Abstract
High biomass production and wide adaptation has made switchgrass (Panicum virgatum L.) an important candidate lignocellulosic bioenergy crop. One major limitation of this and other lignocellulosic feedstocks is the recalcitrance of complex carbohydrates to hydrolysis for conversion to biofuels. Lignin is the major contributor to recalcitrance as it limits the accessibility of cell wall carbohydrates to enzymatic breakdown into fermentable sugars. Therefore, genetic manipulation of the lignin biosynthesis pathway is one strategy to reduce recalcitrance. Here, we identified a switchgrass Knotted1 transcription factor, PvKN1, with the aim of genetically engineering switchgrass for reduced biomass recalcitrance for biofuel production. Gene expression of the endogenous PvKN1 gene was observed to be highest in young inflorescences and stems. Ectopic overexpression of PvKN1 in switchgrass altered growth, especially in early developmental stages. Transgenic lines had reduced expression of most lignin biosynthetic genes accompanied by a reduction in lignin content suggesting the involvement of PvKN1 in the broad regulation of the lignin biosynthesis pathway. Moreover, the reduced expression of the Gibberellin 20-oxidase (GA20ox) gene in tandem with the increased expression of Gibberellin 2-oxidase (GA2ox) genes in transgenic PvKN1 lines suggest that PvKN1 may exert regulatory effects via modulation of GA signaling. Furthermore, overexpression of PvKN1 altered the expression of cellulose and hemicellulose biosynthetic genes and increased sugar release efficiency in transgenic lines. Our results demonstrated that switchgrass PvKN1 is a putative ortholog of maize KN1 that is linked to plant lignification and cell wall and development traits as a major regulatory gene. Therefore, targeted overexpression of PvKN1 in bioenergy feedstocks may provide one feasible strategy for reducing biomass recalcitrance and simultaneously improving plant growth characteristics.
Keywords: gene regulation, lignin, gibberellin, cell walls, cellulose, hemicellulose, sugar release, biofuel
Introduction
Switchgrass, a C4 perennial prairie forage grass indigenous to North American grasslands, is a leading candidate as a lignocellulosic biofuel feedstock owing to its high yield of biomass, stress resistance, high nutrient-use efficiency, and its ability to thrive on marginal land unsuitable for row crops (Yuan et al., 2008). Controlled field studies to determine the net energy efficiency of switchgrass monoculture on marginal lands showed that it can produce as much as 540% more renewable energy than the non-renewable energy consumed for its production (Schmer et al., 2008), highlighting the potential of this feedstock as a renewable and clean energy source. One of the major obstacles for the development of this and other lignocellulosic biomass feedstocks for biofuels is biomass recalcitrance (resistance of the cellulose and hemicellulose in cell walls to breakdown into fermentable sugars). Currently, biomass must be pretreated before saccharification and fermentation, which is expensive (Ragauskas et al., 2006). Reducing recalcitrance to enhance the economic competitiveness of lignocellulosic-based biofuels is an overarching goal in bioenergy research (Lynd et al., 2008).
Lignin is a complex aromatic polymer composed of hydroxyphenyl, guaiacyl, and syringyl monolignols. Transgenic approaches have been used to reduce lignin, one of the major contributors to biomass recalcitrance. Mostly, a gene-by-gene strategy has been used to downregulate individual biosynthetic genes to reduce lignin (Fu et al., 2011a,b; Saathoff et al., 2011; Xu et al., 2011; Baxter et al., 2014). Several enzymes responsible for monolignol biosynthesis have been identified and functionally characterized in switchgrass (Escamilla-Trevino et al., 2010; Fu et al., 2011a,b; Saathoff et al., 2011; Xu et al., 2011; Shen et al., 2013a). Genetically targeting individual lignin biosynthesis genes to block specific branches in the lignin biosynthesis pathway has been successful, but this strategy appears to have limitations as the build-up of low molecular weight phenolic compounds and other fermentation inhibitors may eventually reduce biofuel output (Tschaplinski et al., 2012). The use of transcriptional repressors of the lignin biosynthesis pathway might be a more effective strategy, as shown by the recent work in which the switchgrass gene coding for MYB4 TF was overexpressed (Shen et al., 2012, 2013b).
Lignin biosynthesis is regulated by multi-layered network of TFs including top-tier NAC and second-tier MYB master regulators (Handakumbura and Hazen, 2012; Zhong and Ye, 2014). The NAC TFs, which activate the lignin biosynthesis pathway via activation of downstream MYB TFs have been well documented in dicots such as Arabidopsis and Populus (Zhao Q. et al., 2010; Zhong and Ye, 2014). Recent studies reported the identification of some rice and maize SWN that are functional orthologs of Arabidopsis SWNs (Zhong et al., 2011; Chai et al., 2015). Similarly, several second-tier MYBs that control the activation of downstream MYB TFs specific to the lignin biosynthesis pathway have been characterized. These include: the Arabidopsis MYB46/83 and its homologs in Populus (PtrMYB2/3/20/21), pine (PtMYB4/8), Eucalyptus (EgMYB2), rice (OsMYB46), and maize (ZmMYB46) (Zhong et al., 2011; Zhong and Ye, 2014). The lignin-specific MYBs act either as a repressors or activators of lignin biosynthesis pathway genes. Several MYB TFs that are repressors of the lignin biosynthesis pathway have been identified in dicots including Arabidopsis (MYB4/7/32) and Eucalyptus (MYB1) (Fornale et al., 2010; Shen et al., 2012; Zhong and Ye, 2014). MYB TFs that are activators of the lignin biosynthesis pathway were also identified in several dicots including Arabidopsis (MYB58/63 and MYB85), Populus (MYB26/28/90/152) and pine (MYB1) (Zhou et al., 2009; Zhong and Ye, 2014). However, with the exception of maize MYB31 (Fornale et al., 2010), wheat MYB4 (Ma et al., 2011) and a recently characterized switchgrass MYB4 TFs (Shen et al., 2012, 2013b), there is limited available information about the transcriptional regulation of lignin biosynthesis in monocots (Handakumbura and Hazen, 2012).
Knotted1(KN1)-like homeobox (KNOX) proteins are members of plant-specific TALE family of TFs, which play a crucial role in the maintenance of meristem tissues and the regulation of various morphogenic processes throughout plant development (Hay and Tsiantis, 2010). These TFs are grouped into two major classes based on their HD identity, intron positions and expression patterns (Kerstetter et al., 1994; Mukherjee et al., 2009; Hay and Tsiantis, 2010; Testone et al., 2012). Class I KNOX genes are mainly expressed in the SAM where they redundantly regulate stem cell maintenance, and in vascular cambium. Class II KNOX genes are diversely expressed and their functions are not as well characterized (Jackson et al., 1994; Hake et al., 2004; Du et al., 2009). Members of class I KNOX genes from Arabidopsis (KNAT1 also known as BREVIPEDICELLUS/BP), peach (KNOPE1), Populus (ARBORKNOX2/ARK2), and maize (KN1) (Mele et al., 2003; Du et al., 2009; Testone et al., 2012; Townsley et al., 2013), and a member of class II KNOX genes from Arabidopsis (KNAT7; Li et al., 2012) have been shown to function as repressors of lignin biosynthetic genes. KNAT1 overexpression in Arabidopsis has been shown to decrease lignin deposition as well as the expression of most lignin biosynthetic genes while binding the promoters of at least two lignin biosynthetic genes, COMT and CCoAOMT (Mele et al., 2003). Similarly, a Populus ortholog of KNAT1, ARK2 has also been shown to negatively regulate several genes in the lignin biosynthesis pathway followed by marked reduction of the polymerized lignin in the stem (Du et al., 2009). Interestingly, ARK2 overexpression in Populus was found to be associated with reduced expression of secondary cell wall cellulose synthase genes (Ces) including three xylem-specific (PttCesA1, PttCesA3-1, and PttCesA3-2) and two phloem-specific (PttCesA2 and estExt_Genewise1_v1.C_LG_VI2188) CesA genes (Du et al., 2009). Despite vast information on class I KNOX gene regulation of cell wall biosynthesis pathway in dicots, such reports are limited in monocots with the exception of maize.
A recent study showed that among three members of maize class I KNOX genes investigated for roles in the regulation of lignin biosynthesis, overexpression of KN1 significantly reduced lignin deposition in maize and tobacco while altering the expression of only two of the four lignin biosynthetic genes analyzed in tobacco (Townsley et al., 2013). However, it was not reported whether similar changes in the expression of lignin biosynthetic genes were observed in maize. This gene has also been shown to regulate gibberellin (GA) signaling via the positive transcriptional regulation of maize GA 2-oxidase (GA2ox1; codes for an enzyme that inactivates bioactive GA) (Bolduc and Hake, 2009). Overexpression of KNAT1 has previously been shown to decrease the levels of GA 20-oxidase (GA20ox1) mRNA in Arabidopsis leaves (Hay et al., 2002) as did the overexpression of ARK2 gene in hybrid aspen (P. alba ×P. tremula; Du et al., 2009). However, it remains to be determined whether KNOX genes regulate GA biosynthetic genes in monocots. GA signaling plays vital roles in the regulation of various developmental and growth processes and its downregulation has been shown to alter plant architecture and lignin deposition (Biemelt et al., 2004; Thomas et al., 2005; Zhao X.Y. et al., 2010). Our recent study demonstrated that the overexpression of the switchgrass GA catabolic gene PvGA2ox5 caused reduced lignification and enhanced sugar release efficiency (Wuddineh et al., 2015). It appears, at least in model dicots, that KNOX TFs may regulate the lignification process via modulation of the GA signaling pathways. However, to the best of our knowledge, there are no published reports on whether class I KNOX TFs from monocots regulate the lignification process and any mechanism thereof.
With the exception of maize KN1-induced alteration of plant lignification, the potential of KNOX TFs to alter various growth and biomass characteristics of bioenergy crops is largely untapped. Investigation of the effect of overexpression of KN1 genes in monocots on the lignin biosynthesis pathway and the potential effects on downstream plant growth parameters is important. Therefore, the objective of this study was to identify and characterize the switchgrass class I KNOX gene (PvKN1) and investigate its association with various plant morphological and developmental processes as well as impacts on cell wall chemistry and recalcitrance in switchgrass.
