Abstract
On observing schizophrenia from a clinical point of view up to its molecular basis, one may conclude that this is likely to be one of the most complex human disorders to be characterized in all aspects. Such complexity is the reflex of an intricate combination of genetic and environmental components that influence brain functions since pre-natal neurodevelopment, passing by brain maturation, up to the onset of disease and disease establishment. The perfect function of tissues, organs, systems, and finally the organism depends heavily on the proper functioning of cells. Several lines of evidence, including genetics, genomics, transcriptomics, neuropathology, and pharmacology, have supported the idea that dysfunctional cells are causative to schizophrenia. Together with the above-mentioned techniques, proteomics have been contributing to understanding the biochemical basis of schizophrenia at the cellular and tissue level through the identification of differentially expressed proteins and consequently their biochemical pathways, mostly in the brain tissue but also in other cells. In addition, mass spectrometry-based proteomics have identified and precisely quantified proteins that may serve as biomarker candidates to prognosis, diagnosis, and medication monitoring in peripheral tissue. Here, we review all data produced by proteomic investigation in the last 5 years using tissue and/or cells from schizophrenic patients, focusing on postmortem brain tissue and peripheral blood serum and plasma. This information has provided integrated pictures of the biochemical systems involved in the pathobiology, and has suggested potential biomarkers, and warrant potential targets to alternative treatment therapies to schizophrenia.
Introduction
Schizophrenia is a complex neuropsychiatric disorder that produces severe symptoms and significant lifelong disability, causing massive personal and societal burden.1,2 About 1% of the world’s population is affected by schizophrenia.3 Despite the strong genetic component, showing increasing risks for those related to schizophrenic patients,4 and the known role of environment as a trigger, schizophrenia signs and symptoms have unknown etiology. Currently, the disease diagnosis is essentially clinically defined by observed signs of psychosis, which often include paranoid delusions and auditory hallucinations,5 with onset during late adolescence and/or early adulthood.
Pharmacological treatments are available for schizophrenia; yet, most of the currently used antipsychotic medications were discovered in the 1950s, or are a variation of those medications, and since then no new major drug class has been introduced to the clinic. In addition, efficacy of medication is poor, and only about 40% of schizophrenic patients respond effectively to initial treatment with antipsychotics.6,7 Unfortunately, comprehensive studies on molecular mechanisms of schizophrenia have been scant; hence, current treatments are only partly beneficial to a subset of symptoms. The response to drugs is heterogenous, mainly because of individual variations of the disease, in addition to scarce knowledge on its pathophysiology, impairing both diagnosis and adequate treatment selection.8,9
Heterogenic and multifactorial aspects of schizophrenia have always hindered biochemical characterization studies and delayed the establishment of preclinical models of the disease.10 Several studies, including postmortem, imaging, pharmacological, and genetic studies, reported common traces of the disease, such as synaptic deficits, abnormal neural network, and changes in neurotransmission, involving dopamine, glutamate, and gamma-aminobutyric acid.2,11–13 Additional abnormalities, such as aberrant inflammatory responses, oligodendrocyte alterations, epigenetic changes, mitochondrial dysfunction, and reactive oxygen species (ROS) imbalance, are often described in schizophrenia.14–16
A complex cross talk between genetic and environmental factors during neurogenesis is responsible for promoting differences of gene and protein expression in schizophrenia, causing abnormal processes during neurodevelopment.2 Recent studies found reinforcement of genes associated with the major hypotheses of glutamatergic neurotransmission, such as DRD2 (dopamine receptor D2)—the main target of antipsychotic drugs17—among other potential targets, involving perturbation of specific neurotransmitter systems or pathways, which are yet to be studied. The complexity of schizophrenia reinforces the need to unravel molecular mechanisms, as those insights have been shown to be essential in identifying and validating drug targets and biomarkers.9 Therefore, unraveling models with relevance to the cause and onset of schizophrenia is essential toward improving treatments and outcomes for those with the disorder.
Here we review the advances of proteomics on schizophrenia research, toward a better understanding of disease mechanisms and response to treatment, and the efforts toward the discovery of biomarkers for diagnosis and disease evolution.
