Abstract
Venoms of most Russian viper species are poorly characterized. Here, by quantitative chromato-mass-spectrometry, we analyzed protein and peptide compositions of venoms from four Vipera species (V. kaznakovi, V. renardi, V. orlovi and V. nikolskii) inhabiting different regions of Russia. In all these species, the main components were phospholipases A2, their content ranging from 24% in V. orlovi to 65% in V. nikolskii. Altogether, enzyme content in venom of V. nikolskii reached ~85%. Among the non-enzymatic proteins, the most abundant were disintegrins (14%) in the V. renardi venom, C-type lectin like (12.5%) in V. kaznakovi, cysteine-rich venom proteins (12%) in V. orlovi and venom endothelial growth factors (8%) in V. nikolskii. In total, 210 proteins and 512 endogenous peptides were identified in the four viper venoms. They represented 14 snake venom protein families, most of which were found in the venoms of Vipera snakes previously. However, phospholipase B and nucleotide degrading enzymes were reported here for the first time. Compositions of V. kaznakovi and V. orlovi venoms were described for the first time and showed the greatest similarity among the four venoms studied, which probably reflected close relationship between these species within the “kaznakovi” complex.
Keywords: snake venom, viper, Vipera kaznakovi, Vipera nikolskii, Vipera orlovi, Vipera renardi, proteome, mass-spectrometry
1. Introduction
Venomous snakes inhabit all continents of the globe except Antarctica. They are particularly abundant in tropical areas of Asia, Africa, South America and Australia. Russia, despite its large territory, is inhabited by only a small number of poisonous snake species, which belong to three genera: Gloydius, Macrovipera and Vipera. The Vipera genus is the most speciose in Russia and includes more than ten species, the systematics within this genus being constantly updated [1,2]. The most abundant species is common (or European) adder Vipera berus, which has a very large habitat in Russia, ranging from its western borders to Sakhalin and the Ussuri region. V. berus is also spread throughout Europe—between 68 and 45 degrees north latitude. The venom of this species is fairly well studied. Biological activities of this venom were characterized and proteolytic, fibrinolytic, anticoagulant, and phospholipolytic ones were demonstrated by in vitro experiments [3]. Several toxic proteins were isolated from V. berus venom, including phospholipase A2 (PLA2) [4], metalloproteinase (SVMP) [5], l-amino acid oxidase (LAAO) [6] and several others. Recently, we have partially characterized the steppe viper V. renardi venom, the PLA2s and Kunitz type protease inhibitors were isolated from this venom and sequenced [7]. The isolated PLA2s were studied in more details and found to exert their action both on lipid membranes [8] and on nicotinic acetylcholine receptor [9]. The venom of Nikolsky’s viper was also partially characterized and several proteins including heterodimeric neurotoxic PLA2s were identified [10,11]. The venoms of other Russian viper species are characterized very poorly. Thus, for Caucasian viper V. kaznakovi and Orlov’s viper V. orlovi, only the toxicity of venoms to insects was determined [12]. Here, we used proteomic chromato-mass-spectrometry analysis to obtain more detailed information on the composition of Russian viper venoms.
Modern proteomic analysis allows both qualitative and quantitative characterization of the venom proteins, leading to suggestions about venom biological effects. So far, among Vipera genus, the venoms of only three species, i.e., V. ammodytes, V. anatolica and V. raddei, were thus studied [13,14,15]. Semi-quantitative venom analysis of V. anatolica showed that the most abundant toxin family was SVMPs (41.5%), followed by two cysteine-rich secretory protein (CRISP) isoforms (15.9%); other proteins represented less than 10% per family [13]. SVMPs (31.6%) were also the most abundant in V. raddei venom, followed by PLA2s (23.8%), and, again, the contents of other toxin families did not exceed 10% each [14]. There is no quantitative analysis of the V. ammodytes venom, however monomeric and heterodimeric Group II PLA2s; serine proteinases (SVSPs); Group I, II, and III SVMPs; l-amino acid oxidases (LAAOs); CRISPs; disintegrins (Dis); and growth factors were found [15]. On the whole, the above data indicate that the composition of different viper venoms might be different. It should also be noted that V. raddei in some publications is classified as Montivipera raddei and attributed to Montivipera genus [16], thus some differences might be attributed to the discrepancy in classification. Using quantitative proteomic, we have studied the venoms from four Vipera species (V. kaznakovi, V. nikolskii, V. orlovi and V. renardi) that inhabit different regions of Russia. In contrast to the venoms of earlier studied Vipera species where the SVMP were found to be predominant [13,14,15], we have observed that the main components of the venoms studied are PLA2s, the content of which ranged between 24 and 65%.
2. Results
2.1. Venom Proteins Identification
In this work, venom proteomes and peptidomes for four species of Vipera snakes were analyzed. Venom proteomes were analyzed by LC-MS/MS after in-solution trypsin proteolysis. In total, for the four Vipera species venoms, the search against the Serpentes database resulted in the identification of 210 proteins (Table 1 and Table 2, and Tables S1 and S2): 116 proteins were identified in V. kaznakovi, 124 in V. renardi, 135 in V. orlovi and 111 in V. nikolskii venoms. Most proteins could be matched to previously reported snake toxins. To minimize individual variations, venoms from several individual animals were pooled for analysis [12].
Table 1.
List of proteins identified in Russian viper venoms.
| Protein No. in SuppData | Protein Name | Taxon | Protein Family 1 | MW, KDa | Seq Cov, % | V. nikolskii | V. kaznakovi | V. orlovi | V. renardi | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prot Abun INT, % | Seq Cov, % | Prot Abun INT, % | Seq Cov, % | Prot Abun INT, % | Seq Cov, % | Prot Abun INT, % | Seq Cov, % | ||||||
| 9 | Natriuretic peptide | Pseudonaja textilis | B-NAP | 14 | 9.8 | 0.007 | 9.8 | 0.029 | 9.8 | 0.012 | 9.8 | - | - |
| 2 | Hemoglobin subunit alpha | Vipera aspis | BP | 15 | 6.4 | 0 | 6.4 | - | - | - | - | 0 | 6.4 |
| 3 | Hemoglobin subunit beta-2 | Naja naja | BP | 16 | 7.5 | 0.016 | 7.5 | - | - | - | - | - | - |
| 4 | Hemoglobin subunit beta | Erythrolamprus miliaris | BP | 15 | 23.3 | 0.018 | 19.2 | - | - | - | - | 0 | 11.6 |
| 33 | Alpha globin | Hydrophis melanocephalus | BP | 16 | 19.7 | 0.011 | 9.2 | 0 | 9.2 | - | - | 0.010 | 19.7 |
| 34 | Alpha globin | Elaphe climacophora | BP | 11 | 25.2 | - | - | - | - | - | - | 0.032 | 25.2 |
| 116 | Murinoglobulin-2 | Ophiophagus hannah | BP | 143 | 2.7 | - | - | - | - | 0.008 | 1.8 | 0.002 | 2.2 |
| 124 | Serum albumin | Protobothrops flavoviridis | BP | 69 | 5.4 | 0.013 | 5.4 | - | - | 0.002 | 5.2 | - | - |
| 158 | Hemoglobin subunit alpha | Hydrophis gracilis | BP | 15 | 15.6 | 0.013 | 15.6 | 0.002 | 15.6 | - | - | 0.017 | 15.6 |
| 165 | Hemoglobin subunit alpha | Crotalus horridus | BP | 15 | 11.3 | 0.012 | 11.3 | 0.003 | 11.3 | - | - | 0.013 | 11.3 |
| 171 | Hemoglobin subunit beta-1 | Boiga irregularis | BP | 16 | 21.8 | 0 | 21.8 | - | - | - | - | 0 | 21.8 |
| 177 | Transferrin | Crotalus adamanteus | BP | 77 | 3.1 | 0.006 | 3.1 | - | 0.004 | 3.1 | 0.009 | 3.1 | |
| 199 | Hemoglobin subunit beta-2 | Thamnophis sirtalis | BP | 16 | 24.5 | 0.021 | 18.4 | - | - | - | - | 0.020 | 12.9 |
| 200 | Hemoglobin subunit beta-1 | Thamnophis sirtalis | BP | 16 | 19.7 | 0.002 | 19.7 | - | - | - | - | - | - |
| 206 | Serum albumin-like | Thamnophis sirtalis | BP | 57 | 8.0 | 0.012 | 8.0 | - | - | - | - | - | - |
| 180 | Cathepsin B-like protein | Crotalus adamanteus | CP | 37 | 5.0 | 0 | 5.0 | - | - | - | - | - | - |
| 7 | Cysteine-rich venom protein | Philodryas patagoniensis | CRISP | 1 | 64.3 | - | - | 0.037 | 64.3 | 0.002 | 64.3 | - | - |
