Abstract
Escherichia coli of the O157 serogroup are comprised of a diverse collection of more than 100 O157:non-H7 serotypes that are found in the environment, animal reservoir and infected patients and some have been linked to severe outbreaks of human disease. Among these, the enteropathogenic E. coli O157:non-H7 serotypes carry virulence factors that are hallmarks of enterohemorrhagic E. coli, such as causing attaching and effacing lesions during human gastrointestinal tract infections. Given the shared virulence gene pool between O157:H7 and O157:non-H7 serotypes, our objective was to examine the prevalence of virulence traits of O157:non-H7 serotypes within and across their H-serotype and when compared to other E. coli pathovars. We sequenced six O157:non-H7 genomes complemented by four genomes from public repositories in an effort to determine their virulence state and genetic relatedness to the highly pathogenic enterohemorrhagic O157:H7 lineage and its ancestral O55:H7 serotype. Whole-genome-based phylogenomic analysis and molecular typing is indicative of a non-monophyletic origin of the heterogeneous O157:non-H7 serotypes that are only distantly related to the O157:H7 serotype. The availability of multiple genomes enables robust phylogenomic placement of these strains into their evolutionary context, and the assessment of the pathogenic potential of the O157:non-H7 strains in causing human disease.
Keywords: enteropathogenic E. coli (EPEC), O157:non-H7, pathogenome evolution, genotyping
Whole-genome sequencing and phylogenomic analyses of 10 enteropathogenic Escherichia coli strains of the O157:non-H7 serotypes provide evidence for their non-monophyletic evolutionary origin and identify a heterogeneous virulence complement.
Graphical Abstract Figure.
Whole-genome sequencing and phylogenomic analyses of 10 enteropathogenic Escherichia coli strains of the O157:non-H7 serotypes provide evidence for their non-monophyletic evolutionary origin and identify a heterogeneous virulence complement.
INTRODUCTION
The enteropathogenic Escherichia coli (EPEC) belong to the large diarrheagenic E. coli (DAEC) pathovar (Nataro and Kaper 1998) and are a major cause of infantile diarrhea and the leading cause of morbidity and mortality in infants in developing countries (Trabulsi, Keller and Tardelli Gomes 2002; Afset, Bergh and Bevanger 2003; Gomes et al. 2004; Kaper, Nataro and Mobley 2004; Alikhani, Mirsalehian and Aslani 2006; Behiry et al. 2011). The EPEC O157 serogroup is comprised of more than 100 O157:non-H7 serotypes that have been isolated from different sources including animal reservoirs and infected patients (Stephan et al. 2004; Wani et al. 2006; Feng et al. 2010); additionally, some of the O157:non-H7 strains have been associated with outbreaks of human disease (Makino et al. 1999; Yatsuyanagi et al. 2002; Feng et al. 2010). An array of virulence determinants have been identified in O157:non-H7 strains that are also present in enterohemorrhagic E. coli (EHEC) strains, including the highly pathogenic O157:H7 serotype (Caprioli et al. 2005; Eppinger et al. 2011a,b; Sadiq et al. 2014). One virulence feature of O157:non-H7 strains shared with O157:H7 is the locus for enterocyte effacement (LEE) pathogenicity-associated island (PAI), which mediates the formation of attaching and effacing lesions (A/E) during colonization of the gastrointestinal (GI) tract (Jerse, Gicquelais and Kaper 1991; Phillips and Frankel 2000; Blanco et al. 2006; Coburn, Sekirov and Finlay 2007). The EPEC serotypes are distinguished from Shiga toxin-producing E. coli (STEC), such as EHEC O157:H7, by the lack of stx-encoding bacteriophages (stx1 and/or stx2) (Cornick et al. 2000; Eklund, Leino and Siitonen 2002). The EPEC serotypes can be further classified into typical (tEPEC) and atypical EPEC (aEPEC) strains by the presence or absence of the enteropathogenic adherence factor (EAF) plasmid-borne bfpA gene, which encodes bundlin, a major structural subunit of the bundle-forming pilus (BFP), used by tEPEC for aggregation and bundle formation during colonization (Blank et al. 2000). The production of BFP protein leads to localized adherence and induction of host epithelium cell death (Melo et al. 2005). Another virulence determinant of tEPEC is the EAF plasmid-encoded regulator, perA, which is involved in autoaggregation and elevated expression of BFP, intimin and Tir proteins during infection (Okeke et al. 2001; Iida et al. 2010).