Materials and Methods
Plants and Growth Conditions
Transgenic and non-transgenic control switchgrass plants were grown under the same conditions (16 h day/8 h night light at 24°C, 390 μE m−2 s−1) in growth chambers and watered three times per week, including weekly nutrient supplements with 100 mg/L Peter’s 20-20-20 fertilizer. For quantification of growth parameters, each transgenic and non-transgenic line was propagated from a single tiller to yield three clonal replicates each (Hardin et al., 2013). Leaf blades, leaf sheath, internode sections, and panicle samples pooled from the top two internodes and young roots, were collected from tillers at the R1 developmental stage (Moore et al., 1991; Shen et al., 2009) to assay transcript abundance. Each sample was snap-frozen in liquid nitrogen and pulverized with mortar and pestle in liquid nitrogen. The pulverized samples were used for RNA extraction as described below.
Gene Identification, Vector Construction, and Transgenic Plant Production
TBLASTN was used to identify the homologous KNOX gene sequences from switchgrass EST databases (Zhang et al., 2013) and draft genome (Panicum virgatum v1.1, DOE Joint Genome Institute [DOE-JGI], 2015) at Phytozome using the amino acid sequences of ZmKN1 (gb/AAP21616.1) and KNAT1 (At4g08150) as heterologous probes. Subsequently, the most closely related genes (PvKN1a and PvKN1b) were identified for cloning based on the cluster analysis and multiple sequence alignment analysis. Overexpression cassettes were constructed by isolating target gene ORFs from switchgrass cDNAs of the ST1 clonal genotype of ‘Alamo’ switchgrass using individual gene-specific primers flanking the ORF of each gene and subsequently cloning each into pCR8 entry vector for sequence confirmation. The list of primer pairs used for cloning is shown in Supplementary Table S1. Sequence-confirmed ORFs were then sub-cloned into pANIC-10A expression vector by GATEWAY recombination (Mann et al., 2012) to place each gene of interest under the control of the ZmUbi1 promoter. Embryogenic callus derived from Alamo switchgrass ST1 or SA37 genotypes (King et al., 2014) was transformed with the expression vector construct using Agrobacterium-mediated transformation (Burris et al., 2009). Selection for positive transformants was carried out for approximately 2 months on 30–50 mg/L hygromycin followed by regeneration of OFP reporter-positive callus sections on regeneration medium (Li and Qu, 2011) containing 400 mg/L timentin. Regenerated plants were rooted on MS medium (Murashige and Skoog, 1962) containing 250 mg/L cefotaxime (Grewal et al., 2006) and the transgenic lines were screened based on the presence of the insert and expression of the transgene. The non-transgenic control ST1 and SA37 lines were generated in tissue culture in parallel with the transgenic lines. Rice transformation was performed using callus derived from mature seeds of rice variety TP309 as described before (Nishimura et al., 2006).
RNA Extraction and qRT-PCR
RNA extraction and analysis of transgene transcripts were performed as previously described (Wuddineh et al., 2015). Briefly, total RNA was extracted from the shoot-tips of transgenic and non-transgenic lines at E4 developmental stage using Tri-Reagent (Molecular Research Center, Cincinnati, OH, USA). Each 3 μg of the purified RNA sample was treated with DNase-I (Promega, Madison, WI, USA) to eliminate any potential genomic DNA contaminants. High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) was used for first-strand cDNA synthesis using the DNase-treated RNA. qRT-PCR analysis was conducted using Power SYBR Green PCR master mix (Applied Biosystems) according to the manufacturer’s protocol. All the experiments were conducted in triplicate. Gene-specific forward primers and an AcV5 tag-spanning reverse primer was used for qRT-PCR of the transgenic lines. The list of all primer pairs used for qRT-PCR is shown in Supplementary Table S1. The relative expression was analyzed using the CT method using UBQ (Switchgrass Unitranscript ID: AP13CTG25905) as a reference gene (Shen et al., 2009; Xu et al., 2011). No amplification products were observed with all the primer pairs when using only the RNA samples or water instead of cDNA.
Phloroglucinol Staining
Qualitative lignin analysis was performed as previously described (Wuddineh et al., 2015). Briefly, leaf samples at R1 developmental stage were collected and cleared in a 2:1 solution of ethanol and glacial acetic acid for 5 days (Bart et al., 2010). The third leaves from the base of the stem were used in this experiment as these were the most uniform-looking in terms of maturity. Subsequently, the cleared leaf sample was immersed in 1% phloroglucinol (in 2:1 ethanol/HCl) overnight for staining. Photographs were taken at 1× magnification using infinity X32 digital camera mounted on Fisher Scientific Stereomaster microscope (Pittsburgh, PA, USA).
Determination of Lignin Content and Composition by Pyrolysis-Molecular Beam Mass Spectrometry
Quantitative lignin analysis and S/G lignin monomer ratio were determined using tillers collected at R1 developmental stage and air dried for 3 weeks at room temperature, followed by milling to 1 mm (20 mesh) particle size. Lignin content and composition were determined at NREL using high-throughput pyrolysis-molecular beam mass spectrometry (pyMBMS) on extractives- and starch-free samples (Sykes et al., 2009; Baxter et al., 2014).
Determination of Sugar Release
For enzymatic hydrolysis, tiller samples were collected at R1 developmental stage and air-dried for 3 weeks at room temperature before grinding to 1 mm (20 mesh) particle size. Subsequently, sugar release efficiency was determined using high-throughput sugar release assays on extractives- and starch-free samples (Selig et al., 2010; Studer et al., 2010; Decker et al., 2012). Briefly, the milled samples were prepared in a 96-well plate along with biomass-only controls, sugar standards, enzyme-only, and blank wells. The pretreatment was performed in a steam chamber at 180°C for 40 min. The enzyme cocktails used for hydrolysis comprises Novozymes Cellic CTec2 cellulase (Novozymes North America, Franklinton, NC, USA) at 70 mg protein/g initial biomass and Novo188 β-glucosidase (Novozymes) at 2.5 mg protein/g initial biomass. The mixture was incubated for 72 h at 54°C and glucose and xylose release were determined by colorimetric assays. The total sugar release was defined as the sum of glucose and xylose released. Each sample for sugar release assays were run in triplicate.
Data Analysis
ANOVA with Fisher’s LSD procedure was used to analyze the differences between treatment means while PROC TTEST procedure was used to examine the statistical difference between the expression of target genes in transgenic vs. non-transgenic lines using SAS version 9.3 (SAS Institute Inc., Cary, NC, USA).
Results
KNOX Family of TFs in Switchgrass and the Identification of PvKN1
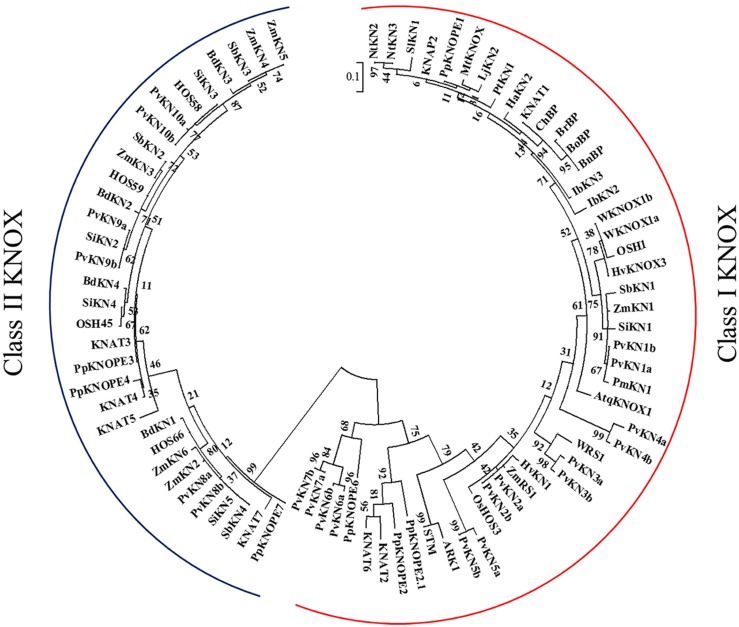
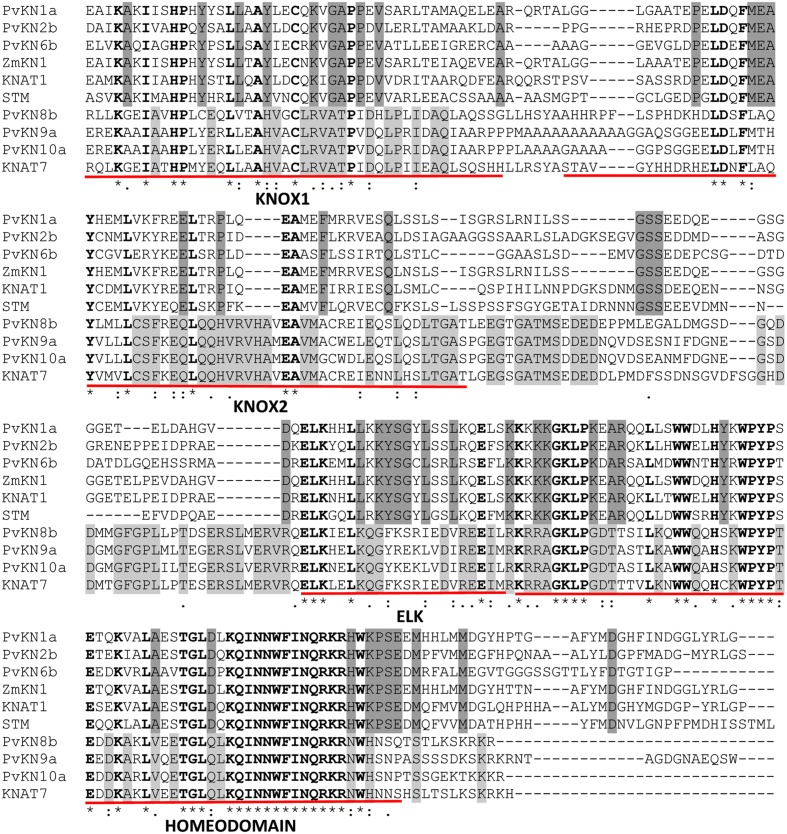
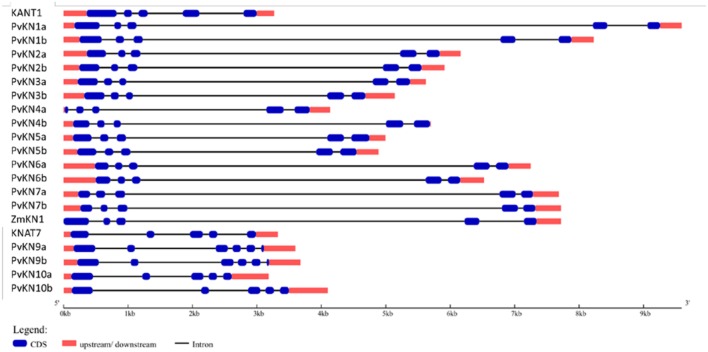
A survey of the publicly available switchgrass genome for the characteristic Meinox domain (KNOX1 and KNOX2) and HD revealed a total of 10 genes belonging to the KNOX family of TFs. The tetraploid switchgrass genome of Alamo AP13 (2n = 4x = 36) contains at least two sub-genomic gene variants representing the ‘A’ and ‘B’ subgenomes (Figure 1). Phylogenetic and sequence analysis showed seven KNOX genes belonging to class I family of KNOX TFs while the remaining three were grouped into the class II family of KNOX TFs (Figures 1 and 2). Analysis of the amino acid sequences of the encoded proteins indicated that all share KNOX1, KNOX2, ELK, and HD conserved domains found in the maize KN1, Arabidopsis KNAT1 and orthologous TFs in dicots (peach and Populus). Moreover, higher identity within members of each class (>44% in class I and >60% in class II) than between the two classes of KNOX genes (≤37%) was observed (Supplementary Table S2). Analysis of gene structure among the two classes of KNOX genes in comparison to the homologs from Arabidopsis (KNAT1 and KNAT7) and maize (KN1) showed a highly conserved pattern in each class, typically with five exons and four introns in the genomic DNA with variable length of introns among the genes (Figure 3). The exception to this pattern was PvKN9, a member of class II KNOX genes family, which had an additional intron inserted in the last exon and, therefore, had a structure of six exons and five introns.
