Abstract
The pathogenesis of celiac disease (CD) has been related to polymorphisms in the regulator of G-protein signaling 1 (RGS1) and interleukin-12 A (IL12A) genes, but the existing findings are inconsistent. Our aim is to investigate the associations of two single-nucleotide polymorphisms (SNPs) (rs2816316 in RGS1 and rs17810546 in IL12A) with CD risk using meta-analysis. We searched PubMed and Web of Science on RGS1 rs2816316 and IL12A rs17810546 with CD risk. Odds ratio (OR) and 95% confidence interval (CI) of each SNP were estimated. All statistical analyses were performed on Stata 12.0. A total of seven studies were retrieved and analyzed. The available data indicated the minor allele C of rs2816316 was negatively associated with CD (C vs. A: OR = 0.77, 95% CI = 0.74–0.80), and a positive association was found for the minor allele G of rs17810546 (G vs. A: OR = 1.37, 95% CI = 1.31–1.43). The co-dominant model of genotype effect confirmed the significant associations between RGS1 rs2816316/IL12A rs17810546 and CD. No evidence of publication bias was observed. Our meta-analysis supports the associations of RGS1 and IL12A with CD and strongly calls for further studies to better understand the roles of RGS1 and IL12A in the pathogenesis of CD.
Keywords: celiac disease, IL12A, meta-analysis, polymorphism, RGS1
1. Introduction
Celiac disease (CD) is a chronic small intestinal immune-mediated enteropathy induced by dietary gluten in genetically predisposed people [1]. Most patients are treated by a life-long gluten-free diet. It has a prevalence of ~1% in many populations worldwide [2]. A genetic linkage study recognized HLA-DQ2 and HLA-DQ8 as the main genetic predisposing factors in the development of CD [3]. However, a genetic-association study by Petronzelli et al. [4] identified the effect of HLA contributing to CD was estimated to be only 36%, so non-HLA genes must be considered [5].
Recently, a two-stage genome-wide association study (GWAS) found the contribution of the regions on chromosome 1q31 and 3q25 containing the regulator of G-protein signaling-1 (RGS1) gene and interleukin-12 A (IL12A) gene in the development of CD in European populations [6]. Both of these genes are involved in the T helper cell type 1 (Th1) pathway, which are responsible for mucosal inflammation in active CD [7]. RGS1, a member of the RGS protein family, activated GTPase by attenuating the signaling activity of G-proteins [8]. Moratz et al. [9] have reported RGS1 regulates chemokine receptor signaling and participates in B cell activation and proliferation. B cell differentiates into plasma cell and produce anti-tTG/antigliadin antibodies [10]. By interacting with mtTG in the basement membrane region, tTG antibody might lead to enterocyte cytoskeleton and epithelial damage in CD [10]. While IL12A encodes the IL12p35 submit, coincides with IL12p40 to form IL12p70. The heterodimeric IL-12 cytokine has a broad range of bioactivities on T and natural killer cells and it induces the production of interferon-γ (INF-γ) [11]. Activated CD4+ T cell produces pro-inflammatory cytokines and results in Th1 response, which is dominantly regulated by INF-γ [12]. Th-1 cytokines promote inflammatory effects and eventually induce enterocyte apoptosis in CD [13].
Several studies have assessed the role for RGS1 rs2816316 (C/A) and IL12A rs17810546 (G/A) in CD development [6,7,14,15,16,17,18]. However, the existing findings were inconsistent even among different ethnic populations in the same study. Thus, we perform a meta-analysis of all available studies to accurately estimate the relationships of RGS1 rs2816316/IL12A rs17810546 polymorphisms with CD risk.
2. Results
2.1. Eligible Studies and Characteristics
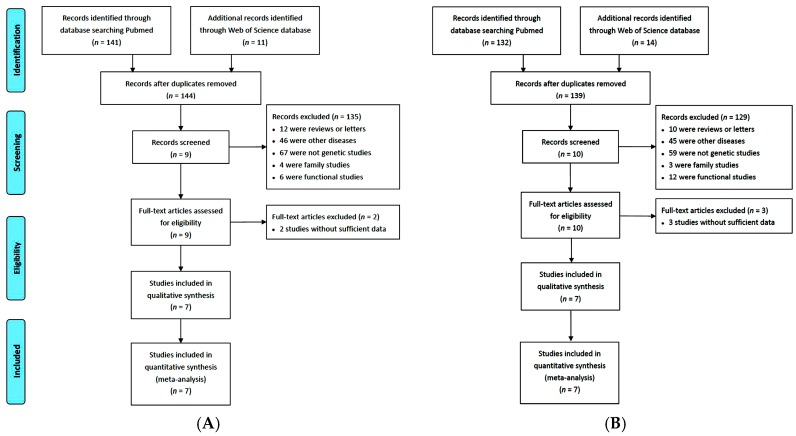
According to the search strategy and inclusion criteria, seven studies involving 20 sub-study collections were identified about the associations of RGS1 and IL12A polymorphisms with CD [6,7,14,15,16,17,18], of which 19 were conducted in Europe and one in America (Table 1). The detailed steps of literature search are shown in Figure 1. Both RGS1 and IL12A polymorphisms were studied in 14,936 CD patients and 24,794 controls.
Table 1.
Characteristics of the included studies.
