Abstract
Molecular Dynamics Flexible Fitting (MDFF) is an established technique for fitting all-atom structures of molecules into corresponding cryo-electron microscopy (cryo-EM) densities. The practical application of MDFF is simple but requires a user to be aware of and take measures against a variety of possible challenges presented by each individual case. Some of these challenges arise from the complexity of a molecular structure or the limited quality of available structural models and densities to be interpreted, while others stem from the intricacies of MDFF itself. The current article serves as an overview of the strategies that have been developed since MDFF's inception to overcome common challenges and successfully perform MDFF simulations.
Keywords: cryo-electron microscopy, molecular dynamics
1. Introduction
Cryo-electron microscopy (cryo-EM) has been playing an increasing role in structure determination in recent years. While the most widely used method for acquiring structures of biomolecules is still X-ray crystallography, crystallization of large biomolecules, macromolecular complexes, and membrane proteins is challenging and limits the method. Cryo-EM single-particle reconstruction does not require the difficult crystallization step and allows structures to be imaged in physiological conditions, though only after rapid freezing of the samples.
X-ray crystallography and cryo-EM often reveal different levels of macromolecular structure. Generally, X-ray crystallography produces atomic-resolution structures (<~3 Å), while cryo-EM densities are resolved typically only at lower resolution (~5-30 Å). However, through recent developments in cryo-EM techniques, high-resolution (~2-5 Å) densities have been obtained [1, 2, 3, 4, 5]. Computational methods that combine information from crystallography and cryo-EM can bridge the resolution gap and hold the promise of generating physiologically accurate, atomic-resolution structures of biomolecular complexes.
Various methods that combine X-ray crystallography structures and cryo-EM densities for structure determination have been developed in recent years. Some of these methods use rigid-fragment fitting [6, 7, 8], while others such as DireX [9], Flex-EM [10], Rosetta [11], and FRODA [12] perform flexible fitting, allowing conformational changes to better shape the structure to the data. Some approaches include the use of low-frequency normal modes [13], deformable elastic networks [9], and cross correlation [14] or least-squares difference between experimental and simulated maps [15] to drive the structure into the cryo-EM density. Some fitting methods use a Monte Carlo-based approach [11], while others such as the present authors’ Molecular Dynamics Flexible Fitting (MDFF) [16, 17] use molecular dynamics (MD) to match structures to multi-modal (crystallography and electron microscopy) data.
The present article concerns MDFF, which has proven to be successful in the hands of its developers as evidenced by applications to solving structural models for the ribosome [18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30], photosynthetic proteins [31, 32], myosin [33], chaperonins [29], bacterial chemosensory array [34], and virus capsids [35, 36, 37], including the first all-atom structure of the HIV capsid [36]. MDFF has played an even greater role in structural modeling efforts outside of its developer group, namely for the ribosome and its substrates [38, 39, 40, 41, 42, 43, 44, 45], the actin-myosin interface [46], the Mot1-TBP complex [47], the 26S proteasome [48, 49], and the HIV-1 virus [50, 51].
The main advantage of MDFF over other fitting methods is that MDFF produces models with improved structural geometries, due to its use of the most advanced MD force fields [52]. Compared to other fitting methods, MDFF has also been shown to produce models with better stereochemistry [53] due to the force fields and stereochemical restraints [54] used during MDFF. However, the emergence of high resolution cryo-EM maps poses a challenge to MDFF which was initially developed for low-resolution densities in the range of 10 – 25 Å. De novo fitting methods, such as the one employed by Rosetta, have been shown to perform better than MDFF at higher resolutions (< 5 Å) [11]. New methodologies are being developed to allow MDFF to overcome the challenges posed by high-resolution maps, as discussed in Section 6.
In this article, we will review the MDFF method and how it has been developed and applied in the years since its creation. A basic protocol will be outlined and employed to highlight specific problems and their solution. An earlier guide [17] covers the basic MDFF workflow; the present article reviews new features and advances introduced since the earlier publication. Details of the application of the features discussed here can also be found in several tutorials listed and discussed in Section 7.
2. Basic MDFF Method
The essence of MDFF is, given an initial all-atom structure (available through crystallography or modeling) and a corresponding cryo-EM density, to match the structure to the density by means of an MD simulation. For this purpose, the structure is first rigidly docked into the density, then flexible fitting is performed by applying to the structure, in addition to the intrinsic (molecular dynamics force field) potential VMD, an external potential VEM(r), obtained by inverting the cryo-EM density and bounding the resultant map Φ(r) from below a threshold Φthr, to prevent fitting to noise, and scaling the potential by a user-defined factor ζ as a control over the strength of the VEM over VMD. Specifically,
| (1) |
where Φmax is the maximum of Φ(r). In addition, an atom-dependent weight wi, where i indexes atoms in the structure, is applied to vary the coupling from atom to atom, such that the potential experienced by atom i at position ri is wiVEM(ri). In practice, wi is commonly set to the atom's atomic mass so that the acceleration due to MDFF forces is uniform amongst all the coupled atoms.
MDFF produces an atomic-resolution structure in the conformation captured by the cryo-EM density by combining all-atom structural information, encapsulated in the initial structure and obtained from structure prediction algorithms or experiments like X-ray crystallography, with EM density information. The MD-based nature of MDFF allows for flexibility and sampling, while maintaining a realistic structural geometry through incorporation of the most advanced force fields in the simulation that define the potential VMD introduced above.
At its simplest, MDFF requires, as initial data, a complete all-atom structure and the corresponding cryo-EM density map. In preparation for the MDFF simulation, the MDFF potential VEM is calculated from the cryo-EM map, and several MDFF-specific parameters are set. These parameters include the subset of atoms within the structure to be coupled to the MDFF potential, the scaling factor ζ, and per-atom weights wi. The user can leave these parameters at their default settings which couple all non-hydrogen protein or nucleic atoms, a scaling factor ζ = 0.3, and per-atom weights wi equal to the atoms’ masses.
The potential energy function is defined on a 3-D grid and incorporated into the MD simulation using the gridForces feature of NAMD [55, 56]. Forces are computed from the added potential and applied (in addition to the intrinsic MD forces) to each atom depending on its position on the grid using an interpolation scheme. The computed VEM-derived forces drive the atoms into regions of high density, producing an atomic-resolution structure in the conformation captured by the cryo-EM density. Restraints imposed during the simulation help preserve the secondary structure, stereochemical correctness [54], and symmetry [57] of the protein investigated. The MDFF plugin and graphical user interface (GUI) (see Section 2.4) in VMD [58] allows one to perform the aforementioned tasks in a user friendly manner to set up an MDFF simulation in NAMD [55].
A thorough description of the basic MDFF technique is given in [16, 17]. Since its inception, MDFF has undergone further developments to address challenging cases. Application of MDFF in such cases is the main focus of the present publication. The MDFF user is instructed to recognize possible difficulties that may arise in their own system demanding structure analysis tasks; available solutions to address these difficulties are outlined.
2.1. Structure Checking
An important step in any modeling workflow, including MDFF, is checking the stereochemistry of the structure for errors. This step is important both at the beginning of modeling, before MDFF is used, to correct errors before they have the potential to propagate, and after the use of MDFF, to ensure the validity of the final fitted structure. One class of stereochemical errors involves the two enantiomer forms (D-/L-) of amino acids around the chiral centers of their Cα (except for glycine) and Cβ (for threonine and isoleucine only) atoms. Most naturally occurring amino acids are found in the L- configuration, however some D-amino acids exist, e.g., in bacterial cell walls [59, 60, 61]. Another class of stereochemical errors involves the conformation of the peptide bond between two amino acids. The dihedral angle ω described by Cα,n, Cn, Nn+1, Cα,n+1, distinguishes between the cis (ω ≈ 0°) or trans (ω ≈ 180°) isomers [62]. The trans isomer is energetically more stable and, therefore, is the one more commonly found in nature, except in some cases, e.g., peptide bonds before a proline residue [63, 64].
