Abstract
Triple negative breast cancer represents a heterogeneous group of breast carcinomas, both at the histologic and genetic level. While recent molecular studies have comprehensively characterized the genetic landscape of these tumors, few have integrated a detailed histologic examination into the analysis. In this study, we defined the genetic alterations in 39 triple negative breast cancers using a high-depth targeted massively parallel sequencing assay and correlated the findings with a detailed morphologic analysis. We obtained representative frozen tissue of primary triple negative breast cancers from patients treated at our institution between 2002 and 2010. We characterized tumors according to their histologic subtype and morphologic features. DNA was extracted from paired frozen primary tumor and normal tissue samples and was subjected to a targeted massively parallel sequencing platform comprising 229 cancer associated genes common across all experiments. The average number of non-synonymous mutations was 3 (range 0–10) per case. The most frequent somatic alterations were mutations in TP53 (74%) and PIK3CA (10%) and MYC amplifications (26%). Triple negative breast cancers with apocrine differentiation less frequently harbored TP53 mutations (25%) and MYC gains (0%), and displayed a high mutation frequency in PIK3CA and other PI3K signaling pathway related genes (75%). Using a targeted massively parallel sequencing platform, we identified the key somatic genetic alterations previously reported in triple negative breast cancers. Furthermore, our findings show that triple negative breast cancers with apocrine differentiation constitute a distinct subset, characterized by a high frequency of PI3K pathway alterations similar to luminal subtypes of breast cancer.
Introduction
Triple negative breast cancer represents a heterogenous group of breast carcinomas that lack expression of estrogen receptor (ER), progesterone receptor (PR) and HER2. As a group, triple negative breast cancers have aggressive clinical course and poor prognosis.1 Morphologically, the majority of the triple negative breast cancers are high-grade invasive ductal carcinomas of no special type, and associated with tumor necrosis, pushing borders, and prominent tumor infiltrating lymphocytes.2, 3 Some special histologic subtypes also show a triple negative phenotype, including most metaplastic carcinomas, a subset of carcinomas with apocrine differentiation, adenoid cystic carcinomas, secretory carcinomas, and acinic cell carcinomas.4–12 Given the lack of expression of ER, PR and HER2, chemotherapy is currently the only option for systemic therapy in patients with triple negative breast cancer.
Recent studies have described the comprehensive molecular genetic landscape of human breast cancers using whole genome/exome sequencing, RNA sequencing, and Affymetrix SNP array analyses.13–15 Recurrent somatic mutations with greater than 10% frequency across all breast cancer subtypes were found in only three genes: TP53, PIK3CA, and GATA3, occurred at 37%, 36%, and 11% respectively.13 The patterns of somatic mutations among the intrinsic breast cancer subtypes are different. Basal-like and triple negative breast cancers showed a high frequency of TP53 mutation (80%), whereas only 12% of luminal A and 29% luminal B tumors harbor TP53 mutations. PIK3CA is the most frequently mutated gene in luminal A (45%) and luminal B (29%) breast cancer. Although PIK3CA is the second most frequently mutated gene in triple negative breast cancers, the frequency of PIK3CA mutation in triple negative breast cancers is lower than that in luminal breast cancer, at 9–10%.13, 14 While such studies have made great strides in cataloguing the genomic diversity inherent in this heterogeneous group of tumors, few have integrated their findings with a detailed histologic evaluation.
In this study, we characterized the genetic alterations in a group of triple negative breast cancers and correlated the results with a detailed morphologic analysis. We utilized the Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) platform,16, 17 a targeted next generation sequencing assay targeting all coding regions and selected regulatory and intronic regions of 229 most common cancer genes.
Materials and Methods
Patient Selection
This study was approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board. Informed patient consent was obtained as appropriate, following the protocols approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board. We identified patients with primary triple negative breast cancer treated at our institution between 2002 and 2010 through a search of the institution database. Triple negative breast cancer was defined as invasive breast carcinoma with ER and PR staining in less than 1% of the tumor cells by immunohistochemistry and no HER2 overexpression [defined as negative (0 to 1+) or equivocal (2+) staining by immunohistochemistry and no amplification by fluorescence in situ hybridization], in accordance with the American Society of Clinical Oncology/College of American Pathologists guidelines.18–21 We retrieved available frozen samples of paired primary tumor and normal tissue for the study. Patients received neoadjuvant chemotherapy were excluded from the study.
Clinicopathological Review
Clinicopathological data for each patient, including age at diagnosis, BRCA1 germline mutation status, tumor characteristics (size, grade, special histologic subtypes and morphologic features – details below), lymph node involvement, distant metastases, length of follow up, and survival status, were recorded. All available slides were reviewed by two pathologists (HYW and REE) to assess the histologic features.
Special histologic subtypes (metaplastic carcinoma and carcinoma with apocrine differentiation) were defined according the 2012 WHO classification.22, 23 Apocrine differentiation was defined as nuclear enlargement with prominent nucleoli and abundant, granular, eosinophilic cytoplasm.23 Metaplastic carcinoma was defined as carcinoma with squamous differentiation, spindle cell morphology, or mesenchymal elements.22 Other morphologic features commonly seen in triple negative breast cancer, such as a large central acellular zone of necrosis or fibrosis and tumor infiltrating lymphocytes were also recorded. Large central acellular zone morphology was defined as the presence of a large, centrally located paucicellular or acellular area occupying >30% of the tumor area and not associated with extensive coagulative necrosis, squamous debris, or overt cartilaginous matrix production.24 Tumor infiltrating lymphocytes were scored as a percentage of the stromal areas occupied by mononuclear cells including lymphocytes and plasma cells, excluding granulocytes and other polymorphonuclear leukocytes.25 Tumor infiltrating lymphocytes were scored according to the recommendations by an International Tumor Infiltrating Lymphocytes Working Group.25 Tumors with ≥50% tumor infiltrating lymphocytes were classified as having prominent tumor infiltrating lymphocytes.26 Triple negative breast cancers with none of these special features were classified as triple negative breast cancer not otherwise specified.
