Abstract
Cell competition is a quality control mechanism that eliminates unfit cells. How cells compete is poorly understood, but it is generally accepted that molecular exchange between cells signals elimination of unfit cells. Here we report an orthogonal mechanism of cell competition, whereby cells compete through mechanical insults. We show that MDCK cells silenced for the polarity gene scribble (scribKD) are hypersensitive to compaction, that interaction with wild-type cells causes their compaction and that crowding is sufficient for scribKD cell elimination. Importantly, we show that elevation of the tumour suppressor p53 is necessary and sufficient for crowding hypersensitivity. Compaction, via activation of Rho-associated kinase (ROCK) and the stress kinase p38, leads to further p53 elevation, causing cell death. Thus, in addition to molecules, cells use mechanical means to compete. Given the involvement of p53, compaction hypersensitivity may be widespread among damaged cells and offers an additional route to eliminate unfit cells.
 Cell competition is a quality control mechanism to eliminate unfit cells. Here the authors show that physical compaction of less fit cells surrounded by healthy neighbours leads to increased expression of tumour suppressor p53 in the compacted cells, causing cell death.
Cell competition is a quality control mechanism to eliminate unfit cells. Here the authors show that physical compaction of less fit cells surrounded by healthy neighbours leads to increased expression of tumour suppressor p53 in the compacted cells, causing cell death.
Cell competition is a remarkable phenomenon, conserved from arthropods to mammals, that causes the elimination of relatively less fit cells from tissues, helping to maintain overall tissue health1,2,3,4,5,6,7,8,9,10. Despite important advances11,12,13,14,15,16, the mechanisms that lead to the elimination of unfit cells are still little understood and it is unclear whether one or multiple pathways lead to cell killing17,18,19,20,21,22.
It has recently been reported that Madin–Darby canine kidney (MDCK) epithelial cells silenced for the polarity gene scribble (scribKD cells) are eliminated in the presence of wild-type MDCK cells23, while they are viable on their own23. However, the mechanisms by which scribKD cells are killed by wild-type cells are largely unknown. We therefore took advantage of this recent observation to investigate the mechanisms of cell competition.
Here we show that scribKD cells are out-competed by wild-type cells through mechanical insults rather than molecular exchange. We find that scribKD cells are hypersensitive to compaction and that this is due to elevation of baseline p53 levels, which is both necessary and sufficient to induce hypersensitivity to crowding and confer a mechanical loser status. We further show that on contact with wild-type cells, scribKD cells become compacted into a high-density arrangement and that compaction is not only required but also sufficient to eliminate scribKD cells. We also delineate the mechano-transduction cascade that leads to scribKD cell death. Specifically, we show that scribKD cells' compaction causes activation of the Rho-associated kinase (ROCK), which in turn activates p38 leading to further p53 elevation and cell death. Overall, this work demonstrates that mechanical forces can be responsible for the elimination of cells during cell competition and that p53 levels play a key role both in instructing the mechanical loser status and in the execution of mechanical cell competition.
Results
Compaction of scrib KD cells induces mechanical competition
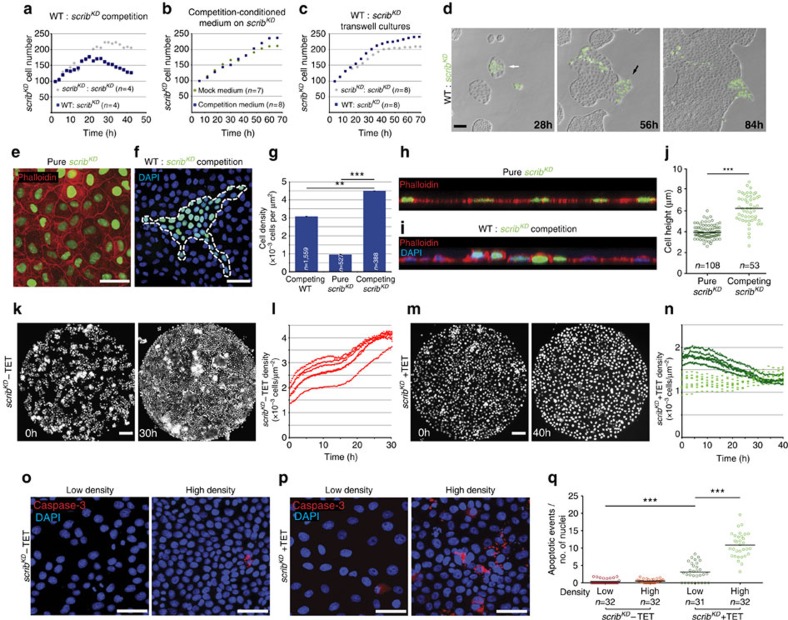
It has previously been shown that scribKD MDCK cells are eliminated when co-cultured with wild-type MDCK cells through cell death and delamination (see ref. 23 and Supplementary Fig. 1a and Supplementary Movie 1, left), while monocultures of scribKD cells are viable (see ref. 23 and Supplementary Fig. 1b and Supplementary Movie 1, right). To investigate the mechanisms of scribKD-mediated cell competition, we first asked whether it is mediated by soluble factors, as in other cases of in vitro cell competition6,24. Growth rate (doubling time) profiles showed that scribKD cells in pure cultures divide, albeit at a reduced rate, to reach a steadily maintained number (Supplementary Fig. 1d), whereas under competing conditions, their numbers collapse following initial growth (Fig. 1a). Interestingly, we found that the growth rate of scribKD cells is not affected by conditioned medium from competing cultures (Fig. 1b and Supplementary Fig. 1c). Similarly, in transwell systems that allow exchange of solutes but prevent cell contact, scribKD cells grown together with co-cultures of competing (wild-type/scribKD) cells grew comparably to scribKD cells grown with other scribKD cells (Fig. 1c and Supplementary Fig. 1c). This indicated that soluble factors are not sufficient to induce cell competition and that cell contact is required. We hypothesized that cell contact enables molecular interactions essential for cell competition, as observed by others11,12. However, to our surprise, we found that sustained contact with wild-type cells is not sufficient for elimination of scribKD cells (Fig. 1d, black arrow and Supplementary Movie 2) and that scribKD clones are efficiently eliminated only when fully surrounded by wild-type cells (Fig. 1d, white arrow and Supplementary Movie 2). This suggested that a type of exchange other than molecular signalling (which would be enabled by contact) may be needed, and prompted us to look for differences between scribKD clones that were surrounded and peripheral clones that were simply contacted.
Figure 1. Compaction of scribKD cells is both required and sufficient for their elimination.
(a–c) Quantification showing growth rate of scribKD cells from time-lapse movies of: competition versus pure cultures (a), pure cultures in mock conditioned versus competition conditioned medium; two biological replicates (b) or transwell experiments, where scribKD cells were co-cultured across transwells with control or with competing cultures; three biological replicates (c). Each dot represents the average of n fields of cells. See also Supplementary Fig. 1c. (d) Time course of cell competition assay between unlabelled wild-type (WT) and GFP-labelled scribKD MDCK cells. Competition is observed in surrounded scribKD cells (white arrow), but not in cells that are only contacted (black arrow), see corresponding Supplementary Movie 2. (e) Confluent GFP-labelled scribKDcells stained with phalloidin. (f) Competing unlabelled WT and GFP-labelled scribKD cells counterstained with DAPI. (g) Quantification showing average (±s.e.m.) cell density values of confluent pure scribKD cells and subconfluent competing WT and scribKDcells as in e and f. (h,i) Confocal xz sections of representative GFP-labelled scribKDcells pure (h) or co-cultured with WT cells (i), stained with phalloidin (h,i) and DAPI (i). (j) Quantifications of cell height from images as in h and i. Black bars=median. (k,m) Representative stills from time lapse of GFP-labelled scribKD cells +/−TET growing on micropatterns (800 μm Ø), see corresponding Supplementary Movie 3. (l,n) Quantifications of cell density over time from movies as in k and m. Each dotted line corresponds to a different movie. (o,p) Cleaved Caspase-3 staining in WT (o) and scribKD (p) cells +/− compression (high density and low density, respectively). (q) Quantification of cell death events (cleaved Caspase-3) from images as in o and p. Data are pooled from three biological replicates. Black bars=mean; three biological replicates across two independent experiments. n=number of fields imaged in a single repeat (a–c,q) or n=number of cells (g,j). Scale bars, 100 μm (movie sequences) and 50 μm (immunofluorescence images) here and throughout all figures. **P<0.005, ***P<0.0005 by KS test.
One striking feature of surrounded clones, which is not shared by peripheral clones, is that they reach a dramatically higher cell density than confluent scribKD pure cultures (Fig. 1e–g and Supplementary Fig. 1e). scribKD cells acquire a flattened morphology upon gene silencing23,25, which at confluence results in a much lower (∼1/3) final density compared with wild-type cells (Fig. 1e–g). However, scribKD clones surrounded by wild-type cells do not flatten and reach a density that is ∼4.5-fold higher than that of pure scribKD cultures (Fig. 1e–g). Furthermore, competing scribKD cells are taller than when grown in single cultures (Fig. 1h–j). Together, this indicates that as a result of their interaction with wild-type cells, scribKD cells become more compacted than in their default state and this correlates with and precedes their elimination.
We next asked whether cell compaction plays a role in scribKD cell elimination, by assessing its effect on scribKD cells in complete absence of wild-type cells. To that end, we used micropatterns to form scribKD microcultures of a defined, homogenous density and size (Ø=800 μm, ref. 26). When plated at high density, control cells that had not undergone scribble silencing (without tetracycline, −TET) continued to grow until they reached a maximal density of 61±4 cells per 10,000 μm2, which they maintained homeostatically (Fig. 1k,l and Supplementary Movie 3, left), as previously shown27,28. Remarkably, scribKD cultures (+TET) instead saw their numbers and densities drop (Fig. 1m,n, dark green and Supplementary Movie 3, right), due to a combination of increased cell death and extrusion, the same two events that lead to the elimination of scribKD cells during cell competition (Supplementary Movie 1, left and ref. 23). Notably, scribKD cells seeded at a lower density maintained that initial density (Fig. 1n, light green) indicating that the drop in cell number observed at higher density is not a general response of scribKD cells to plating. This suggested that scribKD cells are hypersensitive to compaction. To test this directly, we seeded confluent monolayers of control (−TET) or scribKD (+TET) cells on stretched polydimethylsiloxane (PDMS) substrates, which we then released to induce cell compression27. Control cultures showed no increase in apoptosis upon compression (Fig. 1o–q). The scribKD cells, however, displayed a 3-fold increase in apoptosis over their uncompressed baseline cell death, which was already higher than control (Fig. 1p,q). Altogether, these experiments show that scribKD cells display hypersensitivity to cell density and cannot sustain levels of crowding normally reached by wild-type cells. They further indicate that compaction of scribKD cells into higher cell densities, like those imposed on them by wild-type cells during competition, is sufficient to induce cell death. This suggests that wild-type cells eliminate scribKD cells through crowding-induced compaction. We name this new mode of selective elimination of one cell population by another due to differential sensitivity to crowding ‘mechanical cell competition', to contrast it with forms of cell competition that rely on molecular exchange.
