Abstract
The multifaceted field of metabolomics has witnessed exponential growth in both methods development and applications. Owing to the urgent need, a significant fraction of research investigations in the field is focused on understanding, diagnosing and preventing human diseases; hence, the field of biomedicine has been the major beneficiary of metabolomics research. A large body of literature now documents the discovery of numerous potential biomarkers and provides greater insights into pathogeneses of numerous human diseases. A sizable number of findings have been tested for translational applications focusing on disease diagnostics ranging from early detection, to therapy prediction and prognosis, monitoring treatment and recurrence detection, as well as the important area of therapeutic target discovery. Current advances in analytical technologies promise quantitation of biomarkers from even small amounts of bio-specimens using non-invasive or minimally invasive approaches, and facilitate high-throughput analysis required for real time applications in clinical settings. Nevertheless, a number of challenges exist that have thus far delayed the translation of a majority of promising biomarker discoveries to the clinic. This article presents advances in the field of metabolomics with emphasis on biomarker discovery and translational efforts, highlighting the current status, challenges and future directions.
Keywords: Biomarkers, cancer, cardiovascular disease, commercialization, diabetes, diagnostics, inborn errors of metabolism, mass spectrometry, metabolomics, neurological disorder, NMR spectroscopy, statistical analysis, translation, validation
INTRODUCTION
The field of metabolomics, which focuses on the quantitative analysis of large numbers of metabolites in complex specimens including bio-fluids, tissue and cells has grown extremely rapidly [1–3]. Small molecule metabolites (<1000 Da), represent the end products of gene, transcripts and protein function, and provide an instantaneous snapshot of biological status. While vast progress in genomics and proteomics enables understanding of altered genes and proteins under a variety of perturbations, including disease conditions, metabolomics provides an alternative approach to help understand altered metabolic pathways and discover new gene functions [4, 5]. The strong interest in metabolomics is partly based on the fact that subtle changes in genes, transcripts or protein levels can cause substantial changes in the levels and dynamics of metabolites. Hence analysis of metabolites represents a sensitive measure of biological status in health or disease. The altered metabolic fingerprints, which are unique to every individual, offer novel avenues to better understand systems biology, detect or identify potential risks for various diseases and ultimately help achieve the goal of “personalized medicine” [6].
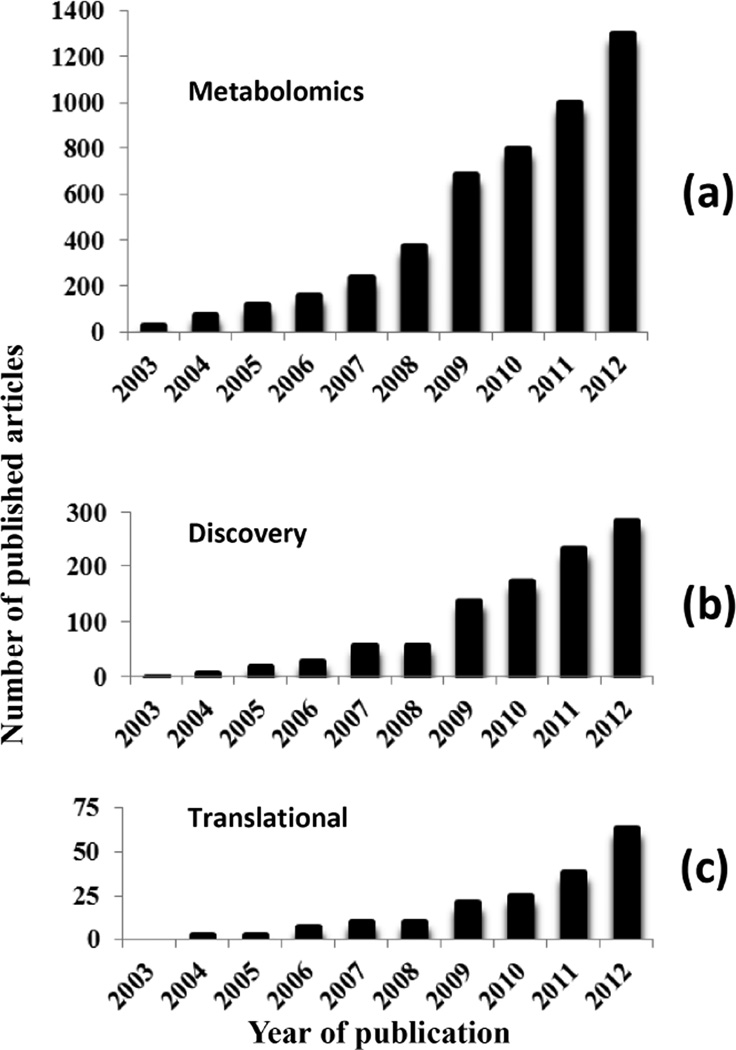
Metabolomics studies have taken advantage of newly advanced instrumentation, an array of mature analytical and statistical methods, and well-established publicly available metabolic pathways and large metabolite databases [7–12]. Over the past 10 years there has been an exponential growth in the number of investigations made in the field of metabolomics. For example, a PubMed search using the keyword “metabolomics” provides nearly 5000 papers published between 2003–2012 ranging from approximately 30 in 2003 to more than 1300 in 2012 (Fig. 1a). Broadly, metabolomics investigations focused on human diseases can be grouped into three categories. A major group of investigations is focused on understanding the molecular basis of pathogenesis of diseases and identifying altered metabolic pathways. The second group is focused on identifying metabolite biomarkers that classify diseases with high sensitivity and specificity. The number of such studies, however, is smaller but increasing exponentially owing to the urgent need to discover sensitive biomarkers for improving disease diagnostics. As an example, Fig. 1b shows the growing number of investigations focused on metabolite based biomarker discovery as obtained from the PubMed search using the key words “metabolomics” and ‘biomarker”. The third group of investigations describes translational opportunities and applications in metabolomics. Such investigations are far fewer, but are also increasing exponentially. Fig. 1c lists the number of published articles obtained from the PubMed search using the key words “metabolomics’ and “translational” and represents a qualitative measure of translational applications. This review will focus on the primary analytical methods and steps in the biomarker discovery process, and emphasize both successes and challenges in translating metabolite profiling to the clinic.
Fig. (1).
The number of metabolomics research studies published over the past 10 years has increased exponentially. The number of papers listed in PubMed using the following keyword searches: (a) “metabolomics”, (b) “metabolomics” and “biomarker” and (c) “metabolomics” and “translational.”
ANALYTICAL METHODS
The two premier analytical methods used in the field of metabolomics are nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). NMR and MS methods are both supplementary and complementary to one another. Numerous techniques within NMR and MS offer multifaceted approaches to measure concentrations of a variety of metabolite classes for disease diagnostics and understanding cellular functions in health and disease.
NMR spectroscopy is rapid, non-destructive, and non-invasive; requires little or no sample preparation; and provides highly quantitative and reproducible results. Peaks in the NMR spectra can be reliably assigned to specific metabolic species, and thus NMR provides a wealth of information on the identity and quantity of a large number of metabolites in parallel, often from a single experiment. Detailed protocols for sample collection, preparation, parameter selection, data acquisition and data analysis for serum/plasma, urine and tissue extract [13], and intact tissue [14] provide step by step procedures along with useful information especially for researchers new to the field. To date NMR-based metabolomics approaches are used to study, among others, the effect of drugs, toxins, and various diseases, to trace metabolic pathways and measure fluxes; numerous reviews provide accounts of NMR based metabolomics research [15, 16] and investigations of diseases including diabetes [17], cardio vascular disease [18], and prostate cancer [19, 20] and toxins [21], drug development [22], and metabolic pathways and flux analysis [23]. Importantly, the non-invasive nature of NMR provides unique opportunities to translate in vitro findings to clinical applications in vivo. The latest technological advancements in NMR such as strong magnetic fields, cryogenic probes and micro-coils, and automation have improved the detection limit, throughput and enabled detection of over hundred quantifiable metabolites from single NMR experiment in biological mixtures [24].
Mass spectrometry (MS) is intrinsically a highly sensitive method and a powerful tool for detection, quantitation and structure elucidation of metabolites. MS’s high sensitivity enables detection of several hundred to over a thousand small molecules from a single experiment, and thus MS applications in metabolomics increasingly dominate the field. Generally, MS methods employ prior separation techniques such as liquid chromatography (LC), gas chromatography (GC), or capillary electrophoresis (CE) to unravel the metabolite complexity, although direct sample infusion and related methods are also used. A majority of the metabolomics studies documented in the literature have used either LC or GC methods. Based on its versatility and recent advances in MS technologies, LC-MS is increasingly the most popular approach for metabolomics applications. The unique characteristic of this approach is that the liquid chromatography allows direct detection of metabolites from biological mixtures with no requirement for chemical modification such as derivatization. GC-MS methods provide a complimentary approach to detect a wide range of metabolites, although chemical derivitization is generally required. Due to the associated gaseous phase and nature of its ionization source, GC-MS achieves better metabolite separation and generally avoids ion suppression, a major challenge faced by LC-MS. Excellent protocols are available for the analysis of urine [25, 26] and serum [27] samples by LC-MS and GC-MS methods. Step by step protocols for animal and human tissue analysis using LC-MS are also reported [28]. More recently, detailed protocols for characterization of unknown peaks using tandem mass spectra and METLIN database of over 10,000 metabolites have been provided [29]. Using a more reliable and quantitative selected reaction monitoring approach, more than 250 targeted metabolites can now be analyzed in a single run of about 15 min with positive/negative ion switching [30]. As with NMR, applications of MS-based methods have spanned numerous areas as reviewed in a number of articles detailing investigations, for example, of breast cancer [31], colorectal cancer [32], prostate cancer [33], oesophago-gastric cancer [34]; cardiovascular [35, 36] and kidney [37] diseases, inborn errors of metabolism [38], toxicology [39], nutrition [40] and metabolic fluxes [41, 42].
