Abstract
Pyrosequencing-based analyses revealed significant effects among low (N50), medium (N80), and high (N100) fertilization on community composition involving a long-term monoculture of lettuce in a greenhouse in both summer and winter. The non-fertilized control (CK) treatment was characterized by a higher relative abundance of Actinobacteria, Acidobacteria, and Chloroflexi; however, the average abundance of Firmicutes typically increased in summer, and the relative abundance of Bacteroidetes increased in winter in the N-fertilized treatments. Principle component analysis showed that the distribution of the microbial community was separated by a N gradient with N80 and N100 in the same group in the summer samples, while CK and N50 were in the same group in the winter samples, with the other N-level treatments existing independently. Redundancy analysis revealed that available N, NO3−-N, and NH4+-N, were the main environmental factors affecting the distribution of the bacterial community. Correlation analysis showed that nitrogen affected the shifts of microbial communities by strongly driving the shifts of Firmicutes, Bacteroidetes, and Proteobacteria in summer samples, and Bacteroidetes, Actinobacteria, and Acidobacteria in winter samples. The study demonstrates a novel example of rhizosphere bacterial diversity and the main factors influencing rizosphere microbial community in continuous vegetable cropping within an intensive greenhouse ecosystem.
Nitrogen (N) is the key limiting nutrient for most crops and terrestrial ecosystems. In order to reach maximal crop yields, excessive N fertilizers are often used, especially in developing countries1. China has been the world’s greatest consumer of N fertilizer since 1985. By 2005, China accounted for 38% of global N application, while possessing only 9% of the world’s arable land2. Despite new information regarding the negative effects of excessive N application, some extension staff and farmers still believe that high yield cropping systems require high amounts of N fertilizer3. Currently, the annual application rate of synthetic N is about 550–600 kg of N per hectare in the summer rice/winter wheat double-cropping systems in the Taihu region. On the North China Plain, the annual application rate of synthetic N has reached 550–600 kg of N per hectare in winter wheat and summer maize rotation systems4,5,6.
Vegetable production in China has increased over the last 20 years. During this period, some of the area devoted to cereal production was converted to intensive vegetable cultivation in plastic greenhouses. The greenhouses are able to produce three or four harvests of more profitable vegetables per year than the traditional rotation of rice/wheat (or rape)7. Compared with cereal production, vegetable production usually requires a greater degree of management and larger input of nutrients and irrigation, and these systems are not sustainable in the long term8. Nitrogen fertilization is as high as 500–1900 kg N ha−1yr−1 in China and can even be more than 3000 kg N ha−1yr−1 in some vegetable cultivation regions in North China9,10,11. In Shouguang, a typical greenhouse vegetable production region in Shandong Province, northern China, excessive fertilizers are commonly combined with organic manure applications to give total nitrogen (N) inputs of >1000 kg N ha−1 for maximum yields of vegetables4.
Large inputs of synthetic N fertilizer have severely disturbed soil quality12 and normal functioning of ecosystems13. Due to their ability to respond quickly to environmental changes, microorganisms are efficient bioindicators of soil biological characteristics14,15. Numerous studies have shown that changes in the size and activity of the soil microbial community are major contributors to soil degradation caused by agricultural management16,17,18. Soil organisms play important roles in maintaining soil health and quality and soil nutrient biogeochemical cycling, particularly in N transformation19,20,21. For example, both the abundance and the diversity of soil microbial communities are altered by increased N fertilization22,23,24,25 or land use intensification26. Recent studies have demonstrated N effects across a range of sites on both bacterial27,28, fungal communities29,30, and ammonia oxidizing bacteria31. Giagnoni et al. also found availability of different N forms was a dominant factor regulating activity and composition of the rhizosphere microbial communities. In addition, N supply rate could change the composition of bacterial, fungal and AOB communities32. However, the effects of excessive N inputs in vegetable production systems on the soil microbial community and their attendant functions have not been fully elucidated. Moreover, little is known about the influences of N management regimes on belowground soil microbial community properties in intensive monocultured greenhouse vegetable systems.
In our study, we established a greenhouse in which lettuce plants were grown continuously for 4 years with six crops per year. The bacterial communities inhabiting the rhizospheric soil were detected using pyrosequencing at different levels of N fertilization. The aims of this work were to: 1) investigate the effect of the N fertilizer application on microbial composition and structure and their changes in different seasons, and 2) explore key bacteria that are significantly correlated with N fertilization application in intensively cultivated greenhouse systems. We hypothesized that the bacterial diversity was different from other ecosystems due to the particular management present in greenhouse vegetable cultivation.
