Abstract
The process of domestication can exert intense trait-targeted selection on genes and regulatory regions. Specifically, rapid shifts in the structure and sequence of genomic regulatory elements could provide an explanation for the extensive, and sometimes extreme, variation in phenotypic traits observed in domesticated species. Here, we explored methylation differences from >24,000 cytosines distributed across the genomes of the domesticated dog (Canis familiaris) and the gray wolf (C. lupus). PCA and model-based cluster analyses identified two primary groups, domestic versus wild canids. A scan for significantly differentially methylated sites (DMSs) revealed species-specific patterns at 68 sites after correcting for cell heterogeneity, with weak yet significant hyper-methylation typical of purebred dogs when compared to wolves (59% and 58%, p<0.05, respectively). Additionally, methylation patterns at eight genes significantly deviated from neutrality, with similar trends of hyper-methylation in purebred dogs. The majority (>66%) of differentially methylated regions contained or were associated with repetitive elements, indicative of a genotype-mediated trend. However, DMSs were also often linked to functionally relevant genes (e.g. neurotransmitters). Finally, we utilized known genealogical relationships among Yellowstone wolves to survey transmission stability of methylation marks, from which we found a substantial fraction that demonstrated high heritability (both H2 and h2>0.99). These analyses provide a unique epigenetic insight into the molecular consequences of recent selection and radiation of our most ancient domesticated companion, the dog. These findings suggest selection has acted on methylation patterns, providing a new genomic perspective on phenotypic diversification in domesticated species.
Keywords: Methylation, genome regulation, domestication, canid
Introduction
As a result of intense trait-targeted selection over a relatively small number of generations, the process of domestication may result in extreme phenotypic diversity (Darwin 1868; Clutton-Brock 1992; Clutton-Brock 1999; Diamond 2002). Recently, the molecular underpinnings of this diversification has been the subject of intense investigation (Ostrander & Wayne 2005; Dobney & Larson 2006; Wiener & Wilkinson 2011; Albert et al. 2012; Andersson 2013; Larson et al. 2014). Various approaches have been used to unravel the genetic changes that occur during domestication, most notably genome-wide SNP and haplotype analyses to map regions under selection and associate those regions with phenotypes (Andersson & Georges 2004; Doebley et al. 2006; Wiener & Wilkinson 2011; Wayne & vonHoldt 2012).
Changes in genomic regulatory elements or transcriptional machinery could explain a significant component of the dramatic evolution of phenotypic diversity in dogs and other domesticated species (Andersson & Georges 2004). Past studies are limited and have generally been transcriptome-based, focusing on differential expression analyses of brain tissues implicated in behavior and brain development (Saetre et al. 2004; Kukekova et al. 2011; Albert et al. 2012), as well as the impact of captivity on genes involved in the stress response (Kennerly et al. 2008). More recently, the use of epigenome-wide association studies (EWAS) have provided a new profile of the epigenomic changes that occur during domestication (Feeney et al. 2014).
Cytosine methylation serves as one mechanism for pre-transcriptional regulatory control and is involved in essential cellular processes, such as cell differentiation, X-inactivation, and genomic imprinting (Bernstein et al. 2007; Suzuki & Bird 2008; Richards et al. 2010). Methylated cytosines in 5’ promoter regions act as transcriptional suppressors by inhibiting the transcriptional complex from binding to the promoter sequence (Bird 2007). New sequencing approaches can now interrogate the methylation state of each cytosine, specifically cytosines in CpG islands (CGIs) comprising putatively regulatory elements and representing a potentially functional fraction of the mammalian genome (Saxonov et al. 2006). In mammals, these islands are often found in association with promoter elements in genes (Deaton & Bird 2011) and epigenetic modifications most frequently occur within the CG dinucleotide motif (Bernstein et al. 2007).
Here, we investigate the methylation variation in the domesticated dog and its wild predecessor, the gray wolf. The dog is considered the oldest domesticated species, providing a rich system for investigation of the evolutionary changes that occur during the domestication process (Sutter & Ostrander 2004; Boyko 2011; Shearman & Wilton 2011; Wayne & vonHoldt 2012; Freedman et al. 2014). Remarkably, the extreme genetic and phenotypic variation observed within dog breeds arose in a short time period, initiating 15,000 to 40,000 years ago (Sablin & Khlopachev 2002; Savolainen et al. 2002; Germonpré et al. 2009; Boyko et al. 2010; Vonholdt et al. 2010; Ovodov et al. 2011; Larson et al. 2012; Freedman et al. 2014; Skoglund et al. 2015). Most modern dog breeds, however, have emerged from controlled breeding practices of the last 200 years involving narrow founding, genetic isolation, and strong phenotypic selection according to breed standards (Parker et al. 2004; Vonholdt et al. 2010). This unique history has created a genetic architecture that is extremely effective for unraveling quantitative phenotypic traits such as body size (Parker et al. 2004; Wayne & Ostrander 2007).
We begin by comparing domestic dogs to the pedigreed population of gray wolves in Yellowstone National Park (YNP) (Vonholdt et al. 2008). Recent studies have revealed many aspects of the Yellowstone population, from their genealogical history and trait evolution (Vonholdt et al. 2008; Anderson et al. 2009), to disease epidemiology (Almberg et al. 2009), life history (MacNulty et al. 2009a; MacNulty et al. 2009b; MacNulty et al. 2011; Stahler et al. 2013; Cubaynes et al. 2014), and population viability (Coulson et al. 2011). We build upon the rich literature on domestic dogs (e.g. heritability estimates, linkage analyses, genome-wide association studies, candidate gene approaches) through the addition of the canine epigenome, an ideal tool to investigate the molecular signature of recent rapid radiation and intense selective pressure. In addition, the samples we analyze include a wide selection of modern and ancient dogs breeds, as well as village dogs, which are dogs whose breeding is not well controlled and represent a mix of modern and ancient lineages (Boyko et al. 2010). Finally, using the Yellowstone wolf genealogy, we estimate the heritability of each methylation mark (Bell et al. 2012).
Methods
Samples and reduced representation bisulfite sequencing
Genomic DNA (1μg) was extracted from whole blood from 50 domestic dogs from 19 recognized American Kennel Club (AKC) purebred dog breeds (further referred to as purebred dogs), two Boz breed dogs from the Turkish Caucasus, two aboriginal island breeds (dingoes, n=2; New Guinea Singing Dogs, ngsd, n=4), and three village dogs. Similarly, we included 35 gray wolves (C. lupus) from YNP (Supplemental Table S1). All DNA was prepared using QIAamp DNA mini kits following manufacturer's protocol (Qiagen, DNeasy blood and Tissue kit). Whole blood is both a reliable proxy for organism-level transcriptional activity (Liew et al. 2006; Kaminsky et al. 2009; Rakyan et al. 2011) and methylation marks are preserved and stable over time in frozen blood, unlike RNA collections (Ferrer et al. 2008; Lofton-Day et al. 2008; Van der Auwera et al. 2009; Wieczorek et al. 2009). DNA was quantified using a Qubit 2.0 Fluorometer and checked on a 2% agarose gel for degradation.
We used reduced representation bisulfite sequencing (RRBS) to explore methylation patterns at CGIs, a single-base resolution methylation analysis from the sequencing of bisulfite converted DNA of CpG-rich regions (Meissner et al. 2005; Saxonov et al. 2006). We constructed RRBS genomic libraries for sequencing on an Illumina HiSeq2500. High purity genomic DNA was digested with Msp1, ends were repaired with adenine on the 3’ ends followed by ligation of adapters with unique 6mer barcode sequence tags using the TruSeq Sample Preparation kit's standard protocol (Illumina Inc). Library purification and size selection were completed using Agencourt AMPure XP beads (Beckman Coulter). We selected fragments between 100-400bp in size then treated them with bisulfite using an EpiTect kit (Qiagen) for conversion of unmethylated cytosines, following the manufacturer's procedure for low DNA input (1 ng-2 μg) with a double incubation time of 10 hours to ensure a high conversion rate. Our converted DNA was then subjected to 12 PCR cycles of amplification for enrichment of adapter-ligated fragments with MyTaq Mix (Bioline Inc). Library quality was assessed with an Agilent 2100 BioAnalyzer (Agilent) and concentration estimated using a Qubit 2.0 Fluorometer. Libraries were then standardized to 10nM, and then pooled on average four samples per lane for sequencing (minimum 2, maximum 6). To avoid generating redundant methylation information, single-end reads were chosen over paired-end reads for sequencing. Single-end DNA sequencing (1×101nt) was performed using the Illumina HiSeq 2500.
RRBS data quality, read mapping, and cytosine methylation levels
We used bowtie2 in BS-Seeker 2 to map our reads to the reference dog genome (canfam3.1, (Lindblad-Toh et al. 2005)) for read lengths bounded from 50-300 bp (Chen et al. 2010). Per barcode pool, there were 3.25 mismatches (min=3, max=5). Therefore, we deplexed pooled sequence data with a custom perl script with a maximum of two mismatches between expected and observed barcode sequence tags. FASTQ files were then trimmed for low quality reads and adapter sequences clipped using Trim_Galore v0.3.7, discarding reads that were <20 bp in length (Krueger 2013). Trimmed reads were mapped to the reference dog genome using bowtie2 (Langmead 2010; Langmead & Salzberg 2012) and methylation called using BS-Seeker2 (Chen et al. 2010). Bowtie aligns C/T converted reads to C/T converted strands. During post processing, the number of user-defined mismatches is counted with the exception of those between read Ts and genomic Cs. Low-quality mappings with user-defined mismatches are then removed.
