Abstract
MicroRNAs (miRNAs) play key roles in plant reproduction. However, knowledge on microRNAome analysis in autotetraploid rice is rather limited. Here, high-throughput sequencing technology was employed to analyze miRNAomes during pollen development in diploid and polyploid rice. A total of 172 differentially expressed miRNAs (DEM) were detected in autotetraploid rice compared to its diploid counterpart, and 57 miRNAs were specifically expressed in autotetraploid rice. Of the 172 DEM, 115 and 61 miRNAs exhibited up- and down-regulation, respectively. Gene Ontology analysis on the targets of up-regulated DEM showed that they were enriched in transport and membrane in pre-meiotic interphase, reproduction in meiosis, and nucleotide binding in single microspore stage. osa-miR5788 and osa-miR1432-5p_R+1 were up-regulated in meiosis and their targets revealed interaction with the meiosis-related genes, suggesting that they may involve in the genes regulation associated with the chromosome behavior. Abundant 24 nt siRNAs associated with transposable elements were found in autotetraploid rice during pollen development; however, they significantly declined in diploid rice, suggesting that 24 nt siRNAs may play a role in pollen development. These findings provide a foundation for understanding the effect of polyploidy on small RNA expression patterns during pollen development that cause pollen sterility in autotetraploid rice.
Keywords: meiosis, microRNAs, polyploidy, pre-meiotic interphase (PMA), single microspore stage (SCP), siRNAs
1. Introduction
The evolutionary history of all angiosperms shows that about 30%–60% of them might be polyploids, whole genome duplication has occurred in 75% of all flowering plants [1,2]. Polyploids could be an important source for plant breeders in the future [3,4] because it offers the following three advantages: increasing the variation in dosage-regulated gene expression evolved to new biological functions, advancing the largest vegetative organs, and high levels of heterosis by polyploidy hybrids, over diploid progenitors [5,6,7,8].
Autotetraploid rice is a useful germplasm resource obtained by chromosome doubling with strong hybrid vigor, ability of resistance against abiotic and biotic stresses, and high biomass production compared with diploid rice [9,10]. However, the poor seed set is the main barrier in commercial utilization of polyploid rice. Pollen sterility is one of the major factors that cause low seed set in polyploid rice [11,12,13]. Previous studies on autotetraploid rice indicated that microtubule organization and abnormal chromosome behavior may result in sterile pollens [11,12]. Microarray analysis revealed that the differential gene expressions altered the meiosis gene network that may result in abnormal chromosome behavior and pollen development in autotetraploid rice [14]. Recently, we conducted a transcriptomic analysis on allelic interactions in autotetraploid rice hybrids, and the results revealed that polyploidy enhanced F1 pollen sterility loci interactions that alter the expression profiles of meiosis-related or meiosis-stage-specific genes, and resulted in low pollen fertility in autotetraploid rice [15]. Though many studies have been conducted to understand the differences between autotetraploid and diploid rice at cytological and molecular levels, microRNAome analysis on developing pollens of autotetraploid rice is still poorly understood.
MicroRNAs (miRNAs) are small (20–25 nucleotides) single stranded non-coding RNAs that modulate plant gene expression by gene silencing through inhibition of target mRNAs. A number of studies have confirmed that miRNAs involved in a series of biological functions, plant growth, reproduction and responses to hormones and stresses [16,17,18,19]. Some pollen-specific miRNAs have been detected after uninucleate microspores stage of developing pollen in rice [20,21,22]. However, little is known about the changes in expression patterns of miRNAs and their association with pollen development in autotetraploid rice compared to diploid counterpart. Therefore, we planned this study to investigate the differentially expressed miRNAs between autotetraploid and diploid rice, and to detect novel miRNAs that may be associated with meiosis or other stages during pollen development in autotetraploid rice. The results of the present study will provide insights into the roles of miRNAs during pollen development in autotetraploid rice and their association with pollen fertility.
2. Results
2.1. Overview of MicroRNAs (miRNAs) Sequencing Datasets in Developing Pollens of Rice
To investigate the miRNAs associated with the pollen development of diploid (CK) and autotetraploid rice, three pollen development stages, PMA (pre-meiotic interphase), MA (meiosis) and SCP (single microspore stage), were selected to construct six libraries and sequenced by Solexa high-throughput sequencing technology. A total of 9,176,755, 9,017,836 and 9,271,861 at PMA, MA and SCP raw reads were obtained in each library of Taichung65-4x, respectively. After removing the low quality sequences, adaptor sequences and RNAs smaller than 18 nt (ACGT101-miR program), we obtained 5,841,382 (63.65%), 5,781,692 (64.11%) and 4,341,659 (46.83%) high quality clean reads, which represented 1,201,066 (53.59%), 1,603,531 (57.64%) and 1,792,394 (55.3%) unique sequences at PMA, MA and SCP libraries ranging from 18 to 30 nt in length in autotetraploid rice, respectively (Table S1). We detected 6,284,253 (70.52%), 6,903,329 (60.76%) and 4,008,705 (34.76%) clean reads that represented 1,362,318 (56.74%), 1,219,793 (55.03%) and 1,191,600 (48.57%) unique reads in Taichung65-2x at PMA-2x, MA-2x and SCP-2x, respectively (Table S1). These high-quality reads were mapped to rice precursors in miRBase to identify known and novel miRNAs for further analysis. Low level of large fragments, such as mRNA (messenger RNA) and rRNA (ribosomal RNA), were also found, which indicated the high-quality and no degradation of RNA samples in the present study. A total of 93.19% and 91.44% small RNAs identified here spanned a size range of 20–24 nt in Taichung65-4x and Taichung65-2x, respectively. After removing the redundant sequences, we found 24 nt sRNA sequences more frequently compared to other sRNAs sequences, and the second highest peak was observed for 21 nt sRNA sequences (Figure S1).
A total of 2336 miRNAs, classified into five categories (Table S2), were detected via BLAST in miRBase. After removing the less expressed miRNAs, i.e., the expression levels were less than 10 after the normalization of dataset, we identified 486 miRNAs including 192 known miRNAs belonging to 112 families and 294 novel miRNAs during pollen development. Of these, 486 miRNAs, 441 and 402 were preferentially expressed in autotetraploid and diploid rice, respectively (Tables S3 and S4, respectively).
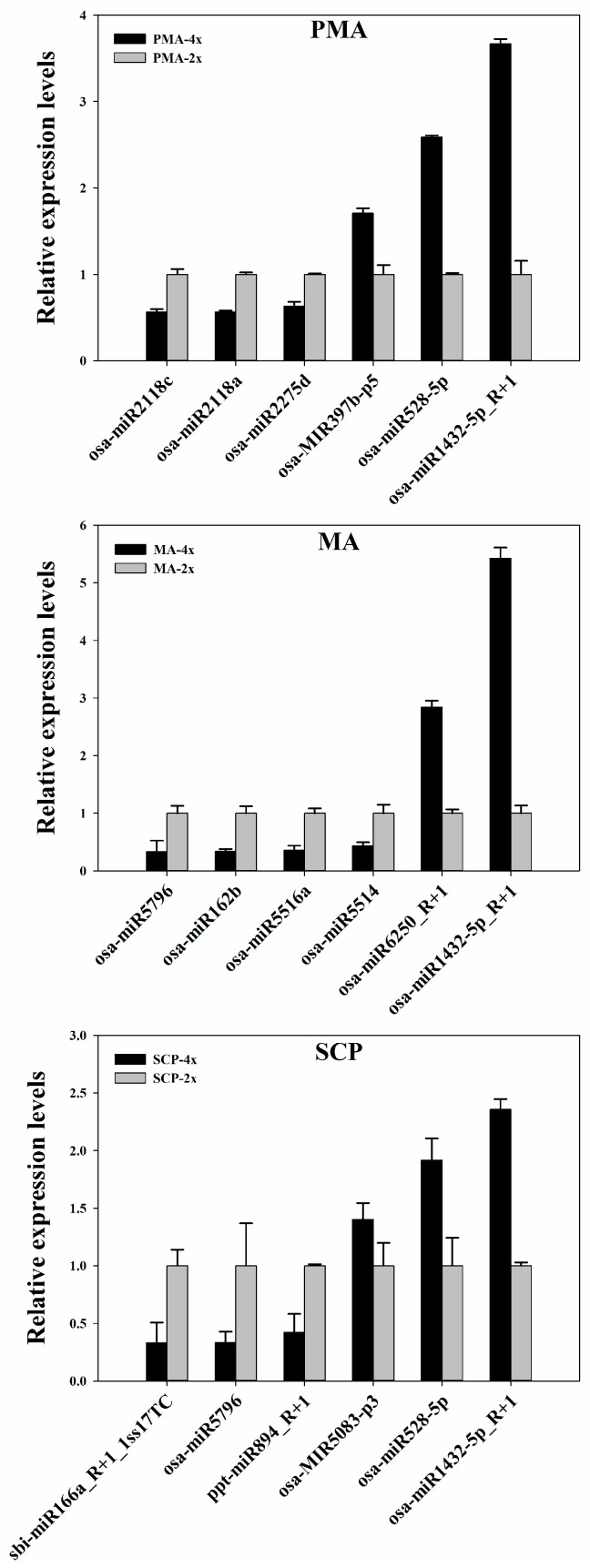
The expression profiles of miRNAs detected in Taichung65-4x and Taichung65-2x during pollen development were compared using principal component analysis (Figure S2). The six samples were grouped into two categories, and the pollen development stages (i.e., PMA, MA and SCP) could be clearly distinguished (Figure S3). These results demonstrated the reliability of the samples used in the present study. Six miRNAs were randomly selected from three pollen development stages for validating the differentially expressed miRNAs (DEM) by quantitative real-time PCR (qRT-PCR). The qRT-PCR results showed that their expression patterns were similar to those by the high-throughput sequencing, demonstrating the accuracy of small RNA sequencing results in the present study (Figure 1).
Figure 1.
Validation of the DEM (differentially expressed miRNAs) in autotetraploid and diploid rice at each pollen development stage (pre-meiotic interphase (PMA), meiosis (MA) and single microspore stage (SCP)). U6 snRNA was used as an internal reference for the qRT-PCR. The x- and y-axis represent the miRNAs and relative expression levels, respectively. Error bars represent the standard deviation (SD) of three biological replicates. “4x” and “2x” represent the autotetraploid and diploid rice, respectively.