The aim of this study was to elucidate the genomic characteristics and plasticity among the different O157:non-H7 serotypes through comparative genomics of six in-house sequenced and four publicly available O157:non-H7 strains as well as a selection of O157:H7, O157:H(-)and O55:H7 strains sequenced in this study or retrieved from public repositories. Comparative genome analysis within each H-serogroup, between different H-serogroups, as well as when compared to other aEPEC (O55:H7) and EHEC (O157:H7/H-) pathovar strains, identified common and unique traits in the pathogenome evolution of O157:non-H7 strains with respect to the aforementioned serotypes. Here we observed that while EHEC O157:H7 and O157:H(-) share certain virulence characteristics with O157:non-H7 serotypes, whole-genome sequence typing approaches, including multilocus sequence typing (MLST) or eae-genotyping, could not establish close genetic relatedness of these O157:non-H7 lineages with EHEC or other EPEC serotypes. These findings are critical in developing a refined phylogenomic framework and assessment for pathogenic potential of the extant O157:non-H7 strains, which are non-monophyletic.
METHODS
Escherichia coli strains used in this study
To capture the genomic plasticity of the O157:non-H7 serotypes, we selected 10 epidemiologically diverse O157:non-H7 strains that were isolated from water, infected patients and processed meat (Table 1) (Feng et al. 2010; Hazen et al. 2013a; Svab et al. 2013). Strains of the O55:H7, O157:H(-) and O157:H7 serotypes were included to study their genetic relatedness to the O157:non-H7 serotypes (Table S1, Supporting Information) (Hayashi et al. 2001; Perna et al. 2001; Eppinger et al. 2011b; Rump et al. 2011; Hazen et al. 2012; Kyle et al. 2012). Representative O157:H7 strains for each of the nine phylogenetic clades (Manning et al. 2008) were used as reference to determine the relationship of O157:non-H7 to the extant O157:H7 strains.
Table 1.
Genomic characteristics of EPEC O157:non-H7 strains.
| Strain | Serotype | Source | Origin | EPEC | MLSTa) | GUD | SOR | eae | tir | bfpA | perA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ARS4.2123 | O157:H16 | Water | USA | T | 79 | + | + | +(ε) | + | +(β1) | +(β2) |
| RN587/1 | O157:H8 | Human | Brazil | T | 725 | + | ND | +(α) | + | +(β6) | +(β2) |
| C844–97 | O157:H45 | Human | Japan | A | 725 | + | ND | +(α) | + | − | − |
| C639–08 | O157:H45 | Human | Denmark | A | 725 | + | ND | +(α) | + | − | − |
| TW00353 | O157:H45 | Human | USA | A | 2 | + | + | +(ε) | + | − | − |
| TW15901 | O157:H16 | Meat | France | A | 10 | + | + | +(ε) | + | − | − |
| N1 | O157:H29 | Meat | Unknown | NA | 2 | + | + | − | − | − | − |
| T22 | O157:H43 | Human | Hungary | NA | 155 | + | ND | − | − | − | − |
| 3006 | O157:H16 | Human | USA | NA | 6 | + | − | − | − | − | − |
| 7798 | O157:H39 | Human | Argentina | A | 2 | + | − | +(κ/δ) | + | − | − |
O157:non-H7 strains were genotyped for prevalence and plasticity in established EHEC- and EPEC-associated phylogenetic and virulence markers (Farmer and Davis 1985; Adu-Bobie et al. 1998; Blank et al. 2000; Okeke et al. 2001; Yoshitomi et al. 2003; Contreras et al. 2010). MLSTa), Achtman ST7 MLST approach (Wirth et al. 2006); NA, Not applicable; SOR, sorbitol fermentation; GUD, β-glucourinidase activity; T, typical EPEC; A, atypical EPEC; ND, No data available.
Whole-genome sequencing and phylogeny
To study the phylogenetic relatedness of diverse O157:non-H7 strains to other E. coli pathotypes, we have sequenced the genomes of 14 E. coli strains of which six belong to O157:non-H7, one to O55:H7, four to O157:H7 and three to O157:H(-) (Table S1, Supporting Information). Genomes were subjected to Illumina sequencing using paired-end libraries with 300-bp inserts on the HiSeq2000 platform. Draft genomes were assembled using Velvet assembler (Zerbino and Birney 2008; Zerbino 2010), and annotated with the IGS Annotation Engine and Manatee for structural and functional genome annotation (Galens et al. 2011). Accession numbers are listed in Table S1 (Supporting Information). Bacterial genomes were aligned using Mugsy (Angiuoli and Salzberg 2011), and the phylogenetic tree was constructed using the maximum-likelihood method in RAxML v8.1 (-f an option) with 100 bootstraps (Stamatakis 2014), and the evolutionary relationship hypothesis was visualized using EvolView (Zhang et al. 2012). For strains G5101 (SRX702352), 493–89 (SRX702359), USDA5905 (SRX702362) and H2687 (SRX702361), the original reads deposited by Rump et al. were retrieved from the short reads archives (SRA) and de novo assembled with Geneious assembler v.7.1.7, (Kearse et al. 2012). Assembled contigs were queried for the presence of stx1 and stx2 genes using VirulenceFinder 1.2 (Joensen et al. 2014).