We calculated the methylation frequency (MF) as the proportion of methylated cytosines out of the total number of methylated and unmethylated cytosine (sequenced as thymine) reads per site (Chen et al. 2010). For a given frequency, we used a 95% confidence interval threshold value of 0.33 to calculate frequency confidence. Our confidence interval threshold (the maximum range we accept for the 95% confidence calculation) was a function of the number of counts at a particular position, so that for every frequency we used, we were 95% confident that our frequency was real within a window of 0.33 around that number. We also excluded cytosines within the CHG and CHH motifs (Zemach et al. 2010). Cytosines were annotated as being either intergenic or in a promoter (within 5Kb of transcriptional start site; (Agirre et al. 2015)), exon, or intron. Additionally, we used RepeatMasker (Smit et al. 1996) to classify cytosines within canine repeat elements (e.g. LTRs, simple repeats).
Conversion to single methylation polymorphism (SMPs)
To convert methylation frequency into a diploid genotype, we discretized methylation values into epigenotypes: sites with little or no methylation (MF<0.33) were annotated as AA epigenotypes, sites with high or fixed methylation (MF>0.66) were annotated as BB epigenotypes, and sites with intermediate methylation levels (0.33≤MF≤0.66) were annotated as AB epigenotypes (Heyn et al. 2013; Wang & Fan 2015). This allowed for the downstream application of traditional population genetic analyses (e.g. Principal Components Analysis, PCA, Bayesian-clustering, association scans) on methylation data through their conversion to diploid genotypes where each epiallele retains some broad level of methylation information (e.g. A epiallele is hypo-methylated; B epiallele is hyper-methylated).
Identification of outlier samples
To assess the samples for quality and general patterns, we employed two methods on our set of 96 samples: 1) hierarchical cluster analysis on methylation frequency data and 2) PCA of SMP data. First, we constructed a Gower pairwise dissimilarity matrix among all samples for autosomes and X chromosome separately with the R function daisy in the cluster package (Gower 1971; Struyf et al. 1997; Kaufman & Rousseeuw 2009). From this matrix, we then performed hierarchical clustering using Ward's method with the hclust function in R (Gordon 1999). Similarly, we conducted a principal component analysis (PCA). However, the nature of RRBS means that we sequence contiguous blocks of nucleotides, thus containing a high degree of redundant genetic information. To avoid violating the assumption of unlinked and independent loci when analyzing SMPs, we chose to use genotypic correlation as a measure of intra-locus linkage through non-random association (e.g. physical linkage). We used PLINK (Purcell et al. 2007) to prune SMPs in linkage disequilibrium (LD) with a genotype correlation of r2>0.2 using the parameter flag --indep-pairwise 50 5 0.2. We used flashPCA (Abraham & Inouye 2014) for PCA as our primary method to identify outlier samples for exclusion in subsequent analyses.
Population genetics of SMPs
To explore the patterns of SMP variation, we analyzed all canid samples after outliers were removed. We surveyed structure using a clustering method with an explicit population genetics model based on Hardy-Weinberg expectations with the program ADMIXTURE and conducted a cross validation with the --cv flag (Alexander et al. 2009). With this method, each individual is assigned an ancestry proportion based on the number of assumed genetic clusters (K=2-8). Additionally, it assumes individuals are unrelated and loci are unlinked. We used ADMIXTURE to explore the relatedness of the YNP wolves. The purebred dogs are known to be unrelated at the grandparent level, while all family history data are lacking for the non-purebred dog samples. To identify loci that are associated with the domesticated dog phenotype, we conducted an association scan using PLINK and 1,307 unlinked SMPs across the purebred dogs (n=47) and YNP wolves (n=33) with the --assoc flag and a case/control phenotype. This is only a small fraction of the methylation dataset due to the nature of the RRBS approach to capture physical clusters of cytosines in CpG islands. Association p-values were converted into -log10(p) scores and displayed as Manhattan plots using the qqman R function (Turner 2014).
Deviations from neutrality with SMP data
We estimated methylation-specific neutrality statistics with a recently developed method based on a Tajima's D framework with the specific application to SMP data. We first estimated the statistic Dm (Wang & Fan 2015) which detects methylation mutations likely under selection at a particular coding locus. For each gene body represented by sites located within exons and/or introns, we calculated a Dm test to assess potential selection on methylation mutations. /standard deviation . and are two estimators of the mutation parameter, . is the value estimated by the average methylation state difference per cytosine site. is the value estimated by the proportion of methylation segregation sites. When there is an excess of intermediate-frequency SMPs, will have a large value, and Dm tends to be positive. When there is an excess of low-frequency SMPs, will have a small value, and Dm tends to be negative. By theoretic analysis and simulation, it has been shown that directional selection in DNA methylation could generate an excess of intermediate-frequency epialleles (positive Dm) or rare epialleles (negative Dm) dependent on epimutation rate, whereas balancing selection in DNA methylation could generate an excess of intermediate-frequency epialleles (positive Dm). Namely, extreme positive Dm could suggest directional or balancing selection on epialleles, while extreme negative Dm may imply directional selection (Wang & Fan 2015).
Purebred dogs and YNP wolves (n=47 and 33, respectively) were analyzed separately for each gene body tested. In order to examine whether gene bodies in purebred dogs and YNP wolves were under different selection pressures, we calculated the difference between purebred dog and YNP wolves’ Dm values (YNP wolves Dm– purebred dog Dm=DmDiff) for each gene body with polymorphism in both groups (nboth=38). We then performed permutation tests to determine p-values for DmDiff. Under the null hypothesis, there should be no difference between dogs and wolves, i.e. DmDiff=0. Therefore, we performed 10,000 permutations, randomly assigning dog and wolf labels without replacement while keeping the vectors of gene bodies intact to preserve linkage disequilibrium. We then performed a gene ontology (GO) analysis as described below.
Finally, we examined the polymorphic gene bodies in purebred dogs to identify deviations from neutrality through a bootstrap analysis with 10,000 replicates, sampling with replacement while keeping the individual gene body vectors intact. This procedure allowed us to estimate the 99.5% bootstrap confidence interval of the Dm range for each gene body in purebred dogs (ngene_bodies=63). We again conducted a GO enrichment analyses on genes that significantly deviated from neutrality as described below.
Detection of differential methylation between wolves and purebred dogs
To measure the genomic signals of purebred dog domestication, we analyzed methylation data from 33 YNP gray wolves and 47 AKC purebred dogs. We report MF in addition to Delta Δ, the absolute difference between the MF of wolves and purebred dogs. All cytosines located within the mitochondrial genome were excluded, as it does not exhibit a strong signal of DNA methylation in vertebrates due to the underrepresentation of CpG motifs in mitochondrial genes (Cardon et al. 1994).
DNA methylation patterns can vary by biological processes, but also artificially by batch and cell heterogeneity (Christensen et al. 2009; Ji et al. 2010; Bock 2012). For example, blood samples consist of heterogeneous population of cell type in varying proportions, each potentially with a distinct DNA methylation signature that may confound association scans (Reinius et al. 2012). To address this issue, we used a reference-free correction approach in the program EWASher, implemented as part of the FaST-LMM Python v 0.2 package (Lippert et al. 2011; Zou et al. 2014). EWASher computes methylome similarity for every pair of samples. These similarities were then used as the covariance in a mixed model as an implicit proxy for cell-type composition. A reference-based cell-type corrected analysis was performed using the estimated cell-type composition as covariates in a linear regression, in addition to other covariates added to the analysis. The analysis begins by converting raw methylation data into β values. We considered a site to be constitutively methylated if the average probe β value across all samples (cases and controls) was MF>0.8 and to be constitutively unmethylated when MF<0.2. Linear regression was used to correct for all covariates, including batch, computing residuals used in the main association analyses. Using the similarity matrix as the covariance component in the LMM, an associated p-value was generated for each site. If the genomic control factor λ was still inflated (i.e. genome-wide type 1 error rate; (Devlin & Roeder 1999)), EWASher computed the PCs across all samples, included the top PC as a covariate, and then repeated the process. Parameters in the linear mixed model were estimated by maximum likelihood (ML). Association P values were obtained by using a 1 degree-of-freedom likelihood-ratio test and all Bonferroni-significant tests were performed at the α = 0.05 significance level.
We observed only small deviations from the 95% error bars in the quantiles of the theoretical null distribution when plotted against the observed, concordant with our expectation that only a small proportion of methylation sites would be significantly different. To control for batch effects, we added batch as a covariate in our regression analysis. In addition, we included sex of the individual and breed as covariates in our analysis. We applied a Bonferroni-adjusted significance threshold of p<0.005. To survey a larger list of sites, we conducted a GO enrichment analysis on sites that had an adjusted p<0.05 and were found in coding regions (n=8). Last, we compared the list of DMSs between the uncorrected SMP list of 1,307 sites (p<0.05) and the corrected methylation (adjusted p<0.05) association scan of 68 sites to determine if there was overlap between the two types of analyses.
Heritability analysis of methylation in a pedigreed wolf population
We utilized the known genealogical relationships among the 35 Yellowstone gray wolves to assess overall heritability of methylation marks and their mode of inheritance (Vonholdt et al. 2008). To limit the analysis to relevant members and thus reduce computation time, the pedigree was reduced to include only wolves with methylation values, along with their relatives by using the statistical genetics software Mendel's pedigree trimming option (Lange & Sinsheimer 2004; Lange et al. 2013). Wolves in singleton pedigrees post trimming were removed from the analysis (three wolves total). Thus, the final pedigree consisted of 63 wolves, 30 of which had methylation data.
To estimate heritability of methylation, we used a variance components analysis, which takes into account known genealogical relationships between wolves by partitioning the phenotypic variance (Lange 2002). The variance components considered in our analyses included additive genetic, dominance, and environmental variance. As discussed in detail in Lange (2002), the variance component model posits that a quantitative trait such as methylation levels is controlled by multiple loci (polygenic model). In this model, the additive genetic variance captures the effect of alleles at multiple loci as if they were acting independently on methylation. Dominance genetic variation results from deviation from this allelic independence. Environmental variation is any residual variation in the trait values after accounting for genetic and shared environmental variation.