2.2. Association between miRNAs Expression Profiles and Pollen Development Stages in Autotetraploid and Diploid Rice
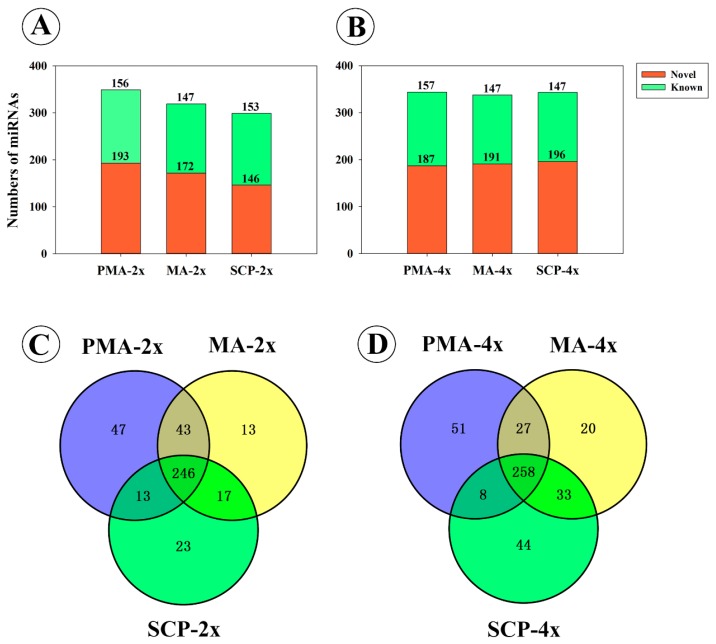
A total of 402 miRNAs (i.e., 349, 319 and 299 at PMA, MA and SCP, respectively), including 178 known and 224 novel miRNAs, were obtained during pollen development of Taichung65-2x by a threshold of >10 (Figure 2A; Table S3). Among 402 miRNAs, 246 miRNAs were co-expressed during the three pollen development stages (i.e., PMA, MA and SCP) (Figure 2C), and 170 miRNAs showed stage-specific differential expression patterns during pollen development stages in the diploid rice (Table S5). Of the 170 differentially expressed miRNAs, 20 and 46 miRNAs in MA compared to PMA stage, and 28 and 96 miRNAs in SCP compared to MA stage were up- and down-regulated, respectively (Table 1 and Table S5). Of the 153 known miRNAs in SCP, 33 miRNAs displayed similar expression patterns as reported by Wei et al. [22], while 120 miRNAs were only found in SCP of Taichung65-2x (Table S6A). Of the 33 miRNAs, 30 miRNAs that belong to 21 miRNAs families were expressed in all development stages (Table S6B), and most of them were pollen-specific miRNAs.
Figure 2.
Classification of miRNAs during pollen development: (A,B) the total number of miRNAs detected during different pollen development stages of Taichung65-2x (A) and Taichung65-4x (B); (C,D) Venn analysis of miRNAs expressed in Taichung65-2x (C) and Taichung65-4x during pollen development (D); and (E,F) specifically up- and down-regulated miRNAs during each adjacent stage of pollen development in Taichung65-2x and Taichung65-4x. PMA, MA and SCP represent pre-meiotic interphase, meiosis and single microspore stage, respectively. “4x” and “2x” represent the autotetraploid and diploid rice, respectively.
Table 1.
Differentially expressed miRNAs (DEM) in autotetraploid rice comparative to diploid rice.
| Material | Description/Stage-Specific Expression | Up-Regulated | Down-Regulated | Total |
|---|---|---|---|---|
| Taichung65-2x | DEM in MA compared to PMA | 20 | 46 | 66 |
| DEM in SCP compared to MA | 28 | 96 | 124 | |
| Taichung65-4x | DEM in MA compared to PMA | 41 | 75 | 116 |
| DEM in SCP compared to MA | 58 | 57 | 115 | |
| Taichung65-4x vs. Taichung65-2x | DEM in PMA | 27 | 19 | 46 |
| DEM in MA | 37 | 21 | 58 | |
| DEM in SCP | 75 | 27 | 102 | |
| DEM specifically in MA compared to PMA of Taichung65-2x | 15 | 34 | 49 | |
| DEM specifically in SCP compared to MA of Taichung65-2x | 19 | 55 | 74 | |
| DEM specifically in MA compared to PMA of Taichung65-4x | 36 | 63 | 99 | |
| DEM specifically in SCP compared to MA of Taichung65-4x | 49 | 16 | 65 |
PMA, MA and SCP represent pre-meiotic interphase, meiosis and single microspore stage, respectively.
A total of 441 miRNAs, including 173 known and 268 novel miRNAs, were detected in Taichung65-4x; of these, 344, 338 and 343 were identified in PMA, MA and SCP by a threshold of >10, respectively (Figure 2B; Table S4). Among 441 miRNAs, 258 miRNAs were co-expressed, and 208 miRNAs were stage-specific and exhibited differential patterns in pollen development stages (i.e., PMA, MA and SCP) in autotetraploid rice (Figure 2D; Table S5). In total, 41 and 75 miRNAs in MA compared to PMA stage, and 58 and 57 miRNAs in SCP compared to MA stage were up- and down-regulated, respectively (Table 1 and Table S5). We also detected 32 known miRNAs in SCP of Taichung65-4x that were reported by Wei et al. [22] (Table S6A), and 29 of these 32 miRNAs were also expressed during the PMA and MA in autotetraploid rice (Table S6B). These 29 known miRNAs were detected in diploid rice as well, but some of them were found to be differentially regulated in both types of rice.
2.3. Differentially Expressed miRNAs during Pollen Development in Autotetraploid and Diploid Rice
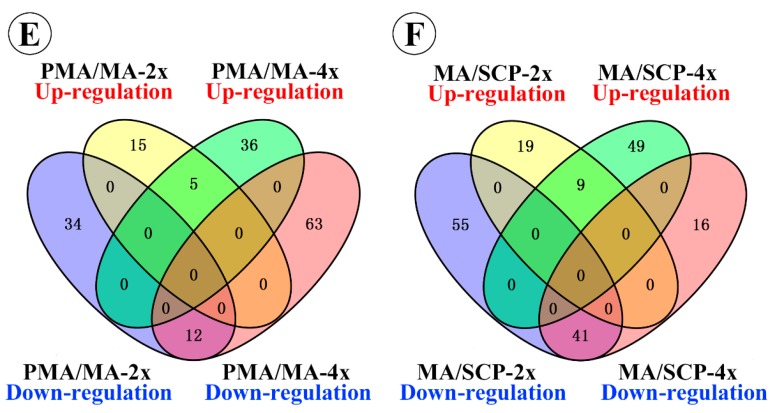
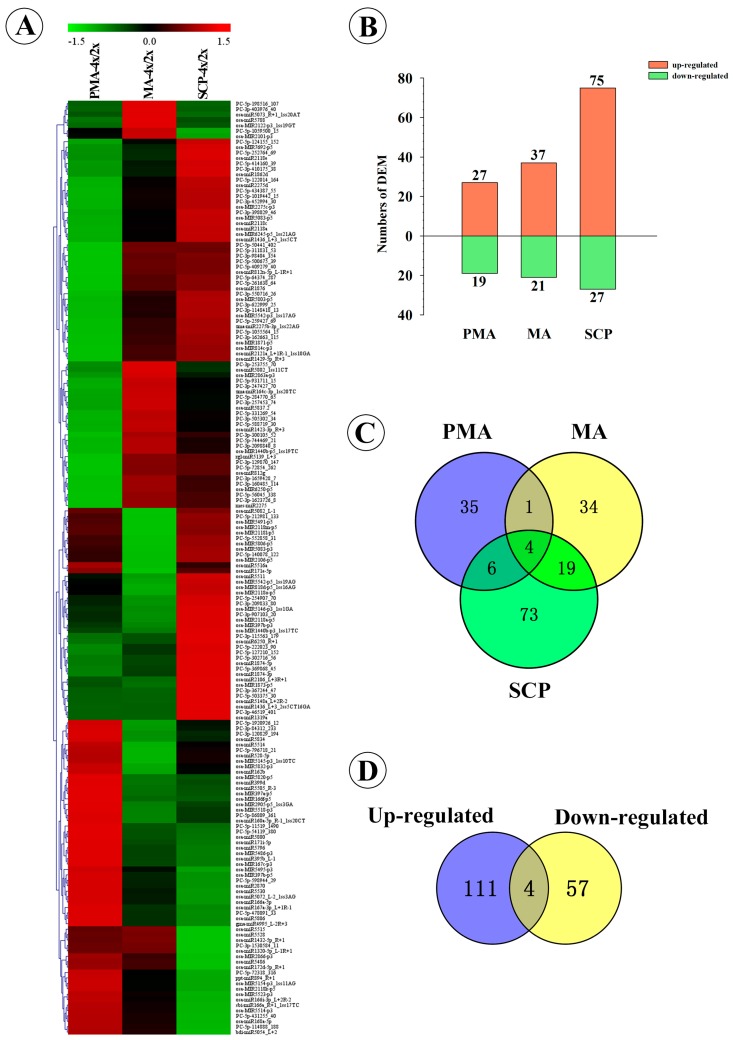
A total of 172 differentially expressed miRNAs (DEM), including 60 known and 112 novel miRNAs, which accounted for 35.39% of the total detected miRNAs, were detected during pollen development between autotetraploid and diploid rice (Figure 3A; Table S7). Among the DEM, 46 miRNAs were differentially expressed in PMA of Taichung65-4x compared with Taichung65-2x, of these 27 and 19 miRNAs were found to be up- and down-regulated, respectively. Fifty-eight miRNAs, including 37 up-regulated and 21 down-regulated, were differentially expressed in MA between Taichung65-4x and Taichung65-2x. One hundred two miRNAs were differentially expressed in SCP, among them 75 and 27 were up- and down-regulated, respectively (Figure 3B; Table 1). The minimum number of DEM was found in PMA and accumulated to maximum in SCP, consistent with the results of principal component analysis (Figure S2). A total of 142 miRNAs were specific/differentially expressed in the three pollen development stages (i.e., 35, 34 and 73 in PMA, MA and SCP, respectively), which accounted for 82.56% of the DEM, and only four miRNAs (osa-MIR5083-p5, osa-miR6250_R+1, osa-miR1320-5p_L-1R+1 and osa-miR1432-5p_R+1) were found to be differentially co-up-regulated in all three pollen development stages (Figure 3C; Table S7). We summed up the differentially expressed miRNAs in each stage, and 111 miRNAs showed up-regulation and 57 revealed down-regulation in autotetraploid rice (Figure 3D). However, we detected four miRNAs that showed the reverse tendency of regulation in different stages (Figure 3D), including osa-MIR1871-p5, osa-miR1429-5p_R+3, osa-MIR2275c-p3 and osa-miR2275d, which were down-regulated in PMA but up-regulated in SCP (Table S7).
Figure 3.
Analysis of DEM (differentially expressed miRNAs) in Taichung65-2x and Taichung65-4x during pollen development. (A) Hierarchical cluster analysis of DEM. The hierarchical clustering tree of 172 DEM in different libraries of pollen development was constructed by MultiExperiment View (version 4.9). Each column represents the difference between Taichung65-2x and Taichung65-4x in each stage. Red and green represent the up- and down-regulated miRNAs, respectively. The scale bar indicates the relative expression levels of miRNAs (log2); (B) The number of DEM at different pollen development stages; (C,D) Venn analysis of DEM during pollen development. PMA, MA and SCP represent pre-meiotic interphase, meiosis and single microspore stage, respectively. “4x” and “2x” represent the autotetraploid and diploid rice, respectively.