FIGURE 1.
Maximum likelihood tree of KNOX family of TFs for the deduced amino acid sequences from switchgrass along with already-characterized proteins from both monocots and dicots. The sequences were aligned using MUSCLE program (Edgar, 2004) and the alignment was curated by Gblocks at the phylogeny.fr (Dereeper et al., 2008). The tree was constructed by maximum likelihood procedure using MEGA6.0 program (Tamura et al., 2013). Analysis using 1000 bootstrap replicates was performed. The scale bar shows 0.1 amino acid substitutions per site. Names of the species and the locus name or GenBank accession numbers of the sequences used in this tree are listed in Supplementary Table S4.
FIGURE 2.
Multiple amino acid sequence alignment of the C-termini of class I: PvKN1a, PvKN2b, PvKN6b, ZmKN1, KNAT1, and STM and class II (PvKN8b, PvKN9a, PvKN10a, and KNAT7) family of KNOX TFs. The conserved domains are underlined in red; the strictly conserved amino acid residues are indicated in bold. The amino acid residues specific to class I KNOX TFs are highlighted in dark gray while those specific to class II are highlighted in light gray. The multiple sequence alignment was constructed using the amino acid sequences of respective genes by MUSCLE program (Edgar, 2004).
FIGURE 3.
Gene structures of class I and II KNOX transcription factor family coding genes from Arabidopsis, switchgrass (Panicum virgatum) and maize (Zea mays). The CDS and the UTR are shown by filled dark blue and pink boxes, respectively. The introns are shown by thick black lines. The gene features were visualized by the gene structure display server (Guo et al., 2007).
Based on amino acid sequence analysis, sub-genomic variants belonging to class I family of KNOX TFs, designated as PvKN1a, PvKN1b, PvKN2a, and PvKN2b, had the highest identity to the maize KN1 and Arabidopsis KNAT1 when compared with the other switchgrass class I family members of KNOX TFs (Figure 2; Supplementary Table S3). However, based on phylogenetic analysis of the KNOX proteins, PvKN1a and PvKN1b showed more similarity to the group containing the previously characterized KNOX TFs from Arabidopsis (KNAT1/BP), maize (ZmKN1), rice (OSH1), wheat (WKNOX1), Populus (ARK2), and peach (PpKNOPE1) while PvKN2a and PvKN2b clustered more closely with maize rough sheath 1 (ZmRS1), rice homeobox (OsHOS3) and wheat rough sheath 1 (WRS1) TFs (Figures 1 and 2). Moreover, the two gene variants of PvKN1 (PvKN1a and PvKN1b) with 95% identity in the predicted amino acid sequences shared over 88% identity with ZmKN1 as opposed to 54% for PvKN2a or PvKN2b (Supplementary Table S3). More importantly, comparison of the HD domain, essential for binding target sequences, showed over 98% identity between the PvKN1 and ZmKN1 proteins versus 89% between PvKN2 and ZmKN1 (Supplementary Table S3). Moreover, PvKN1 displayed the highest conservation at the glycine, serine and glutamic acid-rich motif (also known as GSE) (the stretch of amino acids between the KNOX2 and ELK domains) compared with PvKN2 (Supplementary Table S3).
Expression Patterns of PvKN1 Variants
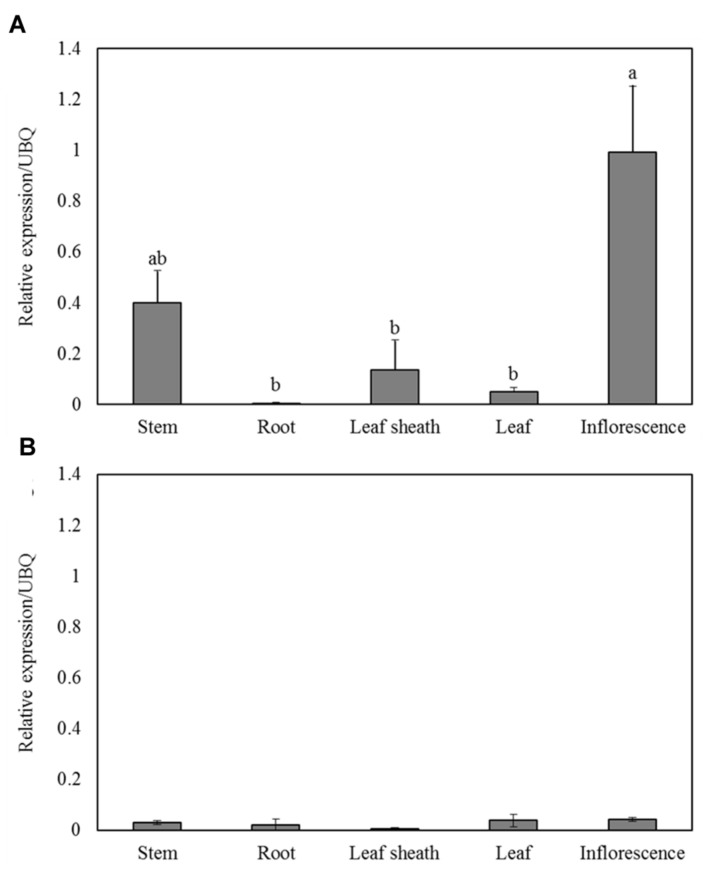
Quantitative reverse transcription polymerase chain reaction analysis showed detectable levels of expression for both PvKN1 gene variants in stems, leaves, leaf sheaths and inflorescences at the R1, the same stage when sugar release, lignin content and growth parameters were analyzed (Figure 4). The relative transcript abundance of PvKN1a appears to be higher than that of PvKN1b in stems, leaf sheath and young inflorescences. Moreover, PvKN1a expression was the highest in young inflorescence tissue followed by stems, with the lowest expression observed in roots and leaves (Figure 4). The expression of PvKN1b was not significantly different among tissues assayed (Figure 4).
FIGURE 4.
Expression patterns of PvKN1a(A) and PvKN1b(B) in different plant tissues as determined by qRT-PCR. Plant samples for RNA extraction used in the qRT-PCR experiments were collected at R1 (reproductive stage 1) developmental stage. The dissociation curve for the qRT-PCR products showed that the primers were gene-specific. The relative levels of transcripts were normalized to UBQ. Bars represent mean values of three replicates ± standard error. Bars represented by different letters are significantly different at P ≤ 0.05 as tested by the LSD method with SAS software (SAS Institute Inc.).
Constitutive Overexpression of PvKN1 in Switchgrass
The switchgrass homologs of maize KN1 gene, PvKN1a and PvKN1b were cloned from cDNA and constitutively overexpressed in switchgrass under the control of the ZmUbi1 promoter. In addition, the same PvKN1a construct was overexpressed in rice to examine its effect on plant development. Only transgenic calli expressing the OFP marker gene were regenerated (Supplementary Figure S1). Five independent transgenic switchgrass lines of ST1 background overexpressing PvKN1a and eight PvKN1b-overexpressing lines were recovered.