| Author | Year | Country | Ethnicity | Genotyping Method | MAF | Sample Size | ||
|---|---|---|---|---|---|---|---|---|
| Control_1 | Control_2 | Case | Control | |||||
| Van Heel [18] | 2007 | UK | European | The Illumina Infinium II assay | 0.174 | 0.123 | 778 | 1422 |
| Hunt [6] | 2008 | UK | European | Infinium assay | 0.188 | 0.127 | 719 | 1561 |
| – | – | Ireland | European | GoldenGate assay | 0.187 | 0.130 | 416 | 957 |
| – | – | Netherlands | European | GoldenGate assay | 0.189 | 0.124 | 508 | 888 |
| Romanos [16] | 2009 | Italy | European | Taqman probe | 0.166 | 0.067 | 538 | 598 |
| Coenen [14] | 2009 | Netherlands | European | Taqman probe | 0.190 | 0.120 | 795 | 1683 |
| Dubois [15] | 2010 | UK1 | European | Illumina Hap300v1-1 | 0.182 | 0.118 | 737 | 2596 |
| – | – | UK2 | European | Illumina 670-QuadCustom_v1 | 0.173 | 0.121 | 1849 | 4936 |
| – | – | Finland | European | Illumina 670-QuadCustom_v1 | 0.143 | 0.101 | 647 | 1829 |
| – | – | Netherlands | European | Illumina 670-QuadCustom_v1 | 0.188 | 0.118 | 803 | 846 |
| – | – | Italy | European | Illumina 670-QuadCustom_v1 | 0.169 | 0.067 | 497 | 543 |
| – | – | USA | American | Illumina GoldenGate | 0.162 | 0.118 | 973 | 555 |
| – | – | Hungary | European | Illumina GoldenGate | 0.158 | 0.113 | 965 | 1067 |
| – | – | Ireland | European | Illumina GoldenGate | 0.182 | 0.133 | 597 | 1456 |
| – | – | Poland | European | Illumina GoldenGate | 0.151 | 0.133 | 564 | 716 |
| – | – | Spain | European | Illumina GoldenGate | 0.188 | 0.095 | 550 | 433 |
| – | – | Italy | European | Illumina GoldenGate | 0.180 | 0.078 | 1010 | 804 |
| – | – | Finland | European | Illumina GoldenGate | 0.162 | 0.113 | 259 | 653 |
| Sperandeo [17] | 2011 | Italy | European | Taqman probe | 0.178 | 0.077 | 637 | 711 |
| Plaza-Izurieta [7] | 2011 | Spain | European | TaqMan probe | 0.199 | 0.094 | 1094 | 540 |
MAF: minor allele frequency; Control_1: controls for RGS1 rs2816316; Control_2: controls for IL12A rs17810546.
Figure 1.
Flow chart showing the literature selection procedure used in this study: (A) RGS1 rs2816316; and (B) IL12A rs17810546.
The detailed characteristics of the seven included studies are described in Table 1. Genotype and allele distributions for each study are listed in Table 2 and Table 3. Studies from Hunt et al. [6] and Dubois et al. [15] included three and twelve independent searches, respectively, which were estimated independently in our meta-analysis. Most of the studies had clear diagnostic criteria and control source, except the Irish Ethnic group by Dubois et al. [15] that did not provide details from citation.
Table 2.
Genotype frequencies of rs2816316 in celiac disease (CD) and control groups and genotype effects in the meta-analysis.
| Author and Year | CD Group | Control Group | C vs. A | AC vs. AA | CC vs. AA | HWE p Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | AA | AC | CC | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||
| Van Heel 2007 UK [18] | 581 | 183 | 14 | 970 | 409 | 43 | 0.74 | 0.63–0.89 | 0.75 | 0.61–0.91 | 0.54 | 0.29–1.00 | 0.988 |
| Hunt 2008 UK [6] | 545 | 162 | 12 | 1029 | 477 | 55 | 0.64 | 0.54–0.77 | 0.64 | 0.52–0.79 | 0.41 | 0.22–0.78 | 0.976 |
| Hunt 2008 Ireland [6] | 293 | 112 | 11 | 633 | 291 | 33 | 0.84 | 0.67–1.04 | 0.83 | 0.64–1.08 | 0.72 | 0.36–1.44 | 0.950 |
| Hunt 2008 Netherlands [6] | 376 | 122 | 10 | 584 | 272 | 32 | 0.70 | 0.56–0.86 | 0.70 | 0.54–0.89 | 0.49 | 0.24–1.00 | 0.962 |
| Romanos 2008 Italy [16] | 409 | 120 | 9 | 416 | 166 | 16 | 0.74 | 0.59–0.94 | 0.74 | 0.56–0.96 | 0.57 | 0.25–1.31 | 0.908 |
| Coenen 2009 Netherlands [14] | 588 | 191 | 16 | 1104 | 518 | 61 | 0.69 | 0.59–0.94 | 0.69 | 0.57–0.84 | 0.49 | 0.28–0.86 | 0.980 |
| Dubois 2010 UK1 [15] | 551 | 172 | 13 | 1737 | 773 | 86 | 0.70 | 0.59–0.82 | 0.70 | 0.58–0.85 | 0.48 | 0.26–0.86 | 0.999 |
| Dubois 2010 UK2 [15] | 1349 | 461 | 39 | 3376 | 1412 | 148 | 0.82 | 0.73–0.91 | 0.82 | 0.72–0.92 | 0.66 | 0.46–0.94 | 0.980 |
| Dubois 2010 Finland [15] | 495 | 142 | 10 | 1343 | 448 | 37 | 0.86 | 0.71–1.04 | 0.86 | 0.69–1.07 | 0.73 | 0.36–1.49 | 0.956 |
| Dubois 2010 Netherlands [15] | 595 | 192 | 16 | 558 | 258 | 30 | 0.70 | 0.58–0.84 | 0.70 | 0.56–0.87 | 0.50 | 0.27–0.93 | 0.978 |
| Dubois 2010 Italy [15] | 376 | 112 | 8 | 375 | 153 | 16 | 0.72 | 0.57–0.92 | 0.73 | 0.55–0.97 | 0.50 | 0.21–1.18 | 0.934 |
| Dubois 2010 USA [15] | 743 | 214 | 15 | 390 | 151 | 15 | 0.74 | 0.60–1.90 | 0.74 | 0.58–0.95 | 0.52 | 0.25–1.08 | 9.933 |
| Dubois 2010 Hungary [15] | 714 | 232 | 19 | 756 | 284 | 27 | 0.86 | 0.73–1.03 | 0.86 | 0.71–1.06 | 0.75 | 0.41–1.35 | 0.957 |
| Dubois 2010 Ireland [15] | 425 | 157 | 15 | 974 | 434 | 48 | 0.83 | 0.70–1.00 | 0.83 | 0.67–1.03 | 0.72 | 0.40–1.29 | 0.967 |
| Dubois 2010 Poland [15] | 412 | 140 | 12 | 516 | 184 | 16 | 0.96 | 0.77–1.19 | 0.95 | 0.74–1.23 | 0.94 | 0.44–2.01 | 0.932 |
| Dubois 2010 Spain [15] | 394 | 143 | 13 | 285 | 132 | 15 | 0.79 | 0.62–1.00 | 0.78 | 0.59–1.04 | 0.63 | 0.29–1.34 | 0.952 |
| Dubois 2010 Italy [15] | 721 | 265 | 24 | 541 | 237 | 26 | 0.84 | 0.70–1.00 | 0.84 | 0.68–1.03 | 0.69 | 0.39–1.22 | 0.994 |
| Dubois 2010 Finland [15] | 199 | 56 | 4 | 459 | 177 | 17 | 0.73 | 0.54–0.99 | 0.73 | 0.52–1.03 | 0.54 | 0.18–1.63 | 0.989 |
| Sperandeo 2011 Italy [17] | 463 | 166 | 8 | 488 | 193 | 30 | 0.77 | 0.62–0.94 | 0.91 | 0.71–1.16 | 0.28 | 0.13–0.62 | 0.054 |
| Plaza-Izurieta 2011 Spain [7] | 787 | 282 | 25 | 346 | 172 | 21 | 0.72 | 0.60–0.87 | 0.72 | 0.57–0.91 | 0.52 | 0.29–0.95 | 0.947 |
| Overall OR | – | – | – | – | – | – | 0.77 | 0.74–0.80 | 0.77 | 0.74–0.81 | 0.57 | 0.50–0.67 | – |
HWE: Hardy–Weinberg equilibrium.