A total of 223 miRNAs were co-expressed in diploid and autotetraploid rice, some of them, such as miR159, miR166, miR396, miR2118 and miR2275 were highly expressed in both types of rice, indicating their conserved and essential roles in pollen development. Moreover, we identified 38 and 57 miRNAs, specifically associated with diploid and autotetraploid rice, respectively. Of these, 21, 7 and 10 miRNAs were expressed in diploid rice at PMA, MA and SCP, while 16, 13 and 28 were expressed during the above mentioned pollen development stages in autotetraploid rice, respectively (Table S8).
We have detected a large number of stage-specific differentially expressed miRNAs from PMA to SCP in autotetraploid rice compared to diploid rice. Among the stage-specific DEM, 36 up- and 63 down-regulated stage-specific DEM were detected from PMA to MA in autotetraploid rice compared to diploid, whereas only 5 up- and 12 down-regulated stage-specific DEM were common in the two lines (Figure 2E; Table 1). We detected 49 up- and 16 down-regulated stage-specific DEM that were specifically found in SCP compared to MA in autotetraploid rice, while 19 up- and 55 down-regulated in diploid rice (Figure 2F; Table 1). A total of nine up- and 41 down-regulated stage-specific DEM were found to be common in the two types of rice (Figure 2F; Table 1). Of these co-expressed stage-specific DEM, novel miRNA PC-5p-11519_1490 showed down-regulation in MA compared to PMA of autotetraploid rice, osa-miR5511 and osa-miR5516a depicted down-regulation in SCP compared to MA in autotetraploid rice (Table S5). These miRNAs were also found to be differentially expressed in autotetraploid rice compared to diploid rice during PMA, SCP and MA (Table S7).
2.4. Target Prediction of Differentially Expressed miRNAs (DEM) and Functional Classification
Identifying regulatory mRNA targets is essential for illustrating the function of miRNAs. To understand the effect of mRNA targets associated with the genetic variation of pollen development in autotetraploid rice, the predicted targets of 172 DEM were analyzed. Of these, 108 DEM were found with 729 predicted targets (Table S9). We found that the 662 of 729 predicted targets were significantly (p ≤ 0.05) categorized into 15 Gene Ontology (GO) terms (Table S10). Among these terms, transport (GO: 0006810) and localization (GO: 0051179) were dominated in the main category of biological processes, and secondary metabolic process (GO: 0019748), signal transduction (GO: 0007165) and catabolic process (GO: 0009056) were also found in biological processes. In the molecular function category, three significant GO terms, including transporter activity (GO: 0005215), lipid binding (GO: 0008289) and receptor activity (GO: 0004872) were identified. Only one term, extracellular region (GO: 0005576) was significant in cellular component category (Table S10).
We found that 111 and 57 miRNAs showed up- and down-regulation in the three stages of pollen development in autotetraploid rice, respectively. Gene Ontology (GO) analysis of the predicted targets of 111 up-regulated DEM showed that they were significantly enriched in transport (GO: 0006810), catabolic process (GO: 0009056), transporter activity (GO: 0005215) and extracellular region (GO: 0005576) (Figure S4). The predicted targets of 57 down-regulated DEM were enriched in multicellular organismal development (GO: 0032501), reproduction (GO: 0000003), signal transduction (GO: 0007165), transcription regulator activity (GO: 0030528) and nucleus (GO: 0005634) (Figure S5).
GO enrichment categories were specifically constructed by the predicted targets of up-regulated DEM in each stage, such as transport (GO: 0006810) and membrane (GO: 0016020) in PMA (Figure S6); response to stimulus (GO: 0050896) and reproduction (GO: 0000003) in MA (Figure S7); and transferase activity (GO: 0016740) and nucleotide binding (GO: 0000166) in SCP (Figure S8). Interestingly, the analysis of the predicted targets of down-regulated DEM revealed the highest number of GO categories in SCP, including multicellular organismal development (GO: 0007275), signal transmission (GO: 0023060), nitrogen compound metabolic process (GO: 0006807), and lipid binding (GO: 0008289) (Figure S9), while nucleotide binding (GO: 0000166) and RNA binding (GO: 0003723) terms were preferentially enriched in PMA and MA, respectively (Figures S10 and S11).
2.5. Functional Classification of Meiosis-Related Predicted Targets of Differentially Expressed miRNAs
Meiosis is a key stage of pollen development in autotetraploid rice; hence, we analyzed the DEM at this stage. We compared the target genes that were predicted by the DEM in autotetraploid rice with the reported microarray data for meiosis-related and stage-specific expression in diploid and autotetraploid rice [14,23,24,25,26,27]. Using the predicted targets of the down-regulated miRNAs, we identified one meiosis-specific gene (i.e., LOC_Os09g39400 gene was predicted by osa-MIR5486-p3 and annotated as histidine-containing phosphotransferase protein) (Table S11). Three meiosis-related genes were identified in this study, including LOC_Os10g17489 (predicted by osa-miR160a-5p_R-1_1ss20CT), LOC_Os11g03300 (predicted by osa-MIR2118l-p5), and LOC_Os08g45160 (predicted by osa-MIR5806-p5), which encode UDP-glucoronosyltransferase and UDP-glucosyltransferase domain containing protein, NAC domain transcription factor, and TENA/THI-4 family protein, respectively. These meiosis-related genes showed up-regulation in autotetraploid compared to the diploid rice during meiosis [14]. Additionally, two meiosis-related genes, LOC_Os10g40420 (predicted by osa-miR5072_L-2_1ss3AG) annotated as LTPL138-Protease inhibitor/seed storage/LTP family protein precursor and LOC_Os07g22930 (predicted by osa-miR160a-5p_R-1_1ss20CT) that encodes starch synthase were also found in this study, which exhibited differential expression patterns in diploid rice during meiosis [24] (Table S11). Of the predicted targets of up-regulated miRNA during meiosis, we identified four genes, including LOC_Os01g64100 (predicted by osa-miR1320-5p_L-1R+1) encoded glycosyl hydrolase, LOC_Os11g02400 (predicted by osa-miR5788) annotated as LTPL8-Protease inhibitor/seed storage/LTP family protein precursor, LOC_Os12g02330 (predicted by osa-miR5788) documented as LTPL13-Protease inhibitor/seed storage/LTP family protein precursor, and LOC_Os08g39000 (predicted by osa-MIR6250-p5) annotated as expressed protein; and these four predicted target genes were up-regulated in autotetraploid compared to diploid rice [14] (Table S11). Moreover, osa-MIR2122-p3_1ss19GT targeted LOC_Os05g33390 (cation-transporting ATPase, annotated as the MALE GAMETOGENESIS IMPAIRED ANTHERS gene in Arabidopsis thaliana [28]) and LOC_Os04g58960 (regulator of chromosome condensation) associated with the meiosis and involved in pollen development in rice, was specifically up-regulated in MA (Table S9A).
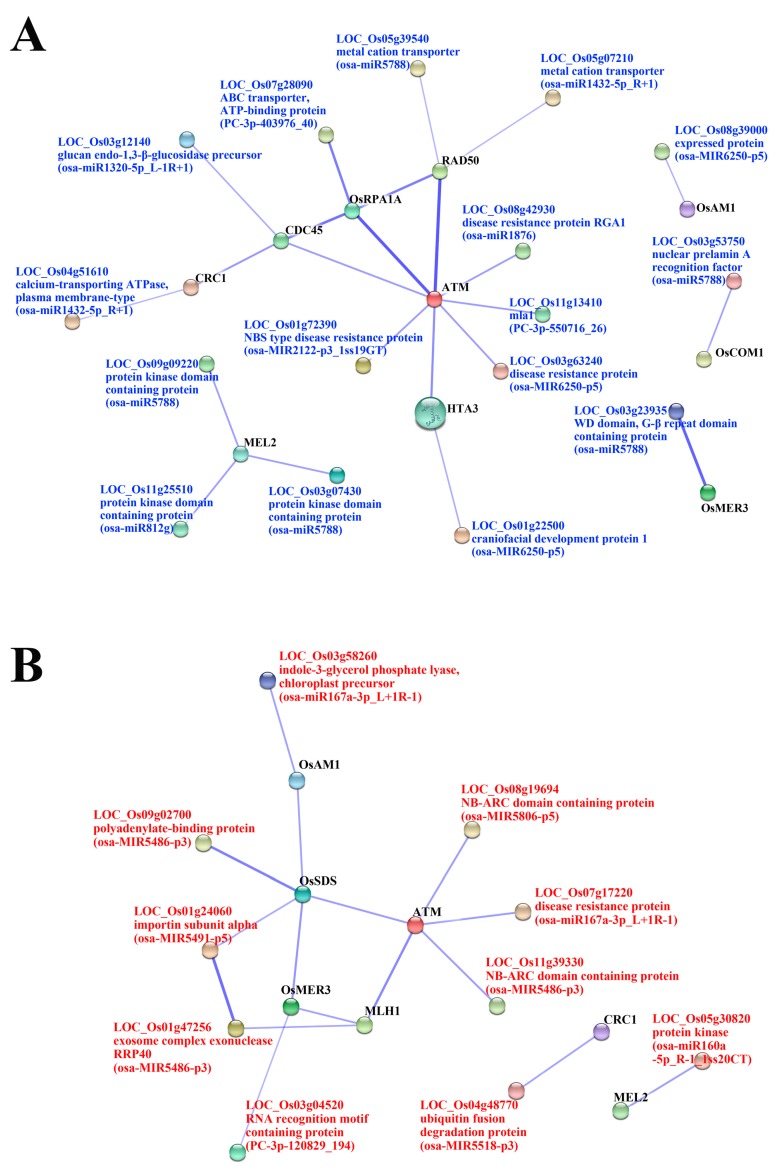
We identified 92 genes with known/proposed meiotic functions (Table S12) based on the putative meiosis-related genes detected by recent transcriptome analyses in rice and Arabidopsis [23,25,29,30]. Then we performed the protein–protein interactions of the important targets (detected at MA division) and 92 meiosis-related genes by using STRING v10. We identified 16 DEM that were associated with the meiosis (Figure 4; Table 2). Among the meiosis-related genes, HTA3, ATM, RAD50, CDC45, CRC1 and OsRPA1A were associated with the targets of up-DEM in MA stage, and constructed a meiosis-related gene-interaction network (Figure 4A). Three predicted targets of osa-MIR6250-p5, including LOC_Os03g63240 (disease resistance protein), LOC_Os01g22500 (craniofacial development protein 1) and LOC_Os08g39000 (expressed protein), were associated with three meiosis-related genes (i.e., ATM, HTA3 and OsAM1), respectively (Table 2). LOC_Os05g07210 (predicted by osa-miR1432-5p_R+1) and LOC_Os05g39540 (predicted by osa-miR5788) annotated as metal cation transporter, and showed interaction with RAD50 (Table 2). Additionally, osa-miR5788 showed the interaction with four meiosis-related genes that were specifically up-regulated in MA of Taichung65-4x, and one of its targets, LOC_Os03g23935 (WD domain, G-β repeat domain containing protein), showed interaction with LOC_Os02g40450 (OsMER3) (Table 2). Novel miRNA, osa-MIR2122-p3_1ss19GT targeted of LOC_Os01g72390 (NBS type disease resistance protein) that related to LOC_Os01g01689 (ATM), and PC-3p-403976_40 targeted of LOC_Os07g28090 (ABC transporter, ATP-binding protein) that related to LOC_Os02g53680 (OsRPA1A) were also up-regulated in MA (Table 2).
