Abstract
This study integrates gene expression, genotype and drug response data in lymphoblastoid cell lines with transcription factor (TF)-binding sites from ENCODE (Encyclopedia of Genomic Elements) in a novel methodology that elucidates regulatory contexts associated with cytotoxicity. The method, GENMi (Gene Expression iN the Middle), postulates that single-nucleotide polymorphisms within TF-binding sites putatively modulate its regulatory activity, and the resulting variation in gene expression leads to variation in drug response. Analysis of 161 TFs and 24 treatments revealed 334 significantly associated TF–treatment pairs. Investigation of 20 selected pairs yielded literature support for 13 of these associations, often from studies where perturbation of the TF expression changes drug response. Experimental validation of significant GENMi associations in taxanes and anthracyclines across two triple-negative breast cancer cell lines corroborates our findings. The method is shown to be more sensitive than an alternative, genome-wide association study-based approach that does not use gene expression. These results demonstrate the utility of GENMi in identifying TFs that influence drug response and provide a number of candidates for further testing.
Introduction
The field of pharmacogenetics aims to understand the relationship between individual variation at the genetic level and variation in cellular or physiological response to a drug. Rapidly emerging genomic technologies have expanded the scope of analysis to genome-wide levels, simultaneously providing a variety of high-quality data to enable the analysis, including genotype, gene expression and proteomic data, as well as functional annotations from the Encyclopedia of Genomic Elements (ENCODE).1 Advances in the burgeoning field of pharmacogenomics2 have the potential to revolutionize health care by guiding personalized health care for patients via genome sequencing.3
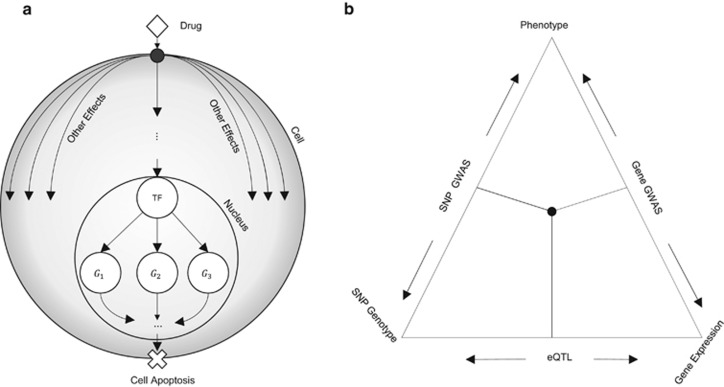
The de facto method for generating biological hypothesis of clinical relevance involves the extraction of pharamacogenomic data from human cell lines that are generalizable and easily manipulated; lymphoblastoid cell lines (LCLs) represent a canonical source with such clear clinical utility.4, 5, 6 A number of studies have analyzed such data sets to relate genotypic variation to drug response,7 revealing important single-nucleotide polymorphisms (SNPs) as well as SNP-carrying genes that are candidates for functional testing. Parallel to the identification of SNPs associated with drug response, there is also considerable interest in characterizing the mechanistic basis of such relationships, that is, pathways and regulatory networks involved in drug response and its variability.8, 9 Identifying systems-level components of the response, such as signaling pathways and transcriptional cascades, can enable discovery of novel drug targets and lead to the realization of ‘precision medicine'.10 Here, we embark upon one such line of enquiry—to identify transcription factors (TFs) whose regulatory activities are associated with cellular response to cytotoxic treatments (Figure 1a), with the expectation that in the future the response may be manipulated by intervening with the function of TF.
Figure 1.
(a) Diagram of how transcription factors (TFs) mediate response to a drug. A drug (diamond) enters the cell and affects multiple cellular processes. One such process involves transport or signal transduction to the nucleus where it alters the transcriptional activity of a TF. Expression of target genes is subsequently altered, potentially resulting in apoptosis. (b) Outline of triangulation procedure proposed in the literature. Each edge of the triangle corresponds to a correlation between two of the three axes of information: drug response, genetic variants and gene expression. Integrative analysis involves intersecting expression quantitative trait locus (eQTL) genes and genome-wide association study (GWAS) genes or eQTL single-nucleotide polymorphisms (SNPs) and GWAS SNPs.
The most widely used statistical method for harnessing pharamacogenomic data to identify biomarkers relevant to drug-induced cellular response is genome-wide association study (GWAS), where SNPs are analyzed for their correlation with drug response across individuals. The multigenic origins of phenotypic variability, correlations between proximally located SNPs and multiple hypothesis correction over millions of candidate markers reduce the ability of GWAS to discover casual SNPs. A number of studies have sought to improve upon the basic GWA approach, for example, by testing subsets of SNPs as opposed to single marker analysis, or combining GWAS associations with prior knowledge of gene networks and pathways.11, 12, 13 We draw inspiration from this emerging paradigm, and attempt to associate drug response variation with multiple SNPs that share a common functional context, viz., that of being located within binding sites of the same TF.
Functional genotypic variants are expected to exert their influence on phenotypic differences at least partly through variation in expression levels of nearby genes.14, 15, 16 A previous study17 argued that if gene expression data are available in addition to genotype and phenotype data on the same cohort, then a statistical approach called ‘mediator analysis' can be employed to discover functional SNPs with greater sensitivity. Gene expression and proteomic data have often been used to predict phenotypes, including drug-induced cellular response.16, 18, 19, 20, 21 Efforts have been made to develop methodologies that integrate both gene expression and genotype information to predict phenotype. One method of achieving this is the following: first conduct a GWAS associating SNPs with the phenotype, and then correlate significant GWAS SNPs with expression of their proximal genes, thereby identifying expression quantitative trait locus (eQTL) SNPs, and finally correlate expression of these eQTL genes with phenotype. This triangulation procedure for integrating genotype, gene expression and phenotype (Figure 1b) data has been used to identify either candidate SNPs or candidate genes for experimentation.4 It has also been used in pharmacogenomics to identify biomarkers and genes related to cisplatin-, etoposide- and radiation-induced cytotoxicity.22, 23, 24 These previous studies motivate us to integrate gene expression data in our search for molecular determinants of drug response variability.
Integration of genotype and expression data in association studies draws our attention to cis-regulatory SNPs that represent a large proportion of individual variability and have been linked to important phenotypes, including diseases.25 A prime difficulty in characterizing regulatory variants is the poor annotation of the noncoding genome, making it difficult to tell neutral from potentially functional cis-variants. Recent community efforts to systematically annotate the regulatory genome, such as the ENCODE project, may alleviate this problem to an extent. However, few existing approaches incorporate ENCODE data, in particular transcription factor-binding site (TFBS) data, into statistical analysis of individual variation at the genotypic and phenotypic levels. The ENCODE consortium analyzed the overlap of disease-associated SNPs from the NHGRI GWAS catalog with TFBS and DNase I hypersensitivity sites.26 A recent study integrated ENCODE data, among other sources of functional data, into a model that selects optimally informative annotation filters to improve SNP association studies.27 Another study concluded that GWAS SNPs embedded in cis-regulatory elements from disease-relevant cell types are likely to function as eQTLs.28 However, these studies do not provide a systematic method for integrating all of the above-mentioned types of genomic data so as to determine candidate regulators of phenotypic variation. Our study aims to address this issue by developing a computational method named GENMi (Gene Expression iN the Middle) that integrates ENCODE TFBS, genotype, gene expression and drug-induced cytotoxicity data in LCLs to quantify the association between a TF and drug response, thereby elucidating putative regulators responsible for observed cellular responses to drugs.