Assignment of the incorrect form for the chiral center or peptide bond configuration results in stereochemical errors. Unfortunately, there has been an increasing occurrence of such errors, particularly non-proline cis-peptide bonds, found in X-ray crystal structures (especially ones solved at low-resolution) [65]. Not only can unchecked errors in the initial structure propagate during MDFF [54], but they can hide worse errors such as incorrect fit to the density which are not always found through other measures such as Ramachandran outliers [65].
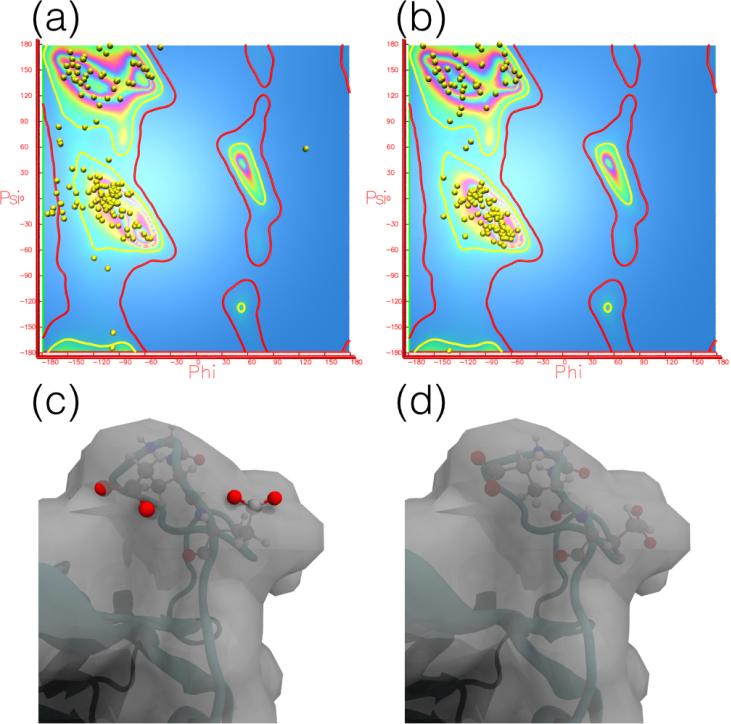
VMD provides plugins, namely cispeptide and chirality, which can be used to detect, visualize, and correct stereochemical errors [54]. A new plugin released in VMD 1.9.3, TorsionPlot (Fig. 1), can similarly be used for marginal and outlier Ramachandran angles. These plugins allow users to easily find errors in their structures and quickly fix them, using short MD simulations to properly equilibrate the structure and improve the geometry, or in more difficult cases, using interactive MD (Section 4) to correct the conformation of the errant regions. Once the errors have been corrected, restraints can be defined for the peptide bond and chirality configurations, as discussed in Section 2.2. Since VMD version 1.9.3, the cispeptide, chirality, and TorsionPlot plugins can work with the newly-developed MDFF Graphical User Interface, discussed in Section 2.4.
Figure 1.
TorsionPlot plugin of VMD for analysing and fixing Ramachandran outliers. Analysis of Ramachandran outliers and marginal residues using the TorsionPlot plugin of VMD can be plotted in the VMD display window (a,b). Results from a Ramachandran analysis on a distorted initial structue of adenylate kinase (PDB 1AKE) (a) demonstrate the reduction in marginal and outlier residues achieved through MDFF (b). An example Ramachandran outlier in residues 150-152 (c) in the initial structure displays a poor fit to the map, with several atoms outside of the density. An MDFF simulation not only improves the fit to the density, but also moves the Ramachandran angles into a favored position (d).
2.2. Restraints
During MDFF, various restraints can be applied to the system, primarily to avoid overfitting the structure to noise in the density map. The most common restraints are applied to a set of internal coordinates relevant to the secondary structure of the macromolecule in its initial conformation, using the ssrestraints plugin of VMD [16, 17]. For proteins, restraints are applied to the ϕ and ψ dihedral angles and hydrogen bonds involving backbone atoms of amino acid residues in helices and β-sheets. For nucleic acids, dihedral angles and interatomic distances between base pairs are restrained. The strength of these restraints can be adjusted during the fitting process. For example, the strength can be decreased as the simulation progresses and large-scale motions have stopped, making way for local refinement of the structure.
A change in isomerism or chirality, as discussed in Section 2.1, during equilibrium simulations at room temperature is unlikely due to a high energy barrier (e.g., 21 kcal/mol in CHARMM22 [66]). However, the fitting forces derived from the potential VEM that are applied during MDFF, especially during initial structure optimization, or other refinement schemes involving high-temperature simulated-annealing, may be strong enough to introduce stereochemical errors into a structure. To avoid such errors, harmonic restraints can be applied to enforce the original isomerism of the peptide bond between residues or the chiral center located at the Cα and Cβ of certain residues using the cispeptide and chirality plugins, respectively, of VMD.
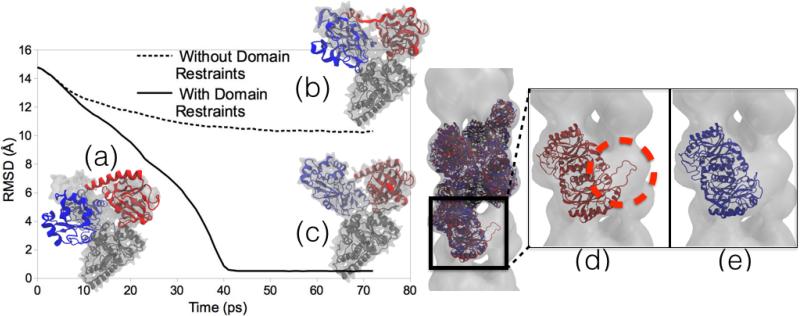
While some form of secondary structure, chirality, and cis-peptide restraints should be used for most MDFF simulations, there are other restraints available which are application-dependent. Domain restraints can be used to maintain non-overlapping, rigid, user-defined domains of a system during MDFF simulations with the Targeted MD (TMD) [67, 68] feature of NAMD. In TMD, a subset of atoms in the simulation is guided towards a final ‘target’ structure by means of steering forces. At each timestep, the root mean-square (RMS) distance between the current coordinates and the target structure is computed (after first aligning the target structure to the current coordinates). The force on each atom is given by the gradient of the potential derived from the RMS distance, such that the atoms are driven towards the selected target. For use as domain restraints, the target coordinates are set to the initial coordinates, thus keeping the domain rigid. Although the restraints keep each domain in the system rigid, they allow for flexibility between domains. Domain restraints are useful when fitting structures that undergo large-scale conformational changes with a ‘hinge-bending’ or ratcheting (as in the case of ribosome translocation [69]) motion where the local structure remains otherwise unchanged (Fig. 2).
Figure 2.
Domain and symmetry restraints for MDFF. Left Image: RMSD plots of MDFF trajectories and representative structures. (a) Initial docking of Acetyl-CoA Synthase structure into cryo-EM density. (b) A structure obtained from MDFF without using domain restraints applied to the two domains (red and blue) to keep the local structure intact is distorted and improperly fit to the density. (c) Result of MDFF with application of domain restraints. The graph shows the RMSD of the structures to a known validation model, with the non-domain-restrained simulation exhibiting a much worse fit. Right Image: A two-turn helix model of a nitrilase from R. rhodochrous J1 [72, 57] fit to a larger cryo-EM density. (d) Final fitted structure from MDFF without using symmetry restraints shows the edge-distortion effect of a subunit being pulled into empty adjacent density (circled in red). (e) Final fitted structure from MDFF using symmetry restraints avoids the edge-distortion effect by restraining the subunits to an average structure.