Figure 4.
Protein interaction between the targets of DEM (differentially expressed miRNAs) and meiosis-related genes. The predicted targets of up-regulated (A) and down-regulated miRNAs (B) showed interaction with the meiosis-related genes (Black font). Blue font and red font represent the targets of up- and down-regulated DEM, respectively. Line thickness represents interaction between proteins/genes.
Table 2.
Association between differentially expressed miRNAs (DEM) and meiosis-related genes in autotetraploid rice during pollen development.
| miRNA | Targets | Genes Annotation | PMA | MA | SCP | Specific in MA | Meiosis-Related Genes |
|---|---|---|---|---|---|---|---|
| osa-miR1320-5p_L-1R+1 | LOC_Os03g12140 | glucan endo-1,3-β-glucosidase precursor, putative, expressed | up | up | up | LOC_Os11g03430 (CDC45) | |
| osa-miR1432-5p_R+1 | LOC_Os04g51610 | calcium-transporting ATPase, plasma membrane-type, putative, expressed | up | up | up | LOC_Os04g40290 (CRC1) | |
| LOC_Os05g07210 | metal cation transporter, putative, expressed | up | up | up | LOC_Os02g29464 (RAD50) | ||
| osa-miR1876 | LOC_Os08g42930 | disease resistance protein RGA1, putative, expressed | ns | up | up | LOC_Os01g01689 (ATM) | |
| osa-MIR2122-p3_1ss19GT | LOC_Os01g72390 | NBS type disease resistance protein, putative, expressed | ns | up | ns | osa-MIR2122-p3_1ss19GT | LOC_Os01g01689 (ATM) |
| osa-miR5788 | LOC_Os03g07430 | protein kinase domain containing protein, expressed | ns | up | ns | osa-miR5788 | LOC_Os12g38460 (MEL2) |
| LOC_Os03g23935 | WD domain, G-β repeat domain containing protein, expressed | ns | up | ns | osa-miR5788 | LOC_Os02g40450 (OsMER3) | |
| LOC_Os03g53750 | nuclear prelamin A recognition factor, putative, expressed | ns | up | ns | osa-miR5788 | LOC_Os06g41050 (OsCOM1) | |
| LOC_Os05g39540 | metal cation transporter, putative, expressed | ns | up | ns | osa-miR5788 | LOC_Os02g29464 (RAD50) | |
| LOC_Os09g09220 | protein kinase domain containing protein, expressed | ns | up | ns | osa-miR5788 | LOC_Os12g38460 (MEL2) | |
| osa-MIR6250-p5 | LOC_Os01g22500 | craniofacial development protein 1, putative, expressed | ns | up | up | LOC_Os12g34510 (HTA3) | |
| LOC_Os03g63240 | disease resistance protein, putative, expressed | ns | up | up | LOC_Os01g01689 (ATM) | ||
| LOC_Os08g39000 | expressed protein | ns | up | up | LOC_Os03g44760 (OsAM1) | ||
| osa-miR812g | LOC_Os11g25510 | protein kinase domain containing protein, expressed | ns | up | up | LOC_Os12g38460 (MEL2) | |
| PC-3p-403976_40 | LOC_Os07g28090 | ABC transporter, ATP-binding protein, putative, expressed | ns | up | up | LOC_Os02g53680 (OsRPA1A) | |
| PC-3p-550716_26 | LOC_Os11g13410 | mla1, putative, expressed | ns | up | ns | PC-3p-550716_26 | LOC_Os01g01689 (ATM) |
| osa-miR160a-5p_R-1_1ss20CT | LOC_Os05g30820 | protein kinase, putative, expressed | ns | down | ns | osa-miR160a-5p_R-1_1ss20CT | LOC_Os12g38460 (MEL2) |
| osa-miR167a-3p_L+1R-1 | LOC_Os03g58260 | indole-3-glycerol phosphate lyase, chloroplast precursor, putative, expressed | ns | down | down | LOC_Os03g44760 (OsAM1) | |
| LOC_Os07g17220 | disease resistance protein, putative, expressed | ns | down | down | LOC_Os01g01689 (ATM) | ||
| osa-MIR5486-p3 | LOC_Os01g47256 | exosome complex exonuclease RRP40, putative, expressed | ns | down | down | LOC_Os01g72880 (MLH1) | |
| LOC_Os09g02700 | polyadenylate-binding protein, putative, expressed | ns | down | down | LOC_Os03g12414 (OsSDS) | ||
| LOC_Os11g39330 | NB-ARC domain containing protein, putative, expressed | ns | down | down | LOC_Os01g01689 (ATM) | ||
| osa-MIR5491-p5 | LOC_Os01g24060 | importin subunit α, putative, expressed | ns | down | ns | osa-MIR5491-p5 | LOC_Os03g12414 (OsSDS) |
| osa-MIR5518-p3 | LOC_Os04g48770 | ubiquitin fusion degradation protein, putative, expressed | ns | down | ns | osa-MIR5518-p3 | LOC_Os04g40290 (CRC1) |
| osa-MIR5806-p5 | LOC_Os08g19694 | NB-ARC domain containing protein, expressed | ns | down | ns | osa-MIR5806-p5 | LOC_Os01g01689 (ATM) |
| PC-3p-120829_194 | LOC_Os03g04520 | RNA recognition motif containing protein, putative, expressed | ns | down | ns | PC-3p-120829_194 | LOC_Os02g40450 (OsMER3) |
ns: non-significant; Up: up-regulated in autotetraploid rice; Down: down-regulated in autotetraploid rice; PMA, MA and SCP represent pre-meiotic interphase, meiosis and single microspore stage, respectively.
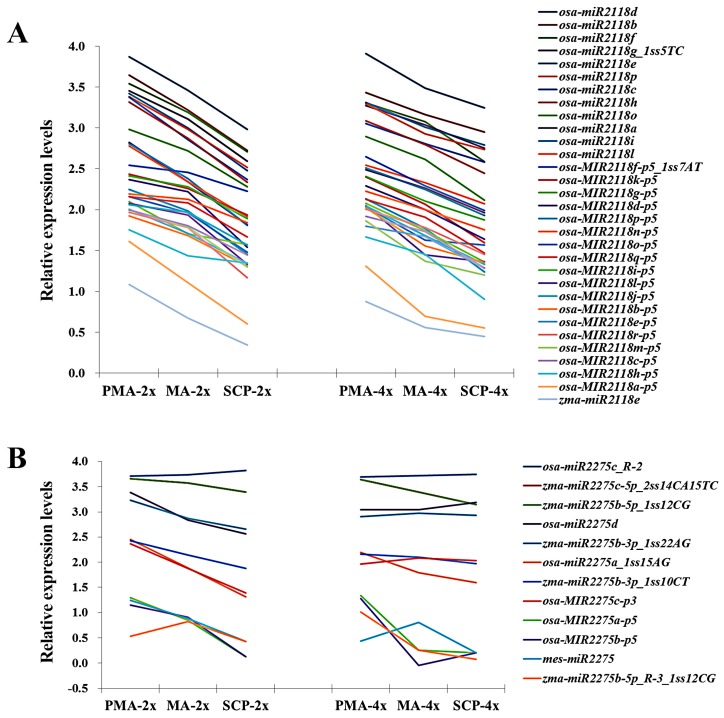
Furthermore, some miRNAs played key roles during meiotic division by generating phasiRNAs [31,32], such as miR2118 and miR2275 that required for biogenesis of phasiRNAs. The expression patterns of miR2118 and miR2275 families from PMA to SCP were almost the same between Taichung65-4x and Taichung65-2x, but with some variations in the members of these families (Figure 5). Eight miRNAs of miR2118 family were differentially expressed and preferentially down-regulated in pollen development stages. Three of them, osa-MIR2118l-p5, osa-MIR2118m-p5 and osa-MIR2118o-p5, were specifically down-regulated in MA. Four members of the miR2275 family also exhibited differential expression patterns in Taichung65-4x. Interestingly, these miRNAs showed similar expression levels in MA, but significantly down-regulated in PMA (Table 3).
Figure 5.
The relative expression levels of miR2118 (A) and miR2275 (B) families between Taichung65-4x and Taichung65-2x. PMA, MA and SCP represent pre-meiotic interphase, meiosis and single microspore stage, respectively. “4x” and “2x” represent the autotetraploid and diploid rice, respectively.
Table 3.
The regulation of miR2118 and miR2275 families in autotetraploid rice.
| MicroRNA_Family | miRNA | Sequence (5’ to 3’) | PMA | MA | SCP |
|---|---|---|---|---|---|
| miR2118 | osa-MIR2118k-p5 | TAAGCTCTATTGTCCCCTCTA | ns | ns | down |
| osa-miR2118e | TTCCCAATGCCTCCCATGCCTA | ns | ns | up | |
| osa-MIR2118l-p5 | TTAGGAAGAGGAAGAAATTGA | ns | down | ns | |
| osa-MIR2118m-p5 | GGAATGGGAACATGAAGGAAAG | ns | down | ns | |
| osa-MIR2118o-p5 | GGCATGGGGACATGAAGGAATG | ns | down | ns | |
| osa-miR2118a | TTCTCGATGCCTCCCATTCCTA | down | ns | ns | |
| osa-miR2118c | TTCCCGATGCCTCCTATTCCTA | down | ns | ns | |
| osa-MIR2118a-p5 | GGACTGGGAACATATGAGAAAG | down | ns | ns | |
| miR2275 | osa-miR2275d | CTTGTTTTTCTCCAATATCTCA | down | ns | up |
| osa-MIR2275c-p3 | ATTGTTTTTCTCCAATATCTCA | down | ns | up | |
| mes-miR2275 | TTTGGTTTCCTCCAATATCTTA | down | ns | ns | |
| zma-miR2275b-3p_1ss22AG | TTCAGTTTCCTCTAATATCTCG | down | ns | ns |
ns: non-significant; Up: up-regulated in autotetraploid rice; Down: down-regulated in autotetraploid rice; PMA, MA and SCP represent pre-meiotic interphase, meiosis and single microspore stage, respectively.