Materials and methods
Data collection
We obtained genotype, gene expression and drug response data on 95 Han-Chinese, 96 Caucasian and 93 African-American lymphoblastoid cell lines from the Coriell Cell Repository (Camden, NJ, USA). Of these 284 individuals, 176 were females and 108 males, with an average age of 33.44 years. The genotype data consisted of 1 344 658 germline SNPs. Quality control analysis had already been performed on these SNPs, removing those that deviated from Hardy–Weinberg equilibrium, were called <95% of the time or had minor allele frequencies <5%. Gene expression data consisted of 54 613 Affymetrix U133 Plus 2.0 Gene-ChIP (Santa Clara, CA, USA) probes assayed for the 284 individuals, with raw expression data being transformed using QUOTE GC robust multiarray averaging. Genotype and gene expression data are available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under SuperSeries accession no. GSE24277. These data were published in a study by Niu et al.24
Drug response data were derived from dosage–response curves of 24 cytotoxic treatments shown in Supplementary Table 1. The phenotype, called EC50, represents the concentration at which the drug reduces the population of LCL cells to half of the initial population. Data for 15 of the 24 treatments have been published in analysis in various studies conducted on these cell lines; in particular, MPA, NAPQI, 6MP, 6TG, ara-C, oxaliplatin, carboplatin, cisplatin, docetaxel, everolimus, gemcitabine, paclitaxel, metformin, radiation and rapamycin have been analyzed in published studies.29, 30, 24, 31, 32, 33, 34, 35, 36 Response data for the following nine drugs have not been published: arsenic, cladribine, doxorubicin, epirubicin, fludarabine, hypoxia, MTX, TCN and TMZ. Cytotoxicity assay was performed for every one of these drugs using the LCL panel. After initial optimization, cells were treated with a range of concentrations for any given drug tested, followed by incubation for 48 to 72 h. MTS cytotoxicity assays were then performed using Cell Titer 96 AQueous Non-Radioactive Cell Proliferation Assay kit (Promega Corporation, Madison, WI, USA), followed by absorbance measurement at 490 nm in a Safire2 microplate reader (Tecan AG, Switzerland). Cytotoxicity phenotypes were determined by the best fitting curve using the R package ‘drc' (dose–response curve) (https://cran.r-project.org/web/packages/drc/drc.pdf) based on a logistic model.
Experimental data on TF binding were retrieved from the ENCODE project;26 specifically, the clustered ChIP (version 3) tracks across 91 were used. ChIP tracks consisted of the clustered ChIP peaks of 161 TFs. TF ChIP high occupancy target regions were removed as described in Supplementary Note 1.
Gene mappings to the Affymetrix arrays were obtained for the Affymetrix Human Genome U133 Plus 2.0 array. ENSEMBL gene symbols were used as the gene reference of choice: we used 55 038 ENSEMBL gene symbols that were annotated with at least one ENSEMBL exon. Of the 54 613 probes assayed on the HG U133 Plus 2.0 array, 37 677 mapped to at least one of the 55 038 ENSEMBL gene symbols.
Human triple-negative breast cancer cell lines, BT549 and MDA-MB231, were obtained from the American Type Culture Collection (Manassas, VA, USA). BT549 cells were cultured in RPMI-1640 containing 10% fetal bovine serum. MDA-MB-231 cells were cultured in L-15 medium containing 10% fetal bovine serum.
Experimental methods
RNA interference and qRT-PCR
The small interfering RNAs (siRNAs) for the candidate transcript factors and negative control siRNA were purchased from Dharmacon (Lafayette, CO, USA). Reverse transfection was performed in 96-well plates. Specifically, 3000–4000 cells were mixed with 0.1 ml of lipofectamine RNAi-MAX reagent (Invitrogen, Grand Island, NY, USA) and 10 nM siRNA for each experiment.
Total RNA was isolated from cultured cells transfected with control or specific siRNAs with the Qiagen RNeasy kit (QIAGEN, Valencia, CA, USA), followed by real-time quantitative reverse transcription-PCR (qRT-PCR) performed with the one-step, Brilliant SYBR Green qRT-PCR master mix kit (Stratagene, Santa Clara, CA, USA). Specifically, primers purchased from QIAGEN were used to perform qRT-PCR using the Stratagene Mx3005P Real-Time PCR detection system (Stratagene). All experiments were performed in triplicate with β-actin as an internal control. Reverse-transcribed Universal Human reference RNA (Stratagene) was used to generate a standard curve. Control reactions lacked RNA template.
MTS cytotoxicity assay
Epirubicin, doxorubicin, paclitaxel and docetaxel were purchased from Sigma-Aldrich (Milwaukee, WI, USA). Drugs were dissolved in dimethyl sulfoxide and aliquots of stock solutions were frozen at −80 °C. Cell proliferation assays were performed in triplicate at each drug concentration. Cytotoxicity assays with the lymphoblastoid and tumor cell lines were performed in triplicate at each dose. Specifically, 90 μl of cells (5 × 103 cells per ml) were plated into 96-well plates (Corning, Corning, NY, USA)37 and were treated with 10 μl of epirubicin or doxorubicin at final concentrations of 0, 0.0156, 0.03125, 0.0625, 0.125, 0.25, 0.55, 1 and 2 μmol l−1. Similarly, cells were treated with paclitaxel or docetaxel at 0, 0.01, 0.1, 1, 10, 50, 100, 1000 and 5000 nmol l−1. After incubation for 72 h, 20 μl of CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay solution (Promega Corporation) was added to each well. Plates were read in a Safire2 plate reader (Tecan AG).
Statistical analysis of cytotoxicity data
Significance of the half-maximal inhibitory concentration (IC50) values between negative control siRNA and gene-specific siRNA was determined by the two-tailed unpaired t-test.
Statistical testing for TF role in varying response to cytotoxicity treatment (GENMi)
We operationally defined the cis-regulatory domain of a gene as the 50-kb sequence upstream of the gene's transcription start site. For any given TF, we assigned a ‘TF-specific cis-eQTL score' to each ENSEMBL gene as follows:
Retain all SNPs located within TF ChIP peaks in the cis-regulatory domain of the gene.
Retrieve all HG U133 Plus 2.0 probes mapped to the gene.
Compute the correlation (eQTL) for each (SNP, probe) combination. This is the correlation coefficient, across all 284 cell lines, between the SNP genotype and the probe's expression value. Also, compute the P-value corresponding to this correlation coefficient.
Use the coefficient of determination of the single best eQTL among all (SNP, probe) combinations as the ‘TF-specific cis-eQTL score' of the gene. Retain probe and SNP identities contributing to the best eQTL for further analysis.
We then considered the set of 400 genes with the strongest TF-specific cis-eQTL scores (we additionally required that a gene included in this set have a TF-specific cis-eQTL P-value ≤0.05, so the cardinality of the set may be ≤400). These may be thought of as genes where genotypic variation in the TFBS correlates with variation in the gene's expression, potentially implicating the TF in their expression variation. Although SNPs outside of TFBS can affect TF regulation of gene regulation38 (for example, SNPs in cofactor binding sites), we limit analysis to eQTL SNPs enveloped within TFBS, as the functional effect of SNPs distant from the TFBS is not well understood. We therefore refer to this gene set as the ‘eQTL gene set' of TF. In addition, we required that in order for us to analyze the role of TF in drug response, there should be at least 15 genes with strong eQTLs within binding sites of that TF, that is, the ‘eQTL gene set' of TF should have at least 15 genes, as per recommendations accompanying the Gene Set Enrichment Analysis (GSEA) tool. This resulted in the analysis being restricted to 114 of the 161 TFs for which ChIP data were available.