The quality of the final model in any fitting method, including MDFF, is highly dependent on the quality of the initial model and the cryo-EM density map. The lower the resolution of a map, the less information it contains. Thus, the greater the degrees of freedom provided for the fitting procedure. Incorporating any additional knowledge of the system into the fitting procedure can reduce the search degrees of freedom and provide better results. One such structural trait to exploit, which is common in many biological systems, is non-crystallographic symmetry. Many large biological macromolecules have inherent structural symmetry, being composed of a few distinct subunits, repeated in a symmetric array. Examples are the poliovirus exhibiting icosahedral symmetry [70] or potassium channels exhibiting 4-fold symmetry [71].
In MDFF, symmetry restraints are available to make use of symmetry information [57]. These restraints are calculated by overlapping selected symmetric subunits, calculating an average structure from them, and applying harmonic forces on the atoms restraining them to that average structure. Similar to the other restraints, the strength of the forces can be adjusted and either be set constant or to increase linearly with time. Increasing the strength only gradually allows for greater flexibility to explore the conformational space of the density map initially before converging to a symmetric structure. In addition to improving the quality of fit and agreement between subunits in low-resolution maps, such as in the case of the bacterial chemosensory array [34], symmetry restraints can also be used to prevent edge-distortion e ects (Fig. 2). Edge-distortion e ects occur when fitting a smaller number of subunits into a much larger EM density. Because the density represents more subunits than are actually being simulated, some subunits will be adjacent to empty density belonging to a subunit not represented in the limited simulation. Such subunits at the edges of the system can be pulled into the adjacent empty density and become distorted if the neighboring subunits, actually present in the real system, are not represented through symmetry restraints. The problem of missing subunits beyond the limited simulation geometry could be solved by cutting the density map to only the regions around the simulated structure, however such a cutting procedure is prone to bias. Another solution is to simulate the entire system, but this solution can be computationally costly and unnecessary, as a small number of subunits could be su cient to model the desired properties. For example, in a nitrilase from R. rhodochrous J1 [72, 57], a two-turn helix model is su cient for determining how interdimer interactions give rise to the stability of a spiral structure. Therefore, only two turns of the helix model needed to be simulated while using symmetry restraints to prevent the edge subunits from distortion [57].
2.3. Simulation Environment
Due to the aqueous environment of the cell in which many biomolecules are found, properly describing the solvent effect on the solutes is an important step in any MD simulation, including MDFF. In the gold standard for accuracy, the explicit solvent method, individual water molecules are represented by atomic models such as TIP3P [73]. While accurate, inclusion of explicit solvent in a MD simulation is computationally demanding because the interactions for all of the solvent molecules must now be calculated in addition to those computed for the solute (e.g. proteins, nucleic acids, and ligands). Furthermore, explicit solvent introduces viscous drag [74] which slows the movement of the solute in the system, causing MDFF simulations to require more simulation time to produce a final fitted structure. Therefore, MDFF simulations are often performed in vacuum, i.e., without any solvent.
While simulations in vacuum are much faster than explicit solvent, they are less accurate, especially for solvent accessible regions on the exterior of a structure [75]. However, the constraints imposed by the cryo-EM derived potential and the additional restraints (e.g., secondary structure) prevent the structure from becoming excessively deformed, so many initial MDFF simulations can still be run in a vacuum environment.
One compromise between the accuracy of the explicit solvent model and the speed of vacuum simulations is use of an implicit solvent model. MDFF can use the generalized Born implicit solvent (GBIS) model of NAMD [75], a fast approximation for calculating the electrostatic interaction between atoms in a dielectric environment described by the Poisson-Boltzmann equation. Not only is this approximation faster to compute than explicit water interactions, but the GBIS algorithms in NAMD are also parallelized for CPUs and GPUs for improved performance. While not as fast as vacuum simulations, MDFF with GBIS produces more accurate final models [75] in approximately the same simulation time due to the absence of viscous drag effects (implicit solvents form a continuum through which model components move as easily as through vacuum). In general, it is adviseable to use at least the implicit solvent model, especially in cases where significant interactions with solvent may affect the shape of the macromolecule, like in case of the 26S proteasome [76].
2.4. Graphical User Interface
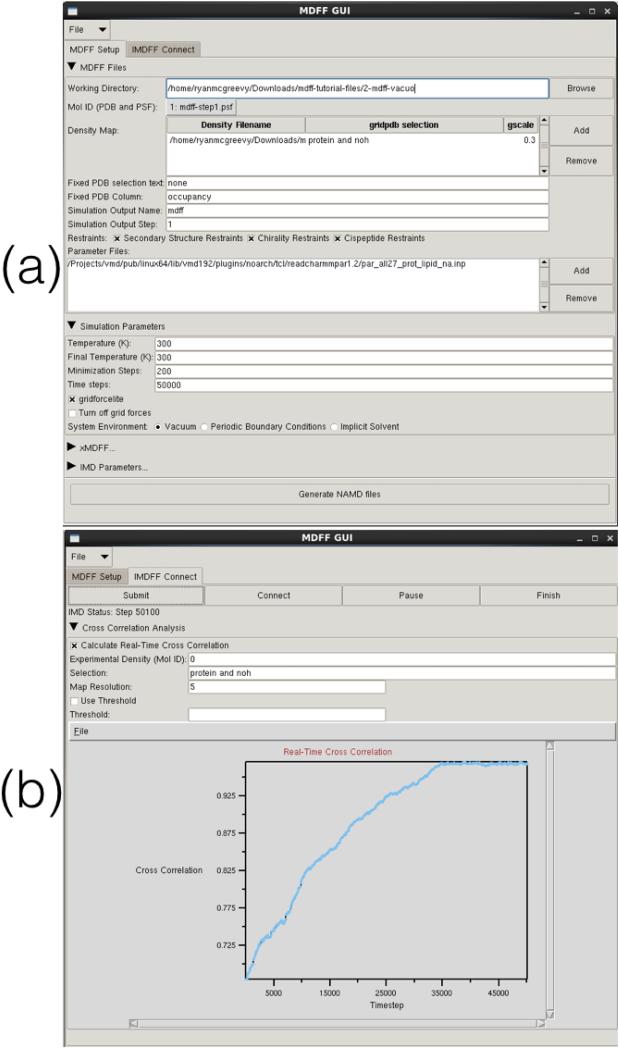
MDFF simulations are set up and analyzed using many different tools in VMD [58]. Many of these tools are plugins (e.g. mdff, cispeptide, and chirality) accessed through VMD's command line interface, the TkConsole. Since VMD version 1.9.2, a new graphical user interface (GUI) for MDFF (Fig. 3) is available, providing a unified gateway for quicker and easier access to all plugins used by MDFF. The GUI, found in the ‘Modeling’ section of VMD's ‘Extensions’ menu, is divided into several sections pertaining to different aspects of MDFF. Both MDFF and xMDFF (Section 3.1) simulations can be set up with a variety of user-defined parameters and all required NAMD files (e.g., restraints and configuration files) are automatically generated. Additionally, the MDFF GUI can be used to set up, run, connect to, and analyze interactive MDFF and xMDFF (Section 3.1) simulations (Section 4). The ‘IMDFF Connect’ tab of the MDFF GUI (Fig. 3B) can be used to monitor the real-time quality of fit of a user-defined selection of the structure, using new e cient cross correlation algorithms (Section 5). This instant feedback on whether or not the structure is fitting well to the target density helps guide a user in manually applying interactive steering forces during the simulations. Since VMD version 1.9.3, the MDFF GUI works in conjunction with the cispeptide, chirality, and TorsionPlot structure checking plugins (Section 2.1). The structure checking plugins can be used to analyze the structure for errors, and then information on the selected residue can be sent directly to the MDFF GUI for automatically setting up a simulation to fix the error.