Results
Overall Diversity of Bacterial Communities
Total microbial communities were characterized by high-throughput pyrosequencing of rhizosphere soil from summer and winter samples in triplicate. Across all 24 samples (12 summer and 12 winter samples), a total of 1,213,034 high-quality sequences (670,908 sequences for summer and 542,126 for winter samples) were obtained after the low quality reads were removed. There were an average of 52,168, 70,601, 39,304 and 61,562 effective sequence reads for CK, N50, N80, and N100 in summer samples, and 42,258, 46,590, 41,373, and 50,485 effective sequence reads in winter samples for CK, N50, N80, and N100, respectively. The effective reads in the summer were more than the winter samples except for the N80 treatment (Table 1). The highest operation taxonomic unit (OTU) number was observed in no fertilizer control (CK) treatment in the winter sample, (2,305 at a distance cutoff level of 3%) (Table 1), while the lowest number of OTUs was obtained for the N50 treatment of the summer sample. There were no significant changes in OTUs across the N gradient in winter samples (P > 0.05). CK in winter showed the highest bacterial diversity among the eight treatments, as shown by the Shannon diversity and Chao 1 index, which are representative of bacterial phylotype richness and evenness levels (Table 1). The richness of CK treatments in either summer or winter samples was higher than other fertilized treatments (Table 1). In the summer samples, there was a significant decrease in Shannon diversity and the Chao 1 index in response to nitrogen addition between N50 and N80 (Duncan analysis, p < 0.05). However, in the winter samples, the N addition treatment did not significantly change Shannon diversity (Table 1).
Table 1. Summary of pyrosequence reads (Mean ± SE, n = 3).
| Samples | Effective Reads |
No. of OTUs* |
Shannon |
Chao1 |
||||
|---|---|---|---|---|---|---|---|---|
| Summer | Winter | Summer | Winter | Summer | Winter | Summer | Winter | |
| CK | 52168 ± 3277ab | 42258 ± 5938a | 1753 ± 34c | 2305 ± 179a | 8.68 ± 0.19c | 9.15 ± 0.03a | 2473 ± 32b | 3033 ± 424a |
| N50 | 70601 ± 9344b | 46590 ± 1751a | 1271 ± 35a | 2063 ± 24a | 7.22 ± 0.08a | 8.76 ± 0.04a | 1892 ± 30a | 2664 ± 42a |
| N80 | 39304 ± 3664a | 41373 ± 7851a | 1333 ± 78ab | 2061 ± 152a | 7.52 ± 0.29ab | 8.74 ± 0.15a | 1941 ± 93a | 2577 ± 260a |
| N100 | 61562 ± 4887b | 50485 ± 457a | 1497 ± 37b | 1959 ± 129a | 7.82 ± 0.04b | 8.57 ± 0.32a | 2269 ± 84b | 2522 ± 157a |
*Operational taxonomic units (OTUs) were defined at 97% sequence identity. 100N: normal application rate of N, P2O5, and K2O was 185, 46, and 185 kg ha−1, respectively; 80N: 0.8 times N a as in the 100N; 50N: 0.5 times N as in the 100N; CK: no N applied. Means followed by the same letter within a column are not significantly different as determined by the LSD test (P = 0.05).
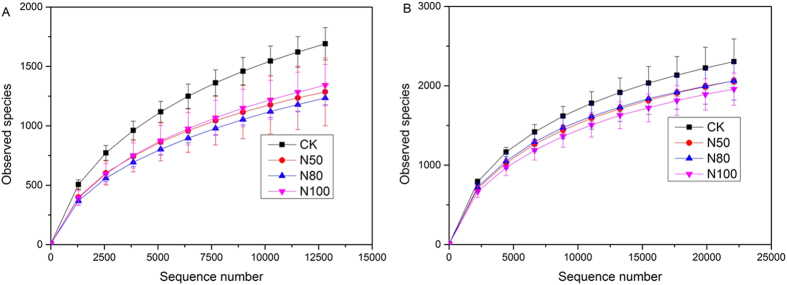
Rarefaction curves were used to estimate and compare bacterial diversity and richness among different treatments in summer and winter samples at a distance cutoff level of 3% as shown in Fig. 1. All amplified rarefaction curves increased rapidly from 0 to 1000 sequences, indicating that sequence-derived diversity and richness in this study were sufficient to characterize the species in each sample. The rarefaction curves showed that the bacterial richness were higher in the winter sample than those of summer samples. Compared to CK, treatments of N50, N80, and N100 consistently decreased the observed OTU richness (observed species) (Fig. 1).
Figure 1. Rarefaction curves of observed species of 16S rRNA genes from the summer.
(A) and winter (B) soil samples based on 97% similarity. The error bars of observed species indicate standard deviations of three replicates. 100N: normal application rate of N, P2O5, and K2O was 185, 46, and 185 kg ha−1, respectively; 80N: 0.8 times N as in the 100N; 50N: 0.5 times N as in the 100N; CK: no N applied.
Bacterial Community Composition in the rhizosphere
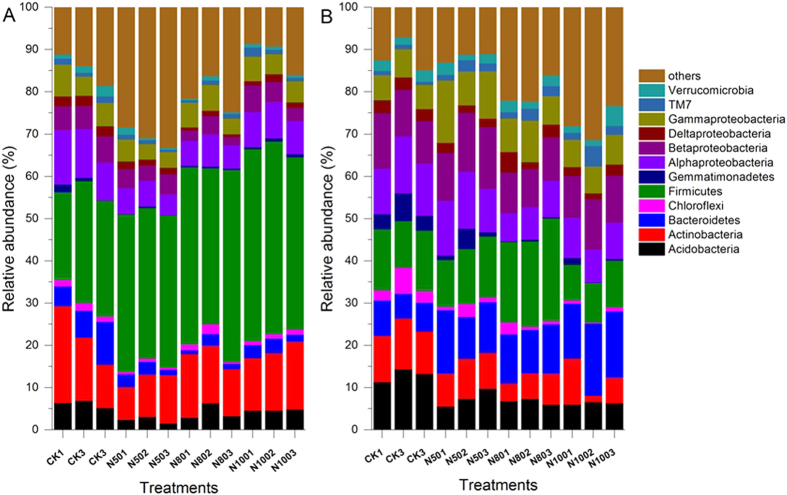
The effective bacterial sequences in the 24 samples were all assigned to corresponding taxonomies using BLAST combined with MEGAN. The dominant phyla across all the samples were Proteobacteria, Firmicutes, Actinobacteria, Acidobacteria, and Bacteroidetes, accounting for more than 65% of the bacterial sequences from each of the soils (Fig. 2, Tables S1 and S2). In addition, Verrucomicrobia, Gemmatimonadetes, Chloroflexi, and TM7 were present in most rhizosphere soils but at relatively low abundances, and 9 other more rare phyla were also identified (Tables S1 and S2).