Table 3.
Genotype frequencies of rs17810546 in CD and control groups and genotype effects in the meta-analysis.
| Author and Year | CD Group | Control Group | G vs. A | AG vs. AA | GG vs. AA | HWE p Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||
| Van Heel 2007 UK [18] | 546 | 211 | 20 | 1094 | 307 | 22 | 1.37 | 1.15–1.63 | 1.38 | 1.12–1.69 | 1.82 | 0.99–3.37 | 0.930 |
| Hunt 2008 UK [6] | 524 | 179 | 15 | 1190 | 346 | 25 | 1.17 | 0.98–1.40 | 1.17 | 0.95–1.45 | 1.36 | 0.71–2.61 | 0.979 |
| Hunt 2008 Ireland [6] | 277 | 125 | 14 | 724 | 216 | 16 | 1.51 | 1.21–1.88 | 1.51 | 1.17–1.96 | 2.29 | 1.10–4.75 | 0.980 |
| Hunt 2008 Netherlands [6] | 349 | 144 | 15 | 681 | 193 | 14 | 1.45 | 1.17–1.80 | 1.46 | 1.13–1.87 | 2.09 | 1.00–4.38 | 0.938 |
| Romanos 2008 Italy [16] | 427 | 105 | 6 | 521 | 75 | 3 | 1.68 | 1.25–2.26 | 1.71 | 1.24–2.36 | 2.44 | 0.61–9.81 | 0.865 |
| Coenen 2009 Netherlands [14] | 548 | 224 | 23 | 1303 | 355 | 24 | 1.50 | 1.27–1.78 | 1.50 | 1.23–1.82 | 2.28 | 1.28–4.07 | 0.974 |
| Dubois 2010 UK1 [15] | 515 | 202 | 20 | 2019 | 540 | 36 | 1.47 | 1.25–1.85 | 1.47 | 1.21–1.77 | 2.18 | 1.25–3.79 | 0.987 |
| Dubois 2010 UK2 [15] | 1339 | 469 | 41 | 3814 | 1050 | 72 | 1.27 | 1.14–1.42 | 1.27 | 1.12–1.44 | 1.62 | 1.10–2.39 | 0.977 |
| Dubois 2010 Finland [15] | 487 | 148 | 11 | 1478 | 332 | 19 | 1.35 | 1.11–1.63 | 1.35 | 1.09–1.68 | 1.76 | 0.83–3.72 | 0.941 |
| Dubois 2010 Netherlands [15] | 555 | 226 | 23 | 658 | 176 | 12 | 1.52 | 1.25–1.73 | 1.52 | 1.21–1.91 | 2.27 | 1.12–4.61 | 0.952 |
| Dubois 2010 Italy [15] | 392 | 99 | 6 | 473 | 68 | 2 | 1.77 | 1.30–2.41 | 1.76 | 1.25–2.46 | 3.62 | 0.73–18.04 | 0.788 |
| Dubois 2010 USA [15] | 720 | 234 | 19 | 432 | 116 | 8 | 1.21 | 0.97–1.51 | 1.21 | 0.94–1.56 | 1.42 | 0.62–3.28 | 0.946 |
| Dubois 2010 Hungary [15] | 674 | 265 | 26 | 839 | 214 | 14 | 1.54 | 1.28–1.84 | 1.54 | 1.25–2.45 | 2.31 | 1.20–4.46 | 0.932 |
| Dubois 2010 Ireland [15] | 398 | 179 | 20 | 1094 | 336 | 26 | 1.46 | 1.22–1.75 | 1.46 | 1.18–1.81 | 2.11 | 1.17–3.83 | 0.972 |
| Dubois 2010 Poland [15] | 390 | 158 | 16 | 538 | 165 | 13 | 1.32 | 1.06–1.64 | 1.32 | 1.02–1.70 | 1.70 | 0.81–3.57 | 0.932 |
| Dubois 2010 Spain [15] | 439 | 105 | 6 | 355 | 74 | 4 | 1.14 | 0.85–1.53 | 1.15 | 0.83–1.59 | 1.21 | 0.34–4.33 | 0.947 |
| Dubois 2010 Italy [15] | 825 | 175 | 9 | 683 | 116 | 5 | 1.24 | 0.98–1.57 | 1.25 | 0.97–1.61 | 1.49 | 0.50–4.47 | 0.975 |
| Dubois 2010 Finland [15] | 200 | 55 | 4 | 514 | 131 | 8 | 1.09 | 0.80–1.50 | 1.08 | 0.76–1.54 | 1.28 | 0.38–4.31 | 0.914 |
| Sperandeo 2011 Italy [17] | 526 | 106 | 5 | 610 | 92 | 9 | 1.20 | 0.91–1.57 | 1.34 | 0.99–1.81 | 0.64 | 0.21–1.93 | 0.029 |
| Plaza-Izurieta 2011 Spain [7] | 847 | 231 | 16 | 443 | 92 | 5 | 1.31 | 1.03–1.67 | 1.31 | 1.00–1.72 | 1.67 | 0.61–4.60 | 0.926 |
| Overall OR | – | – | – | – | – | – | 1.37 | 1.31–1.43 | 1.37 | 1.30–1.44 | 1.82 | 1.55–2.14 | – |
HWE: Hardy–Weinberg equilibrium.