GLOBAL AND TARGETED METABOLIC PROFILING
While it is still impossible to completely profile all metabolites in a complex biological sample, current analytical methods are quite capable of measuring hundreds to a few thousand metabolites, especially when several platforms are combined. Initially, metabolomics investigations primarily used global metabolic profiling methods involving the analysis of all detectable signals, including both known and unidentified metabolite peaks. Very often, metabolites are not identified prior to analysis. Instead, the complex data are subjected to multivariate statistical analysis, after a preprocessing step. For NMR, preprocessing includes apodization and zero-filling, Fourier transformation, phase and baseline correction, peak alignment, solvent peak removal and (optionally) data binning. For MS, spectral peak quantitation and alignment are typically followed by metabolite identification where possible. Typically only 1/3 of metabolite identities are assigned with any confidence. For LC-MS, preprocessing also includes the removal of isotope and adduct peaks resulting in simplified and better quantified spectra. Subsequently, metabolite features that distinguish sample classes are identified and then the structures of distinguishing metabolic features are established where possible. Global profiling approaches are primarily useful for discovery research, and provide an important starting point for further studies to identify distinguishing metabolic changes, or for generating biological hypotheses.
A second approach uses targeted metabolite profiling in which a set of known metabolites are quantitated. The identities of metabolites are initially established based on available databases and using standard compounds; the identified metabolite peaks are then quantified relatively, or sometimes absolutely based on the inclusion of internal or external reference compounds. The resulting data can then be used for pathway analysis to prove or disprove biological hypotheses, or as input variables for statistical analysis. Because of the reliable peak identification and measurement of metabolite intensities or even concentrations, targeted metabolomics provides greater insights into the dynamics and fluxes of metabolites and promises more robust statistical models for distinguishing sample classes with better classification accuracy. As NMR spectroscopy already provides highly reproducible results with a coefficient of variation (CV) of 1–2%, targeted metabolomics can be performed easily using NMR. Targeted metabolomics using tandem mass spectrometry (LC-MS/MS) enables fast MS data acquisition resulting in quantitative data with reasonably good reproducibility (5–30% CV) [43–46]. A variety of internal standardization methods using isotope labeled compounds (2H, 13C, 15N) as well as external standardization methods are also available to provide much better reproducibility. First developed to provide improved quantitation in the field of proteomics, the use of mixtures of metabolites containing light and heavy isotopes is a popular method for relative and absolute quantitation of metabolites using MS methods [47–50]. Targeted methods that use such internal standards compensate ion-suppression effects and provide reliable metabolite quantitation.
A limitation of the analysis of metabolites at steady state is that their concentrations may lack sufficiently specific information to obtain an unambiguous understanding of the biosynthesis of metabolites. This situation occurs because many metabolites are associated with multiple metabolic pathways. In addition, in cells and tissues many metabolic levels are controlled by regulation through homeostasis. Hence the contribution of each metabolite to a specific pathway is masked. Thus it has been increasingly realized that a metabolic pathway-focused, hypothesis testing approach is desirable to further understand pathogenesis and potentially to discover additional disease biomarkers. An approach that promises more specific information on metabolic pathways involves the incorporation of stable isotopes into the downstream metabolites using precursors enriched with stable isotopes such as 2H, 13C or 15N. This approach provides vital clues regarding disease mechanisms through the tracing of isotope enriched metabolite products through the various metabolic pathways and through the measurement of fluxes. Flux analysis allows a better understanding of the dynamics of metabolic pathways, enables quantification of intracellular metabolite levels and determination of the rates at which metabolites are produced or consumed. More specifically, metabolite profiling of cells supplied with 13C-labeled glucose facilitates the determination of the glucose consumption rate as well as the rates at which the downstream metabolic products of glycolysis are produced from the labeled glucose. For example, 13C-labeled lactate produced from the glycolysis of 13C-glucose in cancer development can be distinguished from the same lactate produced from other pathways based on the presence or absence of the embedded 13C isotope [51–52]. Advances in targeted metabolic profiling methods have improved the accuracy in metabolite quantitation and facilitated increased interest in understanding diseases based on altered metabolic pathways. Both NMR and MS methods are used in stable isotope based flux analysis using labeled precursors.
STATISTICAL ANALYSIS
In addition to the analytical methods, advanced statistical and typically multivariate methods play an essential role in metabolite profiling [53–55]. While univariate methods such as p-values or Welch’s t-test calculations are extremely useful in identifying individual biomarker candidates, it has been recognized for some time that individually almost all metabolite biomarkers are likely to be insufficient in terms of performance. Thus, the need to build predictive models based on multiple biomarkers that can improve performance necessitates the use of multivariate methods. Multivariate methods are also very useful for reducing the dimensionality of the NMR/MS data, and to extract the maximum information. Multivariate methods are generally capable of processing several thousand inputs or “variables” and their corresponding intensities; however, most practical applications typically involve a dozen or less biomarkers because of the increased effort needed to develop and validate each marker individually.
In general, multivariate statistical approaches are broadly classified into two categories: “unsupervised” and “supervised” methods. Most predictive models currently rely on supervised models in order to reduce the effects of confounding factors. Supervised methods require a training data set, in which the outcome (i.e., disease or healthy) is known, to build a (hopefully) predictive model. Initially, sample data sets are split into training and test sets, although some practitioners utilize an intermediate set of data that is used to improve the modeling. After training, the model’s accuracy is evaluated by classifying unknown samples in the test set of samples. Supervised methods are very useful for detecting subtle differences between similar samples, however, care must be taken to try to avoid confounding factors and especially overtraining. Typically, cross validation [53] is used to test the robustness of putative biomarker candidates during the training process as well as to identify the best model given the training set of sample data. Supervised techniques can be appropriate to force classification and thereby determine which metabolites distinguish between groups, i.e. for biomarker discovery, or to regress a pattern against a trend (such as correlating a temporal progression with metabolic changes). By far the two most popular methods for supervised pattern recognition include partial least squares discriminant analysis (PLS-DA) [56], which is often combined with orthogonal signal correction [57] and logistic regression. Other methods, including soft independent modeling of class analogies (SIMCA), genetic programming and neural networks are also used. In general, it is extremely important to validate the findings of any of these multivariate methods (including unsupervised methods) using extensive cross validation [58] and, in particular, a second set of samples (preferably blinded and from a second location, vida infra) which is sufficiently powered to yield a statistically significant result. Beyond the statistical aspects of validation, ultimately, biological validation, involving a disease hypothesis specifically related to the discovered biomarkers, may be required before acceptance by the medical and scientific communities can be anticipated.
TRANSLATIONAL PROCESS
As illustrated below, there are 5 main steps along the path of translating a diagnostic biomarker to the clinic (Fig. 2), and these steps are discussed separately in the following sections. The last step of commercialization may also include its own development process, and many of the other steps can have multiple iterations, complicating the picture somewhat. It should also be stated that this process is not unidirectional, but that in practice later steps can inform earlier processes and such feedback can be used to improve the overall process. For example, results from pre-validation may show that additional biomarker discovery is needed, or that development of a particular biomarker candidate is difficult suggesting that another model involving a different mix of biomarkers should be used in the pre-validation stage. The following sections describe the various steps in the chronological order using selected excerpts of studies from the exhaustive literature search.
Fig. (2).
The major steps involved in the translation of a biomarker candidate from the lab to the clinic. In actual practice, some steps may be combined, such as Discovery and Pre-Validation, or there may be multiple Validation steps. Typically, the Development step precedes the final validation step, which is then followed by the commercialization process that may take a number of forms and offered on one or more platforms.
BIOMARKER DISCOVERY
Many major diseases including, but not limited to inborn errors of metabolism, diabetes, cardiovascular disease, neurological diseases and cancer have been studied using MS- or NMR-based metabolomics methods. To date, significant progress has been made in the discovery of metabolite based biomarkers for these diseases [3, 24, 58–66]. A selected set of studies that illustrate this progress and breadth of the investigations are provided below.
Investigations of diseases of inborn errors of metabolism (IEM) are among the early studies focused on metabolite based biomarker discovery, and essentially predate the field of metabolomics [3, 24, 67]. An IEM is generally characterized by a single genetic defect, which deleteriously affects the function of a specific enzyme [68]. It often leads to dysfunction of metabolic pathways and causes accumulation of metabolites associated with the affected enzyme. Such accumulation is reflected in the elevated metabolite levels by several orders of magnitude in body-fluids, particularly in urine. Analytical techniques such as GC-MS and NMR have been widely used to identify and establish metabolite bio-markers for IEMs [60, 61, 69, 70]. Comprehensive screening of urine samples for IEM performed using electrospray tandem mass spectrometry with no need for separation represents a faster and less labor intensive approach for routine applications to a wide variety of IEM [71].