Figure 2. Relative abundances of the dominant bacterial phylotypes in the summer.
(A) and winter (B) soil samples. Relative abundances are based on the proportional frequencies of DNA sequences that could be classified at the phylum level. 100N: normal application rate of N, P2O5, and K2O was 185, 46, and 185 kg ha−1, respectively; 80N: 0.8 times N as in the 100N; 50N: 0.5 times N as in the 100N; CK: no N applied.
In the summer soil samples, Firmicutes was on average the dominant phylum, accounting for 20.79–45.60% of the total 16S rRNA gene reads sequences, and the relative abundance of the Proteobacteria (11.95–28.71%) was second only to the Firmicutes (Fig. 2A, Table S1). However, the winter samples showed an opposite trend, the most dominant phylum was the Proteobacteria, amounting to 28.26–41.83%; and the second was Firmicutes, with the relative abundance of 8.46–24.26% (Fig. 2B, Table S2). The relative abundance of Acidobacteria and Bacteroidetes were lower in summer soil samples than in winter soil samples, but Actinobacteria abundance showed an opposite trend (Fig. 2, Tables S1 and S2).
The distribution of each phylum among the different treatments was evaluated. CK treatment was characterized by a higher average of abundance of Actinobacteria, Acidobacteria, and Chloroflexi (Fig. 2). We also compared the relative abundance in the same treatments of different seasons; the results showed that the dominant phyla were similar in summer and winter, although with different amounts (Fig.S1). In order to further to examine differences in bacterial community composition between summer and winter samples, a multiple-response permutation procedure (MRPP) was employed to compare the difference in the same treatment (with the same N fertilizer application) of summer and winter samples, but there was no significance in the same N level treatment from both seasons (P > 0.05). These results showed that the rhizosphere bacterial composition varied as the season changed and maintained similar trends between N-fertilized and non-N-fertilized CK.
Differences in Bacterial Communities among Fertilizations
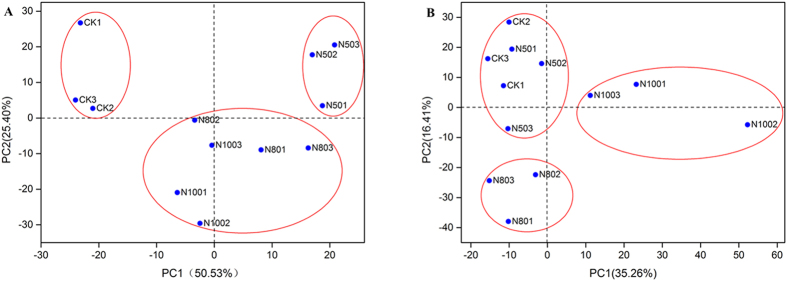
Principal component analyses (PCA) demonstrated clear distinctions in the bacterial communities under different fertilization management regimes in summer and winter soil samples (Fig. 3). In the summer samples, the first principal component (50.53% of contribution rate), which explains the majority of variations in the data, clearly separated all fertilized and unfertilized CK soils. The second principal component axis (25.40% of contribution rate) differentiated bacterial communities in controls and N50s from those in N80s and N100s, which suggests strong variations in bacterial community structure between N-deficient and N-rich fertilization soils (Fig. 3A). In the winter soil samples, both the first and second principal components mainly separated the community composition by differences in the application rates of N fertilizer which was the only difference among the treatments (Fig. 3B). In the summer samples, N-rich treatments (N80s and N100s) harbored similar bacterial community composition, while in the winter samples, the N-deficient treatments (CKs and N50s) formed a separated group (Fig. 3), and the rhizosphere microbial communities were well separated from the medium and high N treatments. These results suggest that the change in seasons also brings changes in the bacterial community structure.
Figure 3. Principal component analyses (PCA) of shifts in soil under different levels of N fertilization for in the summer.
(A) and winter (B) soil samples. 100N: normal application rate of N, P2O5, and K2O was 185, 46, and 185 kg ha−1, respectively; 80N: 0.8 times N as in the 100N; 50N: 0.5 times N as in the 100N; CK: no N applied.