Molecular genotyping and subtyping
We examined the status of the β-glucuronidase enzyme in silico for β-glucuronidase activity (GUD) using BLASTN focusing on position +93 uidA gene (T:G allele) (Altschul et al. 1990; Martins et al. 1993) and functionally examined (when applicable) the ability to ferment sorbitol (SOR) on Sorbitol MacConkey agar (Oxoid, UK) (Farmer and Davis 1985). The eae, perA and bfpA positive O157:non-H7 strains were further subtyped into designated allele profiles (Adu-Bobie et al. 1998; Blank et al. 2000; Blanco et al. 2006; Lacher et al. 2007; Contreras et al. 2010). The Achtman MLST scheme (Wirth et al. 2006) was used for in silico polymorphism profiling of E. coli based on seven housekeeping genes.
RESULTS AND DISCUSSION
The O157:non-H7 strains selected for this study originated from diverse geographical locations and different ecological niches, such as human patients, processed meat, and water (Table S1, Supporting Information). Whole-genome screening of the virulence state and molecular subtyping of known EPEC- and EHEC-associated virulence factors revealed a highly diverse and heterogeneous genomic makeup among the O157:non-H7 serogroups studied (Table 1). Furthermore, the phylogenetic relationship of these O157:non-H7 serotypes to other well-studied pathogenic serotypes, such as EPEC (O55:H7) and EHEC (O157:H7/H(-)), was analyzed (Fig. S1, Supporting Information). Availability of high-quality whole-genome sequences enables the determination of the virulence gene state and phylogenomic grouping according to established genotypic classification methods using both in silico and experimental assays (Farmer and Davis 1985; Yoshitomi, Jinneman and Weagant 2003; Wirth et al. 2006; Manning et al. 2008; Contreras et al. 2010).
Whole-genome phylogeny
The phylogenetic tree confirmed the ancestral status of aEPEC O55:H7 to EHEC serotypes (O157:H7 and O157:H(-)) and exhibited a more distant relationship to the cluster of O157:non-H7 serotypes (Fig. S1, Supporting Information) (Trabulsi, Keller and Tardelli Gomes 2002; Zhou et al. 2010; Eppinger et al. 2011b; Kyle et al. 2012). In agreement with a limited loci-based study by Feng et al. (2010), our whole-genome analysis demonstrated that the O157:non-H7 serotypes are not as closely related to the aEPEC O55:H7 when compared to the descendant EHEC O157:H7 and O157:H(-) serotypes (Feng et al. 2010; Zhou et al. 2010; Kyle et al. 2012). In contrast to the genetically highly homogenous population structure of EHEC O157:H7 and O157:H(-) serotypes (Manning et al. 2008; Eppinger et al. 2011b), whole-genome analysis revealed a heterogeneous genome composition among the O157:non-H7 group of serotypes featuring the same H-serogroup (e.g. O157:H16) (Yatsuyanagi et al. 2002; Feng et al. 2010; Bugarel et al. 2011; Eppinger et al. 2011b). As evident in the tree topology (Figs 1 and S1, Supporting Information), the clustering of the different O157:non-H7 strains did not correlate with the H-serotype, as exemplified by the O157:H16 and O157:H45 strains that are scattered within the O157:non-H7 cluster (Fig. 1).
Figure 1.
