Abstract
Anthracnose, caused by the Colletotrichum species of fungi, is one of the most serious diseases affecting jute in China. The disease causes chlorotic regions with black brown sunken necrotic pits on the surfaces of stems. In late stages of disease, plants undergo defoliation, dieback and blight, which make anthracnose a major threat to jute fiber production and quality in China. In this study, 7 strains of Colletotrichum fungi were isolated from diseased jute stems from Zhejiang, Fujian, Guangxi, and Henan plantations in China. Multi-locus sequence (ACT, TUB2, CAL, GS, GAPDH and ITS) analysis coupled with morphological assessment revealed that C. fructicola, C. siamense and C. corchorum-capsularis sp. nov. were associated with jute anthracnose in southeastern China. C. fructicola and C. siamense were previously not associated with jute anthracnose. C. corchorum-capsularis is a new species formally described here. Pathogenicity tests confirmed that all species can infect jute, causing anthracnose, however the virulence of the 3 species differed. This report is the first associating these three species with jute disease worldwide and is the first description of the pathogens responsible for jute anthracnose in China.
Jute (Corchorus capsularis L.) is an annual bast fiber crop. It is found predominantly in Southeast Asia After cotton, jute is the second cheapest and second most commercially available fiber crop, making it an abundant source of biodegradable and renewable lignocellulose fiber1. Due in large part to its high luster, moisture absorption properties, ability for rapid water loss, and easy degradation, jute fibers have been exploited in various value-added products such as flooring and textiles2. However, jute is affected by a variety of diseases at all stages of its development, from seed germination to the harvested fruits. Anthracnose, caused by Colletotrichum species, has recently become the most serious disease of jute in China. This disease results in sunken necrotic lesions on the surfaces of stems that limit fiber productivity and reduce fiber quality.
Many species of the fungal genera Colletotrichum cause a variety of diseases in a wide range of economically important plants around the world3,4,5. Previously, the identification of Colletotrichum species was based on morphological characteristics3. Cai et al. and Cannon et al. found that such morphological identifications of the species of Colletotrichum depended on experimental methods used, which caused the taxonomy and nomenclature to be inconsistent4,6,7. Recently, Cai et al. recommended a polyphasic approach for accurate identification of Colletotrichum species using multi-locus phylogeny coupled with morphological data6,8. Using this approach, many Colletotrichum strains have been successfully identified and epitypified9,10,11,12,13,14,15,16. This increased understanding of Colletotrichum species can increase the effectiveness of plant disease control interventions7,8,14,17.
Prior to the polyphasic identification of Colletotrichum species, C. gloeosporioides and C. corchorum were generally recognized as the most important jute pathogens worldwide18,19. However, these identifications were based on inadequate techniques including examination of plant symptoms, assessment of the morphology of conidia produced on the infected tissues, or morphology on potato dextrose agar (PDA) cultures. Additionally, following the epitypification of C. gloeosporioides20, Phoulivong et al. reported that C. gloeosporioides sensu stricto was, in fact, not a common pathogen in the tropics21. In China, anthracnose of jute is attributed exclusively to the species C. gloeosporioides and C. corchorum, however there are no studies that perform molecular characterization of Colletotrichum species on jute. Therefore, this study was conducted to unambiguously identify the species of Colletotrichum that cause jute anthracnose by combining morphological and molecular approaches. Further, we aimed to determine the pathogenicity and distribution of the Colletotrichum species associated with jute anthracnose in China.
Materials and Methods
Sampling and spore isolation
From 2011–2012, jute stems showing symptoms of anthracnose were collected from plantations located in Zhejiang, Fujian, Guangxi and Henan provinces of China. 3 pieces (5 × 5 mm) of stem tissue from each plant were surface sterilized in 70% ethanol for 45 s followed by 1% NaClO for 1 min. Samples were then rinsed three times with sterilized water and dried on sterile tissue paper. Samples were placed on PDA and incubated at 25 °C for 2–4 days. Additionally, the leading edge of any fungal hyphae that grew from the tissues was transferred aseptically to PDA. Fungi were monitored for sporulation and spore masses were picked off with a sterilized wire loop and streaked on the surface of water agar. After incubation overnight at 25 °C, single germinated spores were picked up with a sterile needle and transferred to PDA13. Using the procedure described by Cai, et al., single spore cultures were obtained for each Colletotrichum isolate. These pure cultures were stored in sterilized water in Eppendorf tubes at 4 °C and stock cultures were stored in PDA slants at 4 °C in the dark.
Morphological studies of Colletotrichum from jute
Referred to the method described by Cai, et al., characterization of spore morphology and growth in culture were performed6. Mycelial discs (5 mm diameter) were taken from actively sporulating areas near the growing edge of cultures after 5 days of growth and transferred to PDA. Three replicate cultures of each isolate were incubated at 25 °C in the dark. After 7 days, colony diameter was measured and growth rate was calculated as the total growth divided by seven. Colony characteristics of conidial masses and zonation were also recorded6,15.
