Abstract
Bats are the only mammals capable of powered flight, but little is known about the genetic determinants that shape their wings. Here, we generated a genome for Miniopterus natalensis and performed RNA-seq and ChIP-seq (H3K27ac, H3K27me3) on its developing forelimb and hindlimb autopods at sequential embryonic stages to decipher the molecular events that underlie bat wing development. Over 7,000 genes and several lncRNAs, including Tbx5-as1 and Hottip, were differentially expressed between forelimb, hindlimb and different stages. ChIP-seq identified thousands of regions that are differentially modified in forelimb versus hindlimb. Comparative genomics found 2,796 bat-accelerated regions within H3K27ac peaks, several of which cluster near limb-associated genes. Pathway analyses revealed multiple ribosomal proteins and known limb patterning signaling pathways as differentially regulated, and implicated increased forelimb mesenchymal condensations with differential growth. Combined, our work outlines multiple genetic components that contribute to bat wing formation, providing a genomic blueprint for this morphological innovation.
Introduction
The order Chiroptera, commonly known as bats, is the only group of mammals to have evolved the capability of flight. They are estimated to have diverged from their arboreal ancestors ~51 million years ago1. Their adaptions for flight include a radical specialization of the forelimb, characterized by the dramatic extension of digits II to V, a decline in wing bone mineralization along the proximodistal axis, and the retention and expansion of interdigital webbing which is controlled by a novel complex of muscles2,3. Bat hindlimbs are comparatively short, with free symmetrical digits, providing an informative contrast that can be used to highlight genetic processes involved in bat wing formation. Previous studies that examined gene expression in developing bat forelimbs and hindlimbs reported differential expression of several genes, such as Tbx3, Brinp3, Meis2, the 5′ HoxD genes and members of the Shh-Fgf signaling loop, suggesting that multiple genes and processes are involved in generating these morphological innovations4–8. Gene regulatory elements are thought to be important drivers of these changes; for example, the replacement of the mouse Prx1 limb enhancer with the equivalent bat sequence resulted in elongated forelimbs9. However, an integrated understanding of how changes in regulatory elements, various genes and signaling pathways combine to collectively shape the bat wing remains largely unknown.
To characterize the genetic differences that underlie the divergence of bat forelimb and hindlimb development, we used a comprehensive genome-wide strategy. We generated a de novo whole-genome assembly for the vesper bat, Miniopterus natalensis, which has a well characterized stage-by-stage morphological comparison between developing bat and mouse limbs10. In this species, the developing forelimb noticeably diverges from the hindlimb from stages CS15 and CS16 with strong morphological differences seen at a subsequent stage, CS1710. This developmental window is equivalent to embryonic day (E) 12.0 to E13.5 in the mouse4,10. M. natalensis embryos were obtained and transcriptome (RNA-seq) and chromatin immunoprecipitation sequencing (ChIP-seq) for both an active H3K27ac11,12 and a repressive H3K27me313 mark were generated for these three developmental stages (Fig. 1).
Figure 1.
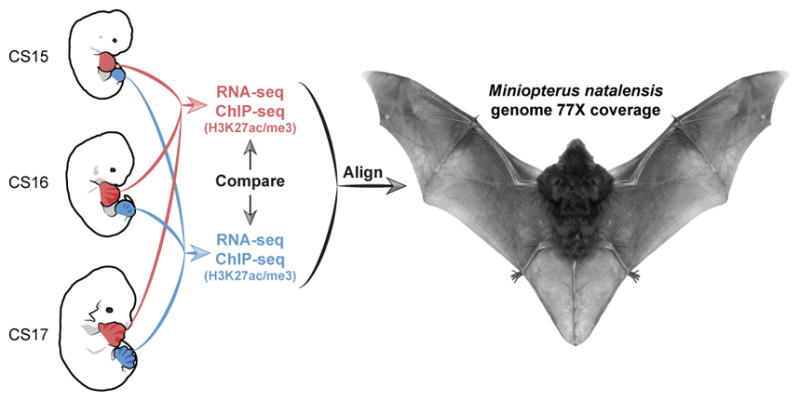
Experimental design. At three developmental stages (CS15, CS16 and CS17) autopods from bat forelimbs (red) and hindlimbs (blue) were analyzed by RNA-seq and ChIP-seq (H3K27ac, H3K27me3) and data aligned to the Miniopterus natalensis genome.
Results
The Miniopterus natalensis genome
High-coverage genomes for three bat species (Pteropus alecto14, Myotis davidii14 Myotis brandtii15) and two low-coverage bat genomes (Myotis lucifugus and Pteropus vampyrus16) have been published. However, the evolutionary distance of these species from M. natalensis (43 million years since the last common ancestor) precludes their use in RNA-seq and ChIP-seq data analyses. We thus generated a draft genome from an adult M. natalensis male at 77X coverage, named Mnat.v1. The quality of Mnat.v1 is comparable to the high coverage bat genomes (Supplementary Table 1). It has an estimated heterozygosity level of 0.13%, with repetitive regions making up 33% of the genome. We annotated 24,239 genes (this includes protein coding genes and long noncoding RNAs) in Mnat.v1. Of the highly conserved genes used by the Core Eukaryotic Genes Mapping Approach (CEGMA)17, 92.7% were found in their entirety with an additional 3.3% partially detected, further confirming Mnat.v1 to be a reliable substrate for subsequent genomic analyses.
Differentially expressed limb transcripts
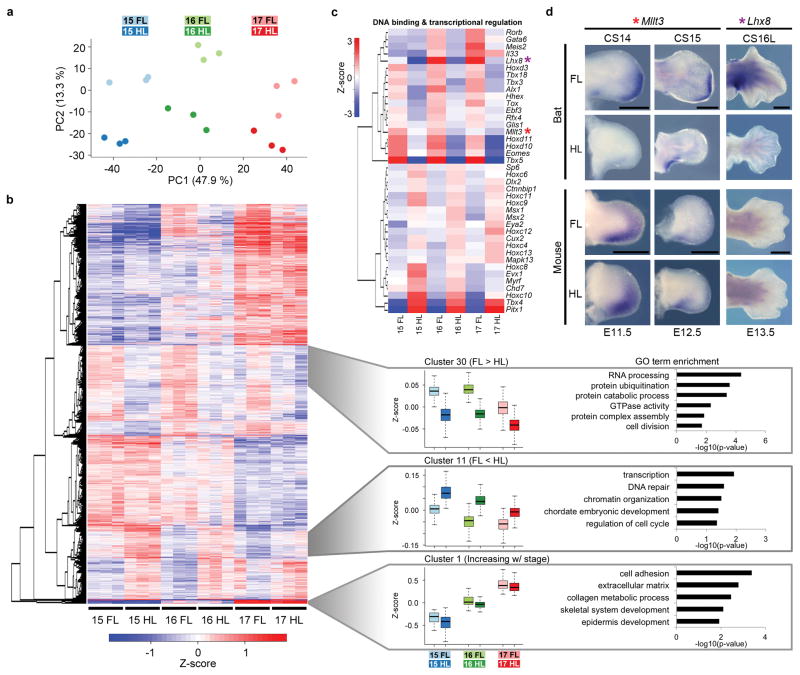
To identify the gene expression differences that could be involved in the morphological divergence of bat limb development, we examined the transcriptomes of whole autopod tissue from the forelimbs and hindlimbs of three sequential developmental stages (CS15, CS16, and CS17). Principle component analysis (PCA) showed an expected segregation pattern, with component one reflecting the developmental stage and component two the tissue type (forelimb or hindlimb; Fig. 2a). We found 2,952 genes differentially expressed between forelimbs and hindlimbs and 5,164 genes differentially expressed between any two sequential stages (adjusted p-value ≤ 0.01; see methods). Pairwise tests for differential expression directly comparing the forelimb and hindlimb at each stage (i.e. CS15 FL vs CS15 HL) contributed an additional 1,596 genes. Combined, these analyses identified 7,172 differentially expressed genes (adjusted p-value ≤ 0.01; Fig. 2b; Supplementary Table 2).