2.2. Risk of Bias Assessment
The results of bias assessment are presented in Table S1. There are risks of bias in terms of ascertainment of CD (“unclear” in 1 out of 20 studies, 5.0%) and ascertainment of control (“unclear” in 1 out of 20 studies, 5.0%).
2.3. Genetic Association between RGS1 rs2816316 and CD Risk
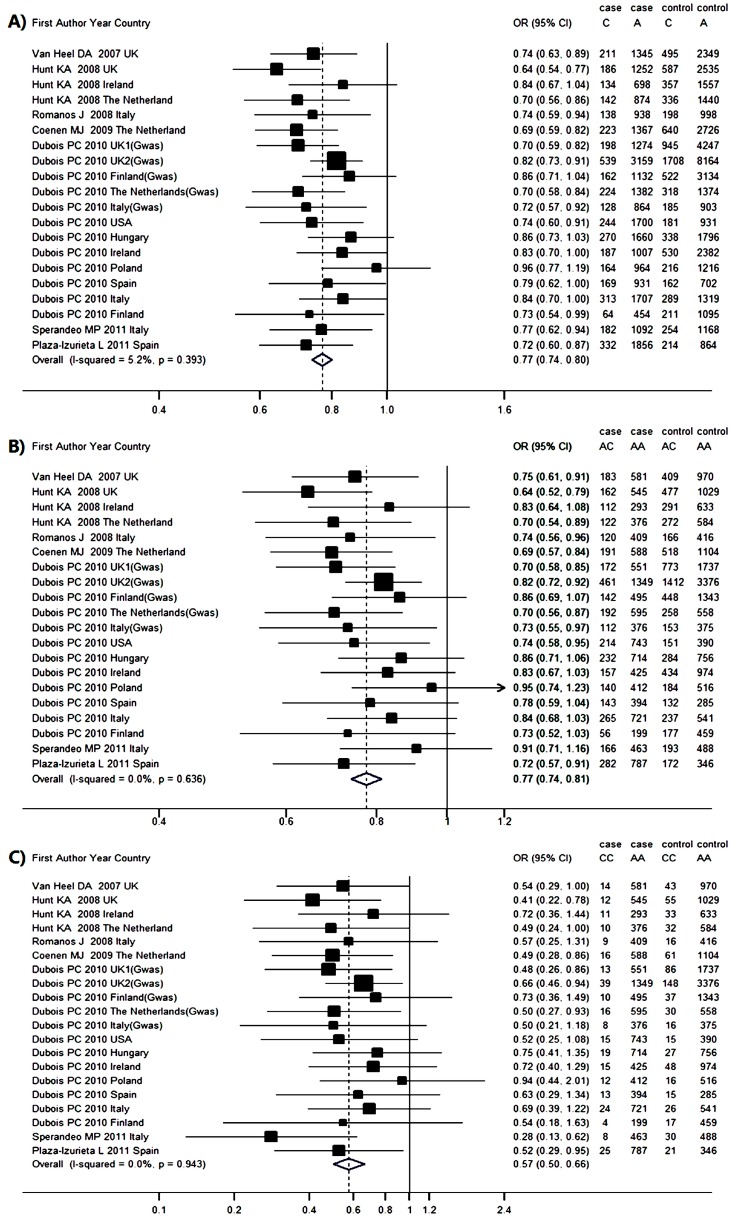
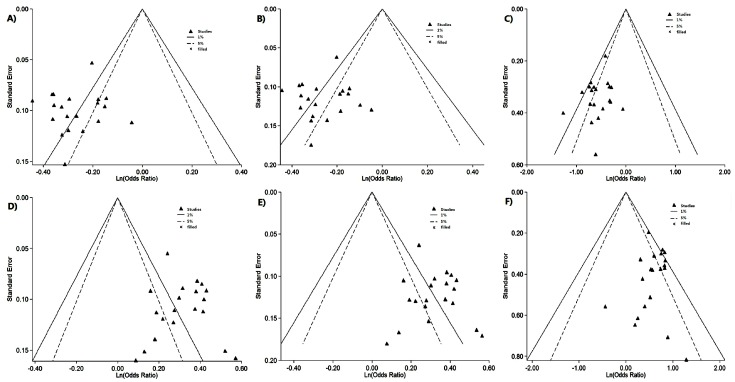
Seven studies reported association between rs2816316 and CD, which involved 14,936 cases and 24,794 controls. Hardy–Weinberg equilibrium (HWE) test showed no study was considered to be disequilibrium. The pooled frequency of the minor allele C was 14.0% (95% CI = 13.6%–14.5%) in the CD groups and 17.5% (95% CI = 16.9%–18.2%) in the control groups. The population-attributable risk (PAR) for allele C was 4.19%. The odds ratio (OR) (C vs. A) was estimated by the fix-effect model with less across-study heterogeneity (p = 0.393, I2 = 5.2%), with a pooled OR of 0.77 (95% CI = 0.74–0.80) (Figure 2A). This result suggests that individuals carrying the C allele have a 23% lower risk of developing CD than those carrying the A allele. Harbord (p = 0.622) and Peters (p = 0.775) tests suggested no existence of publication bias. The results in Figure 3A indicated that the pooled OR (C vs. A) were stable without any publication bias. The sensitive analysis was performed to make sure that no individual study was entirely responsible for the pooled results. In Table S2, none of the individual studies affect the final conclusion obviously.
Figure 2.
Meta-analysis forest plots of the correlations between RGS1 rs2816316 and celiac disease: (A) C vs. A; (B) AC vs. AA; and (C) CC vs. AA. The vertical dotted line and solid line represent the pooled odds ratio (OR) and the OR equals to 1, respectively. The horizontal solid lines mean the confidence intervals (CI) of OR in each population. And if the upper limit of OR exceed the top scale, arrow would replace the solid line. The size of solid squares corresponds to the weight of the study in the meta-analysis. The length of the hollow diamonds represent the CI of the pooled OR.
Figure 3.