Appressoria were obtained by use of a slide culture technique in which 1 cm2 square of agar was inoculated on one side with conidia and then covered with a sterile cover slip6. The shape and size of the appressoria formed across the underside of the cover slip were studied after 5–7 days of incubation at 25 °C. Morphological data were analyzed using analysis of variance (P < 0.05) with Duncan’s Test.
DNA extraction, PCR amplification and Sequencing
Isolates were grown on PDA and incubated at 25 °C for 7 days. Mycelium was scraped from the colony surface using a sterile 10 μl pipette tip. Genomic DNA was extracted from the mycelium using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux®) according to the manufacturer’s protocol. DNA concentrations were estimated visually on a 1% agarose gel by comparing band intensity with a 1 kb DNA ladder (Transgen Biotech®).
Partial actin (ACT), calmodulin (CAL), β-tubulin (TUB2), glutamine synthetase (GS), glyceraldehyde-3-phosphate dehydrogenase (GPDH) genes and the complete rDNA-ITS (ITS) region from 7 Colletotrichum strains were amplified by PCR. Primer pairs for PCR amplifications were referred to the method described by Prihastuti, et al. The PCR products were examined by electrophoresis in 1% agarose gels, purified, and ligated into the pMD18-T vector (Takara, Japan). The vectors containing these gene fragments were transformed into Escherichia coli DH5α and DNA sequencing was performed by BGI Company, Shanghai, China. Sequences derived in this study are deposited in GenBank. The accession numbers of all sequences analyzed in this study are listed in Table 1.
Table 1. Strains of Colletotrichum with details of culture collection, references and GenBank accessions of the sequences generated.
ICMP, International Collection of Microorganisms from Plants (New Zealand); MFLUCC, Mae Fah Luang University Culture Collection (Thailand).
CBS, Centraalbureau voor Schimmelcultures (Netherlands); MAFF, Ministry of Agriculture, Forestry and Fisheries (Japan).
IMI, CABI Genetic Resource Collection (UK); GZAAS, Guizhou Academy of Agriculture Sciences (China);
FAFU, Colletotrichum strains collected in Fujian Agriculture and Forestry University (China); BCC, BIOTEC Culture Collection (Thailand);
*indicate the ex-type culures. New strains and accession numbers produced in this study are bold.
Phylogenetic analysis
Sequences from our isolates, together with reference sequences obtained from GenBank (Table 1), were aligned using ClustalW in MEGA v.522. The multi-locus dataset was subsequently aligned using MAFFT v.623, and manually adjusted using Notepad+ +when necessary. A maximum parsimony (MP) analysis was performed on the multi-locus alignment (ACT, CAL, GAPDH, GS, ITS, TUB2) using PAPU v.4.0b1024. All ambiguously aligned regions were excluded from analyses. Unweighted parsimony (UP) analysis was performed. Trees were inferred using the heuristic search option with Tree Bisection Reconnection (TBR) branch swapping and 1000 random sequence additions. Maxtrees were unlimited, branches of zero length were collapsed and all multiple parsimony trees were saved. Descriptive tree statistics were recorded, including tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI). Robustness of clades was assessed by a bootstrap analysis with 1000 replicates.
In addition, the Markov Chain Monte Carlo (MCMC) algorithm was used to regenerate the phylogenetic trees with Bayesian posterior probabilities in MrBayes v.3.2.125. MrModeltest v.2.326 was used to determine statistical selection of best-fit models of nucleotide substitutions. Two analyses of four MCMC chains were run from random trees for 10 million generations and sampled every 1000 generations. The first 25% of trees generated were discarded because they represented the burn-in phase of each analysis. The remaining trees were used for calculating the posterior probabilities in the majority rule consensus tree.
2 isolates were used in the initial MP analysis using a concatenated alignment for 4 genes: CAL, GAPDH, GS and TUB2. Colletotrichum boninense (MAFF 305972) was used as outgroup in this analysis. A second analysis was carried out to confirm the identity of five isolates with curved conidia based on a concatenated alignment of 6 genes: ACT, CAL, GAPDH, GS, ITS and TUB2. Colletotrichum lindemuthianum (CBS 151.28) was used as the outgroup in this second analysis. Phylogenetic trees were created in Figtree27.