Figure 3.
MDFF GUI: A user-friendly tool in VMD for setting up MDFF simulations in NAMD. (a) The ‘MDFF Setup’ tab in the GUI is used to generate a complete NAMD configuration file from input parameters specified by the user. (b) The ‘IMDFF Connect’ tab provides a quick way to initiate an interactive MDFF (IMDFF) session on VMD, allowing a user to visualize the ongoing simulation and provide manual steering forces if desired. The ‘IMDFF Connect’ tab can also be used to monitor the real-time quality of fit of a user-defined selection of the structure, using new efficient cross correlation algorithms [92] (Section 5)
3. Starting Structure
Successful structural modeling by MDFF depends on the quality of the intial model. If no or only incomplete structural models from experiment are available, computational approaches must be employed to obtain and refine missing structural information. Several options for building initial models are laid out in this section.
3.1. Low-resolution X-ray crystallography
Investigating the structure of large biomolecular complexes poses a serious challenge to traditional crystallography techniques. Inherent flexibility of large systems, presence of disordered solvent and lipids or ligands often cause crystals to diffract at low resolution. Even overall high-resolution X-ray data may contain low-resolution regions that remain unresolved. When low-resolution X-ray data is available for the starting structure, a variant of MDFF for low-resolution X-ray crystallography (xMDFF) [52] can be used to refine the structure and possibly fill in missing pieces. For use with low resolution X-ray crystallography, the MDFF protocol is modified to work with model-phased densities, from the X-ray crystallography program PHENIX [77, 78], which uses the phases ϕ from a tentative model and the amplitudes |F| from the X-ray diffraction data. Next, the tentative model is flexibly fitted into the electron density map using MDFF. The MDFF-fitted structure provides new phases which, together with the experimental diffraction amplitudes, is used to regenerate the electron density. The fitted structure is then used as an updated model to be driven into the new density map. This process is repeated iteratively until the Rwork and Rfree reach a minimum or become lower than a predefined tolerance.
xMDFF has been successfully applied to solve the structure of a voltage-sensing protein, Ci-VSP [79]. In this case xMDFF was used to resolve uncertainty in the low-resolution (3.6, 4, and 7 Å) data in regard to the placement of the important S4 helix involved in the conformational change required for the function of the protein. xMDFF has also been extended to small molecule crystallography, a field not typically explored by macromolecular crystallography programs [80]. Nevertheless, when small molecules come together in multi-molecular assemblies, their structural characteristics closely resemble biomolecules and, thus, can greatly benefit from macromolecular refinement techniques such as xMDFF. A small abiological molecule, cyanostar, exhibited whole molecule disorder and pushed the limits of small-molecule crystallography. However, a hybrid xMDFF-PHENIX approach was able to successfully refine the cyanostar structure and identify multiple conformations contained within the crystal [80]. Importantly, a key contribution to the success of the project was the development of accurate force field parameters using the force field toolkit (ffTK) plugin in VMD [81].
3.2. Homology Modeling and de novo Structure Prediction
If no X-ray or NMR data is available, structure prediction methods can be used to generate the initial model. Homology modeling, which takes direct advantage of data stored in the PDB, is the most widely-employed approach for structure prediction. The application of homology modeling techniques requires, as a prerequisite, the availability of at least one protein structure of similar amino acid sequence [82]. Working under the generalization that structural homology is highly correlated with sequence homology, models are constructed based on alignment of the two sequences and mapping of the homologous template structure to fill the missing regions of the target structure. The quality of models produced by these means are then evaluated based on specific structural and energetic criteria to eliminate erroneous results [83]. There are various programs available, which are suitable for comparative modeling, like MODELLER [84], SWISS MODEL [82], I TASSER [85], and MUFOLD [86]. In particular, MUFOLD was used to supply an initial model for refinement with xMDFF (Section 3.1) to obtain the structure of a voltage-sensing protein [79].
An alternative approach, which also uses knowledge from the PDB, involves building a local fragment library based on the target amino acid residue sequence. Implemented in Rosetta [87], this strategy uses a “Monte Carlo” method guided by a knowledge-based scoring function to exchange and place the fragments into a partial model [88]. Many of the structures deposited in the PDB have missing coordinates for inserted and terminal regions, as they are usually highly flexible or disordered. It has been shown that MDFF can be used to improve sampling and overcome conformational traps that can befall Rosetta by combining Rosetta and MDFF in an iterative protocol [89].
4. Interactive MDFF
In certain situations, automated protocols using MDFF fail to produce a final fitted model with a good quality of fit to the cryo-EM density. For example, an initial model may be required to undergo a large conformational change during the course of the fitting, which will take certain parts of the structure through regions of the density to which it does not belong. Due to the nature of the forces computed from the cyro-EM derived potential, the structure will try to fit to those incorrect regions of the density because the atoms are attracted in the present MDFF procedure to density indiscriminately as the density-induced potential does not incorporate further information, like contextual information. To avoid this problem of becoming trapped, MDFF can be run interactively.
The interactive feature allows a user to manipulate the target structure during the MDFF simulation by manually pulling it to the desired regions of density [90, 22]. Forces are applied to selected atoms or residues using a VMD session that is connected to the running MDFF simulation in NAMD. After selecting the atom or residue in the VMD display window, a user drags the cursor in the desired direction to move the selected component. A force acting on the selected component in the direction denoted by the user with a magnitude relative to the distance the cursor moved is then added to the MD simulation.
Density maps may also contain noisy, washed-out regions in which proper placement of the structure may be ambiguous. Interactive MDFF can be employed to integrate user expertise into the fitting process for ambiguous cases where automated MDFF may fail. For example, interactive MDFF has been used in modeling the ribosome to remove clashes amongst protein and RNA atoms [91]. Particularly helpful in such cases is a new parallel implementation of cross-correlation analysis in VMD [92] (Section 5) that provides a real-time quality of fit estimate during interactive simulations (Fig 3B). A new MDFF graphical user interface released in VMD 1.9.2 (Section 2.4) makes setting up, running, and analyzing MDFF simulations for interactive usage easy.
5. Analysis
An important step in any hybrid fitting method like MDFF is the evaluation of the the final structure. The quality of the strucure itself is commonly measured using MolProbity [93] which assesses structure quality based on various statistics such as Ramachandran and rotamer outliers and steric clashes. Another important factor in judging the quality of an MDFF-derived structure is its fit to the cryo-EM density. One of the most common scoring methods in this regard is the cross correlation coefficient between experimental density map and a density map calculated from the fitted structure.