Effect of Environmental Factors on Bacterial Community at Different Sampling Season
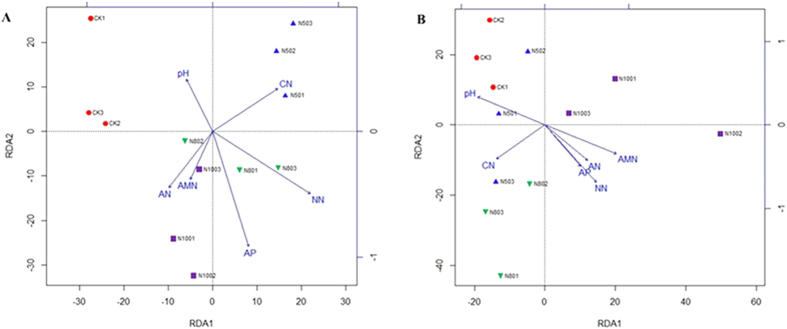
Redundancy analysis (RDA) was used to reveal what environmental factors shifted bacterial assemblages in rhizosphere. Environmental variables included pH value, C/N ratio (CN), available N (AN) and P (AP), nitrate N (NN), and ammonium N (AMN). Statistical analysis showed that environmental variables were different between control and long-term N-fertilized treatments (Fig. 4). In the summer samples, the nitrate N (NN) was the strongest factor (P = 0.002) that was correlated with the distribution of phyla. Available P (AP) also showed marginally significant correlations with community composition (P = 0.039), while the other factors were all non-significant (Fig. 4A). The relationship between environmental variables and microbial composition revealed that controls had higher pH value, while N50s were more related to C/N ratio. The higher contents of AN, AP, NN, and AMN were observed for N80 and N100 treatments, indicating the bacterial distribution in the rhizosphere was correlated with the application rate of N fertilizer (Fig. 4A). In the winter samples, the effect of environmental factors on the microbial community composition resulted in minor changes compared with summer samples. Available N (AN) and ammonium N (AMN) showed marginally significant correlations with community composition (P = 0.018 and P = 0.028, respectively) (Fig. 4B). Besides the CK treatment, pH also influenced the N50 treatment. The bacterial species in the N80s were positively affected by C/N ratio, but negative correlated with N100s (Fig. 4B). All of the results for both summer and winter samples indicated that the N fertilizer content shifted the distribution of the bacterial community, although the main factor influencing the changes was different.
Figure 4. Redundancy analysis (RDA) of the bacterial communities with environmental variables in different N-level fertilized soils for summer.
(A) and winter (B) soil samples. 100N: normal application rate of N, P2O5, and K2O was 185, 46, and 185 kg ha−1, respectively; 80N: 0.8 times N a as in the 100N; 50N: 0.5 times N as in the 100N; CK: no N applied. C/N: C/N ratio; pH: soil pH value; AP: available P; AN: available N; NN: NO3−-N; AMN: NH4+-N.
Relationship between N Gradient and Dominant Phyla
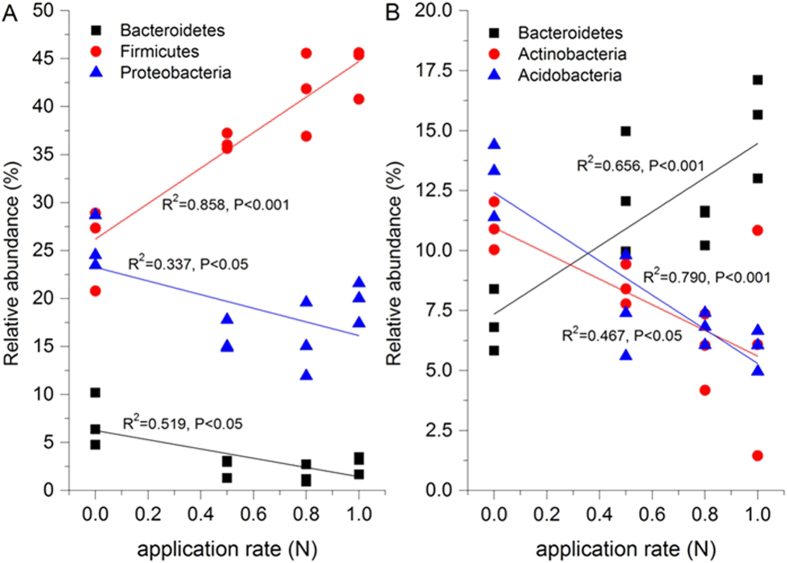
Linear regression analysis was employed to further examine the effect of the N application gradient on the relative abundances of the dominant bacterial phyla (Actinobacteria, Bacteroidetes, Acidobacteria, proteobacteria, and Firmicutes) in both summer and winter samples (Fig. 5). The N application rate was significantly positively correlated with Firmicutes (R2 = 0.858, P < 0.001), while it was also strongly negatively correlated with Bacteroidetes (R2 = 0.519, P < 0.05) and proteobacteria (R2 = 0.337, P < 0.05) in summer samples (Fig. 5A). In the winter samples, we observed a strong positive correlation between N gradient and Bacteroidetes (R2 = 0.656, P < 0.001), but a negative correlation with Actinobacteria (R2 = 0.467, P < 0.05) and Acidobacteria (R2 = 0.790, P < 0.001) (Fig. 5B).
Figure 5. The relationships between relative abundances of dominant bacterial groups and N gradients.
Linear regressions were used to test the correlation between the relative abundances of the taxa and N application. P < 0.05. 100N: normal application rate of N, P2O5, and K2O was 185, 46, and 185 kg ha−1, respectively; 80N: 0.8 times N as in the 100N; 50N: 0.5 times N as in the 100N; CK: no N applied.