Pathogenicity tests
The isolates that were characterized by morphology were also submitted to pathogenicity tests. Single spore isolates were incubated on PDA for 7 days at 28 °C. Conidial suspensions were prepared by adding 10 ml of sterile distilled water to the culture, swirling to isolate the conidia, and filtering through two layers of muslin cloth. Spore concentration was adjusted to 106 conidia/ml with sterile water using a hemocytometer. Jute leaves and stems without symptoms of disease were washed with tap water, surface disinfected in 75% ethanol for 60 sec and 1% sodium hypochlorite for 5 min, and then washed 3 times with sterile distilled water and dried in a fume hood. Spore suspensions, or sterile water for the negative control, were sprayed on the jute leaves. Stems were inoculated by using a sterile scalpel to create superficial wounds in the stem epidermis. The wound was then inoculated with a 5-mm-diameter PDA disk selected from the edge of an actively growing culture. Stems inoculated with sterile PDA were used as a negative control. The inoculated plants were kept in plastic containers, covered with plastic wrap to maintain humidity, and incubated at 28 °C with 12/12 h fluorescent light and darkness. 10 jute seedlings were inoculated for each species; the experiment was performed in triplicate. The incidence of infection was calculated by the formula [Incidence (%) = (infected sites or leaves/inoculated sites or leaves) × 100%] at 12-days post inoculation. The incidence data was analyzed using SPSS software version 20.0 (SPSS Inc., Chicago, USA).
Results
Collection of Colletotrichum species
In total, 7 Colletotrichum strains were isolated from diseased jute samples from the main jute growing regions (Zhejiang, Fujian, Guangxi and Henan provinces) of China. Based on the morphological characterization on PDA, 2 strains produced conidia similar to C. gloeosporioides. 5 strains produced curved conidia, which is typical of fungi in the C. truncatun species complex28.
Phylogenetic analysis
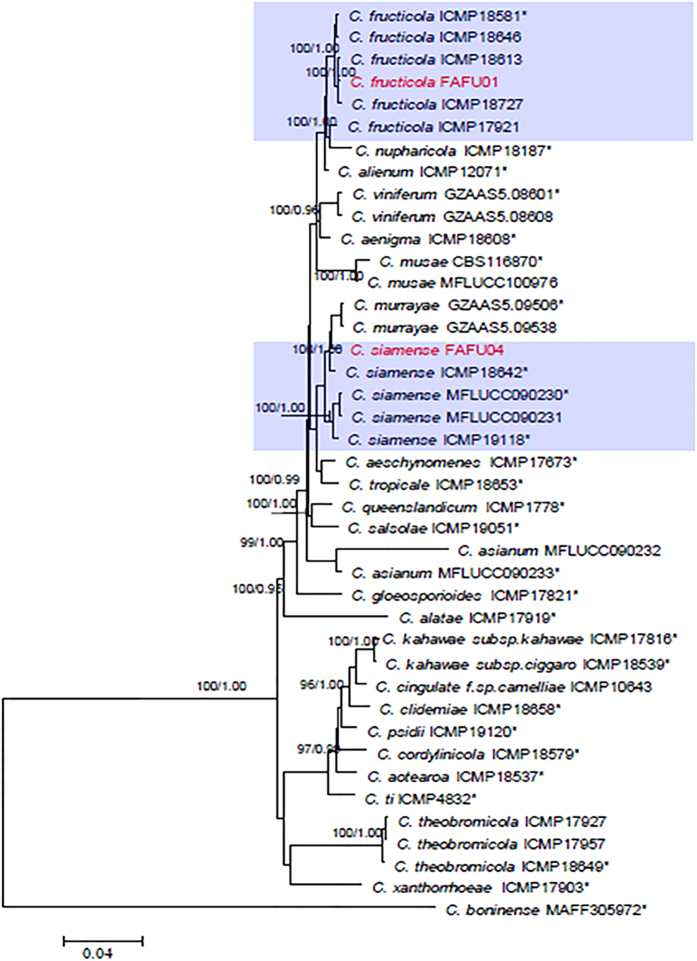
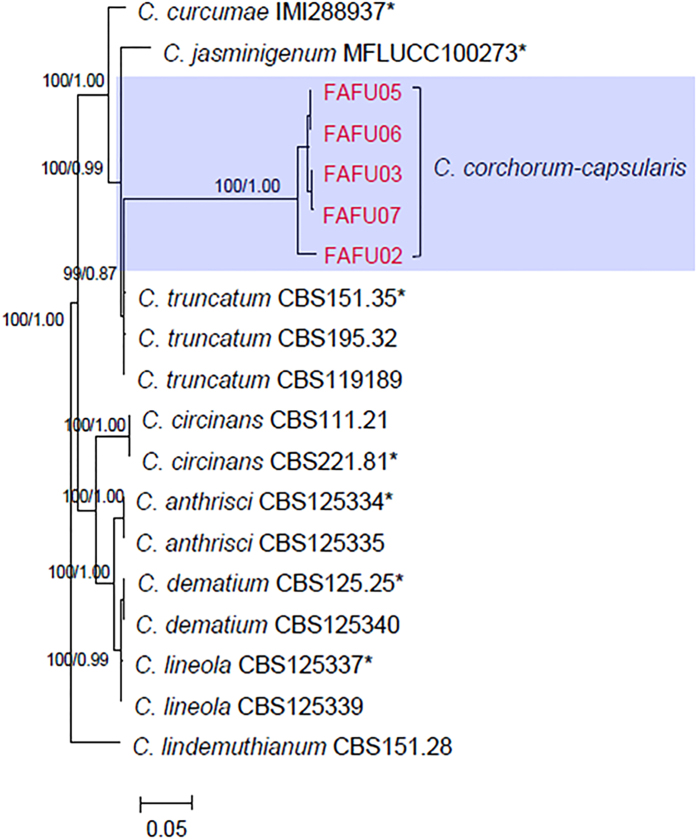