| 18 | Cysteine-rich seceretory protein Dr-CRPK | Daboia russelii | CRISP | 26 | 24.7 | - | - | 0.170 | 24.7 | 0.107 | 21.8 | - | - |
| 19 | Cysteine-rich seceretory protein Dr-CRPB | Daboia russelii | CRISP | 25 | 17.6 | - | - | 0.033 | 17.6 | 0.006 | 15.8 | 0 | 17.6 |
| 89 | Cysteine-rich venom protein | Protobothrops jerdonii | CRISP | 26 | 22.5 | - | - | 0.148 | 22.5 | 0.149 | 22.5 | - | - |
| 90 | Cysteine-rich venom protein | Vipera nikolskii | CRISP | 24 | 83.3 | - | - | 0.112 | 77.4 | 0.050 | 80.1 | 0.116 | 68.3 |
| 91 | Cysteine-rich venom protein | Vipera berus | CRISP | 26 | 77 | 0.345 | 74.1 | 7.916 | 71.5 | 9.924 | 74.1 | 5.125 | 63.2 |
| 145 | Cysteine-rich venom protein triflin | Protobothrops flavoviridis | CRISP | 24 | 23.1 | 0.070 | 23.1 | 2.474 | 23.1 | 2.065 | 23.1 | 3.016 | 23.1 |
| 22 | Snaclec 1 | Sistrurus catenatus edwardsii | CTL | 17 | 6.2 | - | - | 1.404 | 6.2 | 0.168 | 6.2 | - | - |
| 23 | C-type lectin lectoxin-Thr1 | Thrasops jacksonii | CTL | 18 | 7.0 | 0.010 | 6.3 | 0.060 | 7.0 | 0.110 | 7 | 0.069 | 7 |
| 24 | Snaclec A13 | Macrovipera lebetina | CTL | 15 | 33.6 | - | - | 2.042 | 29.0 | 2.555 | 33.6 | - | - |
| 25 | Snaclec A15 | Macrovipera lebetina | CTL | 17 | 46.8 | 0.710 | 46.8 | 0.867 | 46.8 | 0.783 | 46.8 | 0.524 | 46.8 |
| 26 | Snaclec B7 | Macrovipera lebetina | CTL | 15 | 27.2 | - | - | 3.475 | 27.2 | 1.274 | 27.2 | - | - |
| 101 | Snaclec VP12 subunit A | Daboia palaestinae | CTL | 12 | 26.2 | - | - | 0.008 | 26.2 | - | - | - | - |
| 102 | Snaclec VP12 subunit B | Daboia palaestinae | CTL | 15 | 25.6 | - | - | 0.084 | 25.6 | 0.029 | 17.6 | - | - |
| 153 | C-type lectin J | Echis coloratus | CTL | 18 | 14.6 | 0.589 | 14.6 | 0.740 | 14.6 | 0.579 | 14.6 | 0.477 | 14.6 |
| 154 | C-type lectin H | Echis coloratus | CTL | 18 | 10.8 | 0.050 | 10.1 | 0.935 | 10.8 | 0.909 | 10.8 | 0.172 | 10.8 |
| 155 | C-type lectin E | Echis coloratus | CTL | 11 | 12.1 | 0.011 | 7.1 | - | - | 0.008 | 7.1 | 0.045 | 12.1 |
| 156 | C-type lectin B | Echis coloratus | CTL | 13 | 7.1 | - | - | 0.048 | 7.1 | 0.005 | 7.1 | - | - |
| 157 | C-type lectin A | Echis coloratus | CTL | 18 | 10.1 | 0.014 | 10.1 | - | - | - | - | - | - |
| 159 | Snaclec coagulation factor X-activating enzyme light chain 2 | Macrovipera lebetina | CTL | 18 | 6.3 | 0.039 | 6.3 | 0.199 | 6.3 | 0.340 | 6.3 | 0.051 | 6.3 |
| 173 | C-type lectin-like protein 3B | Macrovipera lebetina | CTL | 17 | 42.6 | 2.457 | 42.6 | 2.758 | 42.6 | 2.500 | 42.6 | 1.545 | 42.6 |
| 174 | C-type lectin-like protein 4B | Macrovipera lebetina | CTL | 17 | 14.7 | 0.018 | 12.0 | - | - | 0.017 | 12.0 | 0.353 | 14.7 |
| 175 | Snaclec dabocetin subunit alpha | Daboia siamensis | CTL | 17 | 6.5 | - | - | 0.443 | 6.5 | 0.064 | 6.5 | - | - |
| 181 | Snaclec tokaracetin subunit beta | Protobothrops tokarensis | CTL | 4 | 32.5 | - | - | 0.026 | 32.5 | - | - | - | - |
| 210 | Snaclec anticoagulant protein subunit B | Deinagkistrodon acutus | CTL | 14 | 12.2 | - | - | 0.352 | 12.2 | 0.124 | 12.2 | - | - |
| 14 | Disintegrin VB7A | Vipera berus berus | Dis | 7 | 76.6 | - | - | 0.111 | 23.4 | 0.008 | 23.4 | 12.932 | 76.6 |
| 15 | Disintegrin VB7B | Vipera berus berus | Dis | 6 | 70.3 | - | - | 0.008 | 29.7 | - | - | 0.935 | 70.3 |
| 16 | Disintegrin VLO4 | Macrovipera lebetina obtusa | Dis | 7 | 38.5 | - | - | 0.011 | 24.6 | 0.006 | 24.6 | 0.034 | 38.5 |
| 17 | Disintegrin VA6 | Vipera ammodytes ammodytes | Dis | 7 | 23.4 | - | - | - | - | - | - | 0.133 | 23.4 |
| 191 | Disintegrin lebein-1-alpha | Macrovipera lebetina | Dis | 12 | 30.6 | - | - | 0.389 | 9.0 | 0.578 | 9.0 | 0.006 | 21.6 |
| 86 | Hyaluronidase | Echis ocellatus | Hya | 52 | 13.6 | 0.007 | 13.6 | 0.011 | 7.3 | 0.004 | 2.7 | - | - |
| 176 | Hyaluronidase | Crotalus adamanteus | Hya | 52 | 6.7 | 0.004 | 6.7 | 0.007 | 6.7 | 0.003 | 2.0 | - | - |
| 27 | Inhibitor, chymotrypsin | Vipera ammodytes | Kunitz | 7 | 40.0 | 0.002 | 24.6 | - | - | 0.047 | 29.2 | 0.478 | 29.2 |
| 80 | Protease inhibitor 3 | Walterinnesia aegyptia | Kunitz | 8 | 21.0 | - | - | - | - | 0 | 21.0 | 0.005 | 21.0 |
| 81 | Kunitz-type serine protease inhibitor Vur-KIn | Vipera renardi | Kunitz | 7 | 39.4 | 0.015 | 39.4 | - | - | 0.074 | 39.4 | 0.241 | 39.4 |
| 87 | KP-Sut-1 | Suta fasciata | Kunitz | 13 | 9.4 | 0.072 | 9.4 | - | - | - | - | 0.077 | 9.4 |
| 142 | Kunitz-type serine protease inhibitor ki-VN | Vipera nikolskii | Kunitz | 10 | 34.7 | 0.610 | 34.7 | - | - | - | - | - | - |
| 5 | l-amino-acid oxidase | Macrovipera lebetina | LAAO | 12 | 42.1 | 0.041 | 41.1 | 1.388 | 42.1 | 1.631 | 41.1 | 1.786 | 41.1 |
| 65 | l-amino-acid oxidase | Echis ocellatus | LAAO | 56 | 6.0 | - | - | - | - | 0 | 6.0 | - | - |
| 66 | l-amino-acid oxidase | Vipera ammodytes ammodytes | LAAO | 54 | 13.2 | - | - | 0.009 | 11.2 | 0.012 | 11.2 | 0.007 | 13.0 |
| 75 | l-amino-acid oxidase | Daboia russelii | LAAO | 56 | 11.7 | 0 | 5.8 | 0.316 | 7.7 | 0.045 | 11.7 | 0.049 | 9.9 |
| 92 | Kunitz-type serine protease inhibitor PIVL | Macrovipera lebetina transmediterranea | LAAO | 10 | 8.4 | - | - | - | - | 0 | 8.4 | 0.021 | 8.4 |
| 94 | l-amino-acid oxidase | Gloydius halys | LAAO | 55 | 9.9 | - | - | - | - | 0 | 7.4 | - | - |
| 107 | l-amino acid oxidase | Ovophis okinavensis | LAAO | 58 | 11.0 | 0.001 | 3.9 | 1.893 | 6.8 | 2.106 | 6.8 | 1.378 | 10.9 |
| 144 | l-amino acid oxidase | Protobothrops elegans | LAAO | 57 | 5.7 | - | - | 0 | 5.7 | - | - | - | - |
| 152 | l-amino acid oxidase B variant 1 | Echis coloratus | LAAO | 56 | 17.1 | - | - | 0.425 | 13.1 | 0.213 | 12.9 | 0.216 | 13.9 |
| 164 | l-amino-acid oxidase | Crotalus horridus | LAAO | 58 | 11.2 | 0.001 | 4.5 | 0.042 | 6.6 | 0.005 | 5.8 | 0 | 5.8 |
| 183 | l-amino-acid oxidase | Bothrops moojeni | LAAO | 54 | 12.1 | 0.022 | 6.5 | 0.256 | 7.1 | 0.178 | 9.4 | 0.098 | 9.2 |
| 62 | Venom nerve growth factor 2 | Daboia russelii | NGF | 27 | 14.4 | - | - | 0 | 14.4 | - | - | - | - |
| 63 | Venom nerve growth factor | Vipera ursinii | NGF | 27 | 25.5 | 0.285 | 18.1 | 0.122 | 18.1 | 0.254 | 25.5 | 0.093 | 18.1 |
| 76 | Snake venom 5′-nucleotidase | Gloydius blomhoffii blomhoffii | Nuc | 6 | 27.8 | 0.013 | 27.8 | - | - | - | - | - | - |
| 110 | 5′-nucleotidase | Ovophis okinavensis | Nuc | 55 | 26.6 | 0.091 | 26.6 | 0.012 | 9.1 | 0.018 | 16.5 | 0.057 | 22.4 |
| 125 | Phosphodiesterase | Macrovipera lebetina | Nuc | 96 | 35.4 | 0.243 | 35.4 | 0.103 | 21.4 | 0.088 | 20.7 | 0.038 | 19.5 |
| 126 | 5′-nucleotidase | Macrovipera lebetina | Nuc | 45 | 58.1 | 0.563 | 54.4 | 0.046 | 14.5 | 0.097 | 30.4 | 0.226 | 39.2 |
| 127 | Venom phosphodiesterase 2 | Crotalus adamanteus | Nuc | 91 | 14.0 | 0.007 | 14.0 | - | - | - | - | - | - |
| 132 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3-like | Python bivittatus | Nuc | 93 | 7.5 | 0.004 | 7.5 | 0.002 | 3.5 | 0.003 | 3.5 | - | - |
| 170 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 | Boiga irregularis | Nuc | 100 | 4.5 | - | - | 0.441 | 3.3 | 1.521 | 2.6 | - | - |
| 73 | Proactivator polypeptide-like | Crotalus adamanteus | OP | 58 | 19.7 | 0.036 | 19.7 | 0.011 | 5.2 | 0.024 | 8.7 | 0.006 | 2.5 |
| 104 | ArfGAP with SH3 domain ankyrin repeat and PH domain 3 | Micrurus fulvius | OP | 107 | 2.3 | - | - | 0.027 | 2.3 | 0.033 | 2.3 | - | - |
| 113 | Uncharacterized protein | Ophiophagus hannah | OP | 46 | 3.4 | - | - | 0.040 | 3.4 | 0.015 | 3.4 | - | - |
| 115 | 78 kDa glucose-regulated protein | Ophiophagus hannah | OP | 67 | 6.4 | 0 | 2.8 | 0.004 | 4.4 | 0 | 2.0 | - | - |
| 117 | WD repeat-containing protein 67 | Ophiophagus hannah | OP | 113 | 0.8 | - | - | - | - | - | - | 0.652 | 0.8 |
| 118 | Pituitary adenylate cyclase-activating polypeptide type I receptor | Ophiophagus hannah | OP | 7 | 18.2 | 0.008 | 18.2 | - | - | - | - | - | - |
| 119 | Iron-responsive element-binding protein 2 | Ophiophagus hannah | OP | 90 | 1.8 | - | - | 0.536 | 1.8 | 0.482 | 1.8 | 0.224 | 1.8 |
| 122 | Glutathione peroxidase | Ophiophagus hannah | OP | 29 | 21.6 | 0.078 | 18.9 | 0.047 | 16.7 | 0.095 | 21.6 | 0.081 | 16.7 |
| 128 | PiggyBac transposable element-derived protein 5 | Python bivittatus | OP | 69 | 2.7 | - | - | - | - | - | - | 0.067 | 2.7 |
| 129 | Calmodulin-lysine N-methyltransferase | Python bivittatus | OP | 14 | 12.3 | - | - | - | - | 0 | 12.3 | - | - |