2.6. Functional Classification of Pre-Meiotic Interphase (PMA) and Single Microspore Stage (SCP) Stage-Specific Targets Regulated by Differentially Expressed miRNAs
Pre-meiotic interphase (PMA) is an important step for meiosis initiation, and many genes expressed during this stage. Consequently, the target genes predicted by differentially expressed miRNAs in autotetraploid rice were compared with the previously reported microarray data for pre-meiosis-related and stage-specific expression in diploid and autotetraploid rice during PMA [14,23,24]. Of the 27 up-regulated miRNAs in PMA, 22 predicted target genes were associated with PMA-related and stage-specific expression patterns (Table S11). One gene, LOC_Os01g43870 (predicted by PC-5p-431255_40), encoded NLI interacting factor-like phosphatase, was found in this study, which showed down-regulation in PMA of autotetraploid compared to diploid rice in a previous study [14]. Eight predicted target genes were up-regulated in the same stage of autotetraploid than did in the diploid rice [14]. Another 18 genes were found to be up-regulated during the same stage in diploid rice [24]. Of the 19 down-regulated miRNAs in PMA, only one gene, LOC_Os05g34220 (predicted by osa-miR2118c) was annotated as vrga1 and necessary for PMA in diploid rice, was specifically expressed in PMA (Table S11). In addition, PC-5p-431255_40 was predicted to target the LOC_Os02g07180 (JASON), LOC_Os03g50220 and LOC_Os06g51220 gene encoding an expressed protein, homologous-pairing protein and HMG1/2 (HIGH MOBILITY GROUP), which was specifically up-regulated in autotetraploid rice during PMA (Table S9A). These three genes were involved in male meiosis II [33], chiasma assembly [34] and chromatin assembly or disassembly [35] in Arabidopsis. Here, osa-MIR167c-p3 targeted LOC_Os05g31230 gene that encodes N-acetyltransferase, a homolog of the yeast CTF protein [36] that required for the formation of sister chromatid cohesion, which was also specifically up-regulated in autotetraploid rice during PMA (Table S9A).
Single microspore stage (SCP) development from meiosis owns to haploid chromosome, from which pollen development involves a highly coordinated series of cellular events; therefore, transcriptome is different from the somatic cells. We analyzed the DEM at this stage, and compared the target genes predicted by the DEM in autotetraploid rice with the previously known microarray data for diploid and autotetraploid SCP-related and stage-specific expression patterns [14,24]. Among 75 up-regulated miRNAs in SCP, 25 predicted target genes displayed stage-specific expression patterns. Of these 25 genes, LOC_Os01g64100 (predicted by osa-miR1320-5p_L-1R+1) and LOC_Os01g57610 (predicted by osa-miR1436_L+3_1ss5CT) exhibited up-regulation in SCP of autotetraploid compared to diploid rice [14]. Six predicted target genes of down-regulated miRNAs exhibited SCP-specific expression, and five of them were found to be up-regulated in SCP of diploid rice [24], and LOC_Os01g03330 gene (predicted by PC-5p-72318_316) was up-regulated in SCP of autotetraploid compared to diploid rice [14] (Table S11).
2.7. Correlation between miRNAs and Their Target Genes
To validate the correlations between DEM identified by Illumina sequencing and their potential targets predicted by Targetfinder (MIT, Cambridge, MA, USA), five DEM and their corresponding target genes were taken during different pollen development stages in Taichung65-2x and Taichung65-4x, and examined by qRT-PCR (Figure S12). osa-miR159a.1 targeted a MYB transcription factor family (LOC_Os01g59660), osa-miR5514 and osa-MIR5083-p3 targeted LOC_Os01g49490 and LOC_Os05g05440 gene, respectively. osa-miR528-5p targeted a F-box domain and LRR containing protein (LOC_Os06g06050) and osa-miR1432-5p_R+1 targeted a metal cation transporter (LOC_Os05g07210). The expression patterns of DEM by qRT-PCR were nearly similar to the sequencing data during pollen development. The expression levels of three miRNAs were found to be negatively correlated with their target genes in Taichung65-2x compared to Taichung65-4x rice during SCP (Figure S12). In our study, osa-miR528-5p was up-regulated in PMA and SCP, while osa-miR1432-5p_R+1 showed up-regulation during three pollen development stages in Taichung65-4x. However, osa-miR528-5p and osa-miR1432-5p_R+1 showed negative correlation with their predicted targets in SCP. Different expression levels in Taichung65-4x may be due to the gene dosage effect of polyploidization.
2.8. Differentially Expressed siRNAs in the Autopolyploid Pollen Associated with Transposable Elements
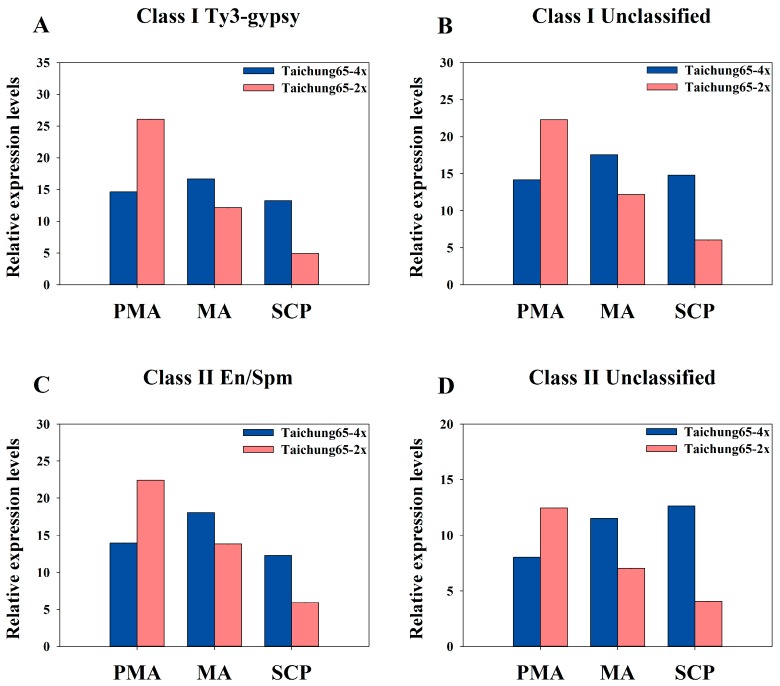
Small RNAs regulates gene expression and directed DNA methylation [37]. The 24 nt siRNA abundance associated with CHG and CHH hypermethylation of class II transposable elements (TEs) inhibited the expression of related genes in autotetraploid rice [38]. Based on the method described by Zhang et al. [38], we identified a total of 1096 siRNAs associated with the transposable elements (Table S13); 21 to 24 nt sRNAs constituted the major portion of siRNAs population (Table S14). We focused on the 24 nt siRNAs (TEs) to investigate the relationship with DNA methylation in autotetraploid compared to diploid rice. We found five types of class I (retrotransposons) and five types of class II (transposon) in 24 nt siRNAs population; Ty3-gypsy and unclassified type with the number of 72 and 171 mainly related to class I (retrotransposons), whereas En/Spm and unclassified type with 84 and 38 in class II (transposon), respectively (Table S15). Thus, we mainly focused on these four main types of siRNAs. Compared to the diploid rice, 24 nt siRNA associated with transposable elements were more abundant during the pollen development in Taichung65-4x, except the PMA stage; very similar changes were detected in class I and class II (Figure 6). The siRNAs with lower expression levels, TEs associated, were found in PMA of polyploid than diploid rice. However, this tendency was reversed during MA and maintained in SCP of polyploid rice (Figure 6). In addition, the expression levels of TEs-associated siRNAs displayed remarkable changes in diploid rice, which gradually declined during the pollen development (Figure 6; Table S16). However, we found irregular changes in the expression levels of siRNAs during the pollen development in polyploid rice, and class I and class II of TEs were enriched in MA compared to PMA in polyploid rice (Figure 6; Table S16).
Figure 6.
Abundance of siRNAs transposable elements associated with Taichung65-4x and Taichung65-2x during pollen development stages: (A) Ty3-gypsy type of class I; (B) unclassified type of class I; (C) En/Spm type of class II; and (D) unclassified type of class II. PMA, MA and SCP represent pre-meiotic interphase, meiosis and single microspore stage, respectively. “4x” and “2x” represent the autotetraploid and diploid rice, respectively.
Furthermore, we compared the predicted targets of differentially expressed miRNAs detected in our study to Zhang et al. [38], and the results revealed that three targets associated with the DNA methylation activity, including LOC_Os01g25450 (predicted by mes-miR2275, the down-regulated DEM in PMA), LOC_Os10g26430 (predicted by osa-miR5796, the down-regulated DEM in MA and SCP) and LOC_Os05g49930 (predicted by bdi-miR5054_L+2, the down-regulated DEM in SCP), were annotated as AIG1 family protein, agenet domain-containing protein and GRAS family transcription factor domain-containing protein, respectively (Table S17).
3. Discussion
3.1. Polyploidy Cause Changes in miRNA Expression Profiles during Pollen Development of Autotetraploid Rice
As negative regulators, miRNAs targets involved in signal transduction, carbohydrate and nitrogen metabolism and hormone homeostasis, indicating that miRNAs play important roles in regulating pollen development in plants [39,40,41] and rice grain filling [42,43]. For example, miR156/7 targets SPL genes required for male fertility in Arabidopsis [44]. The miR159 regulated the GAMYB-like targets (MYB33 and MYB65) during Arabidopsis anther development [45]. The miR167 controlled the expression patterns of ARF6 and ARF8 in Arabidopsis and regulate both male and female reproduction [46]. A recent study on miRNAome analysis of developing rice pollen from the uninucleate microspores (UNM) to tricellular pollen stages (TCP) revealed 202 known miRNAs in developing pollen, many (103) were pollen enriched, such as osa-miR164a/b/f and d, osa-miR169e and n/o, osa-miR171a, osa-miR396a/b, osa-miR399h and osa-miR1881, and more than half of 75 novel miRNAs were expressed in developing pollen [22]. Epigenetic regulation of chromatin assembly and disassembly might be controlled by the pollen development through over-expression of osa-miR820 and osa-miR827, targeted Os03g02010 (DNA cytosine methyl transferase) and Os04g11510 (a methyl-CpG binding domain protein), respectively. Here, we detected significant differences in the abundance of miRNAs during the pollen development stages between autotetraploid and diploid rice; large numbers of specifically up- and down-regulated miRNAs were also found in the same adjacent stages between Taichung65-2x and Taichung65-4x, it may happen due to the effect of polyploidization. Additionally, a total of 223 miRNAs co-expressed in diploid and autotetraploid rice, while only 57 and 38 miRNAs were specifically expressed in autotetraploid and diploid rice, respectively. Autotetraploid rice inherited many miRNAs from the diploid rice; however, some were lost or generated after polyploidization. This phenomenon was also observed in other plants [47] and illustrated as the conserved miRNAs maintained genomic stability and the transcriptionally altered miRNAs might fulfill the need of polyploidy, such as the regulation of pollen fertility.