Altered Growth Phenotypes in Transgenic Plants Overexpressing PvKN1
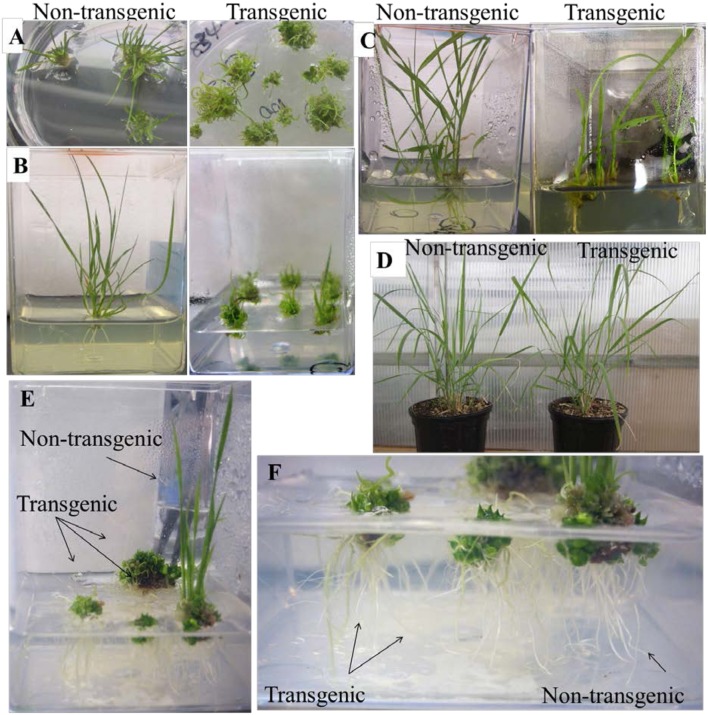
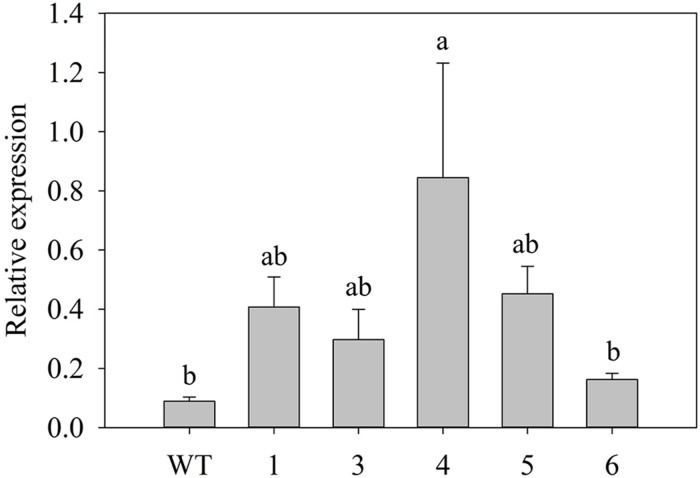
The transgenic lines overexpressing the two PvKN1 gene variants had similar phenotypes, which suggests that each variant may have similar functions upon overexpression (Supplementary Figure S2). Therefore, subsequent work focused on detailed analysis of PvKN1a overexpressing lines. We observed unusually narrow leaf blades with sheath-like tissues and curled leaves just after plants were regenerated. In addition, the PvKN1a overexpression lines had severely inhibited shoot and root elongation as compared with the non-transgenic controls at similar developmental stages (Figures 5A,B). Many of the transgenic lines recovered were dwarfed and some were not fully regenerable. However, several transgenic lines that were regenerated were able to develop into mature plants displaying less severe phenotypes after establishment in the rooting media (Figure 5C); some of these lines eventually grew normally (Figure 5D). The successfully established transgenic lines were confirmed by genomic PCR using transgene and hygromycin-resistance gene specific primers, as well as visualization of OFP in transgenic plants compared with the non-transgenic control lines (Supplementary Figures S1A–S1C). Moreover, there were two to ninefold increases in the PvKN1 transcript levels in transgenics relative to control lines (Figure 6). There were no statistically significant differences in tiller height, tiller number or biomass between transgenic lines and the non-transgenic control (Table 1).
FIGURE 5.
Altered growth phenotypes in transgenic PvKN1a- overexpressing switchgrass (ST1) and rice plants. (A) Transgenic plants overexpressing PvKN1a exhibited reduced shoot elongation and curly leaves compared to the normal phenotype in non-transgenic lines. (B) Transgenic plants with substantially reduced growth compared to the non-transgenic lines at 5 weeks after incubation on the rooting media. (C) Transgenic plants with elongated shoots compared to the non-transgenic lines at 9 weeks after incubation on the rooting media. (D) Morphological phenotypes in 3 months old transgenic lines overexpressing PvKN1a compared to non-transgenic lines. (E) Transgenic rice showing abnormal shoot development compared to the normal phenotype in the non-transgenic plants. (F) Transgenic rice showing normal root development as well as the non-transgenic plants.
FIGURE 6.
Relative transcript levels of the PvKN1a in transgenic and non-transgenic (WT) switchgrass (ST1) plants. The expression analysis was done using RNA derived from the shoot tips at E4 developmental stage. The dissociation curve for the qRT-PCR products showed that the primers were gene-specific. The relative levels of transcripts were normalized to UBQ. Bars represent mean values of three replicates ± standard error. Bars represented by different letters are significantly different at P ≤ 0.05 as tested by LSD method with SAS software (SAS Institute Inc.).
Table 1.
Morphology and biomass yields of transgenic switchgrass lines per plant overexpressing PvKN1 and non-transgenic control (WT) parent plants.
| Lines | Tiller height (cm) | Tiller number | Fresh weight (g) | Dry weight (g) |
|---|---|---|---|---|
| 1 | 50.7 ± 1.6a | 7.5 ± 0.5a | 24.8 ± 6.5a | 7.5 ± 1.2a |
| 3 | 40.5 ± 1.7b | 12.7 ± 4.5a | 24.3 ± 11.8a | 9.3 ± 4.3a |
| 4 | 45.0 ± 3.4ab | 10.3 ± 4.2a | 21.9 ± 3.2a | 8.4 ± 0.4a |
| WT | 45.3 ± 2.5ab | 9.7 ± 1.3a | 27.7 ± 9.3a | 10.7 ± 1.3a |
Tiller height estimates were determined for each plant by taking the mean of the five tallest tillers within each biological replicate. The fresh and dry biomass measurements were taken on 4-mo-old aboveground plant biomass harvested at similar growth stages (reproductive stage R1). Values are means of three biological replicates ± standard errors (n = 3). Values represented by different letters are significantly different at P ≤ 0.05 as tested by the LSD method with SAS software (SAS Institute Inc.).
Overexpression of PvKN1 in a different switchgrass genotype (SA37) background showed abnormal phenotypes at later stages of growth compared to the genotype (ST1) used in this study. These transgenic lines had relatively shorter internodes and twisted or bent culms with altered leaf structure including curly leaves, fused leaf sheath-blade boundary and missing ligules (Supplementary Figures S3A–S3C). The relative expression of the PvKN1 transcript was >50-fold in transgenic compared to the non-transgenic control lines (Supplementary Figure S3D).
When PvKN1 was overexpressed in rice, similar aberrant phenotypes were observed with clumps of vegetative shoots developed from the transgenic callus after incubation on the shoot regeneration medium. In general, shoot elongation was severely reduced in rice even after longer incubation on the growth medium while root growth was apparently unaffected (Figures 5E,F).
Overexpression of PvKN1 in Switchgrass and Subsequent Changes in the Expression of Putative Target Genes in Transgenic Plants
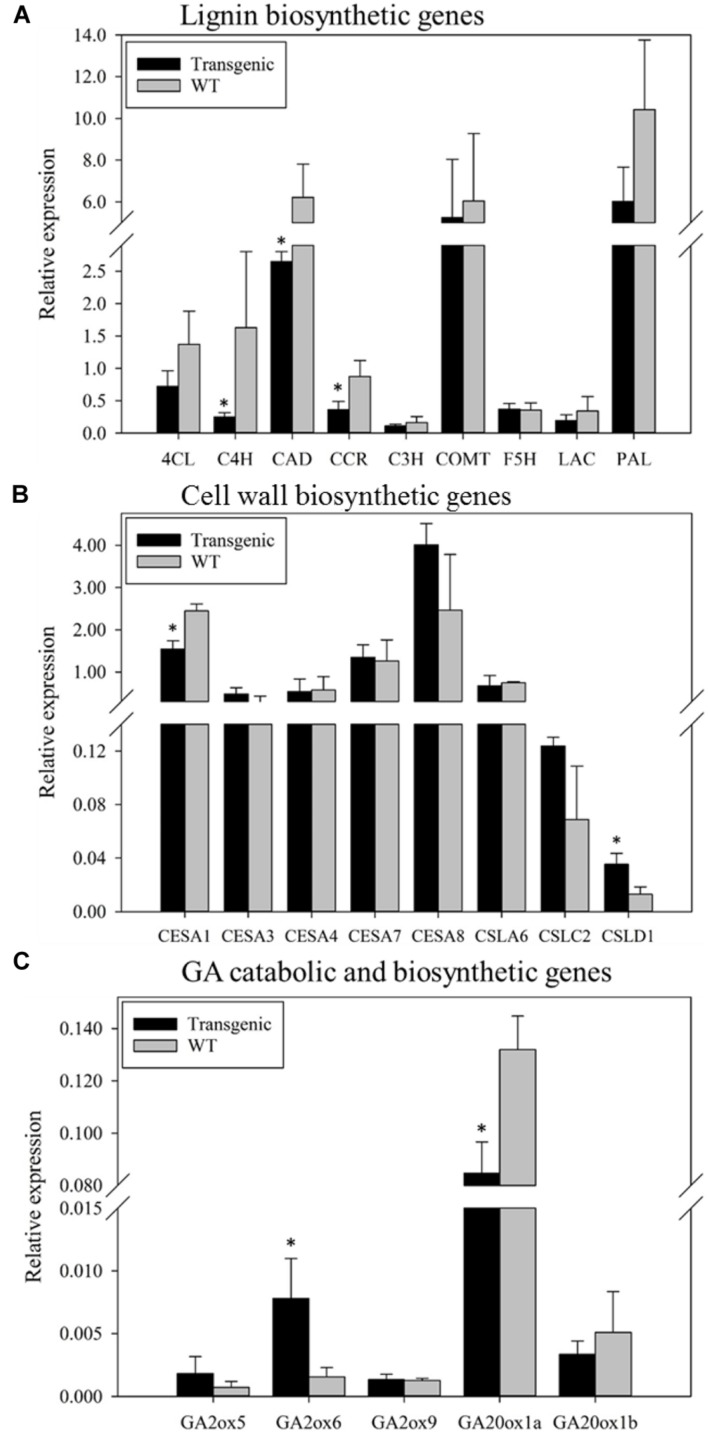
To test whether overexpression of PvKN1 in switchgrass could affect the expression of putative target genes, qRT-PCR analysis was conducted. PvKN1-overexpressing switchgrass displayed significantly reduced expression of lignin biosynthetic genes including PvC4H, PvCAD, and PvCCR1 compared with the control (Figure 7A). The expression of putative cellulose and hemicellulose biosynthetic genes were also evaluated in transgenic and non-transgenic lines. The results showed that the relative transcript level of putative primary cell wall cellulose biosynthetic gene PvCESA1 was significantly reduced, while the expression of other putative primary cell wall biosynthetic gene, PvCESA3 and that of three secondary cell wall associated putative PvCESA genes PvCESA4, PvCESA7, and PvCESA8, were unchanged in transgenic compared with the control lines. A significant increase in the expression level of the putative hemicellulose biosynthetic gene, PvCSLD1 was also observed in transgenic compared to the control lines (Figure 7B). The expression of putative GA signaling pathway genes were also evaluated in transgenic and non-transgenic lines. The results showed that the expression of putative PvGA20ox gene (PvGA20ox1a) was significantly reduced in transgenic compared to the non-transgenic control lines. In contrast, the relative expression level of one (PvGA2ox6) of the three C20 GA2ox genes (responsible for catabolism of the 20-carbon precursors of bioactive GA) evaluated in this study showed a significant sixfold increase in transgenic switchgrass compared with non-transgenic controls (Figure 7C).