| 130 | Dipeptidase 2 | Python bivittatus | OP | 46 | 7.0 | - | - | - | - | - | - | 0 | 7.0 |
| 131 | Serine/threonine-protein phosphatase 6 regulatory subunit 1 isoform X3 | Python bivittatus | OP | 92 | 1.9 | - | - | 0.199 | 1.9 | 0.117 | 1.8 | 0.279 | 1.8 |
| 133 | E3 ubiquitin-protein ligase MARCH8-like isoform X8 | Python bivittatus | OP | 30 | 7.2 | - | - | - | - | - | - | 0.702 | 7.2 |
| 134 | Nucleolar and coiled-body phosphoprotein 1 isoform X5 | Python bivittatus | OP | 104 | 1.4 | - | - | - | - | 0 | 1.4 | - | - |
| 135 | E3 ubiquitin-protein ligase UBR4 | Python bivittatus | OP | 555 | 0.2 | 0 | 0.2 | - | - | - | - | - | - |
| 136 | Extracellular matrix protein 1 | Python bivittatus | OP | 24 | 8.6 | 0.019 | 8.6 | - | - | 0.004 | 8.6 | - | - |
| 137 | Receptor-type tyrosine-protein phosphatase gamma-like | Python bivittatus | OP | 102 | 1.9 | - | - | 0.026 | 1.9 | 0.018 | 1.9 | 0.018 | 1.9 |
| 138 | Nurim-like | Python bivittatus | OP | 32 | 7.7 | 0.010 | 7.7 | - | - | - | - | - | - |
| 162 | Dickkopf-related protein 3-like | Crotalus horridus | OP | 31 | 5.0 | - | - | 0.005 | 5 | 0.007 | 5.0 | - | - |
| 168 | RNA binding motif protein 6 | Boiga irregularis | OP | 59 | 3.5 | 0.007 | 3.5 | - | - | - | - | - | - |
| 172 | Filamin-B isoform 15 | Boiga irregularis | OP | 282 | 0.9 | - | - | 0.010 | 0.9 | - | - | - | - |
| 197 | Leucine-rich repeats and immunoglobulin-like domains protein 1 | Thamnophis sirtalis | OP | 48 | 1.4 | 0.010 | 1.4 | 0.028 | 1.4 | 0.026 | 1.4 | - | - |
| 201 | Peroxiredoxin-4-like | Thamnophis sirtalis | OP | 31 | 8.7 | 0.002 | 8.7 | - | - | - | - | - | - |
| 202 | Obscurin | Thamnophis sirtalis | OP | 1024 | 0.5 | 0.109 | 0.4 | 0.107 | 0.2 | - | - | - | - |
| 203 | CCR4-NOT transcription complex subunit 3 | Callithrix jacchus | OP | 12 | 15.7 | - | - | 0.008 | 15.7 | 0.046 | 15.7 | - | - |
| 205 | Tyrosine-protein phosphatase non-receptor type 20 | Thamnophis sirtalis | OP | 50 | 4.0 | - | - | 0.019 | 4.0 | 0.020 | 4.0 | - | - |
| 208 | Protein BANP | Thamnophis sirtalis | OP | 40 | 4.4 | - | - | 0.064 | 4.4 | 0.076 | 4.4 | 0.119 | 4.4 |
| 209 | Microtubule-associated serine/threonine-protein kinase 1-like | Thamnophis sirtalis | OP | 94 | 0.7 | - | - | - | - | - | - | 0 | 0.7 |
| 10 | Basic phospholipase A2 chain HDP-1P | Vipera nikolskii | PLA2 | 13 | 86.9 | 20.289 | 86.9 | 5.460 | 9.0 | 0.230 | 9.0 | 0.288 | 27.0 |
| 13 | Basic phospholipase A2 B chain | Vipera aspis zinnikeri | PLA2 | 13 | 80.3 | 0.130 | 80.3 | - | - | - | - | - | - |
| 28 | Phospholipase A2 II | Vipera aspis | PLA2 | 5 | 40.4 | 0.018 | 19.2 | - | - | 0.009 | 21.2 | - | - |
| 29 | Phospholipase A2 III | Vipera aspis | PLA2 | 5 | 66.0 | 0.002 | 66.0 | 6.579 | 66.0 | 5.099 | 66.0 | 0 | 66.0 |
| 30 | Acidic phospholipase A2 PLA-1 | Eristicophis macmahoni | PLA2 | 13 | 22.3 | - | - | - | - | - | - | 0.419 | 22.3 |
| 31 | Acidic phospholipase A2 PLA-2 | Eristicophis macmahoni | PLA2 | 13 | 29.8 | 0 | 28.9 | - | - | 0 | 21.5 | 0.388 | 29.8 |
| 32 | Acidic phospholipase A2 homolog vipoxin A chain | Vipera ammodytes meridionalis | PLA2 | 13 | 94.3 | 34.017 | 94.3 | - | - | - | - | 0.048 | 18.9 |
| 56 | Basic phospholipase A2 3 | Daboia russelii | PLA2 | 13 | 27.3 | - | - | 0.003 | 12.4 | 0.002 | 12.4 | 0.048 | 27.3 |
| 57 | Phospholipase A2 | Agkistrodon piscivorus | PLA2 | 13 | 27.6 | 0 | 16.3 | - | - | - | - | - | - |
| 58 | Phospholipase A2 homolog P-elapitoxin-Aa1a beta chain | Acanthophis antarcticus | PLA2 | 3 | 22.6 | - | - | 0.165 | 22.6 | 0.127 | 22.6 | 0.030 | 22.6 |
| 67 | Acidic phospholipase A2 RV-7 | Daboia siamensis | PLA2 | 13 | 45.1 | 1.078 | 45.1 | - | - | - | - | - | - |
| 78 | Basic phospholipase A2 Pla2Vb | Vipera berus berus | PLA2 | 15 | 46.4 | - | - | 0 | 13.8 | 0.056 | 46.4 | - | - |
| 79 | Acidic phospholipase A2 Vur-PL3 | Vipera renardi | PLA2 | 15 | 69.3 | - | - | 12.956 | 67.2 | 12.705 | 67.2 | 7.113 | 58.4 |
| 82 | Acidic phospholipase A2 PL1 | Vipera renardi | PLA2 | 15 | 75.4 | 4.057 | 70.3 | 5.097 | 59.4 | 4.080 | 52.9 | 10.603 | 75.4 |
| 83 | Acidic phospholipase A2 Vur-PL2B | Vipera renardi | PLA2 | 15 | 72.3 | - | - | 0 | 19.7 | 0.041 | 19.7 | 14.999 | 72.3 |
| 84 | Basic phospholipase A2 homolog Vur-S49 | Vipera renardi | PLA2 | 15 | 63.0 | - | - | 0.009 | 18.8 | 0.055 | 18.8 | 7.676 | 63.0 |
| 85 | Basic phospholipase A2 vurtoxin | Vipera renardi | PLA2 | 15 | 50.0 | - | - | - | - | - | - | 4.220 | 50.0 |
| 95 | Ammodytin I1 | Vipera aspis aspis | PLA2 | 15 | 56.5 | 0.032 | 56.5 | 0.048 | 56.5 | 0.040 | 44.9 | 0.026 | 39.9 |
| 96 | Ammodytin I1 | Vipera ammodytes montandoni | PLA2 | 15 | 75.4 | 1.389 | 75.4 | 1.428 | 75.4 | 0.524 | 63.8 | 1.710 | 59.4 |
| 97 | Ammodytin I2 | Vipera aspis aspis | PLA2 | 15 | 19.7 | - | - | - | - | - | - | 0 | 19.7 |
| 98 | Ammodytin I2 | Vipera berus berus | PLA2 | 15 | 36.5 | - | - | 0.021 | 31.4 | - | - | - | - |
| 99 | Ammodytin I2 | Vipera ursinii | PLA2 | 15 | 45.3 | - | - | - | - | - | - | 0.017 | 45.3 |
| 100 | Ammodytin L | Vipera ammodytes ammodytes | PLA2 | 15 | 14.5 | - | - | - | - | - | - | 0.009 | 14.5 |
| 139 | Basic phospholipase A2 vipoxin B chain | Vipera ammodytes meridionalis | PLA2 | 13 | 80.3 | 0.086 | 80.3 | - | - | - | - | - | - |
| 151 | Phospholipase A2 Group IIE | Echis coloratus | PLA2 | 13 | 5.8 | - | - | - | - | - | - | 0.020 | 5.8 |
| 161 | Basic phospholipase A2 | Azemiops feae | PLA2 | 15 | 7.2 | - | - | - | - | 0.012 | 7.2 | 0.016 | 7.2 |
| 193 | Phospholipase A2-III | Daboia russelii | PLA2 | 13 | 13.1 | 0.019 | 13.1 | - | - | - | - | - | - |
| 195 | Phospholipase A2 ammodytin I1 | Vipera nikolskii | PLA2 | 15 | 75.4 | 4.779 | 75.4 | 4.663 | 75.4 | 4.289 | 63.8 | - | - |
| 196 | Basic phospholipase A2 chain HDP-2P | Vipera nikolskii | PLA2 | 15 | 69.6 | 0.068 | 69.6 | - | - | - | - | 0.006 | 31.2 |
| 109 | Phospholipase b | Ovophis okinavensis | PLB | 64 | 20.8 | - | - | 0.015 | 13.6 | 0.037 | 20.1 | 0.057 | 15.0 |
| 114 | Phospholipase B-like 1 | Ophiophagus hannah | PLB | 58 | 16.8 | 0.022 | 7.0 | 0.085 | 12.0 | 0.104 | 16.2 | 0.088 | 10.2 |
| 169 | Phospholipase B | Boiga irregularis | PLB | 64 | 15.6 | - | - | 0.020 | 11.6 | 0.034 | 11.2 | 0.034 | 9.9 |
| 179 | Phospholipase B | Crotalus adamanteus | PLB | 64 | 26.9 | 0.080 | 16.5 | 0.218 | 15.9 | 0.321 | 21.0 | 0.334 | 12.7 |
| 6 | Unassigned | Calloselasma rhodostoma | SP | 24 | 9.4 | - | - | 0.007 | 5.5 | 0.041 | 8.1 | 0.006 | 8.1 |
| 11 | Snake venom serine protease pallase | Gloydius halys | SP | 26 | 13.1 | 0.002 | 9.3 | 0.005 | 10.6 | 0.002 | 11.9 | 0 | 10.6 |
| 12 | Snake venom serine protease ussurase | Gloydius ussuriensis | SP | 26 | 5.6 | 0.077 | 5.6 | - | - | - | - | - | - |
| 21 | Thrombin-like enzyme KN-BJ 2 | Bothrops jararaca | SP | 27 | 11.3 | 1.017 | 11.3 | 0.329 | 11.3 | 1.944 | 11.3 | 0.050 | 11.3 |
| 35 | Serine protease | Echis coloratus | SP | 28 | 8.5 | - | - | 0.043 | 8.5 | 0.172 | 8.5 | 0.087 | 8.5 |
| 36 | Serine protease | Echis ocellatus | SP | 24 | 6.3 | 0.515 | 6.3 | 1.980 | 3.6 | 2.401 | 6.3 | 0.694 | 6.3 |
| 37 | Serine protease | Echis coloratus | SP | 25 | 7.3 | - | - | 0.079 | 4.7 | 0.047 | 7.3 | - | - |
| 38 | Serine protease | Echis coloratus | SP | 25 | 12.0 | 0.004 | 12.0 | 0 | 9.4 | 0.007 | 12.0 | - | - |
| 39 | Serine protease | Echis coloratus | SP | 26 | 5.9 | 0.110 | 5.9 | 0.048 | 3.4 | 0.015 | 5.9 | 0.005 | 5.9 |
| 40 | Serine protease | Echis carinatus sochureki | SP | 25 | 8.1 | 0.021 | 5.5 | - | - | - | - | 0.036 | 5.5 |
| 64 | Factor V activator RVV-V gamma | Daboia siamensis | SP | 25 | 13.7 | 0.006 | 11.1 | 0.473 | 8.1 | 0.075 | 5.6 | - | - |
| 69 | Serine protease VLSP-3 | Macrovipera lebetina | SP | 28 | 15.5 | 3.526 | 15.5 | 2.548 | 12.0 | 3.479 | 14.3 | 1.303 | 14.3 |
| 70 | Beta-fibrinogenase | Macrovipera lebetina | SP | 28 | 18.3 | 0.017 | 18.3 | 0.008 | 9.3 | 0.078 | 11.7 | 0.224 | 11.7 |
| 71 | Chymotrypsin-like protease VLCTLP | Macrovipera lebetina | SP | 28 | 29.6 | 0.069 | 29.6 | - | - | - | - | - | - |
| 74 | Snake venom serine protease nikobin | Vipera nikolskii | SP | 28 | 43.2 | 12.595 | 43.2 | 2.334 | 34.2 | 8.515 | 32.3 | 1.732 | 28.0 |