Molecular analyses were performed on all of the Colletotrichum strains isolated, including 2 strains from the C. gloeosporioides complex and 5 strains with curved conidia. Figure 1 shows the phylogram constructed to identify the strains in the C. gloeosporioides species complex. The strain, FAFU01, could be confidently identified as C. fructicola as it clustered together with the ex-epitype strain ICMP 18581 with 100% bootstrap support. Another strain, FAFU04, clustered with C. siamense strains with 100% bootstrap support, based on the combined datasets of partial CAL, GAPDH, GS and TUB2 sequence analysis. The other 5 strains did not cluster with any currently known species based on these 4 molecular markers. Therefore, a further 6 gene regions (ACT, CAL, GAPDH, GS, ITS and TUB2) of these five strains were sequenced and phylogenetic relationships were predicted using parsimony and Bayesian methods (Fig. 2). However, these 5 strains did not cluster well with any other Colletotrichum species in the 6 gene phylogenetic tree (Fig. 2). The morphological and culture characteristics were closest to the species C. corchorum previously reported18, so these 5 strains are described herein as C. corchorum-capsularis sp. nov.
Figure 1. Maximum parsimony tree obtained from a heuristic search of the combined CAL, GAPDH, GS and TUB2 sequence alignment.
Bootstrap support values ≥ 50% and Bayesian posterior probability values ≥ 0.5 are shown at the nodes. C. boninense was used as the outgroup. * indicates the ex-type strains. Strains isolated in this study are shown in red.
Figure 2. Maximum parsimony tree obtained from a heuristic search of the combined ACT, CAL, GAPDH, GS, ITS and TUB2 sequence alignments, showing the phylogenetic relationships of Colletotrichum species isolated from C. corchorum-capsularis.
Bootstrap support values ≥ 50% and Bayesian posterior probability values ≥ 0.5 are shown at the nodes. C. lindemuthianum was used as the outgroup. * indicates the ex-type strains. Strains isolated in this study are shown in red.
Taxonomy
Colletotrichum corchorum-capsularis
Xiaoping Niu, Hong Gao, Jianmin Qi, Miancai Chen and Jianguang Su, sp. nov. Fig. 3.
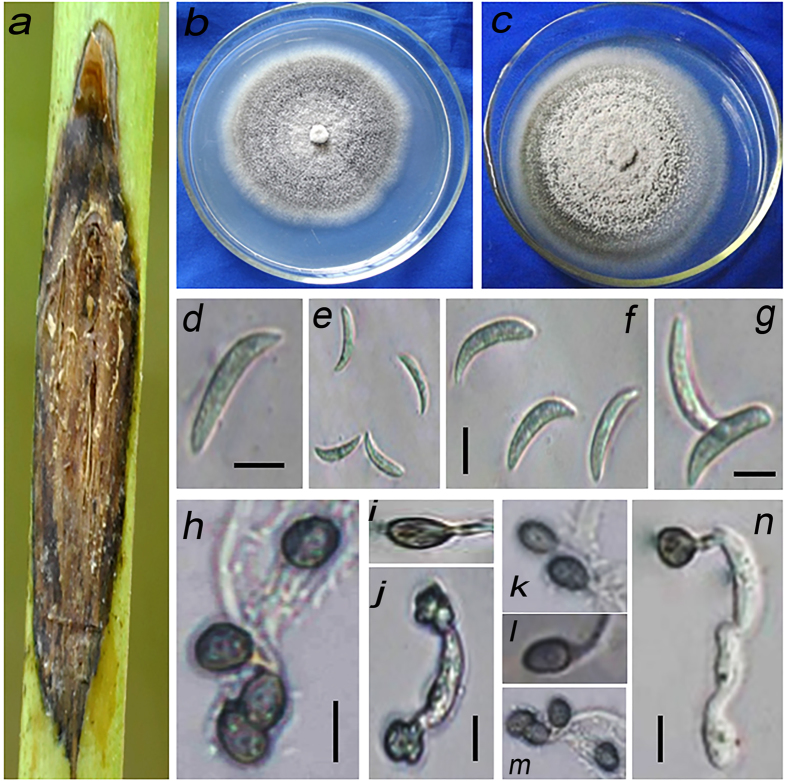
Figure 3. C. corchorum-capsularis (FAFU02).