Ever since the early investigations of altered metabolite levels in serum, plasma and urine from diabetic patients [72], a vast number of studies have focused on discovering additional biomarkers for diabetes beyond glucose and insulin, resulting in the identification of numerous potential bio-markers for both Type 1 and Type 2 diabetes [3,24, 73]. The investigations have utilized NMR and MS, either individually or in combination, to identify biomarkers in blood plasma/serum, urine, and tissue from humans as well as animal models. For example, a multiplatform approach utilizing NMR, LC-MS and GC-MS measured over 400 unique metabolites in blood [74]. This study focused on Type 2 diabetes found a number of lipids, organic acids, carbohydrate molecules and amino acids, including branched chain amino acids that were shown to be distinguishing markers for diabetes. Separately, metabolites associated with lipid and amino acids were found to be linked with Type 1 diabetes [75] and a panel of five branched chain amino acids were shown to be predictive of Type 2 diabetes as early as 5 years before disease onset [76]. An exhaustive metabolomics review on diabetes describes a large number of studies focused on identifying various classes of biomarkers and their association with metabolic pathways linked to Type 1 and Type 2 diabetes [73]. Interestingly, studies indicate that many metabolite biomarkers identified in human patients compare well with the findings from animal models as well as with the studies that investigate altered metabolites during a glucose tolerance test, the gold standard test used to diagnose diabetes. For example, biomarkers in Type 2 diabetes that relate changes in metabolism of N-methylnicotinamide and N-methyl-2-pyridone-5-carboxamide were observed in studies of both human and animal models [77]. Such biomarkers can potentially be of unique value to follow the progression of Type 2 diabetes in clinical settings.
Numerous studies focused on the search for small molecule metabolites as biomarkers for underlying cardio vascular (CVD) and coronary heart diseases have found a number of metabolites to be of potential diagnostic value [3, 24, 59, 78, 79]. To cite a few examples, an MS based metabolomics study of more than 2000 patients undergoing cardiac catheterization finds five baseline metabolic factors to be associated with mortality due to CVD, independent of the standard predictors; they are medium-chain acylcarnitines, short-chain dicarboxylacylcarnitines, long-chain dicarboxylacylcarnitines, branched-chain amino acids and fatty acids [79]. Similarly, a targeted metabolomics study focused on identifying metabolite markers that are predictive of CVD has found 3 phosphocholine metabolites: choline, trimethylamine N-oxide (TMAO) and betaine to be associated with the disease. The study included validation (vida infra) using nearly 2000 patients has confirmed their association [59]. It was inferred that gut flora promote CVD based on the fact that gut bacteria metabolize dietary lipid phosphocholine to form TMAO, which promotes atherosclerosis. Previously, investigations involving the measurement of blood lipids using NMR have led to the identification of certain lipid particles and their prevalence that predict the risk of CVD and have opened avenues for clinical assessment and management of atherosclerotic cardiovascular disease [80, 81]. The methodology involves the multivariate deconvolution of the lipid signals into 15–18 lipoprotein classes and subclasses beyond the normal HDL and LDL designations.
Metabolomics studies focused on identifying distinguishing metabolite biomarkers for neurological disorders including Alzheimer’s disease (AD) and Parkinson’s disease (PD) are steadily increasing [66, 82–85]. Cerebrospinal fluid (CSF) is a rich source of biomarkers for neurological disorders, and therefore there has been increased interest to identify biomarkers for many neurological diseases using CSF [83]. Investigation using blood represents a minimally invasive approach and hence promises a number of transla-tional opportunities. Many studies have used mass spectrometry combined with multivariate statistical methods to identify AD biomarkers in blood serum or plasma. For example, using 20 AD and 20 control plasma samples, ten metabolites including tryptophan, sphingosine and lysophosphatidylcholine were shown to be downregulated in AD [86]. Separately, using 26 AD and 26 control samples, a metabolomics study targeted ceramide and sphingomyelin metabolites in blood and reported downregulation of 8 sphingolipids and upregulation of 2 ceramides [87]. A study that focused on detecting the onset of AD reported upregulation of 2,4-dihydroxybutanoic acid as a major predictor to the progression to AD, using 47 AD, 46 controls and 143 mild cognitive impairment (MCI) serum samples [88]. Targeted analysis of steroid-related metabolites, using 10 AD and 10 control plasma samples, detected decreased desmosterol in AD and the result was consistent when tested using a separate set of 41 AD, 42 controls and 26 MCI samples [89]. Similarly, many metabolomics studies of PD report identification of distinguishing biomarkers. For example, from the metabolic profiling of blood samples from 66 PD patients and 22 controls, decreased uric acid and increased glutathione in PD were reported [90]. A separate study found pyruvate as the key metabolite to distinguish between PD and controls based on 1H NMR analysis of 43 PD and 37 controls [91]. In the same way, many distinguishing metabolites for other neurological disorders such as Amyotrophic lateral sclerosis (ALS) [92, 93], multiple sclerosis [94], Huntington’s disease [95] and schizophrenia [96–98] have been reported.
Major efforts in the metabolomics field are also focused on biomarker identification for improved cancer diagnostics. A vast number of studies have focused on distinguishing cancer patients from matched controls, based on measurements of biofluids and excised or biopsied tumors. These studies have produced a sizable and new body of knowledge on altered metabolite levels and pathways in cancer. Preliminary investigations of almost all major types of cancers have resulted in the identification of numerous potential metabolite biomarkers, many of which are in common. For example, numerous studies have shown elevated choline metabolites, lower glycerophosphocholine and low glucose to have diagnostic value for breast cancer based on investigations of breast tissue metabolites [99–103]. The factors including estrogen receptor, progesterone receptor, and lymph node status have been shown to have links with the altered metabolites in breast cancer [104]. Based on the combined analysis using NMR and MS methods, a panel of biomarkers was identified for the early detection of breast cancer recurrence [105]. Acylcarnitines and metabolites associated with tryptophan metabolism were found to be linked with kidney cancer [106, 107]. It was shown, based on metabolic profiling of seminal fluid using 1H NMR spectroscopy, that low levels of citrate concentration outperforms the conventional prostate specific antigen (PSA) for detecting prostate cancer [108, 109]. Separately, it is shown that the sum of concentrations of choline and creatine, when ratioed to that of citrate, correlates well to prostate cancer aggressiveness [110]. Sarcosine, or N-methyl glycine, has also been shown to be a strong marker of prostate tumor aggressiveness in biopsied tissues [111]. For ovarian carcinoma, a number of potential biomarkers including those involved in purine, pyrimidine and glycerolipid metabolism have been identified [112]. Lower levels of hippurate and trigonelline, and elevated 3-hydroxyisovalerate, α-hydroxyisobutyrate and N-acetylglutamine have been shown to be distinguishing urine markers for lung cancer [113]. Similarly, markers for esophageal cancer have been identified using LC-MS and NMR [64, 65, 114]. Numerous studies in colon cancer have found promising potential biomarkers, including lactate, pyruvate, malic acid and long-chain polyunsaturated fatty acid (GTA-446) identified by LC-MS or GC-MS methods [115–119]; most of these markers are relatively well known metabolites, but some quite promising markers also involve unknown metabolites [115]. The discovery of novel metabolite cancer biomarkers is anticipated to continue at a rapid pace.
PRE-VALIDATION OF BIOMARKERS
A large body of literature documents the identification of putative biomarkers and biomarker panels for numerous diseases. Biomarker identification represents only the starting point in the translation process from lab discovery to clinical diagnostic. The next major step typically involves prevalidation (a.k.a. initial validation) of the putative biomarkers. While this step is sometimes included as part of the discovery, the main function of the pre-validation step is to reduce the number of false positive biomarker candidates, and to assess the overall accuracy of any initial multivariate models. There are a variety of approaches that can be used for this purpose, from simply testing individual markers against a new set of samples using the same analytical platform, to the initial testing of a multivariate statistical model using a cross validation procedure.
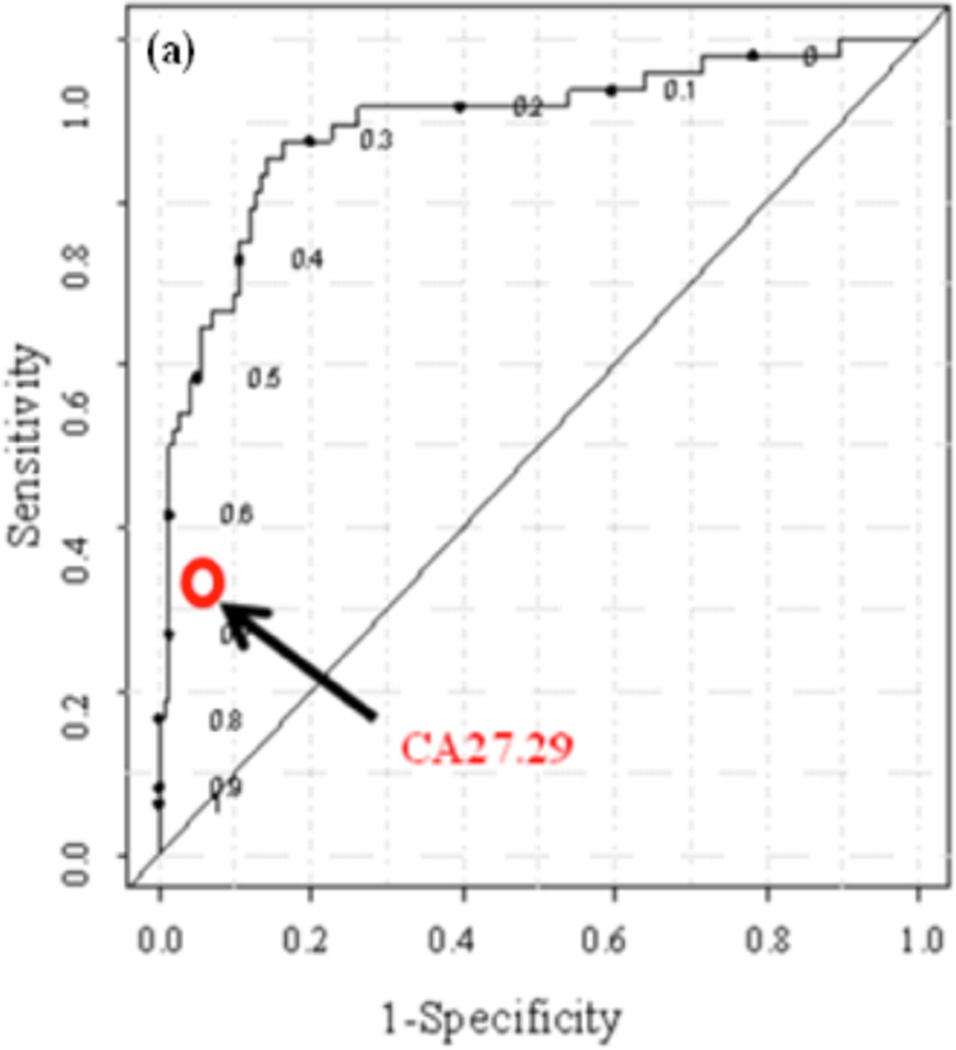
For multivariate models, some practitioners prefer to reduce the complexity of the model by selecting highly ranked metabolites based on regression analysis, p-values, variable influence on projection (VIP) scores, or even the output of multivariate loading plots prior to developing the multivariate model. In any event, multivariate models are first cross-validated using the same data set using a leave-n-out procedure, with n typically chosen between 1 and 20% of the number of samples. The robustness of the model is then evaluated using the sensitivity (true positive rate) and specificity (true negative rate). The sensitivity and specificity, however, depend on the chosen boundary or the cut-off values; by changing the boundary, higher sensitivity can be achieved at the cost of specificity and vice versa. Therefore, the receiver operative characteristic (ROC) curve represents a more general form of representing the performance of a prediction model. The ROC curve enables visualization of specificity and sensitivity at any cut-off value; further, area under the ROC (AUROC) provides a good measure of the overall model performance. An AUROC of 1 represents the ideal performance with a sensitivity and specificity of 100%. An example of an ROC curve is shown in Fig. (3) that depicts the performance of a PLS-DA model using 11 metabolite biomarkers derived from the combination of NMR and MS methods for detecting recurrence of breast cancer [105].
