Abstract
Ants cause a series of accidents involving humans. Such accidents generate different reactions in the body, ranging from a mild irritation at the bite site to anaphylactic shock, and these reactions depend on the mechanism of action of the venom. The study of animal venom is a science known as venomics. Through venomics, the composition of the venom of several ant species has already been characterized and their biological activities described. Thus, the aim of this study was to evaluate the protein composition and biological activities (hemolytic and immunostimulatory) of the venom of Neoponera villosa (N. villosa), an ant widely distributed in South America. The protein composition was evaluated by proteomic techniques, such as two-dimensional electrophoresis. To assess the biological activity, hemolysis assay was carried out and cytokines were quantified after exposure of macrophages to the venom. The venom of N. villosa has a profile composed of 145 proteins, including structural and metabolic components (e.g., tubulin and ATPase), allergenic and immunomodulatory proteins (arginine kinase and heat shock proteins (HSPs)), protective proteins of venom (superoxide dismutase (SOD) and catalase) and tissue degradation proteins (hyaluronidase and phospholipase A2). The venom was able to induce hemolysis in human erythrocytes and also induced release of both pro-inflammatory cytokines, as the anti-inflammatory cytokine release by murine macrophages. These results allow better understanding of the composition and complexity of N. villosa venom in the human body, as well as the possible mechanisms of action after the bite.
Keywords: ant, proteome, venomics, hemolysis, immunostimulatory proteins
1. Introduction
Accidents involving venomous animals, especially insects belonging to the order Hymenoptera, generate a variety of clinical and histopathological manifestations that range from mild irritation at the site of the sting to a severe anaphylactic reaction, often fatal [1,2].
Studies focused on venom, which examine the venom composition and mechanisms of action, have increased and, through these, several bioactive molecules have been isolated and/or identified, often of protein character [3]. We highlight studies on venoms of spiders, snakes, bees, wasps, ants, scorpions, centipedes, and frogs [4,5,6,7,8,9,10,11]. These studies have shown that the bioactivity of venom is related mainly to the release of cytokines as well as inflammatory (e.g., nitric oxide) and lipid (e.g., prostaglandins) mediators [12]. Yi et al. [13] showed that the ant venom from Solenopsis invicta acts on the nitric oxide synthase enzyme of murine macrophages, interfering in the production and release of this mediator. Similar effects were demonstrated in macrophages exposed to the venom from other animals such as scorpions, wasps, bees and spiders [14,15,16,17]. In addition, some venom presents hemolytic and/or cytotoxic activity leading to death, usually by apoptosis in human cells [16,18,19].
In the Neotropical Neoponera genus (Formicidae, Ponerinae), 57 ant species are described [20]. Ants of this genus possess a painful bite, level 2 on a pain scale ranging from 1 to 4 [21] that is attributed to the presence of cyclic dipeptides, as already described for the Neoponera apicalis venom [22]. Although the species of the Ponerinae are very close, several effects inherent to the action of the venom are reported. In insects, the venom initially causes a rapid, reversible, and dose-dependent paralysis, followed by an irreversible paralysis with subsequent permanent paralysis and death. The concentrations that lead to such manifestations vary from 38.7 to 799.2 μg/g, depending on the species of Ponerinae (which includes Neoponera) [23]. The main function of the venom of the genus of these ants is to capture preys and defend the colony, which is the reason why it has a paralytic effect [24].
Protein characterization and biological activity studies have become common for ant venom and, although there are some in this subject focused on the venom from other species of Ponerinae, such as Neoponera goeldii, Brachyponera. chinensis, and Brachyponera. sennaarensis [23,25,26], there are no other studies related to the Neoponera villosa venom, an ant that is found worldwide. In America, it can be found from the southwestern United States of America to southern Brazil [27], and is widely distributed throughout the Brazilian territory and is very common in the state of Bahia [28,29].
Thus, the aim of this study was to evaluate the protein composition and identify the hemolytic activity, cytotoxicity, and cytokine production induced by the N. villosa venom, collected in the south of the state of Bahia, Brazil.
2. Results
2.1. Doses of Protein
The proteins present in venom were quantified using the 2D Quant Kit (GE Healthcare, Little Chalfont, UK) and showed 6.23 µg/µL. By the Bradford method [30], a protein concentration of 2.79 µg/µL was found.
2.2. Protein Profile Obtained by SDS-PAGE
Figure 1 shows the venom protein profile after SDS-PAGE.
Figure 1.

Protein profile of Neoponera villosa (N. villosa) venom by SDS-PAGE. M: molecular weight marker (values are displayed on the left, in kDa); Nv: venom of N. villosa.
The highest density of protein, shown by the largest number of bands, was observed between 20.1 and 97 kDa, revealing a predominance of higher molecular weight proteins.
2.3. Protein Profile Obtained by Two-Dimensional Electrophoresis (2D)
The 2D gels (Figure 2) demonstrated a protein profile composed of 145 spots, from which 139 were identified (Figure 3 and Table 1).
Figure 2.
Two-dimensional gels of Neoponera villosa venom: (A) Replica 1 (Reference gel); (B) Replica 2; and (C) Replica 3.
Figure 3.
Spots corresponding to proteins identified in two-dimensional gel of Neoponera villosa venom.
Table 1.
Identified proteins in Neoponera villosa venom. MW: Molecular weight.
| Spot | Protein/Source | Accession Code | Mw (Da) | pI | Coverage/Score |
|---|---|---|---|---|---|
| 1 | Myosin heavy chain, muscle-like isoform 2/Bombus terrestris (bee) | gi|340718034 | 251,060 | 5.58 | 15%/1509 |
| 2 | Endoplasmin/Harpegnathos saltator (ant) | gi|307192149 | 90,928 | 4.96 | 22%/907 |
| 3 | Transitional endoplasmic reticulum ATPase TER94/Camponotus floridanus (ant) | gi|307174120 | 89,511 | 5.14 | 37%/1213 |
| 4 | Hemocyte protein-glutamine gamma-glutamyltransferase/Harpegnathos saltator (ant) | gi|307215415 | 79,590 | 4.89 | 36%/1291 |
| 5 | Heat shock protein HSP 90-α/Camponotus floridanus (ant) | gi|307186382 | 83,640 | 4.98 | 40%/1410 |
| 6 | Heat shock 70 kDa protein cognate 3/Harpegnathos saltator (ant) | gi|307210158 | 72,646 | 5.02 | 44%/1709 |
| 7 | Transferrin/Harpegnathos saltator (ant) | gi|307215135 | 82,014 | 5.72 | 15%/617 |
| 8 | Transferrin/Harpegnathos saltator (ant) | gi|307215135 | 82,014 | 5.72 | 17%/718 |
| 9 | Vacuolar ATP synthase catalytic subunit A/Harpegnathos saltator (ant) | gi|307196686 | 68,591 | 5.25 | 60%/2338 |
| 10 | Heat shock protein cognate 4 (70 kDa)/Apis mellifera (bee) | gi|229892210 | 71,383 | 5.43 | 50%/2204 |
| 11 | Calreticulin/Harpegnathos saltator (ant) | gi|307191961 | 47,000 | 4.42 | 38%/810 |
| 12 | Tubulin β-1 chain/Harpegnathos saltator (ant) | gi|307201205 | 49,010 | 4.67 | 40%/818 |
| 13 | 60 kDa heat shock protein, mitochondrial/Harpegnathos saltator (ant) | gi|307199045 | 64,756 | 5.32 | 35%/1012 |
| 14 | Tubulin α-1 chain-like/Nasonia vitripennis (wasp) | gi|156548149 | 50,647 | 5.01 | 56%/1598 |
| 15 | Tubulin α-1 chain/Harpegnathos saltator (ant) | gi|307208702 | 52,080 | 5.00 | 51%/1453 |
| 16 | Putative α-tubulin/Maconellicoccus hirsutus (mealybug) | gi|121544009 | 50,633 | 5.01 | 25%/451 |
| 17 | Phosphoglucomutase/Acromyrmex echinatior (ant) | gi|332024861 | 66,241 | 6.20 | 12%/287 |