Immunohistochemistry for Androgen Receptor
Immunohistochemical stain was performed on representative 4 micron-thick sections in cases with available paraffin blocks using antibodies against the androgen receptor (DAKO AR441 clone; 1:75 dilution; pretreatment with citric buffer, pH 6.2; HRP detection; DAB chromogen). Scoring of androgen receptor paralleled the American Society of Clinical Oncology/College of American Pathologists guidelines for ER and PR. Nuclear staining for androgen receptor in ≥1% of tumor cells was considered as positive.
DNA Extraction
DNA was extracted from paired frozen primary tumor (>75% tumor content) and normal tissue samples using the DNeasy Blood and Tissue Kit (QIAGEN). DNA samples were subjected to targeted massively parallel sequencing using the MSK-IMPACT sequencing assay.16 (details below)
The MSK-IMPACT Assay
Deep targeted sequencing of key cancer-associated genes was performed using the MSK-IMPACT assay.16 In this assay, target-specific oligonucleotide probes were designed to capture all protein-coding exons of most common cancer-related genes (229 genes at the time of this study; supplemental table 1) for hybrid selection (Agilent SureSelect or Nimblegen SeqCap) as previously described.17, 27 For 26 samples (13 tumor/normal pairs), barcoded sequence libraries (Illumina TruSeq) were prepared using 500 ng of input tumor or matched normal DNA according to the manufacturer’s instructions. Libraries were pooled at equimolar concentrations (100 ng per tumor library and 50 ng per normal library) for a single exon capture reaction (Agilent SureSelect) as previously described. For the remaining samples, barcoded sequence libraries were prepared using 250 ng of input DNA using a hybrid protocol based on the NEBNext DNA Library Prep Kit (New England Biolabs). Manufacturer’s instructions were followed with two substitutions: we used NEXTflex barcoded adapters (Bio Scientific) and HiFi DNA polymerase (Kapa Biosystems). Libraries were pooled at 100 ng per tumor library and 50 ng per normal library and captured using custom biotinylated DNA probes (Nimblegen SeqCap). To prevent off-target hybridization in all capture reactions, we spiked in a pool of blocker oligonucleotides complementary to the full sequences of all barcoded adaptors (to a final total concentration of 10 μM). Hybridized DNA was sequenced on an Illumina HiSeq 2000 to generate paired-end 75-bp reads.
Data were demultiplexed using CASAVA, and reads were aligned to the reference human genome (hg19) using the Burrows-Wheeler Alignment tool.28 Local realignment and quality score recalibration were performed using the Genome Analysis Toolkit (GATK) according to GATK best practices.29 We achieved mean exon sequence coverage of 507x (678x for all tumor samples and 348x for all normal samples). Deep sequencing ensured sensitivity for detecting mutations in multiclonal and stroma-admixed samples and enabled accurate determination of mutation allele frequencies.
Sequence data were analyzed to identify three classes of somatic alterations: single-nucleotide variants, small insertions/deletions (indels), and copy number alterations, adopting the CLIA-compliant analysis methods as previously published.16 Single-nucleotide variants were called using MuTect,30 and retained if the variant allele frequency in the tumor was >5 times that in the matched normal. Indels were called using the SomaticIndelDetector tool in GATK.29 Somatic single-nucleotide variants and small indels with variant allele frequency <5% were excluded. All candidate mutations and indels were further reviewed manually using the Integrative Genomics Viewer.31 The mean sequence coverage was calculated using the DepthOfCoverage tool in GATK and was used to compute copy number. Copy number gains and losses were determined by calculating the tumor to normal ratio in normalized sequence coverage across all target exons following a loss normalization to adjust for G/C content. Ratios <0.5 were considered as deletions, between 0.5 and 0.7 were considered copy number losses, between 1.3 and 1.5 were considered copy number gains and ratios >1.5 were considered as amplifications.
Mutation Significance Analysis
Mutations resulting in frameshift insertions or deletions (indels) and those involving splice sites or resulting in nonsense mutations were considered pathogenic. We assessed the functional effects of missense mutations using CHASM,32 FATHMM,33 Mutation Assessor,34 Mutation Taster35 and Polyphen-2.36 Mutations predicted to be deleterious or cancer drivers by at least two algorithms were considered potential driver mutations. Deleterious or pathogenic classifications from Mutation Taster and Polyphen-2 were counted as one as these two algorithms frequently returned the same predictions and were underpinned by similar bioinformatic principles. In-frame indels were considered pathogenic if predicted to be deleterious by either PROVEAN37 or Mutation Taster.
Sanger sequencing
Putative somatic mutations in selected genes of interest identified by MSK-IMPACT sequencing were further investigated by Sanger sequencing. Primer sets that amplify mutated exons of the selected genes were designed as previously described38 and are available in Supplementary Table 2. PCR amplification of 5ng of genomic DNA was performed using the AmpliTaq 360 Master Mix Kit (Life Technologies) on a Veriti Thermal Cycler (Life Technologies) as previously described.38 PCR fragments were purified (ExoSAP-IT, Affymetrix) and sequenced on an ABI 3730 capillary sequencer using the ABI BigDye Terminator chemistry (v3.1, Life Technologies). Sequences of the forward and reverse strands were analyzed using MacVector software (MacVector, Inc),38 and all analyses were performed in duplicates. Representative Sanger sequencing chromatograms of validated mutations are shown in supplementary Figure 1. Of the 26 putative somatic mutations selected, 24 were successfully validated (Supplementary Table 3), resulting in an overall validation rate of 92%, providing confidence to include the remaining putative mutations not subjected to Sanger sequencing. The two variants that failed Sanger sequencing were removed from further analyses.
Statistical Analysis
Statistical analysis of clinicopathologic data was performed using a two-tailed student t-test for continuous variables and a Fisher’s exact test for categorical variables. Mutational frequencies and GISTIC2 copy number data for the triple negative breast cancers from the Cancer Genome Atlas were obtained from the publication data portal (https://theCancerGenomeAtlas-data.nci.nih.gov/docs/publications/brca_2012/). For the statistical analysis comparing proportion of cases affected by non-synonymous mutational or copy number alterations (amplifications and deletions, or 2 and −2 in the GISTIC2 copy number data) in a particular gene between the current cohort and triple negative breast cancers from the Cancer Genome Atlas and between subtypes of triple negative breast cancers within the current cohort, Fisher’s exact test for nonparametric variables was employed, with all probabilities reported as two-tailed. P values <0.05 were considered statistically significant. All analyses were performed using R v3.0.2.