Contour-enhanced funnel plot of IL2/IL21 and SH2B3 genes with CD: (A) RGS1 rs2816316 C vs. A; (B) RGS1 rs2816316 AC vs. AA; (C) RGS1 rs2816316 CC vs. AA; (D) IL12A rs17810546 G vs. A; (E) IL12A rs17810546 AG vs. AA; and (F) IL12A rs17810546 GG vs. AA.
Genotype frequency and estimated OR for each study were presented in Figure 2B,C. The OR1 (AC vs. AA) (p = 0.393, I2 = 5.2%) and the OR2 (CC vs. AA) (p = 0.943, I2 = 0.0%) were homogenous. The pooled ORs were estimated by the fix-effects model for OR1 (0.77, 95% CI = 0.74–0.81, p < 0.001) and OR2 (0.57, 95% CI = 0.50–0.66, p < 0.001), both of which were significant. Harbord and Peters tests in OR1 (p = 0.741, 0.910) and OR2 (p = 0.558, 0.553) as well as contour-enhanced funnel plot (Figure 3B,C) showed no publication bias and stable results. The λ = 0.487 (95% CI = 0.319–0.724) suggested a co-dominant model effect was most likely.
2.4. Genetic Association between IL12A rs17810546 and CD Risk
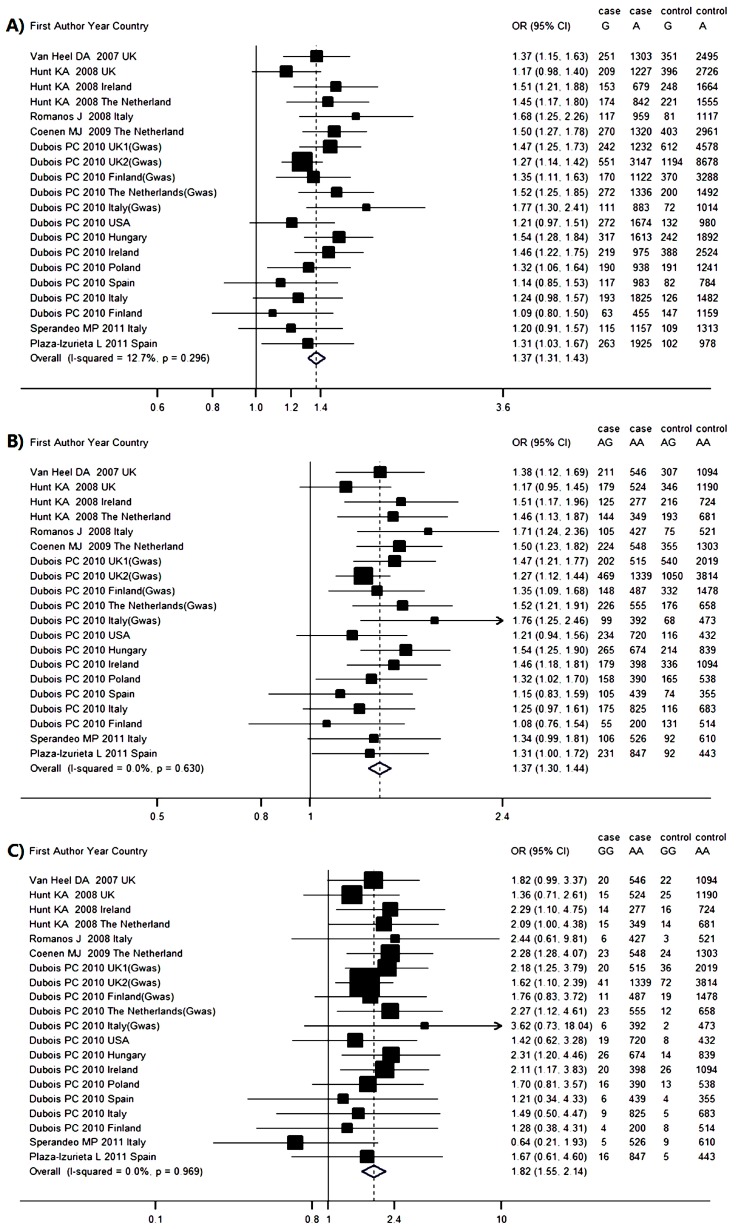
Seven studies reported association between rs17810546 and CD risk, involving 14,936 cases and 24,794 controls. By using HWE test, most studies were considered to be disequilibrium except for Sperandeo MP et al. [17]. The pooled frequency of the minor allele G was 14.3% (95% CI = 13.0%–15.5%) in the CD groups and 10.9% (95% CI = 10.0%–11.7%) in the control groups. The PAR for allele G was 3.87%. The OR (G vs. A) was estimated by the fix-effect model with less across-study heterogeneity (p = 0.296, and I2 = 12.7%), with a pooled OR of 1.37 (95% CI = 1.31–1.43) (Figure 4A). This result suggests that individuals carrying the G allele have a 37% higher risk of developing CD than those carrying the A allele. Harbord and Peters tests (p = 0.281, 0.281) and contour-enhanced funnel plot (Figure 3D) suggested no publication bias and stable pooled results. The sensitive analysis results in Table S3 indicated none of the individual studies affect the final conclusion obviously.
Figure 4.
Meta-analysis forest plots of the correlations between IL12A rs17810546 and celiac disease: (A) G vs. A; (B) AG vs. AA; and (C) GG vs. AA. The vertical dotted line and solid line represent the pooled odds ratio (OR) and the OR equals to 1, respectively. The horizontal solid lines mean the confidence intervals (CI) of OR in each population. And if the upper limit of OR exceed the top scale, arrow would replace the solid line. The size of solid squares corresponds to the weight of the study in the meta-analysis. The length of the hollow diamonds represent the CI of the pooled OR.
Genotype frequency and estimated OR for each study were presented in Figure 4B,C. The OR1 (AG vs. AA) (p = 0.630, I2 = 0.0%) and OR2 (GG vs. AA) (p = 0.969, I2 = 0.0%) were homogenous. The pooled ORs were estimated by the fix-effects model for OR1 (1.37, 95% CI = 1.30–1.44, p < 0.001) and OR2 (1.82, 95% CI = 1.55–2.14, p < 0.001), both of which were significant. Harbord and Peters tests in OR1 (p = 0.678, 0.642) and OR2 (p = 0.971, 0.407) and contour-enhanced funnel plot (Figure 3E,F) showed no publication bias and stable results. The λ = 0.562 (95% CI = 0.398–0.816) also suggested a co-dominant model effect was most likely.