| 18 | Protein disulfide-isomerase A3/Harpegnathos saltator (ant) | gi|307194521 | 56,272 | 5.53 | 11%/260 |
| 19 | Vacuolar ATP synthase subunit B/Camponotus floridanus (ant) | gi|307174076 | 55,263 | 5.37 | 58%/1628 |
| 20 | Eukaryotic initiation factor 4A-II/Camponotus floridanus (ant) | gi|307189936 | 48,514 | 5.30 | 47%/1002 |
| 21 | ATP synthase subunit β, mitochondrial/Harpegnathos saltator (ant) | gi|307195440 | 55,223 | 5.32 | 64%/2075 |
| 22 | Tubulin β-1 chain/Apis mellifera (bee) | gi|48095525 | 50,599 | 4.75 | 65%/1193 |
| 23 | α-N-acetylgalactosaminidase/Harpegnathos saltator (ant) | gi|307213390 | 161,528 | 5.99 | 6%/368 |
| 24 | cAMP-dependent protein kinase type I regulatory subunit isoform 1/Apis mellifera (bee) | gi|48106841 | 41,923 | 4.86 | 34%/611 |
| 27 | Lysosomal aspartic protease/Acromyrmex echinatior (ant) | gi|332024025 | 42,132 | 5.34 | 13%/254 |
| 28 | Actin-5, muscle-specific/Harpegnathos saltator (ant) | gi|307197034 | 42,098 | 5.30 | 68%/1145 |
| 29 | Actin, muscle/Camponotus floridanus (ant) | gi|307166491 | 42,071 | 5.29 | 71%/1252 |
| 30 | Actin, muscle/Camponotus floridanus (ant) | gi|307166491 | 42,071 | 5.29 | 62%/955 |
| 31 | Actin/Aedes aegypti (mosquito) | gi|94468486 | 42,164 | 5.30 | 40%/484 |
| 32 | Rab GDP dissociation inhibitor β/Camponotus floridanus (ant) | gi|307172388 | 50,104 | 5.51 | 36%/757 |
| 33 | Arginine kinase/Acromyrmex echinatior (ant) | gi|332018357 | 40,032 | 5.86 | 18%/313 |
| 34 | Mitochondrial-processing peptidase subunit β/Harpegnathos saltator (ant) | gi|307207091 | 53,918 | 5.77 | 28%/572 |
| 35 | Enolase/Harpegnathos saltator (ant) | gi|307211488 | 47,379 | 5.79 | 40%/650 |
| 36 | Succinyl-CoA ligase (ADP-forming) subunit β, mitochondrial-like/Bombus terrestris (bee) | gi|340726331 | 49,012 | 6.63 | 31%/861 |
| 37 | Arginine kinase/Harpegnathos saltator (ant) | gi|307197996 | 39,996 | 5.75 | 45%/1201 |
| 38 | RecName: Full = Arginine kinase; Short = AK/Homarus gammarus (lobster) | gi|585342 | 40,300 | 6.05 | 4%/88 |
| 39 | Retinal dehydrogenase 1/Acromyrmex echinatior (ant) | gi|332024132 | 53,459 | 6.14 | 9%/283 |
| 40 | Retinal dehydrogenase 1/Acromyrmex echinatior (ant) | gi|332024132 | 53,459 | 6.14 | 9%/333 |
| 41 | α-Tubulin at 84B/Drosophila melanogaster (fly) | gi|17136564 | 50,561 | 5.00 | 25%/402 |
| 42 | Retinal dehydrogenase 1/Acromyrmex echinatior (ant) | gi|332024132 | 53,459 | 6.14 | 9%/333 |
| 43 | Aminoacylase-1/Harpegnathos saltator (ant) | gi|307206409 | 45,783 | 5.66 | 16%/319 |
| 44 | Adenosylhomocysteinase/Harpegnathos saltator (ant) | gi|307206413 | 48,440 | 5.88 | 30%/737 |
| 45 | Enolase/Harpegnathos saltator (ant) | gi|307211488 | 47,379 | 5.79 | 44%/778 |
| 46 | Enolase/Harpegnathos saltator (ant) | gi|307211488 | 47,379 | 5.79 | 16%/307 |
| 47 | Phosphoglycerate kinase/Camponotus floridanus (ant) | gi|307177429 | 45,039 | 6.16 | 35%/1031 |
| 48 | Phosphoglycerate kinase/Camponotus floridanus (ant) | gi|307177429 | 45,039 | 6.16 | 23%/416 |
| 49 | Arginine kinase/Harpegnathos saltator (ant) | gi|307197996 | 39,996 | 5.75 | 47%/1381 |
| 50 | Arginine kinase/Harpegnathos saltator (ant) | gi|307197996 | 39,996 | 5.75 | 41%/1045 |
| 51 | Arginine kinase/Harpegnathos saltator (ant) | gi|307197996 | 39,996 | 5.75 | 45%/637 |
| 52 | Arginine kinase/Harpegnathos saltator (ant) | gi|307197996 | 39,996 | 5.75 | 39%/652 |
| 53 | Arginine kinase/Acromyrmex echinatior (ant) | gi|332018357 | 40,032 | 5.86 | 10%/211 |
| 54 | Malate dehydrogenase, cytoplasmic/Camponotus floridanus (ant) | gi|307166391 | 39,169 | 7.03 | 20%/533 |
| 55 | Glycerol-3-phosphate dehydrogenase (NAD+), cytoplasmic/Acromyrmex echinatior (ant) | gi|332024225 | 37,294 | 7.15 | 44%/1106 |
| 56 | Glycerol-3-phosphate dehydrogenase (NAD+), cytoplasmic/Acromyrmex echinatior (ant) | gi|332024225 | 37,294 | 7.15 | 25%/290 |
| 57 | Superoxide dismutase (Cu-Zn)/Harpegnathos saltator (ant) | gi|307204104 | 14,008 | 6.18 | 43%/213 |
| 58 | Glycogen phosphorylase/Harpegnathos saltator (ant) | gi|307199215 | 121,697 | 6.22 | 17%/837 |
| 59 | Glycogen phosphorylase/Harpegnathos saltator (ant) | gi|307199215 | 121,697 | 6.22 | 20%/1060 |
| 60 | Cytoplasmic aconitate hydratase/Harpegnathos saltator (ant) | gi|307196718 | 98,222 | 6.11 | 11%/319 |
| 61 | 10-formyltetrahydrofolate dehydrogenase/Acromyrmex echinatior (ant) | gi|332029989 | 99,956 | 5.88 | 4%/139 |
| 62 | Elongation factor 2/Camponotus floridanus (ant) | gi|307170298 | 94,303 | 6.11 | 16%/496 |
| 63 | 2-oxoglutarate dehydrogenase E1 component, mitochondrial/Acromyrmex echinatior (ant) | gi|332017156 | 121,347 | 6.78 | 13%/541 |
| 64 | 2-oxoglutarate dehydrogenase E1 component, mitochondrial/Acromyrmex echinatior (ant) | gi|332017156 | 121,347 | 6.78 | 25%/1252 |
| 65 | 2-oxoglutarate dehydrogenase E1 component, mitochondrial/Acromyrmex echinatior (ant) | gi|332017156 | 121,347 | 6.78 | 26%/1262 |
| 66 | Filamin-C/Harpegnathos saltator (ant) | gi|307210403 | 252,199 | 6.12 | 14%/1364 |
| 67 | Filamin-C/Harpegnathos saltator (ant) | gi|307210403 | 252,199 | 6.12 | 17%/1704 |
| 68 | Filamin-C/Harpegnathos saltator (ant) | gi|307210403 | 252,199 | 6.12 | 18%/2261 |
| 69 | Filamin-C/Harpegnathos saltator (ant) | gi|307210403 | 252,199 | 6.12 | 18%/1008 |
| 70 | Aconitate hydratase, mitochondrial/Harpegnathos saltator (ant) | gi|307201595 | 85028 | 8.36 | 25%/839 |
| 71 | Aconitate hydratase, mitochondrial/Harpegnathos saltator (ant) | gi|307201595 | 85028 | 8.36 | 36%/1333 |
| 72 | Succinate dehydrogenase (ubiquinone) flavoprotein subunit, mitochondrial/Acromyrmex echinatior (ant) | gi|332019677 | 73506 | 6.75 | 27%/886 |
| 73 | Putative actin-interacting protein 1/Harpegnathos saltator (ant) | gi|307210939 | 69910 | 5.69 | 36%/1180 |
| 74 | Putative actin-interacting protein 1/Harpegnathos saltator (ant) | gi|307210939 | 69910 | 5.69 | 29%/708 |
| 75 | Glycerol-3-phosphate dehydrogenase, mitochondrial/Harpegnathos saltator (ant) | gi|307212068 | 81972 | 6.51 | 16%/530 |
| 76 | Glycerol-3-phosphate dehydrogenase, mitochondrial/Harpegnathos saltator (ant) | gi|307212068 | 81972 | 6.51 | 13%/292 |
| 77 | Apolipophorins/Harpegnathos saltator (ant) | gi|307201472 | 382,345 | 6.47 | 1%/400 |
| 78 | NADP-dependent malic enzyme/Harpegnathos saltator (ant) | gi|307205633 | 66,782 | 7.19 | 29%/763 |
| 79 | NADP-dependent malic enzyme/Harpegnathos saltator (ant) | gi|307205633 | 66,782 | 7.19 | 13%/207 |
| 80 | NADP-dependent malic enzyme/Harpegnathos saltator (ant) | gi|307205633 | 66,782 | 7.19 | 49%/1238 |
| 81 | Dihydrolipoyl dehydrogenase, mitochondrial/Harpegnathos saltator (ant) | gi|307209020 | 54,499 | 6.87 | 7%/117 |
| 82 | NADP-dependent malic enzyme/Harpegnathos saltator (ant) | gi|307205633 | 66,782 | 7.19 | 54%/1447 |
| 83 | NADP-dependent malic enzyme/Harpegnathos saltator (ant) | gi|307205633 | 66,782 | 7.19 | 12%/195 |
| 84 | Transaldolase/Harpegnathos saltator (ant) | gi|307215256 | 37,393 | 5.27 | 7%/102 |
| 85 | Pyruvate dehydrogenase E1 component subunit β, mitochondrial/Harpegnathos saltator (ant) | gi|307195718 | 39,486 | 5.87 | 34%/475 |
| 86 | Proteasome subunit β type-7/Harpegnathos saltator (ant) | gi|307192825 | 30,622 | 7.00 | 28%/250 |
| 87 | Peroxiredoxin 1/Camponotus floridanus (ant) | gi|307175821 | 21,887 | 6.30 | 32%/423 |
| 88 | Proteasome subunit β type-1/Harpegnathos saltator (ant) | gi|307214019 | 26,343 | 6.81 | 20%/108 |
| 89 | Dihydrolipoyl dehydrogenase, mitochondrial/Harpegnathos saltator (ant) | gi|307209020 | 54,499 | 6.87 | 23%/491 |
| 90 | Glutamate dehydrogenase, mitochondrial/Harpegnathos saltator (ant) | gi|307195623 | 62,095 | 8.40 | 27%/549 |
| 91 | Selenium-binding protein 1-A/Acromyrmex echinatior (ant) | gi|332021867 | 52,719 | 6.99 | 8%/233 |
| 92 | Glutamate dehydrogenase, mitochondrial/Harpegnathos saltator (ant) | gi|307195623 | 62,095 | 8.40 | 52%/1140 |