Results
Clinicopathologic characteristics of the patients
Clinicopathological characteristics of the patients are summarized in Table 1. All patients were female. The median age at diagnosis was 43 years (range 28–78). Fourteen patients had genetic testing for BRCA germline mutations and 9 of which were found to be BRCA1 germline mutation carriers. The median tumor size was 2.9 cm (range 1.2–8.5). Most cases were of high grade (histologic grade III and nuclear grade III). Four (10%) cases had apocrine differentiation, 1 (3%) case was metaplastic carcinoma with chondroid matrix production, and the remaining 34 (87%) triple negative breast cancers were invasive ductal carcinoma of no special type as defined by the WHO classification criteria (Figure 1).39 Of these, 6 (15%) had large central acellular zone and 6 (15%) had prominent tumor infiltrating lymphocytes (Figure 1). Cases with large central acellular zone and prominent tumor infiltrating lymphocytes did not overlap. The remaining 22 triple negative breast cancers (56%) had no special morphologic features (triple negative breast cancer not otherwise specified).
Table 1.
Clinicopathological characteristics of 39 cases
| All cases (n=39) | TNBC NOS (n=22) | Apocrine (n=4) | Metaplastic (n=1) | LCAZ (n=6) | Prominent TILs (n=6) | |
|---|---|---|---|---|---|---|
| Median age (range), years | 43 (28–78) | 37 (28–78) | 64 (52–75) | 51 | 61 (39–67) | 35 (29–58) |
| BRCA1 germline mutation carrier | 9 (23%) | 8 (36%) | 0 | 0 | 0 | 1 (17%) |
| Median tumor size (range), cm | 2.9 (1.2–8.5) | 3.0 (1.2–8.5) | 1.7 (1.4–2.5) | 2.5 | 3.1 (2.1–3.7) | 2.8 (2.5–3.5) |
| Histologic grade* | ||||||
| II | 4 (10%) | 1 (5%) | 2 (50%) | 0 | 1 (17%) | 0 |
| III | 35 (90%) | 21 (95%) | 2 (50%) | 1 (100%) | 5 (83%) | 6 (100%) |
| Nuclear grade** | ||||||
| II | 3 (8%) | 1 (5%) | 2 (50%) | 0 | 0 | 0 |
| III | 36 (92%) | 21 (95%) | 2 (50%) | 1 (100%) | 6 (100%) | 6 (100%) |
| Lymph node metastasis | 19 (49%) | 11 (50%) | 2 (50%) | 0 | 2 (33%) | 4 (67%) |
| Median follow up (range), month | 62 (10–143) | 58 (24–143) | 81 (53–117) | 71 | 47 (10–122) | 66 (32–137) |
| Distant metastasis | 17 (44%) | 9 (41%) | 1 (25%) | 1 (100%) | 4 (67%) | 2 (33%) |
| Lung | 11 | 7 | 0 | 0 | 3 | 1 |
| Bone | 6 | 3 | 1 | 0 | 2 | 0 |
| Brain | 5 | 3 | 0 | 1 | 0 | 1 |
| Liver | 2 | 1 | 1 | 0 | 0 | 0 |
| Other | 1 | 1 | 0 | 0 | 0 | 0 |
| Survival | ||||||
| NED | 22 (56%) | 13 (59%) | 3 (75%) | 0 | 2 (33%) | 4 (67%) |
| AWD | 1 (3%) | 0 | 0 | 0 | 1 (17%) | 0 |
| DOD | 16 (41%) | 9 (41%) | 1 (25%) | 1 (100%) | 3 (50%) | 2 (33%) |
Abbreviations: TNBC, triple negative breast cancer; NOS, not otherwise specified; LCAZ, large central acellular zone; TILs, tumor infiltrating lymphocytes; NED, no evidence of disease; AWD, alive with disease; DOD, died of disease.
No histologic grade I tumor is present in this cohort.
No nuclear grade I tumor is present in this cohort.
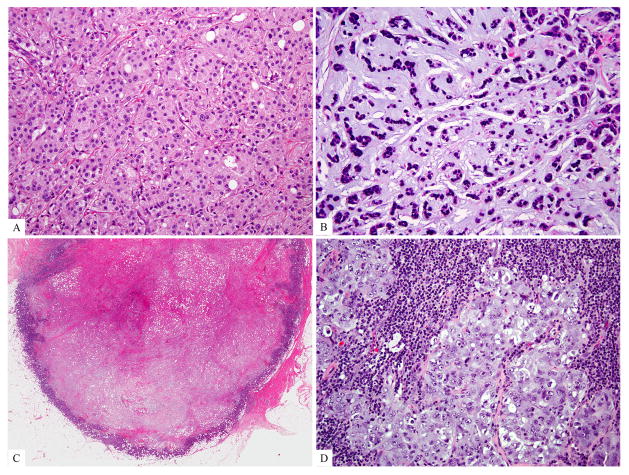
Figure 1. Special histologic subtypes and special morphologic features of triple negative breast cancers.
A) Carcinoma with apocrine differentiation (apocrine triple negative breast cancer); B) metaplastic carcinoma; C) tumor with a large central acellular zone of necrosis or fibrosis; D) tumor with prominent tumor infiltrating lymphocytes.
Lymph node involvement at presentation was seen in 19 (49%) patients and 17 (44%) patients developed distant metastases. The most common site of distant metastasis was lung (n=11), followed by bone (n=6), brain (n=5) and liver (n=2). Six patients had distant metastases to multiple sites. Sixteen (41%) patients died from their breast cancer at a median follow up interval of 62 months (range 10–143).
Immunohistochemistry
Paraffin blocks were available in 30 of 39 cases, including 17 triple negative breast cancers not otherwise specified, 5 tumors with prominent tumor infiltrating lymphocytes, 4 tumors with large central acellular zone, and all 4 triple negative breast cancers with apocrine differentiation. Four tumors were positive for androgen receptor by immunohistochemistry; 3 of these were triple negative breast cancers with apocrine differentiation and the remaining case was triple negative breast cancer not otherwise specified (75% versus 4%; p = 0.0038).