The role of proteomics in schizophrenia research
In the past century, psychiatric research was dedicated to understanding the nature of several disorders, including action of psychotherapeutics. It was also shown that schizophrenia is a highly heritable disease, indicating a strong genetic influence and an estimated heritability of 80–85%,18,19 more likely with a polygenic basis.20 Since the beginning of the twenty-first century, revolution of genomic technologies has allowed a deeper understanding of the genetic basis of diseases, and several genetic findings on psychiatric disorders have been reported,21 unraveling candidate genes linked to risk factors of psychiatric disorders, such as DISC1 (disrupted in schizophrenia 1),22 involved in neuronal development and synapse formation.23,24 In fact, the International Schizophrenia Consortium (ISC) found indication for a polygenic contribution to schizophrenia.25 While candidate gene studies are beneficial, in cases with a not yet well-understood biology, such as schizophrenia, a single gene only adds a small phenotype effect to the multifactorial etiology of the disease.20,26,27
Since 2008, genomic technology innovations have led to a better understanding of psychiatric disorders, providing information about numerous genes that have a role in brain development.21 Recent advances of next-generation sequencing have facilitated a higher coverage and sample throughput of schizophrenia studies.28–30 Furthermore, international collaborations, which increased the number of participant subjects and samples, have combined efforts to provide deeper insight from comprehensive biological data sets, such as the Psychiatric GWAS (genome-wide association studies) Consortium (PGC; http://pgc.unc.edu).31 Most recently, two main studies, reporting comprehensive GWAS analysis, were able to identify 13 (ref. 27) and 108 schizophrenia-associated risk loci,17 the latter being the largest GWAS study on schizophrenia to date, with up to 36,989 cases and 113,075 controls. Unbiased GWAS,17,27,32,33 indicating genetic regions (loci) that contribute to disease susceptibility, and structural variation studies, such as copy number variants,30,34 are the main identification sources of gene variants with small effects on disease phenotype.35 For instance, copy number variants, including deletions and duplications of several DNA segments, confer significant risk increase in alleles of schizophrenia genome up to 10–25-fold.9,34,36 Several of those findings support the leading etiological hypothesis of the disorder, and point to functionally related targets, such as DRD2, miRNA-137, N-methyl-D-aspartate receptor (NMDAR) complex, or calcium channel subunits.17,30,36,37 Information on genetic variations as a base will increase knowledge on mechanisms of schizophrenia and other psychiatric disorders.
Deciphering the human genome was a revolution in genetics, and the anticipated next step was to decode RNA complexity to understand how information was delivered, and its variety between individuals. Development of large-scale transcriptome analyses, such as cDNA microarrays, Serial Analysis of Gene Expression, and the analyses of Expressed Sequence Tag, and more recently the advance of whole transcriptome shotgun sequencing (or RNA-Seq), providing the presence and quantification of RNA at a given time in a genome, allowed a deeper insight into the dynamics of an organism. Transcriptome analyses revealed RNA implication in psychiatric diseases,38,39 including abnormalities resulting from alternative splicing, in addition to messenger RNA transcripts, such as total RNA and small RNA, including micro-RNA.40 Those abnormalities were observed in several biological processes, such as synaptic and mitochondrial/energetic function,41–43 cytoskeleton,44 immune and inflammation response,45–47 and the myelination pathway.48 Although not yet fully understood, the more the pieces of the puzzle discovered, the more comprehensive the pathology network becomes.