| 93 | Glutamate dehydrogenase, mitochondrial/Harpegnathos saltator (ant) | gi|307195623 | 62,095 | 8.40 | 53%/1166 |
| 94 | Glutamate dehydrogenase, mitochondrial/Harpegnathos saltator (ant) | gi|307195623 | 62,095 | 8.40 | 52%/1356 |
| 95 | Catalase/Harpegnathos saltator (ant) | gi|307197480 | 58,234 | 8.58 | 23%/170 |
| 96 | Elongation factor-1 α, partial/Hoplitis ochraceicornis (bee) | gi|426322152 | 40,701 | 8.58 | 41%/226 |
| 97 | Alanine aminotransferase 2/Harpegnathos saltator (ant) | gi|307214462 | 61,520 | 9.26 | 24%/665 |
| 98 | ATP synthase subunit α, mitochondrial/Harpegnathos saltator (ant) | gi|307208992 | 59,687 | 9.12 | 41%/1271 |
| 99 | Elongation factor 1-α/Harpegnathos saltator (ant) | gi|307196337 | 50,620 | 9.13 | 43%/350 |
| 100 | ATP synthase subunit α, mitochondrial/Harpegnathos saltator (ant) | gi|307208992 | 59,687 | 9.12 | 32%/364 |
| 101 | 4-hydroxybutyrate coenzyme A transferase/Harpegnathos saltator (ant) | gi|307203837 | 53,231 | 7.69 | 36%/769 |
| 102 | Probable fumarate hydratase, mitochondrial/Harpegnathos saltator (ant) | gi|307202128 | 47,153 | 8.14 | 24%/478 |
| 103 | Probable citrate synthase 1, mitochondrial/Harpegnathos saltator (ant) | gi|307202019 | 49,316 | 8.94 | 34%/788 |
| 104 | Probable citrate synthase 1, mitochondrial/Harpegnathos saltator (ant) | gi|307202019 | 49,316 | 8.94 | 37%/471 |
| 105 | Isocitrate dehydrogenase (NADP) cytoplasmic-like isoform 1/Bombus terrestris (bee) | gi|340721268 | 46,518 | 8.00 | 9%/146 |
| 106 | Isocitrate dehydrogenase (NADP) cytoplasmic/Harpegnathos saltator (ant) | gi|307210166 | 50,629 | 8.20 | 44%/952 |
| 107 | Isocitrate dehydrogenase (NADP) cytoplasmic/Harpegnathos saltator (ant) | gi|307210166 | 50,629 | 8.20 | 41%/611 |
| 108 | Isocitrate dehydrogenase (NADP) cytoplasmic/Harpegnathos saltator (ant) | gi|307210166 | 50,629 | 8.20 | 19%/422 |
| 109 | Arginine kinase /Harpegnathos saltator (ant) | gi|307197996 | 39,996 | 5.75 | 46%/1213 |
| 110 | Fructose-bisphosphate aldolase-like/Megachile rotundata (bee) | gi|383861513 | 40,009 | 7.00 | 18%/470 |
| 111 | Four and a half LIM domains protein 2/Acromyrmex echinatior (ant) | gi|332021158 | 28,335 | 8.46 | 52%/758 |
| 112 | Isocitrate dehydrogenase (NAD) subunit β, mitochondrial/Harpegnathos saltator (ant) | gi|307210578 | 41,512 | 8.81 | 31%/530 |
| 113 | Sorbitol dehydrogenase/Harpegnathos saltator (ant) | gi|307204829 | 38,256 | 7.56 | 27%/488 |
| 114 | Fructose-bisphosphate aldolase-like/Megachile rotundata (bee) | gi|383861513 | 40,009 | 7.00 | 33%/635 |
| 115 | Aldose reductase/Camponotus floridanus (ant) | gi|307181859 | 35,897 | 6.11 | 17%/212 |
| 116 | Glyceraldehyde-3-phosphate dehydrogenase 2/Harpegnathos saltator (ant) | gi|307198667 | 35,769 | 7.64 | 45%/898 |
| 117 | Glyceraldehyde-3-phosphate dehydrogenase 2/Harpegnathos saltator (ant) | gi|307198667 | 35,769 | 7.64 | 45%/848 |
| 118 | Glyceraldehyde-3-phosphate dehydrogenase 2/Harpegnathos saltator (ant) | gi|307198667 | 35,769 | 7.64 | 42%/603 |
| 119 | Hyaluronoglucosaminidase/Camponotus floridanus (ant) | gi|307180582 | 40,178 | 8.45 | 10%/216 |
| 120 | Succinyl-CoA ligase (GDP-forming) subunit α, mitochondrial/Harpegnathos saltator (ant) | gi|307203732 | 34,525 | 8.70 | 23%/476 |
| 121 | Guanine nucleotide-binding protein subunit β-like protein/Harpegnathos saltator (ant) | gi|307215409 | 36,333 | 7.63 | 68%/913 |
| 122 | Malate dehydrogenase, mitochondrial/Harpegnathos saltator (ant) | gi|307214026 | 36,178 | 9.36 | 46%/670 |
| 123 | Phospholipase A2 isozymes PA3A/PA3B/PA5/Apis mellifera (bee) | gi|328778177 | 26,379 | 7.46 | 6%/103 |
| 124 | Proteasome subunit α type-7-1/Harpegnathos saltator (ant) | gi|307201582 | 28,118 | 6.97 | 32%/385 |
| 125 | Dehydrogenase/reductase SDR family member 11/Harpegnathos saltator (ant) | gi|307212910 | 17,798 | 9.61 | 9%/102 |
| 128 | Phosphoglycerate mutase 2-like/Bombus terrestris (bee) | gi|340726229 | 35,071 | 9.16 | 36%/733 |
| 129 | Glutathione S-transferase sigma 3/Locusta migratoria (locust) | gi|329564873 | 23,498 | 7.63 | 8%/94 |
| 130 | Adenylate kinase isoenzyme F38B2.4/Harpegnathos saltator (ant) | gi|307204436 | 21,291 | 5.42 | 35%/525 |
| 131 | Protein DJ-1/Camponotus floridanus (ant) | gi|307174129 | 20,023 | 5.93 | 7%/110 |
| 132 | RecName: Full = Snake venom metalloproteinase atroxlysin-1; Short = SVMP; AltName: Full = Atroxlysin-I/Bothrops atrox (snake) | gi|353526296 | 23,302 | 5.85 | 12%/135 |
| 134 | Transaldolase/Harpegnathos saltator (ant) | gi|307215256 | 37,393 | 5.27 | 7%/168 |
| 135 | Peroxiredoxin-6/Harpegnathos saltator (ant) | gi|307213913 | 25,248 | 6.65 | 18%/124 |
| 136 | Triosephosphate isomerase/Harpegnathos saltator (ant) | gi|307199049 | 26,956 | 6.00 | 49%/695 |
| 137 | Eukaryotic translation initiation factor 3 subunit I/Acromyrmex echinatior (ant) | gi|332024413 | 36,384 | 6.00 | 25%/210 |
| 138 | Glycine N-methyltransferase/Harpegnathos saltator (ant) | gi|307192516 | 33,820 | 6.26 | 19%/259 |
| 139 | Fumarylacetoacetate hydrolase domain-containing protein 2A/Harpegnathos saltator (ant) | gi|307203489 | 37,311 | 8.54 | 8%/109 |
| 140 | Ribose-phosphate pyrophosphokinase 1/Harpegnathos saltator (ant) | gi|307207320 | 37,856 | 6.26 | 23%/320 |
| 142 | 14-3-3 protein epsilon-like/Nasonia vitripennis (wasp) | gi|156553657 | 29,323 | 4.72 | 4%/123 |
| 143 | Putative 14-3-3 protein/Maconellicoccus hirsutus (mealybug) | gi|121543925 | 28,171 | 4.80 | 9%/83 |
| 144 | 14-3-3 protein ζ/Camponotus floridanus (ant) | gi|307186410 | 35,115 | 5.01 | 11%/149 |
| 145 | Glutathione S-transferase 1, isoform C/Harpegnathos saltator (ant) | gi|307196173 | 24,899 | 7.71 | 18%/175 |
2.4. Hemolytic Activity Induced by Neoponera villosa Venom
Hemolysis was observed up to the fourth largest concentration of venom tested (63 µg/µL). Figure 4 presents the hemolysis rate of the venom in different concentrations. We can see that the hemolytic effect was dose-dependent, and was 50.6%, 40%, 26%, 12.1%, and 2.5% for the concentrations of 500, 250, 125, 63, and 31 µg/mL, respectively. Below these concentrations, it was not possible to observe hemolysis.
Figure 4.
Percentage of hemolysis in different concentrations of Neoponera villosa venom.
2.5. Induction of Cytokines in J774 Macrophages in the Presence of Neoponera villosa Venom
The presence of the venom in cultured murine macrophages was able to stimulate pro- and anti-inflammatory cytokines.
The production of interleukin 6 (IL-6) by macrophages presented significantly higher levels in the presence of venom, especially after 24 h of stimulation. The difference in production was statistically significant at almost all the concentrations of venom used when compared with the negative control (with the exception of the highest and lowest concentrations studied in 2 h). However, within 24 h, the production levels of IL-6 were always lower than the value induced by lipopolysaccharide (LPS) (Figure 5).
Figure 5.
Amount of IL-6 (in ng/mL) released after the stimulus with Neoponera villosa venom. Values expressed in mean + standard deviation. Control: macrophages with supplemented culture medium alone. LPS: macrophages stimulated with 2 µg/mL LPS. * Denotes significantly different (p < 0.05).
There was a larger release of interleukin 12 (IL-12) after 24 h of stimulation, which was statistically significant only at the highest concentrations of venom (500 to 62.5 µg/mL). Although the levels of IL-12 were discreet under 2-h stimulation, they were comparable to the amount found under LPS stimulus (Figure 6).
Figure 6.
Amount of IL-12 (in ng/mL) released after the stimulus with Neoponera villosa venom Values expressed in mean + standard deviation. Control: macrophages with supplemented culture medium alone. LPS: macrophages stimulated with 2 µg/mL LPS. * Denotes significantly different (p < 0.05).