Corralling promotes but is not needed for scrib KD elimination
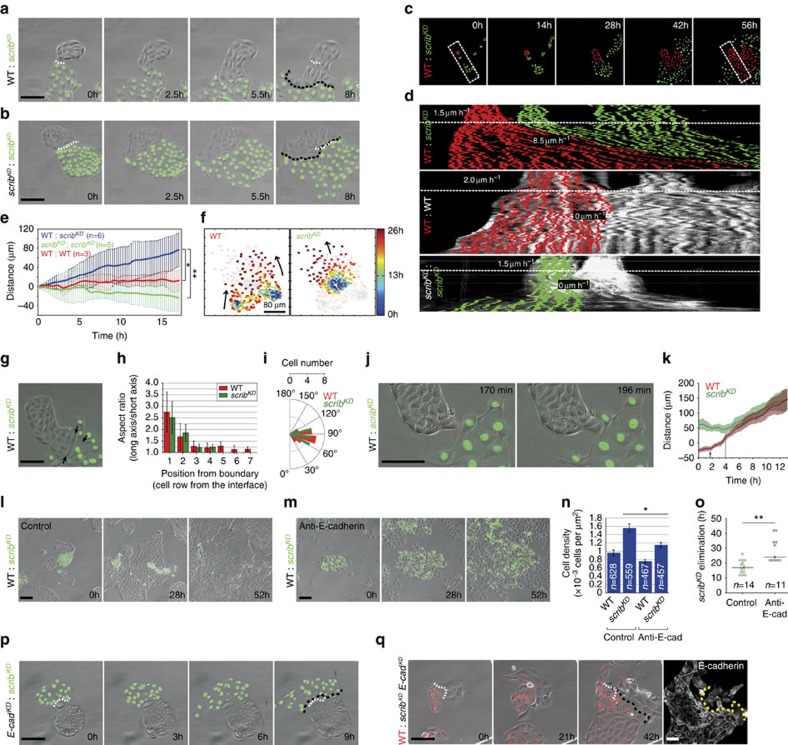
As shown in Fig. 1f,g, in subconfluent competing cultures, the density of scribKD cells surpasses that of surrounding wild-type cells. This indicates that their acquired density does not simply reflect the density of the mixed culture, but is the result of an active process. To understand how scribKD cells become compacted during competition, we plated cells at low density and observed what happens when wild-type and scribKD clones first come into contact. Strikingly, we saw that, on contact, both scribKD and wild-type cells engage in collective cell migration (Fig. 2a and Supplementary Movie 4). This behaviour was specific to wild-type/scribKD encounters, as it was not observed upon homotypic encounters of either cell population (Fig. 2b and Supplementary Movie 5). Interestingly, this migration was highly directional with scribKD cells always at the migrating front and, conversely, wild-type cells always at the back. We next characterized this migratory behaviour and assessed its contribution to cell compaction and elimination. First, we repeated the above experiment using a fluorescent nuclear label in each population to facilitate cell tracking (Fig. 2c). Kymograph analysis showed rapid activation of collective cell migration at the time of contact between wild-type and scribKD cells, with both populations moving synchronously (Fig. 2d, top and Fig. 2e). By contrast, similar kymographs of homotypic collisions did not show significant cell displacement (Fig. 2d, centre and bottom; and Fig. 2e). To further characterize the features of this collective cell migration, we carried out single-cell tracking of both cell populations before and after contact. The analysis of individual cell trajectories shows that single-cell movement is faster and more persistent for both wild-type and scribKD cells upon heterotypic collision (Fig. 2f and Supplementary Fig. 2a–d). We then looked more closely at the dynamic interplay between wild-type and scribKD cells at the onset of migration. Interestingly, analysis of cell shape at the interface between the two populations revealed that both wild-type and scribKD cells become elongated at the site of contact (Fig. 2g,h), indicating that they are under anisotropic stress. Moreover, the cells in both populations move in the direction of their short axis (Fig. 2i), suggesting that they could be locally compressed. This could be the result of wild-type cells beginning migration and locally ‘piling up' against the scribKD cells. In agreement with this, particle image velocimetry (PIV) and single-cell tracking revealed that wild-type cells begin migrating towards scribKD cells on average ∼2 h before the scribKD cells start migrating away (Fig. 2j,k and Supplementary Movie 6). Together, the chronology of these events and the local cellular deformations suggest that cells might engage in a behaviour similar to the ‘chase and run' migration reported for other cell types29. These experiments do not distinguish whether scribKD cells are pushed by wild-type cells or, conversely, whether scribKD cells self-compact to avoid closer interaction wild-type cells. However, it is clear that as migration progresses, scribKD cells become corralled by wild-type cells and are compacted and eliminated (Supplementary Movie 7), indicating that this behaviour may facilitate compaction and outcompetition of scribKD cells.
Figure 2. Contact-induced migration promotes compaction and cell competition.
(a,b) Stills from movies of wild-type (WT) and GFP-labelled scribKD co-cultures (a) or scribKD homotypic cultures (b), see Supplementary Movie 4,5. (c,d) Kymographs (d) from movies as in c. Velocities are shown before (above dashed white line) and after contact (below line). (e) Plot showing displacement of the line of contact between clones from movies as in c. The continuous line is the position of the front average±s.d.; n=number of contact lines averaged. (f) Single-cell tracking of trajectories of WT and scribKD cells during competition. Heat-map representation shows time-resolved position of single cells. (g) Micrograph exemplifying cell shape change (arrows) after contact between WT and scribKD cells. (h) Bar plot representing aspect ratio of WT and scribKD cells as a function of distance from their contact point. n=50 cells of each type from three movies; error bars=s.d. (i) Distribution of angles between a cell's long axis and its direction of motion; n (WT)=18 cells; n (scribKD)=17 cells. (j,k) PIV analysis of images at time of contact (see Supplementary Movie 6) (j); and quantification of cell displacements (k) shows WT cells begin migrating (arrows) before scribKD cells; n=10 cells for each type from three independent movies. Coloured lines=mean; shaded areas=s.d. (l–o) Disrupting cell junctions by E-cadherin blocking antibody and calcium removal prevents contact-induced migration (m), compaction (n) and delays competition (o) compared with control (l), see Supplementary Movie 8; error bars=s.e.m. (p) E-cadherin knockdown in WT cells (E-cadKD) prevents contact-induced migration. (q) E-cadherin knockdown in scribKD cells (scribKD E-cadKD) prevents contact-induced migration, see Supplementary Movie 9. Right panel displays anti-E-cadherin immunofluorescence at end of movie (see Supplementary Fig. 2h,i). Five independent repeats; n=10 events showing absence of directional migration, five were validated for E-cadherin levels and all five had WT levels. White dashed line=initial contact point; black dashed line=final contact point; yellow dashed line separates WT from scribKD E-cadKD cells. *P<0.05, **P<0.005 by KS test.
To directly test the relevance of directional cell migration in the elimination of scribKD cells, we next sought to disrupt this behaviour. Adhesion molecules play an important role in collective cell migration30 and have also been implicated in ‘chase and run' among mesenchymal cells29. In addition, scribble downregulation has previously been shown to induce intracellular accumulation of E-cadherin23,31 and we found that it causes an increase of both total and cell surface E-cadherin levels (Supplementary Fig. 2e–g). We therefore reasoned that targeting E-cadherin-mediated adhesion might disrupt directional migration between the wild-type and scribKD cells. We undertook two separate approaches and found that either blocking E-cadherin function in both populations, through a combination of low calcium and addition of a blocking antibody (Fig. 2l–o and Supplementary Movie 8), or silencing E-cadherin only in wild-type cells (E-cadKD (ref. 32); Fig. 2p) was sufficient to inhibit directional migration. Interestingly, disruption of E-cadherin inhibited active cell compaction (Fig. 2l,m, Supplementary Movie 8 and quantification in Fig. 2n) and resulted in delayed elimination of scribKD cells (Fig. 2l,m, Supplementary Movie 8 and quantification in Fig. 2o). However, it did not rescue the scribKD cells from cell competition, since they were eventually eliminated as the culture became progressively more crowded due to proliferation (Fig. 2l,m and Supplementary Movie 8).
Having established that E-cadherin-mediated adhesion is involved in contact-induced migration between wild-type and scribKD cells, we next asked whether the upregulation of E-cadherin observed in scribKD cells (Supplementary Fig. 2e–g) plays a role in this directional cell movement. We therefore generated an inducible double scribKD E-cadherin knockdown cell line (scribKD E-cadKD) and specifically selected clones that displayed partial silencing, enough to bring E-cadherin down to wild-type levels (Fig. 2q, right panel and Supplementary Fig. 2h,i). Indeed, partial downregulation of E-cadherin inhibited contact-induced migration, suggesting that high E-cadherin levels in the scribKD cells are required for this process (Fig. 2q and Supplementary Movie 9). As expected, these clones were still outcompeted by wild-type cells (Supplementary Fig. 2j). In contrast, we found that E-cadherin upregulation alone is not sufficient to cause contact-induced migration, as cells overexpressing E-cadherin at levels comparable to those of scribKD cells (Supplementary Fig. 2k) did not engage in directional cell migration with wild-type cells upon contact (Supplementary Fig. 2l). Altogether, we conclude that directional cell migration, by enabling corralling and active compaction of scribKD cells, promotes but is not required for mechanical cell competition, and that E-cadherin is necessary for corralling and active compaction but it does not impact on loser cell status.
p53 is activated in scrib KD cells before cell competition
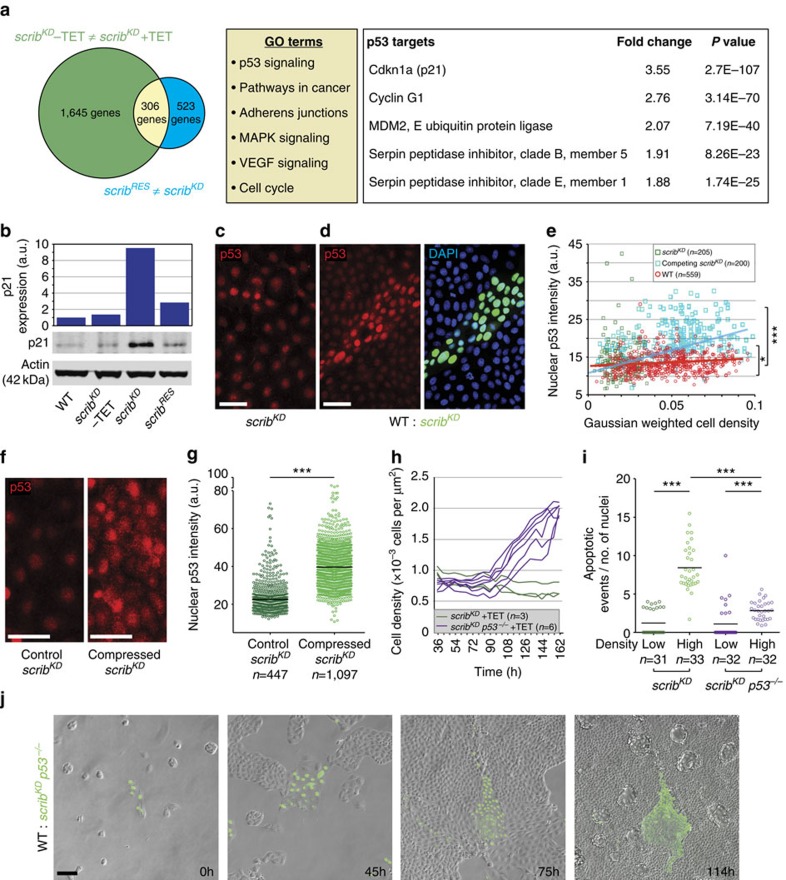
Key to the outcompetition of scribKD cells is their hypersensitivity to compaction (Fig. 1k–q). To identify genes and pathways involved in this behaviour, we carried out transcriptional profiling of scribKD cells (scribKD +TET) and compared it with the transcriptomes of control MDCK cells (scribKD −TET) and, importantly, of an isolate of scribKD cells that is resistant to cell competition (scribRES). Despite maintaining scribble gene knockdown (Supplementary Fig. 3a), the scribRES cells do not display elevated E-cadherin (Supplementary Fig. 3b), do not engage in contact-induced migration with wild-type cells (Supplementary Fig. 3c) and are not outcompeted in cell competition assays (Supplementary Fig. 3d and Supplementary Movie 10). Notwithstanding these fundamental differences, we observed that the scribKD transcriptome is still substantially closer to scribRES (with 523 differentially expressed genes) than to control (scribKD −TET) cells (with 1,645 differentially expressed genes; Fig. 3a, left and Supplementary Data 1 and 2). This allowed us to rule out all genes that are differentially expressed in scribKD versus wild-type cells but are similarly expressed between scribKD and scribRES, as these are clearly not sufficient to induce cell competition. Instead, we focused on the small intersection of 306 genes that are differentially expressed between wild-type and scribKD cells but are also different between scribKD and scribRES cells (Fig. 3a and Supplementary Data 3).
