Abstract
Castration-resistant prostate cancer (CRPC) progression after androgen deprivation therapy shows upregulated expression of androgen receptor (AR) splice variants, induced epithelial-to-mesenchymal transition phenotypes and enhanced stem cell characteristics, all of which are associated with resistance to enzalutamide. Since there is no curative treatment for CRPC, innovative treatments are urgently needed. In our recent study, we found that resistance to enzalutamide was partly due to deregulated expression of microRNAs such as miR-34a, miR-124, miR-27b, miR-320 and let-7, which play important roles in regulating AR and stem cell marker gene expression that appears to be linked with resistance to enzalutamide. Importantly, we found that BioResponse 3,3′-diindolylmethane (BR-DIM) treatment in vitro and in vivo caused downregulation in the expression of wild-type AR. The AR splice variants, Lin28B and EZH2, appear to be deregulated through the re-expression of let-7, miR-27b, miR-320 and miR-34a in human prostate cancer (PCa). BR-DIM administered in clinical trials was well tolerated, and 93% of patients had detectable prostatic DIM levels. The inhibitory effects of BR-DIM on AR and AR target gene such as prostate-specific antigen were also observed in the clinical trial. Our preclinical and clinical studies provide the scientific basis for a ‘proof-of-concept’ clinical trial in CRPC patients treated with enzalutamide in combination with BR-DIM. This strategy could be expanded in future clinical trials in patients with PCa to determine whether or not they could achieve a better treatment outcome which could be partly mediated by delaying or preventing the development of CRPC.
Key Words: Prostate cancer; Micro-RNA; BioResponse 3,3′-diindolylmethane; Clinical trial
Introduction
Prostate cancer (PCa) is the most common noncutaneous epithelial cancer and the second leading cause of cancer-related death in men in the USA with expected 27,540 deaths in 2015 [1]. Most of these deaths are primarily attributed to the acquisition of castration-resistant prostate cancer (CRPC) and subsequent metastasis. The growth of PCa is driven by functional and promiscuous activity of androgen receptor (AR), leading to the aggressiveness, metastasis and recurrence of PCa. Progression to CRPC after androgen ablation therapy (ADT) is predominantly due to deregulated AR signaling [2,3,4,5,6,7,8]. In CRPC, AR functions promiscuously and is associated with deregulation of multiple signaling pathways [2,3,4,5,6,7,8]. More importantly, constitutively active AR splice variants lacking the ligand-binding domain have been found to significantly contribute to the development and progression of CRPC [9,10,11].
In recent years, new drugs targeting AR signaling have been introduced for the treatment of CRPC [12] including abiraterone and enzalutamide [13]. However, patients are developing resistance to these drugs, although abiraterone appears to be effective in previously treated patients with enzalutamide [14]. Enzalutamide resistance has been reported to be mediated by differential expression of AR splice variants [15]. A recent clinical study showed that 39% of enzalutamide-treated patients and 19% of abiraterone-treated patients had detectable AR splice variants in circulating tumor cells [16]. Patients with AR splice variants had lower prostate-specific antigen (PSA) response rates and shorter PSA-free survival, clinical or radiographic progression-free survival, and shorter overall survival compared to patients without AR splice variants [16]. In published reports, it has been suggested that AR splice variants from patients with CRPC were significantly correlated with the drug resistance to enzalutamide and abiraterone [16,17,18]. Therefore, transcriptional inactivation of AR splice variants could be a promising strategy to overcome resistance to enzalutamide or other ADT for achieving better treatment outcomes of PCa patients.