Using the autosomal sites that remain after filtering, methylation levels were standardized and using Mendel, models with different sets of variance components were fit to the data (Lange et al. 2005; Lange et al. 2013). All models included batch and sex as covariates. In addition, the first 5 PCs from EWASher were included in our analysis to correct for cell-type composition. In order to parsimoniously detect evidence for the heritability of methylation marks, we built models in a stepwise fashion. In analysis 1 we fit the simplest model, which includes additive genetic and environmental variance components only. In analysis 2 we reanalyzed those methylation marks that had non-zero estimates of additive genetic variance from analysis 1 by including dominance genetic variance along with the additive genetic and environmental variances. Significance of heritability estimates for all analyses was determined using a 1-sided z-test. Results were termed genome-wide significant if the p-value was less than the Bonferroni-adjusted significance threshold of p<2.08 × 10−6.
Gene ontology enrichment analysis
To test if any gene list was enriched for specific functional categories or ontological types, we used WebGestalt (Zhang et al. 2005; Wang et al. 2013b) on a list of genes that were found to be significant outliers. As a background for our GO analyses, we used all of our unique genes from our RRBS data (n=4580). We applied a Benjamini-Hochberg FDR correction (Benjamini & Hochberg 1995).
Results
Reduced representation bisulfite sequencing
Four samples were dropped due to low coverage, or extremely high variation in coverage (Supplemental Table S1). Of the remaining 92 samples (nwolf=35, npurebred=47, nBoz=2, nsemi=6, nvillage=2), cytosine-specific coverage was calculated by pooling on average four samples per lane (average 27.5× ±4.8 SD) having 17,024,508 ±2,329,082 cytosines, prior to any filtering (Supplemental Table S2). After conservative filtering, 24,205 sites remained (nAuto=23,997; nXchr=208); 33% of the cytosines were located in intergenic regions, 40% were in coding regions and 26% were in promoters (Supplemental Table S3). Further, 19% (n=4,688) of sites were found within repeat elements, with the majority in LINEs (21%, n=978) and SINEs (43%, n=2,002) (Supplemental Table S3). Compared to the 34% TE distribution in the canine genome (Lindblad-Toh et al. 2005), the proportion of methylated loci within all repeat elements for our 24,205 sites was approximately 19% (χ2=0.058, df=1, p=0.81).
Outlier samples and methylation trends
The average methylation (± Standard Deviation; SD) was moderate across 92 samples (MFwolf=0.38 ±0.4; MFpurebred =0.34 ±0.4; MFBoz=0.42 ±0.4; MFsemi=0.35 ±0.4; MFvillage=0.40 ±0.4). However, methylation was significantly higher on the X chromosome than on autosomes (MFautosomes=0.34±0.4; MFXchr=0.75±0.4; 1-tailed t-test unequal variance p<<0.0001). To identify outlier samples, we conducted a PCA on 23,997 autosomal and 208 X-linked cytosines that were converted to SMP genotypes. Using a genotype correlation inclusion threshold of r2<0.2, we retained 1,291 unlinked autosomal and 16 unlinked X chromosome SMPs, of which 8% were found in repeat regions (n=106, where 48 were found within LINEs and SINEs), 32% were within coding regions (n=413), 53% intergenic (n=692), and 15% regulatory (n=202). Across the genome, we identified seven samples that were subsequently excluded from all analyses due to their extremely polarizing effect on the PCA (Supplemental Fig. S1). After removal of outlier samples, the genome-wide average methylation (SD) remained moderate (MFwolf=0.34 ±0.4; MFpurebred =0.34 ±0.4; MFBoz=0.39 ±0.4; MFsemi=0.36 ±0.4; MFvillage=0.38 ±0.4; Supplemental Table S4).
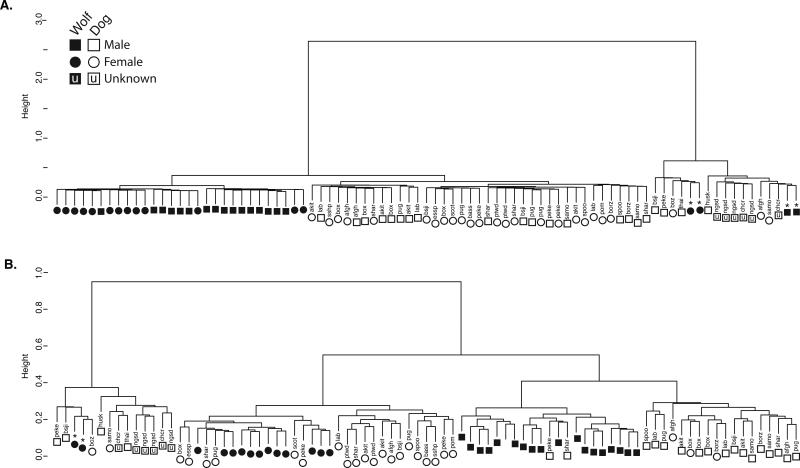
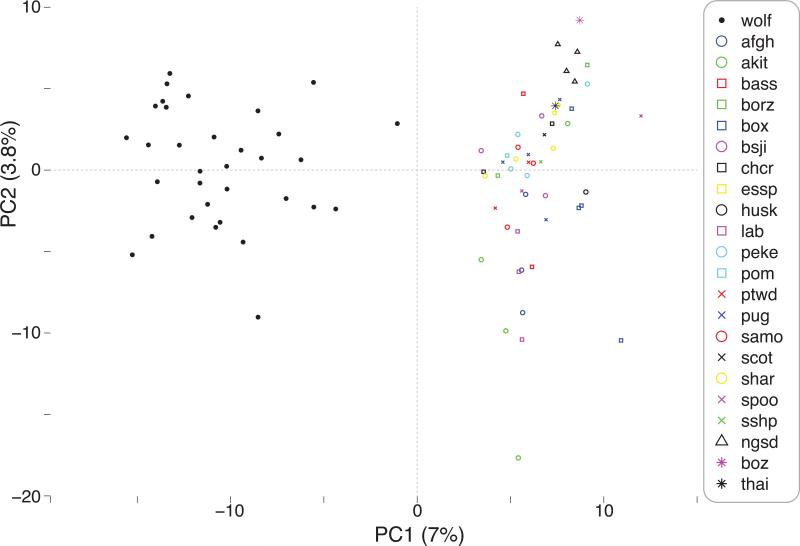
In addition, after removal of outlier samples, a hierarchical cluster analysis of autosomal SMPs (nSMPs=23,997) identified three distinct groups: wolves, dogs, and a “mixed” cluster containing two subgroups of wolves, ancient dogs breeds, and aboriginal island dogs (dingoes removed as outliers) (Fig. 1A). Across the autosomes, the wolves formed a distinct group to that of dogs, with >85% accuracy of species assignment (29/33 wolves; 39/46 purebred dogs) (Fig. 1A). Although there was subdivision of sexes within each cluster, this was less absolute (11/15 female wolves; 10/18 male wolves; 17/25 female purebred dogs; 8/21 male purebred dogs). However, despite a smaller set of X-linked SMPs (nSMPs=208) with variable branch lengths, overall cluster analysis produced five distinct groups: male dogs (15/21), female dogs (14/25), male wolves (18/18), female wolves (12/15), and a “mixed” cluster of aboriginal island/village dogs and wolves (Fig. 1B). The primary axis of divergence for the X-linked SMPs was sex (26/40 females; 33/39 males), followed by species assignments (31/33 wolves; 32/46 purebred dogs). In both dendrograms, there were outlier assignments with wolves processed in a different batch (n=4), aboriginal island dogs (n=4), boz breed dog (n=1), village dogs (n=2), and some putatively ancient dog breeds (bsji, chcr, husk, peke, samo; n=6) (Fig. 1). PC1 also confirmed that the predominant divergence among the samples was a species-specific pattern explaining 7% of the variation (Fig. 2). Relative to the wolves, all dog samples displayed a wider spatial variation along axis 2 (3.8% variation explained).
Figure 1.
Hierarchical cluster analysis of SMP data from 86 samples (excluding outliers) using the Gower dissimilarity matrix and Ward's cluster method for the A) autosomes (nSMPs=23,997); B) X chromosome (nSMPs=208). (See Supplemental Table S1 for codes). Square symbols represent males, circles for females, while “u” are individuals with unknown sex. * indicates wolves that were processed in a different batch.
Figure 2.
PCA (% variance explained) of 1,307 unlinked SMPs after excluding seven outlier samples. See Supplemental Table S1 for breed codes.
We then evaluated population subdivision using the program ADMIXTURE for K=2-8 genetic groups for 1,307 unlinked SMPs (Alexander et al. 2009)(Supplemental Fig. S2; Supplemental Table S5). The best fit at K=2 clearly defined two epigenetic groups, gray wolves and all dogs (purebred, aboriginal island, and village). As K increased, subdivision was found within both groups, though it was not consistent with breed grouping. However, three main groups were distinct at higher K values: wolves, purebred dogs, and the cluster containing the aboriginal island and village dogs (Supplemental Fig. S2).