To determine whether the genotype expression associations identified above are linked to the varying response to a given cytotoxic treatment (drug or radiation), we correlated each probe's expression value with the EC50 value of the treatment, and ranked all genes by this correlation coefficient. (We used the best correlation coefficient among multiple probes for each gene.) Using GSEA,39 we tested for statistical association between this ranked list and the eQTL gene set of TF defined above. The GSEA procedure reported a P-value that served as the basis for inferring a role for the TF in individual variations in response to the specific cytotoxic treatment. As 114 TFs were separately tested in this manner, we relied upon the false discovery rate (FDR) values reported by GSEA to correct for multiple hypothesis testing with each treatment. We refer to this method as GENMi.
Code availability
Scripts used in GENMi, with appropriate documentation, will be freely available upon publication at the following location: veda.cs.uiuc.edu/GENMi.
Results
Integrating genotype, phenotype and expression data with profiles of TF binding
The relationship between genotype and response to cytotoxic treatments is expected to be mediated, at least in part, by regulation of gene expression 17 (Figure 1a). Inclusion of expression data in the LCL data set allows us to investigate this hypothesis by simultaneously examining the correlation between genotype and expression and that between expression and phenotype (Figure 1b). We hypothesized that SNPs manifesting the genotype–expression correlation (eQTLs) should reside within binding sites of TFs that orchestrate the transcriptional programs activated or deactivated by the treatment, and that these SNPs influence phenotypic variation through their effect on gene expression.40
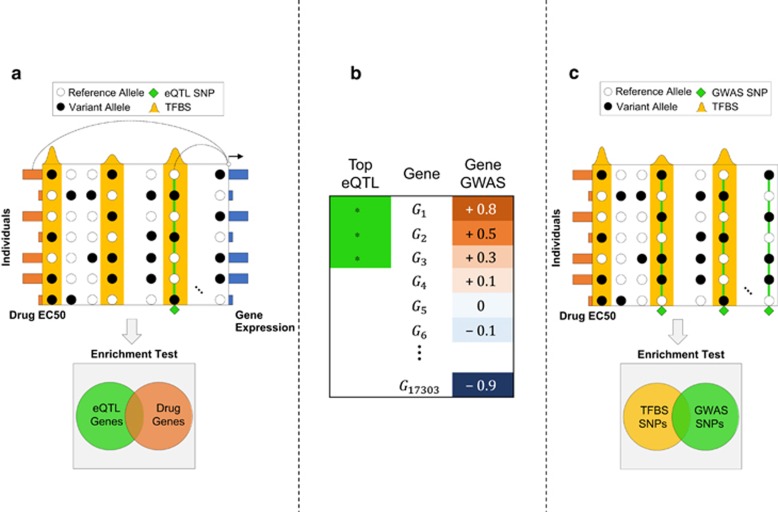
Our goal was to test the possibility that a TF mediates the individual-to-individual variation of gene expression that in turn leads to variation in cytotoxicity across cell lines. To this end, we sought examples (Figure 2a) where a SNP inside the TFBS (ChIP peak) correlates with the neighboring gene's expression (cis-eQTL41), and that gene's expression correlates with drug response. To formalize this idea as a statistical test (Figure 2b), we (1) first ranked genes by the correlation between their expression and the phenotype, (2) separately identified a fixed number of genes (400 in tests reported here) that bear the strongest cis-eQTLs within the TFBS and (3) finally used GSEA39 to test whether the latter set of genes (step 2) is enriched near the top of the former ranked list (step 1). In other words, we asked: when using the TFBS as the context, is genotype-to-expression correlation reflected in expression-to-phenotype correlation? Note that step (1) is performed independently of the TF, and does no hypothesis testing; it simply ranks genes by their (expression) correlation with phenotype. Steps (2) and (3) test whether the cis-eQTLs induced by a TF appear significantly frequently near the top of this phenotype-associated gene list, thus suggesting a role for that TF in the association between genotypic and phenotypic variation, with expression variation in the middle. We call this entire procedure ‘GENMi'.
Figure 2.
(a) The GENMi (Gene Expression iN the Middle) method. Shown is the 50 kb upstream region of a single gene, with transcription factor-binding site (TFBS; ChIP peaks) in yellow, single-nucleotide polymorphisms (SNPs; circles) and their allelic state (black or white) in a sample of seven individuals, as well as gene expression (blue bars on right) and drug response EC50 values (orange bars on left) in these individuals. The gene is scored in two ways : correlation of the best expression quantitative trait locus (eQTL) SNP (green diamond) coincident with a TFBS and correlation of the gene's expression with drug response (these two correlations are illustrated by lines connecting the two correlated variables). Integrating over all genes, testing the overlap between strongest eQTL genes and genes associated with drug response (enrichment test, bottom) quantifies the extent to which a TF is associated with drug response via cis-regulatory mechanisms. (b) Cartoon of Gene Set Enrichment Analysis (GSEA) used as the enrichment test in GENMi. Genes are ranked according to their correlation with drug response (‘gene GWAS'). The analysis looks at the extent to which a given gene set (in this case genes carrying the strongest eQTLs coincident with the TFBS) are enriched near the top or bottom of the ranked list. Here, the gene set is strongly associated with genes positively associated with drug EC50 values. (c) Baseline method that does not use expression data. Shown are SNPs (columns) distributed throughout the genome within TFBS (yellow peaks) and outside. Genome-wide association study (GWAS) SNPs (green diamonds) correlated with drug response across individuals (rows) are tested for enrichment with within-TFBS SNPs to determine whether a TF is associated with drug response.
Identification of TFs with potential role in cytotoxicity variation
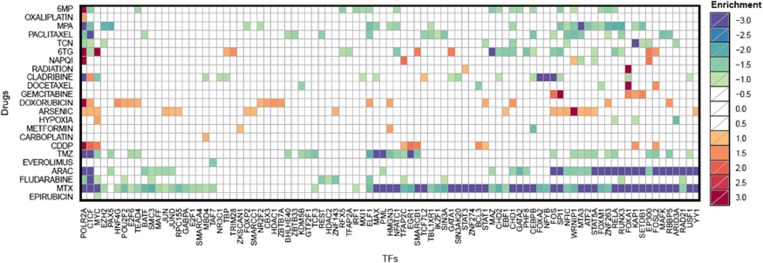
We used the GENMi method to assign statistical significance, that is, P-value and FDR to each (TF, treatment) pair. In total, drug-induced cytotoxicity for 24 drugs were analyzed, of which 9 were prepared specifically for this study (see Materials and Methods). A total of 3864 pairs were tested (114 TFs × 24 treatments, see Supplementary Table 1). There are 334 associations at a threshold of FDR ≤0.10, involving 91 TFs and 23 treatments (Supplementary Table 2). Figure 3 shows all log2 transformed FDR values of any TF and drug with a significant association. The 334 significant associations were distributed unevenly across the treatments, with the drug Methotrexate (MTX) appearing in 70 of the 334 associations (21%), followed by Cytarabine (ara-C) and Medroxyprogesterone Acetate (MPA) as the drugs with most TF associations (Supplementary Table 3). The TFs with the most numbers of associations, as shown in Supplementary Table 4, were POLR2A, the largest subunit of RNA Polymerase II, and CTCF, a versatile regulator involved in gene activation, repression, silencing and chromatin insulation.42 We have reason to believe (Supplementary Figure 1) that these frequent associations involving general TFs that bind the genome extensively are artifacts of our procedure, in conjunction with linkage disequilibrium and the promiscuous DNA binding of these TFs. We ignored such associations in our follow-up investigations. Also included in the six TFs with the most drug associations were MYC, which plays an important role in reversing multidrug resistance,43 and FOS, a member of the AP-1 complex that is linked to chemotreatment resistance.44
Figure 3.