Figure 3. p53 is activated in scribKD cells before competition and further p53 elevation upon compaction causes competition-induced cell death.
(a) Left, transcriptional profiling of scribKD cells without tetracycline (TET) versus scribKD cells with TET (green) and of scribKD cells with TET versus scribKD cells resistant to competition (scribRES) with TET (blue), and corresponding intersection of differentially expressed genes (yellow). Middle, list of pathways functionally enriched in the yellow intersection. Right, list of p53 targets found in this intersection and corresponding fold change difference between scribKD cells −/+TET. Three biological replicates for scribKD cells −/+TET and two biological replicates for scribRES cells +TET. (b) Western blot against p21 with LICOR quantifications and normalization to Actin. (c,d) p53 staining of pure scribKD cells (c) and of co-cultures of GFP-labelled scribKD and wild-type (WT) cells (d). (e) Graph showing single-cell nuclear p53 intensity plotted against cell density from images as in c and d; *P<0.05 by KS test comparing p53 levels in pure scribKD versus WT; ***P<0.0005 by KS test comparing competing scribKD versus WT. Non-parametric Spearman correlation; red line=WT cells; blue line=competing scribKD cells. (f,g) p53 staining in pure scribKD cells on PDMS substrate +/− compression (f) and quantification showing single-cell nuclear p53 intensity (g) from images as in f; black bars=median. (h) Time-resolved density measurement of growing scribKD cells and scribKD p53−/− cells. (i) Quantification of cell death (cleaved Caspase-3) for scribKD cells versus scribKD p53−/− cells on PDMS substrate +/− compression; black bars=mean; data from three biological replicates across two independent experiments. (j) Stills from time-lapse movies of WT and GFP-labelled scribKD p53−/− co-cultures, see corresponding Supplementary Movie 11; n=cell number in e and g or n=number of imaged fields of cells in h and i. a.u.=arbitrary units, here and throughout all the figures. *P<0.05, ***P<0.0005 by KS test.
Gene Ontology term enrichment analysis highlighted p53 signalling as the top functionally enriched pathway (Fig. 3a, middle). A number of known p53 target genes were moderately upregulated in scribKD cells, suggesting p53 activation (Fig. 3a, right). Consistent with this, we found that p21 (a known p53 target33 and the most highly upregulated p53 target in our gene list; Fig. 3a) is specifically upregulated upon scribble knockdown in scribKD cells (Fig. 3b, Supplementary Fig. 3e and Supplementary Fig. 4d,e) but not in scribRES cells (Fig. 3b) and that p53 levels are higher in scribKD cells than in wild-type cells (Fig. 3c,d and Supplementary Fig. 3f), confirming pathway activation. Thus, scribKD cells have high baseline p53 activity and this correlates with their loser status.
p53 is further elevated in scrib KD cells by compaction
Next we asked whether cell competition affects p53 activity. Interestingly, we found that in competing conditions, p53 levels in scribKD cells increase above their already elevated baseline level, in a way that correlates with the degree of cell compaction (Fig. 3c–e; r=0.56 by non-parametric Spearman correlation). This suggested that compaction increases p53 levels or, conversely, that higher p53 levels enable compaction. To distinguish between these two possibilities, we looked at how compression alone affects p53, using deformable PDMS substrates as before (Fig. 1o–q). Importantly, we found that compression alone causes an increase in p53 levels (Fig. 3f,g), as seen during competition. This indicates that cell competition induces further p53 activation in the scribKD cells via compaction-induced mechanical stress.
Given that p53 is upregulated during cell competition, we next sought to ask whether it contributes to this process. Therefore we mutated p53 in scribKD cells by CRISPR-mediated mutagenesis (scribKD p53−/− cells, Supplementary Fig. 4a,b). The scribKD p53−/− cells failed to upregulate p21 following ultraviolet irradiation (Supplementary Fig. 4b) or scribble knockdown (Supplementary Fig. 4c–e), confirming functional p53 inactivation. Strikingly, we found that p53 inactivation in scribKD p53−/− cells was sufficient to partially rescue their low homeostatic cell density (Fig. 3h) and their hypersensitivity to compaction (Fig. 3i). Remarkably, genetic (using scribKD p53−/− cells) or chemical (using the inhibitor Pifithrin-α) inhibition of p53 in scribKD cells was also sufficient to prevent their outcompetition (Fig. 3j and Supplementary Movie 11; Supplementary Fig. 4f). Furthermore, scribKD p53−/− cells maintained high E-cadherin levels (Supplementary Fig. 4g) and still displayed contact-induced migration (Supplementary Fig. 4h), demonstrating again that corralling is not sufficient for competition. Altogether, we conclude that high baseline p53 activity in scribKD cells is associated with their loser status and is required for them to acquire a low homeostatic cell density and hypersensitivity to compaction, two key features of the mechanical loser status.
Interestingly, though it has long been established that scribble−/− cells are eliminated by cell competition in Drosophila34,35, when we tested whether this might happen via mechanical insults in wing imaginal discs we found that, unlike in MDCK cells, scribble−/− wing disc cells did not upregulate E-cadherin (Supplementary Fig. 5a,b) and their outcompetition was not rescued by p53 inhibition (Supplementary Fig. 5c). It is possible that scribble−/− cells are not eliminated by mechanical cell competition in Drosophila or that the function of Scribble or p53 may not be conserved in this process. Alternatively and perhaps more likely, mechanical cell competition may be redundant with other mechanisms of cell competition that have been described to target scribble−/− cells in that system36,37.
ROCK activates p38 leading to p53 elevation and cell death
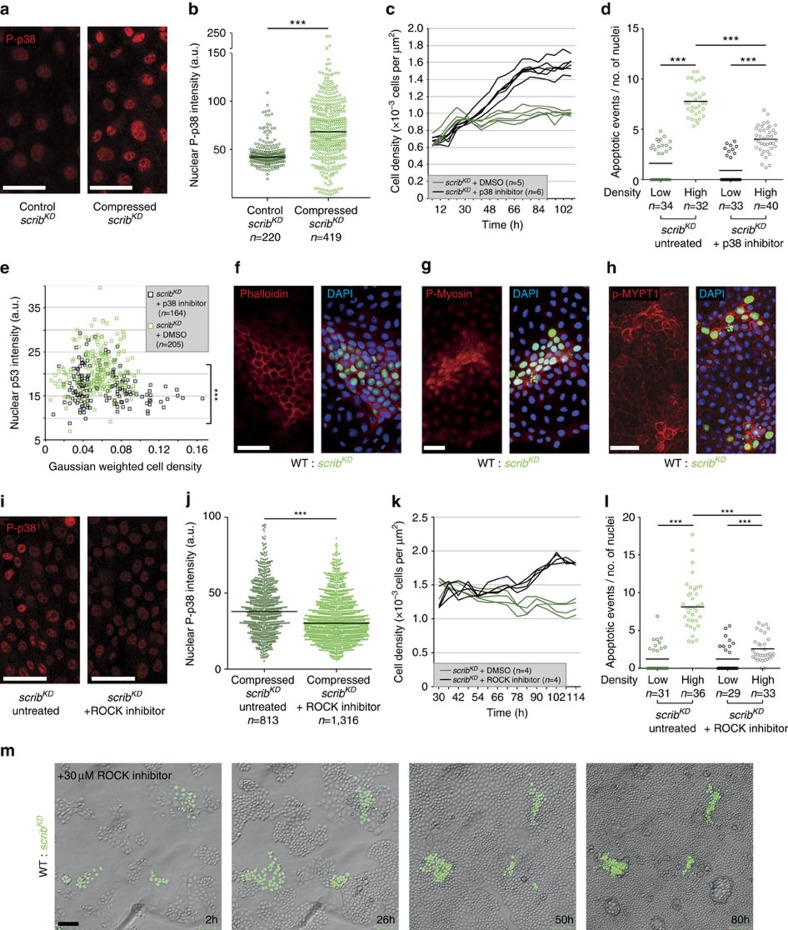
We next wondered how mechanical stress might lead to p53 activation. A potential candidate was p38 signalling, as it is required for scribKD cell competition23, is known to promote p53 activity38,39,40 and has also been involved in the response to mechanical stress41. Consistent with an involvement of p38, we found that compression alone causes an increase in active phosphorylated (T180/Y182) p38 (ref. 38; P-p38) in scribKD cells (Fig. 4a,b) and that chemical inhibition of this pathway partially rescues both the homeostatic density of scribKD cells (Fig. 4c) and their compaction hypersensitivity (Fig. 4d). Moreover, the upregulation of p53 in competing scribKDcells was reduced by p38 inhibition (Fig. 4e). We conclude that in scribKD cells, compression induces p53 via activation of p38.
Figure 4. During mechanical cell competition ROCK activates p38 leading to p53 elevation.
(a) Phosphorylated p38 (P-p38) staining in pure scribKD cells +/− compression. (b) Single-cell nuclear P-p38 intensity from images as in a; black bars=median. (c) Time-resolved density measurement of growing scribKD cells +/− p38 inhibitor. (d) Quantification of cell death (cleaved Caspase-3) of scribKD cells with or without compression and +/− p38 inhibitor; black bars=mean; pooled data from three biological replicates across two independent experiments. (e) Single-cell nuclear p53 signal intensity in competing scribKD cells +/− p38 inhibitor, plotted against cell density. (f–h) F-Actin (phalloidin-stained, f), phosphorylated Myosin-II (P-Myosin, g) and phosphorylated MYPT1 (p-MYPT1, h) are elevated in compacted GFP-labelled scribKD cells compared with wild-type (WT) cells during competition. (i) P-p38 staining in compressed scribKD cells +/− ROCK inhibitor. (j) Single-cell nuclear P-p38 intensity from images as in i; black bars=median. (k) Time-resolved density measurement of growing scribKD cells +/− ROCK inhibitor; two independent repeats. (l) Quantification of cell death (cleaved Caspase-3) in scribKD cells with or without compression and +/− ROCK inhibitor; black bars=mean; pooled data from three biological replicates across two independent experiments. (m) Stills from time-lapse movies of WT and GFP-labelled scribKD co-cultures treated with ROCK inhibitor (30 μM), see corresponding Supplementary Movie 12; n=cell number in b,e and j or n=number of imaged fields of cells in c,d,k and l. ***P<0.0005 by KS test.