Figure 2.
Gene expression profiling during bat wing development by RNA-seq and in situ hybridization. (a) Principle component analysis using 3,000 genes with the highest variances. PC1 is stage dependent, PC2 is tissue dependent (forelimb or hindlimb) and each explain 57.1% and 13.3% of the variance respectively. (b) Gene-wise hierarchical clustering heatmap of all 7,172 genes showing differential expression (adjusted p-value ≤0.01) displays genes primarily segregate into five groups. Z-score scale is mean subtracted regularized log transformed read counts. Cluster 1 (N=64) shows genes with increased expression through stages. Cluster 11 (N=465) displays genes with increased hindlimb expression. Cluster 30 (N=718) highlights genes with increased forelimb expression. The box in the bar chart is the interquartile range (IQR), the line is the median and the whiskers are the furthest data point from the median within 1.5*IQR. Enriched GO terms are shown to the right. (c) Heatmap of genes from the “DNA binding” (GO:0003677) and “regulation of transcription, DNA dependent” (GO:0006355) GO terms that display the most significant differences (adjusted p-value ≤0.01) and greatest fold changes (fold change ≥2) between forelimbs and hindlimbs. Z-score scale is the sample of the mean subtracted average of the regularized log transformed read counts in each sample. Mllt3 and Lhx8 are highlighted by red and purple asterisks respectively. (d) In situ hybridization of Mllt3 and Lhx8 in stage-matched forelimbs and hindlimbs from bat and mouse. Bat Mllt3 expression shows a shift towards the distal autopod in the future location of digits III–V which elongate in bats. Bat expression of Lhx8 is strongest in the most proximal region of the autopod, especially along the anterior and posterior edges of the limb. Scale bars represent 0.5mm.
Differentially expressed genes were grouped by their expression profile across the samples, into 38 manually defined clusters using hierarchical clustering (Supplementary Fig. 1). These clusters were functionally annotated revealing several terms to be correlated with their differential expression (Fig. 2b; Supplementary Table 3). Grouping the top differentially expressed genes based on biological functions of interest (e.g. DNA binding and transcriptional regulation, limb morphogenesis, bone morphogenesis, apoptotic process and others) identified both genes with known roles in limb development and potentially novel functions in bat wing development (Supplementary Fig. 2). For example, genes differentially expressed between forelimbs and hindlimbs involved in DNA binding and transcriptional regulation included Hoxd10, Hoxd11, Meis2, Pitx1, Tbx4 and Tbx5; all genes that were previously shown to be differentially expressed in bats5–7 along with several genes showing higher forelimb expression that have not yet been characterized (Fig. 2c). We also observed hindlimb-specific increased expression for several genes notably Msx1 and Msx2, both key genes involved in apoptotic activity during interdigital tissue regression18.
A limited number of differentially expressed genes of interest were characterized in both mouse and bat embryos using whole mount in situ hybridization (WISH) (Supplementary Fig. 3). Among these, Mllt3 was chosen for its strong forelimb expression at CS15 and CS16 (Fig. 2c) and was found to be uniquely expressed in bat forelimbs in a region restricted to the distal edge where digits III–V are slated to develop (Fig. 2d). Mllt3 is thought to be a Hox gene regulator, with Mllt3-null mouse mutants exhibiting axial defects19, however no gross skeletal limb abnormalities were observed in homozygous knockout mice (Supplementary Fig. 4; see methods). Lhx8, a known regulator of neuronal development20, had higher expression in CS16 and CS17 forelimbs (Fig. 2c). WISH analysis showed localized Lhx8 expression in the posterior portion of the wrist region, specifically in the junction between the base of digit V and the plagiopatagium, while no expression was detected in mouse limbs (Fig. 2d). Together, these experiments support our RNA-seq analyses and highlight genes that have previously uncharacterized roles in limb development.
Bat-specific lncRNAs
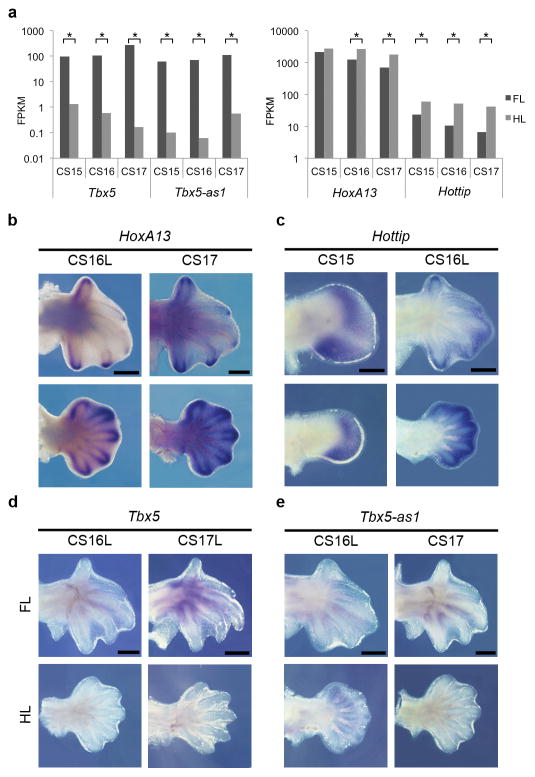
Long noncoding RNAs (lncRNAs) have been shown to be important developmental regulators in several tissues including the limb21. To identify potential lncRNAs associated with bat limb development, we annotated transcripts that did not show similarity to known protein-coding genes, identifying 227 potential lncRNAs (Supplementary Table 4). Amongst these, 188 exhibited some sequence conservation across mammals, 12 of which were similar to characterized lncRNAs in lncRNAdb v2.022. Five putative lncRNAs were identified as being conserved only in bats and 34 were only present in M. natalensis. Within this dataset, 8 known lncRNAs showed differential expression between forelimbs and hindlimbs, including Hottip and an uncharacterized lncRNA, Tbx5-as1 (Fig. 3a). Hottip is thought to be required for the activation of 5′ HoxA genes, important regulators of autopod patterning during limb development21. Both Hottip and HoxA13 showed elevated hindlimb expression in all three stages examined (Fig. 3a). A comparison of their expression patterns revealed both to be more strongly expressed in the interdigital tissue of the bat hindlimb. While Hottip expression was concentrated in the distal interdigital tissue, HoxA13 was more apparent in the digit tips (Fig. 3b,c). The bat Tbx5-as1 transcript maps close to Tbx5 (Fig. 4a), in an antisense direction and shares similarity with human Tbx5 antisense RNA1-as1 transcript (Genbank NR_038440). Tbx5-as1 was the most differentially expressed lncRNA, with elevated expression in the forelimb relative to hindlimb across all stages (Fig. 3a). While its role is unknown, its associated gene Tbx5, is required for forelimb bud initiation with its inactivation in mice abolishing forelimb skeletal formation23,24. In support of a coupled activity for these transcripts, the expression patterns of Tbx5 and Tbx5-as1 were similar, being restricted to the base of digits I to V at CS16L and CS17, with clear expression in the proximal inter-digital tissue (Fig. 3d,e).
Figure 3.
Tbx5-as1 and Hottip expression profiles. (a) RNA-seq fragments per kilobase of exon per million fragments mapped (FPKM) results for Tbx5, Tbx5-as1, Hoxa13 and Hottip. Asterisks display significant forelimb (FL) versus hindlimb (HL) expression changes by stage. (b) In situ hybridization of HoxA13 (b) at CS16 late (L) and CS17 showing reduced expression in the forelimb compared to the hindlimb, but retaining expression in the digit tips in the forelimb. (c) Hottip at CS15 and CS16L, showing expression in the interdigital webbing in both FLs and HLs but with reduced levels in the FLs. (d) Tbx5 at CS16L and CS17L and (e)Tbx5-as1 at CS16L and CS17 shows both transcripts to be restricted to the base of digits I to V, with robust expression in the proximal inter-digital tissue at CS17. Scale bars represent 0.5 mm.