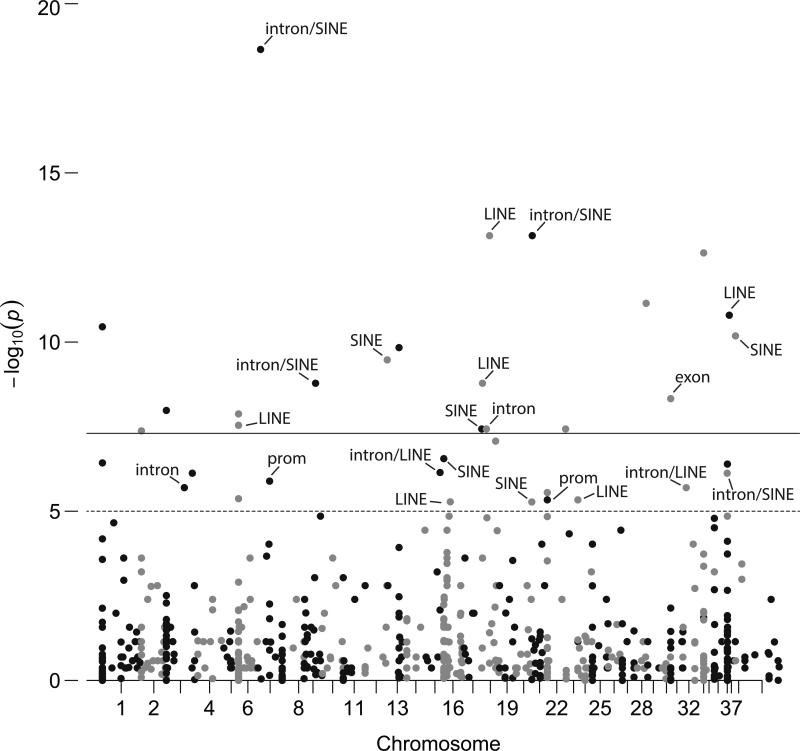
An association scan of 1,307 unlinked SMPs identified 36 significant sites associated with domestication (p<10−5, -log10(p)>5) (Fig. 3; Supplemental Table S6A), of which 47% (17/36) were sites located within a LINE or SINE element, 67% (24/36) intergenic, 25% (9/36) coding, and 6% (2/36) promoter. Both LINES and SINES were significantly enriched compared to the 1,307 unlinked SMPs dataset (Supplemental Table S3). Eleven of the 36 significant sites were located within genes (nexons=1; nintron=8; nprom=2), and the majority of the associated sites (27/36) were hyper-methylated in purebred dogs (average MFwolf=0.62; MFpurebred=0.75). We found an identical pattern of hyper-methylation of transposons in purebred dogs (15/17 hyper-methylated in dogs; average MFwolf=0.61; MFpurebred=0.81). Using the OMIM database (Hamosh et al. 2005), we annotated the significant DMSs associated with genes (Supplemental Table S6A).
Figure 3.
An uncorrected association scan of 1,307 unlinked SMPs across YNP wolves and purebred dogs. Outliers (-log10(p)>5, n=36) are intergenic unless otherwise noted (Abbreviation: prom, promoter). Dashed line (p<10−5) and solid lines (p<10−7) are indicated. All dots above the dashed line are significant outliers. Black dots indicate sites on odd-numbered chromosomes; gray for even-numbered chromosomes.
Differential methylation between purebred dogs and wolves
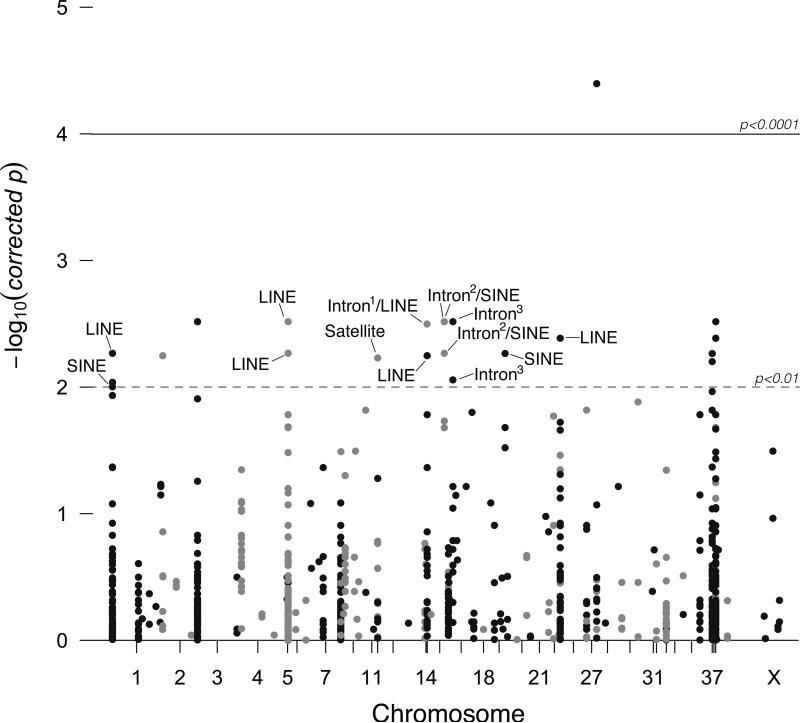
EWASher retained 962 cytosines with 0.2<MF<0.8 between 45 purebred dogs and 33 YNP wolves, of which 68 were significant (corrected p<0.05; Supplemental Table S3) and 14 were highly significant (p<0.005; Fig. 4, Supplemental Table S6B & S7; Supplemental Fig. S3). Out of the 68 significant cytosines, the majority was located within intergenic regions (n=53). The repeat elements were located predominantly in unknown locations (n=33). The majority of the associated sites (36/68) were significantly hyper-methylated in purebred dogs (adjusted p<0.05; average MFwolf=0.58; MFpurebred=0.59). The average methylation frequency difference between purebred dogs and gray wolves was 0.009 (SD±0.018). Among theses significantly associated DMSs, 51% (30/68) consisted of repetitive DNA of which 30 were transposons. We conducted a GO analysis of eight out of 68 sites that were non-overlapping and had an associated annotation. After Benjamini-Hochberg FDR correction, none of our GO categories under Biological Process, Molecular Function, and Cellular Component were significantly enriched (Supplemental Table S8). Further, we found 19 of 68 sites overlapped with the uncorrected SMP association scan, with the majority of sites annotated as intergenic and located within SINES (Table 1). In addition, we found one out of our 14 highly significant, top-ranking sites that overlapped with the uncorrected SMP association scan (Table 1). Out of our 19 overlapping sites, three sites deviated from Hardy-Weinberg Equilibrium (two sites exhibited an excess of heterozygotes and a single site exhibited an excess of homozygotes; Table 1).
Figure 4.
Cell-type and covariate corrected association scan of methylation across YNP wolves and purebred dogs. Manhattan plot for 962 cytosines included in the association scan. Outliers are intergenic unless otherwise noted. Superscripts indicate Ensembl transcript identifiers: 1ENSCAFT00000002993; 2ENSCAFT00000010950; 3ENSCAFT00000042812. All dots above the dashed line are significant outliers. Black dots indicate sites on odd-numbered chromosomes; gray for even-numbered chromosomes.
Table 1.
Sites identified as significant outliers in both the uncorrected SMP association (p<0.05) and corrected methylation scan (p<0.05) between purebred dogs and YNP wolves. (Abbreviations: CFA, canine chromosome)
| CFA | Position | Uncorrected p-value | Adjusted p-value | Gene name | Annotation | In repeat element? | Deviation from Hardy-Weinberg Equilibrium (p<0.05)? |
|---|---|---|---|---|---|---|---|
| 34 | 52849 | <0.000 | 0.005 | Intergenic | |||
| 6 | 49253 | 1.329×10−8 | 0.017 | Intergenic | |||
| 6* | 172430 | 2.890×10−8 | 0.003 | Intergenic | LINE | ||
| 2 | 4609977 | 4.213×10−9 | 0.006 | Intergenic | |||
| 7 | 40156706 | 1.290×10−7 | 0.043 | DEGS1 | Promoter | Dogs (Ho=0.63;He=0.45) | |
| 22 | 48102 | 2.803×10−6 | 0.019 | Intergenic | |||
| 6 | 49272 | 4.261×10−6 | 0.005 | Intergenic | |||
| 24 | 2471 | 4.614×10−8 | 0.015 | Intergenic | LINE | Dogs (Ho=0.02;He=0.08) | |
| 18 | 50124190 | 3.757×10−5 | 0.030 | Intergenic | SINE | Wolves (Ho=0.16);He=(0.12) | |
| 1 | 202655 | 6.568×10−5 | 0.005 | Intergenic | LINE | ||
| 37 | 49946 | 7.712×10−5 | 0.017 | Intergenic | LINE | ||
| 1 | 72422 | 2.655×10−4 | 0.043 | Intergenic | LINE | ||
| 15 | 39451140 | 6.210×10−4 | 0.021 | SLC17A8 | Intron | SINE | |
| 9 | 34354850 | 9.100×10−4 | 0.032 | TUBD1 | Exon | SINE | |
| 11 | 22699 | 9.100×10−4 | 0.006 | Intergenic | Satellite | ||
| 6 | 168151 | 0.001 | 0.003 | Intergenic | |||
| 37 | 28098 | 0.012 | 0.037 | Intergenic | |||
| 37 | 105647 | 0.037 | 0.004 | Intergenic | |||
| 37 | 21264 | 0.045 | 0.021 | Intergenic |
indicates a significant outlier in both uncorrected SMP association scan (p<0.05) and top-ranking corrected methylation scan (p<0.005).
To obtain a perspective at the level of the gene and determine a possible correlation with methylation status, we asked if any repetitive element was within 3Kb of each DMS. We hypothesized that if proximal repetitive elements are epigenetically silenced, their methylation status may spread to nearby sites (Vonholdt et al. 2012). We found that 66% (n=45) of the 68 DMSs were located within or were proximal to repetitive elements. Of the 15 genic DMSs, the majority (67%, n=10) was associated with repeat elements.