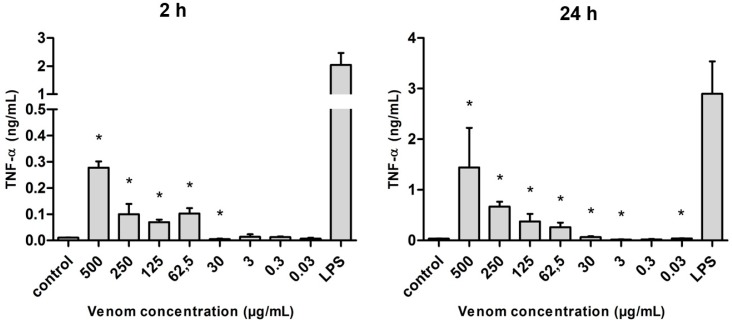
Tumor necrosis factor α (TNF-α) presented the highest levels of production when compared with the other pro-inflammatory cytokines evaluated (IL-6 and IL-12). Regardless of the time of cultivation (2 or 24 h) and when compared with the basal condition, macrophages produced significantly greater amounts of TNF-α under stimulus of the Neoponera villosa venom. The production of this cytokine occurred in a dose-dependent way, as the concentration of the venom decreased, the amount of TNF-α detected also decreased. However, the highest levels of this cytokine under venom stimulus were still lower than those found under the LPS stimulus (Figure 7).
Figure 7.
Amount of TNF-α (in ng/mL) released after the stimulus with Neoponera villosa venom. Values expressed in mean + standard deviation. Control: macrophages with supplemented culture medium alone. LPS: macrophages stimulated with 2 µg/mL LPS. * Denotes significantly different (p < 0.05).
Regarding interleukin 10 (IL-10), the venom was able to stimulate the production of this cytokine at all concentrations used, not demonstrating any relation to the dose of venom used on the cultures. The difference, statistically significant when compared with the control, was observed at all the concentrations used (with the exception of 125 and 62.5 µg/mL). In addition, the levels of IL-10 produced after 24 h of stimulation were slightly higher when compared with the 2-h stimulus, but they were not similar to those observed under LPS stimulus (Figure 8).
Figure 8.
Amount of IL-10 (in ng/mL) released after the stimulus with Neoponera villosa venom. Values expressed in mean + standard deviation. Control: macrophages with supplemented culture medium alone. LPS: macrophages stimulated with 2 µg/mL LPS. * Denotes significantly different (p < 0.05).
3. Discussion
The protein profile of the N. villosa venom showed predominance of proteins with higher molecular weight (between 20.1 and 97 kDa). Similarly, Lee et al. [25] worked with Brachyponera chinensis venom and observed higher protein density between 12 and 85 kDa. Silva et al. [31] analyzed Odontomachus bauri venom, and isolated proteins with molecular weight ranging from 18 to 160 kDa under nonreducing conditions, with more intense staining for bands above 29 kDa. Under reducing conditions, the electrophoretic profile changed, showing a wider range, from 24 to 160 kDa. The profile obtained by two-dimensional electrophoresis was composed of 145 spots. The distribution of these spots showed a higher density in neutral and alkaline areas and molecular weight above 20.1 kDa. To date, there are no data in the literature about protein profile of N. villosa venom; our results are the first ones. However, similar data were demonstrated in other species of the same genus. Costa Manso et al. [32] described, in the 2D profile of N. goeldii venom, higher density of spots between 10 and 66 kDa, and Lee et al. [25], studied B. chinensis venom and reported prevalence of proteins with pI in the neutral and alkaline zones. For the venom from another species of ant, Solenopsis invicta, all the proteins identified in 2D-SDS-PAGE were located above 14.4 kDa and most presented pI in the alkaline lane [33]. Such features are common to the venom of many hymenoptera as well as to ants, bees and wasps [34].
Regarding the 139 proteins identified in this study, it was observed that the venom houses proteins of several functional classes, from structural (e.g., actin), metabolic proteins (e.g., glyceraldehyde 3-phosphate-dehydrogenase), immunostimulatory molecules (e.g., heat shock proteins (HSPs)) to well described allergens (e.g., arginine kinase). Of these, the most biologically and biochemically relevant were: myosin (spot 1), endoplasmin (spot 2), HSPs (spots 5, 6, 10, 13), tubulin (spots 12, 14, 15, 16, 22, 41), α-N-acetylgalactosaminidase (spot 23), aspartyl protease (spot 27), actin (spots 28, 29, 30 and 31), arginine kinase (spots 33, 37, 38, 49, 50, 51, 52, 53, and 109), superoxide dismutase (spot 57), catalase (spot 95), hyaluronoglucosaminidase (spot 119), phospholipase A2 (spot 123), metalloproteinase (spot 132) and peroxiredoxin (spots 87 and 137).
In spots 2, 5, 6, 10, and 13, the following heat shock proteins (HSPs) or stress proteins were identified: HSP 90 (spots 2 and 5), HSP 70 (spots 6 and 10), and HSP 60 (spot 13). These proteins act as chaperones, facilitating the folding of other proteins, and as regulators of signaling, protecting the cell from the event of apoptosis [35]. In addition, the activation of the immune system mediated by HSPs is known. Preparations of purified HSPs, including HSP 60, HSP 70, and HSP 90, from various sources (wild and recombinant bacteria, several mammals, and humans) are potent activators of the innate immune system. These preparations induce the production of pro-inflammatory cytokines, such as TNF-α, IL-1, IL-6, and IL-12, in addition to nitric oxide by monocytes, macrophages and dendritic cells. [36,37]. The presence of such proteins in the venom may be related to the increased release of IL-12 and TNF-α by J774 1.6 macrophages, since, as demonstrated by Asea et al. [38], monocytes derived from human peripheral blood showed high levels of TNF-α, IL-1, and IL-6 release after the exposure to human HSP 70. Similarly, murine peritoneal macrophages and monocytes derived from human peripheral blood showed high expression of mRNA encoder of TNF-α, IL-1, IL-6, and GM-CSF after exposure to HSPs purified from several microorganisms such as Escherichia coli, Mycobacterium tuberculosis, and Legionella pneumophila [39,40].
In spot 23, α-N-acetylgalactosaminidase was identified, which is a hydrolase described for animals and bacteria [41]. It has already been described that this enzyme promotes immunosuppression through the inactivation of macrophages [42].
From spot 27, it was possible to identify an aspartyl protease, an enzyme with cathepsin D activity [43], capable of degrading a variety of substrates, including laminin and fibronectin, constituents of the extracellular matrix, thus contributing to the spread of other venom constituents through the body in which it is inoculated [44].
Several arginine kinase isoforms were identified (spots 33, 37, 38, 49, 50, 51, 52, 53, and 109). This protein has an enzymatic function and is responsible for the energetic metabolism in invertebrates. It is considered a pan-allergen, since it can trigger allergic responses through specific IgE linking [45]. It can be found in many allergenic sources, such as venoms, and has already been isolated from the venom of bees and wasps [35,46], as well as non-venomous animals used as food such as shrimp, octopus, and crab [47,48,49].
In spot 57, a superoxide dismutase (SOD) was identified as well as a catalase in spot 95. Both are responsible for the protecting the venom from the reactive oxygen species produced by the body, such as O2 and H2O2 [46]. In addition, a form of copper/zinc-dependent SOD was identified with the potential to trigger immune responses activation processes in the body where the venom is inoculated [50].
In spot 119, a hyaluronoglucosaminidase (or hyaluronidase) was identified, an enzyme present in several venoms and found in the venom of snakes, scorpions, bees, and wasps [51]. This enzyme acts on the cellular interstitial, downgrading it and is therefore, known as spreading factor [52]. It maximizes the infiltration of the venom as it digests part of the extracellular matrix, facilitating the action of other components [53].
A phospholipase A2 was identified from spot 123. This enzyme, widely distributed in several venoms, destroys phospholipids in biological membranes, leading to the formation of pores, cell lysis and tissue damage [33], inducing inflammation [54], and inhibiting the activity of macrophages [55]. The presence of the enzyme in the venom can be related to hemolytic activity and to inhibition of NO formation by macrophages.
Spot 132 has been identified as a metalloproteinase that is part of a family of enzymes involved in the occurrence of muscle necrosis, tissue damage, swelling and bleeding at the site of the sting, rare coagulopathies, among other related inflammatory reactions [56,57]. In addition, studies with venom from snakes have shown that the metalloproteinases, similar to hyaluronidases, act on the destruction of the extracellular matrix by interfering in cell adhesion, thus providing tissue disorganization [58].
In spots 87 and 137, forms of peroxiredoxin, belonging to a ubiquitous family of antioxidant proteins were identified [59]. Its presence in the venom is related to its own protection from oxidative stress. This function is also performed by glutathione S-transferase protein, found in spots 129 and 145 [33].
Structural proteins were identified in spots 1 (myosin), 12, 14, 15, 16, 22, 41 (tubulin) and 28, 29, 30, and 31 (actin). These proteins originate from muscle or tissues surrounding the venom reservoir. Studies by other authors suggest that the ant cells can rupture when the sting apparatus is removed from the insect with the aid of tweezers, which can cause the addition of these cell proteins to the venom proteins [46].
Although the remaining spots identified in the venom are classified exclusively as intracellular constituents (cytoplasmic or present inside organelles) and their presence in the venom is also related to the lesion of cells surrounding the venom reservoir during the mechanical removal of the glands [46], many of these proteins have been identified in proteomes of several venoms. In Apis mellifera venom, forms of malate dehydrogenase (spots 54 and 122), enolase (spots 35, 45, and 46) and apolipophorine (spot 77) were found [60]. In the venom of the endoparasitoid Pteromalus puparum, forms of vacuolar ATP synthase were found (spots 9, 19, and 21) and glyceraldehyde-3-phosphate dehydrogenase (spots 116, 117, and 118), among others [45]. Furthermore, Torres et al. [61] used transcriptomic approaches, and showed that more than 50% of the genes transcribed in the venom glands of the Dinoponera quadriceps ant were intracellular constituents.
Our study using a proteomic approach showed that N. villosa share many venom proteins with others ant species and even with other hymenopteran venoms. Similar proteomic tools were used by Touchard et al. [62] and they demonstrated that MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight) mass profiles of venom peptides from different species of Neoponera, Pachycondyla and Odontomachus permit taxonomic discrimination in closely related species. This chemotaxonomic tool is important in exploiting the enormous biodiversity of ants and other hymenopteran venoms as sources for novel drugs or biopesticides.