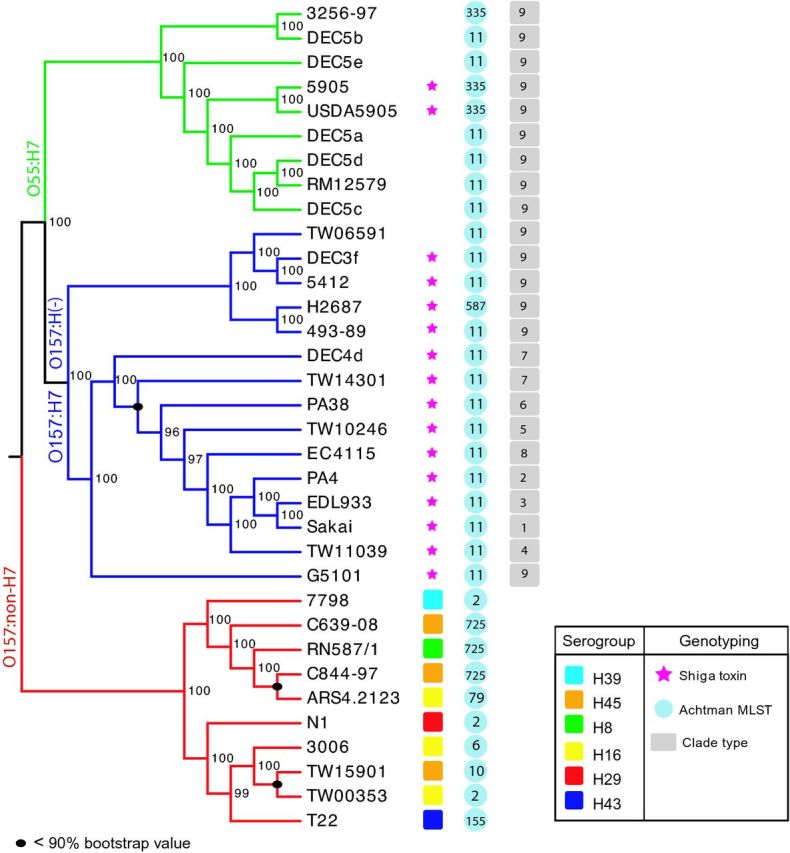
Whole-genome-based phylogeny of O157:non-H7 EPEC strains and select EPEC and EHEC strains. The tree topology demonstrates that O157:non-H7 serogroups form a heterogeneous and non-monophyletic group, as evident from the phylogenetic placement of strains belonging to the same H-serogroup (e.g. H16 and H45). Inferred MLST profiles do not agree with the phylogenetic placement delineated from whole genome alignment. Our data confirm that O157:non-H7 strains are not closely related to the EHEC O157:H7/H(-) lineage or its ancestral aEPEC O55:H7 serotype. For phylogenetic and evolutionary distances, refer to Fig. S1 (Supporting Information).
Using EPEC-associated traits and molecular genotyping, we identified two O157:non-H7 strains as tEPEC (perA/bfpA positive), and the remaining eight strains were classified as aEPEC strains (Table 1). Seven of the studied O157:non-H7 strains were positive for the α, ß or ε eae allele, but none carried the eae- γallele, which is commonly associated with the EHEC O157:H7 serotype (Feng et al. 2010) (Table 1). Overall, these O157:non-H7 strains showed a high degree of variability in their virulence gene complement, which is consistent with the findings reported for other EPEC serotypes (Gomes et al. 2004; Hazen et al. 2013b). As shown in Table 1, seven of the O157:non-H7 strains exhibited an array of EPEC- and/or EHEC-associated virulence determinants with the notable exception of strains N1 (O157:H29), T22 (O157:H43) and 3006 (O157:H16), which lacked these loci and were not classified as tEPEC or aEPEC utilizing the currently established typing schemes (Adu-Bobie et al. 1998; Nataro and Kaper 1998; Blank et al. 2000; Okeke et al. 2001; Dulguer et al. 2003; Contreras et al. 2010), but might originate from an STEC that lost its stx-encoding bacteriophage. Since one of these strains was isolated from a human case with GI symptoms, its pathogenic potential was likely due to carriage of other virulence loci or the result of a coinfection with other pathogenic E. coli strains (Dulguer et al. 2003). The observed combination of virulence determinants likely occurred due to lateral acquisition and cotransfer of genes packaged in virulence plasmids (e.g. EAF-encoded perA and bfpA) (Jerse, Gicquelais and Kaper 1991; Iida et al. 2010) or PAI (e.g. LEE-encoded eae/tir) (Bono et al. 2007; Flockhart et al. 2012). Overall, the diversity observed in these O157:non-H7 genomes can be considered significant.
MLST and phylogenetic placement
The genetically homogenous nature of the E. coli O157:H7 serotype was evidenced by a common Achtman MLST profile of sequence type 11 (ST11) (Wirth et al. 2006; Eppinger et al. 2011b). The non-motile O157:H(-) subgroup (clades 9 and 7) showed the same MLST profile with the exception of strain H2687 (ST587) (Fig. 1). All analyzed non-motile O157:H(-) strains belong to clade 9 and are phylogenetically clustered with the notable exception of the clade 7 strain TW14301 (Fig. 1). We screened for alterations in mobility loci (flhC, flhD or fliC) that have been previously associated with non-motile O157:H(-) strains (Dobbin et al. 2006). Clade 9 O157:H(-) strains exhibit a 12-bp deletion in flhC, consistent with previous findings (Monday, Minnich and Feng 2004), while TW14301 carried the wild-type form of the respective motility genes. Our findings point to loss of motility through independent events in EHEC evolution in the non-motile O157:H(-) serotype (Wang et al. 2003; Dobbin et al. 2006; Kyle et al. 2012). MLST profiling of O55:H7 strains revealed two distinct MLST patterns of ST11 and ST335 (Wirth et al. 2006) based on the Achtman MLST scheme. As shown in Fig. 1, we found instances where MLST profiles of the O157:non-H7 did not corroborate with the corresponding phylogenomic placement. The MLST profile of O157:non-H7 strains (Wirth et al. 2006) was inconclusive in delineating their evolutionary relationship when compared to the whole-genome-based phylogenomic hypothesis (Fig. 1). For example, strains N1 (O157:H29) and TW00353 (O157:H45) or RN587/1 (O157:H8) and C844-97 (O157:H45) feature identical MLST profiles, yet differ in their H-serotype and/or eae-/bfpA-genotype pattern (Table 1), as well as their placement delineated in the evolutionary tree (Fig. S1, Supporting Information).