Figure 4.
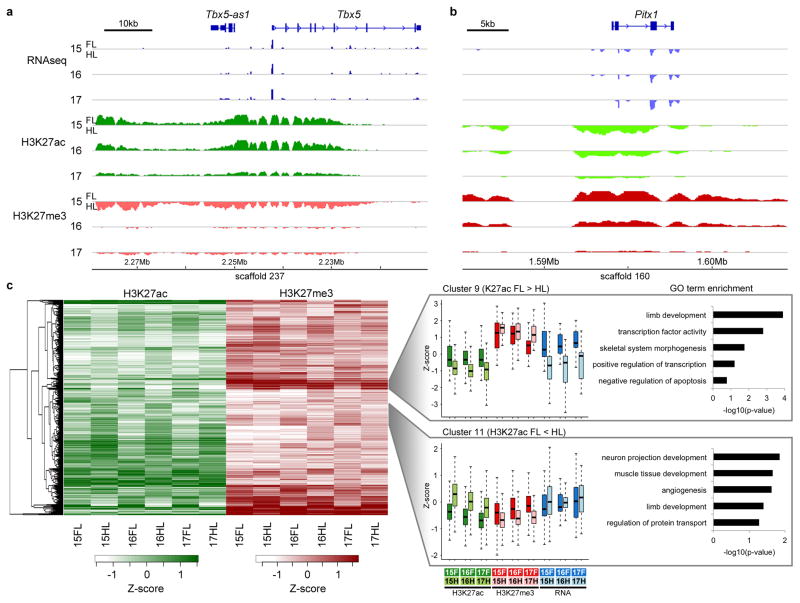
Differing chromatin states between bat forelimbs and hindlimbs during wing development. Schematic of the data tracks for RNA-seq and ChIP-seq for Tbx5 (a) and Pitx1 (b). RNA-seq and ChIP-seq tracks are represented as forelimb (FL) coverage minus hindlimb (HL) coverage in 100 bp intervals for each stage. For RNA-seq, intervals with FL minus HL expression >0 are shown in dark blue and <0 in light blue. Likewise, for ChIP-seq, intervals with FL minus HL enrichment are colored in dark green (H3K27ac) or dark red (H3K27me3) and <0 in light green (H3K27ac) or light red (H3K27me3). (c) Heat map of H3K27ac (green) and H3K27me3 (red) enrichment scores, including a dendrogram of region-wise hierarchical clustering. The heat map shows all regions with differential enrichment between forelimbs and hindlimbs in both marks in at least two stages per mark (2,475 such regions at adjusted p-value ≤ 0.05). The Z-score is the mean subtracted log2 of the signal-to-noise normalized enrichment score plus one. The hierarchical clusters of the regions were segregated into 17 separate clusters; two such clusters are shown as examples, cluster 11 (purple; N=258 peaks) with higher hindlimb H3K27ac and H3K27me3 forelimb and cluster 9 (light blue; N=108 peaks) with higher forelimb H3K27ac and H3K27me3 hindlimb. RNA-seq expression levels of the ChIP-seq peaks neighboring genes are plotted next to the histone marks. Enrichment score distribution is shown as a box plot for each cluster and the enrichment for GO categories of the nearest gene for each region is displayed to the right.
ChIP-seq reveals forelimb regulatory regions
Changes in gene regulatory elements have been shown to be important drivers of morphological adaptations25, including the bat wing9. To identify regulatory elements that could be involved in controlling gene expression in developing bat limbs, we performed low-cell ChIP-seq using antibodies for both H3K27ac (active regions14,15) and H3K27me3 (repressed regions13) on autopods from CS15, CS16 and CS17 forelimbs and hindlimbs, identifying numerous putative regulatory regions (Supplementary Table 5). Using the Genomic Regions Enrichment of Annotations Tool (GREAT26), after converting peaks to the mouse genome, we found significant enrichment for several limb development associated categories for the H3K27ac peaks in GO morphological process, mouse phenotypes and MGI expression (Supplementary Fig. 5). To further validate our ChIP-seq results, we also examined gene loci known to be specifically expressed in the forelimb (Tbx5) or hindlimb (Pitx1) and observed a correspondence with H3K27ac and H3K27me3 peak presence and RNA expression (Fig. 4a,b).
We next set out to analyze differences between forelimb and hindlimb active and repressed ChIP-seq peaks. Differential enrichment analysis was carried out on H3K27ac or H3K27me3 separately identifying 14,553 and 19,352 differentially enriched regions for each mark, respectively (pairwise FDR≤0.05; see methods; Supplementary Table 6). Of these, 2,475 were differentially enriched between forelimbs and hindlimbs for both H3K27ac and H3K27me3 marks. These regions were analyzed using hierarchical clustering based on H3K27ac and H3K27me3 enrichment (Fig. 4c), finding 17 manually defined clusters with distinct H3K27ac and H3K27me3 enrichment patterns (Supplementary Fig. 6). GO term enrichment analysis of the nearest gene showed a strong enrichment for terms associated with limb development. For example, cluster 9 showed an increase in H3K27ac in forelimbs and H3K27me3 in hindlimbs while cluster 11 showed the opposite effect. The regulatory marks of both clusters had a general correspondence with RNA-seq expression levels of the neighboring genes and included fitting GO biological term enrichment for developmental processes (Fig. 4c; Supplementary Table 7).
Bat accelerated regions
To identify genomic changes that may be associated with the innovation of the bat wing, we utilized a comparative genomics approach27 that leveraged the growing number of bat genomes14–16. Whole-genome alignments were generated using repeat masked genomes of eighteen other species, including six bats, nine non-bat mammals and three non-mammal vertebrates. We next used phyloP28 to test for accelerated sequences in the common ancestor of the bat lineage in conserved vertebrate sequences that were marked by H3K27ac in all ChIP-seq experiments. This analysis identified 2,796 bat accelerated regions (BARs; FDR ≤ 0.05) with an average size of 240 bp (Supplementary Table 8). Genomic regions overrepresented for BARs were identified by comparing to vertebrate conserved regions overlapping H3K27ac. Genes contained within these regions were subject to functional annotation clustering, revealing enrichment for categories relating to transcription factors, chromatin conformation and DNA-binding (FDR≤0.05; Supplementary Table 8). The region most highly enriched for BARs included the genes leucine rich repeat neuronal 1 (Lrrn1) and Cereblon (Crbn) (Supplementary Fig. 7a). Lrrn1 is expressed at significantly higher levels in bat hindlimbs compared to forelimbs (Supplementary Table 2) and was also shown to be expressed in the developing mouse limb29. It is important for midbrain-hindbrain boundary formation regulated by Fgf830. Crbn is a known thalidomide target, thought to be important in limb outgrowth by regulating Fgfs31, but did not show significant expression differences between forelimbs and hindlimbs (Supplementary Table 2). Another BAR dense region was around Fgf2 and Spry1 (Supplementary Fig. 7b). Fgf2 is known to have regenerative capabilities in the limb32 and was both the most highly expressed and had the most significant fold change between forelimbs and hindlimbs across all stages amongst Fgf genes in our study (Supplementary Fig. 7c). Spry1, was shown to be involved in limb muscle and tendon development33, but did not have significant expression differences between forelimb and hindlimb (Supplementary Fig. 7c). Combined, our ChIP-seq and BAR analyses highlight specific candidate sequences and genomic regions that may have played a role in the development of the bat wing.