FIGURE 7.
The relative expression of putative target genes of PvKN1 in transgenic switchgrass (line 4) overexpressing PvKN1 vs the non-transgenic (WT) plants as determined by qRT-PCR. (A) Lignin biosynthetic genes. (B) Cell wall biosynthetic genes. (C) Gibberellin (GA) catabolic and biosynthetic genes. The relative levels of transcripts were normalized to UBQ. Asterisks indicate significant differences from non-transgenic control plants at P ≤ 0.05 as determined by PROC TTEST procedure using SAS software (SAS Institute Inc.). Bars represent mean values of three replicates ± standard error. The lignin and GA biosynthetic and catabolic genes were as described before in Wuddineh et al. (2015). The cellulose and hemicellulose biosynthetic genes were labeled according to the naming from the closest rice or Arabidopsis homologs used in Ambavaram et al. (2011) (Supplementary Figure S5). (Pv)4CL (4-coumarate: CoA ligase); (Pv)C3H (coumaroyl shikimate 3-hydroxylase); (Pv)C4H (coumaroyl shikimate 4-hydroxylase); (Pv)CAD (cinnamyl alcohol dehydrogenase); (Pv)CCR1 (cinnamoyl CoA reductase1); (Pv)COMT (caffeic acid 3-O-methyltransferase); (Pv)F5H (ferulate 5-hydroxylase); (Pv)PAL (phenylalanine ammonia-lyase); (Pv)LAC1 (laccase1); (Pv)CESA (cellulose synthase); (Pv)CSL (cellulose synthase-like).
Changes in Lignin Content and Composition in Transgenic Switchgrass
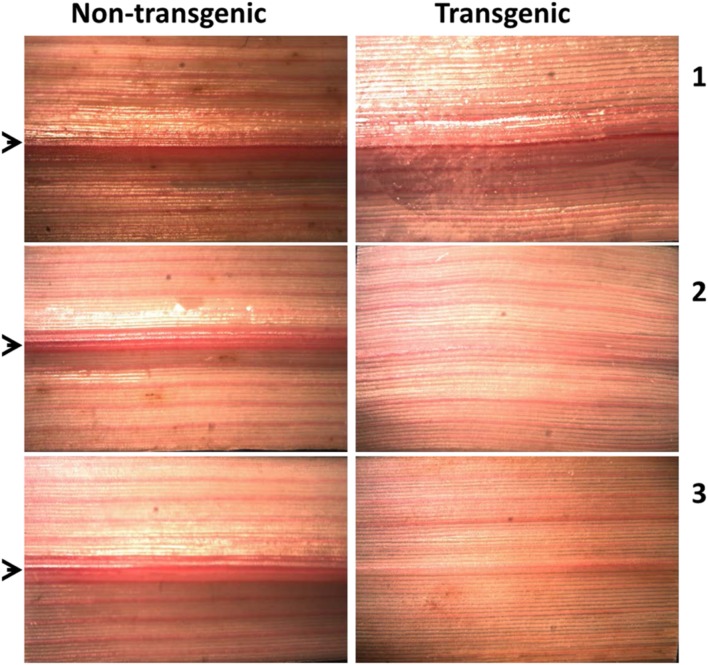
There appeared to be reduced lignin in transgenic leaves stained with phloroglucinol-HCl (Figure 8). Moreover, quantitative analysis of total lignin content in whole tillers by pyMBMS from the R1 developmental stage showed a slight reduction in lignin content by up to 8% in the transgenic lines relative to controls (Supplementary Figure S4A). The S/G lignin monomer ratio in R1 tillers of transgenic lines was not different from that in the non-transgenic control (Supplementary Figure S4B).
FIGURE 8.
Histochemical detection of lignin in leaves of PvKN1a overexpressing and non-transgenic control switchgrass lines using light microscopy. The phloroglucinol-HCl staining was done on leaves from three independent tillers indicated by numbers (1–3). The images were taken at 1× magnification. The arrows indicate midribs of leaves.
Effect of Overexpression of PvKN1 on Sugar Release Efficiency in Switchgrass
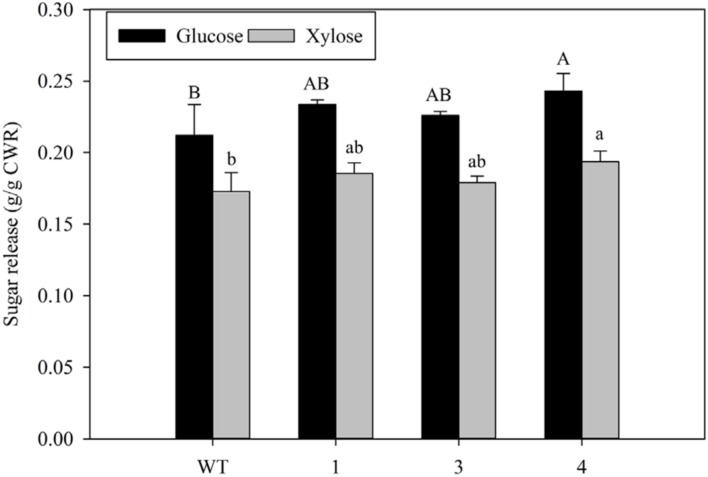
Sugar release in R1 whole tillers was significantly increased in transgenic line 4, which had the highest transgene expression among lines, in which 15% more glucose and 12% more xylose was released as compared with the non-transgenic control. The total sugar release (glucose and xylose combined) in this line was increased by up to 13% relative to the control (Figure 9).
FIGURE 9.
Sugar release by enzymatic hydrolysis in transgenic and non-transgenic control (WT) switchgrass lines. All data are means ± standard deviation (n = 3). Bars represented by different letters are significantly different at P ≤ 0.05 as tested by the LSD method with SAS software (SAS Institute Inc.). CWR, cell wall residues.
Discussion
Structural Conservation in Switchgrass KNOX Genes and Functional Implications
In this study, we identified 10 putative KNOX TF genes in the switchgrass genome, which include seven members of the class I and three of the class II families (Figure 1; Supplementary Table S3). So far, a total of 13 KNOX genes in maize, 12 in rice, 15 in Populus, 8 in Arabidopsis, 10 in peach, 5 in Selaginella moellendorffii, 5 in moss and 1 in Chlamydomonas reinhardtii have been reported (Mukherjee et al., 2009; Hay and Tsiantis, 2010; Testone et al., 2012). Thus, it appears that tetraploid switchgrass could harbor more KNOX genes than we have reported here, which may be revealed as the more complete genome sequences are made available. In general, the identified switchgrass KNOX genes showed high similarity to homologs in other plant species in terms of gene structure, amino acid sequence identity of deduced proteins, with shared specific domains (Figures 1–3, Supplementary Table S3). All members share the characteristic domains found in the maize KN1 including Meinox (essential for suppressing target gene expression and homodimerization), ELK (nuclear localization signal) and HD (Figure 2) (Kerstetter et al., 1994; Nagasaki et al., 2001). Specifically, PvKN1 from class I family KNOX TFs had the highest amino acid sequence identity (>90%) with the well-characterized maize KN1 but even more identity (>98%) in the HD domain (Figure 2, Supplementary Table S3). These remarkable structural resemblances between the KNOX genes among both monocots and dicots perhaps reflect functional conservation among homologs, which is supported by functional studies (Mele et al., 2003; Du et al., 2009; Testone et al., 2012; Townsley et al., 2013).
PvKN1 Gene Expression is Mainly Associated with Meristem Development
Most class I KNOX genes are highly expressed in meristematic tissue in accordance with their proposed function in the maintenance of meristem identity (Kerstetter et al., 1994, 1997; Chuck et al., 1996; Tioni et al., 2003; Testone et al., 2008). Similarly, PvKN1 was also expressed mainly in stem and inflorescence tissues containing meristems, in line with the importance of KNOX homologs in the regulation of apical or intercalary meristem development in young tissues of other species (Kerstetter et al., 1994, 1997). It is interesting to note that PvKN1 transcripts, albeit at low levels, were detected in differentiated lateral organs, including leaves and leaf sheaths, in contrast to the situation in maize, rice, Arabidopsis and Populus where expression of KNOX genes is scantly detected in such tissues (Matsuoka et al., 1993; Kerstetter et al., 1994; Chuck et al., 1996; Du et al., 2009). However, similar expression pattern of KNOX genes in differentiated tissues has been reported in barley, sunflower and tomato (Hareven et al., 1996; Muller et al., 2001; Tioni et al., 2003). KNOX gene expression in tomato leaves was associated with compound leaf development (Hareven et al., 1996). One possible reason for the lack of KNOX gene expression phenotype (ectopic SAM formation) in differentiated switchgrass tissues might be the low transcript level, which might be below the required threshold level to induce the phenotype. Moreover, the involvement of various cell and species-specific factors such as posttranscriptional regulators in the determination of developmental responses to the expression of KN1-like genes might also contribute to this deviation as previously suggested (Tioni et al., 2003; Hay and Tsiantis, 2010). Whether or not PvKN1 has a role in non-meristematic tissues remains to be seen.