(a) Symptom of anthracnose on the stem of jute. (b,c). Colony on PDA of different isolates of FAFU02. (d–g) Conidia. (h–n) Appressoria. Scale: (d–g) = 2 μm; (h–n) = 10 μm.
Fungal Names: FN570235.
Etymology
Named after its host, Corchorus capsularis.
When inoculated on PDA, colonies grew 6.5–10.5 mm/day in diameter at 28 °C. After 7 days, isolates with greyish white to dark gray mycelium and dense, concentric, circular conidia masses were observed.
Conidia
18.3–26.3 × 2.7–4.3 μm (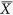 = 22.6 × 3.62 μm), hyaline, non-septate, smooth walled, curved, falcate-fusoid. Appressoria: 6.8–12.5 × 6.0–9.8 μm (
= 22.6 × 3.62 μm), hyaline, non-septate, smooth walled, curved, falcate-fusoid. Appressoria: 6.8–12.5 × 6.0–9.8 μm (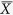 = 8.48 × 7.26 μm) diam, produced from mycelia, brown, ovoid to ellipsoidal. Sexual state was not observed (Table 2; Fig. 3).
= 8.48 × 7.26 μm) diam, produced from mycelia, brown, ovoid to ellipsoidal. Sexual state was not observed (Table 2; Fig. 3).
Table 2. Summary of morphological data of Colletotrichum isolates.
| Species and isolates | Conidia | Appressoria | Growth rate (mm/day)* | |||
|---|---|---|---|---|---|---|
| Length (μm)* | Width (μm)* | Shape | Length (μm)* | Width (μm)* | ||
| C. corchorum-capsularis (FAFU02) | 22.60 (±3.10) | 3.62 (±0.62) | Curved | 8.48 (±0.93) | 7.26 (±1.25) | 6.78 (±0.22) |
| C. corchorum-capsularis (FAFU03) | 22.35 (±3.12) | 3.25 (±0.50) | Curved | 8.33 (±1.87) | 7.23 (±1.36) | 6.76 (±0.38) |
| C. corchorum-capsularis (FAFU05) | 21.75 (±3.50) | 3.50 (±0.50) | Curved | 8.58 (±1.66) | 7.25 (±1.32) | 7.03 (±0.67) |
| C. corchorum-capsularis (FAFU06) | 22.50 (±2.75) | 3.50 (±0.50) | Curved | 8.76 (±1.01) | 7.35 (±0.67) | 6.61 (±0.44) |
| C. corchorum-capsularis (FAFU07) | 22.56 (±2.60) | 3.37 (±0.62) | Curved | 8.75 (±0.90) | 7.36 (±0.63) | 6.84 (±0.10) |
| C. fructicola (FAFU01) | 11.21 (±2.63) | 3.88 (±0.77) | Straight | 8.25 (±0.99) | 5.03 (±0.58) | 6.86 (±0.30) |
| C. siamense (FAFU04) | 11.91 (±2.89) | 3.78 (±0.88) | Straight | 8.75 (±1.75) | 6.38 (±1.97) | 9.02 (±0.34) |
*indicates all figures given in Table 2 are mean values.
Host: FAFU02, FAFU03, FAFU05, FAFU06 and FAFU07 were isolated from the stems of jute (Corchorus capsularis) that was black and withered.
Known distribution: Youxi and Zhaoan, Fujian Province; Xinyang, Henan Province and Xiaoshan, Zhejiang Province, China.
Material examined: CHINA, Fujian Province, Youxi and Zhaoan; Henan Province, Xinyang; Zhejiang Province, Xiaoshan, isolated from stems of Corchorus capsularis, 22–28 June 2013, Xiaoping Niu and Hong Gao, type culture FAFU02.
Notes: All 5 strains form a distinct clade with 100% bootstrap support, indicating that they represent a distinct species. Referring to their colony characteristics, C. corchorum-capsularis is introduced to accommodate this species. This species is similar to C. corchorum by its morphological characteristics and growth in culture. They both produced greyish white and cottony colonies, and grew 6.5–10.5 mm/day in diameter at 28 °C. However, the conidial length was longer (18.3–26.3 μm) than that from C. corchorum (12.0–25.0 μm).
Colletotrichum fructicola
Prihastuti, H., Cai, L. & Hyde, K.D. Fungal Diversity 39:96 (2009).
Material examined: CHINA, Fujian Province, Putian, isolated from stems of Corchorus capsularis, 20 June 2013, Xiaoping Niu and Hong Gao, culture FAFU01 = BPD-I18.