Wing developmental pathways
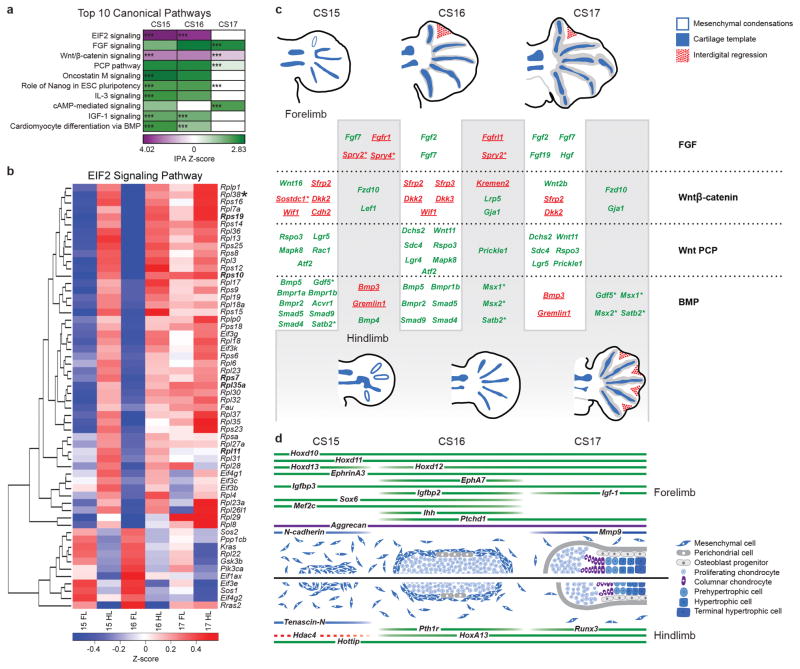
We next used ingenuity pathway analysis (IPA) to identify signaling pathways that were differentially activated across the dataset and could explain the differences in patterning between bat forelimb and hindlimb autopods. Interestingly, the top pathway in our analysis, showing strong hindlimb activation, was the elongation initiation factor 2 (EIF2) signaling pathway (Fig. 5a; Supplementary Table 9), which plays an important role in regulating protein synthesis initiation. A closer inspection shows that 41 ribosomal proteins genes, which are coordinately downregulated in bat forelimbs at CS15 and CS16 (Fig. 5b), were largely responsible for this score. Ribosomal protein expression has been shown to be highly heterogeneous between tissues during embryonic development, including the limb34. Mutations in the ribosomal proteins RPL11, RPL35A, RPS7, RPS10, RPS19 are known to lead to limb malformations in individuals with Diamond-Blackfan anemia35. The Rpl38 gene, which facilitates the translation of several HoxA genes by an IRES-dependent mechanism36 and is mutated in tail short mice which have skeletal patterning defects34, is downregulated in CS15 and CS16 bat forelimbs (Fig. 5b). Rictor, a negative upstream regulator of these ribosomal proteins, had higher expression in forelimbs (Supplementary Table 2). It is a subunit of the mTORC2 complex, playing a role in actin cytoskeleton organization, with conditional deletions in mice resulting in narrower and shorter limb bones37. Combined, our pathway analyses suggest that ribosomal proteins and their regulators could play an important role in bat wing development through the translational control of specific subsets of mRNA transcripts.
Figure 5.
Gene signaling pathways, identified using Ingenuity Pathway Analysis (IPA), are coordinately expressed across the three developmental stages examined (CS15, CS16, CS17). (a) The top ten canonical IPA annotated pathways showing the highest activation scores (p < 0.01 for at least one developmental stage). The Z-score scale is based on the IPA activation Z-score for the pathway. (b) Heatmap showing the RNA-seq expression patterns of gene members of the elongation initiation factor 2 (EIF2) signaling pathway. The Z-score scale is the sample of the mean subtracted average of the regularized log transformed read counts in each sample. Genes that when mutated cause Diamond-Blackfan anemia are highlighted in bold and Rpl38 is highlighted by an asterisk. (c) Differentially expressed genes from the FGF, Wnt/β-catenin, Wnt-PCP and BMP signaling pathways. Forelimb genes are on a white background and hindlimb on a grey background. Activators are highlighted in green and repressors in red. (d) Differentially expressed genes known to be important regulators, or markers of different stages of bone development, including mesenchymal condensation, chondrocyte differentiation, proliferation, maturation and hypertrophy. Genes that are established markers of a particular cell type are indicated in dark blue (mesenchymal condensations), light blue (proliferating chondrocytes) or grey (terminal chondrocytes), while positive regulators are depicted in green and repressors in red. The stages of bone development are aligned to embryonic bat limb developmental stages as described in48.
Several pathways known to play an important role in limb and bone development, including FGF, Wnt, and BMP signaling, were amongst the top ten IPA canonical pathways coordinately activated or repressed in bat forelimb compared to hindlimb (Fig. 5a). FGFs are known to mediate limb patterning by signaling the initial outgrowth of the limb bud from the apical ectodermal ridge38. This pathway showed consistent activation in CS15-CS17 forelimbs (Fig. 5c), with expression of Fgf2, Fgf7, Fgf19, and Hgf in the forelimb at CS16 and CS17 (Fig. 5c). We also observed higher hindlimb expression for several FGF antagonists, including Spry2, Spry4 and Fgfrl1. Wnt ligands are secreted from the limb bud ectoderm and block cartilage formation in the periphery of the limb bud via the β-catenin pathway39. We observed overall suppression of the canonical Wnt/β-catenin pathway in forelimbs versus hindlimbs, with higher levels of several canonical Wnt pathway antagonists in the forelimb and canonical Wnt receptors in the hindlimb (Fig. 5c), including Lef1 which showed strong CS15 hindlimb expression through WISH (Supplementary Fig. 3). The Wnt planar cell polarity (PCP) pathway plays an important role in the elongation of the limb along the proximal-distal axis40 and was activated in bat forelimbs at all stages (Fig. 5c). This upregulation included the PCP pathway ligand, Wnt 11 which has been shown to antagonize the Wnt/β-catenin pathway41.
β-catenin signaling is known to suppress condensation of mesenchymal cells in endochondral bone development39. To test our prediction that β-catenin signaling is diminished and leads to larger fields of condensing mesenchymal cells, we stained sagittal sections of bat forelimb and hindlimb autopods using peanut agglutinin (PNA), a galactose-specific lectin that binds to cell surface markers on condensing pre-cartilage mesenchymal cells42. Haemotoxylin and eosin (H&E) staining of matched sections demarcates the progression from condensing mesenchymal cells (CS15) to differentiation of chondrocytes (CS16) and progression to mature chondrocytes (CS17) in both forelimb and hindlimb autopods (Supplementary Fig. 8). At CS15, PNA staining was far more intense, centered on the emerging digit 4, in sections of forelimb compared to hindlimb autopods (Fig. 6). By CS16, all 5 digits are clearly visible in both bat forelimb and hindlimb sections (Fig. 6, Supplementary Fig. 8). Whereas PNA staining diminishes as chondrocytes differentiate and mature, forelimb digits show more intense, continued recruitment of condensing mesenchymal cells in the distal domain of digits 2 to 5 at both CS16 and CS17 (Fig. 6, Supplementary Fig. 8). These data show that the timing and size of the initial digit condensations and subsequent recruitment of mesenchymal condensations are different in bat forelimb and hindlimb autopods from CS15 onwards and that the foundation for the rapid elongation of forelimb digits could be established far earlier than CS20, as previously proposed43.
Figure 6.