Furthermore, about 40% miRNAs (60 known miRNAs and 112 novel miRNAs) displayed different levels of expression during pollen development stages in autotetraploid and diploid rice. DEM were specifically found during each pollen development stage, suggesting a strong influence of miRNAs on their predicted target genes associated with the regulatory network of pollen development, and most probably resulted in the sterile pollen after the polyploidization. Autotetraploid rice has doubled genomes, more miRNAs should be observed and the expression levels should be increased two-fold compared with diploid rice. However, the expression patterns of miRNAs were not simplified between autotetraploid and diploid rice, some displayed over-expression while some showed down-regulation in autotetraploid rice than did in the diploid rice, as similar behavior was found during the miRNAome analysis in tetraploid Paulownia tomentosa [48]. Ha et al. [49] demonstrated that miRNAs expression levels are highly variable between the allotetraploids and their diploid counterpart (Arabidopsis thaliana and Arabidopsis arenosa), leading to the variations in gene expression, growth, and adaptation. Our data also suggest that miRNA regulation is more complex in autotetraploids than diploids.
GO analysis of the predicted targets of detected miRNAs in Taichung65-4x showed significant GO terms enriched in three stages, whereas some significant GO terms were detected only in one stage (Table S18). Our results showed that transcription factor activity was commonly enriched in three stages of Taichung65-4x. Similar phenomenon was found in the microarray analysis [14], indicating the importance of the transcription factor activity genes in developing pollens of autotetraploid rice. DEM predicted targets showed significant GO terms associated with the transport, transporter activity, signal and other growth and development processes. Among the DEM predicted targets, the predicted targets of up-regulated miRNAs were different from the down-regulated miRNAs. These results demonstrated that the differential expressions of miRNAs may be closely related to the pollen fertility of autotetraploid and diploid rice. In addition, a large number of targets involved in vegetative organ elongation and cell wall thickening were also found in our study. Recently, we have reported the larger PMC and the longer anther in autotetraploid than diploid rice [14], implying that the above mentioned genes have close relationship with cell wall structures of polyploidy.
3.2. Specific miRNA Expression Patterns May Cause Meiosis Abnormalities in Autotetraploid Rice
The 21- and 24-nucleotide phasiRNAs were generated by the target sites of miR2118 and miR2275, recognized as a class of panicle-specific siRNAs [31,32]. Recently, rice Argonaute MEL1 selectively binds 21-nt phasiRNAs generated by miR2118, and the mel1 mutants have abnormal cytoplasm and aberrant PMCs that halt in early meiosis, suggesting that MEL1-phasiRNAs play important role in male fertility [31,50]. In addition, miR2118 family members were abundant at cell fate specification, and then vanished at cell differentiation of maize anther [32]. Similarly, our study also showed that miR2118 was abundant in pre-meiotic and then gradually decreased during other stages of pollen development. MEL1 (Os03g0800200) was significantly up-regulated in Taichuang65-4x [14], closely related to the differential expressions of miR2118. Furthermore, the miR2275 and meiotic-phasiRNAs accumulate preferentially in tapetum and meiocytes to perform their functions [32]. Lower accumulation of miR2275 may cause developmental defects by disrupting the stamen in osdcl4-1 [51]. In our study, the members of miR2275 showed similar variation patterns in diploid rice, while some members displayed contrasting patterns from PMA to SCP, especially in meiosis of autotetraploid that probably give rise to unbalanced regulation of 24-phasiRNAs. These findings were associated with the higher frequency of cytological abnormalities in autotetraploid rice, and we inferred that differential expressions of miR2118 and miR2275 families were associated with meiotic abnormalities in autotetraploid rice.
By the protein–protein interaction analysis, we identified 16 meiosis-related miRNAs whose targets showed interaction with meiosis-related genes. One of predicted target, PC-3p-403976_40, was related to Replication Protein A (RPA1a). OsRPA1a is required for meiosis, the osrpa1a mutants were sterile at the reproductive stage and no embryo sac was found in female meiocytes, and defragmented chromosome were observed in male meiocytes after anaphase I [52]. OsMER3 is necessary for normal meiotic crossing over, and null mutation of MER3 cause complete sterility in rice [53]; OsMER3 was associated with a target predicted by osa-miR5788 in our study. HTA3, ATM and RAD50 associated with each other in the protein interaction network. ATM gene, a chromosome instability disorder in Arabidopsis, might be involved in the telomerase-independent pathway called as alternative lengthening of telomeres [54,55]; target of osa-MIR2122-p3_1ss19GT was related to ATM. ATM contributed to the formation of phosphorylated H2AX (γ-H2AX) [56], which is a meiosis-specific isoform of histone H2A (The histone H2A protein encoded by HTA3). Phosphorylated H2AX accumulated at sites of DNA damage when DNA double-strand breaks (DSBs) happened. Targets predicted by osa-MIR6250-p5 were related to both HTA3 and ATM. Moreover, RAD50 (DNA repair-recombination protein), a component of Mre11/Rad50/Xrs2 (MRX) complex, is essential for meiosis [57,58]. The down-regulation of RAD50 was observed in polyploid rice hybrids during meiosis [15]; the targets predicted by osa-miR1432-5p_R+1 and osa-miR5788 were associated with RAD50 in our study. We speculated that these miRNAs (especially the up-regulated miRNAs) might be involved in the interaction network of meiosis-related genes and accompanied with the abnormal regulation of autotetraploid rice, particularly in meiosis.
osa-miR1432-5p predicted to target the genes of metal cation transporter (LOC_Os05g07210), calcium-transporting ATPase (LOC_Os04g51610) and EF-hand family (LOC_Os03g59790, LOC_Os03g59870, LOC_Os03g59770), and was found to be up-regulated in the early uninucleate stage of developing pollens of MxA (sterile line) which impair the Ca2+-mediated signaling pathway during the rice anther development [59]. In our study, osa-miR1432-5p_R+1 was significantly up-regulated in autotetraploid rice from PMA to SCP and confirmed by the qRT-PCR, and showed the negative correlation with the target genes in SCP. These results displayed that over expression of osa-miR1432 might interrupt the Ca2+ balance in the PMC and played the crucial roles in autotetraploid rice, so we speculated that this miRNA influence tetraploid pollen viability. Another important miRNA, osa-miR528, might be involved in the pollen sterility by targeting the F-box proteins, was also reported by Yan et al. [59]. Notably, a negative tendency was observed in SCP between the levels of osa-miR528-5p and its target LOC_Os06g06050 (F-box protein) in Taichung65-4x and Taichung65-2x. Further, we also found another miRNA, osa-miR172d-5p_R+1, which targeted the LOC_Os06g06050. In the present study, osa-miR172d-5p_R+1 and osa-miR528-5p were abundantly expressed in diploid and autotetraploid rice, respectively. The expression of osa-miR172d-5p_R+1 showed the rising trends gradually from PMA to SCP, while osa-miR528-5p showed the contrary trends over the same period. We speculated that osa-miR528-5p was dominant in the early stage and then taken over by osa-miR172d-5p_R+1 in the later period, which silenced the expression of target genes, and finally resulted in abnormal pollen development in autotetraploid rice. However, the relationship between osa-miR528 and osa-miR172d-5p_R+1 during the rice reproductive development had not been reported, and this required further study.
3.3. Abundance of siRNAs Associated with Transposable Elements May Cause Meiosis Abnormalities in Autotetraploid Rice
Small RNAs are known to be a key component of a signaling network that mediated through DNA methylation and histone modifications [37,60,61,62,63]. Previous study has shown that variation in DNA methylation and activity of class II transposable elements (TEs) play crucial role in the polyploidization of rice to adapt the whole-genome duplication (WGD) [38]. In our study, we abundantly found 24 nt siRNAs associated with both of class I and class II transposable elements, especially Ty3-gypsy in class I and En/Spm type in Class II, in autotetraploid rice during pollen development. Significant decline in 24 nt siRNA TEs associated with developing pollen of diploid rice reflected the activation of gene expression throughout the pollen development and resulted in high seed set. However, 24 nt siRNA maintained its abundance during pollen development in polyploid rice and cause remarkable changes in meiosis compared to diploid rice, suggesting that significant variations during meiosis have a strong influence on the pollen development in polyploid rice. These results are consistent with the previous study, which also found that high chromosomal abnormalities were associated with the altered expression profiles of meiosis related genes in autotetraploid rice [14]. Some 24 nt miRNAs can lead to cytosine DNA methylation at their own loci and target genes, and lead to silencing of gene transcription [64]. LOC_Os10g26430 has hypermethylation of CHG, not only surrounded by DMRs in its flanking 4-kb regions but also in the gene bodies of autotetraploid rice compared with diploid rice [38]. In our study, we found three DEM, especially osa-miR5796 that was down-regulated in MA and SCP of Taichung65-4x and predicted a LOC_Os10g26430 gene, may be associated with the DNA methylation and transposon activity in polyploidization of rice. Taken together, our findings suggest that abundance of siRNAs associated with the transposable elements that involved in DNA methylation in autotetraploid rice, probably influenced the autotetraploid pollen fertility during meiosis.
4. Experimental Section
4.1. Rice Materials
Autotetraploid rice, Taichung65-4x, and its diploid counterpart, Taichung65-2x, were used in the present study. Taichung65-2x was treated with colchicine to develop Taichung65-4x and self-crossed for more than 22 generations in our lab [14]. They were planted at the experimental site of South China Agricultural University (SCAU) under field conditions. Anthers during three stages of pollen development, including pre-meiotic interphase (PMA), meiosis (MA) and single microspore stage (SCP), were collected from Taichung65-4x and Taichung65-2x as described previously [14]. All samples were stored at −80 °C for RNA extraction.
4.2. Small RNA Library Construction, Sequencing and Data Processing
Total RNA was isolated from the anthers using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The total RNA purity and quantity were analyzed by RNA 6000 Nano LabChip Kit (Agilent, Palo Alto, CA, USA) and Bioanalyzer 2100 with RIN number >7.0. About 1 µg of RNA was used to construct small RNA library according to the protocol of Illumina’s TruSeq small RNA sample preparation Kits (San Diego, CA, USA). Then we executed the single-end sequencing (36 bp) on an Illumina Hiseq2500 at the LC-BIO (Hangzhou, China) according to the manufacturer’s protocol.
After the Illumina sequencing, the raw reads were exposed to the Illumina pipeline filter (Solexa 0.3). To remove common RNA families (tRNA, rRNA, snRNA, snoRNA), adapter dimers, low complexity, junk and repeats, the data were further processed with an in-house program, ACGT101-miR (LC Sciences, Houston, TX, USA) [65]. Consequently, the 18–25 nt length unique sequences were BLASTed to rice precursors in miRBase 20.0 [66] to detect known miRNAs. One mismatch inside of the sequence and length variation at both 3′ and 5′ ends were allowed in the alignment. The unique sequences were mapped to rice mature miRNAs in hairpin arms recognized as known miRNAs, and mapped to the other arm of known rice precursor hairpin opposite to the annotated mature miRNA-containing arm considered to be novel 5p- or 3p-derived miRNAs. The remaining sequences were mapped to other plant species in miRBase 20.0 by BLAST search, and the mapped pre-miRNAs were further BLASTed against the rice genomes [67] to identify their genomic positions. The aforementioned miRNAs were considered as known miRNAs. To identify the novel predicted miRNAs, the unmapped sequences were BLASTed against the rice genome database, and the hairpin RNA structures comprising sequences were predicated by using RNAfold software [68].