Carriage of stx-encoding bacteriophages
All analyzed O157:non-H7 strains lack the stx-encoding bacteriophages (stx1 and stx2), which is a virulence hallmark of STEC/EHEC strains (Fig. 1). For some of the strains, the presence of stx1 and stx2 genes had to be assessed from a de novo assembly from the SRA files, as the published contigs were stx negative. However, among the O55:H7 strains we identified two stx2-positive strains, 5905 and USDA5905, which carried a lambda-like stx2d-encoding bacteriophage at the yecE locus. This phage is closely related to the enterobacteria phage BP-4795 found in the EHEC strain RM13516 of the O145:H28 serotype (De Schrijver et al. 2008; Buvens et al. 2011). This particular serotype is related to serotypes O157 and O55 (EHEC1/EPEC1 lineage) (Cooper et al. 2014). We note here that several strains of the O55:H7 serotype have been reported as stx-positive (Feng et al. 1998a,b; Kyle et al. 2012); however, to determine the distinct evolutionary relationship of stx-positive O55:H7 strains compared to EHEC O157:H7 and O157:H(-) serotypes, further phylogenetic studies on larger genomic data sets are needed.
Metabolism
All of the EPEC O157:non-H7 and O55:H7 strains carried the T allele at position +93 uidA and were functionally GUD positive, while all of the O157:H7 and O157:H(-) serotypes carried the G allele and lacked GUD metabolic activity. In analogy to a subset of O157:H7 strains, two of the O157:non-H7 strains (e.g. 3006 and 7798) also did not have the ability to ferment sorbitol (Table 1) (Kyle et al. 2012). Although these two metabolic features are closely associated with the highly pathogenic EHEC O157:H7 lineage, while basal EHEC and ancestral O55:H7 strains are GUD/SOR positive, there are atypical strains of O157:H7 that are GUD positive and the O157:H(-) group is positive for both sorbitol and GUD (Feng et al. 2007).
CONCLUSIONS
The availability of high-quality annotated draft genome sequences for EPEC O157:non-H7 strains provides a critical resource in further understanding the pathogenome evolution and relatedness among different E. coli pathotypes (Dugan et al. 2014; Franz et al. 2014). Our analysis revealed the heterogeneous nature of O157:non-H7 serotypes by studying their genomic inventory and plasticity in the core genome and laterally acquired regions. Overall, the genotypic data for majority of the O157:non-H7 strains we studied (Table 1) did not corroborate with the phylogenomic clustering, as delineated from the whole-genome analysis (Fig. 1), and as suggested by previous publications (Steyert et al. 2012; Hazen et al. 2013b). Even among the same H-serotype strains (e.g. H45 or H16), we observed a non-monophyletic origin that accounts for their diverse genomic content (Fig. 1). Contrary to previous reports of a close relationship of aEPEC and STEC strains in terms of genetic characteristics (Trabulsi, Keller and Tardelli Gomes 2002), we observed a more distant evolutionary relationship between the different tEPEC and aEPEC O157:non-H7 serotypes and the EHEC O157:H7 serotype, as compared to the EPEC O55:H7 (Fig. S1, Supporting Information). In general, aEPEC strains are reported to carry a more heterogeneous virulence profile in comparison to tEPEC strains, and their degree of pathogenicity in the absence of the EAF plasmid remains more elusive (Levine et al. 1978; Trabulsi, Keller and Tardelli Gomes 2002). Here we report that even strains within the same aEPEC O157:non-H7 serotype remain genotypically distinct, and that the reported variability could be attributed to polymorphisms within MLST and established virulence markers. On the other hand, the aEPEC O55:H7 exhibits a more homogenous genotype with few exceptions (Fig. 1), which highlights the degree of diversity found within and among different EPEC serotypes. Further studies are needed to elucidate the evolutionary origin and emergence of EPEC O157:non-H7, which appear to be only distantly related to aEPEC O55:H7 and the highly pathogenic EHEC O157:H7 and H(-) serotypes.