3. Discussion
In the past few years, several GWAS have identified 40 non-HLA genomic regions harboring 64 CD-associated candidate genes [15]. However, non-HLA SNPs investigated in the present study are not regarded as strong markers of CD. To our knowledge, this is the first meta-analysis to reveal the associations of RGS1 (rs2816316) and IL12A (rs17810546) with CD and represents a pooled total of 14,936 cases and 24,794 controls across different ethnic populations. We found significant evidence for a modest decrease in CD risk associated with the minor C allele in rs2816316, and a modest increase in the minor G allele in rs17810546.
CD is a gluten-sensitive enteropathy and the only adopted treatment is a life-long gluten-free diet [19]. However, the treatment renders ineffective in about 5% of cases whom could develop into enteropathy-associated T cell lymphoma [20]. Thus, there is an urgent need to better understand the pathogenesis of CD, which could propel the development of pharmacologic agents in the treatment of CD. Our study identifies that major allele of rs2816316 and minor allele of rs17810546 are risk factors of CD, which suggests the potential role of RGS1 and IL12A in the treatment of CD. This finding should be confirmed by further biological and clinical studies.
The rs2816316 susceptibility allele is located on chromosome 1q31 and maps 8 kb from distal to the 5′ end of RGS1, which is involved in lymphocyte migration and can influence cell trafficking both in immune system development and exogenous infection [21]. Interestingly, RGS1 has been recently shown to be associated with multiple sclerosis (MS) and type 1 diabetes (T1D), both of which are T cell-mediated diseases [12,22]. The minor allele of rs2816316 is shared with MS and T1D, which suggested that the RGS1 might be an important T cell regulator. Study from Gibbons et al. [23] has identified that RGS1 expression is significantly higher in T cells from human gut compared with peripheral blood, especially in intestinal inflammation. Actually, 15%~20% of CD patients suffer from other autoimmune diseases [24], which was strengthened by our study.
rs17810546 locates in a ~70-kb LD block, which is immediately 5′ of IL12A. Similar with the locus RGS1, IL12A has been reported with risk of MS and T1D, confirming the shared genetic factors among different autoimmune diseases [22,25]. In addition, a meta-analysis by Kappen et al. [26] identified the genome wide significance association between IL12A rs17810546 and Behcet’s disease, which was also regarded as a Th1 mediated autoimmune disease. The minor allele of rs17810546 showed higher expression level of SCHIP1 gene, located in the same LD block with IL12A gene [7]. IL12A encodes IL12 and induces the Th1 response in the perspective of the etiology of CD [27]. Paajanen et al. [28] has shown that the children with CD had the lower expression of IL12A mRNA, which suggested that the major allele of these loci might be a protective factor in CD. In our study, the rs17810546 GG homozygote and heterozygote AG have 82% and 37% increased risk of CD compared with the wide genotype AA, without evidence of between-study heterogeneity, which strengthening the pathogenic effect of IL12A minor allele in CD.
Although several previous studies have investigated the associations between CD risk and RGS1 or IL12A polymorphisms, our meta-analysis is more persuasive and well-documented. First, a total of 20 subsets were included to pool the results for each SNP, dramatically improving the power of statistical analysis. Second, the quality of each included study was desirable, since only studies satisfying the risk-of-bias score test could be included. Third, when we removed the studies one by one to detect the influence of a single study on the pooled OR of other studies, none of the individual studies affect the final conclusion obviously, suggesting the stability of our results. However, this retrospective analysis has several limitations. First, the number of included studies for each SNP is limited (seven studies for rs2816316 and 17810546 each). Nevertheless, this limitation is made up for by the relatively large sample sizes (14,936 cases and 24,794 controls). Second, controls in Italian population are not in Hardy–Weinberg equilibrium [17]. In the sensitivity analysis, however, when this study was excluded, the results of disease associations were not altered. Third, since our study includes almost European ethnicities, the associations among other populations still need to be confirmed.
In summary, our meta-analysis indicates that RGS1 and IL12A are both associated with CD, especially in European populations. The associations should be validated by large-scale, well-designed epidemiological studies in the future.
4. Experimental Section
4.1. Search Strategy
Studies concerning the associations of RGS1 and IL12A polymorphisms with CD were searched from PubMed and Wed of Science by two reviewers independently. The updating date was 20 August, 2015. The detailed search strategy was as follows: (celiac disease or CD) and (polymorphism or SNP or rs2816316 or rs17810546) or (G-protein signaling-1 or RGS1) or (interleukin-12 A or IL12A). We only considered studies published in English and Chinese. We scanned the titles and abstracts of all relevant articles, manually examined reference lists for additional relevant publications and obtained the full texts of all possibly relevant studies. For multiple articles published with the same subjects, we selected the latest and most complete one.
4.2. Inclusion Criteria
Any human population-based association study was included only if it met all three criteria: (1) evaluation of the RGS1 (rs2816316) or IL12A (rs17810546) polymorphism and CD; (2) odds ratio (OR) and its 95% confidence interval (CI) was presented or could be calculated; and (3) there was clear diagnosis of CD. We contacted the authors to acquire indispensable information when their data were incomplete. Studies that did not provide data were excluded.
4.3. Data Extraction
Summary data were extracted by two reviewers independently using a standardized form (Table 1). The following information was extracted from each study: (1) name of first author; (2) year of publication; (3) country of population; (4) ethnicity of population; (5) genotyping method; (6) minor allele frequency (MAF) in controls; and (7) sample size of cases and controls. Genotype frequencies of each study and p-values for the Hardy–Weinberg equilibrium HWE test in control groups were extracted (Table 2 and Table 3). If there was no detailed genotype data in a study, the theoretical frequency of genotypes was estimated from the MAF of polymorphisms. Any disagreement was resolved through consensus.
4.4. Risk of Bias Assessment
The quality of each study was assessed by two reviewers based on a risk-of-bias score for genetic association studies [29]. The score considers 6 domains (Table S1): information bias (ascertainment of outcome and gene), quality control for genotyping, population stratification, confounding bias, selective report of outcomes and HWE assessment in the control group. Each item was classified into “yes”, “no” or “unclear”, which represented low risk, high risk and insufficient information, respectively. Disagreement between the two reviewers was solved by a senior reviewer.