Overexpression of PvKN1 Differentially Regulate Plant Growth Phenotypes in Switchgrass and Rice
Ectopic expression of KNOX genes have been shown to modify plant growth and development in both monocots and dicots (Matsuoka et al., 1993; Sentoku et al., 2000; Du et al., 2009; Hay and Tsiantis, 2010). Similarly, ectopic overexpression of PvKN1 in switchgrass dramatically altered growth phenotype, including plant architecture, leaf structure, and shoot elongation in transgenic plants especially, at early developmental stages (Figure 5B; Supplementary Figure S3B). There was a gradation of phenotype severity in different transgenic lines. In the worst-case, growth was arrested at the early seedling stage (Figure 5A), whereas other lines eventually attained normal shoots and roots after prolonged incubation on the rooting medium (Figure 5C) even though PvKN1 transcript level in these lines was up to nine times higher than the control. In contrast, higher levels of PvKN1 transcripts (upward of 50-fold increase) in transgenic lines in the SA37 switchgrass genotype background displayed aberrant phenotypes even at later stages of development (Supplementary Figure S3). The apparent phenotypic differences in these transgenic plants might be attributed to the differences in transcript abundance as well as the genotypic differences in response to PvKN1 overexpression as previously reported for maize (Kerstetter et al., 1997; Foster et al., 1999). The dependence of phenotype severity on KNOX transcript level has been reported in rice and Arabidopsis (Sentoku et al., 2000; Hay et al., 2003).
Interestingly, ectopic expression of PvKN1 failed to induce ectopic meristem development in lateral organs, in contrast to other plants with simple leaves such as maize KN1 dominant gain-of-function mutants (Vollbrecht et al., 1991), rice overexpressing OSH1 (Matsuoka et al., 1993) or Populus overexpressing ARK1 (Groover et al., 2006). However, differences in the effect of KNOX gene overexpression may be related to levels or patterns of ectopic overexpression. Consistent with this idea, no aberrant meristems or shoot phenotypes were observed in dicots such as tobacco that overexpressed the rice OSH1 gene (Kano-Murakami et al., 1993), and Populus that overexpressed ARK2 genes (Du et al., 2009), or in rice overexpressing OSH1 under a weak CaMV 35S promoter (Sentoku et al., 2000). On the other hand, PvKN1 overexpressing plants clearly exhibited altered distal-proximal patterns of leaf growth where the development of distal leaf blades was inhibited, which, in turn, gave rise to the development of proximal leaf sheath-like tissues as previously reported in rice and maize expressing KNOX genes (Figure 5A; Supplementary Figure S3A; Vollbrecht et al., 1991; Sentoku et al., 2000). This observation is in accordance with the maturation schedule hypothesis, which proposes that leaf primordia progress through a series of developmental stages along the proximal to distal axis of the leaf to form distinct tissue types, namely sheath, ligule, auricle and leaf blade, and ectopic expression of KNOX genes causes developmental delays impeding cells from normal progression through subsequent phases of maturation schedule (Freeling, 1992).
Another intriguing observation was the twisted stem and altered leaf phenotypes in SA37 switchgrass plants overexpressing PvKN1, which resembled the phenotypes observed in dominant mutants of maize KNAT4 (Gnarley1; Foster et al., 1999). This specific phenotype was not observed in rice and maize overexpressing the homologs of PvKN1 (Vollbrecht et al., 1991; Sentoku et al., 2000). The diverse effects of ectopic KNOX gene expression in lateral organs may reflect the differential competence of tissues in addition to the transcript level in dictating tissue response to KNOX gene action. Two major factors have been suggested to modulate the competence of leaf tissues in Arabidopsis and tomato, i.e., KNOX-independent regulation of common target genes, and the regulation of tissue differentiation (Hay and Tsiantis, 2010). The interaction between different KNOX proteins and BELL or OVATE proteins might, in turn, be involved in the regulation of these factors (Bolduc et al., 2014). In general, such differences in the phenotypes among homologous gene expression could also indicate divergence in the gene regulatory mechanisms of KNOX genes among plant species as radial adaptation.
Rice with PvKN1 overexpression produced clumps of multiple shoots, but with no aberrant effect on root development. These phenotypes were consistent with the previous findings in rice where class I KNOX genes such as OSH1 and OSH3 were overexpressed (Sentoku et al., 2000; Nagasaki et al., 2001), further supporting the homology between these genes. The difference between switchgrass and rice in response to the expression of PvKN1 might be attributed to differential cis-regulation of the gene by different regulatory elements, as suggested by Hay and Tsiantis (2010). Taken together, these results hinted that the mechanism behind PvKN1-induced regulation of genes and downstream phenotypes is complex and may differ from species to species.
PvKN1 Expression is Associated with the Regulation of Cell Wall Biosynthesis via Regulation of GA and BR Signaling Pathways
PvKN1 overexpression resulted in reduced lignin deposition in the plant biomass accompanied by reduced expression of lignin biosynthetic genes including PvC4H, PvCAD, and PvCCR1 (Figure 7A). This observation is generally in line with the previous reports in Arabidopsis, peach and Populus (Mele et al., 2003; Du et al., 2009; Testone et al., 2012). Interestingly, PvKN1 overexpression appeared to downregulate the expression of a primary cell wall cellulose biosynthetic gene, PvCESA1, while upregulated the expression of hemicellulose biosynthetic gene (PvCSLD1; Figure 7B) indicating that PvKN1 may be involved in the regulation of cell wall biosynthetic genes. Unlike overexpression of ARK2 in Populus, which downregulated the expression of three secondary cell wall cellulose biosynthetic genes (Du et al., 2009), no significant alteration of expression of these genes was observed in PvKN1-overexpressing transgenic switchgrass (Figure 7B). This difference might be attributed to the biological differences between the two species or the differences between the tissues analyzed. However, the regulatory mechanism of cell wall biosynthesis involving KN1 TFs is less clear.
It was recently suggested that the maize KN1 gene regulates the expression of genes responsible for the biosynthesis of lignin, and perhaps other cell wall components indirectly via the regulation of ‘executive’ genes (Bolduc et al., 2012). However, the direct regulation of a few lignin biosynthetic genes including COMT and CCoAOMT in Arabidopsis (Mele et al., 2003), and CCoAOMT in peach (Testone et al., 2012) has also been demonstrated. The GA-signaling pathway has been reported as one of the direct targets of KN1 through which other plant morphological and biochemical characteristics are regulated (Bolduc et al., 2012). In this study, we showed that overexpression of PvKN1 downregulated the expression of putative PvGA20ox (the rate-limiting enzyme in the GA biosynthesis pathway) while upregulated the expression of C20 PvGA2ox genes (Figure 7C) suggesting that PvKN1 might limit shoot elongation and lignification at early growth stages via regulation of GA biosynthetic and catabolic genes and GA signaling.
The mechanism whereby PvKN1 regulates cellulose biosynthetic genes (Figure 7B) is unknown. One possible scenario is that PvKN1 regulates the expression of cellulose biosynthetic genes through modulation of BR signaling, which has been shown to positively regulate the expression of cellulose biosynthetic genes (Xie et al., 2011). This hypothesis is supported by the recent report that maize KN1 could interact with BR biosynthetic and catabolic genes (Bolduc et al., 2012), and OSH1 regulates BR-signaling in rice (Tsuda et al., 2014).
Implications of PvKN1 TF for Improvement of Bioenergy Crops
The prospect of drastically decreasing lignin by ectopic expression of KNOX genes motivated our study in switchgrass. The challenge here, as with the case of transgenic switchgrass in which MYB4 (Shen et al., 2012) and miRNA156 (Fu et al., 2012) gene expression was changed, is to empirically determine the optimal TF transgene expression level that decreases lignin and possibly provides favorable plant architecture changes, without reducing shoot growth. Clearly, if KN1 expression is too high, plants are dwarfed and abnormal. Indeed, relatively low PvKN1 overexpression resulted in reduced lignin deposition/biomass recalcitrance and improved sugar release efficiency without significantly affecting various growth parameters such as tiller height, tiller number, fresh and dry biomass weight (Figure 8 and 9; Table 1). Therefore, the observed increase in sugar release in transgenic switchgrass with reduced lignin content highlight the potential biotechnological application of PvKN1 for the improvement of biomass characteristics for bioenergy feedstocks as well as forage grasses. Previous studies have suggested that expression of KNOX genes may enhance cytokinin levels (Frugis et al., 2001; Jasinski et al., 2005; Yanai et al., 2005) while cytokinin accumulation was shown to increase drought tolerance via coordinated regulation of carbon and nitrogen assimilation (Peleg et al., 2011; Reguera et al., 2013). Whether PvKN1 expression plays similar roles should be the subject of future investigation as such traits may enhance the value of using transgenic lines as improved bioenergy feedstocks.
Conclusion
In summary, we identified a gene coding for class I KNOX TF in switchgrass, PvKN1, as a putative ortholog of maize KN1. Our results demonstrated that PvKN1 may facilitate shoot elongation and lignification via transcriptional regulation of the GA-biosynthesis pathway. Moreover, we showed that PvKN1 may also be involved in the regulation of the biosynthetic genes of other cell wall polymers (cellulose and hemicellulose) and hence play an important role in cell wall biosynthesis. PvKN1-overexpressing lines displaying normal growth phenotypes but with reduced recalcitrance to enzymatic saccharification could potentially be utilized for the improvement of lignocellulosic bioenergy feedstocks for biofuels or for the improvement of forage grass. PvKN1 could also be potentially used in gene-stacking studies along with genes imparting novel traits, such as higher biomass yield, biotic or abiotic stress resistance to develop more improved bioenergy feedstock.
Author Contributions
WW designed and performed the experiments, analyzed the data, and drafted the manuscript. MM participated in experimental design and data analysis, assisted with revisions to the manuscript and coordination of the study. J-YZ and MU assisted with cloning of the target gene and contributed in revision of the manuscript. GT, RS, SD, and MD assisted with performing lignin and sugar release assays and contributed in revision of the manuscript. CNS conceived the study and its design and coordination, and assisted with revisions to the manuscript. All authors read and consented to the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Angela Ziebell, Erica Gjersing, Crissa Doeppke, and Melvin Tucker for their assistance with the cell wall characterization and Susan Holladay for her assistance with data entry into LIMS. We thank Wayne Parrot for providing the switchgrass SA37 clone.