Neutrality estimation
Estimating methylation-specific neutrality statistics using a Dm test revealed that out of 3,229 annotated gene bodies (i.e. the gene bodies in the genome covered by our methylome mapping approach), 121 (ndog= 63; nwolf= 96; nshared= 38) exhibited SMP variation (Supplemental Table S9A). The mean observed gene body Dm values we calculated for purebred dogs was −0.5598 and −0.144571 for YNP wolves, eight of which were significantly different (p<0.05) between dogs and wolves (DmDiff different from 0) as assessed by permutation (Table 2). Of these eight genes, SLC17A8, a glutamate transport gene, overlapped the list of significant outliers from both the uncorrected SMP association and corrected methylation scan (Table 1; Table 2). GO analyses revealed that none of our GO categories under Biological Process, Molecular Function, and Cellular Component were significantly enriched after Benjamini-Hochberg FDR correction (Supplemental Table S8). Further, 75% (6/8) of dog genes that deviated from neutrality contained repeat elements within their boundaries (Table 2).
Table 2.
Genes identified through permutation analyses with significantly different neutrality estimates (Dm) between purebred dogs and YNP wolves (DmDiff). The number of cytosines analyzed per gene is indicated in the parentheses following the gene name.
| Ensembl gene(s) list | Description | Observed YNP wolf Dm | Observed purebred dog Dm | Observed DmDiff | Two tailed p-value | M-Fwolves | M-Fpurebred |
|---|---|---|---|---|---|---|---|
| SLC17A8 (6)* | Solute carrier family 17 (vesicular glutamate transporter), member 8 | 6.3571 | 1.3058 | 5.0512 | 0.0001 | 0.427 | 0.583 |
| EPM2A (2)* | Epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) | 0.9911 | −1.1272 | 2.1183 | 0.0007 | 0.878 | 0.975 |
| MYRIP (5)* | Myosin VIIA and Rab interacting protein | 1.2400 | −0.8429 | 2.0828 | 0.0009 | 0.885 | 0.893 |
| DCAF6 (2)* | DDB1 and CUL4 associated factor 6 | 0.8128 | −1.0148 | 1.8277 | 0.0010 | 0.853 | 0.885 |
| ANXA6 (4)* | Annexin A6 | −1.2512 | 0.1042 | −1.3554 | 0.0063 | 0.905 | 0.884 |
| KLHL26 (2) | Kelch-like family member 26 | −0.4590 | 1.0967 | −1.5556 | 0.0209 | 0.207 | 0.214 |
| ENSCAFT00000045887 (95)* | Uncharacterized protein | −0.9634 | −2.1759 | 1.2124 | 0.0265 | 0.174 | 0.165 |
| ENSCAFT00000012091 (11) | Uncharacterized protein | 2.5544 | −0.1056 | 2.6600 | 0.0472 | 0.905 | 0.888 |
| Averaged Value | 0.654 | 0.698 |
Gene contains repeat elements
We also assessed if gene body methylation in purebred dogs deviated from neutrality through a bootstrap analyses performed on polymorphic SMPs. We found that 11 of 63 gene bodies had 99.5% Bootstrap Confidence Intervals (BCIs) that did not overlap the mean Dm value (Supplemental Table S9B), consistent with substantial deviation from neutrality. From the list of 63 gene bodies, we again found neurotransmitter-related genes (GRBRB1, SLC17A8, and ADCY1). GO analyses did not yield significant results (Supplemental Table S8).
Heritability of methylation
In YNP wolves, pedigree-assisted estimates of heritability were calculated using 23,997 autosomal methylation sites. Analysis 1, which includes fixed effect covariates for batch, sex, and cell-type composition and incorporates variance components for additive genetic and environmental variance, resulted in 4,504 sites with non-zero variance component estimates, 2,282 of which exhibited significant narrow sense heritability after accounting for multiple testing (p<2.08×10−6) (Table 3). Most of the sites exhibited low narrow sense heritability, 81.25% of the methylation marks exhibited virtually zero narrow sense heritability (h2<0.01), whereas a sizable minority (8.89%) had extremely high narrow sense heritability (h2>0.99) with the remaining 2,367 sites showing moderate levels of heritability (median h2=0.536) (Supplemental Fig. S4A; Supplemental Table S6). In addition, the methylation marks with the least variability (<5th percentile) had standard deviations ranging between 0.0004 and 0.0129. After adjusting for multiple testing (p<2.09×10−6), 7% of these 1,200 least variable methylation marks were significantly heritable. When the 1,200 least variable sites were removed, 9.64% of the 22,797 remaining autosomal methylation marks were significantly heritable, demonstrating that the percentage of sites that were significantly heritable differed when considering non-variable sites compared to the rest of the autosomal sites (p= 0.002).
Table 3.
Heritability models (all analyses include covariates for batch, sex, and the first 5 PCs from Fast-LMM-EWASher). Asterisk indicates significance compared to Bonferroni threshold* based on total sites analyzed.
| Analysis Model | Variance components | Total number of sites analyzed | Sites with non-zero variance components# | Sites with non-zero variance components and significant heritability* |
|---|---|---|---|---|
| Analysis 1 | Additive Environmental | 23997 | 4504 | 2282 |
| Analysis 2 | Additive Dominance Environmental | 4504 (from Analysis 1) | 1432 | 1362 |
In analysis 1, both additive and environmental variance components are non-zero. In analysis 2, all three components (additive, dominance and environmental) are non-zero.
The narrow sense heritability significance threshold from analysis 1 and the broad sense heritability significance threshold from analysis 2 equals p<2.08 × 10−6.
Because the wolf pedigree was large and highly detailed, we were able to further explore the broad sense heritability by looking for evidence of dominance genetic variance. In Analysis 2, we included only sites with non-zero additive genetic variance estimates from Analysis 1 (n=4,505) to avoid building on a model in which the additive genetic variance was already estimated to be zero, and extended the model to include dominance variance. As a result, Analysis 2 identified 1,362 sites with significant broad sense heritability using the same fixed effect covariates and genome-wide significance threshold (p=2.08×10−6) (Table 3). In addition, 309 of these methylation marks with significant broad sense heritability also had significant narrow sense heritability. As expected from the narrow sense heritability, the vast majority of methylation marks had near-zero broad sense heritability (81.26% with H2<0.01). This percentage represented only two additional methylation marks than were observed to have near-zero narrow sense heritability. A sizable minority of methylation marks were estimated to have either extremely high narrow or broad sense heritability (14.71% with either h2>0.99 or H2>0.99), an additional 1,397 methylation marks over the number estimated when only allowing for additive genetic variance (Supplemental Fig. S4B). Some of these estimates of high heritability were likely due to the small sample size, making it difficult to accurately estimate the heritability; however, some of these estimates were likely due to methylation marks that were inherited in an essentially Mendelian fashion. The remaining 968 sites had moderate levels of broad sense heritability (median H2=0.50). Manhattan plots confirmed that the methylation marks with significant narrow or broad sense heritability were distributed throughout the genome (Supplemental Fig. S5).
Discussion
From a genome-wide scan of methylation at CpG islands across 33 Yellowstone wolves and 45 American Kennel Club registered purebred dogs, we found that the regions exhibiting differential methylation are enriched for transposons and also contain several intriguing categories of functionally relevant genes (e.g. neurotransmitters). In order to minimize differences in methylation values resulting from cell composition, we used a correction-based approach that estimates the degree of genomic inflation (i.e. genome-wide type 1 error rate) with a subsequent correction for cell-type composition using a linear mixed model conjointly with PCA. In addition, we corrected our analyses for multiple covariates including batch, sex, and breed, when applicable. To measure methylation differences between purebred dogs and YNP wolves, we uncovered the signature of selection for polymorphic genes and determined trends in the overall heritability of methylation, providing a revealing picture on the epigenetic changes that occurred during the domestication of the dog from the wolf.
Role of transposons and phenotype evolution
An association scan revealed that differentially methylated sites were generally hyper-methylated in dogs and frequently found within repetitive DNA, primarily transposons. In canines, transposons make up 34% of the canine genome (Lindblad-Toh et al. 2005), but nearly half of other mammalian genomes (~45%) (Smit 1999; Whitelaw & Martin 2001). Transposons play a variety of roles for genome functioning and stability, which include chromosome breakage and rearrangement, in addition to epigenetic transcriptional regulators (Deininger et al. 2003; Slotkin & Martienssen 2007). Due to their large copy number variation and insertion site polymorphism, transposons can serve as the raw material for evolution and influence local transcriptional patterns as they are targeted by epigenetic mechanisms for silencing (McClintock 1984; Kidwell 2002; Deininger et al. 2003). Insertion sites may also disrupt gene function and alter linked phenotypes, serving as a mechanism that can rapidly generate phenotypic variation, an aspect often noted during species domestication. For example, miniature inverted-repeat transposons (MITEs) experienced a rapid amplification and sequence diversification over a relatively short period of time during rice domestication (Feschotte et al. 2002; Jiang et al. 2003; Naito et al. 2006). After transposition, newly inserted elements are often targeted for epigenetic silencing, which frequently suppresses transcription of linked genes (Kaasik & Lee 2004; Zhanget al. 2006; Lister et al. 2008; Hollister et al. 2011). In maize, a Hopscotch retro-element approximately 60Kb upstream of the tb1 coding region is associated with increased tb1 expression, resulting in apical dominance (i.e. changes in branch structure) in maize compared to teosinte. In addition, molecular dating of the Hopscotch retro-element provides suggestive evidence that it predates the domestication of maize and existed in standing genetic variation of teosinte prior to artificial selection (Studer et al. 2011).