Discussion
Soil microorganisms are vital to the environment due to their role in cycling mineral compounds, decomposing organic materials, and various soil biophysical processes33,34. In turn, soil microbial populations exhibit natural seasonal dynamics for their biodiversity and functions are influenced by various factors including vegetation, soil nutrients, and agricultural management33,35. In China, a large amount of traditional croplands have been recently transformed into high N input greenhouse vegetable production systems. Recently, significant effects have been observed from current soil management practices on soil quality and sustainability in the polytunnel greenhouse vegetable systems7,36. Intense management and excessive N application in the greenhouse fields inevitably creates microbial communities distinct from those found on traditional farming land or other ecosystems.
In the present study, a long-term fertilization experiment was conducted in greenhouse fields, in which the lettuce was mono-cultured six times each year with the same fertilizer regimes for four years. The summer and winter soil samples of the fourth year were collected separately. We observed that the soils from the CK treatment had higher levels of bacterial taxonomic diversity than the N application treatments exhibited by the Chao1 index, Shannon diversity, and rarefaction curve (Table 1; Fig. 1). Reduction in diversity in fertilized plots has also been observed in the arctic tundra27,37, suggesting that nitrogen enrichment may reduce niche space and therefore community diversity. In contrast, a study in another grassland ecosystem (Cedar Creek) observed no reduction in diversity in response to nitrogen enrichment, indicating that species richness responses to nitrogen enrichment may be site or soil specific38. More comprehensive cross-site studies are required to test this hypothesis.
Soil bacterial community composition is altered in response to N addition in both seasons. In the summer, Firmutes and many of the dominant Actinobacteria and Acidobacteria groups increased in relative abundance across the N gradient (Table S1, Fig. 2A); while in the winter, the relative abundance of Actinobacteria and Acidobacteria decreased, but Bacteroidetes increased with the N addition (Table S1, Fig. 2B). Similar taxon shifts have been found in other studies examining bacterial community responses to N addition. For example, Campbell et al.27 noted that Acidobacteria were relatively less abundant in fertilized samples, a pattern also observed by Wessen et al.28. A study of multiple terrestrial ecosystems showed that nitrogen enrichment significantly shifted bacterial communities39, suggesting that this may be a more general effect of N enrichment. One interpretation of this phenomenon may be that there are threshold responses in microbial community changes to enhanced soil nitrogen availability, and the changes associated with high level N application elicit the responses and accordingly drive the shifts in the bacterial community37.
Soil microorganisms play important roles in soil nutrient biogeochemical cycling, particularly in N transformation20. Compared with other cultivation systems, polytunnel greenhouse vegetable land in China is characterized with year-round cultivation and excessive N fertilizer application, which results in significant changes in soil chemical properties with accumulation of soil N, P, and K, as well as lower N use efficiency, and acidification as soil pH sharply decreased26. In our study, results showed that environmental variables were different between control and long-term fertilization treatments (Fig. 4). In the summer samples, controls had higher pH values, indicating pH may also be an important factor controlling soil microbial communities40. While N50s were more related to C/N ratio, the higher contents of available N, NO3−-N, and NH4+-N were observed for N80 and N100 treatments (Fig. 4A). In the winter samples, environmental factors created changes in microbial community composition compared with summer samples (Fig. 4B). The bacterial species in the N80s were affected by C/N ratio, while in the N100s, they were negatively influenced by C/N ratio (Fig. 4B). These results for both summer and winter samples further confirmed that the distribution of the bacterial community mainly induced by N nutrition in the soil originated from N fertilizer. Shen et al.26 also found that both the abundance and the diversity of soil microbial communities were altered by increased N fertilization or land use intensification.
The correlation between the N gradient and dominant bacterial phyla was also investigated to examine the key bacteria that respond to N addition (Fig. 5). Interestingly, we found that the N level exhibited a distinct negative correlation with Bacteroidetes in the summer (R2 = 0.519, P < 0.05, Fig. 5A), but demonstrated a strong positive correlation between N gradient and Bacteroidetes in the winter (R2 = 0.656, P < 0.001, Fig. 5B). This may be due to the distinct differences in climatic conditions, such as temperature and humidity, which influence nutrient cycling and the interaction between plants and microorganisms in the soil ecosystem.
Season was a factor influence microbial community shift in the same plots, however, some phyla in the same N level between summer and winter had the similar shift trend, in which the bacterial communities receiving the highest levels of N addition were significantly different from the communities receiving intermediate levels or no added N (Fig. S1), and there was no significant difference between them when analyzed by the MRPP method (P > 0.05). The bacterial diversity, community composition, and structure variation in winter samples showed distinct characteristics compared with summer samples. These possible mechanisms need further investigation to explain soil microbial responses to N amendments.
The data presented here showed that N addition can significantly alter bacterial community diversity and structure, and successfully identified key bacteria phyla that respond to excessive N input in intensively cultivated greenhouse soils with a long-term monoculture of lettuce. However, a more detailed study would be required to explain the mechanisms in this process.