| 77 | Snake venom serine protease pallabin | Gloydius halys | SP | 28 | 12.3 | 0.002 | 8.8 | - | - | - | - | 0.010 | 10.0 |
| 103 | Kallikrein-CohID-1 | Crotalus oreganus helleri | SP | 28 | 10.8 | - | - | - | - | - | - | 0 | 10.8 |
| 105 | Serine protease | Protobothrops flavoviridis | SP | 18 | 17.4 | 0 | 17.4 | - | - | - | - | - | - |
| 106 | Serine protease | Ovophis okinavensis | SP | 10 | 23.3 | 0.061 | 20.0 | 0.028 | 23.3 | 0.352 | 20.0 | 0.067 | 23.3 |
| 140 | Factor V activator | Macrovipera lebetina | SP | 28 | 18.9 | 0.049 | 13.9 | 0.776 | 11.6 | 0.088 | 11.2 | 0.005 | 5.0 |
| 141 | Venom serine proteinase-like protein 2 | Macrovipera lebetina | SP | 28 | 30.8 | 1.028 | 30.8 | 0.896 | 26.5 | 1.680 | 25.4 | 1.595 | 26.5 |
| 163 | Serine proteinase 1 | Crotalus horridus | SP | 28 | 13.2 | 1.023 | 10.9 | 0.274 | 4.3 | 2.042 | 6.6 | - | - |
| 187 | Snake venom serine protease HS112 | Bothrops jararaca | SP | 27 | 14.9 | - | - | 0.157 | 12.5 | 0.323 | 14.9 | - | |
| 188 | Snake venom serine protease KN6 | Trimeresurus stejnegeri | SP | 28 | 3.5 | 0.036 | 3.5 | - | - | - | - | - | - |
| 189 | Snake venom serine protease 5 | Trimeresurus stejnegeri | SP | 28 | 11.6 | 0 | 11.6 | - | - | - | - | - | - |
| 190 | Snake venom serine protease catroxase-2 | Crotalus atrox | SP | 27 | 17.4 | 0.352 | 17.4 | 0.162 | 8.5 | 0.531 | 10.9 | 0.012 | 10.9 |
| 198 | Rho GTPase-activating protein 28-like | Thamnophis sirtalis | SP | 24 | 3.2 | - | - | 0.811 | 3.2 | 0.822 | 3.2 | 0.207 | 3.2 |
| 41 | Metalloproteinase | Echis carinatus sochureki | SVMP | 69 | 14.6 | - | - | 1.488 | 8.7 | 1.023 | 11.5 | 0.401 | 14.6 |
| 42 | Metalloproteinase | Echis carinatus sochureki | SVMP | 68 | 6.9 | - | - | - | - | - | - | 0.107 | 6.9 |
| 43 | Metalloproteinase | Echis carinatus sochureki | SVMP | 27 | 6.5 | - | - | 2.987 | 6.5 | 2.973 | 6.5 | 1.419 | 6.5 |
| 44 | Metalloproteinase | Echis coloratus | SVMP | 56 | 6.3 | - | - | - | - | - | - | 0.004 | 6.3 |
| 45 | Metalloproteinase | Echis coloratus | SVMP | 56 | 11.2 | - | - | 0.109 | 9.4 | 0.080 | 9.4 | 0.116 | 9.0 |
| 46 | Metalloproteinase | Echis coloratus | SVMP | 66 | 10.7 | - | - | 0 | 9.2 | 0 | 9.2 | 0 | 10.7 |
| 47 | Metalloproteinase | Echis coloratus | SVMP | 69 | 3.1 | - | - | - | - | - | - | 0.008 | 3.1 |
| 48 | Metalloproteinase | Echis coloratus | SVMP | 69 | 7.4 | - | - | 0.208 | 7.4 | 0.435 | 4.7 | 0.058 | 4.9 |
| 49 | Metalloproteinase | Echis coloratus | SVMP | 68 | 8.8 | - | - | - | - | - | - | 0 | 8.8 |
| 50 | Metalloproteinase | Echis coloratus | SVMP | 68 | 13.8 | - | - | 0 | 8.9 | 0 | 5.2 | 0.488 | 4.9 |
| 51 | Metalloproteinase | Echis coloratus | SVMP | 61 | 5.4 | - | - | 2.479 | 5.4 | 2.296 | 5.4 | 1.071 | 5.4 |
| 52 | Metalloproteinase | Echis coloratus | SVMP | 68 | 6.4 | - | - | 0.019 | 6.4 | 0.012 | 5.6 | 0.021 | 6.4 |
| 53 | Metalloproteinase | Echis pyramidum leakeyi | SVMP | 62 | 8.1 | 0.009 | 2.5 | - | - | - | - | - | - |
| 54 | Metalloproteinase | Echis pyramidum leakeyi | SVMP | 46 | 5.4 | - | - | 0.019 | 5.4 | 0.014 | 5.4 | - | - |
| 55 | Metalloproteinase | Echis carinatus sochureki | SVMP | 54 | 5.6 | - | - | - | - | - | - | 0.010 | 5.6 |
| 59 | Group III snake venom metalloproteinase | Echis ocellatus | SVMP | 62 | 10.7 | 0 | 3.1 | 1.254 | 6.0 | 1.744 | 9.0 | 0.651 | 10.7 |
| 61 | Snake venom metalloproteinase VMP1 | Agkistrodon piscivorus leucostoma | SVMP | 46 | 6.8 | - | - | - | - | - | - | 0.833 | 6.8 |
| 72 | Snake venom metalloproteinase | Crotalus adamanteus | SVMP | 68 | 9.0 | - | - | 0.005 | 3.4 | 0.016 | 4.7 | 0.085 | 7.7 |
| 93 | H3 metalloproteinase 1 | Vipera ammodytes ammodytes | SVMP | 68 | 43.5 | 0.611 | 32.0 | 3.415 | 33.3 | 2.970 | 35.9 | 2.737 | 37.3 |
| 108 | P-III metalloprotease | Ovophis okinavensis | SVMP | 16 | 12.1 | - | - | 2.110 | 12.1 | 1.999 | 12.1 | 0.945 | 12.1 |
| 112 | Metalloproteinase H4-A | Vipera ammodytes ammodytes | SVMP | 68 | 14.2 | 0.022 | 11.4 | 0.342 | 4.2 | 0.006 | 4.2 | - | - |
| 143 | Snake venom metalloproteinase lebetase-4 | Macrovipera lebetina | SVMP | 24 | 19.4 | - | - | - | - | 0.008 | 12.4 | 0.091 | 19.4 |
| 146 | Zinc metalloproteinase-disintegrin-like daborhagin-K | Daboia russelii | SVMP | 69 | 6.2 | - | - | - | - | - | - | 0.112 | 6.2 |
| 147 | Coagulation factor X-activating enzyme heavy chain | Daboia siamensis | SVMP | 69 | 10.0 | 0.003 | 2.9 | 0.473 | 7.3 | 0.088 | 7.1 | 0.010 | 7.3 |
| 160 | Coagulation factor X-activating enzyme heavy chain | Macrovipera lebetina | SVMP | 68 | 11.4 | 0.013 | 5.4 | 1.243 | 11.4 | 0.105 | 6.5 | 0.020 | 5.4 |
| 166 | Metalloproteinase F1 | Vipera ammodytes ammodytes | SVMP | 68 | 25.7 | - | - | - | - | 0.001 | 4.1 | 0.498 | 25.7 |
| 182 | Antihemorrhagic factor cHLP-A | Gloydius brevicaudus | SVMP | 36 | 4.0 | - | - | - | - | - | - | 0 | 4.0 |
| 184 | Zinc metalloproteinase/disintegrin | Macrovipera lebetina | SVMP | 53 | 11.9 | - | - | 0.008 | 4.8 | 0.027 | 8.8 | 0.657 | 7.9 |
| 185 | Zinc metalloproteinase-disintegrin-like VLAIP-B | Macrovipera lebetina | SVMP | 68 | 9.3 | 0 | 4.6 | - | - | - | - | 0.052 | 6.5 |
| 186 | Zinc metalloproteinase-disintegrin-like VLAIP-A | Macrovipera lebetina | SVMP | 68 | 25.2 | 0.005 | 17.7 | 0.138 | 14.9 | 0.110 | 17.5 | 0.048 | 19.0 |
| 192 | Group III snake venom metalloproteinase | Echis ocellatus | SVMP | 69 | 10.7 | - | - | 0 | 7.9 | 0 | 10.7 | - | - |
| 194 | Zinc metalloproteinase-disintegrin-like bothrojarin-2 | Bothrops jararaca | SVMP | 24 | 11.9 | - | - | 0.015 | 11.9 | 0.017 | 11.9 | 0.089 | 11.9 |
| 1 | Renin-like aspartic protease | Echis ocellatus | TBP | 43 | 9.4 | 0.009 | 4.6 | 0.040 | 4.8 | 0.039 | 4.8 | 0.055 | 4.8 |
| 8 | Aminopeptidase A | Gloydius brevicaudus | TBP | 110 | 7.6 | 0 | 2.1 | 0.004 | 2.5 | 0.007 | 2.5 | 0.029 | 7.6 |
| 20 | Aminopeptidase N | Gloydius brevicaudus | TBP | 106 | 1.3 | - | - | - | - | - | - | 0 | 1.3 |
| 68 | Glutaminyl-peptide cyclotransferases | Daboia russelii | TBP | 42 | 37.8 | 0.109 | 37.8 | 0.092 | 29.1 | 0.055 | 32.1 | 0.085 | 37.8 |
| 111 | Glutaminyl-cyclase | Ovophis okinavensis | TBP | 40 | 33.2 | 0.012 | 33.2 | - | - | - | - | - | - |
| 120 | Cathepsin D | Ophiophagus hannah | TBP | 30 | 16.3 | 0.013 | 16.3 | - | - | - | - | - | - |
| 121 | Endoplasmic reticulum aminopeptidase 1 | Ophiophagus hannah | TBP | 91 | 1.6 | 0.003 | 1.6 | - | - | - | - | - | - |
| 123 | Renin | Ophiophagus hannah | TBP | 40 | 5.5 | 0.016 | 2.2 | 0.059 | 5.5 | 0.048 | 5.5 | 0.023 | 5.5 |
| 149 | Renin | Echis coloratus | TBP | 12 | 23.9 | - | - | - | - | 0.013 | 23.9 | - | - |
| 167 | Xaa-Pro aminopeptidase 2 | Boiga irregularis | TBP | 76 | 25.7 | 0.420 | 25.7 | 0.041 | 17.3 | 0.114 | 23.6 | 0.042 | 17.3 |
| 178 | Peptidyl-prolyl cis-trans isomerase | Crotalus adamanteus | TBP | 22 | 12.9 | 0.006 | 12.9 | - | - | - | - | - | - |
| 204 | Dipeptidase 2-like | Thamnophis sirtalis | TBP | 33 | 7.4 | 0.002 | 7.4 | - | - | 0.003 | 7.4 | 0.019 | 7.4 |
| 207 | Xaa-Pro aminopeptidase 2-like | Thamnophis sirtalis | TBP | 27 | 19.4 | 0.057 | 19.4 | 0.011 | 11.3 | 0.016 | 19.4 | - | - |
| 60 | Snake venom vascular endothelial growth factor toxin vammin | Vipera ammodytes ammodytes | VEGF | 16 | 49.7 | 5.315 | 31.0 | 4.239 | 44.1 | 5.109 | 36.6 | 2.446 | 32.4 |
| 88 | Snake venom vascular endothelial growth factor toxin HF | Vipera aspis aspis | VEGF | 12 | 65.5 | 0.083 | 65.5 | 0.002 | 58.2 | 0.002 | 48.2 | - | - |
| 148 | Vascular endothelial growth factor A | Echis coloratus | VEGF | 22 | 34.4 | 0.017 | 34.4 | - | - | 0.004 | 17.2 | - | - |
| 150 | Vascular endothelial growth factor F | Echis coloratus | VEGF | 16 | 28.5 | - | - | 0.390 | 28.5 | 0.640 | 28.5 | 0.030 | 18.8 |
1 B-NAP: Bradykinin potentiating and C-type natriuretic peptides; BP: Blood protein; CP: Cysteine Proteases; CRISP: Cysteine-rich secretory protein; CTL: C-type lectin like; Dis: Disintegrin; Hya: Hyaluronidase; Kunitz: Kunitz type proteinase inhibitor; LAAO: l-amino acid oxidase; NGF: Nerve growth factor; Nuc: Nucleic acid degrading enzymes; OP: Other protein; PLA2: Phospholipase A2; PLB: Phospholipase B; SP: Serine proteinase; SVMP: Metalloproteinase; TBP: Toxin biosynthesis proteins (including aminopeptidases); VEGF: Vascular endothelial growth factor.
Table 2.