We next asked how compression of scribKD cells causes p38 activation. We monitored cytoskeletal changes induced by compression and found that both cortical Actin (by phalloidin staining) and active phosphorylated-myosin (P-Myosin) are upregulated in compacted scribKD cells during competition (Fig. 4f,g). Since the cytoskeletal regulator ROCK42 is one of the main kinases responsible for Myosin phosphorylation, this suggested that ROCK might be activated. Indeed P-Myosin upregulation was reduced in the presence of a ROCK inhibitor (Supplementary Fig. 6a,b) and the ROCK target phospho-MYPT1 (ref. 43; p-MYPT1) was also elevated in compacted scribKD cells (Fig. 4h), indicating that ROCK is activated in compacted scribKD cells. This was potentially relevant because ROCK has been shown to phosphorylate p38 (ref. 44). Thus, to ask whether ROCK is upstream of p38 activation, we compressed pure cultures of scribKD cells in the presence or absence of a ROCK inhibitor and looked at P-p38 levels. ROCK inhibition led to a partial reduction of P-p38 levels, thus placing ROCK upstream of p38 signalling (Fig. 4i,j). In addition, ROCK inhibition was sufficient to partially rescue the homeostatic density of scribKD cells (Fig. 4k) and compression-induced cell death (Fig. 4l). Importantly, inhibition of ROCK was also sufficient to prevent the out-competition of scribKD cells, with no appreciable cell death observed even though cells were compacted far beyond standard competition densities (Fig. 4m and Supplementary Movie 12). Altogether, these experiments indicate that mechanical cell competition is caused by compaction-induced ROCK activation, which activates p38, leading to p53 elevation and cell death. Interestingly, ROCK has also been implicated in apical extrusion of dying or crowded MDCK cells45,46. However, in the case of Scribble competition, ROCK has a different function, as it is involved in p53 activation and cell death. In addition, while in that context ROCK was activated by S1P2 and Piezo signalling45,46, inhibition of these pathways had no effect on Scribble cell competition (Supplementary Fig. 7a,b), suggesting a distinct upstream activation mechanism.
p53 activation turns wild-type cells into mechanical losers
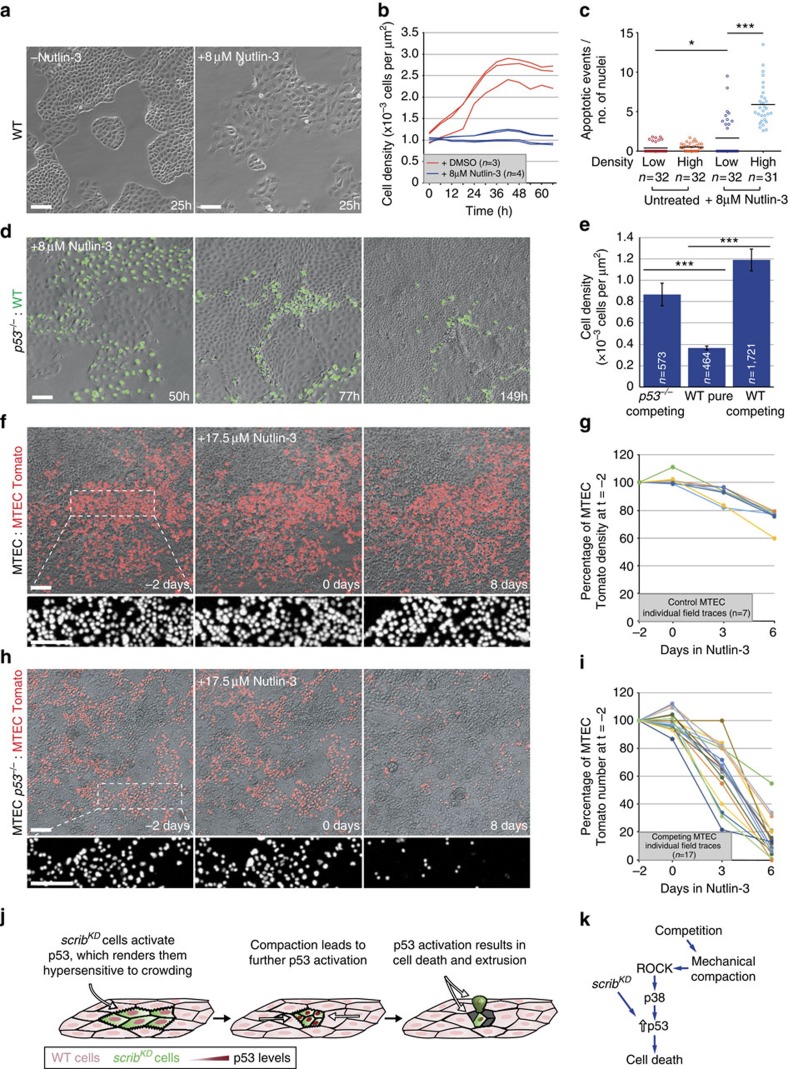
Finally, we decided to address how scribble knockdown earmarks cells as losers. Our data indicated that a loser cell status is not an obligate outcome of scribble silencing, as we could isolate scribKD cells that are competition resistant (scribRES cells; Supplementary Fig. 3d). Instead, our data showed that p53 is necessary for competition and that mild p53 elevation is required for scribKD cells to acquire key features of the mechanical loser status (Fig. 3h,i). This prompted us to investigate whether p53 might actually be sufficient to induce the mechanical loser status and cell competition. Nutlin-3, a chemical inhibitor of the E3 ubiquitin ligase MDM2 (Mouse Double Minute 2 (ref. 47)), activates p53 in a dose-dependent manner. This allowed us to establish conditions to induce mild p53 activation in wild-type MDCK cells. Strikingly, low-level p53 activation was sufficient to induce cell flattening (Fig. 5a) and to lower the homeostatic density of wild-type cells (Fig. 5b). Remarkably, Nutlin-3 treatment was also sufficient to induce compaction hypersensitivity (Fig. 5c). Thus, mild p53 activation is sufficient to induce in wild-type cells all the features that we observed in scribKD cells that are hallmarks of their mechanical loser status. Therefore, we next asked whether differential p53 levels are sufficient to induce mechanical cell competition. To test this, we generated a p53 knockout cell line (p53−/−), which we then co-cultured with wild-type cells. Not surprisingly, simply mixing wild-type and p53−/− cells was not sufficient to induce competition (Supplementary Fig. 8a). Strikingly however, mild p53 activation in wild-type cells, by Nutlin-3 addition at sublethal doses, caused the elimination of these cells specifically in the co-culture, but not when they were cultured alone (Fig. 5d, Supplementary Fig. 8b and Supplementary Movie 13). Importantly, p53-induced cell competition was indistinguishable from scribble cell competition, as it resulted in loser cell compaction (Fig. 5e) and only caused elimination when cells were compacted by p53−/− cells (Fig. 5d). Overall, these results demonstrate that mild elevation of p53 is sufficient to phenocopy Scribble cell competition and induce both hypersensitivity to cell crowding and the mechanical loser status in otherwise wild-type cells.
Figure 5. p53 activation is sufficient to induce crowding hypersensitivity and mechanical cell competition.
(a) Addition of Nutlin-3 (8 μM) causes flattening of wild-type (WT) MDCK cells; n=4 fields per repeat. (b) Time-resolved cell density measurement of growing WT MDCK cells +/− Nutlin-3 (8 μM). (c) Quantification of cell death (cleaved Caspase-3) of WT MDCK cells with and without compression and +/− Nutlin-3 (8 μM); black bars=mean; three biological replicates from two independent experiments. (d) Stills from time-lapse movies of WT and p53−/− MDCK co-cultures with Nutlin-3 (8 μM) see corresponding Supplementary Movie 13. (e) Cell density measurement from movies as in d; mean±s.e.m. (f) Stills from time-lapse movies of primary cultures of unlabelled and Tomato-labelled WT MTECs (see corresponding Supplementary Movie 14). Nutlin-3 (17.5 μM) was added at t=0. (g) Time-resolved cell density measurement from movies as in f of WT MTECs before and after Nutlin-3 (17.5 μM) addition. (h) Stills from time-lapse movies of primary cultures of unlabelled p53−/− and Tomato-labelled WT MTECs (see corresponding Supplementary Movie 15). Nutlin-3 (17.5 μM) was added at t=0. (i) Time-resolved measurement of cell number from movies as in h of WT MTECs before and after Nutlin-3 (17.5 μM) addition. (j,k) Model of mechanical cell competition of scribKD cells; n=cell number in e or n=number of imaged fields of cells in b,c,g and i. For c and e, data are pooled from three biological replicates. *P<0.05, ***P<0.0005 by KS test.
To demonstrate the existence of p53-mediated mechanical cell competition beyond the MDCK experimental paradigm, we turned to primary cultures of epithelial cells, specifically to mouse tracheal epithelial cells (MTECs), and looked for evidence of this process. Under normal conditions, confluent MTEC monolayers show little proliferation or change in cell density (Fig. 5f,g, compare first and second time points, and Supplementary Movie 14, left, before Nutlin-3 addition). Interestingly however, when we induced mild elevation of p53 (by Nutlin-3 treatment) in these cultures, we found that this results in a 26% average reduction in cell density, accompanied by cell extrusion, suggesting acquired hypersensitivity to crowding (Fig. 5f,g, compare before and after Nutlin-3 addition, and Supplementary Movie 14, left, after Nutlin-3 addition). To test whether differential levels of p53 could induce cell competition in MTECs, we mixed Tomato-labelled wild-type MTECs (MTEC Tomato; from Rosa26R-nTomato-nGFP mice48) either with unlabelled wild-type MTECs or with unlabelled p53 null MTECs (MTEC p53−/−; from p53 null mice). Before Nutlin-3 addition, the proportion of wild-type (Tomato) MTECs in the monolayer did not change for several days when co-cultured with either p53−/− cells or more wild-type cells (Fig. 5f–i, first two time points, and Supplementary Movie 14, before Nutlin-3 addition). However and strikingly, Nutlin-3 addition induced robust cell competition specifically in wild-type/p53−/− MTEC co-cultures, causing the number of wild-type cells to plummet within 6 days to ∼17% of their starting number, with pronounced cell death and fragmentation (Fig. 5h,i and Supplementary Movie 14, right). Thus, mild p53 elevation is sufficient to induce crowding hypersensitivity and mechanical cell competition in MTEC cultures.
Discussion
In summary, our work demonstrates that in addition to molecular signals, cells use mechanical means to compete (see model in Fig. 5j,k), a concept that had previously only been speculated about and proposed on theoretical grounds19,49. We show that hypersensitivity to crowding provides an additional route to the loser cell status, leading to mechanical cell competition and the elimination of loser cells. One interesting prediction of our findings is that they suggest that competition could take place between genetically identical cells if they are endowed with differential sensitivity to mechanical stress or if they reside in tissues with varying mechanical properties50,51, a hypothesis that should be investigated. In addition, given the involvement of p53, a general sensor of cell stress, we suggest that mechanical cell competition may be widespread among the damaged cells. Moreover, future work should also examine the implications of our findings on the behaviour of cancer cells. Since p53 is one of the most commonly mutated genes in cancer52, our findings suggest that its loss could enable neoplastic cells (scribble is a tumour-suppressor gene53,54) to evade mechanical cell competition. Identifying the physiological contexts where mechanical cell competition plays a role may help better understand tissue biology and potentially cancer formation.
Note added in proof: Evidence of mechanical competition in Drosophila was reported just prior to acceptance of this work55.