Methods
Site and Experimental Design
The experimental site was in a demonstration garden in Wangting, Jiangsu Province, China (36°26′N, 120°27′E). Average annual rainfall is 1100 mm with mean air temperatures of 2.5 and 28 °C in January and July, respectively. Soil was characterized by a pH of 5.58, and the contents of organic matter, available N and available P were 28.05, 147.06, and 9.82 mg kg−1, respectively, at the beginning of the experiment. The vegetable cultivar tested was lettuce (Lactuca sativa L. var. ramosa Hort.), which was continuously sown from the beginning of 2010 to the end of 2013 with harvests six times each year (The last soil sample was collected in February 2014). Twenty-four crops in total were harvested from the beginning of 2010 to the end of 2013. Each time, the normal application rate of N, P2O5, and K2O was 185, 46, and 185 kg ha−1, respectively (100N). The treatments consisted of four fertilizer regimes in a completely random plot design with three replicates including: (1) 100N (applied as urea, superphosphate, and potassium sulfate, respectively); (2) 80N (0.8 times N and the same rate of PK application as in the 100N treatment); (3) 50N (0.5 times N and the same rate of PK applied as in the 100N treatment); (4) CK (no N applied), Each plot has received the same fertilizer management since 2010. The plot size was 1.8 × 2.4 m2. In each plot, 96 plants of the same variety were planted after each harvest. The fertilizer was a one-time application.
Soil Sample Collection
The soil samples were separately collected twice at the time of the lettuce harvest in August 2013 (summer) and February 2014 (winter). The rhizospheric soil samples were collected as described by Li et al.41. Each soil sample was stored at 4 °C until it was brought to the lab for immediate processing. Soil samples were homogenized by filtering through a 2-mm sieve to remove aboveground plant materials, roots, and stones for downstream processing, including microbial genomic DNA extraction. For soil chemical parameter analysis, a soil auger was inserted to 20 cm soil depth in order to collect soil samples from each plot, and five soil samples were randomly collected from each plot. Soil samples were air dried at room temperature and sieved to pass a 2-mm mesh.
Soil Characteristics Analysis
All three replicates were used for soil chemistry analysis. Soil pH was determined with a glass electrode using a soil-to-water ratio of 1:2.5. Soil organic C (SOC) and total N (TN) were determined by dichromate oxidation42 and Kjeldahl digestion43. Total P was determined by the Vanado-Molybdate phosphoric yellow colorimetric procedure44. Available P was analyzed by the method of Olsen et al.45. Available N was estimated by the alkaline permanganate method suggested by Subbiah and Asija46. The concentrations of NH4+-N and NO3−-N in the soil samples were analyzed as described by Turner and Romero47.
Soil DNA Extraction
Genomic DNA was extracted from 0.5 g of each soil sample using a FastDNA SPIN Kit for soil (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s instructions. The extracted soil DNA was dissolved in 70 μL of TE buffer, quantified by spectrophotometry, and stored at −20 °C for further analysis. To isolate genomic DNA from endophytic microbial communities, tomato roots were washed under running water, and surface sterilized by immersion in 70% ethanol (60 s) followed by treatment in 0.1% (w/v) mercuric chloride for 10 min. Surface sterilization of the plant material was checked by rolling the sterilized plant material on nutrient agar plates, which were then incubated for up to 7 days at 28 °C.
PCR Amplification and Pyrosequencing
The V4 region of the 16S rRNA genes were amplified using the universal primers set 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACVSGGGTATCTAAT-3′) with the barcode48. All PCR reactions were carried out in 30 μL reactions with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM of forward and reverse primers, and about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, elongation at 72 °C for 60 s, and, finally, 72 °C for 5 min. The triplicate PCR amplicons were mixed with the same volume of 1× loading buffer (containing SYBR green) and subjected to electrophoresis on 2% agarose gels for detection. Samples with a bright main band of between 400–450 bp were chosen for further experiments. Then, mixture PCR products were purified with a GeneJET Gel Extraction Kit (Thermo Scientific).
Sequencing libraries were generated using NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations (index codes were added). The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina MiSeq platform and 250 bp/300 bp paired-end reads were generated. Sequence analysis was performed by the UPARSE software package using the UPARSE-OTU and UPARSE-OTUref algorithms49. In-house Perl scripts were used to analyze alpha (within samples) and beta (among samples) diversity. Sequences with ≥97% similarity were assigned to the same OTUs. We chose representative sequences for each OTU and used the RDP classifier to annotate taxonomic information for each representative sequence50. In order to compute alpha diversity, we rarified the OTU table and calculated three metrics: Chao1 estimates of speciesrichness; observed species estimates of the amount of unique OTUs found in each sample, and the Shannon index. Rarefaction curves were generated based on these three metrics.
Data Analysis
Cluster analysis was preceded by principal component analysis (PCA) to evaluate the degree of similarity between microbial communities associated with different samples. Redundancy analysis (RDA) was chosen to examine the relationship between the bacterial community and environmental factors. Multiple-response permutation procedure (MRPP) was performed with Bray-Curtis distances to examine the difference between the same treatment in the summer and winter. The PCA, RDA, and MRPP were calculated used assigned OTUs using R with “vegan” package51, which were performed at the phylum level. The significance of the effects of N level effects on microbial pyrosequencing reads and diversity was analyzed by analysis of variance (ANOVA) for repeated measures with the statistical software package SPSS 20.0 (SPSS Inc.). Statistical analyses were run in Origin 9.0.
Additional Information
How to cite this article: Li, J.-G. et al. Effect of different levels of nitrogen on rhizosphere bacterial community structure in intensive monoculture of greenhouse lettuce. Sci. Rep. 6, 25305; doi: 10.1038/srep25305 (2016).