Transposons, specifically in SINES, have been previously associated with genome evolution of the domestic dog (Wang & Kirkness 2005; Kirkness 2006). SINES were originally characterized as having a genome-wide distribution (Minnick et al. 1992; Wang & Kirkness 2005) with insertion mutation mapped to phenotypes: narcolepsy in Doberman Pinschers (Lin et al. 1999), autosomal recessive centronuclear myopathy in Labrador retrievers (Pele et al. 2005), and merle patterning in multiple breeds (Clark et al. 2006). There are ~170,000 SINEC_Cf elements, a subfamily of canine-specific SINES, with approximately half of all genes containing a SINEC_Cf insertion (Wang & Kirkness 2005). When these elements are transcribed, they can provide splice receptor sites that can result in the addition of novel exons to the genome.
A recent study revealed the evolutionary history of an intronic SINE insertion located within IGF1 of purebred dogs was lacking in a variety of canid species, including the gray wolf (Gray et al. 2010). Taken together, these results provide a unique perspective of canine domestication where ancient transposons present at a low copy number in the ancestral genome could have experienced rapid amplification as a response to strong artificial selective pressures during domestication, and have since contributed towards functional variation. The “stress hypothesis” can provide an intuitive context for transposon expansion and genome restructuring (Capy et al. 2000; Chénais et al. 2012). As it states, under certain conditions of stress that impact fitness, transposons may become activated as a potential genomic mechanism for novel phenotype production. Dogs, unlike their wild counterparts, recently experienced significant genomic stress due to strong artificial selection and line breeding (e.g. inbreeding; Capy et al. 2000). Physiological stress, for example, is easily imagined as changes in temperature, nutrition, pesticides, or infection. Genomic or molecular stress often include selection or inbreeding which elicits responses, either at the genotypic or phenotypic level (Capy et al. 2000; Bijlsma & Loeschcke 2005). This transposon expansion process, however, cannot continue uncontrolled. Methylation may have served as a flexible epigenetic control of transposition, with targeted methylation acting to suppress rapidly expanding transposon families and marking them for subsequent purging (Smit 1999; Matzke et al. 2007; Slotkin & Martienssen 2007). This speculative mechanism may have contributed to the rapid production of phenotypic variation evident in the history of the domestic dog.
Neutrality Estimation
Similar to single nucleotide changes at the genome level, spontaneous epigenetic mutations can occur at the level of the methylome (Becker et al. 2011; Schmitz et al. 2011; Schmitz et al. 2013) and contribute to fitness differences on which natural selection can act (Rapp & Wendel 2005; Geoghegan & Spencer 2012; Hirsch et al. 2012; Geoghegan & Spencer 2013). Specifically, these epialleles can accumulate in an evolutionary lineage in which the linked phenotype shows non-zero heritability and is thus expected to contribute toward individual fitness and the action of natural selection (Wang & Fan 2015).
We identified eight genes with neutrality estimates that are under different selection pressure or demography effects between purebred dogs and YNP wolves, indicating that both selection and demography have played a role in shaping species-specific methylation patterns. Of these eight genes, SLC17A8, a glutamate transport gene, was an outlier in multiple analyses, exhibiting high heritability and containing repetitive elements. In a surveillance of non-neutral methylation patterns within the purebred dog genome, we again found neurotransmitter-related genes (GRBRB1, SLC17A8, and ADCY1). ADCY1 is a neurotransmitter that is believed to play a role in memory acquisition and learning (Wang et al. 2004). Interestingly, ADCY1 is found within the same gene family as ADCY8, a gene previously identified to be under positive selection in the dog genome with a similar role in functional memory (Vonholdt et al. 2010).
Heritability of methylation marks
Epigenetic mechanisms, such as DNA methylation, can directly affect gene expression (Jaenisch & Bird 2003; Szyf et al. 2005; Ong & Corces 2011; Jones 2012). To date, few studies have investigated transgenerational epigenetic variation in natural animal populations (Peaston & Whitelaw 2006; Ho & Burggren 2010; Feil & Fraga 2011). Fitness-related phenotypes have been documented across generations in Yellowstone wolves, including reproductive success, fecundity, lifespan, inbreeding coefficients, behavior, and coat color (Coulson et al. 2011; Stahler et al. 2013; Hedrick et al. 2014). To account for inbreeding effects that can result from a founding bottleneck, we used the Yellowstone wolf genealogy to estimate the heritability of each methylation mark (Bell et al. 2012). We did not have relatedness information for dogs; therefore, dogs were not included in heritability analyses.
More specifically, we examined the heritability of methylation marks using a variance component model, a classic approach used for many years with quantitative phenotype traits (Lange 2002). As with humans, a minority of the methylation marks in the wolves showed highly significant evidence of heritability (Bell et al. 2012; Grundberg et al. 2013; McRae et al. 2014; Shah et al. 2014). When using a simple model where the level of methylation was determined by multiple genes each acting independently, a fraction (9.51%) of 23,997 autosomal methylation marks was significantly heritable after adjusting for multiple testing (p<2.08×10−6).
With the advantage of having an extensive and accurate pedigree for the Yellowstone wolves, we could further examine whether any of these methylation marks showed evidence of dominance genetic variance (broad sense heritability). We found 1,362 methylation marks (5.68%) that showed evidence of significant broad sense heritability. As our sample size for the heritability analysis was too small to reliably dissect the contribution of random household effects from genetic effects, the effects of common environment may inflate our estimates of the percentage of significantly heritable sites. However, they are similar to that observed in humans and, as in humans, these methylation marks seem to be uniformly distributed throughout the chromosomes.
While we detected transgenerational stability of differentially methylated sites, a necessary requirement for evolution, the scope of our study cannot address the functionality of these sites. However, we demonstrate the co-occurrence of such sites with repetitive elements. This relationship may indicate genotype-dependent epigenetic activity and, thus the patterns we describe may actually represent the underlying genotype that is both inherited and selected. The prevalence of repetitive elements found among the outlier sites support the notion that transposons, in particular, experienced a recent expansion during dog domestication with rapid and subsequent silencing through an epigenetic mechanism. Therefore, while we observe the stability of favorable epialleles, we cannot determine the cause of transmission or their association.
Role of neurotransmitters and domestication
In our study, we found both GABA (GABRB1) and glutamate (SLC17A8) to be significantly associated with the differences between dogs and wolves, both of which are essential inhibitory and excitatory neurotransmitters in the brain, respectively. More specifically, GABA receptors are a family of proteins involved in the GABAergic neurotransmission of the mammalian central nervous system (Johnston 1996; Bormann 2000). GABA-A receptors are the site of action of a number of important pharmacologic agents including barbiturates, benzodiazepines, and ethanol (Whiting et al. 1999). Glutamate transporters are located on the surface of the cells expressing them in the central nervous system, as well as peripheral organs and tissues whereby they control glutamate signaling, and affect a wide-array of functions including cognition, memory, learning, and cell migration, differentiation, and death (Danbolt 2001). Due to the fact that GABA is inhibitory and glutamate is excitatory, both neurotransmitters conjointly control many processes, including brain excitation (Petroff 2002). A recent study found two glutamate receptors (GRIK3 and GRIK2) that were differentially expressed in the frontal cortex of dogs and wolves, suggesting a role of increased excitatory plasticity during domestication (Li et al. 2014).
Alterations in neural signaling and pathways due to changes in density of receptors, biosynthetic pathway alteration, or gene expression differences are indicative of neural plasticity, or flexible changes in the neural system, as a result of recent adaptation, such as changes that may occur during the process of domestication (Popova et al. 1997; Cheng 2010; Kukekova et al. 2011). Behavioral selection is thought to be one of the primary factors that contributed to canid domestication, in which selection acted directly on the genes involved in behavioral development (Trut et al. 2004). Not surprisingly, changes in brain gene expression have been shown in studies of domestication. Specifically, wild canids show a distinct signature of gene expression of a number of genes potentially targeted during behavioral selection in the hypothalamus, prefrontal cortex, and other brain regions compared to domestic dogs (Saetre et al. 2004; Li et al. 2013).
Similarly, genetic polymorphisms in neurotransmitters have been observed both between wolves and dogs and within dog breeds (Hejjas et al. 2007). A study using whole genome sequencing in dogs and wolves uncovered 19 genes related to brain function, with approximately half of the genes associated specifically with nervous system development (Axelsson et al. 2013). Similarly, another study using whole genome sequencing generated a candidate list of genes thought to be under positive selection during dog domestication and found multiple genes categorized as being involved in neurological processes (Wang et al. 2013a).
Our results on neurotransmitter-related genes must be interpreted with caution, however, as we did not collect methylation values directly from brain tissue. Although other studies have shown that methylation in blood is a reliable proxy for methylation in brain tissue (Davies et al. 2012), future studies are needed to verify whether our aforementioned neurotransmitter-related genes are differentially methylated in brain-specific tissue, such as tissue originating from the cortex.
In conclusion, our analysis of DNA methylation provides novel insights into the processes (e.g. demography, selection) that shape the epigenetic patterns in the genome of wild and domesticated canids. We found distinct methylation patterns between dogs and wolves, with a prevalence of differentially methylated sites located in transposons. A substantial fraction of these methylated sites demonstrated high heritability. We develop a speculative hypothesis regarding canine methylome evolution that may have enabled phenotypic diversification. Future studies will be needed to measure the effect of specific transposons on gene functionality, particularly genes involved in neurotransmission.
Supplementary Material
Acknowledgements
The authors would like to thank the purebred dog owners for providing blood samples, and Brian Peckinpaugh for providing Boz samples. In addition, we would like to thank Jennifer Listgarten for providing support for the program FaST-LMM-EWASher. We would also like to thank the Princeton University Computational Science & Engineering Support (CSES) group for providing computational assistance for multiple components of our work. This study was supported in part by the National Science Foundation grants DEB-0613730 and DEB-1245373, Yellowstone National Park, and many donors through the Yellowstone Park Foundation. This work was also partially funded through the Intramural Program of the National Human Genome Research Institute and these additional grants: AKC OAK (CHF #1822), NIH T32 HG002536, National Science Foundation DMS-1264153, and NIH GM053275.