Protein families found in the venoms of Russian vipers.
| Protein Family 3 | # of Identified Proteins | Protein Abundance 1 LFQ/INT, % (# of Identified Proteins 2) | |||
|---|---|---|---|---|---|
| V. nikolskii | V. kaznakovi | V. orlovi | V. renardi | ||
| PLA2 | (29) | 64.68/65.96 (14) | 41.03/36.43 (11) | 24.21/27.27 (14) | 44.05/47.64 (18) |
| SVMP | (32) | 0.66/0.66 (8) | 16.15/16.31 (19) | 14.77/13.92 (21) | 11.98/10.53 (28) |
| CTL | (18) | 4.01/3.9 (9) | 12.48/13.44 (15) | 11.2/9.46 (15) | 3.46/3.24 (8) |
| SP | (27) | 19.34/20.51 (20) | 10.79/10.96 (18) | 23.97/22.61 (19) | 7.87/6.03 (15) |
| CRISP | (7) | 0.66/0.41 (2) | 9.72/10.89 (7) | 12.2/12.3 (7) | 7.98/8.26 (4) |
| LAAO | (11) | 0.08/0.07 (5) | 3.99/4.33 (8) | 4.59/4.19 (10) | 4.21/3.56 (8) |
| VEGF | (4) | 7.57/5.42 (3) | 3.96/4.63 (2) | 4.2/5.76 (4) | 2.92/2.48 (2) |
| Dis | (5) | 0/0 (0) | 0.53/0.52 (4) | 0.56/0.59 (3) | 13.43/14.04 (5) |
| OP | (28) | 0.17/0.28 (11) | 0.49/1.13 (13) | 0.95/0.96 (16) | 1.8/2.15 (8) |
| PLB | (4) | 0.12/0.1 (2) | 0.32/0.34 (3) | 0.52/0.5 (3) | 0.54/0.51 (4) |
| Nuc | (7) | 0.88/0.92 (6) | 0.21/0.6 (4) | 2.12/1.73 (5) | 0.47/0.32 (3) |
| TBP | (13) | 0.68/0.65 (11) | 0.17/0.25 (5) | 0.3/0.3 (8) | 0.33/0.25 (7) |
| NGF | (2) | 0.33/0.28 (1) | 0.14/0.12 (2) | 0.25/0.25 (1) | 0.12/0.09 (1) |
| Hya | (2) | 0/0.01 (2) | 0.01/0.02 (2) | 0/0.01 (2) | 0/0 (0) |
| BP | (14) | 0.15/0.12 (12) | 0/0.01 (2) | 0.01/0.01 (3) | 0.06/0.1 (9) |
| B-NAP | (1) | 0.01/0.01 (0) | 0/0.03 (1) | 0/0.01 (1) | 0/0 (0) |
| Kunitz | (5) | 0.66/0.7 (4) | 0/0 (0) | 0.15/0.12 (3) | 0.79/0.8 (4) |
| CP | (1) | 0/0 (1) | 0/0 (0) | 0/0 (0) | 0/0 (0) |
| total | (210) | (111) | (116) | (135) | (124) |
1 Protein abundance was calculated on the basis of peptide abundances for the peptides identified by MS/MS, as well as the peptides identified by MS1 matching between chromatograms. Protein abundances were calculated either on the basis of the MaxLFQ (Label-Free Quantification) algorithm (LFQ) or on the basis of the comparison of total protein intensities (sums of peptide intensities were calculated for each protein) within a single venom (INT). 2 Numbers of proteins were calculated on the basis of the peptides identified by MS/MS only (no MS1 matching hits were used). Therefore the protein might not be listed as identified, but it would still be quantified ”By Matching” with non-zero abundance value (e.g., B-NAP in V. nikolskii venom). 3 B-NAP: Bradykinin potentiating and C-type natriuretic peptides; BP: Blood protein; CP: Cysteine Proteases; CRISP: Cysteine-rich secretory protein; CTL: C-type lectin like; Dis: Disintegrin; Hya: Hyaluronidase; Kunitz: Kunitz type proteinase inhibitor; LAAO: l-amino acid oxidase; NGF: Nerve growth factor; Nuc: Nucleic acid degrading enzymes; OP: Other protein; PLA2: Phospholipase A2; PLB: Phospholipase B; SP: Serine proteinase; SVMP: Metalloproteinase; TBP: Toxin biosynthesis proteins (including aminopeptidases); VEGF: Vascular endothelial growth factor.
The proteins were categorized into 14 known venom protein families (Table 2). The most numerous classes were PLA2, SVMP, C-type lectin like (CTL) and serine protease (SP). Eleven families were represented in all viper venoms, while disintegrins (Dis) were absent in V. nikolskii. There were no Kunitz type proteinase inhibitors in V. kaznakovi, no hyaluronidase (Hya) in V. renardi and no bradykinin potentiating and C-type natriuretic peptides (B-NAP) in V. renardi and V. nikolskii. Besides, in the V. nikolskii venom, a single low abundance protein was identified belonging to Cysteine Proteases (CP), which are not common for snake venoms. Along with venom proteins, several Blood Proteins (BP) (up to 0.15% of the total protein abundance) and proteins with unclear family annotation (Other Proteins (OP)) (up to ~2% of the total protein content) were also found.
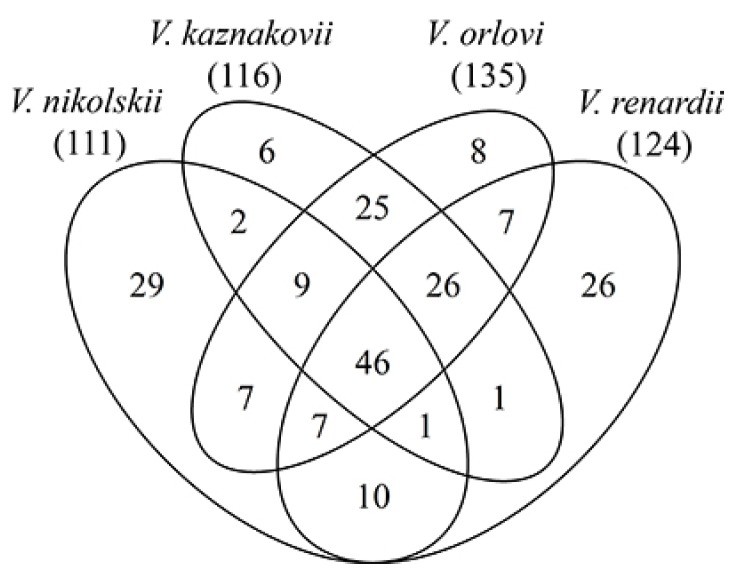
While the most numerous venom protein families were fairly similar in all snakes studied, individual protein composition was quite different (Figure 1). From 210 proteins only 46 were common for all four species and each species featured unique proteins: six in V. kaznakovi, 26 in V. renardi, eight in V. orlovi and 29 in V. nikolskii. These differences did not correlate with the total number of individual proteins identified in each venom.
Figure 1.
The number of common proteins in four Vipera species studied. The number in bracket under each species name indicates the total number of proteins identified in this species venom.
2.2. Composition of Russian Viper Venoms
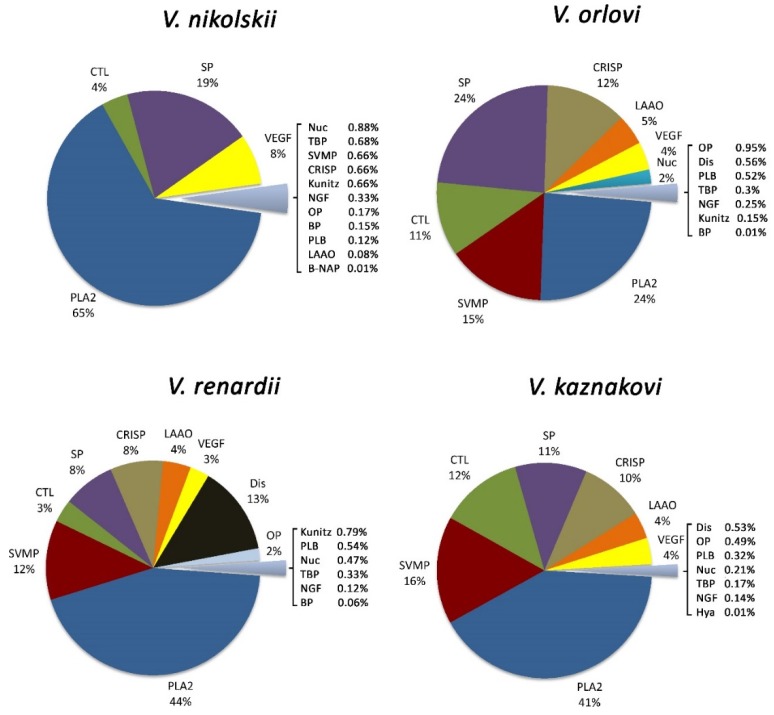
As a result of venom protein quantification, it was found that the main venom components were PLA2s; their content ranged from about 24% in V. orlovi venom to more than 60% in V. nikolskii (Table 2, Figure 2). The overwhelming majority of PLA2s belonged to D49 subgroup of group IIA as it might be expected for the snakes from Viperidae family. The venom of V. nikolskii contained PLA2s only from this group. One PLA2 of S49 subgroup was highly represented in V. renardi. One PLA2 of group IA was observed in small amounts in three venoms and a low quantity of group IIE PLA2 was detected in V. renardi venom.
Figure 2.
Relative abundance of venom proteins that were identified by LC MS/MS in Russian viper venoms. B-NAP: Bradykinin potentiating and C-type natriuretic peptides; BP: Blood protein; CRISP: Cysteine-rich secretory protein; CTL: C-type lectin like; Dis: Disintegrin; Hya: Hyaluronidase; Kunitz: Kunitz type proteinase inhibitor; LAAO: l-amino acid oxidase; NGF: Nerve growth factor; Nuc: Nucleic acid degrading enzymes; OP: Other protein; PLA2: Phospholipase A2; PLB: Phospholipase B; SP: Serine proteinase; SVMP: Metalloproteinase; TBP: Toxin biosynthesis proteins (including aminopeptidases); VEGF: Vascular endothelial growth factor.
Altogether, the enzyme content in venom of V. nikolskii reached about 85%, however this venom was characterized by a very low content of SVMPs (less than 1%) and LAAO (less than 0.1%). PLA2s accounted for more than 40% in V. kaznakovi and V. renardi venoms. While the content of SVMPs was less than 1% in the V. nikolskii venom, they comprised 12%–16% in V. kaznakovi, V. orlovi and V. renardi. The highest content of SPs was in V. orlovi venom (24%) and the lowest in V. renardi (8%). LAAO was at the level of 4%–5% in all the analyzed venoms with the exception of V. nikolskii. Nucleic acid degrading enzymes (Nuc) represented about 2% in V. orlovi venom and less that 1% in all the others. Phospholipase B (PLB) was found in all venoms (less than 1%) and very low amount of Hya (0.01%) was detected in three venoms. Among the non-enzymatic proteins, Dis (13%) in the V. renardi venom, CTL (12%) in V. kaznakovi, CRISPs (12%) in V. orlovi and vascular endothelial growth factors (VEGF, 8%) in V. nikolskii were the most abundant ones in the venoms studied. The total amount of non-enzymatic proteins was about 13% in the V. nikolskii venom and about 27%–28% in all the others. In addition to the proteins mentioned above, nerve growth factor (NGF) and Kunitz were present in all the venoms (less than 1%) with exception of V. kaznakovi, where Kunitz was absent.
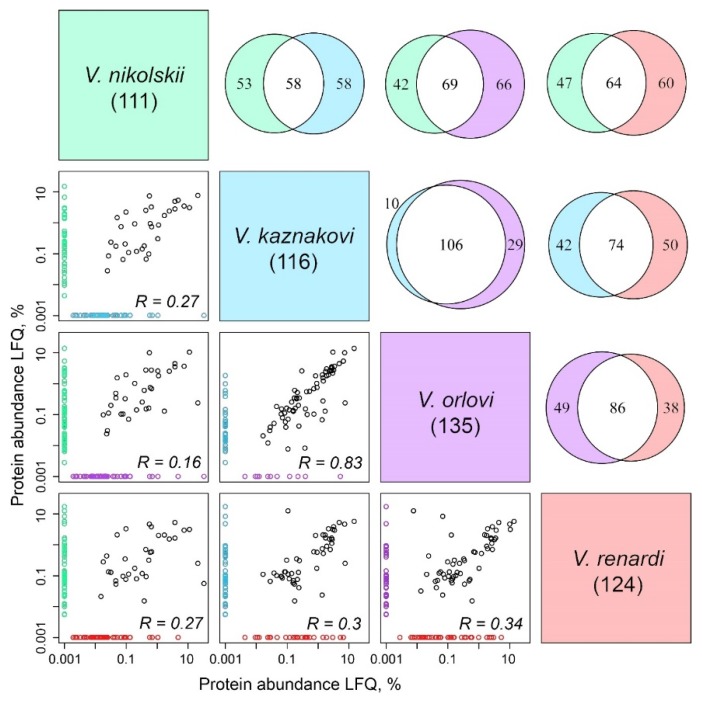
Interestingly, comparison of both the nature of the identified proteins and their abundance showed very close venom compositions for the species V. kaznakovii and V. orlovii. The Pearson correlation coefficient for individual protein abundance LFQ was 0.83, while for the rest of pairs the correlation coefficient varied from 0.16 to 0.34 (Figure 3).
Figure 3.