Fig. (3).
(a) ROC curve generated from the PLS-DA model based on eleven serum metabolite markers for detection of breast cancer recurrence; the model was cross validated using a leave-one-patient-out procedure. The red circle compares the sensitivity and specificity for the conventional breast cancer marker used for recurrence test. The area under the ROC curve is 0.88. The sensitivity and specificity at two selected cutoff values are shown in Table 1 [Reproduced with permission from ref. 105].
Table 1 shows sensitivity and specificity for this model selected at two different cut-off values and compares performance of the metabolite biomarkers with conventional breast cancer recurrence marker CA27.29, which is also shown in the figure at its clinically used threshold value. A multivariate model based on the metabolite biomarkers performed much better than the conventional marker, especially with respect to sensitivity.
Table 1.
Comparison of the Diagnostic Performance of the Breast Cancer Recurrence Metabolite Profile, at cut-off Values of 48 and 54, and CA 27.29
| Sensitivity | Specificity | |
|---|---|---|
| Metabolite biomarkers | 86% | 84% |
| 68% | 94% | |
| CA27.29 | 35% | 96% |
In another example, a recent metabolomics study of pancreatic cancer combining GC-MS and statistical analyses of blood serum metabolites reported a multivariate model that possessed high sensitivity (86.0%) and specificity (88.1%) towards distinguishing pancreatic cancer and healthy controls. Upon validation, the prediction model fared reasonably well when tested using a similar number of independent samples (sensitivity 71.4%; specificity 78.1%) and the same GC-MS instrument. A loss of performance in sensitivity, specificity or both is to be anticipated because of the tendency for multivariate models to be over-trained on even moderately large sample sets. Nevertheless, the model displayed higher sensitivity for detecting patients with resectable pancreatic cancer (sensitivity 77.8%) and lower false positive rate for chronic pancreatitis (17.4%) than conventional tumor markers [120].
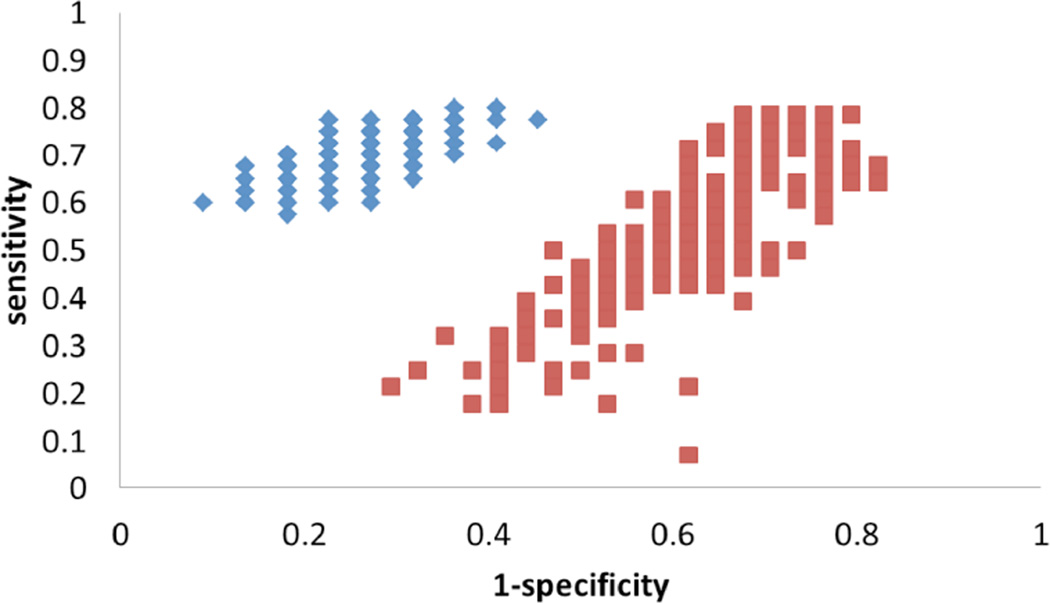
Statistical model robustness can be tested using a technique called Monte Carlo Cross Validation (MCCV) [121]. Typically, several hundred calculations are performed in which the whole dataset is randomly divided into a training set (for example 60% of the whole data set) and a testing set (the remaining 40%). The multivariate statistical model (PLS-DA, for example) is then built using the training set with internal cross-validation. The internal cross-validation prediction on the training set and the external prediction of the testing set are then typically combined to form the prediction result for each MCCV run. The sensitivity and specificity are calculated and compared with the results of a permutation analysis, as shown in (Fig. 4) below. In the permutation, the sample classifications are randomly permuted and several hundred MCCV iterations are typically performed.
Fig. (4).
Results of the MCCV (200 iterations) shown in ROC space for PLS-DA models based on 3 metabolites used to discriminate hepatocellular carcinoma and hepatitis C patients. Each blue diamond represents an iteration of the true model; each red square represents an iteration of the permutation model [Reproduced with permission from ref. 63].
ANALYTICAL DEVELOPMENT
The analytical development stage is normally used to prepare a biomarker or panel of biomarkers for the final validation stage, in which a large number of samples (ideally originating from multiple sites) are carefully tested according to set protocols. A statistical model, if any, is also fixed with respect to its coefficients and parameters ahead of any validation sample measurements. However, in some complex marker studies there may be multiple steps of validation, and therefore the development stage may occur in between those validations. Essentially, the development stage is where the analytical performance of the biomarkers is determined, including the linearity, sensitivity, limit of detection, recovery, robustness, reproducibility, and any matrix effects or interferences are identified and minimized. Often internal or external standards and controls are chosen at this stage to allow careful quantitation. For the development of multiple metabolite biomarkers that are used as a panel, this will involve a significant amount of work, as each biomarker candidate will need to be developed individually and then combined into a panel. The inclusion of good quality control samples and calibrators is also part of the diagnostic development stage as well as the discovery stage [122]. A number of resources exist that describe this development process in general, which fall under the heading of Quality Control [123–126].
VALIDATION OF BIOMARKERS
Most biomarker discovery efforts in the metabolomics field (and elsewhere) typically rely on relatively small numbers of case and control samples due to the cost and effort required to acquire such samples. For the discovery and pre-validation stages discussed above, the biomarker identification process typically involves dividing patient data into training and internal validation sets, developing prediction models using the training sample set, performing cross validation to evaluate the statistical model, and then validating the model using a validation sample set. While such practice is accepted in biomarker discovery efforts, and one might argue necessary at this early stage of biomarker development and translation, it comes with many limitations. For example, the number of samples used in both training and validation is often too small and hence potentially confounding factors including diet, age, gender and subtle differences in pathology can strongly affect biomarker performance and especially selection into biomarker panels. This is particularly true for only moderately strong biomarker candidates, where the concentration differences between disease and control are often small. In addition, as often practiced, randomly dividing the same sample set into two groups, training and validation sets, still may not sufficiently represent independent sample sets, since both groups are often collected, stored and processed using the same procedures and analyzed under identical conditions using the same analytical platform(s). While this approach can reduce confounding factors that arise in the analysis stage and are of a technical nature, a number of other existing biases will likely be carried over to both sets of samples. Therefore, a thorough validation of identified and pre-validated biomarkers is very important, and should involve large cohorts of samples from diverse patient groups. Such validation studies should consider the fact that metabolite profiles are very sensitive to conditions including patient confounding factors such as age, gender, ethnicity, diet, etc., as well as sample processing, chromatographic separation and analytical instrumental settings. Thus validation design should take into account both biological and technical variance, and provide the important information on biomarker sensitivity, specificity and also robustness to the technical aspects of measuring the biomarkers.