Significant (transcription factor (TF), treatment) associations. Shown are the log-transformed false discovery rate (FDR) values for all associations meeting FDR ≤0.1. The green–blue range refers to enrichment for genes whose expression negatively correlates with cytotoxicity, and the yellow–red range indicates enrichment of genes whose expression positively correlates with cytotoxicity. The yellow–red log-transformed FDR values are multiplied by negative 1, creating the −3 to 3 range in the legend enrichment. Anything with an FDR ≥0.1 is shown as white.
We next examined the collection of statistically significant (TF, treatment) associations for prior experimental evidence supporting them. To our knowledge, there is no standard benchmark that can help us with such an assessment, and hence we resorted to surveying the literature for studies implicating a TF in the response to a specific cytotoxic treatment, for example, TFs whose knockdown or overexpression has been shown to affect cytotoxicity, though not necessarily in the lymphoblastoid cell line. We focused on significant (TF, treatment) associations that are relatively unique, that is, the TF is associated with ≤5 (of 24) treatments and the treatment is associated with ≤10 (of 114) TFs. These 20 associations are shown in Table 1. We noted 6 of the 20 associations to be supported by direct experimental evidence involving the drug and the TF. We discuss these below.
Table 1. Literature support for 20 significant (TF, treatment) associations at FDR ≤0.1 where the TF is associated with ≤5 treatments and the treatment is associated with ≤10 TFs.
| Association no. | Treatment | TF | Literature evidence |
|---|---|---|---|
| 1 | Cisplatin | EGR1 | Direct |
| 2 | Cisplatin | STAT1 | Direct |
| 3 | Docetaxel | FOXM1 | Direct |
| 4 | Radiation | STAT3 | Direct |
| 5 | Cisplatin | SMARCB1 | Direct |
| 6 | Cisplatin | BCL3 | Direct |
| 7 | TCN | EZH2 | Indirect |
| 8 | Gemcitabine | SETDB1 | Indirect |
| 9 | Docetaxel | KDM5B | Indirect |
| 10 | Hypoxia | ARID3A | Indirect |
| 11 | Carboplatin | MBD4 | Indirect |
| 12 | Cisplatin | TFAP2C | Indirect |
| 13 | Docetaxel | BCL3 | Indirect |
| 14 | Everolimus | TAF7 | None found |
| 15 | NAPQI | TFAP2C | None found |
| 16 | TCN | ARID3A | None found |
| 17 | TCN | STAT1 | None found |
| 18 | Metformin | HMGN3 | None found |
| 19 | Metformin | ZKSCAN1 | None found |
| 20 | TCN | SETDB1 | None found |
Abbreviations: FDR, false discovery rate; TF, transcription factor.
Observation
FoxM1 (transcription factor forkhead box protein M1) is associated with response to docetaxel. Remarks: overexpression of FoxM1 in gastric cancers was previously shown to mediate resistance to docetaxel and inhibiting FoxM1 was found to reverse docetaxel resistance in gastric cancers.45 Similar conclusions were reached by other studies.46
Observation
EGR-1 (early growth response protein 1) is associated with cisplatin treatment. Remarks: EGR-1 has been shown to regulate cisplatin-induced apoptosis in human esophageal squamous cell carcinoma cell lines (WHCO1).47 The EGR-1 promoter has been shown to be induced by this drug.48, 49
Observation
STAT1, a member of the signal transducer and activator family of transcription factors, is associated with cisplatin. Remarks: overexpression of STAT1 in A2780 human ovarian cancer cells was shown to increase cisplatin resistance.50 Moreover, inhibiting STAT1 expression has been shown to attenuate cisplatin-induced ototoxicity in rats51 and mice.52
Observation
STAT3, a homolog of STAT1 in the signal transducer and activator family of transcription factors, is associated with radiation treatment. Remarks: a previous study found STAT3 blockade to enhance radiosensitivity in Hep-2 cells.53 Other studies have reported that radiation activates STAT3,54 and that targets of STAT3 are upregulated by radiation in a mouse model of glioblastoma.55
Observation
SMARCB1, a core component of the switch/sucrose nonfermentable (SWI/SNF) nucleosome remodeling complex, is associated with cisplatin. Remarks: recent sequencing of various cancer cells have demonstrated frequent mutations in SWI/SNF factors such as ARID1A. Suppression of ARID1A and its paralog ARID1B sensitized the cell to cisplatin as well as radiation. Suppression of SMARCB1 reproduced the same effects.56
Observation
BCL-3 is associated with cisplatin. Remarks: a previous study found BCL-3 overexpression to suppress cisplatin-induced apoptosis in MCF7 breast cancer cell lines.57
In addition to the above six examples of experimental results directly supporting an association, we also noted seven of the statistical associations from Table 1 to be supported by indirect experimental evidence involving transcriptional regulation of the TF in response to the drug or direct experimental evidence involving a protein closely related to the TF. These are described in Supplementary Note 2. For seven of the associations noted in Table 1, we were unable to find strong supporting evidence from the literature, making these promising candidates for future experimental follow-up. To test the effects of imputation on these results, we replicated the GENMi pipeline using both imputed and genotyped SNPs (Supplementary Note 3) to see how many of the literature-supported associations in Table 1 were corroborated. The results are shown in Supplementary Table 5. Overall, imputation did not significantly alter the associations reported in Table 1: we found 10 of the 13 associations with literature support reported in Table 1 to be recovered in this new analysis at the nominal P-value threshold of 0.05 (FDR ≤0.13). In additionally, to test the effect of population stratification, we performed GENMi analysis on each population separately using only genotyped SNPs and genotyped SNPs with imputation. Results are shown in Supplementary Table 6 and discussed in Supplementary Note 4. Of the 13 TF–drug associations in Table 1 that had some form of literature support, 9 were significant in at least one sub-population, at a nominal P-value of 0.05, using either the genotyped SNPs or genotyped as well as imputed SNPs.
An analogous method that does not use expression data reports fewer associations
To determine the utility of our method that integrates genotype, gene expression and phenotype to identify (TF, treatment) pairs, we devised a baseline method agnostic of gene expression. This baseline method (Figure 2c) tests whether GWAS SNPs (P-value ≤10−8) for a given treatment are enriched within peaks of a particular TF (see Materials and Methods), computing a P-value of association for each (TF, treatment) pair. We sought to compare the number of significant associations discovered by GENMi and the baseline method respectively at a fixed false positive rate. For a fair comparison, we devised a procedure that generates randomized data sets and asked whether either method discovers a significant association (a false positive, as the data set is a randomized one) on it. By performing this test repeatedly and counting how frequently each method (GENMi or the baseline method) reported false associations, we were able to control for the false positive rate of each method in exactly the same manner. Details of the randomization procedure are articulated in Supplementary Note 5. The number of (TF, treatment) pairs reported by either method on the real data set, at each false positive rate threshold, is shown in Table 2. The number of significant associations found by the GENMi procedure far outweighs those in the baseline, indicating that utilizing expression information improves the sensitivity of the association study.