SUPPLEMENTARY DATA
FUNDING
This work received support from the South Texas Center of Emerging Infectious Diseases (STCEID), Department of Biology and Computational System Biology Core at the University of Texas at San Antonio, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract HHSN272200900007C, the High Performance Computing Center/Computational Bioinformatics Initiative (HPC/CBI) under contract 2G12RR013646-12. This project is supported by the Army Research Office of the Department of Defense under Contract No. W911NF-11-1-0136. FS is supported by the South Texas Center for Emerging Infectious Diseases (STCEID) and an University Teaching Fellowship (UTF). BR is supported by the Swiss National Science Foundation (SNSF) Early.Postdoc.Mobility Fellowship (P2LAP3-151770).
Conflict of interest. None declared.
Supplementary Material
REFERENCES
- Adu-Bobie J, Frankel G, Bain C, et al. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol. 1998;36:662–8. doi: 10.1128/jcm.36.3.662-668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afset JE, Bergh K, Bevanger L. High prevalence of atypical enteropathogenic Escherichia coli (EPEC) in Norwegian children with diarrhoea. J Med Microbiol. 2003;52:1015–9. doi: 10.1099/jmm.0.05287-0. [DOI] [PubMed] [Google Scholar]
- Alikhani MY, Mirsalehian A, Aslani MM. Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J Med Microbiol. 2006;55:1159–63. doi: 10.1099/jmm.0.46539-0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Angiuoli SV, Salzberg SL. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 2011;27:334–42. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behiry IK, Abada EA, Ahmed EA, et al. Enteropathogenic Escherichia coli associated with diarrhea in children in Cairo, Egypt. ScientificWorldJournal. 2011;11:2613–9. doi: 10.1100/2011/485381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M, Blanco JE, Dahbi G, et al. Typing of intimin (eae) genes from enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea in Montevideo, Uruguay: identification of two novel intimin variants (muB and xiR/beta2B) J Med Microbiol. 2006;55:1165–74. doi: 10.1099/jmm.0.46518-0. [DOI] [PubMed] [Google Scholar]
- Blank TE, Zhong H, Bell AL, et al. Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia coli strains. Infect Immun. 2000;68:7028–38. doi: 10.1128/iai.68.12.7028-7038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JL, Keen JE, Clawson ML, et al. Association of Escherichia coli O157:H7 tir polymorphisms with human infection. BMC Infect Dis. 2007;7:98. doi: 10.1186/1471-2334-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugarel M, Martin A, Fach P, et al. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol. 2011;11:142. doi: 10.1186/1471-2180-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvens G, Posse B, De Schrijver K, et al. Virulence profiling and quantification of verocytotoxin-producing Escherichia coli O145:H28 and O26:H11 isolated during an ice cream-related hemolytic uremic syndrome outbreak. Foodborne Pathog Dis. 2011;8:421–6. doi: 10.1089/fpd.2010.0693. [DOI] [PubMed] [Google Scholar]
- Caprioli A, Morabito S, Brugere H, et al. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- Coburn B, Sekirov I, Finlay BB. Type III secretion systems and disease. Clin Microbiol Rev. 2007;20:535–49. doi: 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras CA, Ochoa TJ, Lacher DW, et al. Allelic variability of critical virulence genes (eae, bfpA and perA) in typical and atypical enteropathogenic Escherichia coli in Peruvian children. J Med Microbiol. 2010;59:25–31. doi: 10.1099/jmm.0.013706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KK, Mandrell RE, Louie JW, et al. Comparative genomics of enterohemorrhagic Escherichia coli O145:H28 demonstrates a common evolutionary lineage with Escherichia coli O157:H7. BMC Genomics. 2014;15:17. doi: 10.1186/1471-2164-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornick NA, Matise I, Samuel JE, et al. Shiga toxin-producing Escherichia coli infection: temporal and quantitative relationships among colonization, toxin production, and systemic disease. J Infect Dis. 2000;181:242–51. doi: 10.1086/315172. [DOI] [PubMed] [Google Scholar]
- De Schrijver K, Buvens G, Posse B, et al. Outbreak of verocytotoxin-producing E. coli O145 and O26 infections associated with the consumption of ice cream produced at a farm, Belgium, 2007. Euro Surveill. 2008;13:pii:8041. doi: 10.2807/ese.13.07.08041-en. [DOI] [PubMed] [Google Scholar]
- Dobbin HS, Hovde CJ, Williams CJ, et al. The Escherichia coli O157 flagellar regulatory gene flhC and not the flagellin gene fliC impacts colonization of cattle. Infect Immun. 2006;74:2894–905. doi: 10.1128/IAI.74.5.2894-2905.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan VG, Emrich SJ, Giraldo-Calderon GI, et al. Standardized metadata for human pathogen/vector genomic sequences. PLoS One. 2014;9:e99979. doi: 10.1371/journal.pone.0099979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulguer MV, Fabbricotti SH, Bando SY, et al. Atypical enteropathogenic Escherichia coli strains: phenotypic and genetic profiling reveals a strong association between enteroaggregative E. coli heat-stable enterotoxin and diarrhea. J Infect Dis. 2003;188:1685–94. doi: 10.1086/379666. [DOI] [PubMed] [Google Scholar]