Data normalization followed the procedures as described in a previous study [69] with minor modification. (1) Find a common set of sequences among all samples; (2) Construct a reference data set. Each data in the reference set is the copy number median value of a corresponding common sequence of all samples; (3) Perform 2-based logarithm transformation on copy numbers (Log2 (copy#)) of all samples and reference data set; (4) Calculate the Log2 (copy#) difference (ΔLog2 (copy#)) between individual sample and the reference data set; (5) Form a subset of sequences by selecting |ΔLog2 (copy#)| < 2, which means less than 4 (22) fold change from the reference set; (6) Perform linear regressions between individual samples and the reference set on the subset sequences to derive linear equations y = aix + bi, where ai and bi are the slop and interception, respectively, of the derived line, x is Log2 (copy#) of the reference set, and y is the expected Log2 (copy#) of sample i on a corresponding sequence; (7) Calculate the mid value xmid = (max(x) − min(x))/2 of the reference set. Calculate the corresponding expected Log2 (copy#) of sample i, yi,mid = aixmid + bi. Let yr,mid = xmid, let Δyi = yr,mid − yi,mid, which is the logarithmic correction factor of sample i. We then derive the arithmetic correction factor fi = 2Δyi sample I; (8) Correct copy numbers of individual samples by multiplying corresponding arithmetic correction factor to original copy numbers.
To find out the siRNAs associated with the transposable elements, we filtered reads that did not match miRNA, rRNA, tRNA, snRNA, or snoRNA, and mapped them against the rice reference genome to select the genome sequences annotated as transposable elements [38]. These subsequent of siRNAs associated with transposable elements (TEs) were used for further analysis. The type of transposable elements (TEs) associated with siRNAs was classified firstly, and then we averaged the expression levels of siRNAs associated with transposable elements in each type to estimate the relative expression levels. These relative expression levels represented the TEs expression levels of each type during each stage in diploid and autotetraploid rice. Small RNA sequencing data have been deposited to NCBI GEO database (Accession Number: GSE79344).
4.3. Analysis of Differentially Expressed miRNAs
MicroRNAs were regarded as differentially expressed based on normalized deep-sequencing levels (with the exclusion of 10 RPM) in Taichung65-2X and Taichung65-4X during pollen development. p-Value was estimated by selectively by using Chi-square (X2) test and Fisher exact test. A significance threshold level was set to be 0.05 in each test. miRNAs with p-value <0.05 and log2 (fold change ratio) >1 were considered as differentially expressed miRNA (DEM). The normalized read count of some miRNAs were set to be 0.01 for further calculation if it has no reads in the library.
4.4. The Prediction and Functional Analysis of Target Genes of miRNAs
To predict the genes targeted by DEM, computational target prediction algorithms and TargetFinder [70] were used to detect miRNA binding sites. The algorithm of TargetFinder was based on miRNA and mRNA complementary pairing principle. The miRNA sequence (5′–3′) from 2 to 13 nt was defined as the seed sequence. The mismatching of Guanine and Uracil will lose 0.5 point and the rest of the mismatches cut off 1 point outside the seed sequence, while doubling the lost point in seed sequence. Finally, the results were obtained with the points less than or equal to 4. GO enrichment analysis was done by using AgriGO [71]. Protein–protein interaction networks were used to predict the meiosis-related targets/genes by using STRING v10 [72].
4.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
The RNA was extracted from anthers of Taichung65-2x and Taichung65-4x at each stage of pollen development, and used as template for reverse transcription with miRNA-specific stem-loop RT primers [73] using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany). The reactions were incubated at 16 °C for 30 min, then pulsed RT of 60 cycles at 30 °C for 30 s, 42 °C for 30 s and 50 °C for 1 s, and a final incubation for 5 min at 85 °C to inactivate the reverse transcriptase [74]. The cDNA template for the miRNA target gene was reverse transcribed using the OligodT20 primer with the Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany).
The qRT-PCRs were performed on Lightcycler480 (Roche) using the SsoAdvanced universal SYBR Green Supermix (Bio-RAD, Hercules, CA, USA). The reaction profile was as follow: 30 s at 95 °C, 40 cycles of 95 °C denaturation for 5 s and 58 °C annealing and extension for 20 s. We performed three biological replications of each reaction, and U6 snRNA and ubiquitin were used as internal control genes for qRT-PCR of DEM and target genes, respectively. The relative expression values of DEM or target genes were calculated based on the 2−ΔΔCt method [75]. The stem-loop RT primers and target gene-specific primers were designed by using the Primer Premier 5.0 software (Palo Alto, CA, USA) (Table S19).
Acknowledgments
The authors thank Chen Zhixiong, Shuhong Yu, Jianning Liu (LC Sciences, Hangzhou, China) and Jie Zhang (Kunming Institute of Botany, Kunming, China) for technical assistance and data analysis. This work was supported by the NSFC to Xiangdong Liu (31270352; 31571625; and 31210103023), Guangdong provincial key platform of university and major research project (Natural Science)—characteristic innovation project (Yue-JK2014 (65) to Xiangdong Liu) and the Guangdong science and technology program (2014A030304055 to Xiangdong Liu).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/4/499/s1.
Author Contributions
Xiangdong Liu, Yonggen Lu and Muhammad Qasim Shahid conceived and designed the experiments. Xiang Li, Muhammad Qasim Shahid and Xiangdong Liu wrote the paper. Xiang Li, Muhammad Qasim Shahid, and Jinwen Wu performed the experiment and analyzed the data. Lan Wang contributed to data analysis. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Soltis D.E., Bell C.D., Kim S., Soltis P.S. Origin and early evolution of angiosperms. Ann. N. Y. Acad. Sci. 2008;1133:3–25. doi: 10.1196/annals.1438.005. [DOI] [PubMed] [Google Scholar]
- 2.Jiao Y., Wickett N.J., Ayyampalayam S., Chanderbali A.S., Landherr L., Ralph P.E., Tomsho L.P., Hu Y., Liang H., Soltis P.S., et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 3.Bennett M.D. Perspectives on polyploidy in plants—Ancient and neo. Biol. J. Linn. Soc. 2004;82:411–423. doi: 10.1111/j.1095-8312.2004.00328.x. [DOI] [Google Scholar]
- 4.Shahid M.Q., Li Y.J., Saleem M.F., Naeem M., Wei C.M., Liu X.D. Yield and yield components in autotetraploid and diploid rice genotypes (indica and japonica) sown in early and late seasons. Aust. J. Crop Sci. 2013;7:632–641. [Google Scholar]
- 5.Osborn T.C., Pires J.C., Birchler J.A., Auger D.L., Chen Z.J., Lee H.S., Comai L., Madlung A., Doerge R.W., Colot V., et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003;19:141–147. doi: 10.1016/S0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- 6.Comai L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 7.Yu Z., Haage K., Streit V.E., Gierl A., Ruiz R.A. A large number of tetraploid Arabidopsis thaliana lines, generated by a rapid strategy, reveal high stability of neo-tetraploids during consecutive generations. Theor. Appl. Genet. 2009;118:1107–1119. doi: 10.1007/s00122-009-0966-9. [DOI] [PubMed] [Google Scholar]
- 8.Shahid M.Q., Liu G., Li J., Naeem M., Liu X. Heterosis and gene action study of agronomic traits in diploid and autotetraploid rice. Acta Agric. Scand. Sect. B Soil Plant Sci. 2011;61:23–32. doi: 10.1080/09064710903428140. [DOI] [Google Scholar]
- 9.Shahid M.Q., Xu H., Lin S., Chen Z., Naeem M., Li Y., Liu X. Genetic analysis and hybrid vigor study of grain yield and other quantitative traits in autotetraploid rice. Pak. J. Bot. 2012;44:237–246. [Google Scholar]
- 10.Tu S., Luan L., Liu Y., Long W., Kong F., He T., Xu Q., Yan W., Yu M. Production and heterosis analysis of rice autotetraploid hybrids. Crop Sci. 2007;47:2356–2363. doi: 10.2135/cropsci2007.01.0058. [DOI] [Google Scholar]
- 11.He J.H., Shahid M.Q., Chen Z.X., Chen X.A., Liu X.D., Lu Y.G. Abnormal PMC microtubule distribution pattern and chromosome behavior resulted in low pollen fertility of an intersubspecific autotetraploid rice hybrid. Plant Syst. Evol. 2011;291:257–265. doi: 10.1007/s00606-010-0386-y. [DOI] [Google Scholar]
- 12.He J.H., Shahid M.Q., Li Y.J., Guo H.B., Cheng X.A., Liu X.D., Lu Y.G. Allelic interaction of F1 pollen sterility loci and abnormal chromosome behaviour caused pollen sterility in intersubspecific autotetraploid rice hybrids. J. Exp. Bot. 2011;62:4433–4445. doi: 10.1093/jxb/err098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shahid M.Q., Sun J.F., Wei C.M., Zhang P., Liu X.D. Studies on the abnormality of embryo sac and pollen fertility in autotetraploid rice during different growing seasons. Pak. J. Bot. 2010;42:7–19. [Google Scholar]
- 14.Wu J., Shahid M.Q., Guo H., Yin W., Chen Z., Wang L., Liu X., Lu Y. Comparative cytological and transcriptomic analysis of pollen development in autotetraploid and diploid rice. Plant Reprod. 2014;27:181–196. doi: 10.1007/s00497-014-0250-2. [DOI] [PubMed] [Google Scholar]
- 15.Wu J., Shahid M.Q., Chen L., Chen Z., Wang L., Liu X., Lu Y. Polyploidy enhances F1 pollen sterility loci interactions that increase meiosis abnormalities and pollen sterility in autotetraploid rice. Plant Physiol. 2015;169:2700–2717. doi: 10.1104/pp.15.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong D.H., Green P.J. The role of rice microRNAs in abiotic stress responses. J. Plant Biol. 2013;56:187–197. doi: 10.1007/s12374-013-0213-4. [DOI] [Google Scholar]
- 17.Kong X., Zhang M., Xu X., Li X., Li C., Ding Z. System analysis of microRNAs in the development and aluminium stress responses of the maize root system. Plant Biotechnol. J. 2014;12:1108–1121. doi: 10.1111/pbi.12218. [DOI] [PubMed] [Google Scholar]
- 18.Meng Y., Shao C., Wang H., Ma X., Chen M. Construction of gene regulatory networks mediated by vegetative and reproductive stage-specific small RNAs in rice (Oryza sativa) New Phytol. 2013;197:441–453. doi: 10.1111/nph.12018. [DOI] [PubMed] [Google Scholar]
- 19.Tang M., Mao D., Xu L., Li D., Song S., Chen C. Integrated analysis of miRNA and mRNA expression profiles in response to Cd exposure in rice seedlings. BMC Genom. 2014;15:835. doi: 10.1186/1471-2164-15-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujioka T., Kaneko F., Kazama T., Suwabe K., Suzuki G., Makino A., Mae T., Endo M., Kawagishi-Kobayashi M., Watanabe M. Identification of small RNAs in late developmental stage of rice anthers. Genes Genet. Syst. 2008;83:281–284. doi: 10.1266/ggs.83.281. [DOI] [PubMed] [Google Scholar]