Table 2. Number of (TF, treatment) associations discovered by the GENMi method and the baseline method that does not use expression data, at varying FPR thresholds.
| FPR | 1 | 0.2 | 0.002 | 0.0002 | 0.00002 |
| Baseline | 1932 | 75 | 16 | 2 | 0 |
| GENMi | 2736 | 943 | 211 | 33 | 14 |
Abbreviations: FPR, false positive rate; GENMi, Gene Expression iN the Middle; TF, transcription factor.
The FPR is estimated by running either method on 5000 randomized data sets where transcription factor-binding site (TFBS) locations have been shuffled genome wide.
In vivo experimental validation of TFS regulating anthracycline and taxane response
We sought to verify whether TFs associated with drug response variation can be linked in vivo to significant changes in cellular sensitivity to drug-induced apoptosis. Though we utilized lymphoblastoid cell lines data for our association analysis, we performed siRNA knockdown experiments in two different cell lines to demonstrate the generalizability of our results and the LCL model system. Specifically, we choose two triple-negative breast cancer cell lines, BTF549 and MDA-MB-231, that are of great clinical significance. In addition, we restricted our analysis to two of the most widely utilized family of drugs used in the treatment of breast cancer: anthracyclines (doxorubicin, epirubicin) and taxanes (docetaxel, paclitaxel). In addition to being clinically relevant, the mechanisms of the drugs within each family are very similar, gifting us with the ability to check self-consistency within a drug family, that is, if TF increases resistance to doxorubicin, it should also increase resistance to epirubicin. Cost and time constraints restricted the experimental validation in this study to these four drugs.
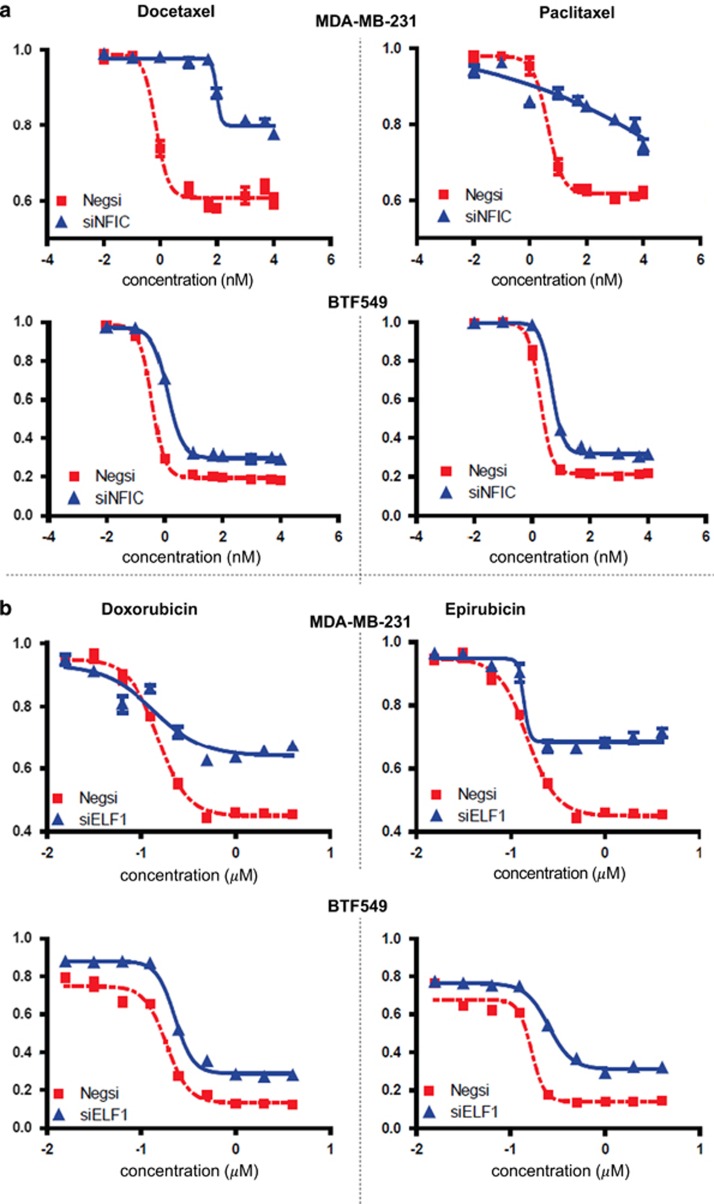
To choose candidate TFs for validation, we restricted ourselves to those TFs that exhibited a P-value of ≤0.05 and FDR of ≤0.1 for at least one drug in the family of interest. For the taxanes, this produced 21 TFs, as shown in Table 3, 4 of which were omitted for various reasons. For the anthracyclines, these criteria yielded 14 TFs, as shown in Table 4. CTCF and POLR2A were original candidates, but omitted because they are ubiquitous activators for expression. The siRNA knockdowns were performed for the 21 taxane- and 14 anthracycline-associated TFs with negative siRNA as a control. Tables 3 and 4 show the results of the assays for the taxanes and anthracyclines, respectively. Even though a TF was tested even if it was predicted to be associated with only one of the two drugs in a family, our definition of successful validation conservatively required a TF to affect significant change in the dosage–response curve for both drugs in the family. For additional stringency, this requirement had to be met in both the tested cell lines. Using these stringent criteria, we found 6 out of the 21 predicted TFs, specifically BCL3, CEBPB, ELF1, MAFK, NFIC and REST, to increase resistance to taxane-induced cytotoxicity. An example of what constitutes significant change (induced by a TF knockdown) in the dosage–response curve for the taxanes is shown in Figure 4a, for NFIC knockdown. For the anthracyclines, we found 4 out of the 14 predicted TFs, namely ELF1, HDAC1, HNF4G and ZNF263, to increase cytotoxic resistance to doxorubicin and epirubicin in both tested cell lines. An example of a validated TF association (ELF1) for the anthracyclines is shown in Figure 4b. The rest of the cytotoxicity curves are shown in Supplementary Figures 2. In addition, the GENMi analysis predicted MYC to be associated only with epirubicin, and the experimental validation supports this as it is associated only with epirubicin (in BTF549).