As there is no single, universally accepted procedure for metabolic profiling, different laboratories often use their own optimized protocols and hence it can be challenging to obtain identical metabolic profiles for the same samples by independent laboratories. Thus well designed cross-over studies are important to validate marker performance across different laboratories. Inter-laboratory differences in measuring metabolite biomarkers contribute to the list of bottlenecks in biomarker validation and translation. And obviously, the proper use of appropriate quality control samples is also very important. A small difference in protocols between discovery and validation steps can affect the outcome and inferences. For example, a GC-MS based metabolomics study reported sarcosine as a potential marker for aggressive prostate cancer [111]. Sarcosine was an especially strong marker in prostate tissue, but less so in urine. Considering its potential implications for clinical use, a validation study was undertaken by an independent group using the same types of bio-specimens and analytical technique, but reported negative results [127]. However, the outcome of such a validation study is somewhat inconclusive as there were fundamental differences in the two studies, as noted by an accompanying editorial [128]. While both studies used GC-MS for analysis, the discovery study used urinary sediment and analyzed the sarcosine to alanine ratio, while the validation study used urine supernatant and determined sarcosine to creatinine ratio for comparison. In sum, validation studies for translational applications should be properly designed and ideally take into account all sources of variation, whether biological or technical, and have standardized protocols for sample acquisition, chemical analysis as well as data analysis. For further confidence, samples should be processed and analyzed using well developed standard protocols to limit the potential bias introduced by different lab personnel. Many researchers believe the samples should be blinded to the analyst, especially for the validation studies. And in appropriate cases, the use of different analytical platforms would provide additional information on biomarker robustness.
Additional, well designed validation studies may be used to further assess the clinical performance and utility of a certain biomarker assay. Finally the use of multisite clinical trials is an increasingly important step in the validation process, since some confounding factors (diet, environment, race, socioeconomic factors) may have a geographic origin. Additionally, SOPs and protocols might be altered by clinical or laboratory personnel in different centers. Despite these challenges efforts to surmount them are increasing. For example, an international team of scientists performed a multisite study to discover a panel of metabolites in blood for the prediction of preeclampsia in early pregnancy. They were able to validate the 14 metabolite model using an independent patient cohort from a participating center in a different country. The metabolite panel was consistently discriminatory for predicting preeclampsia with an ROC area of 0.94 and 0.92 for discovery and validation, respectively [129]. As a second example, major efforts have been focused on discovering and validating reduced blood levels of gastrointestinal tract acids to be associated with the risk of developing colorectal cancer [115, 116]. A large validation trial involving almost 5,000 patients was performed and showed that gastrointestinal tract acid-446, a 28 carbon fatty acid metabolite with m/z 446, was low in patients with high risk for colon cancer [116]. The relative risk of having colorectal cancer for a patient under 50 with low GTA-446 was 10. It is anticipated that additional and 446 was 10. It is anticipated that additional and similarly large scale trials will be performed for a number of important diseases, despite the significant resources required.
COMMERCIALIZATION
Metabolomics investigations, specifically, over the last few years have led to the establishment of putative bio-marker panels for many diseases. Efforts to commercialize such findings for diagnostics, however have generally met with many challenges, which is much the case for many types of biomarker candidates, whether they are genes, transcripts, proteins or other markers. A major challenge is the cost involved to validate diagnostic metabolite profiles using larger patient cohorts. In some cases, and depending on the application, the technology or biomarkers themselves may need to be approved by the FDA. In spite of the challenges, efforts have been made to commercialize metabolic technologies for disease diagnostic applications, and a number of companies are currently focusing on developing metabolite biomarker based diagnostic tests (Table 2). Within this group of companies, specific metabolic profiles for diseases including colon cancer, breast cancer and prostate cancer are being tested, further validated and readied for translation of the technology from the bench to the beside. Metabolon, one of the longest established metabolomics related companies, is focused on providing diagnostic services for several diseases including prostate cancer, and provides a wide range of global and targeted metabolic profiling services to customers from academia and pharma/biotech. In Europe, Metanomics Health GmbH is a large metabolomics service provider which offers metabolite profiling services to healthcare customers in industry and academia. Liposcience, established in 1997, has commercialized its NMR technology that provides a personalized diagnostic test to assess the risk of heart disease through the measurement of lipoprotein particles in the blood. Two Canadian companies are focused on detecting colon cancer. Metabolomic Technologies Inc. (MTI) is developing diagnostic tests to detect adenomatous polyps and colorectal cancer (CRC) using spot urine samples, while Phenomenome has developed a blood test for colon cancer screening. Matrix-Bio is focused on developing blood-based diagnostic test to detect breast cancer recurrence.
Table 2.
List of Commercialization Ventures of Metabolomics Technologies
| Metabolomics Company | Technology | Country |
|---|---|---|
| Metabometrix Ltd. http://www.metabometrix.com/ |
Broad metabolomics services company | UK |
| Metabolomic Discoveries GmbH http://www.metabolomicdiscoveries.com/ |
Broad metabolomics services company | Germany |
| Metabolon, Inc. http://www.metabolon.com/ |
Diagnostic and broad metabolomics services company | USA |
| Matrix-Bio, Inc. http://www.matrix-bio.com/ |
Diagnostic company for screening breast cancer recurrence using blood | USA |
| Metabolomic Technologies Inc http://www.metabolomictechnologies.ca/ |
Diagnostic company for screening colorectal cancer using urine | Canada |
| Lipo Science, Inc. http://www.liposcience.com/ |
Diagnostic company that tests cardiovascular disease risk based on measurement of lipoprotein particles in blood |
USA |
| ClinMet, Inc. http://www.clinmet.com/ |
Metabolomics and clinical solutions to drug discovery and development | USA |
| Metanomics Health GmbH http://www.metanomics-health.de/ |
Metabolite profiling services | Germany |
| Stemina Biomarker Discovery, Inc. http://www.stemina.com/ |
Diagnostic, cardiotoxicity screening and metabolomics services company | USA |
| SiDMAP, Inc. http://www.sidmap.com/ |
Stable isotope tracer based metabolite profiling | USA |
| Phenomenome Discoveries Inc. http://www.phenomenome.com/ |
Diagnostic company for screening colorectal cancer using blood | Canada |
Other companies have focused on providing broad-based metabolic technology for academic and industrial research. For example, the start-up company Metabometrix possesses advanced analytical technology for data processing, and statistics tools for generating data and providing interpretation. ClinMet offers metabolomics services to pharmaceutical companies to improve efficacy of drug discovery and development focusing diabetes and kidney diseases. The bio-marker discovery company Stemina offers a broad range of metabolomics services including diagnostics and cardiotoxicity screening, with a specialization on the use of human derived stem cells. The company SidDMAP offers stable isotope tracer based dynamic metabolic profiling services.
It is foreseen that more commercialization efforts for specific metabolic technologies will emerge as the number of validation studies using large patient cohorts increases. In addition to the analytical and clinical validation studies, biological validation is becoming increasingly important. This is in part driven by the large number of false positive biomarker candidates that arise. While it is reassuring when the altered metabolism can be directly related to the disease biology, it is not always possible at present to understand the biological origin of the metabolic changes, given the somewhat dormant state of metabolism studies over the past 40 years. In addition, some biomarkers that are well used are not too well understood. As a case in point, one could argue that urinary glucose was shown as an accurate indicator of diabetes well before the biological understanding between glucose levels and diabetes. Hence, in our opinion, efforts to commercialize metabolite biomarker panels and to find biological connection between the disease state and the derived biomarker panel should go in parallel. Despite the challenges (Table 3), there continues to be great excitement for the potential for metabolite profiling to provide improved diagnostic biomarkers and targets for drug development.
Table 3.
Some of the Challenges Faced in Biomarker Discovery and Translation in Metabolomics
| Major Steps | Major Challenge |
|---|---|
| Biomarker Discovery |
|
| Pre-validation |
|
| Development | High costs of or inaccessibility to isotope labeled analogues for accurate metabolite quantitation using mass spectrometry. |
| Validation | Limited access to large and independent sample cohorts. |
| Commercialization |
|
Acknowledgments
We acknowledge financial support from NIH (National Institute of General Medical Sciences NIH 2R01GM085291).
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
Daniel Raftery reports holding equity and an executive role in Matrix-Bio, Inc.