Several microRNAs (miRNAs) have been shown to downregulate the level of wild-type AR, AR splice variants, and the epithelial-to-mesenchymal transition (EMT) and stem cell markers which are deregulated in CRPC cells (table 1). These miRNAs are aberrantly expressed in PCa possibly due to epigenetic regulation. Therefore, targeting miRNAs to transcriptionally inactivate AR splice variants and stem cell markers could prevent and inhibit the development and progression of CRPC and drug resistance. Several natural agents including 3,3′-diindolylmethane (DIM), isoflavone and curcumin have been shown to have inhibitory effects on AR signaling [19,20,21,22]. Therefore, these natural agents could be useful for delaying or preventing the development of CRPC. Among these natural agents, DIM showed more potent inhibitory effects on AR signaling [19]. The effects of BR-DIM (formulated DIM manufactured by BioResponse) on cell signal transduction and biological behaviors of PCa cells have been investigated. BR-DIM inhibits PCa cell proliferation and induces apoptotic cell death through the regulation of important signal transduction pathways including NF-κB, AR, Akt-mTOR and others [19,23,24,25,26]. BR-DIM also regulates the expression of miRNAs which modulate cellular signaling pathways and suppress cancer growth [27,28,29,30]. Therefore, BR-DIM inhibits AR signaling and stem cell signatures partly via the regulation of miRNAs. Thus, BR-DIM appears to be a promising agent for the inhibition of CRPC development and drug resistance.
Table 1.
miRNAs in the regulation of AR, EMT and stem cell signaling
| miRNA | Targets | Regulation | [Ref] |
|---|---|---|---|
| let-7 | EZH2, Lin28B, c-MYC, RAS, HMGA2, CDC25a, CDC34, CDK6 | Cell proliferation, EMT, stem cell renewal | 47, 65 – 67 |
| miR-200 | ZEB1, ZEB2, TGF-β | EMT, stem cell renewal | 66, 68, 69 |
| miR-34a | AR, E2F3, NOTCH1, DLL1, NANOG, SOX2 | AR signaling, stem cell renewal | 51, 65, 68, 70 |
| miR-124 | AR, CDK6, ITGB1, FOXA2 | AR signaling, cell proliferation | 66, 71 |
| mi-27b | AR, LIMK1, VEGF, Rac1 | AR signaling, angiogenesis | 72 – 74 |
| miR-320 | β-Catenin, RUNX2, MIB1 | Stem cell renewal | 54, 75 |
The Biological Significance of miRNAs, AR Splice Variants, EMT and Stem Cell Markers in PCa
The ADT is a common therapeutic strategy for the management of PCa. Although ADT initially show efficacy on tumor suppression, most patients with advanced PCa ultimately develop resistance to the therapy and proceed to CRPC. In the investigation of molecular mechanisms underlying the development of CRPC, ADT has been found to upregulate the levels of AR splice variants [31,32,33]. In PCa after ADT, a high rate of C-terminal truncated AR variants has been observed, suggesting that the AR variants could contribute to CRPC development [11,31,32,33,34,35]. Our recent findings showed a higher expression of AR splice variants such as AR3 (also known as ARv7) and AR132b in PCa tissues, especially in PCa with a higher Gleason grade [36]. The human AR gene is comprised of 8 exons and encodes a multidomain protein including an NH2 terminal transactivation domain (NTD encoded by exon 1), a central DNA-binding domain (DBD encoded by exons 2-3), a hinge region (encoded by exon 4), and a COOH-terminal ligand-binding domain (LBD encoded by exons 4-8; fig. 1). The AR belongs to the steroid receptor superfamily. It functions as a transcription factor. The ligand, androgen, binds to AR and activates AR; AR then translocates to the nucleus and interacts with androgen response elements in the promoter of AR target genes. In this way, AR regulates the transcription of its target genes such as PSA, an important biomarker for PCa. The AR splice variant AR3 is encoded by exons 1-3 and 3b [15,35], while AR132b is encoded by exons 1-3 and 2b [37] (fig. 1). Both AR3 and AR132b variants contain intact NTD and DBD but lack the hinge region and LBD. These variants have been found to be constitutively activated without androgen binding and associated with CRPC [9,11,34,35]. Moreover, AR3 expression has been found to be correlated with risk of tumor recurrence after radical prostatectomy [35]. These findings suggest that AR splice variants confer the castration resistance phenotype and drug resistance characteristics in PCa [9,10,11,34,35,38].