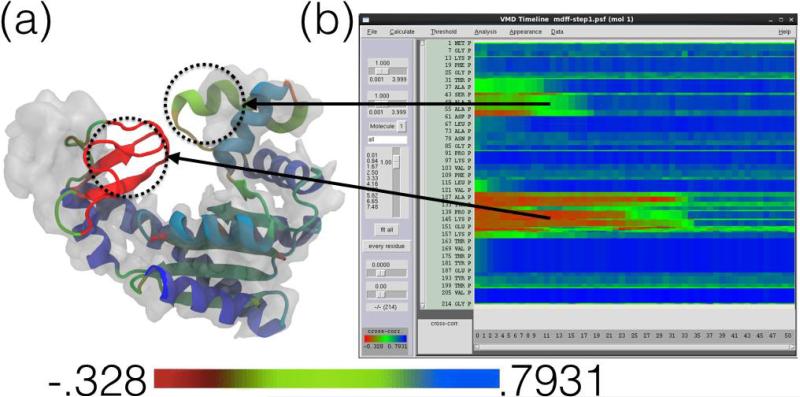
A single global correlation value for the entire protein can be calculated, giving a coarse overall evaluation of the model. However, global cross correlation analysis is prone to producing false-positive information and the result is inherently degenerate: two different structures can be fitted into a density map that produce the same cross-correlation coefficients (ccc). Even though the ccc can be close to 1 (perfect fit), the fitted structure could be locally distorted. Instead of just using the global ccc, it is often useful to calculate the correlation at a finer decomposition, for example per-residue, revealing which parts of the structure are fitted well and which parts might require additional fitting or investigation. Fast parallel CPU and GPU algorithms make the routine computation of local ccc feasible, even for large structures and long fitting trajectories [92]. The Timeline analysis plugin of VMD (Fig. 4) can be used to quickly calculate and visualize local cross correlations of MDFF trajectories [92]. In the default analysis scheme, the structure is split into contiguous sections of secondary structure which are used to calculate local cross correlations for each section independently. The heatmap-style 2-D matrix plot provided by Timeline displays the time dimension horizontally and the structure-component dimension vertically (e.g., residues). The plot is zoomable and connected interactively to the VMD display window, so that when a specific residue at a specific frame has just been selected, VMD will display the neighborhood of the residue in this frame and permit any VMD style viewing of the structure. The features described allow users to quickly inspect the correlation analysis to locate poorly fitted parts of the structure. Additionally, Fourier shell correlation (FSC) which is often used to calculate the resolution of a cryo-EM density, can also be used to judge the quality of a model-to-map-fit. In case of the gold standard method of FSC, which utilizes two independent half maps [94], a cross-validation protocol can be followed during model refinement [95, 96] in order to identify possible overfitting. This protocol involves fitting a model to one half map while calculating the real-space or Fourier shell correlations with respect to the other half-map in a manner that is similar to the Rfree concept in crystallography [97].
Figure 4.
Timeline local cross correlation analysis (adapted from [92]). The Timeline plugin in VMD provides analysis and visualization of local structural properties such as cross correlation [92]. The structure in (a) is in the process of being fitted to the corresponding map in an MDFF simulation and is colored by the per-secondary-structure-component local cross correlation analysis (red is worst correlation, green is intermediate and blue is best). In this analysis scheme, the structure is split into contiguous sections of secondary structure which are used to calculate local cross correlations for each section independently. The Timeline window (b) shows the corresponding plot of local cross correlation of individual residues (y-axis) over time (x-axis) during the simulation. For example, two regions of relatively low correlation (colored red and green) have been highlighted in (a) and matched to the corresponding parts on the Timeline plot in (b). Both regions exhibit low cross correlation due to their varying degrees of distance outside of the density, but are fit as the MDFF simulation progresses, as evidenced in the Timeline plot.
There are many different strategies that can be employed to fix poorly fitting regions of a structure. Such strategies include varying simulation parameters such as lowering the scaling factor or increasing the temperature to improve sampling, using a different fitting methodology such as interactive MDFF or others discussed in the preceding sections, and selectively fitting regions of low local cross correlation while restraining other regions. However, the user is cautioned that poor fitting may be the result of missing or poorly resolved densities in the map, which limits the refinement of the structure.
Even though high-resolution cryo-EM data are becoming more readily obtainable, resolution is not always uniform throughout a map. Flexible regions of the structure still produce local resolutions lower than that of the overall map, as evaluated with tools such as ResMap [98]. MDFF protocols can be adjusted to account for such local variations and better inform the process of model validation. For example, if ResMap analysis shows that certain residues reside in low-resolution regions of the density, the per-atom weighting factor applied to the forces derived from the density can be lowered. Adjusting to a lower weight reduces the strength of the coupling to the map, allowing for greater flexibility and improved conformational sampling of the low-resolution region of density. Local resolution analysis can be especially important for determining the parts of a high-resolution map that realistically contain side chain information and the parts that do not, preventing over-interpretation of the latter.
6. Conclusion
MDFF is a simple and intuitive, yet powerful, method for fitting structures to EM maps while respecting inter-atomic interactions which determine local structure. In this article, we have laid out characteristics of structures and density maps that users must be mindful of in practical applications of MDFF. The corresponding strategies provided are by no means exhaustive and further development of MDFF will likely improve the accuracy and applicability of the method.
As a tool for combining structural and density information, MDFF can easily be adapted to address a variety of circumstances faced in hybrid structure analysis. A large part of MDFF's versatility is inherited from known MD techniques (restraints, solvent models, interactive MD) as well as traditional modelling approaches (homology, de novo, structure prediction). Thus, developments in MD and molecular modelling in general present opportunities to improve MDFF to suit the needs of the structural biology community.
Future development in MDFF will also proceed in tandem with advances in the cryo-EM field. In particular, the increasing resolution of EM maps presents a challenge to MDFF. At the time of MDFF's inception, cryo-EM maps were typically in the low resolution range of 10 – 25 Å. Advances in the cryo-EM field over the years have led to dramatic increases in resolution. Most recently, high-resolution EM maps (< 5 Å) have emerged [1, 2, 3, 4, 5], placing cryo-EM technology alongside X-ray crystallography at the forefront of molecular structure determination. For example, cryo-EM maps of TRPV1 [1](3.4 Å) and β-galactosidase [2, 3](3.2 and 2.2 Å) have been resolved and used to construct atomic structures de novo.
The emergence of high resolution cryo-EM maps poses a challenge to MDFF. If maps have such high resolutions as to enable the construction of accurate atomic structures de novo [99, 100, 101], then structure refinement techniques like MDFF would become unnecessary. However, the production of high resolution maps continues to be a difficult undertaking and the resolutions of produced maps will continue, in the foreseeable future, to fall within a broad spectrum, only a fraction of which being amenable to straight interpretation by atomic resolution structures. Furthermore, EM maps, including high-resolution ones, typically do not have a uniform local resolution throughout the subject molecule, so that low-resolution regions, such as flexible exterior segments, will still require refinement by means of MDFF or other methods.
An adaptation of MDFF for high-resolution EM maps is presently being developed. The protocol for fitting to high-resolution maps involves the use of a low-pass or Gaussian filter that can be applied to the cryo-EM density map to smooth the resulting potential energy function that acts in the MDFF simulation, as previously proposed in [16]. Such a filter removes steep wells commonly found in high-resolution densities in which the structure can become trapped. A structure fit to a smoothed density can then be used in a subsequent MDFF simulation using the original, high-resolution, map for further refinement. The ease of conceptualizing and implementing such adaptations is an important feature that underpins the usefulness of MDFF.
7. Tutorials
Instruction on the practical usage of MDFF and all of the features discussed in this article is available in a series of tutorials. The basic MDFF method (Section 2) in vacuum and in explicit solvent (Section 2.3), application of restraints (Section 2.2), the MDFF graphical user interface (Section 2.4), interactive MDFF (Section 4), xMDFF (Section 3.1), and Timeline analysis (Section 5) are covered in the tutorial found at http://www.ks.uiuc.edu/Training/Tutorials/science/mdff/tutorial_mdff-html/. The tutorial for structure checking (Section 2.1) can be found at http://www.ks.uiuc.edu/Training/Tutorials/science/structurecheck/tutorial_structurecheck-html/. The tutorial for the combined use of ROSETTA and MDFF (Section 3.2) to build complete initial models can be found at http://www.ks.uiuc.edu/Training/Tutorials/science/rosetta-mdff/rosetta-mdff-tutorial-html/. Additional tutorials covering more general topics including use of NAMD, VMD, and Timeline can be found at http://www.ks.uiuc.edu/Training/Tutorials/.