Notes: Colletotrichum fructicola was originally reported as a pathogen of coffee berries in Thailand10. This species was also known as a pathogen of Pyrus pyrifolia (Japan), Persea americana (Australia), Malus domestica (Brazil), Dioscorea (Nigeria), Theobroma and Tetragastris (Panama)15, Vitis (China)14, and Mangifera indica (Brazil)13. Strain FAFU01 in our study was identified as C. fructicola based on morphology and multi-locus (CAL, GAPDH, GS and TUB2) phylogenetic analysis. In the phylogram, the strain clustered with C. fructicola (ICMP 18581) with 100% bootstrap support and posterior probability values of 1.00 (Fig. 1).
Colletotrichum siamense
Prihastuti, H., Cai, L. & Hyde, K.D. Fungal Diversity 39:98 (2009).
Material examined: CHINA, Guangxi Province, Nanning, cultured from stems of jute, 16 June 2013, Xiaoping Niu and Hong Gao, culture FAFU04 = BPD-I2. THAILAND, Chiang Mai, Mae Lod Village, on Coffea arabica berries, 12 December 2007, Prihastuti, H., culture CBS 130417 = ICMP 18642 = MFLUCC 090230 = BPD-I2.
Notes: A detailed description of Colletotrichum siamense was provided by Prihastuti, et al. C. siamense was also reported as a pathogen of Hymenocallis sp. (China), Malus (USA), Jasminum (Vietnam), Dioscoria (Nigeria), Persea and Pistacia (Australia)15, and Proteaceae29. In the present study, C. siamense was isolated from stems of jute. The conidial shape and dimensions match the holotype of C. siamense10. However, the appressoria were (4.8–9.6 μm wide) slightly wider than that from ex-holotype culture (3.5–5.3 μm wide)10. In the phylogram, this strain clustered together with the type strain of C. siamense (ICMP18642) and strain MFLUCC090230 with bootstrap support/posterior probability values of 100%/0.99 and 100%/1.00, respectively (Fig. 1).
Pathogenicity testing
The pathogenicity of the Colletotrichum isolates was tested on both leaves and stems of jute to confirm Koch’s postulates. As shown in Table 3, the 3 species recovered in this study exhibited different virulence. Colletotrichum corchorum-capsularis strain FAFU02 was the most virulent on experimental leaves, with a mean infection incidence of 83%. Colletotrichum fructicola strain FAFU01 was also pathogenic to jute leaves with a mean infection incidence of 62%. Colletotrichum siamense strain FAFU04 infected experimental leaves with a lower mean infection incidence (58%) but this was not significantly different from strain FAFU01. As for lesion size on stems, C. corchorum-capsularis strain FAFU02 produced the largest lesions (mean length = 10.7 ± 1.70 mm, mean width = 6.4 ± 0.27 mm).
Table 3. Pathogenicity testing of Colletotrichum species from C.capsularis.
| Species and isolates | Mean infection incidence (%) | Lesion diameter of stems (mm) | ||
|---|---|---|---|---|
| Leaves | Stems | Mean Length | Mean Width | |
| C. corchorum-capsularis (FAFU02) | 83 | 100 | 10.7 ± 1.70 | 6.4 ± 0.27 |
| C. fructicola (FAFU01) | 62 | 100 | 7.5 ± 0.36 | 3.5 ± 0.25 |
| C. siamense (FAFU04) | 58 | 100 | 6.7 ± 0.51 | 3.6 ± 0.38 |
| control | 0 | 0 | – | – |
Discussion
Colletotrichum species on jute (C. capsularis) have been poorly studied, with reports focusing on C. gloeosporioides and C. corchorum18,19. Previous studies on Colletotrichum species causing disease on jute used morphological and culture characterizations which restrains identification to species complexes rather than individual species. The current study represents the first identification of Colletotrichum species associated with anthracnose of jute in China using a muti-locus phylogenetic approach. In this study, we isolated 7 Colletotrichum strains representing 3 distinct taxa, 2 of which have not been previously associated with jute disease. Although this investigation is limited in the sampling scale and isolations obtained, it appears that jute may harbor more Colletotrichum species than previously expected.
The most striking finding of this study was the absence of the C. gloeosporioides that was previously reported to be one of the main causal agents of jute anthracnose. However, 2 members of the C. gloeosporiodes species complex were newly associated with jute anthracnose, C. fructicola (located in Youxi, Fujian province) and C. siamense (located in Nanning, Guangxi province). Although C. fructicola and C. siamense were isolated only from symptomatic stems, pathogenicity tests showed that both species can also cause anthracnose on jute leaves. This could indicate that C. fructicola and C. siamense begin their lifecycles as endophytes and grow into opportunistic pathogens30.