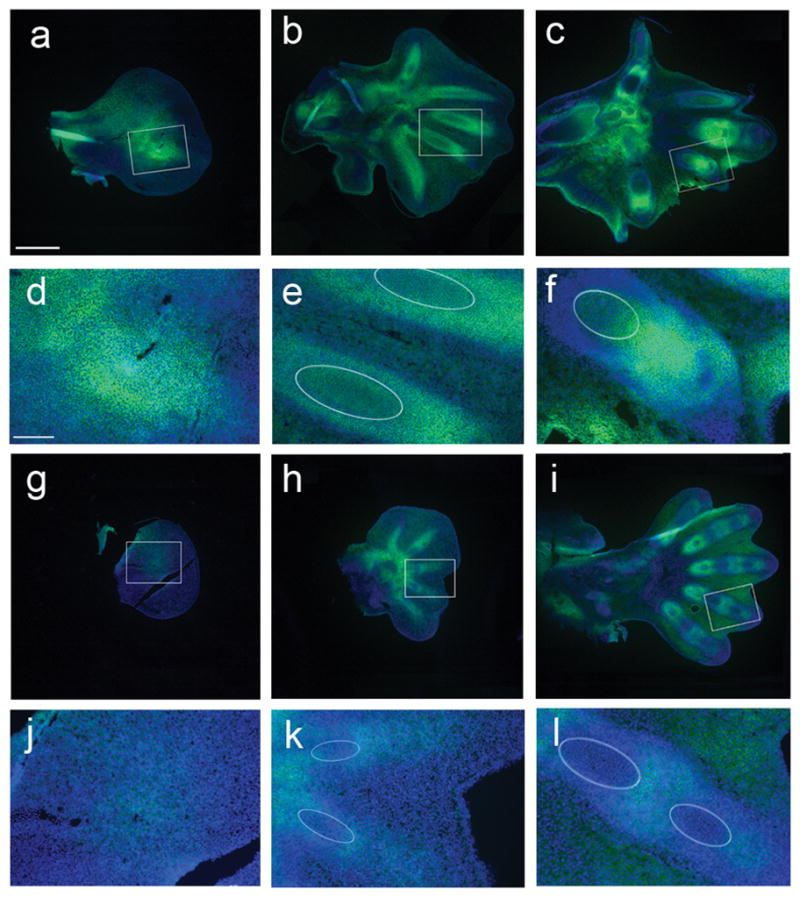
Peanut agglutinin (green) and Hoechst (blue) staining of sagittal sections of bat CS15, CS16 and CS17 forelimb (FL) (a–c) and hindlimb (HL) (g–i) autopods. Higher magnified views of the boxed regions are shown which correspond to the strongest area of PNA staining at CS15 (d and j), digits 3 and 4 at CS16 (e and k) and digit 4 at CS17 (f and l). Ellipses indicate regions of maturing chondrocytes, determined by comparison to H&E staining of adjacent sections (Supplementary Fig. 8). Scale bars of 500 μm (a–c and g–i) and 100 μm (d–f and j–l) are shown.
In the limb, BMP signaling regulates both bone formation and interdigital tissue regression44. We observed two distinct phases of BMP signaling in our datasets (Fig. 5c). During digit initiation and specification (CS15), we observed high levels of BMP inhibitors Gremlin and Bmp3 in the hindlimb (which shows a slight developmental lag at this stage) while BMP receptors (Bmpr1a, Bmpr1b, Bmpr2 and Acvr1) and ligands (Bmp5 and Gdf5) are more abundant in the forelimb. Mutations in Bmp5 were shown to decrease mouse limb width45 and overexpression of Gdf5 in chickens increases skeletal length46. The pattern of BMP signaling starts to switch at CS16, with CS17 forelimbs showing higher levels of Bmp3 and Gremlin. Expression of these BMP antagonists in the forelimb is consistent with the observed decrease in Msx1 and Msx2 expression. A similar suppression of the BMP signaling pathway has been shown to have an important role in interdigital webbing retention in ducks47. Ranking genes from our differentially expressed signaling pathways for consistency across the RNA-seq and ChIP-seq datasets (Supplementary Table 9) found Msx2 (BMP signaling) and Fzd10 (Wnt/β-catenin pathway) to be positively correlated for RNA-seq, H3K27ac and H3K27me3. These genomic regions contain 8 and 12 BARs respectively (within 500kb of their transcription start site) suggesting that they could be important determinants of bat wing development.
Discussion
To identify the genetic components that contribute to bat wing development, we carried out whole-genome sequencing combined with RNA-seq and ChIP-seq for H3K27ac and H3K27me3 on developing bat forelimbs and hindlimbs at three key developmental time points. Overall, we found that multiple genetic components are likely to contribute to the development of the bat wing. These include numerous gene expression changes, both in known limb developmental regulators and newly characterized ones, such as Mllt3 and Lhx8. lncRNAs could also have a strong influence on wing development, with observed forelimb/hindlimb expression differences for Hottip and Tbx5-as1, an uncharacterized lncRNA. A combined pathway analysis found numerous signaling pathways to be differentially activated. These include ribosomal proteins, whose alteration has been shown to result in limb malformations35. Suppression of the Wnt/β-catenin pathway in the forelimb is consistent with the condensation of larger fields of digit mesenchymal cells in the developing bat wing. In contrast, Wnt-PCP signaling, which maintains the polarity of proliferating chondrocytes in the growth plate, was more active in the forelimb (Fig. 6d) and may set the foundation for extended digit growth. Interestingly, the BMP signaling pathway showed two distinct phases with the inhibitors Gremlin and Bmp3 expressed at high levels early in the hindlimb and at later stages in the forelimb with different tissue and temporal identity of BMP activators, fitting with its diverse roles in chondrogenesis, osteogenesis and apoptosis. Combined, the differential activation of these pathways is consistent with the changes in expression of key genes in long bone development, including enhanced expression of chondrogenic markers (e.g. Sox6, Aggrecan, Mmp9) across CS15-CS17 (Fig. 6d). These expression changes could be driven by gene regulatory elements, with potential candidate sequences residing in our ChIP-seq datasets.
Our study obtained unique genomic data from wild-caught non-model organisms. Though restricted sample sizes, biological variation, and gross tissue sampling may have reduced the scope of the experiments and the power of some of the analyses, we were able to generate robust genomic datasets, which identified important regulators of the processes involved in bat limb development. As bats are not currently amenable to transgenic experimentation, future functional characterization of the genes, lncRNAs and regulatory elements identified here could be performed in the mouse, with the potential to further our understanding of their functional importance in the limb. Combined, our results uncover, on a genomic level, the molecular components and pathways that play a role in the formation of the bat wing and provide a foundation of work for studies that examine such unique morphological innovations.
Online Methods
Genome assembly
DNA was extracted from the leg muscle tissue of a single male Miniopterus natalensis using phenol chloroform. The 4 ug protocol of the Nextera Mate Pair Sample Preparation Kit (Illumina) was used to build 2 kb, 5–6 kb and 8–10 kb libraries. For the 5–6 kb and 8–10 kb libraries, multiple reactions were pooled (4, 7 respectively) into one before size selection. The smaller insert libraries were made with the TruSeq DNA LT Sample Preparation Kit (Illumina) following manufacturer instructions. All libraries were sequenced on the Illumina HiSeq2500. The 175 bp and 300 bp paired reads were trimmed on either side to a minimum quality of 17 using Trimmomatic49. Trimmed reads were then used to calculate the 27 bp k-mer frequency using KmerFreq_HA in SOAPdenovo50. The 175 bp paired reads were then merged using their theoretical 25 bp overlap and FLASH51. The remaining read pairs were trimmed to a minimum quality of 17 on the 3′ end before all reads were error corrected using Corrector_HA in SOAPdenovo. K-mers within a read with a frequency of 3 or lower were corrected to a more common k-mer. These changes were limited to two times in the non-overlapping, paired end reads and four times with the 175 bp reads. After these corrections, further erroneous k-mers were removed, to a minimum read length of 60 bp. Duplicate reads were removed using FastUniq52. Combined, the reads totaled over 77x coverage, of which 17.5x was composed of long insert mate pairs. Processed reads were assembled using SOAPdenovo50 and a k-mer size of 49. Pairs with one read mapping to a contig and one read mapping to a gap in a scaffold were used to fill in gaps using GapCloser (SOAPdenovo; submitted to WGS as PRJNA283550). Heterozygosity was estimated using BWA53 and Samtools54. The coherency of the genomic sequence was tested with CEGMA17, using the mammalian optimization.