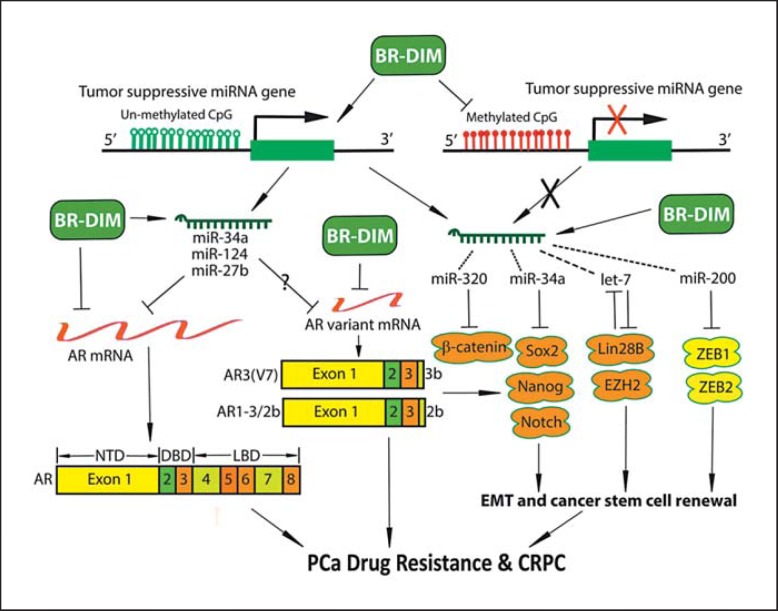
Fig. 1.
The role of miRNA, AR splice variants, EMT and stem cell signaling axis in the progression of CRPC and the effects of BR-DIM on the axis. AR signaling is targeted by miR-34a, miR-124 and miR-27b, while EMT/stem cell signatures are targeted by several miRNAs (miR-43a targeting Sox2, Nanog and Notch1; let-7 targeting Lin28B and EZH2; miR-200 targeting ZEB1 and ZEB2; miR-320 targeting β- catenin).
The EMT was originally recognized as an important differentiation and morphogenetic process in embryogenesis. However, EMT has recently been known as a pathological process during the development of various diseases including inflammation and cancers. Among cancer cells, a small population of cancer cells has been identified as cancer stem cells which retain the capability to self-renew and generate the diverse cell populations that comprise the cancer mass. Recent experimental and clinical evidence suggests that the acquisition of the EMT phenotype and cancer stem cell characteristics significantly contribute to drug resistance and progression of PCa [39,40,41,42,43,44]. Tumor recurrence is also thought to be associated with the EMT and stem cell phenotypes and signatures [45,46]. We found that the stem cell markers Lin28B and EZH2 were highly expressed in PCa tissues, especially in a higher Gleason grade [47]. Importantly, the AR splice variants were positively correlated with the expression levels of stem cell markers such as Lin28B and EZH2 in PCa tissue specimens with a higher Gleason grade [36,48]. Moreover, androgen depletion caused the upregulation of AR and AR splice variants concomitantly with elevated levels of stem cell markers (Lin28B, Nanog, Oct4, Sox2 and CD44) and EMT markers (ZEB1, Snail, Twist, N-cadherin and vimentin) in various PCa cell lines including LNCaP, C4-2B, PC-3, DU145, CWR22Rv1 and VCaP cells [36,49]. These findings strongly suggest that higher expression of AR and AR splice variants after ADT contributes to the stem cell characteristics and induction of EMT in PCa, which will likely lead to the drug resistance and the development of CRPC (fig. 1).