- Eklund M, Leino K, Siitonen A. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J Clin Microbiol. 2002;40:4585–93. doi: 10.1128/JCM.40.12.4585-4593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinger M, Mammel MK, Leclerc JE, et al. Genome signatures of Escherichia coli O157:H7 isolates from the bovine host reservoir. Appl Environ Microb. 2011a;77:2916–25. doi: 10.1128/AEM.02554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinger M, Mammel MK, Leclerc JE, et al. Genomic anatomy of Escherichia coli O157:H7 outbreaks. P Natl Acad Sci USA. 2011b;108:20142–7. doi: 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer JJ, 3rd, Davis BR. H7 antiserum-sorbitol fermentation medium: a single tube screening medium for detecting Escherichia coli O157:H7 associated with hemorrhagic colitis. J Clin Microbiol. 1985;22:620–5. doi: 10.1128/jcm.22.4.620-625.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Lampel KA, Karch H, et al. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998a;177:1750–3. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- Feng P, Sandlin RC, Park CH, et al. Identification of a rough strain of Escherichia coli O157:H7 that produces no detectable O157 antigen. J Clin Microbiol. 1998b;36:2339–41. doi: 10.1128/jcm.36.8.2339-2341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng PC, Keys C, Lacher D, et al. Prevalence, characterization and clonal analysis of Escherichia coli O157: non-H7 serotypes that carry eae alleles. FEMS Microbiol Lett. 2010;308:62–7. doi: 10.1111/j.1574-6968.2010.01990.x. [DOI] [PubMed] [Google Scholar]
- Feng PC, Monday SR, Lacher DW, et al. Genetic diversity among clonal lineages within Escherichia coli O157:H7 stepwise evolutionary model. Emerg Infect Dis. 2007;13:1701–6. doi: 10.3201/eid1311.070381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockhart AF, Tree JJ, Xu X, et al. Identification of a novel prophage regulator in Escherichia coli controlling the expression of type III secretion. Mol Microbiol. 2012;83:208–23. doi: 10.1111/j.1365-2958.2011.07927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz E, Delaquis P, Morabito S, et al. Exploiting the explosion of information associated with whole genome sequencing to tackle Shiga toxin-producing Escherichia coli (STEC) in global food production systems. Int J Food Microbiol. 2014;187C:57–72. doi: 10.1016/j.ijfoodmicro.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Galens K, Orvis J, Daugherty S, et al. The IGS standard operating procedure for automated prokaryotic annotation. Stand Genomic Sci. 2011;4:244–51. doi: 10.4056/sigs.1223234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes TA, Irino K, Girao DM, et al. Emerging enteropathogenic Escherichia coli strains? Emerg Infect Dis. 2004;10:1851–5. doi: 10.3201/eid1010.031093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Hazen TH, Sahl JW, Fraser CM, et al. Draft genome sequences of three O157 enteropathogenic Escherichia coli isolates. Genome Announc. 2013a;193:2058–9. doi: 10.1128/genomeA.00516-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen TH, Sahl JW, Fraser CM, et al. Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. P Natl Acad Sci USA. 2013b;110:12810–5. doi: 10.1073/pnas.1306836110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen TH, Sahl JW, Redman JC, et al. Draft genome sequences of the diarrheagenic Escherichia coli collection. J Bacteriol. 2012;194:3026–7. doi: 10.1128/JB.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida M, Okamura N, Yamazaki M, et al. Classification of perA sequences and their correlation with autoaggregation in typical enteropathogenic Escherichia coli isolates collected in Japan and Thailand. Microbiol Immunol. 2010;54:184–95. doi: 10.1111/j.1348-0421.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- Jerse AE, Gicquelais KG, Kaper JB. Plasmid and chromosomal elements involved in the pathogenesis of attaching and effacing Escherichia coli. Infect Immun. 1991;59:3869–75. doi: 10.1128/iai.59.11.3869-3875.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen KG, Scheutz F, Lund O, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52:1501–10. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle JL, Cummings CA, Parker CT, et al. Escherichia coli serotype O55:H7 diversity supports parallel acquisition of bacteriophage at Shiga toxin phage insertion sites during evolution of the O157:H7 lineage. J Bacteriol. 2012;194:1885–96. doi: 10.1128/JB.00120-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacher DW, Steinsland H, Blank TE, et al. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J Bacteriol. 2007;189:342–50. doi: 10.1128/JB.