Genomic and transcriptomic studies generated significant data, although these changes cannot yet be translated into biomarkers. The main limitation of genetic approaches in schizophrenia is extrapolation to functional protein expression, as proteins undergo several modifications from transcription to posttranslation, and transcript abundance cannot really predict protein levels either in normal conditions or in response to stress, such as diseases.49,50 Therefore, proteomic techniques are being increasingly used in screening for identification of biomarkers in schizophrenia,51,52 providing several insights into the pathophysiology of the disease. Proteomics can show global expression of proteins or protein groups, and is more complex than genomics as it can change from each cell type at any given time or state.49 Also a high-throughput method, proteomic studies detect fewer expressed proteins than a transcriptomic detects expressed genes, but protein expression provides a precise functional profile and presents an unbiased current physiological state as a reflex of the complex interaction of gene versus environment. The importance of those interactions has been increasing in the research of schizophrenia and other neurological diseases.10,41,53
Regarding research into schizophrenia, numerous studies have investigated the proteome of postmortem brain tissue, including several brain regions such as the dorsolateral prefrontal cortex,41,54,55 frontal cortex,56 thalamus,57 anterior cingulate cortex,58–60 hippocampus,61,62 corpus callosum,63 and insular cortex.64 Postmortem brain tissue has yielded many valuable insights into the pathophysiology of schizophrenia, but less information on disease onset and development. Thus, other tissues and cells have been tested, providing data from naive patients as well, such as from cerebrospinal fluid (CSF),65–68 blood serum and plasma,69–73 liver,68,74 and fibroblasts,75 which can be biopsied from living patients, among others,76,77 aiming to reveal more about potential biomarkers of discovery and monitoring of the disease.
Proteomic methodologies used in schizophrenia research
A proteome comprises the entire set of proteins in a biological system (cell, tissue, or organism) in a particular state, at a given time.78 The need to understand all proteins derived from almost 20,000 genes identified by the Human Genome Project turns molecular biology studies toward proteomics. Because of the progress of mass spectrometry techniques, more fine and high-throughput methods are available, supporting the identification of hundreds (or thousands) of proteins in a single biological sample. In 2014, two major consortiums have delivered a draft of the human proteome,79,80 with a large-scale data set covering 84–92% of the protein-coded genes annotated for the human genome. The more information annotated on protein knowledge databases, the more unknown causes of diseases and biomarker identification can be performed.
In the first decade of proteomics, the main quantitative methods used were gel-based, such as two-dimensional gel electrophoresis (2DE), including the fluorescent two-dimensional differential gel electrophoresis (2D-DIGE). Despite its recognized usefulness,81 gel-based techniques have been consistently replaced by gel-free techniques with the introduction of the concept of shotgun proteomics, which employs basically liquid chromatography followed by mass spectrometry (LC/MS).82 The large scale was possible only because of the development of proteomics based on mass spectrometry, which offers insights into protein abundance, expression profiles according to cell type, posttranslational modifications, and protein–protein interactions, and the possibility to study modifications at the protein level.83
2DE was first described in 1975,84 and after intense enhancements in the 1980s85,86 it became widely used in the separation of complex protein mixtures according to their isoelectric point, in the first dimension by isoelectric focusing, and according to their molecular weight (MW), in the second dimension, by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. This separation leads to a protein profile comprising several spots, each of which, in theory, represents a single protein, providing information about intact proteins and isoforms. Protein visualization techniques include common post-run methods, such as Coomassie blue or silver staining, and also pre-labeling of samples with fluorescent dies, such as in the 2D-DIGE.87 Image analysis of the latter provides a more sensitive quantification method, as up to 10-fold lower amount of samples can be applied. In addition, 2D-DIGE allows co-running of different samples in the same gel, labeled with distinct fluorescent dies (i.e., Cy3, Cy2, Cy5), and might also include an internal control for cross-gel comparison purposes. Those techniques have significantly improved in the previous years with respect to reproducibility and robustness, allowing better comparison between samples and across different laboratories.