Colletotrichum fructicola was previously found to be an important pathogen on a variety of hosts13,14,15, and was also found as a leaf endophyte in several plants13,31. However, this is the first report of C. fructicola causing jute anthracnose. Similarly, Colletotrichum siamense is another species that had not been thought to cause anthracnose in jute in southeastern China. This species was originally isolated from coffee berries in Thailand, and was biologically and geographically diverse15,29. Pathogenicity tests showed that this species can cause disease of both the leaves and stems of jute. Interestingly, a recent study by Sharma et al. of ApMat sequence data recognized several clades within C. siamense, suggesting C. siamense may be a species complex32,33. Although the strain FAFU04 resembles the type strain of C. siamense (ICMP18642) with bootstrap support/posterior probability values of 100%/0.99 (Fig. 1), further collections and investigations need to be conducted to gain a better understanding of its phylogenetic relationships and infraspecific variation. Colletotrichum corchorum-capsularis (FAFU02, FAFU03, FAFU05, FAFU06 and FAFU07) produced curved conidia (Fig. 3), which have similarity to species in the C. truncatum species complex. Phylogenetic analysis showed that these 5 strains with curved conidia formed a distinct clade with 100% bootstrap support, indicating that they represent a distinct species. The morphological characteristics of these 5 strains were most closely related to those of C. corchorum, as they both produced colonies of same color and growth rate at 28 °C. However, the conidial length of the former is significantly (P < 0.05) longer (18.3–26.3 μm) than that from C. corchorum (12–25 μm)18. Thus, Colletotrichum corchorum-capsularis is introduced by this study to accommodate this species.
Pathogenicity tests showed that 3 species were pathogenic to jute leaves and stems, and the virulence was significantly different. C. corchorum-capsularis was the most virulent species with a mean incidence of disease of 83% on leaves, while C. fructicola and C. siamense showed mild virulence (Table 3). Symptom development may vary considerably with factors such as species, inoculation conditions, humidity, temperature, and the concentration of the inoculum34,35. Therefore, this result may not reflect the true virulence potential of these species. Additional research should be conducted to determine the virulence potential of Colletotrichum species in natural infections rather than artificial inoculations.
In the present study, we have combined morphological and molecular data to identify the species of Colletotrichum that cause disease of jute (C. capsularis) in the most important jute producing areas of China. The most important causal agent was C. corchorum-capsularis. C. corchorum-capsularis encompasses the most virulent strains and appears to be responsible for most jute anthracnose in China (Fujian, Henan and Zhejiang provinces). C. corchorum-capsularis shows phylogenetic divergence and is probably a species complex; further work with more discerning genes is required to characterize the new species. This is the first report to link C. fructicola and C. siamense to jute anthracnose. Both caused disease in Fujian and Guangxi provinces. Pathogenicity tests showed that both species could cause disease at similar frequencies. C. gloeosporioides, which is reported to be the main pathogen for jute anthracnose, was not found in this study, possibly because we did not survey in the whole vegetative period, and the collected strains were only from the infected stems.
Additional Information
How to cite this article: Niu, X. et al. Colletotrichum species associated with jute (Corchorus capsularis L.) anthracnose in southeastern China. Sci. Rep. 6, 25179; doi: 10.1038/srep25179 (2016).
Acknowledgments
We are grateful to Prof. Yusen Chen who gave valuable suggestions to our work on Colletotrichum taxonomy. We also thank Dr. Fangluan Gao, who provided us with the PAPU v.4.0b10 software. This work was financed by the National Bast Fiber Research System of China (nycytx-19-E05). This work was also supported by a grant from the National Natural Science Foundation of China (31471549) and the National Bast Fiber Germplasm Resources Project of China (2013BAD01B03-13).
Footnotes
Author Contributions Conceived and designed the experiments: X.N., H.G., J.Q., M.C. and J.S. Performed the experiments: X.N., H.G. and A.T. Analyzed the data: X.N., A.T., J.X. and Z.D. Wrote the paper: X.N., M.C. and J.Q. Prepared tables and figures: X.N. Revised and approved the final version of the paper: X.N., J.Q. and J.S.
