Abstract
The wound healing assay (or scratch assay) is a technique frequently used to quantify the dependence of cell motility—a central process in tissue repair and evolution of disease—subject to various treatments conditions. However processing the resulting data is a laborious task due its high throughput and variability across images. This Robust Quantitative Scratch Assay algorithm introduced statistical outputs where migration rates are estimated, cellular behaviour is distinguished and outliers are identified among groups of unique experimental conditions. Furthermore, the RQSA decreased measurement errors and increased accuracy in the wound boundary at comparable processing times compared to previously developed method (TScratch).
Availability and implementation: The RQSA is freely available at: http://ophid.utoronto.ca/RQSA/RQSA_Scripts.zip. The image sets used for training and validation and results are available at: (http://ophid.utoronto.ca/RQSA/trainingSet.zip, http://ophid.utoronto.ca/RQSA/validationSet.zip, http://ophid.utoronto.ca/RQSA/ValidationSetResults.zip, http://ophid.utoronto.ca/RQSA/ValidationSet_H1975.zip, http://ophid.utoronto.ca/RQSA/ValidationSet_H1975Results.zip, http://ophid.utoronto.ca/RQSA/RobustnessSet.zip, http://ophid.utoronto.ca/RQSA/RobustnessSet.zip). Supplementary Material is provided for detailed description of the development of the RQSA.
Contact: juris@ai.utoronto.ca
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Since cell migration is significant in repair processes and disease advancement such as metastasis and cancer angiogenesis, it can be used as a parameter to describe the state of health of various cell lines (Gebäck et al., 2009; Zordan et al., 2011). Furthermore, quantification of cell migration can be used to measure the effect of drug treatments on the same cell line. The Wound Healing Assay (WHA) is used to quantify motility by monitoring the evolution of a wounded monolayer of cells. The experimental set-up consists of growing a confluent monolayer of cells under specific conditions (i.e. treated or not treated with a drug), then creating a wound or scratch on each layer/well, and then imaging all wounds multiple times over a period of time. Since the rate how wounds close may depend on conditions used, measuring wound areas from individual images are used to estimate migration rates, and thus quantify condition-specific cell motility. (Jonkman et al. 2014) provide extensive insight on experimental design considerations and workflow of the WHA measurements. In order to obtain reliable results, it is necessary to measure wound areas across multiple replicates of the same cell condition, and record multiple frames per replicate to determine whether changes are statistically significant. To make this task feasible, an automated system is required to process the high volumes of data and make measurements independent of image artefacts such as uneven illuminations, smudges or scratches on the well plate lid, in addition to some variation of initial wound width and shape. Sample images of wound areas are shown in Supplementary Material.
While previous methods by Gebäck et al. and Zordan et al. support automated WHA image analysis, they lack a final statistical output estimating the migration rates per condition, and lack an image set for validation, therefore depending on the acquisition parameters, these methods may or may not work. Therefore, we have developed the Robust Quantitative Scratch Assay (RQSA) algorithm using MATLAB 7.11.0 to address the challenges imposed by the large datasets, variability in image illuminations and implemented a statistical output for an improved quantification of cell motility using the wound healing assay. Furthermore, we provide image datasets for reproducibility and have developed a set of guidelines for future WHA image acquisitions.
2 Methods
The algorithm requires MATLAB 7.11.0 (or GNU Octave) and uses the Statistics and Image Processing Toolboxes. In order to design and validate the RQSA algorithm a training and testing image datasets were acquired and are described in detail in Section 1.1 of the Supplementary Material. The image and data processing comprises measuring individual open wound areas for each well using a sequence of morphological and spatial filters (Gonzales, 2003; http://www.mathworks.com). Then the resulting open wound area vs. frame curve is fitted to a polynomial curve using the Least Squares method in order to calculate migration rates, coefficients of determination (R2), and to label the temporal behavior for each well. The estimated migration rates for all wells in a single condition are grouped, then outliers are flagged and discarded before calculating the average migration rate for each experimental condition. Last, the average migration rates are compared across all experimental conditions. The statistical analysis is output as a text file; Section 1.3 in Supplementary Material shows sample open wound area curves, sample output text file for a single condition and describes the process for outlier detection.