Table 3. Shown are the functional validation results for 21 TFs enriched (at FDR ≤0.1 and P-value ≤0.05) for either taxane, paclitaxel (PAX) or docetaxel (DOC).
| Number | TF |
GENMi enrichments |
Cell lines |
||||||
|---|---|---|---|---|---|---|---|---|---|
|
PAX |
DOC |
BTF549 |
MDA-MB-231 |
||||||
| P-value | FDR | P-value | FDR | PAX | DOC | PAX | DOC | ||
| 1 | BATF | 0.0142 | 0.07 | 0.5160 | 1.00 | UP | UP | ||
| 2 | BCL3 | 0.4112 | 0.56 | 0.0020 | 0.03 | UP | UP | UP | UP |
| 3 | BHLHE40 | 0.0088 | 0.06 | 0.0487 | 0.30 | UP | UP | ||
| 4 | CEBPB | 0.0024 | 0.02 | 0.3199 | 0.86 | UP | UP | UP | UP |
| 5 | ELF1 | 0.0020 | 0.03 | 0.0147 | 0.20 | UP | UP | UP | UP |
| 6 | FOS | 0.0127 | 0.09 | 0.2007 | 0.63 | UP | UP | ||
| 7 | FOSL2 | 0.0285 | 0.12 | 0.0012 | 0.03 | UP | UP | ||
| 8 | FOXM1 | 0.2754 | 0.44 | 0.0052 | 0.07 | UP | UP | UP | |
| 9 | IKZF1 | 0.0213 | 0.09 | 0.1422 | 0.78 | UP | UP | ||
| 10 | KDM5B | 0.0016 | 0.02 | 0.0004 | 0.04 | UP | UP | ||
| 11 | MAFK | 0.0016 | 0.02 | 0.6006 | 1.00 | UP | UP | UP | UP |
| 12 | MTA3 | 0.0027 | 0.03 | 0.1556 | 0.62 | UP | UP | UP | |
| 13 | NFIC | 0.0020 | 0.02 | 0.0131 | 0.18 | UP | UP | UP | UP |
| 14 | RBBP5 | 0.0012 | 0.02 | 0.3613 | 0.69 | UP | |||
| 15 | REST | 0.0008 | 0.04 | 0.2343 | 0.57 | UP | UP | UP | UP |
| 16 | SIN3A | 0.0028 | 0.03 | 0.5193 | 0.78 | UP | |||
| 17 | TBL1XR1 | 0.0071 | 0.04 | 0.5347 | 1.00 | ||||
| 18 | TCF7L2 | 0.0056 | 0.04 | 0.1004 | 0.45 | UP | |||
| 19 | WRNIP1 | 0.0004 | 0.02 | 0.2273 | 0.59 | UP | UP | UP | |
| 20 | ZBTB33 | 0.0044 | 0.05 | 0.9282 | 0.95 | UP | UP | ||
| 21 | ZNF263 | 0.0168 | 0.09 | 0.0255 | 0.26 | UP | UP | ||
Abbreviations: FDR, false discovery rate; GENMi, Gene Expression iN the Middle; TF, transcription factor.
Validation was performed in two triple-negative breast cancer cell lines (BTF549 and MDA-MB-231). For each cell line, drug and TF, a small interfering RNA (siRNA) knockdown experiment was performed, followed by an MTA assay for the drug. Comparisons were made to negative siRNA experiments to determine whether the TF decreased, increased or did not affect the sensitivity of the cell to the drug. ‘UP' in the table refers to decreased sensitivity or desensitization of the cell to the drug, that is, the TF knockdown increased cell resistance/survivability to increasing concentrations of the apoptotic drug. In no case was the knockdown found to decrease cell resistance. Cells with P-value ≤0.05 or FDR ≤ 0.1 are colored gray, and represent the drug (PAX or DOC or both) for which the TF was predicted to influence response.
Table 4. Shown are the functional validation results for 14 TFs enriched for either anthracycline drug, epirubicin (EPI) or doxorubicin (DOX) at FDR ≤0.1 and P-value ≤0.05.
| Number | TF |
GENMi enrichments |
Cell lines |
||||||
|---|---|---|---|---|---|---|---|---|---|
|
DOX |
EPI |
BTF549 |
MDA-MB-231 |
||||||
| P-value | FDR | P-value | FDR | DOX | EPI | DOX | EPI | ||
| 1 | CBX3 | 0.0033 | 0.05 | 0.4734 | 0.77 | UP | UP | ||
| 2 | CHD1 | 0.0036 | 0.04 | 0.5706 | 0.82 | ||||
| 3 | E2F6 | 0.0051 | 0.06 | 0.0944 | 0.76 | ||||
| 4 | ELF1 | 0.0008 | 0.05 | 0.3030 | 0.79 | UP | UP | UP | UP |
| 5 | HDAC1 | 0.0020 | 0.04 | 0.9026 | 0.89 | UP | UP | UP | UP |
| 6 | HMGN3 | 0.0077 | 0.05 | 0.4534 | 0.76 | UP | UP | ||
| 7 | HNF4G | 0.0004 | 0.03 | 0.5844 | 0.82 | UP | UP | UP | UP |
| 8 | MYC | 0.1112 | 0.32 | 0.0028 | 0.09 | UP | UP | UP | |
| 9 | NR2F2 | 0.0112 | 0.08 | 0.8199 | 0.82 | UP | UP | ||
| 10 | POU2F2 | 0.0048 | 0.06 | 0.1192 | 0.72 | UP | UP | ||
| 11 | RBBP5 | 0.0004 | 0.04 | 0.7434 | 0.83 | ||||
| 12 | STAT1 | 0.0065 | 0.07 | 0.7122 | 0.87 | UP | UP | ||
| 13 | TEAD4 | 0.0101 | 0.07 | 0.8462 | 0.98 | ||||
| 14 | ZNF263 | 0.0071 | 0.05 | 0.1045 | 0.72 | UP | UP | UP | UP |
Abbreviations: FDR, false discovery rate; GENMi, Gene Expression iN the Middle; TF, transcription factor.
Validation was performed in two triple-negative breast cancer cell lines (BTF549 and MDA-MB-231). For each cell line, drug and TF, a small interfering RNA (siRNA) knockdown experiment was performed, followed by an MTA assay for the drug. Comparisons were made to negative siRNA experiments to determine whether the TF decreased, increased or did not affect the sensitivity of the cell to the drug. ‘UP' in the table refers to decreased sensitivity or desensitization of the cell to the drug. In other words, it means that the TF knockdown increased cell resistance/survivability to increasing concentrations of the apoptotic drug. Cells with P-value ≤0.05 or FDR ≤0.1 are colored gray.
Figure 4.
(a) Dosage–response curves for the transcription factor (TF) NFIC across two drugs, docetaxel (left) and paclitaxel (right), and two cell lines, MDA-MB-231 and BTF549. Each plot shows significant increase in resistance to the drug upon knockdown of NFIC compared with normal response of the cells, using a two-tailed paired t-test. (b) Dosage–response curves for the TF ELF1 across two drugs, doxorubicin (left) and epirubicin (right), and two cell lines, MDA-MB-231 and BTF549. Each plot shows significant increase in resistance to the drug upon knockdown of NFIC compared with normal response of the cells, using a two-tailed paired t-test.
Although several of GENMi associations were not corroborated experimentally in both drugs within a family and in both cell lines, this is expected to an extent as the selection of TF knockdowns was based on GENMi predictions for either one of the drugs in the family and made from a different cell line, that is, the experimental test was more stringent than what the statistical association suggests. In total, we find the hit rates of 6/21 and 4/14 as significant evidence that GENMi identifies TFs that truly regulate cellular response to drug-induced apoptosis.