References
- Niu X. et al. Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in jute (Corchorus capsularis) under stress treatments. Front. Plant Sci. 6, 848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. Y. et al. Overexpression of UDP-glucose pyrophosphorylase gene could increase cellulose content in jute (Corchorus capsularis L.). Biochem. Biophy. Res. Co. 442, 153–158 (2013). [DOI] [PubMed] [Google Scholar]
- Hyde K. D. et al. Colletotrichum: a catalogue of confusion. Fungal Divers. 39, 1–17 (2009). [Google Scholar]
- Cannon P. F., Damm U., Johnston P. R. & Weir B. S. Colletotrichum - current status and future directions. Stud. Mycol. 73, 181–213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoulivong S. Colletotrichum, naming, control, resistance, biocontrol of weeds and current challenges. Curr. Res. Environ. Appl. Mycol. 1, 53–73 (2011). [Google Scholar]
- Cai L. et al. A polyphasic approach for studying Colletotrichum. Fungal Divers. 39, 183–204 (2009). [Google Scholar]
- Yan J.-Y. et al. Diverse species of Colletotrichum associated with grapevine anthracnose in China. Fungal Divers. 71, 233–246 (2014). [Google Scholar]
- Cai L. et al. The need to carry out re-inventory of plant pathogenic fungi. Trop. Plant Pathol. 36, 205–213 (2011). [Google Scholar]
- Moriwaki J. & Tsukiboshi T. Colletotrichum echinochloae, a new species on Japanese barnyard millet (Echinochloa utilis). Mycosci. 50, 273–280 (2009). [Google Scholar]
- Prihastuti H., Cai L., Chen H., McKenzie E. H. C. & Hyde K. D. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 39, 89–109 (2009). [Google Scholar]
- Shivas R. & Yu Y. A taxonomic re-assessment of Colletotrichum acutatum, introducing C. fioriniae comb. et stat. nov. and C. simmondsii sp. nov. Fungal Divers. 39, 111 (2009). [Google Scholar]
- Huang F. et al. Colletotrichum species associated with cultivated citrus in China. Fungal Divers. 61, 61–74 (2013). [Google Scholar]
- Lima N. B. et al. Five Colletotrichum species are responsible for mango anthracnose in northeastern Brazil. Fungal Divers. 61, 75–88 (2013). [Google Scholar]
- Peng L. J. et al. Colletotrichum species on grape in Guizhou and Yunnan provinces, China. Mycosci. 54, 29–41 (2013). [Google Scholar]
- Weir B. S., Johnston P. R. & Damm U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 73, 115–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikee S. et al. Colletotrichum species from jasmine (Jasminum sambac). Fungal Divers. 46, 171–182 (2010). [Google Scholar]
- Damm U., Cannon P. F., Woudenberg J. H. & Crous P. W. The Colletotrichum acutatum species complex. Stud. Mycol. 73, 37–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikata S. & Yoshida M. A new anthracnose of jute Plant. Ann. Phytopath. Soc. Japan 10, 141–149 (1940). [Google Scholar]
- Purkayastha R. & Sen-Gupta M. Studies on Colletotrichum gloeosporioides inciting anthracnose of jute. Indian phytopathol. 26, 650–653 (1975). [Google Scholar]
- Cannon P. F., Buddie A. G. & Bridge P. D. The typification of Colletotrichum gloeosporioides. Mycota. 104, 189–204 (2008). [Google Scholar]
- Phoulivong S. et al. Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Divers. 44, 33–43 (2010). [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. & Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinform. 26, 1899–1900 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0 b10. Sinauer Associates, Sunderland, Massachusetts. URL http://paup.csit.fsu.edu (2002). [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J. A. A. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, 2 (2004). [Google Scholar]
- Rambaut A. & Drummond A. FigTree v1. 3.1: Tree figure drawing tool. Institute of Evolutionary Biology, Edinburgh, UK. URL http://tree.bio.ed.uk/software/figtree. (2009) [Google Scholar]
- Damm U., Woudengerg J. H. C., Cannon P. F. & Crous P. W. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Divers. 39, 45–87 (2009). [Google Scholar]
- Liu F., Damm U., Cai L. & Crous P. W. Species of the Colletotrichum gloeosporioides complex associated with anthracnose diseases of Proteaceae. Fungal Divers. 61, 89–105 (2013). [Google Scholar]
- Promputtha I., Hyde K. D., McKenzie E. H. C., Peberdy J. F. & Lumyong S. Can leaf degrading enzymes provide evidence that endophytic fungi becoming saprobes? Fungal Divers. 41, 89–99 (2010). [Google Scholar]
- Rojas E. I. et al. Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panama: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycol. 102, 1318–1338 (2010). [DOI] [PubMed] [Google Scholar]
- Sharma G., Kumar N., Weir B. S., Hyde K. D. & Shenoy B. D. The ApMat marker can resolve Colletotrichum species: a case study with Mangifera indica. Fungal Divers. 61, 117–138 (2013). [Google Scholar]
- Sharma G., Pinnaka A. K. & Shenoy B. D. Resolving the Colletotrichum siamense species complex using ApMat marker. Fungal Divers. 71, 247–264 (2014). [Google Scholar]
- Simmonds J. A study of the species of Colletotrichum causing ripe fruit rots in Queensland. Queensland J. Agr. Anim. Sci. 22, 437–459 (1965). [Google Scholar]
- Freeman S., Katan T. & Shabi E. Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant dis. 82, 596–605 (1998). [DOI] [PubMed] [Google Scholar]