Genome Annotation
The M. natalensis genome was annotated using the Maker2 pipeline55. Repetitive regions comprised 33% of the genome and were soft masked using RepeatMasker56. Several transcriptome assemblies were used to annotate genes. This included a draft assembly of the M. natalensis forelimb and hindlimb RNA-seq data for each of the three time points (6 assemblies) and a pooled assembly of all the RNA-seq data. Combined, these resulted in 6.1 million transcripts that were aligned to the genome using BLAST57. In addition, 960,000 M. brandtii RNA-seq transcripts, from the liver, kidney and brain15, were aligned using relaxed blastn settings (75% coverage, 80% identity and an e-value cut off of 5e−9) and 51,778 mouse proteins from the RefSeq protein database were aligned using blastp. After alignment, Exonerate58 was used to clear up intron-exon boundaries. Ab initio gene prediction was performed by SNAP59, which was trained off the earlier annotation, and AUGUSTUS60, which was run using the Human optimization. Once complete, gene predictions with poor evidence (AED > 0.75) were ignored. Finally, PASA61 was used to identify and confirm alternatively spliced transcripts.
RNA extraction, sequencing and analysis
RNA was extracted from paired forelimbs and hindlimbs from three individuals (biological replicates) at three developmental stages (CS15, CS16, CS17) using the RNeasy Midi (Qiagen) kit. All bat embryos were staged according to Hockman et al10. Total RNA samples were enriched for poly-A containing transcripts using the Oligotex mRNA Mini kit (Qiagen) and strand-specific RNA-seq libraries62 were generated using PrepX RNA library preparation kits (IntegenX) following the manufacturer’s protocol. After clean up with AMPure XP beads (Beckman Coulter) and amplification with Phusion High-Fidelity polymerase (NEB), RNA libraries were sequenced on a HiSeq 2500 to a depth of at least 30 M reads (submitted to SRA as SRP051253). For de novo transcriptome analysis, raw reads were quality trimmed and adapters sequences removed using Trimmomatic49. Two de novo assembly strategies were employed (Supplementary Fig. 9). First, all three replicates from each tissue/stage combination were pooled and assembled separately using Trinity63. Second, reads from all stages and tissues were pooled and went through digital down sampling and assembly using the Trinity pipeline63. All de novo assemblies were then used in the Maker2 pipeline55 to improve gene annotations. The sequences of 436 transcripts from 227 genes, which did not have a match to the Mammalian Uniprot database, were compared to sequences in the Long Noncoding RNA Database v2.022 and the GENCODE v7 Long non-coding RNA gene annotation database64. These non-coding transcripts were also compared using BLAST57 to the mouse, human, dog, horse, cat and other bat genomes to identify novel lncRNA transcripts that were conserved either in bats, or in a subset of mammals. The coding potential calculator was used to score whether the transcript is likely to be coding or non-coding65. For differential expression, raw sequencing reads were mapped to the M. natalensis draft genome using Tophat66. Read counts for each gene were calculated for each replicate using HTSeq67 and differential expression tests done using DESeq268. Following differential expression testing, genes with p-values adjusted for multiple testing (FDR) less than 0.01 in any of the five differential expression tests were clustered for similar expression using the R package hclust and displayed in the heat map. Additionally, genes found to be differential expressed between forelimbs and hindlimbs across all stages were grouped based on specific GO categories and subject to analysis by clustered heat maps.
In situ hybridization
Mus musculus embryos (C57BL6-StrainUCT3) were supplied by the Animal Research Facility, University of Cape Town (UCT). M. natalensis embryos were collected from a maternity roost at De Hoop Nature Reserve, South Africa (Cape Nature Conservation permit number: AAA007-00133-0056) as previously described69. Ethical approval by the University of Cape Town for the use of these embryos was granted by the UCT Animal Ethics Approval committee, code: 2014/V14/NI and protocol 014-017). Bat and mouse embryos of equivalent stages were matched as described by Hockman et al10. Fixation and storage of embryos, WISH probe synthesis and conditions were done as previously described6. Primers to generate WISH probes are summarized in Supplementary Table 11.
Mllt3 mouse skeletal preps
Skeletons of newborn Mllt3 homozygous knockouts19 and wild-type littermates were stained for cartilage with Alcian blue and for bone with Alizarin red as previously described70. Briefly, newborn mice were sacrificed, skinned, eviscerated, fixed in 95% ethanol for several days and then incubated at 37°C for 2 days in Alcian blue stain (15 mg Alcian blue, 80 ml 95% ethanol, 20 ml glacial acetic acid). Samples were rinsed twice in 95% ethanol for 2 hour each. Specimens were cleared in 1% KOH for 4–5 hours and counterstained overnight with Alizarin red stain (50 mg Alizarin red, 1 liter 2% KOH). Finally, samples were cleared in 20% glycerol, 1% KOH followed by 50% glycerol, 1% KOH for several days each and then stored in 80% glycerol.
ChIP sequencing and analysis
Developing bat forelimbs and hindlimbs (dissected from CS15, CS16 and CS17 embryos) were cross-linked with 1% formaldehyde for 10 minutes, quenched with glycine and flash frozen in the field. Cross-linked limbs were then combined in the lab into pools of 4–7 pairs per stage for chromatin sheering using a Covaris S2 sonicator. Sheared chromatin was then used for chromatin immunoprecipitation (ChIP) with antibodies against active (anti-H3K27ac; Abcam ab4729) or repressed (anti-H3K27me3; Millipore 07-449) chromatin marks using the Diagenode LowCell# ChIP kit following the manufacture’s protocol. Libraries were prepared using the Rubicon ThruPLEX-FD Prep Kit following the manufacturer’s protocol and sequenced on an Illumina HiSeq2500 using single end 50 bp reads to a sequencing depth of at least 25 M (submitted to SRA as SRP051267). Uniquely mapping raw reads were aligned using bowtie71 with default settings. Peak regions for each histone mark were called using SICER72, informed by the estimated average fragmentation size of the chromatin after shearing, as measured by the Agilent 2100 BioAnalyzer. Peaks from all samples were merged using BEDTools73 and then partitioned using BEDOPS74 (Supplementary Fig. 10). Differentially enriched regions between forelimbs and hindlimbs for each stage and histone mark were then obtained following a similar methodology as MAnorm, which uses a linear model that assumes peaks shared between samples can serve to normalize ChIP-seq datasets for differing signal to noise. In this methodology, from the partitioned regions, genomic regions not appearing as a peak in any sample were obtained using BEDTools and used to normalize the background noise present in each sample. Furthermore, a set of common regions for each histone mark was obtained and these regions were used to normalize the ChIP-seq signal between all samples by creating a scaling factor based on the average signal in shared peaks minus the average noise in non-peak regions. After removing duplicate reads with PICARD MarkDuplicates, read counts were obtained with BEDTools coverage command. The average noise from each region based on its genomic size was then subtracted from each region’s read counts. Noise subtracted read counts were then normalized by multiplying each by the signal scaling factor and a read depth scaling factor, to create an enrichment score for each portioned region. Pairwise differential enrichment tests were then carried out using a Bayesian model75, which is also used by MAnorm, followed by adjusting for multiple testing using the R package p.adjust.