4.5. Statistical Analysis
All statistical analyses were performed on Stata 12.0. The association strength of RGS1 rs2816316 and IL12A rs17810546 with CD risk was assessed by OR and its 95%CI. p < 0.05 was considered statistically significant in all tests, except for the heterogeneity test in which p < 0.10 was used. The HWE of the genotype distribution in a control group was assessed by the χ2 goodness-of-fit test.
The statistical significance of the pooled OR was determined by the Mantel-Haenszel method. The across-study heterogeneity of allele effects was checked using a Q test and was quantified by I2 (I2 < 25%, no; 25%–50%, moderate; 50%–75%, large; >75%, extreme) [30]. If heterogeneity was present (i.e., Q test was significant), the cause of heterogeneity was explored by sensitivity analysis. A random-effect model was used if I2 > 50%, otherwise, a fixed-effect model was used. The population-attributable risk PAR for mutant allele was calculated based on the results from a discrete-time model [31].
We supposed that A and a were wild-type and mutant alleles, respectively; and AA, Aa and aa were common homozygous, heterozygous, and minor homozygous, respectively. The model-free approach was used to estimate the genotype effect, and two types of ORs for each study: aa vs. AA (OR1) and Aa vs. AA (OR2). The model of genetic effect, measured by the parameter lambda (λ: the ratio of logOR2 to logOR1), was then estimated by the model-free Bayesian approach. The λ represents the heterozygote effect as a proportion of the homozygote variant effect. Furthermore, the λ-value ranges from 0 to 1. We get information about the genetic mode of action as follows: λ = 0 suggests a recessive model (Aa + aa vs. AA); λ = 1 suggests a dominant model (AA + Aa vs. aa); λ = 0.5 suggests a co-dominant model (AA vs. aa; Aa vs. aa). λ > 1 or λ < 0 indicates a homozygous or heterosis model, although this is rare. The best genetic model, once identified, is used to collapse the three genotypes into two groups and re-pool the results. WinBugs 1.4.2 was used with vague prior to distributions for parameter estimation (i.e., λ and OR). The models were run with a burn-in of 1000 iterations, followed by 10,000 iterations for parameter estimation.
The Harbord and Peters test were adopted to assess and quantify the publication bias. Additionally, a novel Contour-enhanced meta-analysis funnel plot method was used to combat publication biases [32]. In the sensitive analysis, we removed the studies one by one to identify the influence of a single study on the pooled OR from other studies.
5. Conclusions
In conclusion, our study identified RGS1 rs2816316 was negatively associated with CD. In the other hand, IL12A rs17810546 was positively associated with CD. Further studies are required to elucidate how these variants contribute to the susceptibility of CD.
Acknowledgments
This work was supported in part by Training Program of the Major Research Plan of the National Natural Science Foundation of China (Grant numbers: 91543132), National Natural Science Foundation of China (Grant numbers: 30901249, 81101267 and 81541070), Guangdong Natural Science Foundation (Grant numbers: 10151063201000036 and S2011010002526), Guangdong Province Medical Research Foundation (Grant number: A2014374 and A2015310) and Project from Jinan university (Grant number: 21612426 and 21615427).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/4/457/s1.
Author Contributions
Chun-Xia Jing and Guang Yang contributed to study conception and design; Cong-Cong Guo and Man Wang acquired the data; Wei-Huang Huang, Di Xiao, Xing-Guang Ye, Mei-Ling Ou, Na Zhang, Bao-Huan Zhang and Yang Liu performed the analyses and the interpretation of the results; Cong-Cong Guo and Man Wang wrote the first draft and Feng-Di Cao revised it critically for important intellectual content. All authors approved the final version to be published.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Meresse B., Malamut G., Cerf-Bensussan N. Celiac disease: An immunological jigsaw. Immunity. 2012;36:907–919. doi: 10.1016/j.immuni.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Kelly C.P., Bai J.C., Liu E., Leffler D.A. Advances in diagnosis and management of celiac disease. Gastroenterology. 2015;148:1175–1186. doi: 10.1053/j.gastro.2015.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sollid L.M., Markussen G., Ek J., Gjerde H., Vartdal F., Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ α/β heterodimer. J. Exp. Med. 1989;169:345–350. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petronzelli F., Bonamico M., Ferrante P., Grillo R., Mora B., Mariani P., Apollonio I., Gemme G., Mazzilli M.C. Genetic contribution of the HLA region to the familial clustering of coeliac disease. Ann. Hum. Genet. 1997;61:307–317. doi: 10.1017/S0003480097006258. [DOI] [PubMed] [Google Scholar]
- 5.Wolters V.M., Wijmenga C. Genetic background of celiac disease and its clinical implications. Am. J. Gastroenterol. 2008;103:190–195. doi: 10.1111/j.1572-0241.2007.01471.x. [DOI] [PubMed] [Google Scholar]
- 6.Hunt K.A., Zhernakova A., Turner G., Heap G.A., Franke L., Bruinenberg M., Romanos J., Dinesen L.C., Ryan A.W., Panesar D., et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plaza-Izurieta L., Castellanos-Rubio A., Irastorza I., Fernandez-Jimenez N., Gutierrez G., Bilbao J.R., CEGEC Revisiting genome wide association studies (GWAS) in coeliac disease: Replication study in spanish population and expression analysis of candidate genes. J. Med. Genet. 2011;48:493–496. doi: 10.1136/jmg.2011.089714. [DOI] [PubMed] [Google Scholar]
- 8.Han S.B., Moratz C., Huang N.N., Kelsall B., Cho H., Shi C.S., Schwartz O., Kehrl J.H. Rgs1 and Gnai2 regulate the entrance of B lymphocytes into lymph nodes and B cell motility within lymph node follicles. Immunity. 2005;22:343–354. doi: 10.1016/j.immuni.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 9.Moratz C., Kang V.H., Druey K.M., Shi C.S., Scheschonka A., Murphy P.M., Kozasa T., Kehrl J.H. Regulator of G protein signaling 1 (RGS1) markedly impairs gi alpha signaling responses of B lymphocytes. J. Immunol. 2000;164:1829–1838. doi: 10.4049/jimmunol.164.4.1829. [DOI] [PubMed] [Google Scholar]