Neoponera villosa venom presented hemolytic activity which was dose-dependent, and the percentage of hemolysis was 50.6%, 40%, 26%, 12.1%, and 2.5% for concentrations of 500, 250, 125, 63, and 31 µg/mL, respectively. Below these concentrations, it was not possible to observe hemolysis. It has been shown that venoms from many organisms present hemolytic activity (in crude or fractionated form), among these organisms are: jellyfish, spiders, scorpions, and snakes [63,64,65,66].
Venoms from various species of hymenoptera also present hemolytic activity, such as Pogonomyrmex barbatus (P. barbatus) [18], Paraponera clavata (P. clavata) [21], Myrmecia pilosula (M. pilosula) [67], Ectatomma tuberculatum [68] ants, and wasps of the genus Vespa and Polistes [69]. The minimum concentrations of hemolysis vary and, within the same genus, there may be a great discrepancy. For example, it takes 500 U/mg dried venom from P. barbatus to provide 50% hemolysis [18], but it only takes 10 U/mg dried venom from P. clavata to obtain the same value of hemolysis [21]. Values close to those of the present study were found by Matuszek et al. [67], who took a concentration of 65 µg/mL venom from the M. pilosula ant to generate 12.5% hemolysis. The Polistes lanio lanio wasp requires a concentration equal to or greater than 50 µg/mL to obtain minimal hemolysis. Hemolysis may be the result of one or more mechanisms of action, and is usually linked to enzyme action. There may be direct hemolysis, in which one or more venom components act on the erythrocytic membrane, yielding pores and leading them to lysis, for example, there is the action of phospholipases that cleave the phospholipids of the membrane forming channels, where the intracellular content pours through [70] and proteins/peptides with surfactant activity [64]. Indirect hemolysis can be mediated by the action of complement, in which venom components, such as metalloproteinases, facilitate the action of cascade-signaling proteins [71].
Regarding N. villosa venom, a synergistic effect is suggested between the phospholipase and the metalloproteinase activities in the venom (spots 123 and 132). As these components are at low concentration in the venom, higher doses are required to obtain hemolysis (in this case, over 31 µg/mL).
The dosage of cytokines (IL-6, IL-10, IL-12, and TNF-α) released in the culture supernatant by macrophages exposed to the venom was performed by ELISA technique and, despite the significant increase in all pro-inflammatory cytokines (IL-6, IL-12, and TNF-α) in 2 and 24 h, there was also an increase of IL-10, a cytokine of anti-inflammatory and immunomodulatory character in both the periods evaluated. The induction pattern of pro-inflammatory cytokine has been observed for venom of several hymenoptera. Lam et al. [72] demonstrated that A. mellifera venom presents pro-inflammatory properties, inducing, in vitro, the release of cytokine IL-1 by macrophages through the activation of the p38 MAPK pathway. Tambourgi et al. [73] showed that, in vivo, the venom from the Loxosceles intermedia spider, is able to increase the IL-6 and TNF-α serum levels. Similar effect was observed by Malaque et al. [74], who stimulated primary culture human keratinocytes with venom from the Loxosceles gaucho spider and noted an increase in TNF-α. The IL-6 cytokine, as well as some chemokines, are also related to inflammation at the site of the sting induced by the venom from the same spider, as presented by Barbaro et al. [75]. The venom from the Tityus serrulatus scorpion generates local inflammation with high serum levels of IL-6 and TNF-α [12]. The same was observed, in vitro, when macrophages were exposed to the venom from the same species of scorpion and showed an increase in the release of IL-6 and TNF-α [76]. This pro-inflammatory pattern of the venom can be explained by the presence of some proteins known to induce macrophages to produce and release cytokines of this character, such as the heat shock proteins (HSPs), phospholipases, and metalloproteinases identified in this study. However, we also observed the production of IL-10, a cytokine that is anti-inflammatory and immunomodulatory in character able to inhibit the secretion of pro-inflammatory cytokines and regulate the differentiation and proliferation of some cells [77]. Some studies with venom also demonstrate increased IL-10 after stimulation. Fialho et al. [12] showed that in addition to the pro-inflammatory cytokines IL-6 and TNF-α, the venom from the scorpion T. serrulatus also induced an increase in serum IL-10. Andrade et al. [78] demonstrated that the stimulus with venom from the same scorpion raises the increased mRNA expression of both pro- (IL-1, IL-6, IFN-γ, and TNF-α) and anti-inflammatory (IL-10) cytokines. Ribeiro et al. [79] noted a similar fact and showed that after stimulating peripheral blood mononuclear cells with the venom from the snake Crotalus durissus collilineatus, there was an increase in the release of IL-10 and TNF-α as well. This fact could be related to the presence of components that stimulate both pro- and anti-inflammatory response in cells stimulated by the venom. Petricevich et al. [80] suggest that the production of IL-10 has a protective character in the process of poisoning by venom from snakes of the Bothrops genus, preventing the systemic shock condition.
4. Materials and Methods
4.1. Venom Extraction
The ants were collected while foraging on the campus of the State University of Santa Cruz (UESC) and on the Executive Committee of the Cocoa Crop (CEPLAC), in Ilhéus, Bahia, Brazil (14°47′20″S and 39°02′58″W). The insects were killed by freezing at −20 °C, identified in stereoscopic microscope and stored at −20 °C until use. We used 350 individuals (250 for protein characterization and 100 for biological activity tests). The crude venom, obtained by breaking the bag attached to the stinger after the ants’ dissection, was diluted in phosphate buffered saline (PBS) 100 mM pH 7.8.
4.2. Protein Extraction
To precipitate the proteins from the crude venom, the same amount of 20% (v/v) trichloroacetic acid (TCA) in acetone was added to each microtube containing venom. The microtube was packaged at −20 °C for 60 min and centrifuged at 16,100× g for 45 min to form the pellet. The supernatant was discarded. The pellet was washed with 500 µL cold acetone and then sonicated on ice for 21 s in cycles of 7 s with 70% range, and centrifuged for 10 min at 16,100× g, forming the pellet again. The supernatant was discarded and the pellet washed four times with acetone. The acetone was evaporated at room temperature and then rehydration buffer was added (7 M urea, 2 M thiourea, 2% CHAPS, and 0.002% bromophenol blue) to solubilize the proteins.
4.3. Protein Quantification
A 2D Quant Kit (GE Healthcare) was used to quantify proteins dissolved in the rehydration buffer, according to the manufacturer’s recommendations.
For the tests of biological activity (venom dissolved in PBS), dosage was used according to the Bradford protocol [30], using bovine serum albumin as standard.
4.4. Electrophoresis (SDS-PAGE)
For electrophoresis, 20 µg protein were subjected to SDS-PAGE (12.5% acrylamide). Aliquots of 7 µL of standard molecular weight (Amersham, Little Chalfont, UK) were also submitted to electrophoresis for comparison. After running, the gels were fixed (40% ethanol and 10% acetic acid) for 10 min, and then the solution was replaced by Coomassie Blue G-50 (Sigma Aldrich, St. Louis, MO, USA) dye, which was maintained for 12 h while stirring. Then, the dye was removed and the gels bleached with distilled water.
4.5. Two-Dimensional Electrophoresis (2D-SDS-PAGE)
Gel strips (Immobiline Dry Strip) of 13 cm pH 3-10 NL (GE Healthcare) were rehydrated for 12 h in rehydration buffer containing 350 µg protein. They were focused using a multiple-stage protocol (1 h at 500 V, 1 h at 1000 V and 6 h at 8000 V). Before the second dimension, each strip was balanced for 15 min in buffer containing 50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue, supplemented with 10 mg/mL DTT. After this stage, strips were balanced for 15 min in the buffer described above, but the DTT was replaced by 25 mg/mL of iodoacetamide. The second dimension electrophoresis was performed at 150 V using polyacrylamide gels (12.5%) in triplicate.
4.6. Protein Visualization and Image Analysis
Polyacrylamide gels were fixed (40% ethanol and 10% acetic acid) for 30 min, and then stained with Coomassie Blue G50 (Sigma) for five days while stirring. Subsequently, the gels were bleached with distilled water. Images of the gels were scanned and analyzed by ImageMaster 2D Platinum 7.0 (GE Healthcare) software to assess the number of spots, and characterize the values of pI and molecular weight.
4.7. Spot Excision and Triptych Digestion
The spots were excised from the gels and arranged separately in microtubes. Later, they were added to the gel fragments 200 µL NH4HCO3 (ammonium bicarbonate), 25 mM, pH 8.0 in 50% acetonitrile to discolor and remove SDS. This solution was kept in the microtubes for 24 h. After this period, the spots were washed with 200 µL ultrapure water, 100 µL acetonitrile were added to dehydrate the gel, and the fragments were incubated for 5 min at room temperature. The excess acetonitrile was removed, and the microtubes were vacuum dried for 20 min. Then, the proteins were partially digested with trypsin (5 µL at the concentration of 20 ng/µL) for 10 min at 4 °C. Later, 200 µL NH4HCO3, 25 mM were added, followed by incubation at 37 °C for 24 h to complete the digestion of the peptides. To extract the peptides, the remaining solution was transferred to a differente and clean microtube, and 50 µL 50% acetonitrile were added to the first microtube containing 0.1% formic acid. These microtubes containing a mixture of acetonitrile, formic acid, and residual peptides were agitated slightly for 30 min. The solution was pipetted to the new microtubes, and this process was repeated. In the end, the microtubes containing fully digested peptides were vacuum dried to the volume of 5–10 µL.
4.8. Peptide Sequencing and Protein Identification
The peptides obtained as described in the previous item were separately applied in the nanoAcquity UPLC (Waters, Milford, MA, USA) in two C18 columns (“trapping” of 5 µm, 180 µm × 20 mm; and the second of 1.7 µm, 100 µm × 100 mm), under a stream of 0.6 µL/min in a 50-min run. The peptides were separated according to acetonitrile gradient grade, transferred to the Mass Spectrometer (Micromass Q-TOFmicro, Waters) and ionized in a capillary under 3000 V. They were fragmented in the positive mode with selection of minimum relative intensity of 10 counts, and the three most intense ions were analyzed per every 1 s sweep, with collision energy ranging from 20 to 95 V according to the mass/charge (m/z) of peptides. The spectra obtained in each run were analyzed by MASCOT Server Online (Matrix Science: http://www.matrixscience.com/), and compared with the NCBI database. On hydrolysis, by trypsin, the possible loss of a cleavage site was considered. The tolerance of the peptide masses was ±0.3 Da, and the tolerance of the fragment masses was ±0.1 Da.