Footnotes
Author Contributions
IJK and BMV designed the study, performed analyses, and wrote the manuscript. BMV and LR constructed the RRBS libraries. MJT analyzed RRBS data for quality, and assisted with mapping and computing cytosine methylation levels. MMC and JSS conducted heritability analyses and contributed to the discussion. LD assisted with heritability analyses. KDM and JW conducted neutrality analyses and contributed to the discussion; KDM also assisted with mapping samples to the dog genome. JSS provided statistical approaches to assess the neutrality analyses. GEG and ELM conducted initial analyses of differential methylation in dogs and wolves. DRS, EAO, and RKW provided blood samples and manuscript edits. MP, DRS, EAO, and RKW were an integral part of interpretation of analyses and discussion of results.
Data Accessibility
BS-Seeker2 log files, epigenotype variant files (used for ADMIXTURE, dendrograms, and PCA), and GO results/input files are accessioned onto Dryad (doi:10.5061/dryad.q2g6h). Raw Illumina Reads are accessioned onto NCBI's Sequence Read Archive (SRA) SRP065319: BioSample Accessions numbers 4217607-4217698. Bam files are accessioned onto SRA SRP065666: BioSample Accessions numbers 4217607-4217698.
References
- Abraham G, Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS One. 2014;9:e93766. doi: 10.1371/journal.pone.0093766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre X, Castellano G, Pascual M, et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res. 2015 doi: 10.1101/gr.180240.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert FW, Somel M, Carneiro M, et al. A comparison of brain gene expression levels in domesticated and wild animals. PLoS Genet. 2012;8:e1002962. doi: 10.1371/journal.pgen.1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almberg ES, Mech LD, Smith DW, Sheldon JW, Crabtree RL. A serological survey of infectious disease in Yellowstone National Park's canid community. PLoS One. 2009;4:e7042. doi: 10.1371/journal.pone.0007042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TM, vonHoldt BM, Candille SI, et al. Molecular and Evolutionary History of Melanism in North American Gray Wolves. Science. 2009;323:1339–1343. doi: 10.1126/science.1165448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L. Molecular consequences of animal breeding. Curr Opin Genet Dev. 2013;23:295–301. doi: 10.1016/j.gde.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- Axelsson E, Ratnakumar A, Arendt ML, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- Becker C, Hagmann J, Muller J, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- Bell JT, Tsai P-C, Yang T-P, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995:289–300. [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bijlsma R, Loeschcke V. Environmental stress, adaptation and evolution: an overview. Journal of evolutionary biology. 2005;18:744–749. doi: 10.1111/j.1420-9101.2005.00962.x. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- Bormann J. The ‘ABC’of GABA receptors. Trends in Pharmacological Sciences. 2000;21:16–19. doi: 10.1016/s0165-6147(99)01413-3. [DOI] [PubMed] [Google Scholar]
- Boyko AR. The domestic dog: man'best friend in the genomic era. Genome Biol. 2011;12:216. doi: 10.1186/gb-2011-12-2-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Quignon P, Li L, et al. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8:e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P, Gasperi G, Biémont C, Bazin C. Stress and transposable elements: co-evolution or useful parasites? Heredity. 2000;85:101–106. doi: 10.1046/j.1365-2540.2000.00751.x. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Burge C, Clayton DA, Karlin S. Pervasive CpG suppression in animal mitochondrial genomes. Proceedings of the National Academy of Sciences. 1994;91:3799–3803. doi: 10.1073/pnas.91.9.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chénais B, Caruso A, Hiard S, Casse N. The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene. 2012;509:7–15. doi: 10.1016/j.gene.2012.07.042. [DOI] [PubMed] [Google Scholar]
- Cheng HW. Breeding of tomorrow's chickens to improve well-being. Poult Sci. 2010;89:805–813. doi: 10.3382/ps.2009-00361. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LA, Wahl JM, Rees CA, Murphy KE. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1376–1381. doi: 10.1073/pnas.0506940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock J. A natural history of domesticated mammals. Cambridge University Press; 1999. [Google Scholar]
- Clutton-Brock J. The process of domestication. Mammal Review. 1992;22:79–85. [Google Scholar]
- Coulson T, MacNulty DR, Stahler DR, et al. Modeling effects of environmental change on wolf population dynamics, trait evolution, and life history. Science. 2011;334:1275–1278. doi: 10.1126/science.1209441. [DOI] [PubMed] [Google Scholar]
- Cubaynes S, MacNulty DR, Stahler DR, et al. Density-dependent intraspecific aggression regulates survival in northern Yellowstone wolves (Canis lupus). J Anim Ecol. 2014 doi: 10.1111/1365-2656.12238. [DOI] [PubMed] [Google Scholar]
- Danbolt NC. Glutamate uptake. Progress in neurobiology. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Darwin C. The variation of animals and plants under domestication. 1868 [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Moran JV, Batzer MA, Kazazian HH. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic Control for Association Studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- Dobney K, Larson G. Genetics and animal domestication: new windows on an elusive process. Journal of Zoology. 2006;269(2):261–71. [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Feeney A, Nilsson E, Skinner MK. Epigenetics and transgenerational inheritance in domesticated farm animals. Journal of Animal Science and Biotechnology. 2014;5:48. doi: 10.1186/2049-1891-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Martinez A, Boluda S, Parchi P, Barrachina M. Brain banks: benefits, limitations and cautions concerning the use of post-mortem brain tissue for molecular studies. Cell and Tissue Banking. 2008;9:181–194. doi: 10.1007/s10561-008-9077-0. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- Freedman AH, Gronau I, Schweizer RM, et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JL, Spencer HG. Population-epigenetic models of selection. Theor Popul Biol. 2012;81:232–242. doi: 10.1016/j.tpb.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Geoghegan JL, Spencer HG. Exploring epiallele stability in a population-epigenetic model. Theor Popul Biol. 2013;83:136–144. doi: 10.1016/j.tpb.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Germonpré M, Sablin MV, Stevens RE, et al. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. Journal of Archaeological Science. 2009;36:473–490. [Google Scholar]
- Gordon AD. Classification. Chapman & Hall/CRC Monographs on Statistics & Applied Probability; 1999. [Google Scholar]
- Gower JC. A general coefficient of similarity and some of its properties. Biometrics. 1971:857–871. [Google Scholar]
- Gray MM, Sutter NB, Ostrander EA, Wayne RK. The IGF1 small dog haplotype is derived from Middle Eastern grey wolves. BMC Biol. 2010;8:16. doi: 10.1186/1741-7007-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Meduri E, Sandling JK, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. The American Journal of Human Genetics. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW, Stahler DR, Dekker D. Heterozygote advantage in a finite population: black color in wolves. Journal of Heredity. 2014;105:457–465. doi: 10.1093/jhered/esu024. [DOI] [PubMed] [Google Scholar]
- Hejjas K, Vas J, Kubinyi E, et al. Novel repeat polymorphisms of the dopaminergic neurotransmitter genes among dogs and wolves. Mamm Genome. 2007;18:871–879. doi: 10.1007/s00335-007-9070-0. [DOI] [PubMed] [Google Scholar]
- Heyn H, Moran S, Hernando-Herraez I, et al. DNA methylation contributes to natural human variation. Genome Res. 2013;23:1363–1372. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Baumberger R, Grossniklaus U. Epigenetic variation, inheritance, and selection in plant populations. Cold Spring Harb Symp Quant Biol. 2012;77:97–104. doi: 10.1101/sqb.2013.77.014605. [DOI] [PubMed] [Google Scholar]
- Ho DH, Burggren WW. Epigenetics and transgenerational transfer: a physiological perspective. J Exp Biol. 2010;213:3–16. doi: 10.1242/jeb.019752. [DOI] [PubMed] [Google Scholar]
- Hollister JD, Smith LM, Guo Y-L, et al. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proceedings of the National Academy of Sciences. 2011;108:2322–2327. doi: 10.1073/pnas.1018222108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, et al. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- Johnston GAR. GABA C receptors: relatively simple transmitter-gated ion channels? Trends in Pharmacological Sciences. 1996;17:319–323. [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding groups in data: an introduction to cluster analysis. John Wiley & Sons; 2009. [Google Scholar]
- Kennerly E, Ballmann A, Martin S, et al. A gene expression signature of confinement in peripheral blood of red wolves (Canis rufus). Mol Ecol. 2008;17:2782–2791. doi: 10.1111/j.1365-294X.2008.03775.x. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- Kirkness EF. 12 SINEs of Canine Genomic Diversity. Cold Spring Harbor Monograph Archive. 2006;44:209–219. [Google Scholar]
- Krueger F. Trim Galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) libraries. 2013 [Google Scholar]
- Kukekova AV, Johnson JL, Teiling C, et al. Sequence comparison of prefrontal cortical brain transcriptome from a tame and an aggressive silver fox (Vulpes vulpes). BMC Genomics. 2011;12:482. doi: 10.1186/1471-2164-12-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K. Mathematical and statistical methods for genetic analysis. Springer Science & Business Media; 2002. [Google Scholar]
- Lange K, Papp JC, Sinsheimer JS, et al. Mendel: the Swiss army knife of genetic analysis programs. Bioinformatics. 2013;29:1568–1570. doi: 10.1093/bioinformatics/btt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Sinsheimer JS. The pedigree trimming problem. Human heredity. 2004;58:108–111. doi: 10.1159/000083031. [DOI] [PubMed] [Google Scholar]