Somatic mutations and copy number alterations
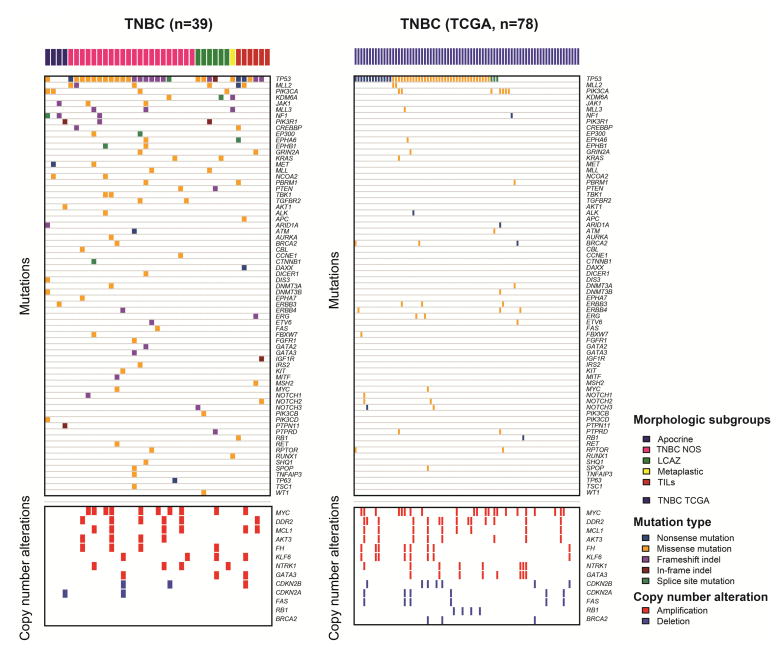
The average depth of sequencing was 678x for tumor samples and 348x for normal samples. Among the 229 genes profiled across our entire cohort, the average number of non-synonymous mutations per case was 3.33 (range 0–10), significantly higher than that of the triple negative breast cancers in the Cancer Genome Atlas data set (mean 2.37, range 0–7, p = 0.008737, Mann-Whitney U test). The most frequently mutated genes in our cohort were TP53 (29 cases, 74%), followed by KMT2D (also known as MLL2, 6 cases, 15%) and PIK3CA (4 cases, 10%) (Figure 2). While the frequencies of TP53 and PIK3CA mutations are similar to those of triple negative breast cancers reported by the Cancer Genome Atlas and other studies,13, 14 mutations in MLL2 were significantly more frequent in our cohort than in triple negative breast cancers from the Cancer Genome Atlas (MLL2: 6/39 (15%) vs 2/78 cases (3%), p = 0.0162, Fisher’s exact test). Other statistically significant differences between our cohort and the cohort of triple negative breast cancers from the Cancer Genome Atlas include the frequency of KDM6A, JAK1 and PIK3R1 mutations, each present in 3 (8%) in our cohort in contrast to none of the cases in the Cancer Genome Atlas cohort (p = 0.0351, Fisher’s exact tests).
Figure 2. The landscape of somatic genetic alterations in triple negative breast cancers.
Top panels: the repertoire of somatic non-synonymous mutations in our triple negative breast cancer cohort as compared to the corresponding genes in triple negative breast cancers in the Cancer Genome Atlas Cohort. Bottom panels: the genes amplified in at least 10% of our triple negative breast cancer cohort or deleted in at least 5% of our triple negative breast cancer cohort, as compared to the same genes in triple negative breast cancers in the Cancer Genome Atlas cohort.
Abbreviations: TNBC, triple negative breast cancer; TCGA, the Cancer Genome Atlas; NOS, not otherwise specified; LCAZ, large central acellular zone; TILs, prominent tumor infiltrating lymphocytes.
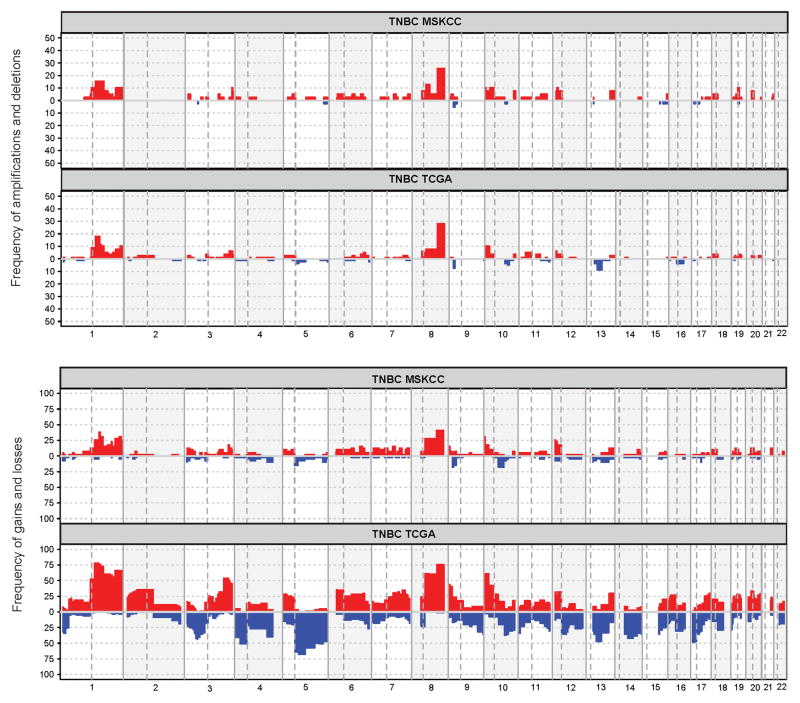
The overall pattern of copy number alterations in this cohort is similar to that in the triple negative breast cancers from the Cancer Genome Atlas breast cancer study (Figure 3). The most frequent copy number alterations in this cohort were amplifications of MYC (10 cases, 26%), MCL1 (6 cases, 15%), and DDR2 (6 cases, 15%) and deletions of CDKN2A/CDKN2B (2 cases, 5%) (Figure 2). Amplifications of PIK3C2G were more frequent in our cohort than that of the triple negative breast cancers in the Cancer Genome Atlas cohort (4/39, 10% vs 0/78, 0%, p=0.011, Fisher’s exact test).