4.9. Hemolytic Activity
To perform the test, a 96-well microplate with a U-shaped bottom was used. The initial venom concentration was 500 µg/mL and was performed a serial dilution to the concentration of 0.2 µg/mL venom. Then, 100 µL red blood cell suspension was added to 1% (v/v). For the reference value of zero, 100 µL saline solution and 100 µL red blood cell suspension were added. For the reference value of 100% hemolysis, 100 µL saline solution containing 1% (v/v) Triton X-100 (Sigma) and 100 µL red blood cell suspension were added. The plates were incubated at 25 °C for 120 min. Then, the microplates were centrifuged for 5 min at 1440× g. The supernatant of each well was collected and transferred to the wells of a new plate with a flat bottom. The rate of hemolysis was taken by the absorbance reading at 540 nm and the hemolysis was calculated by the following equation:
4.10. Murine Macrophages Culture and Stimulus with Neoponera villosa Venom
J774 1.6 murine macrophages (Rio de Janeiro Cell Bank—BCRJ, access No. 0273) were cultured for 24 h until they reached the confluence amid DMEM (Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% antibiotics (penicillin and streptomycin) (Gibco), at 37 °C, 5% CO2 in the concentration of 1 × 106 cells/mL, and the viability was monitored with trypan blue dye at each bouncing.
For the tests with venom stimulation, the macrophages were cultured for 48 h at the concentration of 1 × 106 cells/mL in 24-well plates containing DMEM, 10% FBS and 1% antibiotics as well as different concentrations of venom (ranging from 500 to 0.03 μg/mL). As positive stimulus control the cells were grown in the presence 2 μg/mL of bacterial lipopolysaccharide (LPS) (Sigma) and as negative control (basal condition) only with the culture supplemented with FBS and antibiotic.
4.11. Cytokines Doses
Five hundred milliliter aliquots from culture supernatants described in item 4.10 were removed, in periods of 2 and 24 h after the stimulus. Cytokines IL-6, IL-10, IL-12, and TNF-α were evaluated by ELISA kits (Peprotech, Ribeirão Preto, São Paulo, Brazil), according to the manufacturer’s recommendations.
The statistical analysis of cytokine concentration was carried out via the GraphPad Prism 5 software, using the Mann–Whitney non-parametric test, considering p < 0.05 and confidence interval (CI) of 95%.
5. Conclusions
The venom of Neoponera villosa has a complex protein composition, which consists of 145 proteins in the neutral and alkaline lanes of pH and above 20.1 kDa. In addition, N. villosa venom presents hemolytic activity for concentrations exceeding 31 µg/µL and this fact is related to the presence of phospholipase and metalloproteinase enzymes in the venom (spots 123 and 132, respectively). N. villosa venom induces an increase in the release of pro-inflammatory cytokines (IL-6, IL-8, and TNF-α), and this increase is related to the presence of the following proteins in the venom: HSP 90 (spots 2 and 5), HSP 70 (spots 6 and 10), and HSP 60 (spot 13), since it is known that HSPs are potent activators of the innate immune system, inducing the release of pro-inflammatory cytokines. In addition to the pro-inflammatory cytokines, there was slight increase in IL-10 production by macrophages, and this fact could be related to the presence of components that stimulate both a pre- and anti-inflammatory response in the venom or with a cellular compensatory mechanism aimed at preventing damages caused by the high concentration of pro-inflammatory cytokines. These results allow a better understanding of the composition and complexity of N. villosa venom in the human body, as well as the venom’s possible mechanisms of action after the bite.
Acknowledgments
Research supported by grants from National Council for Scientific and Technological Development (CNPq), Coordination of Improvement of Higher Education Personnel (CAPES) and Research Foundation of the State of Bahia (Fapesb).
Abbreviations
| 2D | Two-dimensional |
| BCRJ | Rio de Janeiro Cell Bank |
| CEPLAC | Executive Committee of the Cocoa Crop |
| CHAPS | 3-((3-Cholamidopropyl)dimethylammonio)-1-propanesulfonate |
| CI | Confidence interval |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DTT | Dithiothreitol |
| ELISA | Enzyme linked immuno sorbent assay |
| FBS | Fetal bovine serum |
| HSP | Heat shock proteins |
| IL | Interleukin |
| LPS | Lipopolysaccharide |
| MALDI-TOF | Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight |
| MAPK | Mitogen-activated protein kinase |
| MW | Molecular weight |
| NCBI | National Center for Biotechnology Information |
| NO | Nitric oxide |
| PAGE | Polyacrylamide gel electrophoresis |
| PBS | Phosphate buffered saline |
| pI | Isoelectric point |
| SOD | Superoxide dismutase |
| SDS | Sodium dodecyl sulfate |
| TCA | Trichloroacetic acid |
| TNF | Tumor necrosis factor |
| UESC | State University of Santa Cruz |
| UPLC | Ultra performance liquid chromatography |
Author Contributions
Wallace Felipe Blohem Pessoa performed the experiments, analyzed data and prepared the manuscript; Ludimilla Carvalho Cerqueira Silva and Leila de Oliveira Dias helped in 2D electrophoresis; Jacques Hubert Charles Delabie identified the ants and contributed the reagents and materials; and Helena Costa and Carla Cristina Romano conceived and designed the experiments. All the authors have contributed with the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Steen C.J., Janniger C.K., Schutzer S.E., Schwartz R.A. Insect sting reactions to bees, wasps, and ant. Int. J. Dermatol. 2005;44:91–94. doi: 10.1111/j.1365-4632.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald K.T., Flood A.A. Hymenoptera Stings. Clin. Tech. Small Anim. Pract. 2006;21:194–204. doi: 10.1053/j.ctsap.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Escoubas P., King G.F. Venomics as a drug discovery platform. Expert Rev. Proteom. 2009;6:221–224. doi: 10.1586/epr.09.45. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn-Nentwig L., Stöcklin R., Nentwig W. Venom composition and strategies in spiders: Is everything possible? Adv. Insect Physiol. 2011;40:1–86. [Google Scholar]
- 5.Georgieva D., Seifert J., Öhler M., von Bergen M., Spencer P., Arni R.K., Genov N., Betzel C. Pseudechis australis venomics: Adaptation for a defense against microbial pathogens and recruitment of body transferrin. J. Proteome Res. 2011;10:2440–2464. doi: 10.1021/pr101248e. [DOI] [PubMed] [Google Scholar]
- 6.De Graaf D.C., Aerts M., Danneels E., Devreese B. Bee, wasp and ant venomics pave the way for a component-resolved diagnosis of sting allergy. J. Proteom. 2009;72:145–154. doi: 10.1016/j.jprot.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Santos L.D., Pieroni M., Menegasso A.R.S., Pinto J.R.A.S., Palma M.S. A new scenario of bioprospecting of Hymenoptera venoms through proteomic approach. J. Venom. Anim. Toxins Incl. Trop. Dis. 2011;17:364–377. [Google Scholar]
- 8.Hoffman D.R., Sakell R.H., Schmidt M. Sol i 1, the phospholipase allergen of imported fire ant venom. J. Allergy Clin. Immunol. 2005;115:611–616. doi: 10.1016/j.jaci.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz E.F., Camargos T.S., Zamudio F.Z., Silva L.P., Bloch C., Jr., Caixeta F., Schwartz C.A., Possani L.D. Mass spectrometry analysis, amino acid sequence and biological activity of venom components from the Brazilian scorpion Opisthacanthus cayaporum. Toxicon. 2008;15:1499–1508. doi: 10.1016/j.toxicon.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 10.Rates B., Bemquerer M.P., Richardson M., Borges M.H., Morales R.A., de Lima M.E., Pimenta A.M. Venomic analyses of Scolopendra viridicornis nigra and Scolopendra angulata (Centipede, Scolopendromorpha): Shedding light on venoms from a neglected group. Toxicon. 2007;49:810–826. doi: 10.1016/j.toxicon.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Rates B., Silva L.P., Ireno I.C., Leite F.S., Borges M.H., Bloch C., Jr., de Lima M.E., Pimenta A.M. Peptidomic dissection of the skin secretion of Phasmahyla jandaia (Bokermann and Sazima, 1978) (Anura, Hylidae, Phyllomedusinae) Toxicon. 2011;57:35–52. doi: 10.1016/j.toxicon.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Fialho E.M., Maciel M.C., Silva A.C., Reis A.S., Assunção A.K., Fortes T.S., Silva L.A., Guerra R.N., Kwasniewski F.H., Nascimento F.R. Immune cells recruitment and activation by Tityus serrulatus scorpion venom. Toxicon. 2011;58:480–485. doi: 10.1016/j.toxicon.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Yi G.B., McClendon D., Desaiah D., Goddard J., Lister A., Moffitt J., Meer R.K., de Shazo R., Lee K.S., Rockhold R.W. Fire ant venom alkaloid, isosolenopsin A, a potent and selective inhibitor of neuronal nitric oxide synthase. Int. J. Toxicol. 2003;22:81–86. doi: 10.1080/10915810305090. [DOI] [PubMed] [Google Scholar]