Supplementary Material
Acknowledgments
This research was supported by national natural science foundation of China (No. 41201241), Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB15030301), Knowledge Innovation Program of the Institute of Soil Science, Chinese Academy of Sciences (Grant No. ISSASIP1634), China postdoctoral science foundation (2013 M531412).
Footnotes
Author Contributions J.-G.L. and Y.-H.D. designed the research. J.-G.L., L.L., J.-F.H. and J.-X.W. performed the experiments, J.-G.L. and M.-C.S. analyzed the data, J.-G.L. and Y.-H.D. wrote the manuscript.
References
- Good A. G. & Beatty P. H. Fertilizing Nature: A Tragedy of Excess in the Commons. Plos Biol. 9, e1001124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO. FAOSTAT. Food and Agriculture Organization of the United Nations (FAO), FAO Statistical Databases. Agriculture Data. (2006) FAO Rome, Italy. accessed 20 October 2006. (Available at: http://www.fao.org). [Google Scholar]
- Hou P. et al. Grain yields in relation to N requirement: Optimizing nitrogen management for spring maize grown in China. Field Crops Res. 129, 1–6 (2012). [Google Scholar]
- Ju X. T., Liu X. J., Zhang F. S. & Roelcke M. Nitrogen fertilization, soil nitrate accumulation, and policy recommendations in several agricultural regions of China. Ambio 33, 300–305 (2004). [DOI] [PubMed] [Google Scholar]
- Zhao R. F. et al. Fertilization and nitrogen balance in a wheat maize rotation system in North China. Agron J. 98, 938–945 (2006). [Google Scholar]
- Norse D., Powlson D. S. & Lu Y. Integrated nutrient management as a key contributor to China’s low-carbon agriculture. In Climate change mitigation in agriculture (eds Wollenberg E., Nihart A., Tapio-Bistrom M.-L. & Grieg-Gran M.) 347–359 (Earthscan, Abingdon, 2012). [Google Scholar]
- Shen W. S. et al. Higher rates of nitrogen fertilization decrease soil enzyme activities, microbial functional diversity and nitrification capacity in a Chinese polytunnel greenhouse vegetable land. Plant Soil 2, 137–150 (2010). [Google Scholar]
- Hao Z. P. et al. Excessive Nitrogen Inputs in Intensive Greenhouse Cultivation May Influence Soil Microbial Biomass and Community Composition. Commun. Soil Sci. Plant. Anal. 40, 2323–2337 (2009). [Google Scholar]
- Zhu J. H., Li X. L., Christie P. & Li J. L. Environmental implications of low nitrogen use efficiency in excessively fertilized hot pepper (Capsicum frutescens L.) cropping systems. Agric Ecosyst Environ 111, 70–80 (2005). [Google Scholar]
- Ju X. T., Kou C. L., Zhang F. S. & Christie P. Nitrogen balance and ground water nitrate contamination: comparison among three intensive cropping systems on the North China Plain. Environ Pollut 143, 117–125 (2006). [DOI] [PubMed] [Google Scholar]
- He F., Chen. Q., Jiang R., Chen X. & Zhang F. S. Yield and nitrogen balance of greenhouse tomato (Lycopersicum esculentum Mill.) with conventional and site-specific nitrogen managenment in Northern China. Nutr Cycl Agroecosyst 77, 1–14 (2007). [Google Scholar]
- Mäder P. A. et al. Soil fertility and biodiversity in organic farming. Science 296, 1694–1697 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang W. L., Wu S. X., Ji H. J. & Kolbe H. The current situation and controlling manures of non-point source pollutions in China (in Chinese with English abstract). Agri Sci China 37, 1008–1017 (2004). [Google Scholar]
- Avidano L., Gamalero E., Cossa G. P. & Carraro E. Characterization of soil health in an Italian polluted site by using microorganisms as bioindicators. Appl. Soil Ecol. 30, 21–33 (2005). [Google Scholar]
- Bell C. W. et al. Linking microbial community structure and function to seasonal differences in soil moisture and temperature in a Chihuahuan Desert grassland. Microbial Ecol. 58, 827–842 (2009). [DOI] [PubMed] [Google Scholar]
- Courty P., Smith P., Koegel S., Redecker D. & Wipf D. Inorganic Nitrogen Uptake and Transport in Beneficial Plant Root-Microbe Interactions. Crit Rev Plant Sci. 34, 4–16 (2015). [Google Scholar]
- Bell C. W., Asao C. S., Woik, F. B. & Wallenstein M. D. Plant nitrogen uptake drives rhizosphere bacterial community assembly during plant growth. Soil Biol Biochem. 85, 170–182 (2015). [Google Scholar]
- Ciccazzo S. et al. Safe-site effects on rhizosphere bacterial communities in a high-Altitude Alpine environment. Bio Med Res Int. 2014, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich J. J. & Myrold D. D. Community composition and activities of denitrifying bacteria from adjacent agricultural soil, riparian soil, and creek sediment in Oregon, USA. Soil Biol Biochem 36, 1431–1441 (2004). [Google Scholar]
- Leininger S. et al. Archaea predominate among ammoniaoxidizing prokaryotes in soils. Nature 442, 806–809 (2006). [DOI] [PubMed] [Google Scholar]
- Janvier C. et al. Soil health through soil disease suppression: which strategy from descriptors to indicators? Soil Biol Biochem 39, 1–23 (2007). [Google Scholar]