Figure 3. Frequency of copy number aberrations in our triple negative breast cancer cohort as compared to triple negative breast cancers in the Cancer Genome Atlas cohort.
Abbreviations: TNBC, triple negative breast cancer; MSKCC, Memorial Sloan Kettering Cancer Center; TCGA, the Cancer Genome Atlas.
Genetic alterations by histologic subtypes and morphologic features
We next performed an exploratory, hypothesis generating analysis of the repertoire of somatic genetic alterations found in triple negative breast cancers according to histologic subtypes and specific morphologic features (Table 2 and Figure 4).
Table 2.
Comparison of genetic alterations by histologic subtypes and morphologic features
| All cases (n=39) | TNBC NOS (n=22) | Apocrine (n=4) | Metaplastic (n=1) | LCAZ (n=6) | Prominent TILs (n=6) | |
|---|---|---|---|---|---|---|
| Average number of mutations per case (range) | 3.33 (0–10) | 3.41 (0–10) | 4.5 (3–8) | 3 | 2.5 (1–6) | 3.17 (0–6) |
| Most frequent mutations |
TP53 (74%) MLL2 (15%) PIK3CA (10%) |
TP53 (82%) MLL2 (14%) |
PIK3CA (50%) NF1 (50%) |
TP53 KDM6A MLL3 |
TP53 (67%) KDM6A (33%) |
TP53 (83%) MLL2 (33%) |
| Most frequent copy number alterations (Amplifications) |
MYC (26%) MCL1 (15%) DDR2 (15%) |
MYC (36%) MCL1 (23%) |
None | - | ATM (33%) | CCNE1 (33%) |
Abbreviations: TNBC, triple negative breast cancer; NOS, not otherwise specified; LCAZ, large central acellular zone; TILs, tumor infiltrating lymphocytes
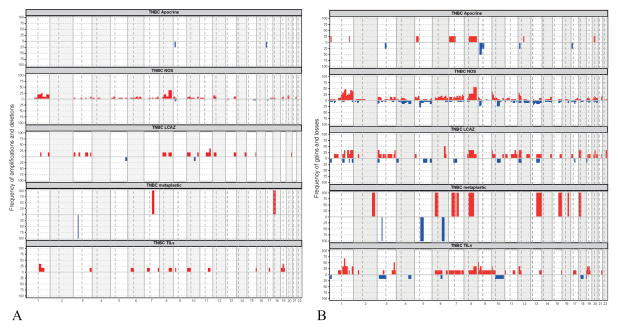
Figure 4. Frequency of copy number aberrations in our triple negative breast cancer cohort, by morphologic group.
A) amplifications and deletions; B) gains and losses
Abbreviations: TNBC, triple negative breast cancer; NOS, not otherwise specified; LCAZ, large central acellular zone; TILs, prominent tumor infiltrating lymphocytes.
Triple Negative Breast Cancer Not Otherwise Specified
Triple negative breast cancer not otherwise specified comprised the majority of the cases in our study (22/39, 56%). No statistically significant differences in age, tumor size, grade, lymph node involvement, distant metastases, or survival were seen between the triple negative breast cancer not otherwise specified group and the overall cases in our cohort. The average number of non-synonymous mutations per case in the triple negative breast cancer not otherwise specified group was 3.41 (range 0–10), significantly higher than that of the triple negative breast cancers in the Cancer Genome Atlas cohort (mean 2.37, range 0–7) (p = 0.01567, Fisher’s exact test). The most frequently mutated genes in this group were TP53 (18 cases, 82%) followed by MLL2 (3 cases, 14%), mirroring the findings in our overall cohort. PIK3CA mutations, however, were infrequent in this group, limited to only one case (5%). None of the above mutations was significantly different in frequency from that of the triple negative breast cancers in the Cancer Genome Atlas cohort. Mutations that reached statistical significance (despite having a lower mutation frequency than TP53 or MLL2) include JAK1, TBK1, and TGFBR2, each present in 2 (9%) cases in our study and in none of the cases in the triple negative breast cancers in the Cancer Genome Atlas cohort (p = 0.047). The vast majority of the highest frequency copy number alterations (amplifications of MYC, MCL1, and DDR2) were seen in this group (Figure 4).
Triple Negative Breast Cancer with Apocrine Differentiation
Triple negative breast cancer with apocrine differentiation (apocrine triple negative breast cancer) comprised a small subset of the cases in our cohort (4/39, 10%). Patients with apocrine triple negative breast cancer were older (mean age 64 years) than patients with non-apocrine triple negative breast cancer (mean age 45 years) (p = 0.01303, two-tailed student t-test). Apocrine triple negative breast cancers had lower histologic and nuclear grade than non-apocrine triple negative breast cancers (Table 1). Specifically, 50% of the apocrine triple negative breast cancers had histologic grade II, versus only 5% of the non-apocrine triple negative breast cancers (p = 0.04512, Fisher’s exact test); similarly 50% of apocrine triple negative breast cancers had nuclear grade II, versus only 3% of the non-apocrine triple negative breast cancers (p=0.02342, Fisher’s exact test). Patients with apocrine triple negative breast cancer also appeared to have smaller tumor size (median size 1.7 cm, range 1.4–2.5 cm), a lower rate of distant metastasis (1 case, 25%), and a lower rate of death from breast cancer (1 case, 25%); however, none of these differences reached statistical significance.