The miRNAs are small single-stranded RNAs. They do not code for proteins or peptides at all. Nevertheless, a single miRNA controls the expression of many target genes through several regulatory processes. The critical step of the regulation is that miRNA imperfectly binds to the 3′-untranslated region of its target mRNAs, leading to the translational repression or target mRNA cleavage. By regulating gene expression, miRNAs control many physiological and pathological processes including cancer development and progression. The miRNAs are emerging as master regulators of specific genes such as AR during PCa progression, and are critically involved in the regulation of stem cell and EMT phenotypes (table 1) [50]. In PCa cells, several tumor suppressive miRNAs have been found to be downregulated. We found that loss of let-7 and miR-34a expression in human PCa tissues, especially those with higher Gleason grades, was associated with increased expression of Lin28B, EZH2, AR and AR variants [36,47,51]. The miR-34 family [51,52], miR-124 [53] and miR-27b have been shown to inhibit the expression of AR and AR splice variants (table 1; fig. 1); however, the levels of those miRNAs are reduced in PCa, which appears to be responsible for higher expression of AR. Moreover, the expression of let-7 and miR-320 families has been found to suppress the expression of EMT and stem cell markers [54]; their expression has been found to be decreased in CRPC cells. Therefore, loss of expression of specific miRNAs appears to increase the levels of both wild-type AR and AR splice variants in PCa, resulting in an increased expression of EMT and stem cell markers which confer drug resistance and leads to further progression of CRPC to metastasis.
The Effects of BR-DIM on PCa
The DIM is a dimeric product of indole-3-carbinol (I3C). I3C is mainly found from naturally occurring glucosinolates that exist in a lot of vegetables including members of the family Cruciferae, and particularly members of the genus Brassica. Under the acidic conditions of the stomach, I3C proceeds to broad and fast self-condensation reactions to form different derivatives including DIM, which appears to be the main derivative and condensed product of I3C. Epidemiological studies found that human exposure to indoles through consumption of cruciferous vegetable could decrease cancer risk [55]. The molecular effects and mechanisms of action of DIM in various cancers have been investigated [19,56,57]. We conducted cDNA microarray experiments to reveal the gene expression profiles altered by DIM treatment in PC-3 cells [58]. We found that DIM regulated the expression of many genes that are involved in the control of carcinogenesis, cell survival and physiological behaviors. In addition, DIM also regulates signal transduction in several important signaling pathways such as NF-κB, Akt-mTOR, AR and others in various PCa cells including LNCaP, C4-2B and PC-3 cells. DIM significantly inhibited NF-κB DNA binding activity and suppressed the activation of Akt with induction of apoptosis [23,25]. Importantly, we found that BR-DIM, a formulated DIM with higher bioavailability [59], significantly inhibited cell proliferation and induced apoptosis with the downregulation of AR and its downstream gene PSA in LNCaP and C4-2B cells [26]. BR-DIM also significantly inhibited Akt activation, FOXO3a phosphorylation, Wnt activation, NF-κB activation, AR phosphorylation, and AR nuclear translocation [19,26]. These results suggest that BR-DIM inhibits the growth of PCa through regulation of Akt/FOXO3a/Wnt/NF-κB/AR signaling.
Furthermore, we observed that BR-DIM treatment suppressed the expression of AR, EZH2 and Lin28, caused AR nuclear exclusion, and induced re-expression of let-7 and miR-34a in LNCaP and CWR22Rv1 cells [47,51]. In addition, PCa tissue specimens from patients prior to radical prostatectomy clearly showed strong nuclear AR staining before BR-DIM intervention, whereas after BR-DIM intervention the level of AR expression was decreased. There was nuclear exclusion of the AR in PCa tissue specimens after BR-DIM intervention. BR-DIM administered to patients with PCa before radical prostatectomy inhibited the expression of the cancer stem cell marker Lin28 [36]. More interestingly, BR-DIM decreased the expression of wild-type AR, AR variants and PSA in CWR22Rv1 and LNCaP cells and sphere-forming cells from CWR22Rv1 and LNCaP cells. BR-DIM also inhibited the expression of stem cell markers including Lin28B, Nanog, Oct4, Sox2 and CD44 in sphere-forming cells from CWR22Rv1 cells [36]. Moreover, we also observed that BR-DIM treatment led to the re-expression of miR-34a, miR-27b, miR-320, miR-200 and let-7 significantly, which are regulators of AR, AR splice variants, and stem cell and EMT markers [36,47,51]. By inhibiting the expression of AR splice variants, BR-DIM suppressed cell migration and prostasphere-forming ability with the downregulation of EMT and stem cell markers in LNCaP and CWR22Rv1 cells [36]. These results suggest that BR-DIM could induce re-expression of several specific miRNAs, leading to the downregulation of AR, AR splice variants, EMT and stem cell markers, suggesting an inhibitory effect of BR-DIM on the development of drug resistance and CRPC (fig. 1).