Comparative genomics
Whole genome alignments were carried out using Lastz76 with soft-masked genome assemblies from 18 species (E. fuscus, M. brandti, M. davidii, M. lucifugus, P. alecto, P. vampyrus, B. taurus, C. familiaris, E. caballus, F. catus, H. sapiens, L. africana, M. domestica, M. musculus, S. scrofa, D. rerio, A. carolinensis, G. gallus) using the repeat masked M. natalensis genome as a reference. If no publically available repeat masked genome was available, RepeatMasker56 was run using the mammal repeat database and default conditions. Alignment files were then chained, netted and converted to MAFs using UCSC utilities. Individual MAF files from each pairwise species alignments were then combined into a multiple MAF file using the roast command, which is part of the Multiz-TBA package77. A tree model for both conserved and non-conserved sequences was then created for the species used in the multiple MAF file using phyloFit78. These tree models were then used inside PhastCons78 to identify vertebrate conserved sequences in the M. natalensis genome and generate base-by-base conservation scores to be displayed in genome browsers. Bat accelerated regions (BARs) were identified by using phyloP28,78 to test for acceleration in the common ancestor of the bat lineage over regions identified as vertebrate conserved sequences after filtering for quality alignments. Genomic regions enriched for BARs were identified by scanning the genome using a 100 kb sliding window with a step size of 50 kb while counting BARs and phyloP tested regions within them. On average phyloP found acceleration in 0.812% of sequences tested. The expected number of BARs in each region was then set to be the number of sequences tested by phyloP in that region multiplied by 0.00812. Regions enriched with BARSs were then identified by comparing the average expected number of BARs to the observed number of BARs using a Poisson test. After correction for multiple testing, genes contained in or overlapping the genomic regions with significant over-representation of BARs were analyzed for functional annotation clustering using DAVID79,80 with the background set to the genes contained in regions with valid multiple sequence alignments and H3K27ac peaks.
Ingenuity pathway analysis
Pairwise differential expression testing between forelimbs and hindlimbs at each stage identified a total of 3,140 bat genes (FDR < 0.05). This list was filtered for genes that had an average FPKM value greater than 2, and which had been mapped to a human Entrez GeneID, generating 2,751 genes. Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City) was used to analyze this set of 2,751 genes to identify whether specific canonical signaling pathways and their upstream regulators were coordinately regulated across three developmental stages (CS15, CS16 and CS17) using fold change values. A Fisher’s exact right tailed test identified significantly enriched pathways, while a Z-score was computed to determine whether the pathway was activated or inhibited at each stage. IPA was also used to predict upstream regulators that would explain the patterns of differential gene expression observed across the dataset.
Coherency (marked with a 1) was tested by comparing significant differences between forelimb and hindlimb ChIP and RNAseq signals for genes differentially expressed in the top 10 canonical IPA pathways (Fig. 5a). Significantly different acetylation marks were required to be antagonist to their equivalent methylation marks, with at least a single mark being significantly different between the forelimb and hindlimb. RNA-seq levels between the forelimb and hindlimb were also required to be positively correlated with any significantly different acetylation marks.
Histochemistry
Bat embryos were fixed in 4% paraformaldehyde for 3 hours at room temperature, washed in phosphate buffered saline (PBS), and stored in 30% sucrose/PBS at 4°C for 5–6 days. Whole limbs were dissected from these embryos and were embedded in tissue freezing medium (Leica Biosystems). These were sectioned at 8 μm, using a Leica CM1850 cryotome at −17°C, collected on Superfrost Plus (Thermoscientific) slides and stored at −70°C. Serial sections were stained with either haemotoxylin and eosin, or peanut agglutinin (PNA). Slides containing sections were fixed in phosphate-buffered formalin for 5 minutes, washed in distilled water for 1 minute, and then stained with haemotoxylin for 30 seconds. Slides were rinsed in running water, acid alcohol, running water and incubated in Scott’s water for 1 minute. Following rinses in running water, and 80% alcohol, slides were stained with acid-based eosin for 2.5 minutes. Slides were dehydrated through alcohol, dipped in xylene, and coverslips were secured with DPX mountant (Sigma).
For PNA staining, bat autopod sections were fixed for 10 minutes in acetone. Slides were washed three times in PBS and blocked in 3% bovine serum albumin (BSA) in PBS for 1 hour at room temperature. Sections were incubated with 100 μg/ml FITC-conjugated PNA (Sigma L7381) in 3% BSA/PBS at 4°C overnight. Control slides were incubated in 3% BSA/PBS only. All slides were washed in PBS, stained for 10 minutes in 1 μg/ml Hoechst nuclear stain, before another three PBS washes. ProLong Gold antifade reagent (Life Technologies) was used to mount coverslips. Sections were photographed on a Nikon Ti-E inverted fluorescent microscope using the same standardized camera setting for all sections.
Supplementary Material
Acknowledgments
We thank the Broad Institute Genomics Platform, Vertebrate Genome Biology group and Kerstin Lindblad-Toh for making the data for Eptesicus fuscus available. We would also like to thank Dr. Matthew Kelley and Elizabeth Driver (NIDCD) for providing us with Mllt3 newborn mice and Morea Petersen for assistance with sectioning bat autopods. This work was supported in part by the NICHD grant 1R01HD059862. N.A. is also supported in part by the NIDDK award number 1R01DK090382, NINDS award number 1R01NS079231 and NCI award number 1R01CA197139. N.I. is supported by National Research Foundation (NRF) (South Africa) grants (NBIG#86932) while C.N.M was supported by NRF grant #85207. L.C. and K.N. are supported by the office of the director, National Institutes of Health under Award Number P51OD011092.
Footnotes
Author Contributions
W.L.E, S.A.S, M.K.M., J.E.V., J.D.W. N.I. and N.A. conceived key aspects of the project and planned experiments. S.A.S, M.K.M., Z.G., A.V.P., C.M-N. and N.I. collected bat embryos. S.A.S and C.M-N. extracted genomic DNA and RNA from bats. E.T., K.N. and L.C. generated genome sequencing libraries. W.L.E, S.A.S, M.K.M., Z.G., A.V.P., B.M.B., S.N., N.M., J.E.V. and N.I. performed experiments. T.F. and K.S.P. developed statistical methods. W.L.E, S.A.S, M.K.M., Z.G., T.F., L.C., K.S.P, J.D.W., N.I. and N.A. analyzed data. W.L.E, S.A.S, M.K.M., L.C., N.I. and N.A. wrote the manuscript. All authors commented on and revised the manuscript.
Competing Financial Interests
The authors have no competing financial interests.