Protein number and abundance distributions for four Vipera species. ( The panels under the diagonal showing the species names) Individual protein abundance label-free quantification (LFQ) pairwise comparison. Proteins unique for a single species in a pair are highlighted in the corresponding color and for better visualization in logarithmic scale are assigned 0.001% abundance instead of real 0%. (The diagrams above the diagonal showing the species names) Pairwise Venn diagrams showing the number of common and unique proteins for each pair of the venoms.
2.3. Identification of Endogenous Peptides in the Venoms
It is well known that snake venoms may contain various peptides: several peptide families including bradykinin-potentiating peptides, natriuretic peptides, sarafotoxin, etc. were identified [17]. Moreover, the venoms studied in this work contain proteases, therefore their proteins may undergo proteolysis leading to generation of peptides. To study endogenously generated peptides in the venoms of interest, high molecular weight (MW) proteins were separated by ultrafiltration (10 KDa cut-off). The peptide fractions obtained were analyzed by LC-MS/MS in the same fashion as proteins, but without preliminary proteolysis. Peptides were searched at first against a full NCBI Serpentes database by Mascot search engine with 10% protein FDR (False Discovery Rate). A fusion database containing the peptidogenic proteins from Mascot search and the SwissProt Serpentes database was used for the final search in MaxQuant. Full NCBI database search with unspecific digestion failed in MaxQuant due to internal software limitations. In summary, 512 endogenous peptides from 80 proteins belonging to 13 protein families were found (Table S3). As expected, most of the peptides (462 peptides) belonged to proteins (48 proteins) which were earlier found in proteome, thus most likely representing venom protein degradation in vivo as a result of proteases and peptidases activity. At the same time, 50 peptides (Table S4) belonged to 32 unique proteins from nine protein families (Table 3). Among these, proteins in six families mostly had one peptide per protein, which can explain their identification only in the peptidome analysis as a result of a very low concentration of original proteins before degradation, so they were missed in the shotgun MS/MS selection in proteome analysis (or were beyond the taken FDR cut off). The largest number of unique peptides was found in proteins belonging to Dis and SVMP/Dis families: 25 peptides from 14 proteins were found. The peptides identified were mainly fragments of larger venom proteins. However, we found 12 peptides from proteins belonging to B-NAP family (Figure 4). These peptides may represent real endogenous peptides and possess their own biological activity. This is the first indication for the presence of bradykinin-potentiating and natriuretic peptides in venoms of vipers from the Pelias group.
Table 3.
Snake venom protein families for which peptides were found in peptidome only.
| Family | Number of Proteins | Number of Peptides |
|---|---|---|
| CTL | 4 | 5 |
| Dis | 3 | 7 |
| Kunitz | 2 | 2 |
| LAAO | 2 | 3 |
| NAP | 6 | 14 |
| PLA2 | 1 | 1 |
| SP | 2 | 2 |
| SVMP | 11 | 22 |
| VEGF | 1 | 1 |
Figure 4.
Bradykinin-potentiating and natriuretic peptides found in four viper venoms. P85169 (BPPDB_BOTJA)—Bradykinin-potentiating peptide 13b from Bothrops jararaca, P0DJK3 (BPPAE_BOTCO)—Bradykinin-potentiating peptide 10e from Bothrops cotiara, Q90Y12 (BNP_CRODU)—Bradykinin potentiating and C-type natriuretic peptides from Crotalus durissus terrificus, P83231 (VNPC_OXYSA)—Natriuretic peptide TNP-c from Oxyuranus scutellatus canni (Papuan taipan), and P0DMD5 (VNP_BUNMU)—amino acid sequence fragment 81–100 of Natriuretic peptide BM026 from Bungarus multicinctus. Vn, Vk, Vo, and Vr indicate V. nikolskii, V. kaznakovi, V. orlovi, and V. renardi, respectively.
3. Discussion
We have analyzed venom proteomes and peptidomes for four species of Vipera, for which there is no genomic or transcriptomic data published. For each species, the venoms of at least 15 individual animals were pooled for the analysis. Protein identification for such “non-sequenced” species is problematic for inherently database oriented bottom-up LC-MS/MS-based proteomics [18]. A possible solution is to use the protein sequences of closely related species, based on the assumption of their high homology level [18]. Thus, when the exact protein sequence is missing in the database, the protein might still be identified by partial/full homology with a known protein of another species. Here, we searched LC-MS/MS data against the database containing all the proteins from the taxon Serpentes in the NCBI database on the date of the experiment (the results are given in Table 1).
In the bottom-up proteomics, there are two major approaches for the quantitative analysis: (a) relative quantification of a single protein across samples; and (b) comparison of different proteins within a single sample. Principle (a) is based on the measurement of all the peptides belonging to a protein in several samples (and pair-wise peptide Fold Changes estimation) followed by protein Fold Change calculation as, e.g., mean or median value of the peptide fold changes. Principle (b) is based on the assumption that the sum of peptide peak areas (either all or just some of them, like in the top 3 theory [19,20]) for a given protein is proportional to its absolute abundance. Thus, comparison of these sums for two proteins is supposed to give the difference in their content within one sample. What is the most important, when making a comparison between several samples, the two approaches (a) and (b) are supposed to give consistent results.
In case of protein analysis of “non-sequenced” species, both these approaches encounter significant albeit different problems arising from the incomplete peptide identification due to the lack of adequate protein amino acid sequences in the search database. Principle (a) works best when as many as possible shared peptides per protein are identified and quantified for a pair of samples, since individual peptide measurements are prone to err due to possible post-translational modifications or isoforms. When it comes to different species, the number of shared peptides between samples goes down just because of different protein sequences. Besides, this approach works only when there are shared peptide sequences identified and quantified in both samples (recommended number of shared peptides for a reliable quantitation is 2). Thus, if a protein is unique for a sample, it cannot be quantified this way at all. Besides, it provides no data for concentration comparison between different proteins within a single sample.
Principle (b) was developed and verified for systems (artificial protein mixtures) where all the best flyer peptides for a protein (the peptides which have the best proportion between peptide concentration and intensity and thus have the maximum impact on the summed protein intensity) can be easily identified and quantified [19]. For “non-sequenced” species it would mess the final results through protein abundance underestimation if the missed peptides were among the best flyers for the given protein of some particular species but were overlooked because their amino acid sequence was missing in the database. For that, peptide MS/MS sequencing de novo might help a bit, but many peptides would still be missed for the reasons that are not clarified.
Here, we used two approaches to quantify proteins. First, we used MaxLFQ approach [21] which is basically principle (a), but it also uses absolute peptide intensities in addition to peptide FC comparison between samples (such results are labeled LFQ in Table 2). Second, we used direct comparison of sums of peptide intensities to make quantitation within each sample (such results are labeled INT in Table 2). The results of protein quantitation made by different methods gave quite similar results (Table 2, Table S1), especially when potential errors in individual protein contents were compensated by consolidation of proteins into families.
There is also a question of which types of peptides should be used for protein quantitation. Protein identification process deals not with separate proteins, but with protein groups, which are sets of individual proteins (at least partially homologous) sharing a set of identified peptide sequences. In the absence of unique specific peptides, no distinction between these proteins within a group can be made. A standard approach is to take as a hit the protein from a protein group which has a maximum number of assigned identified peptides. Thus, there are three types of peptides for a single protein group in the identification list: unique, razor and other (shared) peptides (MaxQuant terminology) [22,23]. Usually, protein groups have some unique peptides to pinpoint them as “correct” hits, but it is also possible that the number of unique peptides for a protein is zero. Absence of unique peptides means that all the peptides from the current protein group are shared and can be just as successfully assigned to some other protein groups. In such situation, the final set of protein groups shown in the identification list is the minimal one sufficient to explain all the identified peptides (Occam’s razor principle). Shared peptides are named “razor” when they belong to the protein group with the maximum total number of peptides among other possible protein groups. These razor peptides are used for quantitation (along with unique peptides), both LFQ and intensity based [24]. A shared peptide, which is “razor” for some particular group, is counted in “all peptides” in all the protein groups to which it can be potentially assigned, and “all peptides” list is used to calculate Sequence Coverage.
Importantly, any analytical method may prove only that the amount of the compound under investigation is below the method sensitivity, rather than show the absolute absence of the compound in the sample. This is specifically applicable for the LC-MS/MS-based shotgun identification principle which selects peptide ions pseudo-randomly, sometimes missing the peptides with very low intensities just because of a wrong choice. Thus, quantitation is much more reliable for showing the absence of the compound (or, more accurate, the concentration being lower than its Low Limit of Detection). MaxQuant features chromatogram alignment and the possibility to quantify peptides on the basis of similarity of their retention time and m/z in the sample, where they were identified by MS/MS and in another sample where this particular m/z signal got lost during the shot-gun selection for the MS/MS analysis (proteins with such peptides are marked “By matching” in “Identity Type” column in Tables S1 and S2). In this work the protein is considered to be identified (and considered as present) in the sample only if it has an MS/MS spectrum identified in this particular sample. However, for quantitation both MS/MS identified peptides and the peptides identified on the basis of the above described similarity were used. This might lead to apparent contradictions when there are no proteins identified, but the protein abundance is non-zero (like natriuretic peptides (B-NAP) in the V. nikolskii venom—0.01/0.01 (0) in Table 2).
At the present time, the genus Vipera includes 22 species, however it is not homogenous. Molecular phylogeny studies showed that this genus comprises the V. aspis group, the V. ammodytes complex, and the Pelias group as separate clades [25]. Of these clades, only snakes from the Pelias group inhabits Russia. The Pelias was further classified into two subgroups, one comprising V. dinniki, V. kasnakovi, and V. ursinii, and another including V. berus, V. barani, V. nikolskii, and V. seoanei [25]. The first subgroup was further subdivided into the “kaznakovi” complex, including V. kasnakovi, V. orlovi and some other closely related species, and the “ursinii” complex, in which V. renardi was included [26,27]. Earlier, for the vipers of the Pelias group, we have studied the venom toxicity towards crickets Gryllus assimilis [12] and found that it differed depending on feeding preferences. The snakes from the V. renardi, V. lotievi, V. kaznakovi, and V. orlovi species feed on a wide range of animals including insects, whereas the snakes from V. berus and V. nikolskii species do not include insects in their diet. The venom from vipers which hunt insects was found to possess a greater toxicity towards crickets. This suggests that the venom composition may greatly differ among these species. As concerns the toxicity to other animals, it was shown that the venom of V. nikolskii was more toxic than that of V. berus to frogs (9–11 µg/g vs. 30–52 µg/g) and mice (0.93 vs 1.58 µg/g) at intraperitoneal injection [28]. The venom of V. renardi was less toxic to mice (2.96 µg/g) than that of V. berus [28]. We were not able to find any data about toxicity of V. orlovi and V. kaznakovi venoms.
Regarding the danger to humans, the data about bites by these snakes are sparse. Most of the documented cases refer to steppe viper V. renardi and report that it usually has calm and timid behavior, is reluctant to bite, and seeks to escape. This viper bites only when it is in danger, for example, if the snake is suddenly stepped on or picked up. V. renardi is considered less dangerous to humans than common adder. The human fatalities as a consequence of steppe viper bites are not reliably known [29], though there are some cases of the death of horses and small ruminants. A picture of human envenomation is characterized mainly by local signs which include severe pain at the site of the bite, redness, swelling that spreads far beyond the site of the biting. In severe cases, drowsiness, dizziness, nausea, increase of heart rate, and reduction in body temperature may be observed [30].
Records of the bites of humans by the Caucasian viper V. kaznakovi and Nikolsky’s viper V. nikolskii are practically absent. However, V. kaznakovi may be dangerous. Solitary human deaths and livestock losses after Caucasian viper bites were mentioned [30]. We were able to find only one report about human fatalities after the Nikolsky’s viper bites [31]. No information on the V. orlovi bites is available.
It should be noted that the venoms of not all Pelias species were studied equally well. The venom of V. berus is the best characterized. As mentioned earlier, the V. berus venom displayed in vitro proteolytic, fibrinolytic, anticoagulant, and phospholipolytic activities. In mice, significant local tissue-damaging effects, including edema, hemorrhage and myonecrosis were observed for this venom [3]. Several proteins involved in manifestation of those effects were isolated from V. berus venom. These proteins included basic PLA2 [4], SVMP [5], LAAO [6] and some others.