The induction of miRNA expression by BR-DIM could be mediated through epigenetic regulation. Histone deacetylation and methylation of the CpG region in the promoter regions of miRNA genes are frequently associated with loss of miRNA expression [53,60,61,62,63]. We observed that BR-DIM caused re-expression of miR-34a by demethylating the miR-34a promoter [51]. We also found that BR-DIM induced acetylation of lysine 9 on histone 3 (Ac-H3) in LNCaP and C4-2B cells. Thus, BR-DIM could cause the re-expression of miRNAs, such as miR-34a, miR-124, miR-27b, miR-320 and let-7 by modifying promoter methylation and histone acetylation, suggesting that BR-DIM has the capacity for epigenetic regulation of miRNAs (fig. 1).
More importantly, we have conducted a dose escalation, phase I clinical study of BR-DIM with objectives to determine the maximum tolerated dose, toxicity profile and pharmacokinetics of BR-DIM, and to assess its effects on serum PSA and quality of life [64]. The results showed that BR-DIM was well tolerated and that modest efficacy was demonstrated. We have subsequently conducted a phase II clinical trial (NCT00888654; https://clinicaltrials.gov/ct2/show/NCT00888654?term=DIM&rank=2) for assessing the value of BR-DIM in the treatment of PCa, and the results were promising. The results will be published soon in a clinical journal. Our preclinical and clinical studies provide the scientific basis for a ‘proof-of-concept’ clinical trial in CRPC patients treated with enzalutamide in combination with BR-DIM, which is being planned. This strategy could also be expanded in further future clinical trials in PCa patients to determine whether the patients could achieve a better treatment outcome which could in part be mediated by delaying or preventing the development of CRPC.
Conclusion and Future Perspectives
The emerging evidence has demonstrated that ADT in PCa induces the expression of AR and AR variants, which lead to the upregulation of EMT and stem cell markers including ZEB1, EZH2, Lin28, Nanog, Oct4, Sox2 and CD44. The upregulated AR, AR splice variants, EMT and stem cell markers may contribute to the development of CRPC and drug resistance. Importantly, the in vitro and in vivo investigations showed that BR-DIM upregulates the expression of several miRNAs including miR-34a, miR-124, miR-27b, miR-320, miR-200 and let-7 in human PCa tissue specimens and cell lines, leading to the downregulation of AR, AR splice variants, EMT and stem cell markers. Clinical trials showed safety and bioavailability of BR-DIM administration and the inhibitory effects of BR-DIM on AR signaling were clearly seen. These results demonstrate that BR-DIM could be a promising agent for overcoming enzalutamide (or other ADT) resistance by regulating the miRNA/AR splice variant/stem cell signature axis.
Based on these preclinical and clinical data, we are planning to conduct a ‘proof-of-concept’ clinical trial using enzalutamide together with BR-DIM treatment in metastatic CRPC patients to investigate whether BR- DIM in vivo in this patient population could inactivate AR and AR splice variants by regulating the expression of miRNAs, resulting in decreased expression of EMT and stem cell markers, and increased sensitivity of PCa to enzalutamide. This clinical strategy would likely lead to better treatment outcomes of PCa patients which may in part be due to delaying or preventing the development of CRPC and subsequent metastasis (having EMT and cancer stem cell characteristics). Such studies would likely have a significant impact on the management of PCa patients in the future.