URLs
Ingenuity® Pathway Analysis, www.qiagen.com/ingenuity; PICARD MarkDuplicates, http://broadinstitute.github.io/picard; Supplementary Table 6. ChIP-seq differential enrichment analysis.https://dl.dropboxusercontent.com/u/923636/Supplementary_Table_6_ChIPseq_Differential_Enrichment.xlsx
Accession codes
Whole Genome Shotgun:
Genomic Assembly PRJNA283550
Sequence Read Archive:
RNAseq: PRJNA270639 (SRP051253)
ChIPseq: PRJNA270665 (SRP051267)
References
- 1.Gunnell GF, Simmons NB. Evolutionary History of Bats: Fossils, Molecules and Morphology. Cambridge Univ. Press; 2012. [Google Scholar]
- 2.Cooper LN, Cretekos CJ, Sears KE. The evolution and development of mammalian flight. Wiley Interdiscip Rev Dev Biol. 2012;1:773–779. doi: 10.1002/wdev.50. [DOI] [PubMed] [Google Scholar]
- 3.Swartz SM, Middleton KM. Biomechanics of the bat limb skeleton: scaling, material properties and mechanics. Cells Tissues Organs. 2008;187:59–84. doi: 10.1159/000109964. [DOI] [PubMed] [Google Scholar]
- 4.Hockman D, et al. A second wave of Sonic hedgehog expression during the development of the bat limb. Proc Natl Acad Sci U S A. 2008;105:16982–16987. doi: 10.1073/pnas.0805308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, et al. Unique expression patterns of multiple key genes associated with the evolution of mammalian flight. Proc Biol Sci. 2014;281:20133133. doi: 10.1098/rspb.2013.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason MK, et al. Retinoic acid-independent expression of Meis2 during autopod patterning in the developing bat and mouse limb. Evodevo. 2015;6:6. doi: 10.1186/s13227-015-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, et al. Digital gene expression tag profiling of bat digits provides robust candidates contributing to wing formation. BMC Genomics. 2010;1:619. doi: 10.1186/1471-2164-11-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weatherbee SD, Behringer RR, Rasweiler JJt, Niswander LA. Interdigital webbing retention in bat wings illustrates genetic changes underlying amniote limb diversification. Proc Natl Acad Sci U S A. 2006;103:15103–15107. doi: 10.1073/pnas.0604934103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cretekos CJ, et al. Regulatory divergence modifies limb length between mammals. Genes Dev. 2008;22:141–151. doi: 10.1101/gad.1620408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hockman D, Mason MK, Jacobs DS, Illing N. The role of early development in mammalian limb diversification: a descriptive comparison of early limb development between the Natal long-fingered bat (Miniopterus natalensis) and the mouse (Mus musculus) Dev Dyn. 2009;238:965–979. doi: 10.1002/dvdy.21896. [DOI] [PubMed] [Google Scholar]
- 11.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Zhang G, et al. Comparative Analysis of Bat Genomes Provides Insight into the Evolution of Flight and Immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seim I, et al. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat Commun. 2013;4:2212. doi: 10.1038/ncomms3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–82. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 18.Lallemand Y, Bensoussan V, Cloment CS, Robert B. Msx genes are important apoptosis effectors downstream of the Shh/Gli3 pathway in the limb. Dev Biol. 2009;331:189–198. doi: 10.1016/j.ydbio.2009.04.038. [DOI] [PubMed] [Google Scholar]
- 19.Collins EC, et al. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol Cell Biol. 2002;22:7313–7324. doi: 10.1128/MCB.22.20.7313-7324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, et al. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci U S A. 2003;100:9005–10. doi: 10.1073/pnas.1537759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quek XC, et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwal P, et al. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–633. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- 24.Rallis C, et al. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 2003;130:2741–2751. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- 25.Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotech. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Booker BM, et al. Bat Accelerated Regions Identify a Bat Forelimb Specific Enhancer in the HoxD Locus. PLoS Genet. 2016;12:e1005738. doi: 10.1371/journal.pgen.1005738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Homma S, Shimada T, Hikake T, Yaginuma H. Expression pattern of LRR and Ig domain-containing protein (LRRIG protein) in the early mouse embryo. Gene Expr Patterns. 2009;9:1–26. doi: 10.1016/j.gep.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Tossell K, et al. Lrrn1 is required for formation of the midbrain-hindbrain boundary and organiser through regulation of affinity differences between midbrain and hindbrain cells in chick. Dev Biol. 2011;352:341–352. doi: 10.1016/j.ydbio.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–50. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 32.Taylor GP, Anderson R, Reginelli AD, Muneoka K. FGF-2 induces regeneration of the chick limb bud. Dev Biol. 1994;163:282–284. doi: 10.1006/dbio.1994.1144. [DOI] [PubMed] [Google Scholar]
- 33.Eloy-Trinquet S, Wang H, Edom-Vovard F, Duprez D. Fgf signaling components are associated with muscles and tendons during limb development. Developmental dynamics : an official publication of the American Association of Anatomists. 2009;238:1195–1206. doi: 10.1002/dvdy.21946. [DOI] [PubMed] [Google Scholar]
- 34.Kondrashov N, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ball S. Diamond Blackfan anemia. Hematology Am Soc Hematol Educ Program. 2011;2011:487–91. doi: 10.1182/asheducation-2011.1.487. [DOI] [PubMed] [Google Scholar]
- 36.Xue S, et al. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Holguin N, Shi Y, Silva MJ, Long F. mTORC2 signaling promotes skeletal growth and bone formation in mice. J Bone Miner Res. 2015;30:369–378. doi: 10.1002/jbmr.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin GR. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- 39.Kozhemyakina E, Lassar AB, Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142:817–831. doi: 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao B, Yang Y. Planar cell polarity in vertebrate limb morphogenesis. Curr Opin Genet Dev. 2013;23:438–444. doi: 10.1016/j.gde.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bisson JA, Mills B, Paul Helt JC, Zwaka TP, Cohen ED. Wnt5a and Wnt11 inhibit the canonical Wnt pathway and promote cardiac progenitor development via the Caspase-dependent degradation of AKT. Dev Biol. 2015;398:80–96. doi: 10.1016/j.ydbio.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Hall BK, Miyake T. Divide, accumulate, differentiate: cell condensation in skeletal development revisited. Int J Dev Biol. 1995;39:881–893. [PubMed] [Google Scholar]
- 43.Sears KE, Behringer RR, Rasweiler JJt, Niswander LA. Development of bat flight: morphologic and molecular evolution of bat wing digits. Proc Natl Acad Sci U S A. 2006;103:6581–6586. doi: 10.1073/pnas.0509716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pignatti E, Zeller R, Zuniga A. To BMP or not to BMP during vertebrate limb bud development. Semin Cell Dev Biol. 2014;32:119–127. doi: 10.1016/j.semcdb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Kingsley DM, et al. The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell. 1992;71:399–410. doi: 10.1016/0092-8674(92)90510-j. [DOI] [PubMed] [Google Scholar]
- 46.Francis-West PH, et al. Mechanisms of GDF-5 action during skeletal development. Development. 1999;126:1305–1315. doi: 10.1242/dev.126.6.1305. [DOI] [PubMed] [Google Scholar]
- 47.Merino R, et al. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development. 1999;126:5515–5522. doi: 10.1242/dev.126.23.5515. [DOI] [PubMed] [Google Scholar]
- 48.Hockman D, Mason MK, Jacobs DS, Illing N. The role of early development in mammalian limb diversification: a descriptive comparison of early limb development between the Natal long-fingered bat (Miniopterus natalensis) and the mouse (Mus musculus) Dev Dyn. 2009;238:965–979. doi: 10.1002/dvdy.21896. [DOI] [PubMed] [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H, et al. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS ONE. 2012;7:e52249. doi: 10.1371/journal.pone.0052249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2010. [Google Scholar]
- 57.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 58.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borodina T, Adjaye J, Sultan M. A strand-specific library preparation protocol for RNA sequencing. Methods Enzymol. 2011;500:79–98. doi: 10.1016/B978-0-12-385118-5.00005-0. [DOI] [PubMed] [Google Scholar]
- 63.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Pyl PT, Huber W. HTSeq A Python framework to work with high-throughput sequencing data. 2014 doi: 10.1093/bioinformatics/btu638. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love MI, Huber H, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. 2014 doi: 10.1186/s13059-014-0550-8. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mason MK, Hockman D, Jacobs DS, Illing N. Evaluation of maternal features as indicators of asynchronous embryonic development in Miniopterus natalensis. Acta Chiropterologica. 2010;12:161–171. [Google Scholar]
- 70.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo: A laboratory manual. Vol. 764. Cold Spring Harbor; New York: 2002. [Google Scholar]
- 71.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neph S, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012;28:1919–20. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 76.Harris, R.S. Pennsylvania State University (2007).
- 77.Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hubisz MJ, Pollard KS, Siepel A. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform. 2011;12:41–51. doi: 10.1093/bib/bbq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 80.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.