The V. nikolskii species is phylogenetically very close to V. berus and is included in the same subgroup within the Pelias group. It is regarded as a V. berus subspecies in some publications. However, the analysis of the V. nikolskii venom has shown it to differ greatly from that of V. berus. Thus, two heterodimeric PLA2s were isolated from the V. nikolskii venom [10], but similar proteins are absent in V. berus. The data obtained in the present work are in good agreement with the published results; the basic and acidic PLA2 subunits forming heterodimeric enzymes account for more than 50% of the V. nikolskii venom (Table 1). Earlier, cDNA encoding SP nikobin and Kunitz type inhibitor in the V. nikolskii venom gland was cloned and sequenced [11]. In this study we have found that nikobin is the main SP in the V. nikolskii venom (more than 12% of the total protein content, Table 1) and Kunitz-type serine protease inhibitor ki-VN was also the main protein of the Kunitz family in this venom (about 0.6%, Table 1). CRISP, the sequence of which was also deduced from cDNA analysis [32], was found in the venom in fairly low amount (0.66%, Figure 2). Interestingly, the content of CRISPs was much higher in other venoms studied and accounted for 8%, 10% and 12% in V. renardi, V. kaznakovi and V. orlovi venoms, respectively (Figure 2).
The steppe viper V. renardi is included in the “ursinii” complex [33] while the other two vipers, V. kaznakovi and V. orlovi, belong to the “kaznakovi” complex. Among these vipers only the composition of the V. renardi venom was in some way studied [7]. The amino acid sequences for several PLA2s and Kunitz-type inhibitor were deduced from the cloned cDNA of venom gland. Some PLA2s and Kunitz protein were isolated from the venom. The most abundant was ammodytin I2d analogue. In this work we have found all the PLA2s described by Tsai et al. [7] in the V. renardi venom, Vur-PL2 having the highest content (Table 1). Interestingly, this viper venom has very high content of disintegrins which accounts for about 13% of total protein, while the V. kaznakovi and V. orlovi venoms contain less than 1% and in the V. nikolskii venom no disintegrins were detected.
There are no published data on the composition of the V. kaznakovi and V. orlovi venoms and they are characterized for the first time in this work. These two venoms have the highest similarity among the four ones studied (Figure 3) that confirms the inclusion of V. orlovi in the “kaznakovi” complex. They have a fairly high content of SVMPs (15%–16%), CTL (11%–12%) and CRISPs (11%–12%) (Figure 3). The V. orlovi venom has the highest amount of SP (24%) among the four venoms studied (Figure 3) and only V. kaznakovi contains a small quantity of hyaluronidase (Hya) at the level of 0.01%. However no Kunitz type proteins were detected in the latter venom.
Although a limited number of B-NAP proteins (one in V. kaznakovi and one in V. orlovi, Table 2) were detected in the proteome analysis, several peptides derived from proteins of this family were found in the peptidomes of all the venoms studied (Figure 4). The mature bradykinin-potentiating peptide QGGLPRPGPEIPP was observed in the V. nikolskii venom and several fragments of similar peptides were detected in the other analyzed venoms. Several fragments of C-type natriuretic peptides were found in all four venoms as well (Figure 4). It should be noted that no bradykinin-potentiating and C-type natriuretic peptides from the vipers of Pelias group were reported so far.
In total, 210 proteins (Table 1) and 512 endogenous peptides (Table S3) were identified in four viper venoms. The overwhelming majority of the proteins (98%–99% of the total protein content) and the peptides represented 14 snake venom protein families (Table 2). The comparison of our results with those for other snakes of the Vipera genus shows higher representation of venom protein families in our data (Table 4). For example, while Nuc and PLB were found in all venoms studied in this work, no proteins of these families were reported for other venoms from the Vipera species (Table 4).
Table 4.
Snake venom protein families represented in viper venoms.
| Snake Venom Protein Family | V. kaznakovi Venom | V. renardi Venom | V. orlovi Venom | V. nikolskii Venom | V. anatolica Venom 1 | V. raddei Venom 2 | V. a. ammodytes Venom 3 | V. a. meridionalis Venom 3,4 |
|---|---|---|---|---|---|---|---|---|
| PLA2 | + | + | + | + | + | + | + | + 3 |
| SP | + | + | + | + | + | + | + | + 3 |
| Dis | + | + | + | − | + | + | + | + 3 |
| CRISP | + | + | + | + | + | + | + | − |
| Kunitz | − | + | + | + | + | + | − | + 4 |
| LAAO | + | + | + | + | − | + | + | + 3 |
| SVMP | + | + | + | + | + | + | + | + 3 |
| NGF | + | + | + | + | − | + | + | + 3 |
| CTL | + | + | + | + | + | + | − | − |
| PLB | + | + | + | + | − | − | − | − |
| VEGF | + | + | + | + | − | + | + | + 3 |
| Nuc | + | + | + | + | − | − | − | − |
| B-NAP | + | + | + | + | − | + | − | + 4 |
| Hya | + | − | + | + | − | − | − | − |
Hya was observed in three of the studied venoms and this is also the first indication for the presence of this enzyme in the venoms of the Vipera species. We have found that the main components of all venom studied are PLA2s, while SVMPs were prevailing in venoms of V. anatolica [13] and V. raddei [14].
4. Conclusions
In this work, quantitative proteomic and peptidomic characterization of venoms from four vipers inhabiting Russia was done; the compositions of the venoms from V. kaznakovi and V. orlovi, which showed the highest similarity among the four studied species, were analyzed for the first time.
More than 200 proteins and over 500 peptides were detected in total in all four venoms. They represented 14 snake venom protein families. In all venoms studied, over 70% of the total proteins were enzymes, the highest enzyme content (85.7%) being in the V. nikolskii venom. The main components of the venoms were PLA2s, which accounted for 65% of total protein content in the V. nikolskii venom. For the first time, bradykinin-potentiating and C-type natriuretic peptides were reported for vipers of the Pelias group. Nucleic acid degrading enzymes and phospholipase B were found in the venoms of Vipera species for the first time.
Due to the low toxicity of the steppe viper, or a limited habitat of the Caucasian and Orlov’s vipers, these snakes do not pose an epidemiological threat to Russian population. However, the envenomation by Nikolsky’s viper, the venom of which was shown in this study to contain a considerable amount of neurotoxic phospholipase A2, may represent certain danger. An antiserum “Antigadyuka” (“Antiviper”) produced by Russian company “Allergen” is based on the venom of the common viper and may not be effective against the Nikolsky’s viper bites due to strong differences in the composition of the venoms. The need to consider the differences in the composition of the venoms in the antivenom production is discussed in recent publications [28,35] and should be taken into account by antiserum manufacturers.
5. Materials and Methods
The venoms of V. kaznakovi, V. nikolskii, V. orlovi and V. renardi vipers were obtained as described earlier [12]. The venoms from several individual animals were pooled as described in [12]. Snakes were captured in their natural habitat: V. kaznakovi in Krasnodar Territory near Adler, V. nikolskii in Penza region near Zubrilovo village, V. orlovi in Krasnodar Territory at Mikhaylovskiy mountain pass and V. renardi in Krasnodar Territory near Beysugskiy firth.
5.1. In-Solution Trypsin Digestion of Venom Samples
Lyophilized venom sample (100 μg each) was dissolved in 10 μL of a buffer containing 100 mM ammonium bicarbonate (ABC), 5% sodium deoxycholate (SDC) and 5 mM dithiothreitol (DTT) and incubated for 40 min at 60 °C to reduce cysteine residues. Then, 5 μL of 50 mM iodoacetamide (IAA) water solution was added and the mixture was incubated 30 min at RT, in the dark. Residual IAA was neutralized by 5 μL of 50 mM DTT and sample was diluted with 50 μL 50 mM ABC and trypsin was added in a 1:100 (enzyme/protein) ratio to the final volume 100 μL and the protein concentration ~1 mg/mL. Samples were incubated overnight at 37 °C. Trypsin was deactivated by addition of 5 μL of 10% TFA. Tryptic peptides were desalted using reverse-phase solid extraction cartridges Discovery DSC-18 (100 mg) (Supelco, Bellefonte, PA, USA) according to the manufacturer protocol. Final peptide solution was dried in vacuum and stored at −80 °C prior to LC-MS/MS analysis.
5.2. Endogenous Venom Peptides Isolation
Endogenous peptides from venom samples were isolated using C18 StageTips [36]. To make StageTips, two pieces of C18 Empore extraction disk were cut using blunt-ended 16-gauge needle and packed into a P200 pipette tip. Membranes were conditioned by 20 μL of methanol and equilibrated by 20 μL of 0.1% aqua TFA. Venom solutions were applied onto the conditioned tips, followed by membrane washing with 20 μL of 0.1% aqua TFA. Peptides were eluted by 20 μL of 80% ACN, 0.1% TFA. Eluates were dried in vacuum and stored at −80 °C prior to LC-MS/MS analysis.
5.3. LC-MS/MS Analysis
Analysis was performed on the QExactive HF mass-spectrometer (Thermo Scientific, Waltham, MA, USA) coupled to the Dionex 3000 RSLCnano HPLC system (Thermo Scientific, Waltham, MA, USA). The HPLC system was configured in a trap-elute mode. An analytical column (75 μm × 150 mm) and a precolumn (100 μm × 10 mm) were in-house packed with Aeris Peptide C18 2.6 μm sorbent (Phenomenex, Torrance, CA, USA). Samples were loaded on the precolumn for 10 min at 3 mL/min with buffer A (3% AcN, 96.9% H2O, 0.1% FA), followed by separation at 300 nL/min with the 4%–55% gradient of buffer B (80% AcN, 19.9% H2O, 0.1% FA).
Mass-spectrometer experiment consisted of one full survey MS1 scan followed by 20 dependent MS2 scans for the most intense ions. MS1 spectra were acquired in the profile mode in mass range 350–1400 m/z, maximum IT time 100 ms, AGC target 3e6, resolution 60000. Dependent MS2 scan were performed at resolution 15000 for 200–2000 m/z mass range, AGC target 1e5, maximum IT 25 ms, isolation window 1.4 m/z. Dynamic exclusion was set to 30 s.
5.4. LC-MS/MS Data Analysis
Data analysis was performed in the MaxQuant software (V. 1.5.3.30, Max Planck Institute of Biochemistry, Martinsried, Germany, 2016). Proteomic LC-MS/MS data was searched with the Andromeda search engine incorporated in the MaxQuant software against NCBI Serpentes DataBase exported from the NCBI web-site [37] for the Taxon Serpentes 2015/11/17 and containing 134677 entries with the following parameters: digestion Trypsin/P; max number of miscleavages 2; include contaminants; fixed modification: carbamidomethyl (Cys); variable modifications: Oxidation (Met), Acetylation (N-term), Deamidation (Asn, Gln); min peptide length 6; max peptide MW 5500; PSM FDR 0.01; protein FDR 0.05; decoy mode: revert; min number of peptides for identification 1; razor protein FDR; second peptide; match between runs; LFQ quantitation with minimum 2 peptide pairs; and stabilize large LFQ ratios. Full set of MaxQuant parameters for the analysis can be found in the Supplementary Data file mqpar_proteins.xml.
Peptidomic LC-MS/MS data were searched with the Mascot search engine against the same full NCBI Serpentes data base with the following parameters: MS tolerance 5 ppm; MS/MS tolerance 0.01 Da; charge: +1, +2, +3; fixed modification: carabamidomethyl (Cys); variable modifications: Oxidation (Met), Deamidation (Asn, Gln); enzyme none. Mascot results were reprocessed in the Scaffold software and identified peptidogenic proteins (protein FDR 10%) for all four venoms were added to the SwissProt Serpentes database exported from the NCBI web-site on 2015/11/30 and contains 2567 sequences to generate a fused database. This database was used for Andromeda search in MaxQuant software with the digestion parameter set to unspecific. Peptide length for unspecific digestion search was from 6 to 50 amino acids. The rest of the parameters were the same as for proteome data analysis, however in the peptidogenic protein features in the in the “Number of Unique and Razor Peptides (NoURP)” column, the number of peptides corresponds to that before PEP-based filtration.