Furthermore, mass spectrometry (MS) revolutionized proteomic studies when combined with the 2DE/2D-DIGE workflow, improving sensitivity for identification of differentially expressed proteins, by measuring molecular mass-to-charge ratio of ions (m/z).88 Protein spots, excised from the gel, are digested (i.e., trypsin) and masses of these peptides measured on MS instruments, providing a peptide mass fingerprint of each protein, which is then compared with an in silico-digested database. Further fragmentation of each peptide, performed on an MS/MS instrument, provides the sequence of that peptide, assisting in protein identification. Disadvantages of 2DE/MS combination include the incompatibility to very low or very high molecular weight or isoelectric point, in addition to those proteins with low abundance, which will not be spotted.89 Nevertheless, several proteome studies in schizophrenia were performed using proteomic screening approaches such as 2DE/2D-DIGE, providing large-scale data on the pathophysiology of the disease.41,55,56,58,61,66,90
Hence, schizophrenia and other psychiatric disorder studies have intensively used shotgun proteomics for the analysis of peptides and proteins for profiling, and for quantification of protein modification analysis.54,59,72,75,77,91 For shotgun proteomics, proteins are first digested into peptides (i.e., using trypsin, as previously), which are next separated by high-performance liquid chromatography online-connected to a hybrid MS, providing a gel-free proteomic system (LC-MS/MS). Shotgun proteomics lead to the possibility of identifying more proteins, increasing sampling of low abundant and extreme-sized proteins.82 Most proteomic studies on schizophrenia have used label-free methods for quantification,59,75,92 which assume chromatographic peak areas correlated to the concentration of peptides,93 and is one of the simplest ways to compare proteomics, allowing comparison among several samples at once.94 Nevertheless, both in vitro and in vivo stable isotopic labeling methods are available in shotgun proteomics, for quantification accuracy of protein concentrations simultaneously in several biological samples. In vitro approaches include isobaric tags for relative and absolute quantification95 and isotope-coded protein labeling, whereas in vivo metabolic methods, such as stable isotope labeling by amino acids in cell culture96 and stable isotope labeling in mammals, have been used in proteomic quantification.97 Some have been applied to neuropsychiatric disorders, in postmortem brains and CSF,54,57,98 and in animal and cell studies99,100 on proteomic research.
The power to identify and quantify proteins and protein sets at high resolution, among multiple samples, is essential to understand large case studies in biomedical research. Recent advances have been made in MS-based techniques, such as selected reaction monitoring/multiple reaction monitoring, which has just emerged as a promising technology for a more precise MS-based quantification of targeted protein,101,102 and was awarded Nature’s Method of the Year in 2012 on biological research methods.103 Selected reaction monitoring is specific, accurate, and sensitive, as it selects proteotypic peptides—those that uniquely identify the targeted protein—for its analysis, which might overcome several current validation issues, such as semiquantitative western blotting techniques, availability, and specificity.104 Furthermore, this ability to quantify specific proteins across several samples is particularly interesting with regard to biomarkers, as clinical validation of biomarker signatures for a given disease must be tested over a large sample set to achieve satisfactory statistical power. Indeed, proteomic studies in psychiatric disorders slowly start to validate pathways and biological functions that were found differentially expressed by selected reaction monitoring.105–108
Likewise, other proteomic methodologies have been extensively applied to schizophrenia research in order to discover and validate biomarkers, such as multiplex immunoassays,69 which use multiplexed dye-coded microspheres of selected protein sets, thus providing profile studies of cytokines, growth factors, or metabolic pathways, from blood serum or CSF samples.70,109–111 Aiming to reach the broader spectrum of protein visualization, concerns regarding the possibility to obtain sub-proteomes (using fractionation methods)112 by depleting high-abundant proteins or enriching a group of proteins in a sample should be part of the design and technique choice. Protein separation and quantification using SELDI-TOF-MS ProteinChip analysis or metal ion affinity chromatography to select proteins from a mixture have been used in schizophrenia research lately.65,68,72 Regardless of the protein analysis method, study design and sample preparation choice are crucial steps in proteomic studies. Platforms using reduced number of analytes, but a broader number of clinical samples, provide a precise statistical interpretation.
Indeed, statistics and bioinformatics are of extreme importance for proteomic studies, as different types of assays (2DE, shotgun-MS, or multiplex immunoassays) are required to precisely quantify changes in expression of hundreds (or thousands) of proteins. Therefore, those fields are improving, together with the development of new tools and methods for proteomic analysis, offering better algorithms and image analysis tools, in order to provide a more robust analysis from the growing number of data generated.
What do proteomics tell us about schizophrenia?
Proteomic technologies, mostly focusing on mass spectrometric analysis, are a valuable tool in psychiatric research. A simple search on PubMed using the terms ‘proteomics or proteome and schizophrenia’ provides a total of 218 articles since the first article on proteomics of schizophrenia in the beginning of the 2000s.56 Out of them, 124 articles (and growing) were published within the last 5 years (2010–2014) on human and animal studies, including some reviews, showing considerable increase in awareness of the importance of proteomics in the study of schizophrenia. We have focused, for the purpose of the review, on proteomic studies on human samples of schizophrenia patients compared with controls, from the last 5 years.