REFERENCES
- 1.Djukovic D, Nagana Gowda GA, Raftery D. Mass Spectrometry and NMR Spectroscopy Based Quantitative Metabolomics. In: Isssaq HJ, Veenstra TD, editors. Proteomic and Metabolomic Approaches to Biomarker Discovery. Amsterdam: Academic Press; 2013. pp. 279–298. Ch 18. [Google Scholar]
- 2.Gu H, Nagana Gowda GA, Raftery D. Metabolic profiling: are we en route to better diagnostic tests for cancer? Future Oncology. 2012;8:1207–1210. doi: 10.2217/fon.12.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature. 2012;491:384–392. doi: 10.1038/nature11708. [DOI] [PubMed] [Google Scholar]
- 4.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. (USA) 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature Genetics. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. HMDB: a knowledgebase for the human metabolome. Nucleic. Acids. Res. 2009:D603–D610. doi: 10.1093/nar/gkn810. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markley JL, Anderson ME, Cui Q, Eghbalnia HR, Lewis IA, Hegeman AD, Li J, Schulte CF, Sussman MR, Westler WM, Ulrich EL, Zolnai Z. New bioinformatics resources for metabolomics. Pac. Symp. Biocomput. 2007:157–168. [PubMed] [Google Scholar]
- 9.Tautenhahn R, Cho K, Uritboonthai W, Zhu ZJ, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 11.Kind T, Wohlgemuth G, Lee DY, Lu L, Palazoglu M, Shahbaz S, Fiehn O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckonert O, Keun HC, Ebbels TMD, Bundy J, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007;2(11):2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 14.Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TMD, Holmes E, Lindon JC, Nicholson JK. High-resolution magicangle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat. Protoc. 2010;5(6):1019–1032. doi: 10.1038/nprot.2010.45. [DOI] [PubMed] [Google Scholar]
- 15.Gebregiworgis T, Powers R. Application of NMR metabolomics to search for human disease biomarkers. Comb. Chem. High Throughput Screen. 2012;15(8):595–610. doi: 10.2174/138620712802650522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barding GA, Jr, Salditos R, Larive CK. Quantitative NMR for bioanalysis and metabolomics. Anal. Bioanal. Chem. 2012;404(4):1165–1179. doi: 10.1007/s00216-012-6188-z. [DOI] [PubMed] [Google Scholar]
- 17.Maher AD, Lindon JC, Nicholson JK. (1)H NMR-based metabonomics for investigating diabetes. Fut. Med. Chem. 2009;1:737–747. doi: 10.4155/fmc.09.54. [DOI] [PubMed] [Google Scholar]
- 18.Griffin JL, Atherton H, Shockcor J, Atzori L. Metabolomics as a tool for cardiac research. Nat. Rev. Cardiol. 2011;8:630–643. doi: 10.1038/nrcardio.2011.138. [DOI] [PubMed] [Google Scholar]
- 19.DeFeo EM, Wu CL, McDougal WS, Cheng LL. A decade in prostate cancer: from NMR to metabolomics. Nat. Rev. Urol. 2011;8:301–311. doi: 10.1038/nrurol.2011.53. [DOI] [PubMed] [Google Scholar]
- 20.Kumar V, Dwivedi DK, Jagannathan NR. High-resolution NMR spectroscopy of human body fluids and tissues in relation to prostate cancer. NMR Biomed. 2013 Jul 4; doi: 10.1002/nbm.2979. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Lindon JC, Keun HC, Ebbels TMD, Pearce JMT, Holmes E, Nicholson JK. The Consortium for Metabonomic Toxicology (COMET): aims, activities and achievements. Pharmacogenomics. 2005;6:691–699. doi: 10.2217/14622416.6.7.691. [DOI] [PubMed] [Google Scholar]
- 22.Lindon JC, Holmes E, Nicholson JK. Nicholson, Metabonomics in pharmaceutical R & D. FEBS J. 2007;274:1140–1151. doi: 10.1111/j.1742-4658.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- 23.Fan TW, Lane AN. NMR-based stable isotope resolved metabolomics in systems biochemistry. J. Biomol. NMR. 2011;49(3–4):267–280. doi: 10.1007/s10858-011-9484-6. Erratum in: J Biomol NMR. 2011, 49(3–4), 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye T, Zhang S, Nagana Gowda GA, Raftery D. Nuclear magnetic resonance and statistical analysis. In: Meyers RA, editor. Encyclopedia of Analytical Chemistry. Hoboken: John Wiley; 2010. [Google Scholar]
- 25.Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockcor J, Holmes E, Nicholson JK. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010;5(6):1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- 26.Chan EC, Pasikanti KK, Nicholson JK. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat. Protoc. 2011;6(10):1483–1499. doi: 10.1038/nprot.2011.375. [DOI] [PubMed] [Google Scholar]
- 27.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011;6(7):1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 28.Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, Plumb RS, Shockcor J, Loftus N, Holmes E, Nicholson JK. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013;8(1):17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- 29.Zhu ZJ, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, Siuzdak G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013;8(3):451–460. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012;7(5):872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connell TM. Recent advances in metabolomics in oncology. Bioanalysis. 2012;4:431–451. doi: 10.4155/bio.11.326. [DOI] [PubMed] [Google Scholar]
- 32.Williams MD, Reeves R, Resar LS, Hill HH., Jr Metabolomics of colorectal cancer: past and current analytical platforms. Anal. Bioanal. Chem. 2013;405(15):5013–5030. doi: 10.1007/s00216-013-6777-5. [DOI] [PubMed] [Google Scholar]
- 33.Trock BJ. Application of metabolomics to prostate cancer. Urol. Oncol. 2011;29(5):572–581. doi: 10.1016/j.urolonc.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbassi-Ghadi N, Kumar S, Huang J, Goldin R, Takats Z, Hanna GB. Metabolomic profiling of oesophago-gastric cancer: A systematic review. Eur. J. Cancer. 2013 doi: 10.1016/j.ejca.2013.07.004. S0959-8049(13)00548-0 (in print) [DOI] [PubMed] [Google Scholar]
- 35.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126(9):1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee EP, Gerszten RE. Metabolomics and cardiovascular biomarker discovery. Clin. Chem. 2012;58(1):139–147. doi: 10.1373/clinchem.2011.169573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss RH, Kim K. Metabolomics in the study of kidney diseases. Nat. Rev. Nephrol. 2011;8(1):22–33. doi: 10.1038/nrneph.2011.152. [DOI] [PubMed] [Google Scholar]
- 38.Mamas M, Dunn WB, Neyses L, Goodacre R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch. Toxicol. 2011;85(1):5–17. doi: 10.1007/s00204-010-0609-6. [DOI] [PubMed] [Google Scholar]
- 39.Robertson DG, Watkins PB, Reily MD. Metabolomics in toxicology: preclinical and clinical applications. Toxicol. Sci. 2011;(Suppl 1):S146–S170. doi: 10.1093/toxsci/kfq358. [DOI] [PubMed] [Google Scholar]
- 40.Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal BS, van Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5:435–458. doi: 10.1007/s11306-009-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reaves ML, Rabinowitz JD. Metabolomics in systems microbiology. Curr. Opin. Biotechnol. 2011;22(1):17–25. doi: 10.1016/j.copbio.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zamboni N, Fendt SM, Rühl M, Sauer U. (13)C-based metabolic flux analysis. Nat. Protoc. 2009;4(6):878–892. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- 43.Bajad SU, Lu WY, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatographytandem mass spectrometry. J. Chromatogr. A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 44.Melamud E, Vastag L, Rabinowitz JD. Metabolomic Analysis and Visualization Engine for LC-MS Data. Anal. Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei R, Li GD, Seymour AB. High-Throughput and Multiplexed LC/MS/MRM Method for Targeted Metabolomics. Anal. Chem. 2010;82:5527–5533. doi: 10.1021/ac100331b. [DOI] [PubMed] [Google Scholar]
- 46.Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding Coverage of the Metabolome for Global Metabolite Profiling. Anal. Chem. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo K, Bamforth F, Li L. Qualitative metabolome analysis of human cerebrospinal fluid by 13C-/12C-isotope dansylation labeling combined with liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass. Spectrom. 2011;22:339–347. doi: 10.1007/s13361-010-0033-4. [DOI] [PubMed] [Google Scholar]
- 48.Huang X, Regnier FE. Differential metabolomics using stable isotope labeling and two-dimensional gas chromatography with time-of-flight mass spectrometry. Anal. Chem. 2008;80:107–114. doi: 10.1021/ac071263f. [DOI] [PubMed] [Google Scholar]
- 49.Yang WC, Adamec J, Regnier FE. Enhancement of the LC/MS analysis of fatty acids through derivatization and stable isotope coding. Anal. Chem. 2007;79:5150–5157. doi: 10.1021/ac070311t. [DOI] [PubMed] [Google Scholar]
- 50.Yang WC, Regnier FE, Sliva D, Adamec J. Stable isotope-coded quaternization for comparative quantification of estrogen metabolites by high-performance liquid chromatography-electrospray ionization mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008;870:233–240. doi: 10.1016/j.jchromb.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lloyd SG, Zeng H, Wang P, Chatham JC. Lactate isotopomer analysis by 1H NMR spectroscopy: consideration of long-range nuclear spin-spin interactions. Magn. Reson. Med. 2004;51:1279–1282. doi: 10.1002/mrm.20075. [DOI] [PubMed] [Google Scholar]