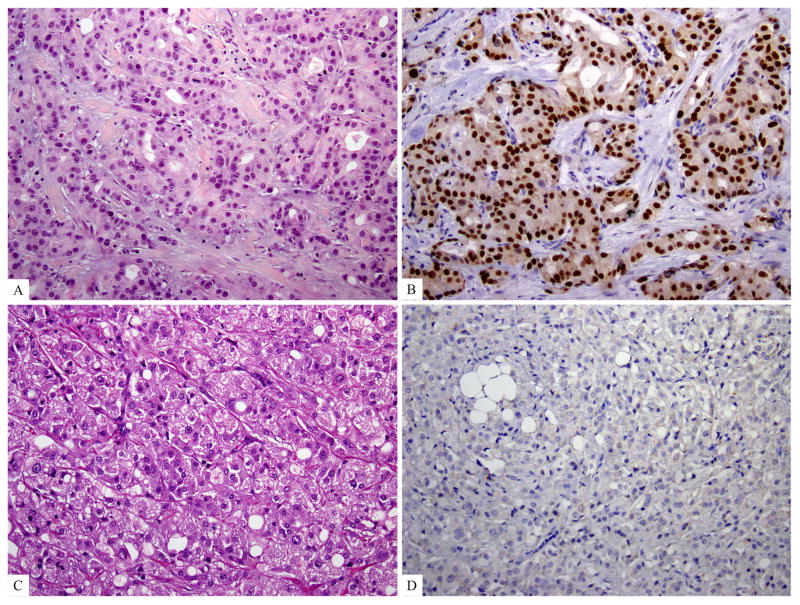
The average number of non-synonymous mutations per case in the apocrine triple negative breast cancer group was 4.5 (range 3–8), significantly higher than that of the triple negative breast cancers of the Cancer Genome Atlas cohort (mean 2.37, range 0–7, p = 0.03145, Mann-Whitney U test). Apocrine triple negative breast cancers displayed a distinctive repertoire of somatic genetic alterations, characterized by less frequent TP53 mutations (one of four cases, 25%) than in other morphologic subgroups of triple negative breast cancers from our cohort and from triple negative breast cancers from the Cancer Genome Atlas (p = 0.027, Fisher’s exact test). The most commonly mutated genes in the apocrine triple negative breast cancer group were PIK3CA and NF1, each mutated in 2 cases (50%). While the relative frequencies of PIK3CA mutations in the apocrine triple negative breast cancer group and the triple negative breast cancers of the Cancer Genome Atlas cohort did not reach statistical significance, a trend was observed toward higher frequency of PIK3CA mutations in the apocrine triple negative breast cancer group (2 cases (50%) in apocrine triple negative breast cancer; 7 cases (9%) triple negative breast cancers of the Cancer Genome Atlas cohort, p = 0.058, Fisher’s exact test). In addition, an apocrine triple negative breast cancer without PIK3CA mutation in our study showed a mutation in PIK3R1. Accordingly, if all activating mutations in PI3K pathway components are considered, apocrine triple negative breast cancers displayed significantly more frequent mutations affecting canonical genes of the PI3K pathway, including PIK3CD, PIK3R1 and AKT1, than triple negative breast cancers from the Cancer Genome Atlas study (3 cases (75%) of apocrine triple negative breast cancers vs 8 cases (10%) triple negative breast cancers of the Cancer Genome Atlas cohort, p = 0.0069, Fisher’s exact test). In the immunohistochemical analysis of AR expression in apocrine triple negative breast cancers, all but one case expressed androgen receptor (Figure 5). The case lacked androgen receptor expression was re-evaluated morphologically and the apocrine histologic features were confirmed (Figure 5 C &D). This case harbored a PIK3CA mutation.
Figure 5. Immunohistochemical staining for androgen receptor in triple negative breast cancer with apocrine differentiation (apocrine triple negative breast cancer).
A and B) An example of androgen receptor positive apocrine triple negative breast cancer, H&E and androgen receptor immunohistochemical stain; C and D) The apocrine triple negative breast cancer that is androgen receptor negative but harboring a PIK3CA mutation, H&E and androgen receptor immunohistochemical stain.
NF1 mutations were more frequently observed in the apocrine triple negative breast cancer group in our cohort than in triple negative breast cancers of the Cancer Genome Atlas cohort (2/4, 50% vs 2/78, 3%, p = 0.010, Fisher’s exact test). Other genes with significantly higher mutation frequency in the apocrine triple negative breast cancer group in our study included known cancer genes such as DIS3, JAK1, MET, NCOA2, and PTPN11, all of which were mutated in a single case each (25%) in our apocrine triple negative breast cancer group and in none of the triple negative breast cancer cases in the Cancer Genome Atlas data set (all p = 0.047, Fisher’s exact tests).
Apocrine triple negative breast cancers had a lower level of genetic instability, with few amplifications and deletions (Figure 4). No recurrent copy number alterations were identified in this group.
Metaplastic Carcinoma
There was a single case of metaplastic carcinoma (matrix producing type) in this study. The tumor measured 2.5 cm and was high grade. Lymph nodes were not involved at presentation, but the patient developed brain metastasis and died of disease 71 months following breast cancer diagnosis. The number of non-synonymous mutations in this case was 3, not different from that of the triple negative breast cancers from the Cancer Genome Atlas study (mean 2.37, range 0–7, p = 0.4572, Mann-Whitney U test). This tumor harbored mutations affecting TP53, KDM6A and MLL3. A separate study of genetic alterations in a larger cohort of metaplastic carcinoma is ongoing.
Triple Negative Breast Cancer with Large Central Acellular Zone
Six (15%) triple negative breast cancers had large central acellular zone. Patients with large central acellular zone tumors demonstrated a trend toward higher rates of distant metastasis (67%) and death from disease (50%), although did not reached statistical significance. We found no statistically significant differences in age, tumor size, grade, or frequency of lymph node involvement between triple negative breast cancer with large central acellular zone group and other patients in our cohort. The average number of non-synonymous mutations per case in the large central acellular zone group was 2.5 (range 1–6), not significantly different from that of the triple negative breast cancers from the Cancer Genome Atlas study (mean 2.37. range 0–7, p = 0.2691, Mann-Whitney U test). The most commonly mutated gene in the large central acellular zone group were TP53 (4 cases, 67%) followed by KDM6A (2 cases, 33%). We observed a trend toward a higher percentage of KDM6A mutations in the triple negative breast cancer with large central acellular zone group than in the triple negative breast cancer not otherwise specified group (triple negative breast cancer with large central acellular zone: 2/6, 33%; triple negative breast cancer not otherwise specified: 2/22, 9%), although this difference did not reach statistical significance (p = 0.191, Fisher’s exact test). In both groups, the frequency of KDM6A mutations was significantly higher than of that of triple negative breast cancers from the Cancer Genome Atlas study which did not show any KDM6A mutations (large central acellular zone vs triple negative breast cancers in the Cancer Genome Atlas cohort, p = 0.004; triple negative breast cancer not otherwise specified vs triple negative breast cancers in the Cancer Genome Atlas cohort, p = 0.047, Fisher’s exact tests). Two (33%) of the cases in the large central acellular zone group had amplification of the ATM gene – a significantly higher frequency than that of triple negative breast cancers in the Cancer Genome Atlas cohort (1 case, 1%; p=0.012, Fisher’s exact test).