These studies, which are summarized in Table 1, have been using proteomic screening approaches such as shotgun-MS (10/23), 2DE/DIGE (7/23), and multiplex immunoassays (10/23), alone or combined. Although postmortem brains are the main studied tissue in schizophrenia research,57–59,61,113 influences of chronic medication or sample heterogeneity and age have impaired some interpretation of the molecular differences found in postmortem brain tissue of schizophrenia patients compared with control subjects.114 Thus, current studies have been mostly focusing on more accessible peripheral tissues, with a preference for blood serum and plasma,72,73,109,115 and CSF,65,66 although there are studies on skin fibroblasts75 and saliva as well.76 Those have become the main tissues used in proteomic studies of schizophrenia because of the possibility of multiple sampling, thus providing better characterization of disease onset, development, and response to treatment. This broader characterization could lead to a more complete understanding of the disease and to development of diagnostic/prognostic biomarkers. Indeed, an analysis of proteins that are common to brain, CSF, and blood samples from at least two studies presented in Table 1, using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Qiagen, Redwood, CA, USA; www.ingenuity.com—Figure 1), shows biomarker candidates of psychiatric disorders and their interactions, and is further discussed.
Table 1. Human proteomic studies from the last 5 years of different tissues and cells in schizophrenic patients.
| Reference | Tissue/Cells | Samples | Technique used | Antipsychotics | Major findings | |
|---|---|---|---|---|---|---|
| 2010 | 73 | Plasma | 229 SCZ, 245 MDD, 254 CTR | Multiplex immunoassay | Naive and treated | Disease signatures using multi-analyte profiling showed growth factors and neurotrophin family differences in schizophrenic patients. |
| 110 | Blood serum | 66 SCZ, 10 BPD, 78 CTR | Multiplex immunoassay | Naive | Circulating levels of insulin-related peptides and the secretory protein chromogranin A significantly elevated in first-onset schizophrenic subjects. | |
| 77 | PBMC | 19 SCZ, 19 CTR | Shotgun (label-free) | 12 Naive, 7 treated | Small clusters of glycolysis pathway proteins can be used to discriminate schizophrenia response with high precision. | |
| 57 | Brain (thalamus) | 11 SCZ, 8 CTR | 2DE/Shotgun (iTRAQ) | Treated, CPE calculated | Differentially expressed proteins on neural transmission and signaling, calcium homeostasis, proteasome metabolism, glycolysis, oligodendrocyte metabolism, and cytoskeleton assembly. | |
| 58 | Brain (ACC) | 11 SCZ, 8 CTR | 2DE | NA | Shows for the first time sex-specific differential expression of proteins in SCZ. | |
| 66 | CSF | 17 SCZ, 10 CTR | 2DE | Naive | Disturbed cholesterol and phospholipid metabolism in SCZ, and potential biomarkers. | |
| 69 | Blood serum | 110 BPD, 827 SCZ, 69 CTR | Multiplex immunoassay | First-onset naive/drug-free >6 weeks | Validation of a biomarker panel for onset schizophrenia molecular signature. | |
| 75 | Skin fibroblast | 11 SCZ, 11 CTR | Shotgun (label-free) | Treated | Alterations in the expression of mRNA and proteins of the cell cycle and growth response of fibroblasts from schizophrenic patients. Fibroblasts are a fit model for SCZ studies. | |
| 2011 | 61 | Brain (HC) | 35 SCZ, 35 BPD, 35 CTR | 2D-DIGE | Treated | Differential expression of hippocampal proteins, involving cytoskeletal and metabolic changes, in addition to clathrin-mediated endocytosis. |
| 90 | Blood serum | 230 SCZ, 36 CTR | Multiplex immunoassay/2D-DIGE | First-onset naive/drug-free >6 weeks | Seven analytes showed significant differences in schizophrenia, supporting the hypothesis of metabolic unbalance such as insulin resistance. | |