- Avrahami S. et al. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl Environ Microbiol 68, 5685–5692 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enwall K., Conrad R. & Braker G. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl Environ Microbiol 71, 8335–8343 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H. Y. et al. Community structure of ammonia-oxidizing bacteria under long-term application of mineral fertilizer and organic manure in a sandy loam soil. Appl Environ Microbiol 73, 485–491 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. Z. et al. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9, 2364–2374 (2007). [DOI] [PubMed] [Google Scholar]
- Shen W. S. et al. Land use intensification affects soil microbial populations, functional diversity and related suppressiveness of cucumber Fusarium wilt in China’s Yangtze River Delta. Plant Soil 306, 117–127 (2008). [Google Scholar]
- Campbell B. J., Polson S. W., Hanson T. E., Mack M. C. & Schuur E. A. G. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ Microbiol 12, 1842–1854 (2010). [DOI] [PubMed] [Google Scholar]
- Wessen E., Hallin S. & Philippot L. Differential responses of bacterial and archaeal groups at high taxonomical ranks to soil management. Soil Biol Biochem 42, 1759–1765 (2010). [Google Scholar]
- Frey S. D., Knorr M., Parrent J. L. & Simpson R. T. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. Forest Ecol Man 196, 159–171 (2004). [Google Scholar]
- Egerton-Warburton L. M., Johnson N. C. & Allen E. B. Mycorrhizal community dynamics following nitrogen fertilization: a cross-site test in five grasslands. Ecol Monogr 77, 527–544 (2007). [Google Scholar]
- Zhou X. G., Guan S. N. & Wu F. Z. Composition of soil microbial communities in the rhizosphere of cucumber cultivars with differing nitrogen acquisition efficiency. Appl Soil Ecol 95, 90–98 (2015). [Google Scholar]
- Giagnoni L. et al. Availability of different nitrogen forms changes the microbial communities and enzyme activities in the rhizosphere of maize lines with different nitrogen use efficiency. Appl Soil Ecol 98, 30–38 (2016). [Google Scholar]
- Critter S. A. M., Freitas S. S. & Airoldi C. Comparison of microbial activity in some Brazilian soils by microcalorimetric and respirometric methods. Thermochim Acta 410, 35–46 (2004). [Google Scholar]
- Wardle D. A. & Ghani A. Why is the strength of relationships between pairs of methods for estimating soil microbial biomass often so variable? Soil Biol. Biochem. 27, 821–828 (1995). [Google Scholar]
- Griffiths R. I., Bailey M. J., McNamara N. P. & Whiteley A. S. The functions and components of the Sourhope soil micrbiota, Appl. Soil Ecol. 33, 114–126 (2006). [Google Scholar]
- Shi W. M., Yao J. & Yan F. Vegetable cultivation under greenhouse conditions leads to rapid accumulation of nutrients, acidification and salinity of soils and groundwater contamination in South-Eastern China. Nutr Cycl Agroecosyst 83, 73–84 (2009). [Google Scholar]
- Coolon J. D. et al. Long-Term Nitrogen Amendment Alters the Diversity and Assemblage of Soil Bacterial Communities in Tallgrass Prairie. Plos ONE 8, e67884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Jones K. L., Todd T. C., Blair J. M. & Herman M. A. Comparative metagenomic, phylogenetic, and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez K. S., Craine J. M. & Fierer N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Change Biol 18, 1918–1927 (2012). [Google Scholar]
- Wallenstein M. D., McNulty S., Fernandez I. J., Boggs J. & Schlesinger W. H. Nitrogen Fertilization Decreases Forest Soil Fungal and Bacterial Biomass in Three Long-Term Experiments. For. Ecol. Manage. 222, 459–468 (2006). [Google Scholar]
- Li J. G., Gai D. R., Jiang Z. J. & Dong Y. H. Composition and Activity of Rhizosphere Microbial Communities Associated with Healthy and Diseased Greenhouse Tomatoes. Plant soil 380, 337–347 (2014). [Google Scholar]
- Mebius L. J. A rapid method for the determination of organic carbon in soil. Anal. Chim. Acta 22, 120–124 (1960). [Google Scholar]
- Nelson D. W. & Sommers L. E. Total carbon, organic carbon and organic matter. In Methods of Soil Analysis Part 2 539–580 (American Society of Agronomy, Madison, WI, 1982). [Google Scholar]
- Jackson M. L. Soil chemical analysis 151–154 (Prentice Hall Inc., Englewood Cliffs, New Jersey, 1960). [Google Scholar]
- Olsen S. R. et al. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate 18 (Circular No. 939, US Department of Agriculture, Washington, DC, 1954). [Google Scholar]
- Subbiah B. V. & Asija G. L. A. rapid procedure for estimation of available nitrogen in soils. Curr Sci 25, 259–260 (1956). [Google Scholar]
- Turner B. L. & Romero T. E. Short-term changes in extractable inorganic nutrients during storage of tropical rain forest soils. Soil Sci Soc Am J 73, 1972–1979 (2009). [Google Scholar]
- Caporaso J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Nati Acade Sci USA. 108, 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Meth 10, 996–998 (2013). [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J. et al. The vegan package. Community ecology package 631–637 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.