3 Results
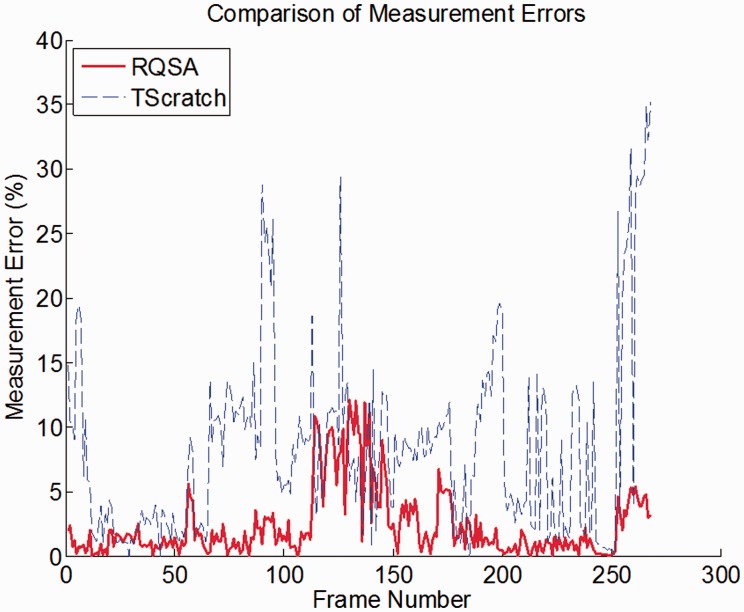
The robustness and improved performance of the RQSA algorithm was evaluated over a ‘Robustness set’—a set of images (N = 268) with various features, such as high/low image illumination, resolution, cell confluence, and stray cells in the wound area. Open Wound Areas and Wound Estimate were measured with the RQSA algorithm and TScratch were compared to manual measurements, Figure 1 shows the error in Open Wound Area for this subset. Section 2.1 in Supplementary Material describes these metrics of comparison with more detail and discusses the robustness of the algorithm. A paired t-test was performed on the errors in Open wound area and errors in Wound estimate using both methods, where resulting p-values (P < 0.05) determined that there is a statistical difference between both methods. Overall RQSA showed better representation of the wound boundaries resulting in a more accurate estimate of migration rates over all conditions.
Fig. 1.
Measurement errors resulting from TScratch (dashed line) and RQSA (solid line)
Both methods were tested using a validation set, and results were compared by determining R2 from the fitted curves in both experimental conditions. The non-treated condition with RQSA shows an R2 value centered at 1.0, suggesting that the open wound areas vs. frame from the RQSA algorithm yield a better description of the temporal behavior of the wound, and shows less variability across time points. Meanwhile, the lower R2 values of the TScratch polynomial fits suggest that measurements fluctuate highly from one frame to the other.
The RQSA was run using an additional independent set with a different cell line and treatments, to demonstrate its robustness. Results of both datasets with statistical analysis including average migrations rates, uncertainties, identified possible well outliers, and are further discussed in Section 2.2 of Supplementary Material.
Future directions for the RQSA include the incorporation of cell segmentation routines to provide a morphological and proliferation assessment on individual cells, and implementation of parallel computing techniques to decrease processing times and increase efficiency.
Conflict of Interest: none declared.
Supplementary Material
References
- Gebäck T. et al. (2009) TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques, 46, 265–274. [DOI] [PubMed] [Google Scholar]
- Gonzalez R. et al. (2003) Digital Image Processing Using MATLAB. New Jersey: Prentice Hall. [Google Scholar]
- Jonkman J. et al. (2014) An introduction to the wound healing assay using live-cell microscopy. Cell Adhesion Migration, 8, 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan M.D. et al. (2011) A high throughput, interactive imaging, bright-field. Cytometry A, 79, 227–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.