Acknowledgment
The authors’ work cited in this article was in part funded by grants from the National Cancer Institute, NIH (R01CA164318 awarded to F.H.S.). We would like to thank all the colleagues who either directly or indirectly participated in the clinical trial in PCa patients with BR-DIM, and who helped in the acquisition of prostate tissue specimens.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Culig Z, Santer FR. Molecular aspects of androgenic signaling and possible targets for therapeutic intervention in prostate cancer. Steroids. 2013;78:851–859. doi: 10.1016/j.steroids.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Nadiminty N, Tummala R, Liu C, et al. NF-kappaB2/p52 induces resistance to enzalutamide in prostate cancer: role of androgen receptor and its variants. Mol Cancer Ther. 2013;12:1629–1637. doi: 10.1158/1535-7163.MCT-13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt LJ, Tindall DJ. Androgen receptor: past, present and future. Curr Drug Targets. 2013;14:401–407. doi: 10.2174/1389450111314040002. [DOI] [PubMed] [Google Scholar]
- 5.Sharifi N. Minireview: androgen metabolism in castration-resistant prostate cancer. Mol Endocrinol. 2013;27:708–714. doi: 10.1210/me.2013-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh S, Lin HK, Kang HY, et al. From HER2/Neu signal cascade to androgen receptor and its coactivators: a novel pathway by induction of androgen target genes through MAP kinase in prostate cancer cells. Proc Natl Acad Sci USA. 1999;96:5458–5463. doi: 10.1073/pnas.96.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asangani IA, Dommeti VL, Wang X, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornberg E, Ylitalo EB, Crnalic S, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu R, Lu C, Mostaghel EA, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Morrissey C, Sun S, et al. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PLoS One. 2011;6:e27970. doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung PH, Gayed BA, Thoreson GR, et al. Emerging drugs for prostate cancer. Expert Opin Emerg Drugs. 2013;18:533–550. doi: 10.1517/14728214.2013.864635. [DOI] [PubMed] [Google Scholar]
- 13.Dhingra R, Sharma T, Singh S, et al. Enzalutamide: a novel anti-androgen with prolonged survival rate in CRPC patients. Mini Rev Med Chem. 2013;13:1475–1486. doi: 10.2174/13895575113139990003. [DOI] [PubMed] [Google Scholar]
- 14.Noonan KL, North S, Bitting RL, et al. Clinical activity of abiraterone acetate in patients with metastatic castration-resistant prostate cancer progressing after enzalutamide. Ann Oncol. 2013;24:1802–1807. doi: 10.1093/annonc/mdt138. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Chan SC, Brand LJ, et al. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakazawa M, Antonarakis ES, Luo J. Androgen receptor splice variants in the era of enzalutamide and abiraterone. Horm Cancer. 2014;5:265–273. doi: 10.1007/s12672-014-0190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao B, Qi Y, Zhang G, et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646–1656. doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Wang Z, Kong D, et al. Regulation of FOXO3a/beta-catenin/GSK-3beta signaling by 3,3′-diindolylmethane contributes to inhibition of cell proliferation and induction of apoptosis in prostate cancer cells. J Biol Chem. 2007;282:21542–21550. doi: 10.1074/jbc.M701978200. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Wang Z, Kong D, et al. Regulation of Akt/FOXO3a/GSK-3beta/AR signaling network by isoflavone in prostate cancer cells. J Biol Chem. 2008;283:27707–27716. doi: 10.1074/jbc.M802759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fajardo AM, MacKenzie DA, Ji M, et al. The curcumin analog ca27 down-regulates androgen receptor through an oxidative stress mediated mechanism in human prostate cancer cells. Prostate. 2012;72:612–625. doi: 10.1002/pros.21464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi HY, Lim JE, Hong JH. Curcumin interrupts the interaction between the androgen receptor and Wnt/beta-catenin signaling pathway in LNCaP prostate cancer cells. Prostate Cancer Prostatic Dis. 2010;13:343–349. doi: 10.1038/pcan.2010.26. [DOI] [PubMed] [Google Scholar]
- 23.Kong D, Li Y, Wang Z, et al. Inhibition of angiogenesis and invasion by 3,3′-diindolylmethane is mediated by the nuclear factor-kappaB downstream target genes MMP-9 and uPA that regulated bioavailability of vascular endothelial growth factor in prostate cancer. Cancer Res. 2007;67:3310–3319. doi: 10.1158/0008-5472.CAN-06-4277. [DOI] [PubMed] [Google Scholar]