Methods
Antibodies and materials
For immunofluorescence we used:
Rabbit anti-p53 antibody (1:750, Cell Signaling Technology #9382), rabbit anti-p21 antibody (1:200, Santa Cruz sc-397). In functional tests, both antibodies show an increase in staining intensity +ultraviolet-C treatment and a reduction or complete absence of staining in scribKD p53−/− and p53−/− cells (Supplementary Fig. 4b). In addition, the anti-p21 antibody is documented to cross-react in canine. Mouse anti-E-cadherin antibody (1:600 total stain or 1:200 surface stain56), rabbit anti-E-cadherin antibody (1:500 total stain or 1:50 surface stain57), both documented to cross-react in canine. Rabbit anti-P-p38 MAPK (T180/Y182) antibody (1:50, Cell Signaling Technology #9215) was used as in Norman et al.23 Rabbit anti-P-Myosin light chain (phospho S20) antibody (1:100, Abcam ab2480), predicted to cross-react with all mammals. Goat p-MYPT1 (Thr 853) (1:50, Santa Cruz sc-17432) antibody, documented to cross-react in canine. Rabbit anti-cleaved Caspase-3 antibody (1:200, Cell Signaling Technology #9661s), predicted to cross-react in canine based on 100% sequence homology. DAPI (1 μg ml−1 Invitrogen); Alexa Fluor conjugated secondary antibodies (1:1,000, Invitrogen); Alexa Fluor-568 and Alexa Fluor-647 conjugated phalloidin (1:40, Invitrogen).
For western blotting we used:
Rabbit anti-p53 antibody (1:1000, Cell Signaling Technology #9382), rabbit anti-p21 antibody (1:500, Santa Cruz sc-397). Goat anti-Scribble antibody (1:500, Santa Cruz sc-11048), functional tests show a decrease in signal intensity in scribKD cells +TET and it is documented to cross-react in canine (Supplementary Fig. 3a). Mouse anti-E-cadherin antibody (1:1,000; (ref. 56)), rabbit anti-β-tubulin antibody (1:50,000, Abcam ab6046), HRP conjugated secondary antibodies (1:3,000, Bio-Rad) and IRDye infrared fluorescent dyes (1:10,000, LI-COR).
Inhibitors and treatments were used at the following concentrations: Pifithrin-α (Sigma) 10 μM, p38 inhibitor SB202190 (Calbiochem) 10 μM, ROCK inhibitor Y27632 (Sigma) 30 μM, Piezo inhibitor gadolinium III chloride (Sigma) 100 μM, S1P2 inhibitor JTE013 (Tocris Bioscience) 10 μM, tetracycline (Sigma) 5 μg ml−1 (except for pre-treating scribKD cells for RNA-seq analysis where it was used at 10 μg ml−1), doxycycline (Sigma) 1 μg ml−1, Nutlin-3 (Cayman Chemicals) was added as specified.
Cell culture and plasmids
The cell lines used in this publication are not listed in the database of misidentified cell lines maintained by ICLAC. MDCK cell lines were authenticated in our laboratory by RNA sequencing, which confirmed their canine origin. All cell lines used in this publication have been tested in our laboratory and were found to be negative for mycoplasma infection (EZ PCR Mycoplasma Test Kit, Geneflow).
Wild-type MDCK, MDCK-pTR E-cadherin shRNA (EcadKD)32 and MDCK-pTR scribble shRNA (scribKD) cells23 were a kind gift from Yasuyuki Fujita. Wild-type and E-cadKD MCDK cells were cultured in DMEM (21885; Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) in a humidified incubator at 37 °C with 5% CO2. The scribKD cells were cultured as described for wild-type cells with the addition of blasticidin 50 μg ml−1 (Sigma) and G418 800 μg ml−1 (Invitrogen) to the culture media. To establish MDCK cell lines that stably express a nuclear green fluorescent protein (GFP) or red fluorescent protein (RFP) marker, we used a modified pGIPZ-turboGFP-Puro (Thermo Scientific) plasmid where we replaced turbo-GFP with either GFP-NLS or RFP-NLS. Selection following infection was carried out with puromycin (0.65 μg ml−1, Sigma).
Resistant scribKD MDCK cells (scribRES) were generated by culturing MDCK-pTR scribble shRNA cells in 5 μg ml−1 tetracycline for 20 days. The resulting final population was expanded in the absence of tetracycline and tested for efficient Scribble knockdown by western blotting.
The MDCK p53−/− and scribKD p53−/− cells were generated using Cas9 D10A CRISPR technology. sgRNAs against canine TP53 were manually designed following published guidelines58
(p53_CRISPR#1_Fw: 5′-GCAGAAGTGGCTGGCATCCT-3′, p53_CRISPR#2_Fw: 5′-CCCTGGACCGGCCCCCTCC-3′).
sgRNAs were individually cloned into the PX461 vector58 and the pair was co-transfected into recipient cells. Pools of both p53−/− and scribKD p53−/− cells were generated by functional selection with Nutlin-3 (20–30 μM) for 5–7 days and either used immediately or expanded from single cells. p53 knockout was verified by functional tests.
To knock down E-cadherin expression in MDCK scribble shRNA cells (scribKD E-cadKD), the recipient cells were stably transfected with a modified version (pSUPERIOR.hygro+gfp E-cadherin shRNA) of a plasmid provided by Yasuyuki Fujita (pSUPERIOR.neo+gfp E-cadherin shRNA32). Selection was carried out with Hygromycin B (75 μg ml−1, Invitrogen). To overexpress E-cadherin in wild-type MDCK cells, the E-cadherin-GFP cDNA59 was introduced into a modified pTRIPZ-turboRFP-Puro plasmid (Thermo Scientific) in which turboRFP was replaced by RFP-NLS (pTRIPZ-RFP-NLS-Puro). The cells infected with the pTRIPZ ECAD-GFP-P2A-RFP-NLS construct express doxycycline-inducible E-cadherin-GFP and RFP-NLS as a single transcript. Selection following infection was carried out with puromycin (0.65 μg ml−1, Sigma).
Primary mouse tracheal epithelial cells (MTECs) were obtained from 5-month-old animals from 26R-Tomato (Gt(ROSA)26Sortm1(CAG-tdTomato*,-EGFP*)Ees) and p53-null (Trp53tm1tyj) strains, both of C57BL/6 background, using a protocol adapted from published methods60. Tracheas were dissected from the larynx to the bronchial main branches and collected in ice-cold DMEM:F12 (11330-32; Invitrogen) supplemented with a solution of 100 units ml−1 penicillin and 100 μg ml−1 streptomycin (Invitrogen). The muscle, vascular tissue and glands were then removed and the trachea cut into three to four rings. Each fragment was washed in phosphate-buffered saline (PBS) and then incubated in Dispase (BD Biosciences) at 7.5 Caseinolytic Units in PBS (total volume 450 μl per trachea) for 25 min at room temperature (RT). Tracheal fragments were then transferred into ice-cold DMEM:F12 and the sheets of epithelial tissue were peeled off. The epithelial sheets and medium were transferred to an ice-cold 1.5 ml tube, and pelleted twice at 500 g for 3 min with a PBS wash in between. The pellets were resuspended in 0.05% TE (Invitrogen) supplemented with 5 mM EDTA for 30 min at 37 °C. Five hundred microlitres of DMEM:F12 supplemented with 5% FBS was added to stop the reaction. The cells were pelleted (500 g, 3 min), resuspended in MTEC/Plus media and plated on collagen-coated (50 μg ml−1 rat tail collagen I (BD Biosciences)/0.02 M acetic acid) 24-well tissue culture inserts (BD Falcon) in MTEC/Plus media at approximately 5 × 104 cells per insert. The MTEC cells were cultured in MTEC/Plus media consisting of: DMEM:F12 basal media supplemented with a solution of 100 units ml−1 penicillin and 100 μg ml−1 streptomycin, 10 μg ml−1 insulin (Invitrogen), 5.5 μg ml−1 transferrin (Invitrogen), 6.7 μg ml−1 selenium (Invitrogen), 0.1 μg ml−1 cholera toxin (Sigma), 25 ng ml−1 epidermal growth factor (R&D Systems), 30 μg ml−1 bovine pituitary extract (Invitrogen), 5% FBS, 15 mM HEPES and 0.01 μM freshly added retinoic acid (Sigma) in a humidified incubator at 37 °C with 5% CO2.
Conditioned media and transwell assays
For competition-conditioned medium experiments, media were conditioned for 48 h by cultures of GFP-labelled scribKD and wild-type cells with (competition condition) or without (mock condition) tetracycline. Recipient GFP-labelled scribKD cells were pre-treated with tetracycline for 24 h and 10,000 cells per well were seeded into 24-well plates with 1 ml of conditioned medium. For transwell assays, GFP-labelled scribKD cells (10,000 per well) were seeded at the bottom of a 24-well plate. Co-cultures at a ratio of 1:10 (GFP-labelled scribKD: wild-type or GFP-labelled scribKD: scribKD) were plated onto a transwell insert (3.0 μm pore size-polyester membrane, Corning) at 1,500 cells per insert). All scribKD and GFP-labelled scribKD cells were pre-treated for 24 h with tetracycline before plating.
Fences system
Where applicable, immunofluorescence of cell competition was carried out in a 24-well plate using ‘fences' (Aix Scientifics, http://www.aix-scientifics.co.uk/en/fences.html). The system allows two different populations of cells to be seeded on either side of a silicone barrier, thus allowing experimental and control conditions to be cultured, treated, imaged and (where applicable) stained within the same well/coverslip.
Cell competition and contact-induced migration assays
Cell competition assays on MDCK cells were carried out in 24-well plate fences. Control cultures were plated in the centre of the fence (1,000 cells per fence) at a ratio of 1:10 GFP-labelled scribKD: scribKD. Competition cultures were seeded on the outside of the barrier (8,000 cells per fence) at a ratio of 1:10, scribKD: wild-type cells. The fences were removed approximately 5 h after plating and the culture medium was replaced with or without addition of tetracycline. Twenty hours later, the culture medium was replaced with phenol red-free DMEM (+10% FBS and 1% L-glutamine, Invitrogen) with or without tetracycline. Where specified, the chemical inhibitors were also added at this point, except for the ROCK, Piezo and S1P2 inhibitors, which were added after an additional 24 h. Live imaging was started 2–4 h after the final media change and was continued for at least 50 h with regular media changes every 2–3 days.
For cell migration assays, the cells were seeded as for competition assays, except that they were plated at a lower density (1,000 cells in the centre of the fence, 2,000–5,000 cells in the outer chamber) and imaged more frequently (every 10 min to every 2 h). Alternatively, where specified, cells for competition or migration assays were plated on gridded tissue culture plates (μ-Dish 35 mm Grid-500, Ibidi) at 6,250 cells per plate. This allowed us to find specific clones of cells for immunofluorescence analysis after live imaging had finished. Co-cultures, unless otherwise specified, were plated at a ratio of 1:10 (scribKD: wild-type).
The MTEC cells were plated in collagen-coated tissue culture inserts and allowed to grow for approximately 2 weeks until they reached homeostatic density before commencing experiments. In control cultures, wild-type (unlabelled) cells were plated with wild-type Rosa26R-Tomato (nuclear red) cells (MTEC Tomato)48 cells at a 2:1 ratio. In competition cultures, unlabelled p53-null cells (MTEC p53−/−) and wild-type Rosa26R-Tomato cells (MTEC Tomato) were plated at a 2:1 ratio. Nutlin-3 was added at 17.5 μM on day 3 of live imaging. The medium was changed every 2 days.
Homeostatic cell density assays
These assays were carried out in 96-well plates. GFP-labelled scribKD and scribKD p53−/− cells (8,000–12,000 cells per plate) were pre-treated with tetracycline for 12 h before plating where applicable, in the presence or absence of inhibitors. Where applicable, Nutlin-3 was added 24 h after plating. The whole well was then imaged and the individual images were stitched together for further analysis to obtain data on total cell number and density. The plates were imaged every 6 h and cell culture medium and drug treatments were changed every 2 days.