- 52.Lane AN, Fan TW. Quantification and identification of isotopomer distributions of metabolites in crude cell extracts using 1H TOCSY. Metabolomics. 2007;3:79–86. [Google Scholar]
- 53.Johnson RA, Wichern DW. Applied Multivariate Statistical Analysis. New Jersey: Prentice Hall, Upper Saddle River; 1999. [Google Scholar]
- 54.Lindon JC, Nicholson JK. Spectroscopic and Statistical Techniques for Information Recovery in Metabonomics and Metabolomics. Ann. Rev.Anal. Chem. 2008;1:45–69. doi: 10.1146/annurev.anchem.1.031207.113026. [DOI] [PubMed] [Google Scholar]
- 55.Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J. Proteome Res. 2007;6(2):469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 56.Barker M, Rayens W. Partial least squares for discrimination. J. Chemom. 2003;17:166–173. [Google Scholar]
- 57.Beckwith-Hall BM, Brindle JT, Barton RH, Coen M, Holmes E, Nicholson JK, Antti H. Application of orthogonal signal correction to minimise the effects of physical and biological variation in high resolution 1H NMR spectra of biofluids. Analyst. 2002;127:1283–1288. doi: 10.1039/b205128c. [DOI] [PubMed] [Google Scholar]
- 58.Xia J, Broadhurst DI, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9(2):280–299. doi: 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engelke UFH, Oostendorp M, Wevers RA. NMR spectroscopy of body fluids as a metabolomics approach to inborn errors of metabolism. In: Lindon JC, Nicholson JK, Holmes E, editors. The handbook of metabonomics and metabolomics. Amsterdam: Elsevier; 2007. pp. 375–412. [Google Scholar]
- 61.Moolenaar SH, Engelke UFH, Wevers RA. Proton nuclear magnetic resonance spectroscopy of body fluids in the field of inborn errors of metabolism. Ann. Clin. Biochem. 2003;40:16–24. doi: 10.1258/000456303321016132. [DOI] [PubMed] [Google Scholar]
- 62.Wei S, Liu L, Zhang J, Bowers J, Nagana Gowda GA, Seeger H, Fehm T, Neubauer HJ, Vogel U, Clare SE, Raftery D. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol Oncol. 2013;7(3):297–307. doi: 10.1016/j.molonc.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei S, Suryani Y, Nagana Gowda GA, Skill N, Maluccio M, Raftery D. Differentiating Hepatocellular Carcinoma from Hepatitis C Using Metabolite Profiling. Metabolites. 2012;2:701–716. doi: 10.3390/metabo2040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Bowers J, Liu L, Wei S, Nagana Gowda GA, Hammoud Z, Raftery D. Esophageal Cancer Metabolite Biomarkers Detected by LC-MS and NMR Methods. PlosOne. 2012;7:e30181. doi: 10.1371/journal.pone.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Liu L, Wei S, Nagana Gowda GA, Hammoud Z, Kessler KA, Raftery D. Metabolomics Study of Esophageal Adenocarcinoma. J. Thoracic Cardiovas. Surg. 2011;141:469–475. doi: 10.1016/j.jtcvs.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 66.Kaddurah-Daouk R, Krishnan KR. Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacolog. 2009;34(1):173–186. doi: 10.1038/npp.2008.174. [DOI] [PubMed] [Google Scholar]
- 67.Arn PH. Newborn screening: current status. Health AV (Millwood) 2007;26(2):559–566. doi: 10.1377/hlthaff.26.2.559. [DOI] [PubMed] [Google Scholar]
- 68.Jones PM, Bennett MJ. The changing face of newborn screening: diagnosis of inborn errors of metabolism by tandem mass spectrometry. Clin. Chim. Acta. 2002;324:121–128. doi: 10.1016/s0009-8981(02)00238-3. [DOI] [PubMed] [Google Scholar]
- 69.Chace DH. Mass spectrometry in the clinical laboratory. Chem. Rev. 2001;101(2):445–477. doi: 10.1021/cr990077+. [DOI] [PubMed] [Google Scholar]
- 70.Wilcken B, Wiley V, Hammond J, Carpenter K. Screening newborns for inborn errors of metabolism by tandem mass spectrometry. New Eng. J. Med. 2003;348(23):2304–2312. doi: 10.1056/NEJMoa025225. [DOI] [PubMed] [Google Scholar]
- 71.Pitt JJ, Eggington M, Kahler SG. Comprehensive screening of urine samples for inborn errors of metabolism by electrospray tandem mass spectrometry. Clin Chem. 2002;48(11):1970–1980. [PubMed] [Google Scholar]
- 72.Nicholson JK, O'Flynn MP, Sadler PJ, Macleod AF, Juul SM, Sönksen PH. Proton-nuclear-magnetic-resonance studies of serum, plasma and urine from fasting normal and diabetic subjects. Biochem. J. 1984;217:365–375. doi: 10.1042/bj2170365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Friedrich N. Metabolomics in diabetes research. J. Endocrinol. 2012;215:29–42. doi: 10.1530/JOE-12-0120. [DOI] [PubMed] [Google Scholar]
- 74.Suhre K, Meisinger C, Döring A, Altmaier E, Belcredi P, Gieger C, Chang D, Milburn MV, Gall WE, Weinberger KM, Mewes HW, Hrabé de Angelis M, Wichmann HE, Kronenberg F, Adamski J, Illig T. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5(11):e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oresic M, Simell S, Sysi-Aho M, Näntö-Salonen K, Seppänen-Laakso T, Parikka V, Katajamaa M, Hekkala A, Mattila I, Keskinen P, Yetukuri L, Reinikainen A, Lähde J, Suortti T, Hakalax J, Simell T, Hyöty H, Veijola R, Ilonen J, Lahesmaa R, Knip M, Simell O. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J. Exp. Med. 2008;205:2975–2984. doi: 10.1084/jem.20081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nature Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salek RM, Maguire ML, Bentley E, Rubtsov DV, Hough T, Cheeseman M, Nunez D, Sweatman BC, Haselden JN, Cox RD, Connor SC, Griffin JL. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol Genomics. 2007;29(2):99–108. doi: 10.1152/physiolgenomics.00194.2006. [DOI] [PubMed] [Google Scholar]
- 78.Griffin JL, Atherton H, Shockcor JP, Atzori L. Metabolomics as a tool for cardiac research. Nature Rev. Cardiol. 2011;8:630–643. doi: 10.1038/nrcardio.2011.138. [DOI] [PubMed] [Google Scholar]
- 79.Shah SH, Sun JL, Stevens RD, Bain JR, Muehlbauer MJ, Pieper KS, Haynes C, Hauser ER, Kraus WE, Granger CB, Newgard CB, Califf RM, Newby LK. Baseline metabolomic profiles predict cardiovascular events in patients at risk for coronary artery disease. Am. Heart J. 2012;163:844–850. doi: 10.1016/j.ahj.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 80.Otvos JD, Jeyarajah EJ, Bennett DW. Quantification of plasma lipoproteins by proton nuclear magnetic resonance spectroscopy. Clin. Chem. 1991;37:377–386. [PubMed] [Google Scholar]
- 81.Jeyarajah EJ, Cromwell WC, Otvos JD. Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy. Clin Lab Med. 2006;26(4):847–870. doi: 10.1016/j.cll.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 82.Kaddurah-Daouk R, Zhu H, Sharma S, Bogdanov M, Rozen SG, Matson W, Oki NO, Motsinger-Reif AA, Churchill E, Lei Z, Appleby D, Kling MA, Trojanowski JQ, Doraiswamy PM, Arnold SE. Alterations in metabolic pathways and networks in Alzheimer's disease. Transl Psychiatry. 2013;3:e244. doi: 10.1038/tp.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang AH, Sun H, Wang XJ. Recent advances in metabolomics in neurological disease, and future perspectives. Anal Bioanal Chem. 2013 May 30; doi: 10.1007/s00216-013-7061-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 84.Hassan-Smith G, Wallace GR, Douglas MR, Sinclair AJ. The role of metabolomics in neurological disease. J Neuroimmunol. 2012;248(1–2):48–52. doi: 10.1016/j.jneuroim.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 85.Trushina E, Dutta T, Persson XM, Mielke MM, Petersen RC. Identification of Altered Metabolic Pathways in Plasma and CSF in Mild Cognitive Impairment and Alzheimer's Disease Using Metabolomics. PLoS One. 2013;8(5):e63644. doi: 10.1371/journal.pone.0063644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li NJ, Liu WT, Li W, Li SQ, Chen XH, Bi KS, He P. Plasma metabolic profiling of Alzheimer's disease by liquid chromatography/mass spectrometry. Clin Biochem. 2010;43(12):992–997. doi: 10.1016/j.clinbiochem.2010.04.072. [DOI] [PubMed] [Google Scholar]
- 87.Han X, Rozen S, Boyle SH, Hellegers C, Cheng H, Burke JR, Welsh-Bohmer KA, Doraiswamy PM, Kaddurah-Daouk R. Metabolomics in early Alzheimer's disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS One. 2011;6(7):e21643. doi: 10.1371/journal.pone.0021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Orešič M, Hyötyläinen T, Herukka SK, Sysi-Aho M, Mattila I, Seppänan-Laakso T, Julkunen V, Gopalacharyulu PV, Hallikainen M, Koikkalainen J, Kivipelto M, Helisalmi S, Lötjönen J, Soininen H. Metabolome in progression to Alzheimer's disease. Transl Psychiatry. 2011;1:e57. doi: 10.1038/tp.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sato Y, Suzuki I, Nakamura T, Bernier F, Aoshima K, Oda Y. Identification of a new plasma biomarker of Alzheimer's disease using metabolomics technology. J. Lipid Res. 2012;53:567–576. doi: 10.1194/jlr.M022376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bogdanov M, Matson WR, Wang L, Matson T, Saunders-Pullman R, Bressman SS, Beal MF. Metabolomic profiling to develop blood biomarkers for Parkinson's disease. Brain. 2008;131:389–396. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- 91.Ahmed SS, Santosh W, Kumar S, Christlet HT. Metabolic profiling of Parkinson's disease: evidence of biomarker from gene expression analysis and rapid neural network detection. J Biomed Sci. 2009;16:63. doi: 10.1186/1423-0127-16-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kumar A, Bala L, Kalita J, Misra UK, Singh RL, Khetrapal CL, Babu GN. Metabolomic analysis of serum by (1) H NMR spectroscopy in amyotrophic lateral sclerosis. Clin Chim Acta. 2010;411(7–8):563–567. doi: 10.1016/j.cca.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 93.Blasco H, Corcia P, Moreau C, Veau S, Fournier C, Vourc’h P, Emond P, Gordon P, Pradat PF, Praline J, Devos D, Nadal-Desbarats L, Andres CR. 1H–NMR-based metabolomics profiling of CSF in early amyotrophic lateral sclerosis. PLoS One. 2010;5(10):e13223. doi: 10.1371/journal.pone.0013223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lutz NW, Cozzone PJ. Metabolic profiling in multiple sclerosis and other disorders by quantitative analysis of cerebrospinal fluid using nuclear magnetic resonance spectroscopy. Curr Pharm Biotechnol. 2011;12(7):1016–1025. doi: 10.2174/138920111795909122. [DOI] [PubMed] [Google Scholar]