* Cryo-EM is an important tool for structure determination of biomolecular complexes.
* Hybrid computational methods combine data from cryo-EM and X-ray crystallography.
* MDFF is a highly adaptable method for obtaining atomic structures from cryo-EM data.
* New MDFF features developed to overcome challenges faced in molecular modeling.
Acknowledgements
This work has been supported by grants NIH 9P41GM104601, NIH 5R01GM098243-02, and NIH U54GM087519 from the National Institutes of Health. The authors also acknowledge the Beckman Postdoctoral Fellowship program supporting A. Singharoy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. doi:10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S. Structure of β-galactosidase at 3.2 Å resolution obtained by cryo-electron microscopy. Proc. Natl. Acad. Sci. USA. 2014;111(32):11709–11714. doi: 10.1073/pnas.1402809111. doi:10.1073/pnas.1402809111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JL, Subramaniam S. 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348(6239):1147–1151. doi: 10.1126/science.aab1576. doi:10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H. Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by cs-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. doi:10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 5.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. NatureIn press. doi: 10.1038/nature14896. doi:10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman MS. Restrained real-space macromolecular atomic refinement using a new resolution-dependent electron-density function. Acta Cryst. A. 1995;51:69–80. doi: 10.1107/S0108767394007130. [Google Scholar]
- 7.Roseman AM. Docking structures of domains into maps from cryo-electron microscopy using local correlation. Acta Cryst. D. 2000;56:1332–1340. doi: 10.1107/s0907444900010908. doi:10.1107/S0907444900010908. [DOI] [PubMed] [Google Scholar]
- 8.Wriggers W, Chacón P. Modeling tricks and fitting techniques for multiresolution structures. Structure. 2001;9:779–788. doi: 10.1016/s0969-2126(01)00648-7. doi:10.1016/S0969-2126(01)00648-7. [DOI] [PubMed] [Google Scholar]
- 9.Schröder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. doi:10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. doi:10. 1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMaio F, Song Y, Li X, Brunner MJ, Xu C, Conticello V, Egelman E, Marlovits TC, Cheng Y, Baker D. Atomic-accuracy models from 4.5 Å cryoelectron microscopy data with density-guided iterative local refinement. Nat. Methods. doi: 10.1038/nmeth.3286. doi:10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jolley CC, Wells SA, Fromme P, Thorpe MF. Fitting low-resolution cryo-EM maps of proteins using constrained geometric simulations. Biophys. J. 2008;94:1613–1621. doi: 10.1529/biophysj.107.115949. doi:10.1529/biophysj.107.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tama F, Miyashita O, Brooks CL., III Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J. Struct. Biol. 2004;147:315–326. doi: 10.1016/j.jsb.2004.03.002. doi:10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Orzechowski M, Tama F. Flexible fitting of high-resolution X-ray structures into cryo electron microscopy maps using biased molecular dynamics simulations. Biophys. J. 2008;95(12):5692–5705. doi: 10.1529/biophysj.108.139451. doi:10.1529/biophysj.108.139451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman MS, Trzynka A, Chapman BK. Atomic modeling of cryo-electron microscopy reconstructions - Joint refinement of model and imaging parameters. J. Struct. Biol. 2013;182:10–21. doi: 10.1016/j.jsb.2013.01.003. doi:10.1016/j.jsb.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. doi:10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular Dynamics Flexible Fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. doi:10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl. Acad. Sci. USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. doi:10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gumbart J, Trabuco LG, Schreiner E, Villa E, Schulten K. Regulation of the protein-conducting channel by a bound ribosome. Structure. 2009;17:1453–1464. doi: 10.1016/j.str.2009.09.010. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache J-P, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. doi:10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trabuco LG, Schreiner E, Eargle J, Cornish P, Ha T, Luthey-Schulten Z, Schulten K. The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. J. Mol. Biol. 2010;402:741–760. doi: 10.1016/j.jmb.2010.07.056. doi:10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gumbart J, Schreiner E, Trabuco LG, Chan K-Y, Schulten K. Viewing the mechanisms of translation through the computational microscope. In: Frank J, editor. Molecular Machines in Biology. Cambridge University Press; 2011. pp. 142–157. Ch. 8. doi:10.1017/CBO9781139003704.010. [Google Scholar]
- 24.Gumbart J, Schreiner E, Wilson DN, Beckmann R, Schulten K. Mechanisms of SecM-mediated stalling in the ribosome. Biophys. J. 2012;103:331–341. doi: 10.1016/j.bpj.2012.06.005. doi: 10.1016/j.bpj.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frauenfeld J, Gumbart J, van der Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. doi:10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agirrezabala X, Schreiner E, Trabuco LG, Lei J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. Structural insights into cognate vs. near-cognate discrimination during decoding. EMBO J. 2011;30:1497–1507. doi: 10.1038/emboj.2011.58. doi:10.1038/emboj.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Trabuco LG, Schulten K, Frank J. Molecular dynamics of EF-G during translocation. Proteins: Struct., Func., Bioinf. 2011;79:1478–1486. doi: 10.1002/prot.22976. doi:10.1002/ prot.22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Atkinson GC, Thakor NS, Allas Ü, Lu C, Chan K-Y, Tenson T, Schulten K, Wilson KS, Hauryliuk V, Frank J. Mechanism of tetracycline resistance by ribosomal protection protein Tet(O) Nat. Commun. 2013;4:1477. doi: 10.1038/ncomms2470. doi:10.1038/ncomms2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang K, Wang L, Liu Y, Chan K-Y, Pang X, Schulten K, Dong Z, Sun F. Flexible interwoven termini determine the thermal stability of thermosomes. Protein & Cell. 2013;4:432–444. doi: 10.1007/s13238-013-3026-9. doi:10.1007/s13238-013-3026-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickles S, Singharoy A, Andreani J, Seemayer S, Bischoff L, Berninghausen O, Soeding J, Schulten K, van der Sluis E, Beckmann R. structural model of the active ribosome-bound membrane protein insertase YidC. A. eLife. 3:e03035, 17. doi: 10.7554/eLife.03035. doi:10.7554/eLife.03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsin J, Gumbart J, Trabuco LG, Villa E, Qian P, Hunter CN, Schulten K. Protein-induced membrane curvature investigated through molecular dynamics flexible fitting. Biophys. J. 2009;97:321–329. doi: 10.1016/j.bpj.2009.04.031. doi:10.1016/j.bpj.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sener MK, Hsin J, Trabuco LG, Villa E, Qian P, Hunter CN, Schulten K. Structural model and excitonic properties of the dimeric RC-LH1-PufX complex from Rhodobacter sphaeroides. Chem. Phys. 2009;357:188–197. doi: 10.1016/j.chemphys.2009.01.003. doi:10.1016/j. chemphys.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H, Hsin J, Liu Y, Selvin PR, Schulten K. Formation of salt bridges mediates internal dimerization of myosin VI medial tail domain. Structure. 2010;18:1443–1449. doi: 10.1016/j.str.2010.09.011. doi:10.1016/j.str.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassidy CK, Himes BA, Alvarez FJ, Ma J, Zhao G, Perilla JR, Schulten K, Zhang P. CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling. eLife. doi: 10.7554/eLife.08419. 10.7554/eLife.08419, (50 pages). doi: 10.7554/eLife.08419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Xu F, Liu J, Gao B, Liu Y, Zhai Y, Ma J, Zhang K, Baker TS, Schulten K, Zheng D, Pang H, Sun F. Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS Pathog. 9:e1003132, 14. doi: 10.1371/journal.ppat.1003132. doi:10.1371/journal.ppat.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. doi:10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C, Perilla JR, Ning J, Lu M, Hou G, Ramalho R, Bedwell G, Byeon I-J, Ahn J, Shi J, Gronenborn A, Prevelige P, Rousso I, Aiken C, Polenova T, Schulten K. Cyclophilin A stabilizes HIV-1 capsid through a novel non-canonical binding site-Submitted. doi: 10.1038/ncomms10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R. α-helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2010;17:313–317. doi: 10.1038/nsmb.1756. doi:10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- 39.Guo Q, Yuan Y, Xu Y, Feng B, Liu L, Chen K, Sun M, Yang Z, Lei J, Gao N. Structural basis for the function of a small GTPase RsgA on the 30S ribosomal subunit maturation revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. USA. 2011;108:13100–13105. doi: 10.1073/pnas.1104645108. doi:10.1073/pnas.1104645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dinda G, Franckenber S, Marquz V, Mielk T, Thomm M, Berninghausen O, Beatrix B, Södina J, Westho E, Wilso DN, Beckmann R. Localization of eukaryote-specific ribosomal proteins in a 5.5 Åcryo-EM map of the 80S eukaryotic ribosome. Proc. Natl. Acad. Sci. USA. 2011;107:19754–19759. doi: 10.1073/pnas.1010005107. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker T, Armache J-P, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. doi:10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 42.Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, III, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. doi:10.1126/ science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache J-P, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, Karcher A, Thomm M, Hopfner K-P, Green R, Beckmann R. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482(7386):501–506. doi: 10.1038/nature10829. doi:10.1038/ nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park E, Ménétret J-F, Gumbart JC, Ludtke SJ, Li W, Whynot A, Rapoport TA, Akey CW. Structure of the SecY channel during initiation of protein translocation. Nature. 2014;506:102–106. doi: 10.1038/nature12720. doi:10.1038/nature12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gogala M, Becker T, Beatrix B, Armache J-P, Barrio-Garcia C, Berninghausen O, Beckmann R. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature. 2014;506(7486):107–110. doi: 10.1038/nature12950. doi: 10.1038/nature12950. [DOI] [PubMed] [Google Scholar]
- 46.Lorenz M, Holmes KC. The actin-myosin interface. Proc. Natl. Acad. Sci. USA. 2010;107:12529–12534. doi: 10.1073/pnas.1003604107. doi:10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wollmann P, Cui S, Viswanathan R, Berninghausen O, Wells MN, Moldt M, Witte G, Butryn A, Wendler P, Beckmann R, Auble DT, Hopfner K-P. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature. 2011;475:403–407. doi: 10.1038/nature10215. doi:10.1038/nature10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lasker K, Förster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26s proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. USA. 2012;109(5):1380–1387. doi: 10.1073/pnas.1120559109. doi:10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unverdorben P, Beck F, Śledź P, Schweitzer A, Pfeifer G, Plitzko JM, Baumeister W, Förster F. Deep classification of a large cryo-em dataset defines the conformational landscape of the 26s proteasome. Proc. Natl. Acad. Sci. USA. 2014;111(15):5544–5549. doi: 10.1073/pnas.1403409111. doi:10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bharat TAM, Castillo Menendez LR, Hagen WJH, Lux V, Igonet S, Schorb M, Schur FKM, Kräusslich H-G, Briggs JAG. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc. Natl. Acad. Sci. USA. 2014;111(22):8233–8238. doi: 10.1073/pnas.1401455111. doi: 10.1073/pnas.1401455111. URL http://www.pnas.org/content/111/22/8233.abstract . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schur FKM, Hagen WJH, Rumlova M, Ruml T, Muller B, Krausslich H-G, Briggs JAG. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature. 2015;517:505–508. doi: 10.1038/nature13838. URL http://dx.doi.org/10.1038/nature1383810.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 52.McGreevy R, Singharoy A, Li Q, Zhang J, Xu D, Perozo E, Schulten K. xMDFF: Molecular dynamics flexible fitting of low-resolution X-Ray structures. Acta Cryst. D. 2014;70:2344–2355. doi: 10.1107/S1399004714013856. doi:10.1107/S1399004714013856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan K-Y, Trabuco LG, Schreiner E, Schulten K. Cryo-electron microscopy modeling by the molecular dynamics flexible fitting method. Biopolymers. 2012;97:678–686. doi: 10.1002/bip.22042. doi:10.1002/bip.22042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schreiner E, Trabuco LG, Freddolino PL, Schulten K. Stereochemical errors and their implications for molecular dynamics simulations. BMC Bioinform. 2011;12:190. doi: 10.1186/1471-2105-12-190. doi:10.1186/1471-2105-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. doi:10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells D, Abramkina V, Aksimentiev A. Exploring transmembrane transport through alpha-hemolysin with grid-steered molecular dynamics. J. Chem. Phys. 2007;127:125101–125101–10. doi: 10.1063/1.2770738. doi:10.1063/1.2770738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan K-Y, Gumbart J, McGreevy R, Watermeyer JM, Sewell BT, Schulten K. Symmetry-restrained flexible fitting for symmetric EM maps. Structure. 2011;19:1211–1218. doi: 10.1016/j.str.2011.07.017. doi:10.1016/j.str.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. doi:10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 59.Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lam H, Oh D-C, Cava F, Takacs CN, Clardy J, de Pedr MA, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;235(5947):1552–1555. doi: 10.1126/science.1178123. doi:10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. doi:10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 62.on Biochemical Nomenclature I-IC. Abbreviations and symbols for the description of the conformation of polypeptide chains. J. Mol. Biol. 1970;52:1–17. doi:10.1016/ 0022-2836(70)90173-7. [PubMed] [Google Scholar]
- 63.Pal D, Chakrabarti P. Cis peptide bonds in proteins: residues involved, their conformations, interactions and locations. J. Mol. Biol. 1999;294:271–288. doi: 10.1006/jmbi.1999.3217. doi:doi: 10.1006/jmbi.1999.3217. [DOI] [PubMed] [Google Scholar]
- 64.Jabs A, Weiss MS, Hilgenfeld R. Non-proline cis peptide bonds in proteins. J. Mol. Biol. 1999;286:291–304. doi: 10.1006/jmbi.1998.2459. doi:doi:10.1006/jmbi.1998.2459. [DOI] [PubMed] [Google Scholar]
- 65.Croll TI. The rate of cis–trans conformation errors is increasing in low-resolution crystal structures. Acta Cryst. D. 2015;71(3):706–709. doi: 10.1107/S1399004715000826. doi:10.1107/S139900471500082. [DOI] [PubMed] [Google Scholar]
- 66.MacKerell AD, Jr., Bashford D, Bellott M, Dunbrack RL, Jr., Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher IWE, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. doi:10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 67.Schlitter J, Engels M, Krüger P, Jacoby E, Wollmer A. Targeted molecular dynamics simulation of conformational change – application to the T ↔ R transition in insulin. Mol. Sim. 1993;10(2–6):291–308. doi:10.1080/08927029308022170. [Google Scholar]
- 68.Schlitter J, Engels M, Krüger P. Targeted molecular dynamics: A new approach for searching pathways of conformational transitions. J. Mol. Graph. 1994;12:84–89. doi: 10.1016/0263-7855(94)80072-3. doi:10.1016/0263-7855(94)80072-3. [DOI] [PubMed] [Google Scholar]