Results were processed in the Perseus (V. 1.5.2.6, Max Planck Institute of Biochemistry, Martinsried, Germany, 2016) and Excel software (V. 12.06743.5000, Microsoft Corporation, Redmond, WA, USA, 2007) and with the use of R.
Acknowledgments
This study was supported in part by the Russian Foundation for Basic Research (project No. 15-04-01843) and the Russian Science Foundation (project No. 16-14-00215).
Abbreviations
The following abbreviations are used in this manuscript:
- FC
Fold Changes
- FDR
False Discovery Rate
- B-NAP
Bradykinin potentiating and C-type natriuretic peptides
- BP
Blood protein
- CP
Cysteine Proteases
- CRISP
Cysteine-rich secretory protein
- CTL
C-type lectin like
- Dis
Disintegrin
- Hya
Hyaluronidase
- Kunitz
Kunitz type proteinase inhibitor
- LFQ
label-free quantification
- LAAO
l-amino acid oxidase
- NGF
Nerve growth factor
- Nuc
Nucleic acid degrading enzymes
- OP
Other protein
- PLA2
Phospholipase A2
- PLB
Phospholipase B
- SP
Serine proteinase
- SVMP
Snake venom metalloproteinase
- TBP
Toxin biosynthesis proteins (including aminopeptidases)
- VEGF
Vascular endothelial growth factor
Supplementary Materials
The following materials are available online at www.mdpi.com/2072-6651/8/4/105/s1, Table S1: The detailed list of proteins identified in Russian viper venoms; Table S2: The detailed peptide identification list for proteome results (contains all peptide-related data from MaxQuant proteome analysis); Table S3: Endogenous peptides found in four viper venoms: “endogPeptidesWithProteins”—combined peptide and protein data, “endogenousPeptides”—peptide only data, “endogenousPeptidogenicProteins”—protein only data; Table S4: Endogenous peptides identified in proteins unique for peptidome analysis.
Author Contributions
S.I.K., R.H.Z. and Y.N.U. conceived and designed the experiments; S.I.K. and R.H.Z. performed the experiments; S.I.K., R.H.Z., V.I.T. and Y.N.U. analyzed the data; V.G.S. contributed materials; and S.I.K., R.H.Z., V.I.T. and Y.N.U. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tuniyev B.S., Ostrovskikh S. Two new species of vipers of “kaznakovi” complex (Ophidia, Viperinae) from the Western Caucasus. Russ. J. Herpetol. 2001;8:117–126. [Google Scholar]
- 2.Tuniyev S.B., Orlov N.L., Tuniyev B.S., Kidov F.F. On the taxonomical status of steppe viper from foothills of the south macroslope of the East Caucasus. Russ. J. Herpetol. 2013;20:129–146. [Google Scholar]
- 3.Calderón L., Lomonte B., Gutiérrez J.M., Tarkowski A., Hanson L.Å. Biological and biochemical activities of Vipera berus (European viper) venom. Toxicon. 1993;31:743–753. doi: 10.1016/0041-0101(93)90380-2. [DOI] [PubMed] [Google Scholar]
- 4.Križaj I., Siigur J., Samel M., Cotič V., Gubenšek F. Isolation, partial characterization and complete amino acid sequence of the toxic phospholipase A2 from the venom of the common viper, Vipera berus berus. Biochim. Biophys. Acta. 1993;1157:81–85. doi: 10.1016/0304-4165(93)90081-I. [DOI] [PubMed] [Google Scholar]
- 5.Samel M., Vija H., Subbi J., Siigur J. Metalloproteinase with factor X-activating and fibrinogenolytic activities from Vipera berus berus venom. Comp. Biochem. Physiol. B. 2003;135:575–582. doi: 10.1016/S1096-4959(03)00171-4. [DOI] [PubMed] [Google Scholar]
- 6.Samel M., Vija H., Rönnholm G., Siigur J., Kalkkinen N., Siigur E. Isolation and characterization of an apoptotic and platelet aggregation inhibiting l-amino acid oxidase from Vipera berus berus (common viper) venom. Biochim. Biophys. Acta. 2006;1764:707–714. doi: 10.1016/j.bbapap.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 7.Tsai I.H., Wang Y.M., Cheng A.C., Starkov V., Osipov A., Nikitin I., Makarova Y., Ziganshin R., Utkin Y. cDNA cloning, structural, and functional analyses of venom phospholipases A2 and a Kunitz-type protease inhibitor from steppe viper Vipera ursinii renardi. Toxicon. 2011;57:332–341. doi: 10.1016/j.toxicon.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Ghazaryan N.A., Ghulikyan L., Kishmiryan A., Andreeva T.V., Utkin Y.N., Tsetlin V.I., Lomonte B., Ayvazyan N.M. Phospholipases A2 from Viperidae snakes: Differences in membranotropic activity between enzymatically active toxin and its inactive isoforms. Biochim. Biophys. Acta. 2015;1848:463–468. doi: 10.1016/j.bbamem.2014.10.037. [DOI] [PubMed] [Google Scholar]
- 9.Vulfius C.A., Kasheverov I.E., Starkov V.G., Osipov A.V., Andreeva T.V., Filkin S.Y., Gorbacheva E.V., Astashev M.E., Tsetlin V.I., Utkin Y.N. Inhibition of nicotinic acetylcholine receptors, a novel facet in the pleiotropic activities of snake venom phospholipases A2. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0115428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramazanova A.S., Zavada L.L., Starkov V.G., Kovyazina I.V., Subbotina T.F., Kostyukhina E.E., Dementieva I.N., Ovchinnikova T.V., Utkin Y.N. Heterodimeric neurotoxic phospholipases A2—The first proteins from venom of recently established species Vipera nikolskii: Implication of venom composition in viper systematics. Toxicon. 2008;51:524–537. doi: 10.1016/j.toxicon.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Ramazanova A.S., Fil’kin S.Iu., Starkov V.G., Utkin Iu.N. Molecular cloning and analysis of cDNA sequences encoding serine proteinase and Kunitz type inhibitor in venom gland of Vipera nikolskii viper. Bioorg. Khim. 2011;37:374–385. doi: 10.1134/s1068162011030149. [DOI] [PubMed] [Google Scholar]
- 12.Starkov V.G., Osipov A.V., Utkin Y.N. Toxicity of venoms from vipers of Pelias group to crickets Gryllus assimilis and its relation to snake entomophagy. Toxicon. 2007;49:995–1001. doi: 10.1016/j.toxicon.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Goçmen B., Heiss P., Petras D., Nalbantsoy A., Süssmuth R.D. Mass spectrometry guided venom profiling and bioactivity screening of the Anatolian Meadow Viper, Vipera anatolica. Toxicon. 2015;107:163–174. doi: 10.1016/j.toxicon.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Sanz L., Ayvazyan N., Calvete J.J. Snake venomics of the Armenian mountain vipers Macrovipera lebetina obtusa and Vipera raddei. J. Proteomics. 2008;71:198–209. doi: 10.1016/j.jprot.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Georgieva D., Risch M., Kardas A., Buck F., von Bergen M., Betzel C. Comparative analysis of the venom proteomes of Vipera ammodytes ammodytes and Vipera ammodytes meridionalis. J. Proteome Res. 2008;7:866–886. doi: 10.1021/pr070376c. [DOI] [PubMed] [Google Scholar]
- 16.Stümpel N., Joger U. Recent advances in phylogeny and taxonomy of near and middle eastern vipers—An update. ZooKeys. 2009;31:179–191. doi: 10.3897/zookeys.31.261. [DOI] [Google Scholar]
- 17.McCleary R.J., Kini R.M. Non-enzymatic proteins from snake venoms: A gold mine of pharmacological tools and drug leads. Toxicon. 2013;62:56–74. doi: 10.1016/j.toxicon.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Waridel P., Frank A., Thomas H., Surendranath V., Sunyaev S., Pevzner P., Shevchenko A. Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics. 2007;7:2318–2329. doi: 10.1002/pmic.200700003. [DOI] [PubMed] [Google Scholar]
- 19.Silva J.C., Gorenstein M.V., Li G.Z., Vissers J.P., Geromanos S.J. Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Grossmann J., Roschitzki B., Panse C., Fortes C., Barkow-Oesterreicher S., Rutishauser D., Schlapbach R. Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J. Proteomics. 2010;73:1740–1746. doi: 10.1016/j.jprot.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J.V., Mann M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 23.Nesvizhskii A.I., Aebersold R. Interpretation of shotgun proteomic data: The protein inference problem. Mol. Cell. Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 25.Garrigues T., Dauga C., Ferquel E., Choumet V., Failloux A.-B. Molecular phylogeny of Vipera Laurenti, 1768 and the related genera Macrovipera (Reuss, 1927) and Daboia (Gray, 1842), with comments about neurotoxic Vipera aspis aspis populations. Mol. Phylogenet. Evol. 2005;35:35–47. doi: 10.1016/j.ympev.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Kalyabina-Hauf S., Schweiger S., Joger U., Mayer W., Orlov N., Wink M. Phylogeny and systematics of adders (Vipera berus complex) Mertensiella. 2004;15:7–15. [Google Scholar]
- 27.Zinenko O., Stumpel N., Mazanaeva L., Bakiev A., Shiryaev K., Pavlov A., Kotenko T., Kukushkin O., Chikin Yu., Duisebayeva T., et al. Mitochondrial phylogeny shows multiple independent ecological transitions and northern dispersion despite of Pleistocene glaciations in meadow and steppe vipers (Vipera ursinii and Vipera renardi) Mol. Phylogenet. Evol. 2015;84:85–100. doi: 10.1016/j.ympev.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Bakiev A.G., Garanin V.I., Gelashvili D.B., Gorelov R.A., Doroni I.V., Zaytseva O.V., Zinenko F.I., Klyonina A.A., Makarova T.N., Malenyov A.L., et al. Vipers (Reptilia: Serpentes: Viperidae: Vipera) of the Volga basin. Part. 1. Cassandra; Togliatti, Russia: 2015. p. 234. (In Russian) [Google Scholar]
- 29.Bakiev А.G., Garanin V.I., Pavlov А.V., Shurshina I.V., Malenyov А.L. East steppe viper Vipera renardi (Reptilia, Viperidae) in Volga river basin: Materials on biology, ecology and toxicology. Samar. Luka. 2008;17:817–845. (In Russian) [Google Scholar]
- 30.Orlov B.N., Gelashvili D.B., Ibrahimov A.K. Venomous Animals and Plants of USSR. Vysshaya Shkola; Moscow, Russia: 1990. pp. 111–117. (In Russian) [Google Scholar]
- 31.BelPressa. [(accessed on 14 February 2016)]. Available online: http://www.belpressa.ru/news/news/osoba-holodnyh-krovej08494/ (In Russian)
- 32.Ramazanova A.S., Starkov V.G., Osipov A.V., Ziganshin R.H., Filkin S.Y., Tsetlin V.I., Utkin Y.N. Cysteine-rich venom proteins from the snakes of Viperinae subfamily—molecular cloning and phylogenetic relationship. Toxicon. 2009;53:162–168. doi: 10.1016/j.toxicon.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Nilson G., Andren C. The meadow and steppe vipers of Europe and Asia—The Vipera (Acrodiphaga) ursinii complex. Acta Zool. Acad. Sci. Hung. 2001;47:87–267. [Google Scholar]
- 34.Munawar A., Trusch M., Georgieva D., Spencer P., Frochaux V., Harder S., Arni R.K., Duhalov D., Genov N., Schluter H., et al. Venom peptide analysis of Vipera ammodytes meridionalis (Viperinae) and Bothrops jararacussu (Crotalinae) demonstrates subfamily-specificity of the peptidome in the family Viperidae. Mol. Biosyst. 2011;7:3298–3307. doi: 10.1039/c1mb05309d. [DOI] [PubMed] [Google Scholar]
- 35.Zaitseva О.V., Маlenyov А.L., Bakiev А.G. Studies of properties of the common viper’s venom in the Volga river basin: Practical value of the results obtained. Samar. Luka. 2011;20:180–184. (In Russian) [Google Scholar]
- 36.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, prefractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 37.NCBI (National Center for Biotechnology Information) [(accessed on 17 November 2015)]; Available online: http://www.ncbi.nlm.nih.gov/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.