Discussion
We have presented a methodology for interrogating the extent to which specific TFs are associated with individual variations in drug-induced cytotoxicity. We employ a statistical approach that assumes cis-regulatory variants embedded within TFBS affect proximal genes, whose varying expression is then reflected in drug response. Our approach is fundamentally different from the aforementioned triangulation approach in that we integrate TF–DNA-binding data and are able to associate TFs with drug response; also, we do not require the direct association of SNPs with drug-induced phenotype. Focusing on drugs and TFs that feature in a limited number of associations, we noted the statistically significant (TF, treatment) associations to be frequently supported by the literature, in the form of experiments where knockdown or overexpression of the TF changes drug response. Stringent control of the randomization procedure illuminated the benefit of GENMi over a simple GWAS–TFBS overlap approach where expression data are not used. Although our results showcase the true positive rate of GENMi, they do not yet allow us to determine false negative rates or sensitivity because of the absence of a comprehensive benchmark of true (TF, treatment) associations. Nevertheless, our methods represent the first comprehensive methodology for assessing regulatory associations with drug response.
There are a number of areas in which the GENMi method could be improved. For one, differences in allele frequencies between stratified populations has been shown to induce spurious associations;58 adjusting for this confounding factor in a more principled framework may reduce the number of false positives in our results. Second, although filtering for ChIP high occupancy target regions helps eliminate regions where it is difficult to assign function to any one TF, one may not assume that all bound TFs are nonfunctional,59 and future extensions of our method will be cognizant of this. The literature also indicates the existence of eQTL hot spots—eQTLs associated with a large number of genes—as a result of various confounding factors; elimination of these factors would aid in the discovery of true eQTL signals.60 Another way of improving GENMi would be to exploit prior knowledge of the relationship between drugs; relevant methods already exist, to an extent.61 GENMi could also be improved at the level of determining gene targets for a TF. At a statistical level, GENMi reduces to a two-step procedure of enriching top cis-eQTL genes under a given transcriptional regulatory context with genes whose expression correlates with a particular drug. Ideas from a recent study that employs a two-step regressive framework to a similar end (but without the integration of TF ChIP data)17 may be adapted to eliminate the arbitrary threshold of the GENMi method in determining transcription gene targets. In addition, GENMi could be improved by considering more elegant eQTL models, such as the methods employed by Sudarsanam and Cohen;62 however, exploiting more complex methods capturing multi-additive and epistatic interactions at the genome-wide level carries a heavy computational price that is not easy to circumnavigate. Another area of investigation involves the determination of the cis-regulatory region: although cis-regulatory eQTLs are replicated better across studies than trans-eQTLs,63 definitions of cis-regulatory regions differ widely.64 In our analysis, we use an operational regulatory region size of 50 kb upstream of the gene's transcription start site; a size that has been used in many other studies.28, 65 In fact, studies have even used regions up to 100 kb.66 In additionally, we denote the entire region upstream of the gene as the de facto regulatory region of the gene. Together, these assumptions carry the risk that the regulatory region of one gene may contain regulatory sequences for other genes; a more conservative regulatory size would dilute this effect, at the expense of sensitivity. It is not known the extent to which different regulatory sizes and schemes affect the GENMi analysis and more work needs to be conducted on this front. Enhancer–promoter interaction data from chromatin capture-based technologies such as Hi-C67 will help obviate this problem to a certain degree, though such data have to be obtained from the cell type of interest. Furthermore, the GENMi method only considers single TFBS for filtering eQTL SNPs; associations may be more conspicuous when considering combinations of transcriptional contexts. Though this is hard to compute greedily, there are methods for finding combinations of TFs overrepresented in cis-regulatory regions.68 Finally, future work will benefit from analysis of protein QTLs—SNPs correlated with protein abundance—as opposed to mRNA abundance,6, 63 as the activity of the gene at the protein level is hypothesized to implement the target cellular response; however, genome-wide protein data are not readily available.
Our approach utilizing single TF contexts with eQTLs estimated from basal gene expression data in LCLs corresponds to a logical entry point analysis into the pharmacological effects of TFs on drug response. To our knowledge, the GENMi approach is novel in its direct interrogation of transcriptional regulation on drug-induced cellular response. Although many improvements can be made, the fruit of the existing GENMi analysis in both our literature review and control experiments illustrates the remarkable utility of the method.
Acknowledgments
Funding for this work was provided in part by the NIH (Grant U54 GM114838 to SS, U19 GM61388 Pharmacogenomics Research Network and R01 CA138461 to LW) and in part by the Mayo Clinic-UIUC Alliance and by Grant 1U54GM114838 awarded by NIGMS through funds provided by the trans-NIH Big Data to Knowledge (BD2K) initiative (www.bd2k.nih.gov). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Encode Project Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007; 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Weinshilboum RM. Pharmacogenomics: candidate gene identification, functional validation and mechanisms. Hum Mol Genet 2008; 17: R174–R179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med 2011; 364: 1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen EL, Godley LA, Zhang W, Dolan ME. Pharmacogenomics of chemotherapeutic susceptibility and toxicity. Genome Med 2012; 4: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler HE, Dolan ME. Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation. Pharmacogenomics 2012; 13: 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark AL, Hause RJ Jr, Gorsic LK, Antao NN, Wong SS, Chung SH et al. Protein quantitative trait loci identify novel candidates modulating cellular response to chemotherapy. PLoS Genet 2014; 10: e1004192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther 2007; 81: 328–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg Y, Gottlieb A, Kupiec M, Ruppin E, Sharan R. Large-scale elucidation of drug response pathways in humans. J Comput Biol 2012; 19: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Wallqvist A, Thanki N, Covell DG. Linking pathway gene expressions to the growth inhibition response from the National Cancer Institute's anticancer screen and drug mechanism of action. Pharmacogenomics J 2005; 5: 381–399. [DOI] [PubMed] [Google Scholar]
- National Research Council (US) Committee on A Framework for Developing a New Taxonomy of DiseaseToward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease, National Academies Press (US): Washington (DC), 2011. [PubMed]
- Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am J Hum Genet 2010; 86: 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L, Macciardi F, Subramanian A, Guffanti G, Potkin SG, Yu Z et al. SNP-based pathway enrichment analysis for genome-wide association studies. BMC Bioinformatics 2011; 12: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ et al. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet 2010; 86: 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JL, Taylor JM, Richards JB, Watts T, Pettersson FH, Broxholme J et al. The use of genome-wide eQTL associations in lymphoblastoid cell lines to identify novel genetic pathways involved in complex traits. PLoS One 2011; 6: e22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kabesch M, Bouzigon E, Demenais F, Farrall M, Moffatt MF et al. Using eQTL weights to improve power for genome-wide association studies: a genetic study of childhood asthma. Front Genet 2013; 4: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet 2008; 4: e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SD, Cai TT, Li H. More powerful genetic association testing via a new statistical framework for integrative genomics. Biometrics 2014; 70: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Ding Z, Qian Y, Shi X, Castranova V, Harner EJ et al. Predicting cancer drug response by proteomic profiling. Clin Cancer Res 2006; 12: 4583–4589. [DOI] [PubMed] [Google Scholar]