- 69.Agirrezabala X, Liao H, Schreiner E, Fu J, Ortiz-Meoz R, Schulten K, Green R, Frank J. Structural characterization of mRNA-tRNA translocation intermediates. Proc. Natl. Acad. Sci. USA. 2012;109:6094–6099. doi: 10.1073/pnas.1201288109. doi:10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358. doi: 10.1126/science.2994218. doi:10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 71.Doyle DA, Cabral JM, Pfuetzer RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. doi:10.1126/science.280. 5360.69. [DOI] [PubMed] [Google Scholar]
- 72.Thuku RN, Weber BW, Varsani A, Sewell BT. Post-translational cleavage of recombinantly expressed nitrilase from Rhodococcus rhodochrous J1 yields a stable, active helical form. FEBS J. 2007;274:2099–2108. doi: 10.1111/j.1742-4658.2007.05752.x. doi:10.1111/j.1742-4658.2007. 05752.x. [DOI] [PubMed] [Google Scholar]
- 73.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79(2):926–935. doi:10.1063/1.445869. [Google Scholar]
- 74.Rhee YM, Pande VS. Solvent viscosity dependence of the protein folding dynamics. J. Phys. Chem. B. 2008;112:6221–6227. doi: 10.1021/jp076301d. doi:10.1021/jp076301d. [DOI] [PubMed] [Google Scholar]
- 75.Tanner DE, Chan K-Y, Phillips J, Schulten K. Parallel generalized Born implicit solvent calculations with NAMD. J. Chem. Theor. Comp. 2011;7:3635–3642. doi: 10.1021/ct200563j. doi: 10.1021/ct200563j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, et al. Near-atomic resolution structural model of the yeast 26s proteasome. Proc. Natl. Acad. Sci. USA. 2012;109(37):14870–14875. doi: 10.1073/pnas.1213333109. doi:10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adams P, Afonine P, Bunkóczi G, Chen V, Davis I, Echols N, Headd J, Hung L, Kapral G, Grosse-Kunstleve R, McCoy A, Moriarty N, Oeffner R, Read R, Richardson D, Richardson J, Terwilliger T, Zwart P. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. doi:10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adams PD, Afonine PV, Bunkczi G, Chen VB, Echols N, Headd JJ, Hung L-W, Jain S, Kapral GJ, Kunstleve RWG, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. The phenix software for automated determination of macromolecular structures. Methods. 2011;55(1):94–106. doi: 10.1016/j.ymeth.2011.07.005. doi:10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Q, Wanderling S, Paduch M, Medovoy D, Singharoy A, McGreevy R, Villalba- Galea C, Hulse RE, Roux B, Schulten K, Kossiakoff A, Perozo E. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat. Struct. Mol. Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. doi:10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singharoy A, Venkatakrishnan B, Liu Y, Mayne CG, Chen C-H, Zlotnick A, Schulten K, Flood AH. Macromolecular crystallography for synthetic abiological molecules: Combining xMDFF and PHENIX for structure determination of cyanostar macrocycles. J. Am. Chem. Soc. 2015;137:8810–8818. doi: 10.1021/jacs.5b04407. doi:10.1021/jacs.5b04407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mayne CG, Saam J, Schulten K, Tajkhorshid E, Gumbart JC. Rapid parameterization of small molecules using the Force Field Toolkit. J. Comp. Chem. 2013;34:2757–2770. doi: 10.1002/jcc.23422. doi:10.1002/jcc.23422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arnold K, Bordoli L, Kopp J, Schwede T. The swiss-model workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. doi:10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 83.Eswar N, Webb B, Marti-Renom MA, Madhusudhan M, Eramian D, Shen M.-y., Pieper U, Sali A. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinf. 2006:5–6. doi: 10.1002/0471250953.bi0506s15. http://dx.doi.org/10.1002/0471140864.ps0209s50 doi:10. 1002/0471140864.ps0209s50. [DOI] [PMC free article] [PubMed]
- 84.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. doi:10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 85.Roy A, Kucukural A, Zhang Y. I-tasser: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. doi:10. 1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang J, Wang Q, Barz B, He Z, Kosztin I, Shang Y, Xu D. MUFOLD: A new solution for protein 3D structure prediction. Proteins: Struct., Func., Bioinf. 2010;78:1137–1152. doi: 10.1002/prot.22634. doi:10.1002/prot.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. Rosetta3: an object-oriented software suite for the simulation and design of macromolecules. Methods in enzymology. 2011;487:545. doi: 10.1016/B978-0-12-381270-4.00019-6. doi:10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaufmann KW, Lemmon GH, DeLuca SL, Sheehan JH, Meiler J. Practically useful: what the rosetta protein modeling suite can do for you. Biochemistry. 2010;49(14):2987–2998. doi: 10.1021/bi902153g. doi:10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lindert S, McCammon JA. Improved cryoem-guided iterative molecular dynamicsrosetta protein structure refinement protocol for high precision protein structure prediction. J. Chem. Theor. Comp. 2015;11(3):1337–1346. doi: 10.1021/ct500995d. doi:10.1021/ct500995d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stone JE, Gullingsrud J, Grayson P, Schulten K. A system for interactive molecular dynamics simulation. In: Hughes JF, Séquin CH, editors. ACM Symposium on Interactive 3D Graphics. ACM SIGGRAPH; New York: 2001. pp. 191–194. 2001. doi: 10.1145/364338.364398. [Google Scholar]
- 91.Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Söding J, Westhof E, Wilson DN, Beckmann R. Cryo-em structure and rrna model of a translating eukaryotic 80s ribosome at 5.5-åresolution. Proc. Natl. Acad. Sci. USA. 2010;107(46):19748–19753. doi: 10.1073/pnas.1009999107. doi:10.1073/pnas. 1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stone JE, McGreevy R, Isralewitz B, Schulten K. GPU-accelerated analysis and visualization of large structures solved by molecular dynamics flexible fitting. Faraday Discuss. 2014;169:265–283. doi: 10.1039/c4fd00005f. doi:10.1039/C4FD00005F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen VB, W. B. A., Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, Jiang W, Ludtke SJ, Medalia O, Penczek PA, Rosenthal PB, Rossmann MG, Schmid MF, Schrder GF, Steven AC, Stokes DL, Westbrook JD, Wriggers W, Yang H, Young J, Berman HM, Chiu W, Kleywegt GJ, Lawson CL. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20–330(2):205–214. doi: 10.1016/j.str.2011.12.014. doi:10.1016/j. str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.DiMaio F, Zhang J, Chiu W, Baker D. Cryo-EM model validation using independent map reconstructions. Prot. Sci. 2013;22(6):865–868. doi: 10.1002/pro.2267. doi:10.1002/pro.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9(9):853–854. doi: 10.1038/nmeth.2115. doi:10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brünger A. The free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. doi:doi:10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 98.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11(1):63–65. doi: 10.1038/nmeth.2727. doi: http://dx.doi.org/10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang RY-R, Kudryashev M, Li X, Egelman EH, Basler M, Cheng Y, Baker D, DiMaio F. De novo protein structure determination from near-atomic-resolution cryoem maps. Nat. Methods. 2015;12(4):335–338. doi: 10.1038/nmeth.3287. doi:10.1038/nmeth.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kudryashev M, Wang RY-R, Brackmann M, Scherer S, Maier T, Baker D, DiMaio F, Stahlberg H, Egelman EH, Basler M. Structure of the type VI secretion system contractile sheath. Cell. 2015;160(5):952–962. doi: 10.1016/j.cell.2015.01.037. doi:10.1016/j.cell.2015. 01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lindert S, Alexander N, Wötzel N, Karakaş M, Stewart PL, Meiler J. Em-fold: De novo atomic-detail protein structure determination from medium-resolution density maps. Structure. 2012;20(3):464–478. doi: 10.1016/j.str.2012.01.023. doi:10.1016/j.str.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]