- 14.Petricevich V.L., Hernández Cruz A., Coronas F.I., Possani L.D. Toxin gamma from Tityus serrulatus scorpion venom plays an essential role in immunomodulation of macrophages. Toxicon. 2007;50:666–675. doi: 10.1016/j.toxicon.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Ponvert C., le Courvoisier C., Weill B., Bloch E., Paupe J., Scheinmann P. Kinetics of plasma cytokine levels in children hyposensitized with wasp venom. Cytokine. 2001;15:229–231. doi: 10.1006/cyto.2001.0926. [DOI] [PubMed] [Google Scholar]
- 16.Jang H.S., Kim S.K., Han J.B., Ahn H.J., Bae H., Min B.I. Effects of bee venom on the pro-inflammatory responses in RAW264.7 macrophage cell line. J. Ethnopharmacol. 2005;99:157–160. doi: 10.1016/j.jep.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 17.Zanchet E.M., Longo I., Cury Y. Involvement of spinal neurokinins, excitatory amino acids, proinflammatory cytokines, nitric oxide and prostanoids in pain facilitation induced by Phoneutria nigriventer spider venom. Brain Res. 2004;1021:101–111. doi: 10.1016/j.brainres.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 18.Bernheimer A.W., Avigad L.S., Schmidt J.O. A hemolytic polypeptide from the venom of the red harvester ant, Pogonomyrmex barbatus. Toxicon. 1980;18:271–278. doi: 10.1016/0041-0101(80)90005-7. [DOI] [PubMed] [Google Scholar]
- 19.De Souza A.L., Malaque C.M., Sztajnbok J., Romano C.C., Duarte A.J., Seguro A.C. Loxosceles venom-induced cytokine activation, haemolysis, and acute kidney injury. Toxicon. 2008;51:151–156. doi: 10.1016/j.toxicon.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt C.A., Shattuck S.O. The higher classification of the ant subfamily ponerinae (Hymenoptera: Formicidae), with a review of ponerine ecology and behavior. Zootaxa. 2014;3817:1–242. doi: 10.11646/zootaxa.3817.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Schimidt J.O., Blum M., Overal W. Hemolytic activities of stinging insect venoms. Arch. Insect Biochem. Physiol. 1984;1:155–160. doi: 10.1002/arch.940010205. [DOI] [Google Scholar]
- 22.Lopez L.C., Morgan E.D. Explanation of bitter taste of venom of ponerine ant, Pachycondyla apicalis. J. Chem. Ecol. 1997;23:705–712. doi: 10.1023/B:JOEC.0000006405.26872.ef. [DOI] [Google Scholar]
- 23.Orivel J., Dejean A. Comparative effect of the venoms of ants of the genus Pachycondyla (Hymenoptera: Ponerinae) Toxicon. 2001;39:195–201. doi: 10.1016/S0041-0101(00)00113-6. [DOI] [PubMed] [Google Scholar]
- 24.Mill A.E. Predation by the ponerine ant Pachycondyla commutata on termites of the genus Syntermes in Amazonian rain forest. J. Nat. Hist. 1982;18:405–410. doi: 10.1080/00222938400770341. [DOI] [Google Scholar]
- 25.Lee E.K., Jeong K.Y., Lyu D.P., Lee Y.W., Sohn J.H., Lim K.J., Hong C.S., Park J.W. Characterization of the major allergens of Pachycondyla chinensis in ant sting anaphylaxis patients. Clin. Exp. Allergy. 2009;39:602–607. doi: 10.1111/j.1365-2222.2008.03181.x. [DOI] [PubMed] [Google Scholar]
- 26.Dkhil M.A., Abdel-Baki A.S., Al-Quraishi S., Al-Khalifa M. Anti-inflammatory activity of the venom from samsum ants Pachycondyla sennaarensis. Afr. J. Pharmacol. 2010;4:115–118. [Google Scholar]
- 27.Mackay W.P., Mackay E.E. The Systematics and Biology of the New World Ants of the Genus Pachycondyla (Hymenoptera: Formicidae) 1st ed. The Edwin Mellen Press; Lewiston, NY, USA: 2010. pp. 571–579. [Google Scholar]
- 28.Lucas C., Fresneau D., Kolmer K., Heinze J., Delabie J.H.C., Pho D.B. A multidisciplinary approach to discriminating different taxa in the species complex Pachycondyla villosa (Formicidae) Biol. J. Linn. Soc. 2002;65:249–259. doi: 10.1111/j.1095-8312.2002.tb01425.x. [DOI] [Google Scholar]
- 29.Mariano C.S.F., Delabie J.H.C., Santos J.R.M., Pompolo S.G. Karyotype evolution in Neotropical Pachycondyla spp. (Ponerinae) Biológico. 2007;69:409–412. [Google Scholar]
- 30.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Silva M.F., Mota C.M., Miranda V.S., Cunha A.O., Silva M.C., Naves K.S.C., Oliveira F., Silva D.A.O., Mineo T.W.P., Santiago F.M. Biological and enzymatic characterization of proteases from crude venom of the ant Odontomachus bauri. Toxins. 2015;7:5114–5128. doi: 10.3390/toxins7124869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa Manso E., Croce M., Pinto J.R., Souza Santos K., Delazari Santos L., Baptista Dias N., Palma M.S. Anaphylaxis due to Pachycondyla goeldii ant: A case report. J. Investig. Allergol. Clin. Immunol. 2010;20:352–353. [PubMed] [Google Scholar]
- 33.Dos Santos Pinto J.R., Fox E.G., Saidemberg D.M., Santos L.D., da Silva Menegasso A.R., Costa-Manso E., Machado E.A., Bueno O.C., Palma M.S. Proteomic view of the venom from the fire ant Solenopsis invicta Buren. J. Proteome Res. 2012;11:4643–4653. doi: 10.1021/pr300451g. [DOI] [PubMed] [Google Scholar]
- 34.Leluk J., Schmidt J., Jones D. Comparative studies on the protein composition of hymenopteran venom reservoirs. Toxicon. 1989;27:105–114. doi: 10.1016/0041-0101(89)90410-8. [DOI] [PubMed] [Google Scholar]
- 35.Peiren N., de Graaf D.C., Vanrobaeys F., Danneels E.L., Devreese B., Van Beeumen J., Jacobs F.J. Proteomic analysis of the honey bee worker venom gland focusing on the mechanisms of protection against tissue damage. Toxicon. 2008;52:72–83. doi: 10.1016/j.toxicon.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Tsan M.F., Gao B. Cytokine function of heat shock proteins. Am. J. Physiol. Cell Physiol. 2004;286:739–744. doi: 10.1152/ajpcell.00364.2003. [DOI] [PubMed] [Google Scholar]
- 37.Tsan M.F., Gao B. Heat shock protein and innate immunity. Cell. Mol. Immunol. 2004;1:274–279. [PubMed] [Google Scholar]
- 38.Asea A., Kraeft S.K., Kurt-Jones E.A., Stevenson M.A., Chen L.B., Finberg R.W., Koo G.C., Calderwood S.K. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat. Med. 2000;6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- 39.Retzlaff C., Yamamoto Y., Hoffman P.S., Friedman H., Klein T.W. Bacterial heat shock proteins directly induce cytokine mRNA and interleukin-1 secretion in macrophage cultures. Infect. Immun. 1994;62:5689–5693. doi: 10.1128/iai.62.12.5689-5693.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galdiero M., de l’Ero G.C., Marcatili A. Cytokine and adhesion molecule expression in human monocytes and endothelial cells stimulated with bacterial heat shock proteins. Infect. Immun. 1997;65:699–707. doi: 10.1128/iai.65.2.699-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu A., Wang Z.K., Beavis R. Structural studies of α-N-acetylgalactosaminidase: Effect of glycosylation on the level of expression, secretion efficiency, and enzyme activity. Arch. Biochem. Biophys. 1998;352:1–8. doi: 10.1006/abbi.1998.0575. [DOI] [PubMed] [Google Scholar]
- 42.Mohamad S.B., Nagasawa H., Uto Y., Hori H. Tumor cell α-N-acetylgalactosaminidase activity and its involvement in GcMAF-related macrophage activation. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002;132:1–8. doi: 10.1016/S1095-6433(01)00522-0. [DOI] [PubMed] [Google Scholar]
- 43.Cho W., Dhadialla T.S., Raikhel A.S. Purification and characterization of a lysosomal aspartic protease with cathepsin D activity from the mosquito. Insect Biochem. 1991;21:165–176. doi: 10.1016/0020-1790(91)90047-I. [DOI] [Google Scholar]
- 44.Lkhider M., Castino R., Bouguyon E., Isidoro C., Ollivier-Bousquet M. Cathepsin D released by lactating rat mammary epithelial cells is involved in prolactin cleavage under physiological conditions. J. Cell Sci. 2004;117:5155–5164. doi: 10.1242/jcs.01396. [DOI] [PubMed] [Google Scholar]
- 45.Zhu J.Y., Fang Q., Wang L., Hu C., Ye G.Y. Proteomic analysis of the venom from the endoparasitoid wasp Pteromalus puparum (Hymenoptera: Pteromalidae) Arch. Insect Biochem. Physiol. 2010;75:28–44. doi: 10.1002/arch.20380. [DOI] [PubMed] [Google Scholar]
- 46.Dos Santos L.D., Santos K.S., Pinto J.R., Dias N.B., de Souza B.M., dos Santos M.F., Perales J., Domont G.B., Castro F.M., Kalil J.E., et al. Profiling the proteome of the venom from the social wasp Polybia paulista: A clue to understand the envenoming mechanism. J. Proteome Res. 2010;9:3867–3877. doi: 10.1021/pr1000829. [DOI] [PubMed] [Google Scholar]
- 47.García-Orozco K.D., Aispuro-Hernández E., Yepiz-Plascencia G., Calderón-de-la-Barca A.M., Sotelo-Mundo R.R. Molecular characterization of arginine kinase, an allergen from the shrimp Litopenaeus vannamei. Int. Arch. Allergy Immunol. 2007;144:23–28. doi: 10.1159/000102610. [DOI] [PubMed] [Google Scholar]
- 48.Shen H.W., Cao M.J., Cai Q.F., Ruan M.M., Mao H.Y., Su W.J., Liu G.M. Purification, cloning, and immunological characterization of arginine kinase, a novel allergen of Octopus fangsiao. J. Agric. Food Chem. 2012;60:2190–2199. doi: 10.1021/jf203779w. [DOI] [PubMed] [Google Scholar]