- 21.Peng H., Chun J., Ai T.B., Tong Y.A., Zhang R., Zhao M.M., Chen F., Wang S.H. MicroRNA profiles and their control of male gametophyte development in rice. Plant Mol. Biol. 2012;80:85–102. doi: 10.1007/s11103-012-9898-x. [DOI] [PubMed] [Google Scholar]
- 22.Wei L.Q., Yan L.F., Wang T. Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol. 2011;12:R53. doi: 10.1186/gb-2011-12-6-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita M., Horiuchi Y., Ueda Y., Mizuta Y., Kubo T., Yano K., Yamaki S., Tsuda K., Nagata T., Niihama M., et al. Rice expression atlas in reproductive development. Plant Cell Physiol. 2010;51:2060–2081. doi: 10.1093/pcp/pcq165. [DOI] [PubMed] [Google Scholar]
- 24.Deveshwar P., Bovill W.D., Sharma R., Able J.A., Kapoor S. Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biol. 2011;11:78. doi: 10.1186/1471-2229-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yant L., Hollister J.D., Wright K.M., Arnold B.J., Higgins J.D., Franklin F.C., Bomblies K. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 2013;23:2151–2156. doi: 10.1016/j.cub.2013.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright K.M., Arnold B., Xue K., Surinova M., O’Connell J., Bomblies K. Selection on meiosis genes in diploid and tetraploid Arabidopsis arenosa. Mol. Biol. Evol. 2015;32:944–955. doi: 10.1093/molbev/msu398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Y., Yang H., Wei Z., Ma H., Ge X. Rice male development under drought stress: Phenotypic changes and stage-dependent transcriptomic reprogramming. Mol. Plant. 2013;6:1630–1645. doi: 10.1093/mp/sst067. [DOI] [PubMed] [Google Scholar]
- 28.Jakobsen M.K., Poulsen L.R., Schulz A., Fleurat-Lessard P., Moller A., Husted S., Schiott M., Amtmann A., Palmgren M.G. Pollen development and fertilization in Arabidopsis is dependent on the MALE GAMETOGENESIS IMPAIRED ANTHERS gene encoding a type V P-type ATPase. Genes Dev. 2005;19:2757–2769. doi: 10.1101/gad.357305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J.X., Liu Y.G. Molecular control of male reproductive development and pollen fertility in rice. J. Integr. Plant Biol. 2012;54:967–978. doi: 10.1111/j.1744-7909.2012.01172.x. [DOI] [PubMed] [Google Scholar]
- 30.Luo Q., Li Y., Shen Y., Cheng Z. Ten years of gene discovery for meiotic event control in rice. J. Genet. Genom. 2014;41:125–137. doi: 10.1016/j.jgg.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Komiya R., Ohyanagi H., Niihama M., Watanabe T., Nakano M., Kurata N., Nonomura K. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014;78:385–397. doi: 10.1111/tpj.12483. [DOI] [PubMed] [Google Scholar]
- 32.Zhai J., Zhang H., Arikit S., Huang K., Nan G.L., Walbot V., Meyers B.C. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA. 2015;112:3146–3151. doi: 10.1073/pnas.1418918112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Storme N., Geelen D. The Arabidopsis mutant jason produces unreduced first division restitution male gametes through a parallel/fused spindle mechanism in meiosis II. Plant Physiol. 2011;155:1403–1415. doi: 10.1104/pp.110.170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uanschou C., Ronceret A., Von Harder M., de Muyt A., Vezon D., Pereira L., Chelysheva L., Kobayashi W., Kurumizaka H., Schlogelhofer P., et al. Sufficient amounts of functional HOP2/MND1 complex promote interhomolog DNA repair but are dispensable for intersister DNA repair during meiosis in Arabidopsis. Plant Cell. 2013;25:4924–4940. doi: 10.1105/tpc.113.118521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkle T., Grasser K.D. Unexpected mobility of plant chromatin-associated HMGB proteins. Plant Signal Behav. 2011;6:878–880. doi: 10.4161/psb.6.6.15255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Block-Schmidt A.S., Dukowic-Schulze S., Wanieck K., Reidt W., Puchta H. BRCC36A is epistatic to BRCA1 in DNA crosslink repair and homologous recombination in Arabidopsis thaliana. Nucleic Acids Res. 2011;39:146–154. doi: 10.1093/nar/gkq722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon S.A., Meyers B.C. Small RNA-mediated epigenetic modifications in plants. Curr. Opin. Plant Biol. 2011;14:148–155. doi: 10.1016/j.pbi.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J., Liu Y., Xia E.H., Yao Q.Y., Liu X.D., Gao L.Z. Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. USA. 2015;112:E7022–E7029. doi: 10.1073/pnas.1515170112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Y., Zhang Z., Lin H., Liu H., Chen J., Peng H., Cao M., Rong T., Pan G. Cytoplasmic male sterility-regulated novel microRNAs from maize. Funct. Integr. Genom. 2011;11:179–191. doi: 10.1007/s10142-010-0202-3. [DOI] [PubMed] [Google Scholar]
- 40.Wei M., Wei H., Wu M., Song M., Zhang J., Yu J., Fan S., Yu S. Comparative expression profiling of miRNA during anther development in genetic male sterile and wild type cotton. BMC Plant Biol. 2013;13:66. doi: 10.1186/1471-2229-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J., Liu X., Xu B., Zhao N., Yang X., Zhang M. Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines of Brassica juncea. BMC Genom. 2013;14:9. doi: 10.1186/1471-2164-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng T., Sun H., Du Y., Zhang J., Li J., Liu Y., Zhao Y., Zhao Q. Characterization and expression patterns of microRNAs involved in rice grain filling. PLoS ONE. 2013;8:499. doi: 10.1371/journal.pone.0054148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi R., Zhu Z., Hu J., Qian Q., Dai J., Ding Y. Identification and expression analysis of microRNAs at the grain filling stage in rice (Oryza sativa L.) via deep sequencing. PLoS ONE. 2013;8:499. doi: 10.1371/journal.pone.0057863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing S., Salinas M., Hohmann S., Berndtgen R., Huijser P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 2010;22:3935–3950. doi: 10.1105/tpc.110.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millar A.A., Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M.F., Tian Q., Reed J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- 47.Xie F., Zhang B. microRNA evolution and expression analysis in polyploidized cotton genome. Plant Biotechnol. J. 2015;13:421–434. doi: 10.1111/pbi.12295. [DOI] [PubMed] [Google Scholar]
- 48.Fan G., Zhai X., Niu S., Ren Y. Dynamic expression of novel and conserved microRNAs and their targets in diploid and tetraploid of Paulownia tomentosa. Biochimie. 2014;102:68–77. doi: 10.1016/j.biochi.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Ha M., Lu J., Tian L., Ramachandran V., Kasschau K.D., Chapman E.J., Carrington J.C., Chen X., Wang X.J., Chen Z.J. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA. 2009;106:17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nonomura K., Morohoshi A., Nakano M., Eiguchi M., Miyao A., Hirochika H., Kurata N. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell. 2007;19:2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song X., Li P., Zhai J., Zhou M., Ma L., Liu B., Jeong D.H., Nakano M., Cao S., Liu C., et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012;69:462–474. doi: 10.1111/j.1365-313X.2011.04805.x. [DOI] [PubMed] [Google Scholar]
- 52.Chang Y., Gong L., Yuan W., Li X., Chen G., Li X., Zhang Q., Wu C. Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiol. 2009;151:2162–2173. doi: 10.1104/pp.109.142877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang K., Tang D., Wang M., Lu J., Yu H., Liu J., Qian B., Gong Z., Wang X., Chen J., et al. MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sci. 2009;122:2055–2063. doi: 10.1242/jcs.049080. [DOI] [PubMed] [Google Scholar]
- 54.Vespa L., Warrington R.T., Mokros P., Siroky J., Shippen D.E. ATM regulates the length of individual telomere tracts in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104:18145–18150. doi: 10.1073/pnas.0704466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vespa L., Couvillion M., Spangler E., Shippen D.E. ATM and ATR make distinct contributions to chromosome end protection and the maintenance of telomeric DNA in Arabidopsis. Genes Dev. 2005;19:2111–2115. doi: 10.1101/gad.1333805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friesner J.D., Liu B., Culligan K., Britt A.B. Ionizing radiation-dependent γ-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell. 2005;16:2566–2576. doi: 10.1091/mbc.E04-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clerici M., Mantiero D., Lucchini G., Longhese M.P. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 58.Gallego M.E., Jeanneau M., Granier F., Bouchez D., Bechtold N., White C.I. Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J. 2001;25:31–41. doi: 10.1046/j.1365-313x.2001.00928.x. [DOI] [PubMed] [Google Scholar]
- 59.Yan J., Zhang H., Zheng Y., Ding Y. Comparative expression profiling of miRNAs between the cytoplasmic male sterile line MeixiangA and its maintainer line MeixiangB during rice anther development. Planta. 2015;241:109–123. doi: 10.1007/s00425-014-2167-2. [DOI] [PubMed] [Google Scholar]
- 60.Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:499. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao Z., Liu H.L., Daxinger L., Pontes O., He X., Qian W., Lin H., Xie M., Lorkovic Z.J., Zhang S., et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Law J.A., Ausin I., Johnson L.M., Vashisht A.A., Zhu J.K., Wohlschlegel J.A., Jacobsen S.E. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr. Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lister R., O’Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y. DNA methylation mediated by a microRNA pathway. Mol. Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Han X., Yin H., Song X., Zhang Y., Liu M., Sang J., Jiang J., Li J., Zhuo R. Integration of small RNAs, degradome and transcriptome sequencing in hyperaccumulator Sedum alfredii uncovers a complex regulatory network and provides insights into cadmium phytoremediation. Plant Biotechnol. J. 2016 doi: 10.1111/pbi.12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.miRBase 20.0. [(accessed on 20 July 2015)]. Available online: ftp://mirbase.org/pub/mirbase/CURRENT/
- 67.Rice Genomes. [(accessed on 15 July 2015)]. Available online: ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Osativa.
- 68.RNAfold Software. [(accessed on 10 August 2015)]. Available online: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi.
- 69.Cer R.Z., Herrera-Galeano J.E., Anderson J.J., Bishop-Lilly K.A., Mokashi V.P. miRNA Temporal Analyzer (miRNATA): A bioinformatics tool for identifying differentially expressed microRNAs in temporal studies using normal quantile transformation. Gigascience. 2014;3:20. doi: 10.1186/2047-217X-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fahlgren N., Carrington J.C. miRNA target prediction in plants. Methods Mol. Biol. 2010;592:51–57. doi: 10.1007/978-1-60327-005-2_4. [DOI] [PubMed] [Google Scholar]
- 71.AgriGO. [(accessed on 25 July 2015)]. Available online: http://bioinfo.cau.edu.cn/agriGO/
- 72.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.