Abbreviations
- ANOVA
analysis of variance
- BELL
BEL1-like homeodomain
- BR
brassinosteroid
- CAD
cinnamyl alcohol dehydrogenase
- CDS
coding DNA sequence
- CCoAOMT
caffeoyl CoA 3-O-methyltransferase
- CCR
cinnamoyl CoA reductase
- CESA
cellulose synthase
- COMT
caffeic acid 3-O-methyltransferase
- CSL
cellulose synthase-like
- CWR
cell wall residues
- C3H
coumaroyl shikimate 3-hydroxylase
- C4H
coumaroyl shikimate 4-hydroxylase
- F5H
ferulate 5-hydroxylase
- GA2ox
Gibberellin 2-oxidase
- GA20ox
Gibberellin 20-oxidase
- HD
homeodomain
- KN1
Knotted1
- KNAT1
KN1-like from Arabidopsis
- KNOX
KN1-like homeobox
- LAC
laccase
- LSD
least significant difference
- NAC NAM
ATAF1/2 and CUC2
- OFP
orange fluorescent protein
- PAL
phenylalanine ammonia-lyase
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- R1
reproductive stage 1
- SAM
shoot apical meristems
- S/G
syringyl/guaiacyl
- STM
SHOOT MERISTEMLESS
- SWN
secondary wall NAC
- TALE
three amino acid loop extension
- TF
transcription factor
- UBQ
ubiquitin
- UTR
untranslated regions
- ZmUbi1
maize ubiquitin 1
- 4CL
4-coumarate: CoA ligase.
Footnotes
Funding. This work was supported by funding from the BioEnergy Science Center (DE-PS02-06ER64304). The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. We also thank Tennessee Agricultural Experiment Station for providing partial financial support for WW. We also thank DOE-JGI for the prepublication access to the switchgrass genome.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00520
References
- Ambavaram M. M., Krishnan A., Trijatmiko K. R., Pereira A. (2011). Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 2011 916–931. 10.1104/pp.110.168641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R. S., Chern M., Vega-Sanchez M. E., Canlas P., Ronald P. C. (2010). Rice Snl6 a cinnamoyl-CoA reductase-like gene family member, is required for NH1-mediated immunity to Xanthomonas oryzae pv. oryzae. PLoS Genet. 6:e1001123 10.1371/journal.pgen.1001123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter H. L., Mazarei M., Labbe N., Kline L. M., Cheng Q., Windham M. T., et al. (2014). Two-year field analysis of reduced recalcitrance transgenic switchgrass. Plant Biotechnol. J. 12 914–924. 10.1111/pbi.12195 [DOI] [PubMed] [Google Scholar]
- Biemelt S., Tschiersch H., Sonnewald U. (2004). Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol. 135 254–265. 10.1104/pp.103.036988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene GA2ox1. Plant Cell 21 1647–1658. 10.1105/tpc.109.068221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Tyers R. G., Freeling M., Hake S. (2014). Unequal redundancy in maize knotted1 homeobox genes. Plant Physiol. 164 229–238. 10.1104/pp.113.228791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra M. K., Morohashi K., O’connor D., Grotewold E., et al. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26 1685–1690. 10.1101/gad.193433.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris J., Mann D. J., Joyce B., Stewart C. N., Jr. (2009). An improved tissue culture system for embryogenic callus production and plant regeneration in switchgrass (Panicum virgatum L.). Bioenerg. Res. 2 267–274. 10.1007/s12155-009-9048-8 [DOI] [Google Scholar]
- Chai M., Bellizzi M., Wan C., Cui Z., Li Y., Wang G.-L. (2015). The NAC transcription factor OsSWN1 regulates secondary cell wall development in Oryza sativa. J. Plant Biol. 58 44–51. 10.1007/s12374-014-0400-y [DOI] [Google Scholar]
- Chuck G., Lincoln C., Hake S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289. 10.1105/tpc.8.8.1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker S. R., Carlile M., Selig M. J., Doeppke C., Davis M., Sykes R., et al. (2012). Reducing the effect of variable starch levels in biomass recalcitrance screening. Methods Mol. Biol. 908 181–195. 10.1007/978-1-61779-956-3_17 [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36 W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOE Joint Genome Institute [DOE-JGI] (2015). Panicum virgatum v1.1. Available at: http://www.phytozome.net/panicumvirgatum [Google Scholar]
- Du J., Mansfield S. D., Groover A. T. (2009). The Populus homeobox gene ARBORKNOX2 regulates cell differentiation during secondary growth. Plant J. 60 1000–1014. 10.1111/j.1365-313X.2009.04017.x [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla-Trevino L. L., Shen H., Uppalapati S. R., Ray T., Tang Y., Hernandez T., et al. (2010). Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties. New Phytol. 185 143–155. 10.1111/j.1469-8137.2009.03018.x [DOI] [PubMed] [Google Scholar]
- Fornale S., Shi X., Chai C., Encina A., Irar S., Capellades M., et al. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 64 633–644. 10.1111/j.1365-313X.2010.04363.x [DOI] [PubMed] [Google Scholar]
- Foster T., Yamaguchi J., Wong B. C., Veit B., Hake S. (1999). Gnarley1 is a dominant mutation in the knox4 homeobox gene affecting cell shape and identity. Plant Cell 11 1239–1252. 10.2307/3870746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (1992). A conceptual framework for maize leaf development. Dev. Biol. 153 44–58. 10.1016/0012-1606(92)90090-4 [DOI] [PubMed] [Google Scholar]
- Frugis G., Giannino D., Mele G., Nicolodi C., Chiappetta A., Bitonti M. B., et al. (2001). Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 126 1370–1380. 10.1104/pp.126.4.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Mielenz J. R., Xiao X., Ge Y., Hamilton C. Y., Rodriguez M., et al. (2011a). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 108 3803–3808. 10.1073/pnas.1100310108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Sunkar R., Zhou C., Shen H., Zhang J. Y., Matts J., et al. (2012). Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 10 443–452. 10.1111/j.1467-7652.2011.00677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Xiao X., Xi Y., Ge Y., Chen F., Bouton J., et al. (2011b). Downregulation of Cinnamyl Alcohol Dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. Bioenerg. Res. 4 153–164. 10.1007/s12155-010-9109-z [DOI] [Google Scholar]
- Grewal D., Gill R., Gosal S. S. (2006). Influence of antibiotic cefotaxime on somatic embryogenesis and plant regeneration in indica rice. Biotechnol. J. 1 1158–1162. 10.1002/biot.200600139 [DOI] [PubMed] [Google Scholar]
- Groover A. T., Mansfield S. D., Difazio S. P., Dupper G., Fontana J. R., Millar R., et al. (2006). The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 61 917–932. 10.1007/s11103-006-0059-y [DOI] [PubMed] [Google Scholar]
- Guo A. Y., Zhu Q. H., Chen X., Luo J. C. (2007). GSDS: a gene structure display server. Yi Chuan 29 1023–1026. 10.1360/yc-007-1023 [DOI] [PubMed] [Google Scholar]
- Hake S., Smith H. M., Holtan H., Magnani E., Mele G., Ramirez J. (2004). The role of KNOX genes in plant development. Annu. Rev. Cell Dev. Biol. 20 125–151. 10.1146/annurev.cellbio.20.031803.093824 [DOI] [PubMed] [Google Scholar]
- Handakumbura P. P., Hazen S. P. (2012). Transcriptional regulation of grass secondary cell wall biosynthesis: playing catch-up with Arabidopsis thaliana. Front. Plant Sci. 3:74 10.3389/fpls.2012.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin C. F., Fu C., Hisano H., Xiao X., Shen H., Stewart C. N., et al. (2013). Standardization of switchgrass sample collection for cell wall and biomass trait analysis. Bioenerg. Res. 6 755–762. 10.1007/s12155-012-9292-1 [DOI] [Google Scholar]
- Hareven D., Gutfinger T., Parnis A., Eshed Y., Lifschitz E. (1996). The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84 735–744. 10.1016/S0092-8674(00)81051-X [DOI] [PubMed] [Google Scholar]
- Hay A., Jackson D., Ori N., Hake S. (2003). Analysis of the competence to respond to KNOTTED1 activity in Arabidopsis leaves using a steroid induction system. Plant Physiol. 131 1671–1680. 10.1104/pp.102.017434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12 1557–1565. 10.1016/S0960-9822(02)01125-9 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137 3153–3165. 10.1242/dev.030049 [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413. [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15 1560–1565. 10.1016/j.cub.2005.07.023 [DOI] [PubMed] [Google Scholar]
- Kano-Murakami Y., Yanai T., Tagiri A., Matsuoka M. (1993). A rice homeotic gene, OSH1 causes unusual phenotypes in transgenic tobacco. FEBS Lett. 334 365–368. [DOI] [PubMed] [Google Scholar]
- Kerstetter R., Vollbrecht E., Lowe B., Veit B., Yamaguchi J., Hake S. (1994). Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 6 1877–1887. 10.1105/tpc.6.12.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R. A., Laudencia-Chingcuanco D., Smith L. G., Hake S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1 are defective in shoot meristem maintenance. Development 124 3045–3054. [DOI] [PubMed] [Google Scholar]
- King Z. R., Bray A. L., Lafayette P. R., Parrott W. A. (2014). Biolistic transformation of elite genotypes of switchgrass (Panicum virgatum L.). Plant Cell Rep. 33 313–322. 10.1007/s00299-013-1531-1 [DOI] [PubMed] [Google Scholar]
- Li E., Bhargava A., Qiang W., Friedmann M. C., Forneris N., Savidge R. A., et al. (2012). The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 194 102–115. 10.1111/j.1469-8137.2011.04016.x [DOI] [PubMed] [Google Scholar]
- Li R., Qu R. (2011). High throughput Agrobacterium-mediated switchgrass transformation. Biomass Bioenerg. 35 1046–1054. 10.1016/j.biombioe.2010.11.025 [DOI] [Google Scholar]
- Lynd L. R., Laser M. S., Bransby D., Dale B. E., Davison B., Hamilton R., et al. (2008). How biotech can transform biofuels. Nat. Biotechnol. 26 169–172. 10.1038/nbt0208-169 [DOI] [PubMed] [Google Scholar]