Micropattern assays
For assays in micropatterns, GFP-labelled scribKD cells with or without tetracycline pre-treatment (72 h) were plated onto circular adhesive patterns (Ø 800 μm) within a PEG cell-repellent surface26,61. The cells were flushed with culture medium 5 h after plating and the medium was replaced with phenol red-free DMEM (+10% FBS and 1% L-glutamine, Invitrogen) with or without tetracycline. Cell culture medium and drug treatments were changed every 2 days.
PDMS-based cell compression assays
The GFP-labelled scribKD cells were plated onto a stretched flexible silicone substrate (Gel pak PF-60-X4, 150 μm thickness, Teltek), held in a custom-made chamber (GREM; http://www.jove.com/video/51193/stretching-micropatterned-cells-on-a-pdms-membrane). Before plating, the clamped membranes were coated with 25 μg ml−1 fibronectin/PBS (Sigma) for 1 h at 37 °C. The membranes were stretched precisely by 2 cm, which provided a 57% stretch over the resting length (unless otherwise specified). A PDMS rectangular chamber, with two compartments (6.6 × 13 mm each) was attached to the membrane with Baysilone paste (GE Bayer). Two densities (low and high) of tetracycline pre-treated or Nutlin-3 pre-treated (48 h) GFP-labelled scribKD cells were plated, one in each compartment. High-density cells were plated between 75,000 and 120,000 cells per compartment, forming a confluent monolayer; low-density cells were plated at 25,000–35,000 cells per compartment. The cells were allowed to adhere for 24 h and then the membrane was released to induce compression. The cells were fixed in 4% PFA/PBS (3 h after release for p53 staining, 1.5 h for P-p38 staining and 5 h for cleaved Caspase-3 staining) and processed for immunofluorescence. As per design, low- and high-density cells were stained and imaged from the same stretcher avoiding sample-to-sample variability. The p38 inhibitor was added one day before plating; the ROCK inhibitor was added 1 h before releasing the membrane.
E-cadherin-blocking experiments
To block E-cadherin-dependent junctions, 20 h after the addition of tetracycline, the cells were incubated in 10 mM EDTA/PBS for 5 min, followed by a 20 min incubation in calcium-free DMEM (Invitrogen). The cells were then cultured in calcium-free DMEM (+10% FBS and 1% L-glutamine, Invitrogen), supplemented with an anti-E-cadherin-blocking antibody (1:200; ref. 57). Live-image analysis was started 2–4 h after the final media change and continued for at least 50 h.
Immunofluorescence
For immunofluorescence, the cells were cultured on glass coverslips or on gridded dishes (μ-Dish 35 mm Grid-500, Ibidi). The cells were fixed for 10 min in 4% PFA/PBS, quenched for 10 min in 50 mM NH4Cl/PBS and then permeabilized for 10 min with 0.1% Triton X-100/PBS. The cells were blocked in 2% BSA, 2% FBS/PBS for 30 min. Primary and secondary antibodies were diluted in blocking solution diluted 1:1 in PBS. The primary antibodies were incubated for a minimum of 1 h at RT, followed by washes in PBS; secondary antibodies were incubated for a minimum of 30 min at RT followed by washes in PBS. Coverslips were mounted with FluorSave (Millipore). For immunostaining against phosphorylated proteins, fixing solution was supplemented with PhosSTOP (1 tablet per 10 ml, Sigma), all PBS solutions were substituted with TBS, and blocking solution was substituted with 5% BSA/TBS. For surface immunostaining, the cells were washed in ice-cold phenol red-free DMEM (Gibco) and incubated with primary antibody diluted in ice-cold phenol red-free DMEM at 4 °C for 45 min. The cells were then washed in ice-cold PBS before fixation at RT in 4% PFA/PBS for 10 min. Secondary antibody staining was then carried out as outlined previously.
Western blotting
The cells were lysed in 1% SDS/PBS and 10–20 μg of protein was separated on a 4–12% gradient gel (Invitrogen) and transferred onto PVDF membrane for standard ECL blots (Anachem) or Immobilon FL PVDF for LI-COR blots (Millipore, 0.45 μm pore size). The membranes were blocked with 5% Marvel/0.05% Tween-20/PBS (PBST) for 1 h at RT, incubated in primary antibodies (in 2% Marvel/PBST) overnight at 4 °C, washed in PBST and incubated in secondary HRP-conjugated antibodies for ECL blots or Infrared fluorescent dyes (IRDye-800CW and IRDye-680RD) for LI-COR blots, diluted in 2% Marvel/PBST for 1 h at RT. After washes in PBST, the membranes were developed using standard ECL (GE Healthcare) or scanned with a LI-COR Odyssey CLx near-infrared fluorescence imaging system. Quantifications for LI-COR blots were carried out using Image studio lite (http://www.licor.com/bio/products/software/image_studio_lite/?gclid=COWG_Yze98MCFYvpwgodfR8Adg), using Actin to normalize the samples for loading. For original full blots, see Supplementary Fig. 9.
RNA-seq and differential expression analysis
RNA-seq libraries were prepared with the TrueSeq RNA sample preparation V2 kit (Illumina) according to the manufacturer's instructions, and sequenced on an Illumina HiSeq 2000 instrument in single-read mode at 36 or 40 base length. The resulting fastq files were filtered for low-quality reads (<Q20) and low-quality bases were trimmed from the ends of the reads (<Q20).
Genome-based RNA-seq mapping was carried out using Canis lupus familiaris 3.1 (NCBI/Dog Sequencing Consortium) as a reference genome. Transcript sequences were assigned to genome using BLAT62. The resulting mappings were filtered by a mismatch threshold (2%), as well as requiring 90% of the transcript to match the genome and all exons to match a single chromosome. This resulted in 21,571 transcripts mapping to the genome. This mapping was used as a junction file for Tophat 2 (ref. 63), which was used to map the RNA-seq reads to the genome. To provide gene names, transcript sequences were downloaded from the NCBI RefSeq database in March 2013 (24,538 sequences). Orthologues were found against the Mus musculus proteome (downloaded in January 2013—NCBI RefSeq) using Inparanoid64. The sequences were further annotated using InterProScan65 to provide both InterPro Domains and Panther ontology terms66. For differential expression, read counts were generated by quantifying overlaps with transcript locations. These were then used to generate RPKMs. Comparisons were made between pairs of conditions, each with at least four replicates. For a transcript to be included, counts per million had to be above 10 for all samples in at least one condition and within 2-fold between replicates. Differentially expressed transcripts were then called using EdgeR67. Hits were selected applying the following thresholds: P>0.05 log FC (fold change) >0.5. Gene Ontology terms over-represented among these lists were found using David Bioinformatics Resources68, in particular, KEGG pathway analysis.
Experiments in Drosophila wing discs
Flies were raised on a standard fly food containing yeast at 25 °C. The larvae were collected at wandering third instar stage. Clones in wing discs were induced either with en-FLP (Supplementary Fig. 5a,b) or with hs-FLP (with a 10-min heat-shock at 37 °C, 48–72 h before dissection; Supplementary Fig. 5c). For immunofluorescence, late third instar larvae were dissected in PBS followed by fixation in 4% formaldehyde/PBS solution for 20 min, permeabilization in 0.25% Triton X-100 PBS for 20 min and blocking in 4% FBS/PBS for 30 min. The primary and secondary antibodies were diluted in blocking solution. The primary antibodies were incubated overnight at 4 °C, followed by washes in 0.25% Triton X-100/PBS; secondary antibodies were incubated for a minimum of 1 h at RT followed by washes in 0.25% Triton X-100/PBS. The wing discs were mounted in Vectashield (Vector Laboratories) and imaged on Leica SP5 or SP8 confocal microscopes. Optical sections were acquired with 0.8μm steps. The images were processed in ImageJ and Photoshop. Genotypes: Supplementary Fig. 5a,b en-flp, Act>STOP>Gal4, UAS-GFP/+; UAS-Scrib-RNAi, Supplementary Fig. 5c (left) hs-FLP, tub-Gal4, UAS-GFP/+; +/+, FRT82, tub-Gal80/ FRT82, scrib2 and Supplementary Fig. 5c (right) hs-FLP, tub-Gal4, UAS-GFP/+; UAS-p53DN/+, FRT82, tub-Gal80/ FRT82, scrib2. Antibodies used: anti-Scribble (1:2000, from Chris Doe lab), anti-E-Cadherin (DSHB DCAD2, 1:200). Both the antibodies were raised against Drosophila antigens and have been previously used in Drosophila.
Imaging and image analysis
Quantifications shown in the figures are from a single representative experiment of a minimum of three independent repeats per experiment, unless otherwise specified in the figure legend.
Fixed samples were imaged with a Leica SP5 or SP8 confocal microscope. For live imaging, the cells (kept at 37 °C and 5% CO2) were imaged using a Nikon BioStation CT with a × 10 air objective with imaging frequency between every 10 min and every 6 h (as indicated in the movie time stamps), with media changes every 2–3 days. For each live imaging experiment, at least five fields were imaged by time lapse and analysed. Movie sequences or individual field time points that lost focus during the experiment were discarded from further analysis, as this precluded accurate cell counting.
For Fig. 1a–c and Supplementary Fig. 1d, quantifications of cell number over time were carried out using the open-source image analysis software Cell Profiler (http://www.cellprofiler.org/), using the nuclear GFP signal to segment cells.
To measure cell density in competition assays, the number of nuclei was manually counted using DAPI and/or nuclear GFP and divided by surface area, as calculated in Fiji (http://fiji.sc/Fiji; Fig. 1g and 2n, Fig. 5e, Supplementary Fig. 1e).
For Fig. 1k–n, cell number over time was quantified with the open-source image analysis software Icy69, using Icy Protocols. The nuclei were detected using the wavelet-based spot detection plugins. Areas with cysts, where nuclei were not visible due to overexposure or because they were out of focus, and areas without cells were excluded from both cell counting and from area measurements on a frame by frame basis. For Fig. 3h, Fig. 4c,k and Fig. 5b, entire wells of a 96-well plate were imaged by tiling. Individual images were then stitched and processed as above.
Cell height (μm, Fig. 1h–j) was measured from apical to basal membranes using the open image analysis software Fiji.
Cell death in compression assays was quantified as the number of activated caspase-3 positive death events (Fig. 1o–q, Fig. 3i, Fig. 4d,l and Fig. 5c) divided by total number of DAPI-positive nuclei.
Kymographs were generated by first registering the hyperstack containing the different channels (bright-field, RFP and GFP) to remove any global motion or drift over time of the cells (Fig. 2d). The position of the centroid for each nucleus at each time point was extracted and the RFP and GFP channels were then projected on the same mean direction of motion by re-slicing the stacks and then projecting the maximum intensity of the centroid. For the control kymographs (GFP-labelled scribKD: scribKD; RFP-labelled WT: WT), this was done only using the bright-field channel and one fluorescent channel.
The directionality index of migrating cells (D=Euclidean distance/total distance, Supplementary Fig. 2b,d) was calculated after manual tracking of individual cell nuclei (>40 cells and >5 movies per condition) with the aid of ImageJ (http://imagej.nih.gov/ij/). Data analysis was performed with Matlab (MathWorks Inc.).
Analysis of cell shape and migratory features (Fig. 2g–k). The aspect ratio, which is defined as the ratio of the long axis to the short axis of cells, was measured from three independent movies by manually fitting the best fit ellipses to single cells. To plot the distribution of angles between the cell's long axis and the cell's direction of motion, we determined for each cell the orientation of the cell's long axis and calculated its angular deviation with respect to the cell's direction of motion (data were binned at 40° interval), by manual tracking. To measure the displacement of wild-type and scribKD cells, we carried out PIV analysis, using a custom algorithm based on the MatPIV software package for MATLAB, and measured the displacement of individual cells within the first row of contact. Trajectories across different movies were adjusted so that the first time point where both populations formed a broad interface and moved concertedly was set at t=5 h. Distance is the projected distance along the axis that joins both cell populations.