- 10.Di Sabatino A., Corazza G.R. Coeliac disease. Lancet. 2009;373:1480–1493. doi: 10.1016/S0140-6736(09)60254-3. [DOI] [PubMed] [Google Scholar]
- 11.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 12.International Multiple Sclerosis Genetics Conssortium IL12A, MPHOSPH9/CDK2AP1 and RGS1 are novel multiple sclerosis susceptibility loci. Genes Immun. 2010;11:397–405. doi: 10.1038/gene.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahdenpera A., Ludvigsson J., Falth-Magnusson K., Hogberg L., Vaarala O. The effect of gluten-free diet on Th1-Th2-Th3-associated intestinal immune responses in celiac disease. Scand. J. Gastroenterol. 2011;46:538–549. doi: 10.3109/00365521.2011.551888. [DOI] [PubMed] [Google Scholar]
- 14.Coenen M.J., Trynka G., Heskamp S., Franke B., van Diemen C.C., Smolonska J., van Leeuwen M., Brouwer E., Boezen M.H., Postma D.S., et al. Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum. Mol. Genet. 2009;18:4195–4203. doi: 10.1093/hmg/ddp365. [DOI] [PubMed] [Google Scholar]
- 15.Dubois P.C., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A., Adany R., Aromaa A., et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romanos J., Barisani D., Trynka G., Zhernakova A., Bardella M.T., Wijmenga C. Six new coeliac disease loci replicated in an Italian population confirm association with coeliac disease. J. Med. Genet. 2009;46:60–63. doi: 10.1136/jmg.2008.061457. [DOI] [PubMed] [Google Scholar]
- 17.Sperandeo M.P., Tosco A., Izzo V., Tucci F., Troncone R., Auricchio R., Romanos J., Trynka G., Auricchio S., Jabri B., et al. Potential celiac patients: A model of celiac disease pathogenesis. PLoS ONE. 2011;6:457. doi: 10.1371/journal.pone.0021281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Heel D.A., Franke L., Hunt K.A., Gwilliam R., Zhernakova A., Inouye M., Wapenaar M.C., Barnardo M.C., Bethel G., Holmes G.K., et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat. Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trynka G., Wijmenga C., van Heel D.A. A genetic perspective on coeliac disease. Trends Mol. Med. 2010;16:537–550. doi: 10.1016/j.molmed.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Al-Toma A., Verbeek W.H., Hadithi M., von Blomberg B.M., Mulder C.J. Survival in refractory coeliac disease and enteropathy-associated T-cell lymphoma: Retrospective evaluation of single-centre experience. Gut. 2007;56:1373–1378. doi: 10.1136/gut.2006.114512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Z., Xiong K., Issekutz T.B. Regulation of chemokine-induced transendothelial migration of T lymphocytes by endothelial activation: Differential effects on naive and memory T cells. J. Leukoc. Biol. 2000;67:825–833. doi: 10.1002/jlb.67.6.825. [DOI] [PubMed] [Google Scholar]
- 22.Smyth D.J., Plagnol V., Walker N.M., Cooper J.D., Downes K., Yang J.H., Howson J.M., Stevens H., McManus R., Wijmenga C., et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbons D.L., Abeler-Dorner L., Raine T., Hwang I.Y., Jandke A., Wencker M., Deban L., Rudd C.E., Irving P.M., Kehrl J.H., et al. Cutting edge: Regulator of g protein signaling-1 selectively regulates gut T cell trafficking and colitic potential. J. Immunol. 2011;187:2067–2071. doi: 10.4049/jimmunol.1100833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cosnes J., Cellier C., Viola S., Colombel J.F., Michaud L., Sarles J., Hugot J.P., Ginies J.L., Dabadie A., Mouterde O., et al. Incidence of autoimmune diseases in celiac disease: Protective effect of the gluten-free diet. Clin. Gastroenterol. Hepatol. 2008;6:753–758. doi: 10.1016/j.cgh.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 25.Mowry E.M., Carey R.F., Blasco M.R., Pelletier J., Duquette P., Villoslada P., Malikova I., Roger E., Kinkel R.P., McDonald J., et al. Association of multiple sclerosis susceptibility variants and early attack location in the CNS. PLoS ONE. 2013;8:457. doi: 10.1371/journal.pone.0075565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kappen J.H., Medina-Gomez C., van Hagen P.M., Stolk L., Estrada K., Rivadeneira F., Uitterlinden A.G., Stanford M.R., Ben-Chetrit E., Wallace G.R., et al. Genome-wide association study in an admixed case series reveals IL12A as a new candidate in behcet disease. PLoS ONE. 2015;10:457. doi: 10.1371/journal.pone.0119085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Sabatino A., Pickard K.M., Gordon J.N., Salvati V., Mazzarella G., Beattie R.M., Vossenkaemper A., Rovedatti L., Leakey N.A., Croft N.M., et al. Evidence for the role of interferon-α production by dendritic cells in the Th1 response in celiac disease. Gastroenterology. 2007;133:1175–1187. doi: 10.1053/j.gastro.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Paajanen L., Kokkonen J., Karttunen T.J., Tuure T., Korpela R., Vaarala O. Intestinal cytokine mRNA expression in delayed-type cow’s milk allergy. J. Pediatr. Gastroenterol. Nutr. 2006;43:470–476. doi: 10.1097/01.mpg.0000233160.35786.35. [DOI] [PubMed] [Google Scholar]
- 29.Thakkinstian A., McKay G.J., McEvoy M., Chakravarthy U., Chakrabarti S., Silvestri G., Kaur I., Li X., Attia J. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: A huge review and meta-analysis. Am. J. Epidemiol. 2011;173:1365–1379. doi: 10.1093/aje/kwr025. [DOI] [PubMed] [Google Scholar]
- 30.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossman M.D., Thompson B., Frederick M., Maliarik M., Iannuzzi M.C., Rybicki B.A., Pandey J.P., Newman L.S., Magira E., Beznik-Cizman B., et al. HLA-DRB1*1101: A significant risk factor for sarcoidosis in blacks and whites. Am. J. Hum. Genet. 2003;73:720–735. doi: 10.1086/378097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters J.L., Sutton A.J., Jones D.R., Abrams K.R., Rushton L. Contour-enhanced meta-analysis funnel plots help distinguish publication bias from other causes of asymmetry. J. Clin. Epidemiol. 2008;61:991–996. doi: 10.1016/j.jclinepi.2007.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.