- Ma Q. H., Wang C., Zhu H. H. (2011). TaMYB4 cloned from wheat regulates lignin biosynthesis through negatively controlling the transcripts of both cinnamyl alcohol dehydrogenase and cinnamoyl-CoA reductase genes. Biochimie 93 1179–1186. 10.1016/j.biochi.2011.04.012 [DOI] [PubMed] [Google Scholar]
- Mann D. G., Lafayette P. R., Abercrombie L. L., King Z. R., Mazarei M., Halter M. C., et al. (2012). Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol. J. 10 226–236. 10.1111/j.1467-7652.2011.00658.x [DOI] [PubMed] [Google Scholar]
- Matsuoka M., Ichikawa H., Saito A., Tada Y., Fujimura T., Kano-Murakami Y. (1993). Expression of a rice homeobox gene causes altered morphology of transgenic plants. Plant Cell 5 1039–1048. 10.1105/tpc.5.9.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele G., Ori N., Sato Y., Hake S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17 2088–2093. 10.1101/gad.1120003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. J., Moser L. E., Vogel K. P., Waller S. S., Johnson B. E., Pedersen J. F. (1991). Describing and quantifying growth stages of perennial forage grasses. Agron. J. 83 1073–1077. 10.2134/agronj1991.00021962008300060027x [DOI] [Google Scholar]
- Mukherjee K., Brocchieri L., Burglin T. R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26 2775–2794. 10.1093/molbev/msp201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J., Wang Y., Franzen R., Santi L., Salamini F., Rohde W. (2001). In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of KNOX gene function. Plant J. 27 13–23. 10.1046/j.1365-313x.2001.01064.x [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Nagasaki H., Sakamoto T., Sato Y., Matsuoka M. (2001). Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell 13 2085–2098. 10.1105/tpc.13.9.2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A., Aichi I., Matsuoka M. (2006). A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 1 2796–2802. 10.1038/nprot.2006.469 [DOI] [PubMed] [Google Scholar]
- Peleg Z., Reguera M., Tumimbang E., Walia H., Blumwald E. (2011). Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol. J. 9 747–758. 10.1111/j.1467-7652.2010.00584.x [DOI] [PubMed] [Google Scholar]
- Ragauskas A. J., Williams C. K., Davison B. H., Britovsek G., Cairney J., Eckert C. A., et al. (2006). The path forward for biofuels and biomaterials. Science 311 484–489. 10.1126/science.1114736 [DOI] [PubMed] [Google Scholar]
- Reguera M., Peleg Z., Abdel-Tawab Y. M., Tumimbang E. B., Delatorre C. A., Blumwald E. (2013). Stress-induced cytokinin synthesis increases drought tolerance through the coordinated regulation of carbon and nitrogen assimilation in rice. Plant Physiol. 163 1609–1622. 10.1104/pp.113.227702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saathoff A. J., Sarath G., Chow E. K., Dien B. S., Tobias C. M. (2011). Downregulation of cinnamyl-alcohol dehydrogenase in switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PLoS ONE 6:e16416 10.1371/journal.pone.0016416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmer M. R., Vogel K. P., Mitchell R. B., Perrin R. K. (2008). Net energy of cellulosic ethanol from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 105 464–469. 10.1073/pnas.0704767105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig M. J., Tucker M. P., Sykes R. W., Reichel K. L., Brunecky R., Himmel M. E., et al. (2010). Biomass recalcitrance sceening by integrated high throughput hydrothermal pretreatment and enzymatic saccharification. Ind. Biotechnol. 6 104–111. 10.1007/978-1-61779-956-3_17 [DOI] [Google Scholar]
- Sentoku N., Sato Y., Matsuoka M. (2000). Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev. Biol. 220 358–364. 10.1006/dbio.2000.9624 [DOI] [PubMed] [Google Scholar]
- Shen H., Fu C., Xiao X., Ray T., Tang Y., Wang Z., et al. (2009). Developmental control of lignification in stems of lowland switchgrass variety Alamo and the effects on saccharification efficiency. Bioenerg. Res. 2 233–245. 10.1007/s12155-009-9058-6 [DOI] [Google Scholar]
- Shen H., He X., Poovaiah C. R., Wuddineh W. A., Ma J., Mann D. G., et al. (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193 121–136. 10.1111/j.1469-8137.2011.03922.x [DOI] [PubMed] [Google Scholar]
- Shen H., Mazarei M., Hisano H., Escamilla-Trevino L., Fu C., Pu Y., et al. (2013a). A genomics approach to deciphering lignin biosynthesis in switchgrass. Plant Cell 25 4342–4361. 10.1105/tpc.113.118828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Poovaiah C. R., Ziebell A., Tschaplinski T. J., Pattathil S., Gjersing E., et al. (2013b). Enhanced characteristics of genetically modified switchgrass (Panicum virgatum L.) for high biofuel production. Biotechnol. Biofuels 6 71 10.1186/1754-6834-6-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M. H., Demartini J. D., Brethauer S., Mckenzie H. L., Wyman C. E. (2010). Engineering of a high-throughput screening system to identify cellulosic biomass, pretreatments, and enzyme formulations that enhance sugar release. Biotechnol. Bioeng. 105 231–238. 10.1002/bit.22527 [DOI] [PubMed] [Google Scholar]
- Sykes R., Yung M., Novaes E., Kirst M., Peter G., Davis M. (2009). High-throughput screening of plant cell-wall composition using pyrolysis molecular beam mass spectroscopy. Methods Mol. Biol. 581 169–183. 10.1007/978-1-60761-214-8_12 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testone G., Bruno L., Condello E., Chiappetta A., Bruno A., Mele G., et al. (2008). Peach [Prunus persica (L.) Batsch] KNOPE1 a class 1 KNOX orthologue to Arabidopsis BREVIPEDICELLUS/KNAT1 is misexpressed during hyperplasia of leaf curl disease. J. Exp. Bot. 59 389–402. 10.1093/jxb/erm317 [DOI] [PubMed] [Google Scholar]
- Testone G., Condello E., Verde I., Nicolodi C., Caboni E., Dettori M. T., et al. (2012). The peach (Prunus persica L. Batsch) genome harbours 10 KNOX genes, which are differentially expressed in stem development, and the class 1 KNOPE1 regulates elongation and lignification during primary growth. J. Exp. Bot. 63 5417–5435. 10.1093/jxb/ers194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. G., Rieu I., Steber C. M. (2005). Gibberellin metabolism and signaling. Vitam. Horm. 72 289–338. 10.1016/s0083-6729(05)72009-4 [DOI] [PubMed] [Google Scholar]
- Tioni M. F., Gonzalez D. H., Chan R. L. (2003). Knotted1-like genes are strongly expressed in differentiated cell types in sunflower. J. Exp. Bot. 54 681–690. 10.1093/jxb/erg077 [DOI] [PubMed] [Google Scholar]
- Townsley B. T., Sinha N. R., Kang J. (2013). KNOX1 genes regulate lignin deposition and composition in monocots and dicots. Front. Plant Sci. 4:121 10.3389/fpls.2013.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaplinski T. J., Standaert R. F., Engle N. L., Martin M. Z., Sangha A. K., Parks J. M., et al. (2012). Down-regulation of the caffeic acid O-methyltransferase gene in switchgrass reveals a novel monolignol analog. Biotechnol. Biofuels 5 71 10.1186/1754-6834-5-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Kurata N., Ohyanagi H., Hake S. (2014). Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26 3488–3500. 10.1105/tpc.114.129122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Veit B., Sinha N., Hake S. (1991). The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350 241–243. 10.1038/350241a0 [DOI] [PubMed] [Google Scholar]
- Wuddineh W. A., Mazarei M., Zhang J., Poovaiah C. R., Mann D. G. J., Ziebell A., et al. (2015). Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol. J. 13 636–647. 10.1111/pbi.12287 [DOI] [PubMed] [Google Scholar]
- Xie L., Yang C., Wang X. (2011). Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 62 4495–4506. 10.1093/jxb/err164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Escamilla-Trevino L. L., Sathitsuksanoh N., Shen Z., Shen H., Zhang Y. H., et al. (2011). Silencing of 4-coumarate:coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol. 192 611–625. 10.1111/j.1469-8137.2011.03830.x [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., et al. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15 1566–1571. 10.1016/j.cub.2005.07.060 [DOI] [PubMed] [Google Scholar]
- Yuan J. S., Tiller K. H., Al-Ahmad H., Stewart N. R., Stewart CN., Jr (2008). Plants to power: bioenergy to fuel the future. Trends Plant Sci. 13 421–429. 10.1016/j.tplants.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Zhang J. Y., Lee Y. C., Torres-Jerez I., Wang M., Yin Y., Chou W. C., et al. (2013). Development of an integrated transcript sequence database and a gene expression atlas for gene discovery and analysis in switchgrass (Panicum virgatum L.). Plant J. 74 160–173. 10.1111/tpj.12104 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Gallego-Giraldo L., Wang H., Zeng Y., Ding S. Y., Chen F., et al. (2010). An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 63 100–114. 10.1111/j.1365-313X.2010.04223.x [DOI] [PubMed] [Google Scholar]
- Zhao X. Y., Zhu D. F., Zhou B., Peng W. S., Lin J. Z., Huang X. Q., et al. (2010). Over-expression of the AtGA2ox8 gene decreases the biomass accumulation and lignification in rapeseed (Brassica napus L.). J. Zhejiang Univ. Sci. 11 471–481. 10.1631/jzus.B1000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Mccarthy R. L., Reeves C. K., Jones E. G., Ye Z. H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52 1856–1871. 10.1093/pcp/pcr123 [DOI] [PubMed] [Google Scholar]
- Zhong R., Ye Z.-H. (2014). Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci. 229 193–207. 10.1016/j.plantsci.2014.09.009 [DOI] [PubMed] [Google Scholar]
- Zhou J., Lee C., Zhong R., Ye Z. H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21 248–266. 10.1105/tpc.108.063321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.