Tumors with Prominent Tumor Infiltrating Lymphocytes
Six (15%) triple negative breast cancers had prominent tumor infiltrating lymphocytes. While patients in the prominent tumor infiltrating lymphocytes group were younger than patients without prominent tumor infiltrating lymphocytes (mean age 38.5 years vs 48.36 years; p = 0.1319), this figure did not reach statistical significance. No statistically significant differences in tumor size, grade, lymph node involvement, distant metastases, or death from breast cancer were seen between the prominent tumor infiltrating lymphocytes group and the other groups in this cohort. The average number of non-synonymous mutations per case in the tumors with prominent tumor infiltrating lymphocytes group was 3.17 (range 0–6), not significantly different from that of the triple negative breast cancers from the Cancer Genome Atlas study (mean 2.37, range 0–7, p = 0.3691, Mann-Whitney U test) The genes most frequently mutated in triple negative breast cancer with prominent tumor infiltrating lymphocytes group were TP53 (5 cases, 83%) and MLL2 (2 cases, 33%). None of the mutations in triple negative breast cancers with prominent tumor infiltrating lymphocytes reached statistical significance as compared to the triple negative breast cancers in the Cancer Genome Atlas cohort. Recurrent amplifications in the prominent tumor infiltrating lymphocytes group were seen in CCNE1 (2 cases, 33%) and MCL1 (2 cases, 33%). Of these, the amplification of CCNE1 was significantly more frequent in triple negative breast cancers with prominent tumor infiltrating lymphocytes as compared to the triple negative breast cancers in the Cancer Genome Atlas cohort (2 cases (33%) and 2 cases (2.6%), respectively; p = 0.024, Fisher’s exact test).
Genetic Alterations in BRCA1 Germline Mutation Carriers
Nine patients were BRCA1 germline mutation carriers. BRCA1 germline mutation carriers were younger than the other patients in our cohort (mean age 34.2 years vs 50.6 years; p = 0.002). No statistically significant differences in tumor size, grade, lymph node involvement, distant metastases, or death from breast cancer were seen between the BRCA1 germline mutation carriers and the other patients in our study. Most (8/9, 89%) of BRCA1 germline mutation carriers had triple negative breast cancer not otherwise specified, one patient (1/9, 11%) had triple negative breast cancer with prominent tumor infiltrating lymphocytes. The average number of non-synonymous mutations per case in BRCA1 germline mutation carriers’ tumors was 4.44 (range 1–10) and was significantly different from that of the triple negative breast cancers from the Cancer Genome Atlas study (mean 2.37, range 0–7, p = 0.0186). The most frequently mutated gene in tumors from BRCA1 germline mutation carriers was TP53 (8 cases, 89%), a frequency similar to that reported for triple negative breast cancers from the Cancer Genome Atlas cohort (64 cases, 82%; p= 1.00).
Discussion
While the genomic landscape of triple negative breast cancers is heterogeneous and complex, here we show that many of the key somatic genetic alterations in triple negative breast cancers elucidated by studies using large scale genomic analysis techniques can be detected using a targeted next-generation sequencing assay (MSK-IMPACT) in routine clinical use at our institution.16 Moreover, we show that the histologic analysis of triple negative breast cancers can provide useful information regarding the predicted somatic genetic landscape of individual triple negative breast cancer tumors. For instance, mutations in TP53 and PIK3CA are known to predominate in triple negative breast cancers, being reported in approximately 80% and 10% of triple negative breast cancers, respectively.13, 14 We found similar mutation frequencies in our cohort, in which TP53 and PIK3CA mutations were present in 74% and 10% of cases. However, these findings did not apply to all morphologic subtypes of triple negative breast cancer. For example, activating mutations in both PIK3CA as well as other key PI3K pathway components were enriched within the apocrine triple negative breast cancer group (75% of cases), a subset of triple negative breast cancers found to express androgen receptor more frequently than other forms of triple negative breast cancer. Lehmann et al40 also found PIK3CA mutations in 40% of their luminal androgen receptor tumors, a molecular subtype thought to be enriched in triple negative breast cancer with apocrine morphology, but did not correlate the findings with histologic analysis. While our study did not include transcriptomic profiling analysis, we speculate that triple negative breast cancers that express androgen receptor in our study would strongly correlated with the luminal androgen receptor (LAR) subtype described by Lehmann and colleagues 41 and Burstein and colleagues42, since it has been reported that androgen receptor mRNA was highly expressed in the LAR subtype, much greater than all other subtypes.41 To the best of our knowledge, our study is the first comprehensive analysis of apocrine triple negative breast cancers as defined by histologic features. Our study demonstrated that, relative to the triple negative breast cancers from the Cancer Genome Atlas dataset, apocrine triple negative breast cancers displayed a significantly higher rate of PI3K pathway mutations, a significantly higher rate of NF1 mutations, a significant lower rate of TP53 mutations, and a significantly lower rate of copy number aberrations. The high frequency of PI3K pathway mutations in apocrine triple negative breast cancer is more similar to that in luminal types of breast cancers.
For the mutations targeted by our assay, we observed a significantly higher number of somatic non-synonymous mutations per case in our triple negative breast cancer cohort as compared to that reported in the triple negative breast cancers in the Cancer Genome Atlas dataset. This is likely due to the higher depth of coverage and more sensitive detection using MSK-IMPACT than in the samples subjected to whole exome sequencing analysis in the Cancer Genome Atlas study, given that we achieved a coverage of at least 77% (tumors: median 98%, range 84%–99%; normal: median 96%, range 77%–98%) at 100x depth compared to the Cancer Genome Atlas requirement of including samples with at least 70% coverage at 20x depth and we used MuTect for calling SNVs, which has been shown to be more sensitive than VarScan 2 and SomaticSniper used by the Cancer Genome Atlas. 43–45 As the genes targeted by our assay include actionable and potentially actionable targets, these results are of particular interest with regard to the treatment of triple negative breast cancers as a lack of recurrent actionable targets is the basis for the reliance on cytotoxic agents as a means for systemic therapy.