- 95.Underwood B, Broadhurst D, Dunn WB, Ellis DI, Michell AW, Vacher C, Mosedale DE, Kell DB, Barker RA, Grainger DJ, Rubinsztein DC. Huntington disease patients and transgenic mice have similar pro-catabolic serum metabolite profiles. Brain. 2006;129:877–886. doi: 10.1093/brain/awl027. [DOI] [PubMed] [Google Scholar]
- 96.Holmes E, Tsang TM, Huang JT, Leweke FM, Koethe D, Gerth CW, Nolden BM, Gross S, Schreiber D, Nicholson JK, Bahn S. Metabolic profiling of CSF: evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLoS Med. 2006;3:e327. doi: 10.1371/journal.pmed.0030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kaddurah-Daouk R, McEvoy J, Baillie RA, Lee D, Yao JK, Doraiswamy PM, Krishnan KR. Metabolomic mapping of atypical antipsychotic effects in schizophrenia. Mol. Psychiatry. 2007;12:934–945. doi: 10.1038/sj.mp.4002000. [DOI] [PubMed] [Google Scholar]
- 98.Tsang TM, Huang JT, Holmes E, Bahn S. Metabolic profiling of plasma from discordant schizophrenia twins: correlation between lipid signals and global functioning in female schizophrenia patients. J. Proteome Res. 2006;5:756–760. doi: 10.1021/pr0503782. [DOI] [PubMed] [Google Scholar]
- 99.Spratlin JL, Serkova NJ, Eckhardt SG. Clinical Applications of Metabolomics in Oncology: A Review. Clin. Cancer. Res. 2009;15(2):431–440. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gribbestad IS, Sitter B, Lundgren S, Krane J, Axelson D. Metabolite composition in breast tumors examined by proton nuclear magnetic resonance spectroscopy. Anticancer Res. 1999;19:1737–1746. [PubMed] [Google Scholar]
- 101.Sitter B, Lundgren S, Bathen TF, Halgunset J, Fjosne HE, Gribbestad IS. Comparison of HR MAS MR spectroscopic profiles of breast cancer tissue with clinical parameters. NMR Biomed. 2006;19:30–40. doi: 10.1002/nbm.992. [DOI] [PubMed] [Google Scholar]
- 102.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 103.Bartella L, Thakur SB, Morris EA, Dershaw DD, Huang W, Chough E, Cruz MC, Liberman L. Enhancing nonmass lesions in the breast: evaluation with proton (1H) MR spectroscopy. Radiology. 2007;245:80–87. doi: 10.1148/radiol.2451061639. [DOI] [PubMed] [Google Scholar]
- 104.Giskeodegard GF, Grinde MT, Sitter B, Axelson DE, Lundgren S, Fjøsne HE, Dahl S, Gribbestad IS, Bathen TF. Multivariate Modeling and Prediction of Breast Cancer Prognostic Factors Using MR Metabolomics. J. Proteome Res. 2010;9(2):972–979. doi: 10.1021/pr9008783. [DOI] [PubMed] [Google Scholar]
- 105.Asiago VM, Alvarado LZ, Shanaiah N, Nagana Gowda GA, Owusu-Sarfo K, Ballas RA, Raftery D. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 2010;70(21):8309–8318. doi: 10.1158/0008-5472.CAN-10-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ganti S, Weiss RH. Urine metabolomics for kidney cancer detection and biomarker discovery. Urol. Oncol. 2011;29:551–557. doi: 10.1016/j.urolonc.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ganti S, Taylor S, Kim K, Hoppel CL, Guo L, Yang J, Evans C, Weiss RH. Urinary acylcarnitines are altered in kidney cancer. Int. J. Cancer. 2011;130:2791–2800. doi: 10.1002/ijc.26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kline EE, Treat EG, Averna TA, Davis MS, Smith AY, Sillerud LO. Citrate concentrations in human seminal fluid and expressed prostatic fluid determined via 1H nuclear magnetic resonance spectroscopy outperform prostate specific antigen in prostate cancer detection. J. Urol. 2006;176:2274–2279. doi: 10.1016/j.juro.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 109.DeFeo EM, Wu CL, McDougal WS, Cheng LL. A decade in prostate cancer: from NMR to metabolomics. Nature Rev. Urol. 2011;8(6):301–311. doi: 10.1038/nrurol.2011.53. [DOI] [PubMed] [Google Scholar]
- 110.Jordan KW, Cheng LL. NMR-based metabolomics approach to target biomarkers for human prostate cancer. Expert Rev. Proteomics. 2007;4:389–400. doi: 10.1586/14789450.4.3.389. [DOI] [PubMed] [Google Scholar]
- 111.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 112.Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, Niesporek S, Könsgen D, Dietel M, Fiehn O. Mass spectrometry based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 2006;66:10795–10804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 113.Carrola J, Rocha CM, Barros AS, Gil AM, Goodfellow BJ, Carreira IM, Bernardo J, Gomes A, Sousa V, Carvalho L, Duarte IF. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J. Proteome Res. 2011;10:221–230. doi: 10.1021/pr100899x. [DOI] [PubMed] [Google Scholar]
- 114.Djukovic D, Baniasadi HR, Kc R, Hammoud Z, Raftery D. Targeted Serum Metabolite Profiling of Nucleosides in Esophageal Adenocarcinoma. Rapid Comm. Mass Spectrom. 2010;24:3057–3062. doi: 10.1002/rcm.4739. [DOI] [PubMed] [Google Scholar]
- 115.Ritchie SA, Ahiahonu PW, Jayasinghe D, Heath D, Liu J, Lu Y, Jin W, Kavianpour A, Yamazaki Y, Khan AM, Hossain M, Su-Myat KK, Wood PL, Krenitsky K, Takemasa I, Miyake M, Sekimoto M, Monden M, Matsubara H, Nomura F, Goodenowe DB. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: implications for early screening and detection. BMC Med. 2010;8:13. doi: 10.1186/1741-7015-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ritchie SA, Tonita J, Alvi R, Lehotay D, Elshoni H, Myat S, McHattie J, Goodenowe DB. Low-serum GTA-446 anti-inflammatory fatty acid levels as a new risk factor for colon cancer. Int J Cancer. 2013;132(2):355–362. doi: 10.1002/ijc.27673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S, Jia W. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J. Proteome Res. 2009;8(10):4844–4850. doi: 10.1021/pr9004162. [DOI] [PubMed] [Google Scholar]
- 118.Qiu Y, Cai G, Su M, Chen T, Liu Y, Xu Y, Ni Y, Zhao A, Cai S, Xu LX, Jia W. Urinary metabonomic study on colorectal cancer. J. Proteome Res. 2010;9(3):1627–1634. doi: 10.1021/pr901081y. [DOI] [PubMed] [Google Scholar]
- 119.Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, Kind T, Niesporek S, Noske A, Buckendahl A, Dietel M, Fiehn O. Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover. Mol. Cancer. 2008;7:72. doi: 10.1186/1476-4598-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kobayashi T, Nishiumi S, Ikeda A, Yoshie T, Sakai A, Matsubara A, Izumi Y, Tsumura H, Tsuda M, Nishisaki H, Hayashi N, Kawano S, Fujiwara Y, Minami H, Takenawa T, Azuma T, Yoshida M. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiol. Biomarkers Prev. 2013;22(4):571–579. doi: 10.1158/1055-9965.EPI-12-1033. [DOI] [PubMed] [Google Scholar]
- 121.Xu QS, Liang YZ. Monte Carlo cross validation. Chemom. Intell. Lab. Syst. 2001;56(1):1–11. [Google Scholar]
- 122.Dunn WB, Wilson ID, Nicholls AW, Broadhurst D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis. 2012;4(18):2249–2264. doi: 10.4155/bio.12.204. [DOI] [PubMed] [Google Scholar]
- 123.Westgard JO, Ehrmeyer SS, Darcy PT. CLIA Final Rules for Quality Systems. Westgard QC: Madison WI; 2004. p. 220. [Google Scholar]
- 124.Westgard JO. Assuring the Right Quality Right: Good laboratory practices for verifying the attainment of the intended quality of test results. Westgard QC: Madison WI; 2007. p. 288. [Google Scholar]
- 125.Westgard JO, Westgard SA. The quality of laboratory testing today: An assessment of sigma-metrics for analytical quality using performance data from proficiency testing surveys and the CLIA criteria for acceptable performance. Am. J. Clin. Pathol. 2006;125:343–354. [PubMed] [Google Scholar]
- 126.Westgard JO, Barry PL. Cost-Effective Quality Control: Managing the quality and productivity of analytical processes. Washington, DC: AACC Press; 1986. Improving quality control by use of multirole control procedures; pp. 92–117. [Google Scholar]
- 127.Jentzmik F, Stephan C, Miller K, Schrader M, Erbersdobler A, Kristiansen G, Lein M, Jung K. Sarcosine in urine after digital rectal examination fails as a marker in prostate cancer detection and identification of aggressive tumours. Eur Urol. 2010;58(1):12–18. doi: 10.1016/j.eururo.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 128.Schalken J. Is urinary sarcosine useful to identify patients with significant prostate cancer? The trials and tribulations of biomarker development. Eur Urol. 2010;58(1):19–21. doi: 10.1016/j.eururo.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 129.Kenny LC, Broadhurst DI, Dunn W, Brown M, North RA, McCowan L, Roberts C, Cooper GJS, Kell DB, Baker PN on behalf of the Screening for Pregnancy Endpoints Consortium. Robust Early Pregnancy Prediction of Later Preeclampsia Using Metabolomic Biomarkers. Hypertension. 2010;56:741–749. doi: 10.1161/HYPERTENSIONAHA.110.157297. [DOI] [PMC free article] [PubMed] [Google Scholar]