In Figs 3c–e and 4e, local density was measured for each cell by taking the sum of the Gaussian-weighted distances to all other cells within 50 μm using a Gaussian function with σ=20.
Nuclear p53 (Fig. 3f,g), nuclear phospho-p38 (Fig. 4a,b,i,j) and nuclear p21 (Supplementary Fig. 4d) mean intensity was measured using Volocity (http://www.perkinelmer.co.uk/pages/020/cellularimaging/products/volocity.xhtml), using DAPI as a mask to segment the nuclei.
For Fig. 5f–I, quantifications of cell number over time were carried out in ImageJ (http://imagej.nih.gov/ij/), using the nuclear Tomato signal to segment cells.
In Supplementary Fig. 2h–i, E-cadherin staining intensity was measured using Fiji. The individual cells were manually selected on an average z stack projection and their integrated density values were recorded.
Statistical analysis
No statistical methods were used to predetermine sample size. Every experimental condition and treatment was carried out alongside a complete control set of experiments or no treatment control. The sample size was chosen to see a statistical difference between data sets. In the few instances where no difference was observed, sample size was at least as big as in conditions that had shown a difference. The experiments were not randomized and there was no blinding during experiments or analysis, as samples were marked. We carried out a minimum of independent three repeats for each experiment, unless otherwise specified in the figure legend.
The t-Test was used in Supplementary Fig. 1e and Supplementary Fig. 2b,d, where data were normally distributed and with equal variance. The Wicoxon rank-sum test was used in Supplementary Fig. 3f. Non-parametric Spearman correlation was used in Fig. 3e. Otherwise, the non-parametric KS test was used for all the statistical tests, removing the requirement for normally distributed data and equal variance. Throughout: *P<0.05, **P<0.005, ***P<0.0005.
Additional information
Accession codes: The RNAseq data generated in this study have been deposited into the GEO (Gene Expression Omnibus) database under the accession code GSE79042.
How to cite this article: Wagstaff, L. et al. Mechanical cell competition kills cells via induction of lethal p53 levels. Nat. Commun. 7:11373 doi: 10.1038/ncomms11373 (2016).
Supplementary Material
Supplementary Figures 1-9
Supplementary Figures 1-9
Genes differentially expressed upon silencing of scribble. List of differentially expressed genes between control cells (scribKD -TET) and scribKD cells +TET.
Genes differentially expressed between scribKD cells that are sensitive and scribKD cells that are resistant to out-competition. List of differentially expressed genes between scribRES cells and scribKD cells.
Intersection of the list of genes that are both differentially expressed between scribKD and control cells and between scribKD and scribRES cells. List of genes that are present in both Supplementary Data 1 and 2 i.e. genes that are differentially expressed between scribKD cells with respect to both control (scribKD -TET) and scribRES cells.
scribKD cells are out-competed by their wild-type neighbours but are viable as pure cultures. Left: Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells. Right: Time-lapse of co-culture of unlabelled and GFP labelled scribKD cells.
Only clones of scribKD cells that become fully surrounded by wild-type cells are eliminated through cell competition. Time-lapse of cell competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells. Competition is observed in surrounded scribKD cells (white arrow), but not in cells that are only contacted (black arrow).
Forcing scribKD cells above their natural density at confluency is sufficient to induce death and live cell extrusion. Time-lapse of GFP labelled scribKD cells growing on micropatterns (800μm ø), with (right) or without (left) the addition of tetracycline (TET). Movies show GFP labelled nuclei.
Upon contact, scribKD and wild-type cells engage in contact mediated migration. Time-lapse of co-culture of unlabelled wild-type (WT) and GFP labelled scribKD cells. Black asterisk marks the non-migrating end of the wild-type clone; magenta asterisk marks the initial point of contact between the two populations.
Homotypic cultures, of scribKD or wild-type MDCK cells, do not engage in contact mediated migration. Time-lapse of homotypic co-cultures of MDCK cells showing absence of contact mediated migration. Left: Co-culture of unlabelled and GFP labelled scribKD cells. Right: Co-culture of unlabelled and GFP labelled wild-type (WT) cells. Black lines mark the initial point of contact between the cell populations.
Particle image velocimetry shows wild-type cells migrating towards scribKD cells upon contact, before scribKD cells migrate away. Time-lapse of co-culture of unlabelled wild-type (WT) and GFP labelled scribKD cells, analysed with particle image velocimetry (PIV; shown by red arrows). Upon contact, WT cells begin migrating towards scribKD cells before scribKD cells start migrating away.
Contact mediated migration between scribKD and wild-type cells results in compaction and elimination of the scribKD cells. Time-lapse of co-culture of unlabelled wild-type (WT) and GFP labelled scribKD cells. Extended imaging time shows compaction and elimination of scribKD cells is a result of contact mediated migration.
Inhibiting E-cadherin-mediated cell contacts prevents contact mediated migration and delays the elimination of scribKD cells. Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells. Disrupting cell junctions by calcium withdrawal and an E-cadherin blocking antibody (right) prevents contact mediated migration and delays competition compared to control (left).
Reducing E-cadherin expression in scribKD cells down to wild-type levels prevents contact mediated migration. Time-lapse of co-culture of RFP labelled wild-type (WT) cells and unlabelled scribKD cells with E-cadherin knockdown (scribKD E-cadKD) showing absence of contact mediated migration. Black line represents the initial point of contact between the two populations.
Competition-resistant scribble cells (scribRES) are not eliminated by wild-type cells. Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribRES cells.
CRISPR knockout of p53 in scribKD cells protects them from out-competition by wild-type cells. Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells with knockout of p53 (scribKD p53-/-). The clone of scribKD p53-/- cells remaining at the end of the assay is outlined in white.
Inhibition of ROCK activity during competition prevents elimination of scribKD cells. Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells in presence of the ROCK inhibitor Y27632 (30μM).
Mild, sub-lethal elevation of p53 in wild-type MDCK cells is sufficient to induce loser status and activate cell competition. Left: Time-lapse of competition assay between GFP labelled wild-type (WT) cells and unlabelled WT cells with knockout of p53 (p53-/-) in presence of Nutlin-3 (8μM). Right: Homotypic culture of GFP labelled wild-type cells in presence of Nutlin-3 (8μM).
Wild-type MTECs with mild, sub-lethal p53 activation are eliminated specifically in the presence of p53-/- MTECs. Left: Time-lapse of co-cultures of unlabelled mouse tracheal epithelial cells (MTECs) and Tomato labelled MTECs. Right: Time-lapse of co-cultures of unlabelled p53-/- MTECs and Tomato labelled wild-type (WT) MTEC cells. Nutlin-3 (17.5μM) was added to both co-cultures when indicated.
Acknowledgments
This work was supported by a Cancer Research UK Programme Grant (E.P. and L.W. A12460), a Royal Society University Research fellowship to E.P. (UF0905080), a Wellcome Trust PhD studentship to I.K., a Cambridge Cancer Centre PhD studentship to M.G., a European Research Council (ERC) grant to R.E.C.S. (243283-SYSGRO), a BBSRC grant (R.E.C.S. and A.C. BB/K006320/1) and Core grant funding from the Wellcome Trust (092096) and CRUK (C6946/A14492). The ‘Biology-Inspired Physics at Mesoscales' group is a member of the IPGG Labex. The laboratoire PhysicoChimie Curie is member of the Labex CelTisPhyBio Labex. We thank: Yasuyuki Fujita for providing scribKD and E-cadKD MDCK cells and an E-cadherin shRNA construct; Richard Butler, Nicola Lawrence and Alex Sossick for help with microscopy and image analysis; Nicolas Carpi and Matthieu Piel for advice on the use of stretchers; James Nelson for E-cadherin constructs and discussions; Silvia Vivarelli for cloning pGIPZ-GFP-NLS and pGIPZ-RFP-NLS and researchers cited in methods for reagents. We apologize to the researchers whose work could not be cited due to space limitations.
Footnotes
Author contributions E.P. conceived and led the project. E.P. and LW designed the experimental strategy with help from RECS. M.G. characterized the role of ROCK and S1P2 and carried out assays in MTECs with help from E.L.R. M.G. and L.W. carried out the PDMS assays. K.K. carried out all cloning and, with help from M.G. and L.W., characterized the contribution of E-cadherin to contact-induced migration and loser status. M.G., C.R.B. and G.E.A. carried out RNAseq and data analyses. I.K. and L.W. carried out conditioned media and transwell experiments. I.K. carried out all Drosophila work. G.D. and P.S. generated micropatterns and analysed directional cell migration. A.C. and R.E.C.S. developed an algorithm for time-resolved cell density measurements of cell density. L.H.-O. and L.W. characterized scribRES cells and p21 expression. All the other experiments were carried out by L.W. E.P. and L.W. prepared the manuscript with help from K.K., M.G., R.E.C.S. and other co-authors.