This study has several limitations. First, the relatively small number of cases in each morphologic group renders the analysis performed exploratory and hypothesis generating. Second, we have surveyed the presence of somatic genetic alterations affecting 229 genes. Hence, we cannot exclude that additional differences between specific subtypes of triple negative breast cancer may be present if whole exome or whole genome sequencing was performed. In fact, rare subtypes of triple negative breast cancer, such as adenoid cystic carcinomas have been shown to be driven by recurrent MYB-NFIB fusion genes.9 Nevertheless, as a proof-of-concept study, our findings demonstrate that a next-generation sequencing panel (MSK-IMPACT) can reproduce findings similar to those obtained by genomic studies done on a much larger scale13, 14 and that a careful morphologic analysis of triple negative breast cancers can provide useful information regarding the somatic genetic composition of individual tumors. Furthermore, our findings suggest that triple negative breast cancers with apocrine differentiation likely display a landscape of somatic genetic alterations distinct from that of other triple negative breast cancers. Given these observations, further studies dissecting the genomic landscape of specific subsets of triple negative breast cancers are warranted.
Supplementary Material
Acknowledgments
This study was supported by Farmer Family Foundation. SP is funded by a Susan G Komen Postdoctoral Fellowship Grant (PDF14298348). Research reported in this publication was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (Grant No. P30CA008748). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Dr. Kiran Jakate for her assistance.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Part of this work was presented at United States and Canadian Academy of Pathology (USCAP) 103rd Annual Meeting in San Diego March 2014.
References
- 1.Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 2.Livasy CA, Karaca G, Nanda R, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 3.Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH. Triple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol. 2010;23:123–133. doi: 10.1038/modpathol.2009.145. [DOI] [PubMed] [Google Scholar]
- 4.Weigelt B, Kreike B, Reis-Filho JS. Metaplastic breast carcinomas are basal-like breast cancers: a genomic profiling analysis. Breast Cancer Res Treat. 2009;117:273–280. doi: 10.1007/s10549-008-0197-9. [DOI] [PubMed] [Google Scholar]
- 5.Geyer FC, Weigelt B, Natrajan R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220:562–573. doi: 10.1002/path.2675. [DOI] [PubMed] [Google Scholar]
- 6.Vranic S, Tawfik O, Palazzo J, et al. EGFR and HER-2/neu expression in invasive apocrine carcinoma of the breast. Mod Pathol. 2010;23:644–653. doi: 10.1038/modpathol.2010.50. [DOI] [PubMed] [Google Scholar]
- 7.Farmer P, Bonnefoi H, Becette V, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 8.Wetterskog D, Lopez-Garcia MA, Lambros MB, et al. Adenoid cystic carcinomas constitute a genomically distinct subgroup of triple-negative and basal-like breast cancers. J Pathol. 2012;226:84–96. doi: 10.1002/path.2974. [DOI] [PubMed] [Google Scholar]
- 9.Martelotto LG, De Filippo MR, Ng CK, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015 doi: 10.1002/path.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diallo R, Schaefer KL, Bankfalvi A, et al. Secretory carcinoma of the breast: a distinct variant of invasive ductal carcinoma assessed by comparative genomic hybridization and immunohistochemistry. Hum Pathol. 2003;34:1299–1305. doi: 10.1016/s0046-8177(03)00423-4. [DOI] [PubMed] [Google Scholar]
- 11.Piscuoglio S, Hodi Z, Katabi N, et al. Are acinic cell carcinomas of the breast and salivary glands distinct diseases? Histopathology. 2015;67:529–537. doi: 10.1111/his.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerini-Rocco E, Hodi Z, Piscuoglio S, et al. The repertoire of somatic genetic alterations of acinic cell carcinomas of the breast: an exploratory, hypothesis-generating study. J Pathol. 2015 doi: 10.1002/path.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng CK, Schultheis AM, Bidard FC, Weigelt B, Reis-Filho JS. Breast cancer genomics from microarrays to massively parallel sequencing: paradigms and new insights. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv015. [DOI] [PubMed] [Google Scholar]
- 16.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Won HH, Scott SN, Brannon AR, Shah RH, Berger MF. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp. 2013;80:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med. 2010;134:907–922. doi: 10.5858/134.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138:241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 22.Reis-Filho JS, Lakhani SR, Gobbi H, Sneige N. Metaplastic carcinoma. In: Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ, editors. WHO Classification of Tumours of the Breast. Lyon: International Agency for Research on Cancer (IARC); 2012. pp. 48–52. [Google Scholar]
- 23.O’Malley F, Eusebi V, Lakhani SR. Carcinomas with apocrine differentiation. In: Lakhani S, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ, editors. WHO Classification of Tumours of the Breast. Lyon: International Agency for Research on Cancer (IARC); 2012. pp. 53–54. [Google Scholar]
- 24.Tsuda H, Takarabe T, Hasegawa F, Fukutomi T, Hirohashi S. Large, central acellular zones indicating myoepithelial tumor differentiation in high-grade invasive ductal carcinomas as markers of predisposition to lung and brain metastases. Am J Surg Pathol. 2000;24:197–202. doi: 10.1097/00000478-200002000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loi S, Sirtaine N, Piette F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. J Clin Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 27.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–21. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter H, Chen S, Isik L, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 36.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinreb I, Piscuoglio S, Martelotto LG, et al. Hotspot activating PRKD1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–1169. doi: 10.1038/ng.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellis I, Collins L, Ichihara S, MacGrogan G. Invasive carcinoma of no special type. In: Lakhani S, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ, editors. WHO Classification of Tumours of the Breast. Lyon: International Agency for Research on Cancer (IARC); 2012. pp. 34–38. [Google Scholar]
- 40.Lehmann BD, Bauer JA, Schafer JM, et al. PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res. 2014;16:406. doi: 10.1186/s13058-014-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burstein MD, Tsimelzon A, Poage GM, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Jia P, Li F, et al. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med. 2013;5:91. doi: 10.1186/gm495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alioto TS, Buchhalter I, Derdak S, et al. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat Commun. 2015;6:10001. doi: 10.1038/ncomms10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H, DiCarlo J, Satya RV, Peng Q, Wang Y. Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genomics. 2014;15:244. doi: 10.1186/1471-2164-15-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.