- 49.Rosmilah M., Shahnaz M., Zailatul H.M., Noormalin A., Normilah I. Identification of tropomyosin and arginine kinase as major allergens of Portunus pelagicus (blue swimming crab) Trop. Biomed. 2012;29:467–478. [PubMed] [Google Scholar]
- 50.Guarneri F., Guarneri C., Guarneri B., Benvenga S. In silico identification of potential new latex allergens. Clin. Exp. Allergy. 2006;36:916–919. doi: 10.1111/j.1365-2222.2006.02516.x. [DOI] [PubMed] [Google Scholar]
- 51.Marković-Housley Z., Miglierini G., Soldatova L., Rizkallah P.J., Müller U., Schirmer T. Crystal structure of hyaluronidase, a major allergen of bee venom. Structure. 2000;8:1025–1035. doi: 10.1016/S0969-2126(00)00511-6. [DOI] [PubMed] [Google Scholar]
- 52.Gmachl M., Kreil G. Bee venom hyaluronidase is homologous to a membrane protein of mammalian sperm. Proc. Natl. Acad. Sci. USA. 1993;90:3569–3573. doi: 10.1073/pnas.90.8.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffman D.R. Hymenoptera venom allergens. Clin. Rev. Allergy Immunol. 2006;30:109–128. doi: 10.1385/CRIAI:30:2:109. [DOI] [PubMed] [Google Scholar]
- 54.Marques P.P., Esteves A., Lancellotti M., Ponce-Soto L.A., Marangoni S. Novel acidic phospholipase A2 from Porthidium hyoprora causes inflammation with mast cell rich infiltrate. Biochem. Biophys. Rep. 2015;1:78–84. doi: 10.1016/j.bbrep.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sampaio S.C., Rangel-Santos A.C., Peres C.M., Curi R., Cuty Y. Inhibitory effect of phospholipase A2 isolated from Crotalus durissus terrificus venom on macrophage function. Toxicon. 2011;45:671–676. doi: 10.1016/j.toxicon.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 56.Gutiérrez J.M., Rucavado A. Snake venom metalloproteinases: Their role in the pathogenesis of local tissue damage. Biochimie. 2000;82:841–850. doi: 10.1016/S0300-9084(00)01163-9. [DOI] [PubMed] [Google Scholar]
- 57.Koya S., Crenshaw D., Agarwal A. Rhabdomyolysis and acute renal failure after fire ant bites. J. Gen. Intern. Med. 2007;22:145–147. doi: 10.1007/s11606-006-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gutiérrez J.M., Rucavado A., Escalante T., Díaz C. Hemorrhage induced by snake venom metalloproteinases: Biochemical and biophysical mechanisms involved in microvessel damage. Toxicon. 2005;45:997–1011. doi: 10.1016/j.toxicon.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 59.Rhee S.G., Woo H.A., Kil I.S., Bae S.H. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 2012;287:4403–4410. doi: 10.1074/jbc.R111.283432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li R., Zhang L., Fang Y., Han B., Lu X., Zhou T., Feng M., Li J. Proteome and phosphoproteome analysis of honeybee (Apis mellifera) venom collected from electrical stimulation and manual extraction of the venom gland. BMC Genom. 2013;14 doi: 10.1186/1471-2164-14-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torres A.F.C., Huang C., Chong C.M., Leung S.W., Prieto-da-Silva A.R.B., Havt A., Quinet Y.P., Martins A.M.C., Lee S.M.Y., Rádis-Baptista G. Transcriptome analysis in venom gland of the predatory giant ant Dinoponera quadriceps: Insights into the polypeptide toxin arsenal of hymenopterans. PLoS ONE. 2014;9:513. doi: 10.1371/journal.pone.0087556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Touchard A., Dauvois M., Arguel M.J., Petitclerc F., Leblanc M., Dejean A., Orivel J., Nicholson G.M., Escoubas P. Elucidation of the unexplored biodiversity of ant venom peptidomes via MALDI-TOF mass spectrometry and its application for chemotaxonomy. J. Proteom. 2014;105:217–231. doi: 10.1016/j.jprot.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 63.Li R., Yu H., Xing R., Liu S., Qing Y., Li K., Li B., Meng X., Cui J., Li P. Isolation and in vitro partial characterization of hemolytic proteins from the nematocyst venom of the jellyfish Stomolophus meleagris. Toxicol. Vitro. 2013;27:1620–1625. doi: 10.1016/j.tiv.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 64.Chaves-Moreira D., Chaim O.M., Sade Y.B., Paludo K.S., Gremski L.H., Donatti L., de Moura J., Mangili O.C., Gremski W., da Silveira R.B., et al. Identification of a direct hemolytic effect dependent on the catalytic activity induced by phospholipase-D (dermonecrotic toxin) from brown spider venom. J. Cell. Biochem. 2009;107:655–666. doi: 10.1002/jcb.22148. [DOI] [PubMed] [Google Scholar]
- 65.Borchani L., Sassi A., Shahbazzadeh D., Strub J.M., Tounsi-Guetteti H., Boubaker M.S., Akbari A., van Dorsselaer A., El Ayeb M. Heminecrolysin, the first hemolytic dermonecrotic toxin purified from scorpion venom. Toxicon. 2011;58:130–139. doi: 10.1016/j.toxicon.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 66.Biardi J.E., Coss R.G. Rock squirrel (Spermophilus variegatus) blood sera affects proteolytic and hemolytic activities of rattlesnake venoms. Toxicon. 2011;57:323–331. doi: 10.1016/j.toxicon.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 67.Matuszek M.A., Hodgson W.C., Sutherland S.K., King R.G. Pharmacological studies of jumper ant (Myrmecia pilosula) venom: Evidence for the presence of histamine, and haemolytic and eicosanoid-releasing factors. Toxicon. 1992;30:1081–1091. doi: 10.1016/0041-0101(92)90053-8. [DOI] [PubMed] [Google Scholar]
- 68.Pluzhnikov K., Nosyreva E., Shevchenko L., Kokoz Y., Schmalz D., Hucho F., Grishin E. Analysis of ectatomin action on cell membranes. Eur. J. Biochem. 1999;262:501–506. doi: 10.1046/j.1432-1327.1999.00426.x. [DOI] [PubMed] [Google Scholar]
- 69.Bernheimer A.W., Avigad L.S., Schmidt J.O., Ishay J.S. Proteins in venoms of two wasps, Polistes comanchus navajoe and Wasp orientalis. Comp. Biochem. Physiol. C. 1982;71:203–207. doi: 10.1016/0306-4492(82)90037-5. [DOI] [PubMed] [Google Scholar]
- 70.Hink W.F., Pappas P.W., Jaworski D.C. Partial biochemical characterization of venom from the ant, Pseudomyrmex triplarinus. Toxicon. 1994;32:763–772. doi: 10.1016/0041-0101(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 71.Tambourgi D.V., Morgan B.P., de Andrade R.M., Magnoli F.C., van Den Berg C.W. Loxosceles intermedia spider envenomation induces activation of an endogenous metalloproteinase, resulting in cleavage of glycophorins from the erythrocyte surface and facilitating complement-mediated lysis. Blood. 2000;95:683–691. [PubMed] [Google Scholar]
- 72.Lam P.D., Mandal P.K., Hak S.Y., Hwang S.G. Study of the molecular mechanism of anti-inflammatory activity of bee venom in lipopolysaccharide stimulated RAW 264.7 macrophages. Trop. J. Pharm. Res. 2010;10:19–26. doi: 10.4314/tjpr.v9i1.52104. [DOI] [Google Scholar]
- 73.Tambourgi D.V., Petricevich V.L., Magnoli F.C., Assaf S.L., Jancar S., Dias Da Silva W. Endotoxemic-like shock induced by Loxosceles spider venoms: Pathological changes and putative cytokine mediators. Toxicon. 1998;36:391–403. doi: 10.1016/S0041-0101(97)00063-9. [DOI] [PubMed] [Google Scholar]
- 74.Malaque C.M.S., Ori M., Santos S.A., Andrade D.H. Production of TNF-α by primary cultures of human keratinocytes challenged with Loxosceles gaucho venom. Rev. Inst. Med. Trop. Sao Paulo. 1999;41:179–182. doi: 10.1590/S0036-46651999000300009. [DOI] [PubMed] [Google Scholar]
- 75.Barbaro K.C., Lira M.S., Araújo C.A., Pareja-Santos A., Távora B.C., Prezotto-Neto J.P., Kimura L.F., Lima C., Lopes-Ferreira M., Santoro M.L. Inflammatory mediators generated at the site of inoculation of Loxosceles gaucho spider venom. Toxicon. 2010;56:972–979. doi: 10.1016/j.toxicon.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 76.Zoccal K.F., Bitencourt C.S., Secatto A., Sorgi C.A., Bordon K.C., Sampaio S.V., Arantes E.C., Faccioli L.H. Tityus serrulatus venom and toxins Ts1, Ts2 and Ts6 induce macrophage activation and production of immune mediators. Toxicon. 2011;57:1101–1108. doi: 10.1016/j.toxicon.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 77.Moore K.W., O’Garra A., de Waal Malefyt R., Vieira P., Mosmann T.R. Interleukin-10. Annu. Rev. Immunol. 1993;11:165–190. doi: 10.1146/annurev.iy.11.040193.001121. [DOI] [PubMed] [Google Scholar]
- 78.Andrade M.V., Lisboa F.A., Portugal A.L., Arantes R.M., Cunha-Melo J.R. Scorpion venom increases mRNA expression of lung cytokines. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2007;146:581–587. doi: 10.1016/j.cbpa.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 79.Ribeiro C.B., Dos Santos J.C., Silva J.M., de Godoi P.H., Magalhães M.R., Spadafora-Ferreira M., Fonseca S.G., Pfrimer I.A. Crotalus durissus collilineatus venom induces TNF-α and IL-10 production in human peripheral blood mononuclear cells. ISRN Inflamm. 2014;2014 doi: 10.1155/2014/563628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petricevich V.L., Teixeira C.F., Tambourgi D.V., Gutiérrez J.M. Increments in serum cytokine and nitric oxide levels in mice injected with Bothrops asper and Bothrops jararaca snake venoms. Toxicon. 2000;38:1253–1266. doi: 10.1016/S0041-0101(99)00227-5. [DOI] [PubMed] [Google Scholar]