- 24.Kong D, Banerjee S, Huang W, et al. Mammalian target of rapamycin repression by 3,3′-diindolylmethane inhibits invasion and angiogenesis in platelet-derived growth factor-D-overexpressing PC3 cells. Cancer Res. 2008;68:1927–1934. doi: 10.1158/0008-5472.CAN-07-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Chinni SR, Sarkar FH. Selective growth regulatory and pro-apoptotic effects of DIM is mediated by AKT and NF-kappaB pathways in prostate cancer cells. Front Biosci. 2005;10:236–243. doi: 10.2741/1523. [DOI] [PubMed] [Google Scholar]
- 26.Bhuiyan MM, Li Y, Banerjee S, et al. Down-regulation of androgen receptor by 3,3′-diindolylmethane contributes to inhibition of cell proliferation and induction of apoptosis in both hormone-sensitive LNCaP and insensitive C4-2B prostate cancer cells. Cancer Res. 2006;66:10064–10072. doi: 10.1158/0008-5472.CAN-06-2011. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, VandenBoom TG, Wang Z, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Kong D, Ahmad A, et al. Targeting bone remodeling by isoflavone and 3,3′-diindolylmethane in the context of prostate cancer bone metastasis. PLoS One. 2012;7:e33011. doi: 10.1371/journal.pone.0033011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soubani O, Ali AS, Logna F, et al. Re-expression of miR-200 by novel approaches regulates the expression of PTEN and MT1-MMP in pancreatic cancer. Carcinogenesis. 2012;33:1563–1571. doi: 10.1093/carcin/bgs189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, VandenBoom TG, Kong D, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Hwang TH, Oseth LA, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31:4759–4767. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu LL, Xie N, Sun S, et al. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinestel J, Luedeke M, Arndt A, et al. Detecting predictive androgen receptor modifications in circulating prostate cancer cells. Oncotarget. 2015 doi: 10.18632/oncotarget.3925. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun S, Sprenger CC, Vessella RL, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Z, Yang X, Sun F, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong D, Sethi S, Li Y, et al. Androgen receptor splice variants contribute to prostate cancer aggressiveness through induction of EMT and expression of stem cell marker genes. Prostate. 2015;75:161–174. doi: 10.1002/pros.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dehm SM, Schmidt LJ, Heemers HV, et al. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. 2012;287:19736–19749. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JM, Dedhar S, Kalluri R, et al. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins AT, Maitland NJ. Prostate cancer stem cells. Eur J Cancer. 2006;42:1213–1218. doi: 10.1016/j.ejca.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 41.Kasper S. Stem cells: the root of prostate cancer? J Cell Physiol. 2008;216:332–336. doi: 10.1002/jcp.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasper S. Identification, characterization, and biological relevance of prostate cancer stem cells from clinical specimens. Urol Oncol. 2009;27:301–303. doi: 10.1016/j.urolonc.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawson DA, Xin L, Lukacs R, et al. Prostate stem cells and prostate cancer. Cold Spring Harb Symp Quant Biol. 2005;70:187–196. doi: 10.1101/sqb.2005.70.003. [DOI] [PubMed] [Google Scholar]
- 44.Lawson DA, Witte ON. Stem cells in prostate cancer initiation and progression. J Clin Invest. 2007;117:2044–2050. doi: 10.1172/JCI32810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brabletz T, Jung A, Reu S, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brabletz T, Jung A, Spaderna S, et al. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 47.Kong D, Heath E, Chen W, et al. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One. 2012;7:e33729. doi: 10.1371/journal.pone.0033729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Kruithof-de Julio M, Economides KD, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y, Wang BE, Leong KG, et al. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 2012;72:527–536. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]
- 50.Cano A, Nieto MA. Non-coding RNAs take centre stage in epithelial-to-mesenchymal transition. Trends Cell Biol. 2008;18:357–359. doi: 10.1016/j.tcb.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Kong D, Heath E, Chen W, et al. Epigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR-DIM treatment. Am J Transl Res. 2012;4:14–23. [PMC free article] [PubMed] [Google Scholar]