References
- Morata G. & Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev. Biol. 42, 211–221 (1975). [DOI] [PubMed] [Google Scholar]
- Moreno E. & Basler K. dMyc transforms cells into super-competitors. Cell 117, 117–129 (2004). [DOI] [PubMed] [Google Scholar]
- la Cova, de C., Abril M., Bellosta P., Gallant P. & Johnston L. A. Drosophila myc regulates organ size by inducing cell competition. Cell 117, 107–116 (2004). [DOI] [PubMed] [Google Scholar]
- Oertel M., Menthena A., Dabeva M. D. & Shafritz D. A. Cell competition leads to a high level of normal liver reconstitution by transplanted fetal liver stem/progenitor cells. Gastroenterology 130, 507–520 (2006). [DOI] [PubMed] [Google Scholar]
- Rhiner C. et al. Persistent competition among stem cells and their daughters in the Drosophila ovary germline niche. Development 136, 995–1006 (2009). [DOI] [PubMed] [Google Scholar]
- Sancho M. et al. Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev. Cell 26, 19–30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavería C., Giovinazzo G., Sierra R. & Torres M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature 500, 39–44 (2013). [DOI] [PubMed] [Google Scholar]
- Tamori Y. & Deng W.-M. Tissue repair through cell competition and compensatory cellular hypertrophy in postmitotic epithelia. Dev. Cell 25, 350–363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins V. C. et al. Cell competition is a tumour suppressor mechanism in the thymus. Nature 509, 465–470 (2014). [DOI] [PubMed] [Google Scholar]
- Villa Del Campo C., Clavería C., Sierra R. & Torres M. Cell competition promotes phenotypically silent cardiomyocyte replacement in the Mammalian heart. Cell Rep. 8, 1741–1751 (2014). [DOI] [PubMed] [Google Scholar]
- Rhiner C. et al. Flower forms an extracellular code that reveals the fitness of a cell to its neighbors in Drosophila. Dev. Cell 18, 985–998 (2010). [DOI] [PubMed] [Google Scholar]
- Meyer S. N. et al. An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science 346, 1258236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino M. M. et al. Elimination of unfit cells maintains tissue health and prolongs lifespan. Cell 160, 461–476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levayer R., Hauert B. & Moreno E. Cell mixing induced by myc is required for competitive tissue invasion and destruction. Nature 524, 476–480 (2015). [DOI] [PubMed] [Google Scholar]
- Mamada H., Sato T., Ota M. & Sasaki H. Cell competition in mouse NIH3T3 embryonic fibroblasts is controlled by the activity of Tead family proteins and Myc. J. Cell Sci. 128, 790–803 (2015). [DOI] [PubMed] [Google Scholar]
- Li W. & Baker N. E. Engulfment is required for cell competition. Cell 129, 1215–1225 (2007). [DOI] [PubMed] [Google Scholar]
- de Beco S., Ziosi M. & Johnston L. A. New frontiers in cell competition. Dev. Dyn. 241, 831–841 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivarelli S., Wagstaff L. & Piddini E. Cell wars: regulation of cell survival and proliferation by cell competition. Essays Biochem. 53, 69–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J.-P., Fletcher A. G. & Baena-Lopez L. A. Mechanisms and mechanics of cell competition in epithelia 14, 581–591Nature Publishing Group (2013). [DOI] [PubMed] [Google Scholar]
- Baillon L. & Basler K. Reflections on cell competition. Semin. Cell Dev. Biol. 32, 137–144 (2014). [DOI] [PubMed] [Google Scholar]
- Moreno E. Cancer: Darwinian tumour suppression. Nature 509, 435–436 (2014). [DOI] [PubMed] [Google Scholar]
- Levayer R. & Moreno E. Mechanisms of cell competition: themes and variations. J. Cell Biol. 200, 689–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman M. et al. Loss of Scribble causes cell competition in mammalian cells. J. Cell Sci. 125, 59–66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo-Matsuda N. & Johnston L. A. Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc. Natl Acad. Sci. USA 104, 18543–18548 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Capaldo C., Gumbiner B. M. & Macara I. G. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J. Cell Biol. 171, 1061–1071 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deforet M., Hakim V., Yevick H. G., Duclos G. & Silberzan P. Emergence of collective modes and tri-dimensional structures from epithelial confinement. Nat. Commun. 5, 3747 (2014). [DOI] [PubMed] [Google Scholar]
- Eisenhoffer G. T. et al. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 484, 546–549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinari E. et al. Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature 484, 542–545 (2012). [DOI] [PubMed] [Google Scholar]
- Theveneau E. et al. Chase-and-run between adjacent cell populations promotes directional collective migration 15, 763–772Nature Publishing Group (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E. & Mayor R. Cadherins in collective cell migration of mesenchymal cells. Curr. Opin. Cell Biol. 24, 677–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohia M., Qin Y. & Macara I. G. The Scribble polarity protein stabilizes E-cadherin/p120-catenin binding and blocks retrieval of E-cadherin to the Golgi. PLoS ONE 7, e51130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C. et al. Characterization of the interface between normal and transformed epithelial cells 11, 460–467Nature Publishing Group (2009). [DOI] [PubMed] [Google Scholar]
- Zilfou J. T. & Lowe S. W. Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 1, a001883 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby A. M. & Richardson H. E. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 22, 5769–5779 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T., Pagliarini R. A. & Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr. Biol. 16, 1139–1146 (2006). [DOI] [PubMed] [Google Scholar]
- Igaki T., Pastor-Pareja J. C., Aonuma H., Miura M. & Xu T. Intrinsic tumor suppression and epithelial maintenance by endocytic activation of Eiger/TNF signaling in Drosophila. Dev. Cell 16, 458–465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa S. et al. Elimination of oncogenic neighbors by JNK-mediated engulfment in Drosophila. Dev. Cell 20, 315–328 (2011). [DOI] [PubMed] [Google Scholar]
- Cuadrado A. & Nebreda A. R. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 429, 403–417 (2010). [DOI] [PubMed] [Google Scholar]
- Huang C., Ma W. Y., Maxiner A., Sun Y. & Dong Z. p38 kinase mediates UV-induced phosphorylation of p53 protein at serine 389. J. Biol. Chem. 274, 12229–12235 (1999). [DOI] [PubMed] [Google Scholar]
- Bulavin D. V. et al. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 18, 6845–6854 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M. et al. Mechanical pressure-induced phosphorylation of p38 mitogen-activated protein kinase in epithelial cells via Src and protein kinase C. Biochem. Biophys. Res. Commun. 316, 673–679 (2004). [DOI] [PubMed] [Google Scholar]
- Amano M., Nakayama M. & Kaibuchi K. Rho-kinase/ROCK: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton 67, 545–554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S., Paterson H. F. & Marshall C. J. Cdc42–MRCK and Rho–ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat. Cell Biol. 7, 255–261 (2005). [DOI] [PubMed] [Google Scholar]
- Nakagawa K. et al. Cyclic compression-induced p38 activation and subsequent MMP13 expression requires Rho/ROCK activity in bovine cartilage explants. Inflamm. Res. 61, 1093–1100 (2012). [DOI] [PubMed] [Google Scholar]
- Rosenblatt J., Raff M. C. & Cramer L. P. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol. 11, 1847–1857 (2001). [DOI] [PubMed] [Google Scholar]
- Eisenhoffer G. T. & Rosenblatt J. Bringing balance by force: live cell extrusion controls epithelial cell numbers. Trends Cell Biol. 23, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004). [DOI] [PubMed] [Google Scholar]
- Prigge J. R. et al. Nuclear double-fluorescent reporter for in vivo and ex vivo analyses of biological transitions in mouse nuclei. Mamm. Genome 24, 389–399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shraiman B. I. Mechanical feedback as a possible regulator of tissue growth. Proc. Natl Acad. Sci. USA 102, 3318–3323 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Mouw J. K. & Weaver V. M. Forcing form and function: biomechanical regulation of tumor evolution. Trends Cell Biol. 21, 47–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P. & Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147, 992–1009 (2011). [DOI] [PubMed] [Google Scholar]
- Muller P. & Vousden K. H. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder D. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113–116 (2000). [DOI] [PubMed] [Google Scholar]
- Zhan L. et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 135, 865–878 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levayer R., Dupont C. & Moreno E. Tissue crowding induces caspase-dependent competition for space. Curr. Biol. 26, 670–677 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B. & Simons K. A functional assay for proteins involved in establishing an epithelial occluding barrier: identification of a uvomorulin-like polypeptide. J. Cell Biol. 102, 457–468 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D. & Kemler R. Rabbit antiserum against a purified surface glycoprotein decompacts mouse preimplantation embryos and reacts with specific adult tissues. Exp. Cell Res. 152, 169–178 (1984). [DOI] [PubMed] [Google Scholar]
- Ran F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams C. L. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J. Cell Biol. 142, 1105–1119 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Richer E. J., Huang T. & Brody S. L. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. Lung Cell Mol. Physiol. 283, L1315–L1321 (2002). [DOI] [PubMed] [Google Scholar]
- Tourovskaia A., Barber T., Wickes B. T., Hirdes D. & Grin B. Micropatterns of chemisorbed cell adhesion-repellent films using oxygen plasma etching and elastomeric masks. Langmuir 19, 4754–4764 (2003). [Google Scholar]
- Kent W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyenko A., Tamas I., Liu G. & Sonnhammer E. L. L. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics 22, e9–e15 (2006). [DOI] [PubMed] [Google Scholar]
- Zdobnov E. M. & Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848 (2001). [DOI] [PubMed] [Google Scholar]
- Thomas P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2008). [DOI] [PubMed] [Google Scholar]
- de Chaumont F. et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods 9, 690–696 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-9
Supplementary Figures 1-9
Genes differentially expressed upon silencing of scribble. List of differentially expressed genes between control cells (scribKD -TET) and scribKD cells +TET.
Genes differentially expressed between scribKD cells that are sensitive and scribKD cells that are resistant to out-competition. List of differentially expressed genes between scribRES cells and scribKD cells.
Intersection of the list of genes that are both differentially expressed between scribKD and control cells and between scribKD and scribRES cells. List of genes that are present in both Supplementary Data 1 and 2 i.e. genes that are differentially expressed between scribKD cells with respect to both control (scribKD -TET) and scribRES cells.
scribKD cells are out-competed by their wild-type neighbours but are viable as pure cultures. Left: Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells. Right: Time-lapse of co-culture of unlabelled and GFP labelled scribKD cells.
Only clones of scribKD cells that become fully surrounded by wild-type cells are eliminated through cell competition. Time-lapse of cell competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells. Competition is observed in surrounded scribKD cells (white arrow), but not in cells that are only contacted (black arrow).
Forcing scribKD cells above their natural density at confluency is sufficient to induce death and live cell extrusion. Time-lapse of GFP labelled scribKD cells growing on micropatterns (800μm ø), with (right) or without (left) the addition of tetracycline (TET). Movies show GFP labelled nuclei.
Upon contact, scribKD and wild-type cells engage in contact mediated migration. Time-lapse of co-culture of unlabelled wild-type (WT) and GFP labelled scribKD cells. Black asterisk marks the non-migrating end of the wild-type clone; magenta asterisk marks the initial point of contact between the two populations.
Homotypic cultures, of scribKD or wild-type MDCK cells, do not engage in contact mediated migration. Time-lapse of homotypic co-cultures of MDCK cells showing absence of contact mediated migration. Left: Co-culture of unlabelled and GFP labelled scribKD cells. Right: Co-culture of unlabelled and GFP labelled wild-type (WT) cells. Black lines mark the initial point of contact between the cell populations.
Particle image velocimetry shows wild-type cells migrating towards scribKD cells upon contact, before scribKD cells migrate away. Time-lapse of co-culture of unlabelled wild-type (WT) and GFP labelled scribKD cells, analysed with particle image velocimetry (PIV; shown by red arrows). Upon contact, WT cells begin migrating towards scribKD cells before scribKD cells start migrating away.
Contact mediated migration between scribKD and wild-type cells results in compaction and elimination of the scribKD cells. Time-lapse of co-culture of unlabelled wild-type (WT) and GFP labelled scribKD cells. Extended imaging time shows compaction and elimination of scribKD cells is a result of contact mediated migration.
Inhibiting E-cadherin-mediated cell contacts prevents contact mediated migration and delays the elimination of scribKD cells. Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells. Disrupting cell junctions by calcium withdrawal and an E-cadherin blocking antibody (right) prevents contact mediated migration and delays competition compared to control (left).
Reducing E-cadherin expression in scribKD cells down to wild-type levels prevents contact mediated migration. Time-lapse of co-culture of RFP labelled wild-type (WT) cells and unlabelled scribKD cells with E-cadherin knockdown (scribKD E-cadKD) showing absence of contact mediated migration. Black line represents the initial point of contact between the two populations.
Competition-resistant scribble cells (scribRES) are not eliminated by wild-type cells. Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribRES cells.
CRISPR knockout of p53 in scribKD cells protects them from out-competition by wild-type cells. Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells with knockout of p53 (scribKD p53-/-). The clone of scribKD p53-/- cells remaining at the end of the assay is outlined in white.
Inhibition of ROCK activity during competition prevents elimination of scribKD cells. Time-lapse of competition assay between unlabelled wild-type (WT) and GFP labelled scribKD cells in presence of the ROCK inhibitor Y27632 (30μM).
Mild, sub-lethal elevation of p53 in wild-type MDCK cells is sufficient to induce loser status and activate cell competition. Left: Time-lapse of competition assay between GFP labelled wild-type (WT) cells and unlabelled WT cells with knockout of p53 (p53-/-) in presence of Nutlin-3 (8μM). Right: Homotypic culture of GFP labelled wild-type cells in presence of Nutlin-3 (8μM).
Wild-type MTECs with mild, sub-lethal p53 activation are eliminated specifically in the presence of p53-/- MTECs. Left: Time-lapse of co-cultures of unlabelled mouse tracheal epithelial cells (MTECs) and Tomato labelled MTECs. Right: Time-lapse of co-cultures of unlabelled p53-/- MTECs and Tomato labelled wild-type (WT) MTEC cells. Nutlin-3 (17.5μM) was added to both co-cultures when indicated.