- Robert J, Vekris A, Pourquier P, Bonnet J. Predicting drug response based on gene expression. Crit Rev Oncol Hematol 2004; 51: 205–227. [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhang XS, Zhang S. Predicting cooperative drug effects through the quantitative cellular profiling of response to individual drugs. CPT Pharmacometrics Syst Pharmacol 2014; 3: e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci USA 2007; 104: 13086–13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Duan S, Shukla SJ, Kistner EO, Clark TA, Chen TX et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet 2007; 81: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Duan S, Bleibel WK, Kistner EO, Zhang W, Clark TA et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci USA 2007; 104: 9758–9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N, Qin Y, Fridley BL, Hou J, Kalari KR, Zhu M et al. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res 2010; 20: 1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet 2010; 11: 533–538. [DOI] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE et al. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 2011; 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen ES, Lipton G, Clyde MA, Monteiro AN. Functional annotation signatures of disease susceptibility loci improve SNP association analysis. BMC Genomics 2014; 15: 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CD, Mangravite LM, Engelhardt BE. Integrative modeling of eQTLs and cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet 2013; 9: e1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY, Fridley BL, Jenkins GD, Batzler A, Wang L, Weinshilboum RM. Mycophenolic acid response biomarkers: a cell line model system-based genome-wide screen. Int Immunopharmacol 2011; 11: 1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer AM, Fridley BL, Jenkins GD, Batzler AJ, Pelleymounter LL, Kalari KR et al. Acetaminophen-NAPQI hepatotoxicity: a cell line model system genome-wide association study. Toxicol Sci 2011; 120: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley BL, Batzler A, Li L, Li F, Matimba A, Jenkins GD et al. Gene set analysis of purine and pyrimidine antimetabolites cancer therapies. Pharmacogenet Genomics 2011; 21: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Fridley BL, Kalari K, Niu N, Jenkins G, Batzler A et al. Discovery of genetic biomarkers contributing to variation in drug response of cytidine analogues using human lymphoblastoid cell lines. BMC Genomics 2014; 15: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan XL, Moyer AM, Fridley BL, Schaid DJ, Niu N, Batzler AJ et al. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res 2011; 17: 5801–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley BL, Abo R, Tan XL, Jenkins GD, Batzler A, Moyer AM et al. Integrative gene set analysis: application to platinum pharmacogenomics. OMICS 2014; 18: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N, Schaid DJ, Abo RP, Kalari K, Fridley BL, Feng Q et al. Genetic association with overall survival of taxane-treated lung cancer patients - a genome-wide association study in human lymphoblastoid cell lines followed by a clinical association study. BMC Cancer 2012; 12: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Fridley BL, Feng Q, Abo RP, Brisbin A, Batzler A et al. Genome-wide association study for biomarker identification of rapamycin and everolimus using a lymphoblastoid cell line system. Front Genet 2013; 4: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res 2008; 68: 7050–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle JN, Liu M, Wickerham DL, Schaid DJ, Wang L, Mushiroda T et al. Selective estrogen receptor modulators and pharmacogenomic variation in ZNF423 regulation of BRCA1 expression: individualized breast cancer prevention. Cancer Discov 2013; 3: 812–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur I, Tuupanen S, Whitington T, Aaltonen LA, Taipale J. Lessons from functional analysis of genome-wide association studies. Cancer Res 2013; 73: 4180–4184. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet 2012; 8: e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 2001; 17: 520–527. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim MJ, Kim HB, Lee JW, Bae JH, Kim DW et al. Suppression of multidrug resistance by treatment with TRAIL in human ovarian and breast cancer cells with high level of c-Myc. Biochim Biophys Acta 2011; 1812: 796–805. [DOI] [PubMed] [Google Scholar]
- Bonovich M, Olive M, Reed E, O'Connell B, Vinson C. Adenoviral delivery of A-FOS, an AP-1 dominant negative, selectively inhibits drug resistance in two human cancer cell lines. Cancer Gene Ther 2002; 9: 62–70. [DOI] [PubMed] [Google Scholar]
- Li X, Yao R, Yue L, Qiu W, Qi W, Liu S et al. FOXM1 mediates resistance to docetaxel in gastric cancer via up-regulating Stathmin. J Cell Mol Med 2014; 18: 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Fujiwara Y, Takahashi T, Nakamura Y, Takiguchi S, Nakajima K et al. Overexpression of forkhead box M1 transcription factor (FOXM1) is a potential prognostic marker and enhances chemoresistance for docetaxel in gastric cancer. Ann Surg Oncol 2013; 20: 1035–1043. [DOI] [PubMed] [Google Scholar]
- Dong Q, Zhang J, Hendricks DT, Zhao X. GRObeta and its downstream effector EGR1 regulate cisplatin-induced apoptosis in WHCO1 cells. Oncol Rep 2011; 25: 1031–1037. [DOI] [PubMed] [Google Scholar]
- Park JO, Lopez CA, Gupta VK, Brown CK, Mauceri HJ, Darga TE et al. Transcriptional control of viral gene therapy by cisplatin. J Clin Invest 2002; 110: 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WD, Li R, Chen ZT, Li DZ, Duan YZ, Cao ZH. Cisplatin-controlled p53 gene therapy for human non-small cell lung cancer xenografts in athymic nude mice via the CArG elements. Cancer Sci 2005; 96: 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D, Schick J, Conway S, Biade S, Laub PB, Stevenson JP et al. Identification of genes associated with platinum drug sensitivity and resistance in human ovarian cancer cells. Br J Cancer 2005; 92: 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T, Mukherjea D, Sheehan K, Jajoo S, Rybak LP, Ramkumar V. Short interfering RNA against STAT1 attenuates cisplatin-induced ototoxicity in the rat by suppressing inflammation. Cell Death Dis 2011; 2: e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt NC, Rubel EW, Nathanson NM. Cisplatin-induced hair cell death requires STAT1 and is attenuated by epigallocatechin gallate. J Neurosci 2009; 29: 3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang H, Lu X, Di B. STAT3 blockade with shRNA enhances radiosensitivity in Hep-2 human laryngeal squamous carcinoma cells. Oncol Rep 2010; 23: 345–353. [PubMed] [Google Scholar]
- Gao L, Li FS, Chen XH, Liu QW, Feng JB, Liu QJ et al. Radiation induces phosphorylation of STAT3 in a dose- and time-dependent manner. Asian Pac J Cancer Prev 2014; 15: 6161–6164. [DOI] [PubMed] [Google Scholar]
- Halliday J, Helmy K, Pattwell SS, Pitter KL, LaPlant Q, Ozawa T et al. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Nl Acad Sci USA 2014; 111: 5248–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R, Ui A, Kanno S, Ogiwara H, Nagase T, Kohno T et al. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res 2014; 74: 2465–2475. [DOI] [PubMed] [Google Scholar]
- Kashatus D, Cogswell P, Baldwin AS. Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev 2006; 20: 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909. [DOI] [PubMed] [Google Scholar]
- modENCODE Consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 2010; 330: 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JW, Sul JH, Han B, Ye C, Eskin E. Effectively identifying regulatory hotspots while capturing expression heterogeneity in gene expression studies. Genome Biol 2014; 15: r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet 2014; 10: e1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P, Cohen BA. Single nucleotide variants in transcription factors associate more tightly with phenotype than with gene expression. PLoS Genet 2014; 10: e1004325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause RJ, Stark AL, Antao NN, Gorsic LK, Chung SH, Brown CD et al. Identification and validation of genetic variants that influence transcription factor and cell signaling protein levels. Am J Hum Genet 2014; 95: 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen A, Alvarez C, DeClaire S, Tintle NL. Assessing methods for assigning SNPs to genes in gene-based tests of association using common variants. PLoS One 2013; 8: e62161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DJ, Veyrieras JB, Degner JF, Pique-Regi R, Pai AA, Crawford GE et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol 2012; 13: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, McCormick C et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res 2013; 24: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Boyle AP, Wu L, Zhai J, Kawli T, Snyder M. Dynamic trans-acting factor colocalization in human cells. Cell 2013; 155: 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.