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MM, Bergquist EJ, Nalin DR, et al. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;1:1119–22. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- Makino S, Asakura H, Shirahata T, et al. Molecular epidemiological study of a mass outbreak caused by enteropathogenic Escherichia coli O157:H45. Microbiol Immunol. 1999;43:381–4. doi: 10.1111/j.1348-0421.1999.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Manning SD, Motiwala AS, Springman AC, et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. P Natl Acad Sci USA. 2008;105:4868–73. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins MT, Rivera IG, Clark DL, et al. Distribution of uidA gene sequences in Escherichia coli isolates in water sources and comparison with the expression of beta-glucuronidase activity in 4-methylumbelliferyl-beta-D-glucuronide media. Appl Environ Microb. 1993;59:2271–6. doi: 10.1128/aem.59.7.2271-2276.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo AR, Lasunskaia EB, de Almeida CM, et al. Expression of the virulence factor, BfpA, by enteropathogenic Escherichia coli is essential for apoptosis signalling but not for NF-kappaB activation in host cells. Scand J Immunol. 2005;61:511–9. doi: 10.1111/j.1365-3083.2005.01626.x. [DOI] [PubMed] [Google Scholar]
- Monday SR, Minnich SA, Feng PC. A 12-base-pair deletion in the flagellar master control gene flhC causes nonmotility of the pathogenic German sorbitol-fermenting Escherichia coli O157:H- strains. J Bacteriol. 2004;186:2319–27. doi: 10.1128/JB.186.8.2319-2327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeke IN, Borneman JA, Shin S, et al. Comparative sequence analysis of the plasmid-encoded regulator of enteropatho-genic Escherichia coli Strains. Infect Immun. 2001;69:5553–64. doi: 10.1128/IAI.69.9.5553-5564.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, 3rd, Burland V, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–33. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Phillips AD, Frankel G. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J Infect Dis. 2000;181:1496–500. doi: 10.1086/315404. [DOI] [PubMed] [Google Scholar]
- Rump LV, Strain EA, Cao G, et al. Draft genome sequences of six Escherichia coli isolates from the stepwise model of emergence of Escherichia coli O157:H7. J Bacteriol. 2011;193:2058–9. doi: 10.1128/JB.00118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadiq SM, Hazen TH, Rasko DA, et al. EHEC genomics: past, present, and future. Microbiol Spectrum. 2014;2:1–13. doi: 10.1128/microbiolspec.EHEC-0020-2013. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan R, Borel N, Zweifel C, et al. First isolation and further characterization of enteropathogenic Escherichia coli (EPEC) O157:H45 strains from cattle. BMC Microbiol. 2004;4:10. doi: 10.1186/1471-2180-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyert SR, Sahl JW, Fraser CM, et al. Comparative genomics and stx phage characterization of LEE-negative Shiga toxin-producing Escherichia coli. Front Cell Infect Microbiol. 2012;2:133. doi: 10.3389/fcimb.2012.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab D, Horvath B, Szucs A, et al. Draft genome sequence of an Escherichia coli O157:H43 strain isolated from cattle. Genome Announc. 2013;1:e00263-13. doi: 10.1128/genomeA.00263-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8:508–13. doi: 10.3201/eid0805.010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Rothemund D, Curd H, et al. Species-wide variation in the Escherichia coli flagellin (H-antigen) gene. J Bacteriol. 2003;185:2936–43. doi: 10.1128/JB.185.9.2936-2943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani SA, Samanta I, Munshi ZH, et al. Shiga toxin-producing Escherichia coli and enteropathogenic Escherichia coli in healthy goats in India: occurrence and virulence properties. J Appl Microbiol. 2006;100:108–13. doi: 10.1111/j.1365-2672.2005.02759.x. [DOI] [PubMed] [Google Scholar]
- Wirth T, Falush D, Lan R, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsuyanagi J, Saito S, Sato H, et al. Characterization of enteropathogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreaks. J Clin Microbiol. 2002;40:294–7. doi: 10.1128/JCM.40.1.294-297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitomi KJ, Jinneman KC, Weagant SD. Optimization of a 3′-minor groove binder-DNA probe targeting the uidA gene for rapid identification of Escherichia coli O157:H7 using real-time PCR. Mol Cell Probes. 2003;17:275–80. doi: 10.1016/j.mcp.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics. 2010;31:11.5.1–11.5.12. doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao S, Lercher MJ, et al. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012;40:W569–72. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Li X, Liu B, et al. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS One. 2010;5:e8700. doi: 10.1371/journal.pone.0008700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