- Lange K, Sinsheimer JS, Sobel E. Association testing with Mendel. Genetic epidemiology. 2005;29:36–50. doi: 10.1002/gepi.20073. [DOI] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Current protocols in bioinformatics. 2010:11.17, 11–11.17, 14. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Karlsson EK, Perri A, et al. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proceedings of the National Academy of Sciences. 2012;109:8878–8883. doi: 10.1073/pnas.1203005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Piperno DR, Allaby RG, et al. Current perspectives and the future of domestication studies. Proc Natl Acad Sci U S A. 2014;111:6139–6146. doi: 10.1073/pnas.1323964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Vonholdt BM, Reynolds A, et al. Artificial selection on brain-expressed genes during the domestication of dog. Mol Biol Evol. 2013;30:1867–1876. doi: 10.1093/molbev/mst088. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang G-D, Wang M-S, et al. Domestication of the dog from the wolf was promoted by enhanced excitatory synaptic plasticity: a hypothesis. Genome biology and evolution. 2014;6:3115–3121. doi: 10.1093/gbe/evu245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, et al. FaST linear mixed models for genome-wide association studies. Nat Meth. 2011;8:833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, et al. Highly Integrated Single-Base Resolution Maps of the Epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- MacNulty DR, Smith DW, Mech LD, Eberly LE. Body size and predatory performance in wolves: is bigger better? J Anim Ecol. 2009a;78:532–539. doi: 10.1111/j.1365-2656.2008.01517.x. [DOI] [PubMed] [Google Scholar]
- MacNulty DR, Smith DW, Mech LD, Vucetich JA, Packer C. Nonlinear effects of group size on the success of wolves hunting elk. Behavioral Ecology. 2011;23:75–82. [Google Scholar]
- MacNulty DR, Smith DW, Vucetich JA, et al. Predatory senescence in ageing wolves. Ecol Lett. 2009b;12:1347–1356. doi: 10.1111/j.1461-0248.2009.01385.x. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJM. Targets of RNA-directed DNA methylation. Current Opinion in Plant Biology. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- McRae AF, Powell JE, Henders AK, et al. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol. 2014;15:R73. doi: 10.1186/gb-2014-15-5-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Gnirke A, Bell GW, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnick MF, Stillwell LC, Heineman JM, Stiegler GL. A highly repetitive DNA sequence possibly unique to canids. Gene. 1992;110:235–238. doi: 10.1016/0378-1119(92)90654-8. [DOI] [PubMed] [Google Scholar]
- Naito K, Cho E, Yang G, et al. Dramatic amplification of a rice transposable element during recent domestication. Proceedings of the National Academy of Sciences. 2006;103:17620–17625. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Wayne RK. The canine genome. Genome Res. 2005;15:1706–1716. doi: 10.1101/gr.3736605. [DOI] [PubMed] [Google Scholar]
- Ovodov ND, Crockford SJ, Kuzmin YV, et al. A 33,000-year-old incipient dog from the Altai Mountains of Siberia: evidence of the earliest domestication disrupted by the Last Glacial Maximum. PLoS One. 2011;6:e22821. doi: 10.1371/journal.pone.0022821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Kim LV, Sutter NB, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Peaston AE, Whitelaw E. Epigenetics and phenotypic variation in mammals. Mamm Genome. 2006;17:365–374. doi: 10.1007/s00335-005-0180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pele M, Tiret L, Kessler JL, Blot S, Panthier JJ. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet. 2005;14:1417–1427. doi: 10.1093/hmg/ddi151. [DOI] [PubMed] [Google Scholar]
- Petroff OAC. Book Review: GABA and Glutamate in the Human Brain. The Neuroscientist. 2002;8:562–573. doi: 10.1177/1073858402238515. [DOI] [PubMed] [Google Scholar]
- Popova NK, Kulikov AV, Avgustinovich DF, Voĭtenko NN, Trut LN. [Effect of domestication of the silver fox on the main enzymes of serotonin metabolism and serotonin receptors]. Genetika. 1997;33:370–374. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp RA, Wendel JF. Epigenetics and plant evolution. New Phytol. 2005;168:81–91. doi: 10.1111/j.1469-8137.2005.01491.x. [DOI] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CL, Bossdorf O, Pigliucci M. What Role Does Heritable Epigenetic Variation Play in Phenotypic Evolution? BioScience. 2010;60:232–237. [Google Scholar]
- Sablin M, Khlopachev G. The earliest Ice Age dogs: evidence from Eliseevichi 11. Current Anthropology. 2002;43:795–799. [Google Scholar]
- Saetre P, Lindberg J, Leonard JA, et al. From wild wolf to domestic dog: gene expression changes in the brain. Brain Res Mol Brain Res. 2004;126:198–206. doi: 10.1016/j.molbrainres.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Savolainen P, Zhang Y-p, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A Genome-Wide Analysis of CpG Dinucleotides in the Human Genome Distinguishes Two Distinct Classes of Promoters. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, et al. Patterns of population epigenomic diversity. Nature. 2013;495:193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, McRae AF, Marioni RE, et al. Genetic and environmental exposures constrain epigenetic drift over the human life course. Genome Res. 2014;24:1725–1733. doi: 10.1101/gr.176933.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman JR, Wilton AN. Origins of the domestic dog and the rich potential for gene mapping. Genet Res Int. 2011;2011:579308. doi: 10.4061/2011/579308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Ersmark E, Palkopoulou E, Dalén L. Ancient Wolf Genome Reveals an Early Divergence of Domestic Dog Ancestors and Admixture into High-Latitude Breeds. Current Biology. 2015 doi: 10.1016/j.cub.2015.04.019. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Smit AFA. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996 [Google Scholar]
- Stahler DR, MacNulty DR, Wayne RK, vonHoldt B, Smith DW. The adaptive value of morphological, behavioural and life-history traits in reproductive female wolves. J Anim Ecol. 2013;82:222–234. doi: 10.1111/j.1365-2656.2012.02039.x. [DOI] [PubMed] [Google Scholar]
- Struyf A, Hubert M, Rousseeuw PJ. Integrating robust clustering techniques in S-PLUS. Computational Statistics & Data Analysis. 1997;26:17–37. [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Ostrander EA. Dog star rising: the canine genetic system. Nat Rev Genet. 2004;5:900–910. doi: 10.1038/nrg1492. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Szyf M, Weaver IC, Champagne FA, Diorio J, Meaney MJ. Maternal programming of steroid receptor expression and phenotype through DNA methylation in the rat. Front Neuroendocrinol. 2005;26:139–162. doi: 10.1016/j.yfrne.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Trut LN, Plyusnina IZ, Oskina IN. An experiment on fox domestication and debatable issues of evolution of the dog. Russian Journal of Genetics. 2004;40:644–655. [PubMed] [Google Scholar]
- Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014 [Google Scholar]
- Van der Auwera I, Elst HJ, Van Laere SJ, et al. The presence of circulating total DNA and methylated genes is associated with circulating tumour cells in blood from breast cancer patients. Br J Cancer. 2009;100:1277–1286. doi: 10.1038/sj.bjc.6605013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonholdt BM, Pollinger JP, Lohmueller KE, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonholdt BM, Stahler DR, Smith DW, et al. The genealogy and genetic viability of reintroduced Yellowstone grey wolves. Mol Ecol. 2008;17:252–274. doi: 10.1111/j.1365-294X.2007.03468.x. [DOI] [PubMed] [Google Scholar]
- Vonholdt BM, Takuno S, Gaut BS. Recent retrotransposon insertions are methylated and phylogenetically clustered in japonica rice (Oryza sativa spp. japonica). Mol Biol Evol. 2012;29:3193–3203. doi: 10.1093/molbev/mss129. [DOI] [PubMed] [Google Scholar]
- Wang GD, Zhai W, Yang HC, et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun. 2013a;4:1860. doi: 10.1038/ncomms2814. [DOI] [PubMed] [Google Scholar]
- Wang H, Ferguson GD, Pineda VV, Cundiff PE, Storm DR. Overexpression of type-1 adenylyl cyclase in mouse forebrain enhances recognition memory and LTP. Nat Neurosci. 2004;7:635–642. doi: 10.1038/nn1248. [DOI] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B. Web-based gene set analysis toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013b;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fan C. A Neutrality Test for Detecting Selection on DNA Methylation Using Single Methylation Polymorphism Frequency Spectrum. Genome biology and evolution. 2015;7:154–171. doi: 10.1093/gbe/evu271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kirkness EF. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005;15:1798–1808. doi: 10.1101/gr.3765505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne RK, Ostrander EA. Lessons learned from the dog genome. Trends Genet. 2007;23:557–567. doi: 10.1016/j.tig.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Wayne RK, vonHoldt BM. Evolutionary genomics of dog domestication. Mamm Genome. 2012;23:3–18. doi: 10.1007/s00335-011-9386-7. [DOI] [PubMed] [Google Scholar]
- Whitelaw E, Martin DIK. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- Whiting PJ, Bonnert TP, McKernan RM, et al. Molecular and Functional Diversity of the Expanding GABA-A Receptor Gene Family. Annals of the New York Academy of Sciences. 1999;868:645–653. doi: 10.1111/j.1749-6632.1999.tb11341.x. [DOI] [PubMed] [Google Scholar]
- Wieczorek G, Asemissen A, Model F, et al. Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer research. 2009;69:599–608. doi: 10.1158/0008-5472.CAN-08-2361. [DOI] [PubMed] [Google Scholar]
- Wiener P, Wilkinson S. Deciphering the genetic basis of animal domestication. Proc Biol Sci. 2011;278:3161–3170. doi: 10.1098/rspb.2011.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zou J, Lippert C, Heckerman D, Aryee M, Listgarten J. Epigenome-wide association studies without the need for cell-type composition. Nat Methods. 2014;11:309–311. doi: 10.1038/nmeth.2815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.