- 52.Kashat M, Azzouz L, Sarkar SH, et al. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am J Transl Res. 2012;4:432–442. [PMC free article] [PubMed] [Google Scholar]
- 53.Shi XB, Xue L, Ma AH, et al. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene. 2012;32:4130–4138. doi: 10.1038/onc.2012.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsieh IS, Chang KC, Tsai YT, et al. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–538. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 55.Higdon JV, Delage B, Williams DE, et al. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol Res. 2007;55:224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarkar S, Dubaybo H, Ali S, et al. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res. 2013;3:465–477. [PMC free article] [PubMed] [Google Scholar]
- 57.Fan S, Meng Q, Xu J, et al. DIM (3,3′-diindolylmethane) confers protection against ionizing radiation by a unique mechanism. Proc Natl Acad Sci USA. 2013;110:18650–18655. doi: 10.1073/pnas.1308206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Li X, Sarkar FH. Gene expression profiles of I3C- and DIM-treated PC3 human prostate cancer cells determined by cDNA microarray analysis. J Nutr. 2003;133:1011–1019. doi: 10.1093/jn/133.4.1011. [DOI] [PubMed] [Google Scholar]
- 59.Anderton MJ, Manson MM, Verschoyle R, et al. Physiological modeling of formulated and crystalline 3,3′-diindolylmethane pharmacokinetics following oral administration in mice. Drug Metab Dispos. 2004;32:632–638. doi: 10.1124/dmd.32.6.632. [DOI] [PubMed] [Google Scholar]
- 60.Gebauer K, Peters I, Dubrowinskaja N, et al. HSA-miR-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br J Cancer. 2013;108:131–138. doi: 10.1038/bjc.2012.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 62.Pinto R, Pilato B, Ottini L, et al. Different methylation and microRNA expression pattern in male and female familial breast cancer. J Cell Physiol. 2013;228:1264–1269. doi: 10.1002/jcp.24281. [DOI] [PubMed] [Google Scholar]
- 63.Wang P, Chen L, Zhang J, et al. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2014;33:514–524. doi: 10.1038/onc.2012.598. [DOI] [PubMed] [Google Scholar]
- 64.Heath EI, Heilbrun LK, Li J, et al. A phase I dose-escalation study of oral BR-DIM (BioResponse 3,3′-diindolylmethane) in castrate-resistant, non-metastatic prostate cancer. Am J Transl Res. 2010;2:402–411. [PMC free article] [PubMed] [Google Scholar]
- 65.Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 66.Spizzo R, Nicoloso MS, Croce CM, et al. SnapShot: microRNAs in cancer. Cell. 2009;137:586. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 67.Kong D, Banerjee S, Ahmad A, et al. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS One. 2010;5:e12445. doi: 10.1371/journal.pone.0012445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ceppi P, Peter ME. MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene. 2014;33:269–278. doi: 10.1038/onc.2013.55. [DOI] [PubMed] [Google Scholar]
- 69.Kong D, Li Y, Wang Z, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ostling P, Leivonen SK, Aakula A, et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–1967. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- 71.Shi XB, Xue L, Ma AH, et al. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene. 2013;32:4130–4138. doi: 10.1038/onc.2012.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wan L, Zhang L, Fan K, et al. MiR-27b targets LIMK1 to inhibit growth and invasion of NSCLC cells. Mol Cell Biochem. 2014;390:85–91. doi: 10.1007/s11010-013-1959-1. [DOI] [PubMed] [Google Scholar]
- 73.Ye J, Wu X, Wu D, et al. miRNA-27b targets vascular endothelial growth factor C to inhibit tumor progression and angiogenesis in colorectal cancer. PLoS One. 2013;8:e60687. doi: 10.1371/journal.pone.0060687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ishteiwy RA, Ward TM, Dykxhoorn DM, et al. The microRNA-23b/-27b cluster suppresses the metastatic phenotype of castration-resistant prostate cancer cells. PLoS One. 2012;7:e52106. doi: 10.1371/journal.pone.0052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamam D, Ali D, Vishnubalaji R, et al. microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells. Cell Death Dis. 2014;5:e1499. doi: 10.1038/cddis.2014.462. [DOI] [PMC free article] [PubMed] [Google Scholar]