| 70 | Blood serum | 250 SCZ, 35 MDD, 32 BPD 329 CTR | Multiplex immunoassay | First-onset naive/drug-free >6 weeks | Biomarker signature underlying the onset or development of SCZ, though present in MDD and BPD. Blood can be used to identify biomarker signatures in schizophrenia. | |
| 2012 | 65 | CSF | 11 SCZ, 20 AD, 20 CTR, | SELDI-TOF-MS | NA | SCZ patients show an overall reduction of CSF Aβ species, not only Aβ1–42, similarly to AD. |
| 72 | Blood serum | 20 SCZ, 20 CTR | IMAC/Shotgun (label-free) | First-onset naive | Changes in 59 phosphoproteins at the phosphorylation level but not at total protein levels. | |
| 92 | Blood serum | 10+47 SCZ, 10+53 CTR proteomics/validation | Shotgun (label-free) | Naive (proteomics) Treated (validation) | Identified 27 proteins as being schizophrenia-related proteins. Dysregulation of the complement pathway and immune system. | |
| 118 | Eccrine sweat | 23 SCZ, 55 CTR | Shotgun (label-free) | Treated (R, O, Q, C, A) | Eccrine sweat protein set is distinct from serum with sensitivity for MRM analyses. It is a rich source of functionally important proteins for biomarker studies. | |
| 132 | Blood serum | 75 SCZ, 110 BPD, 185 CTR | Multiplex immunoassay | Naive | Identification of 20 molecules (i.e., cortisol, CTGF, SAP, TFF3, IL-17) significantly altered prior clinical manifestations. | |
| 111 | Blood serum | 77 SCZ | Multiplex immunoassay | 36 naive, 41 drug-free >6 weeks | Molecular signatures of symptom severity and response, including biomarkers related to insulin and leptin. | |
| 2013 | 134 | Blood serum | 18 SCZ, 22 BPD, 25 VD, 36 AD, 60+77 CTR | 2D-DIGE | NA | Suggests expression of 14-3-3γ as normalization factor on biomarker research. |
| 158 | Pituitary | 14 SCZ, 13 BPD, 14 MD, 15 CTR | Shotgun (label-free)/2D-DIGE/Multiplex immunoassay | FME measurement | Differential molecular profile of the pituitary gland, and suggestion of translation of profile markers to serum proteins. | |
| 115 | Blood serum | 180 SCZ, 398 CTR | Multiplex immunoassay | Naive | Subgroups of SCZ patients based on distinct differences in their molecular serum profiles. | |
| 113 | Brain (DLPFC) | 10 SCZ, 10 SCZ | Shotgun (label-free)/1H-NMR | FME measurement | Combined metabolome/proteome profiling, differential expression of calcium metabolism, cytoskeleton remodeling pathways. | |
| 2014 | 133 | Blood plasma | 26 SCZ, 26 CTR | GC-MS/shotgun (label-free) | Treated | Combined metabolome/proteome profiling, suggesting molecules such as cholesterol, bioactive lipids, and apoliprotein A as biomarkers. |
| 76 | Saliva | 32 SCZ, 17 BPD, 31 CTR | Shotgun (label-free) | NA | Human saliva as a precious body fluid to characterize putative biomarkers of systemic and multifactorial diseases. | |
| 59 | Brain (ACC) | 10 SCZ, 10 CTR | Shotgun (label-free) | NA | Using PSD-enriched samples confirmed changes in SCZ within this group of proteins. | |
| 109 | Blood serum | 180 SCZ, 350 CTR | Multiplex immunoassay | Naive | Identification of pro- and anti-inflammatory cytokines as biomarkers, and modulation after antipsychotic treatment. |
Abbreviations: A, antidepressants; ACC, Anterior cingulate cortex; AD, Alzheimer’s disease; BPD, bipolar disorder; C, clozapine; CSF, cerebrospinal fluid; CTR, controls; 2DE, two-dimensional gel electrophoresis; 2D-DIGE, two-dimensional differential gel electrophoresis; HC, hippocampus; IMAC, metal ion affinity chromatography; iTRAQ, isobaric tags for relative and absolute quantification; MAP, multiplex analyte profile; MDD, major depression disorder; MRM, multiple reaction monitoring; NA, non available information; O, olanzapine; PD, Parkinson disease; PSD, postsynaptic density; Q, quetiapine; R, risperidone; SCZ, schizophrenia; VD, vascular dementia.
Figure 1.
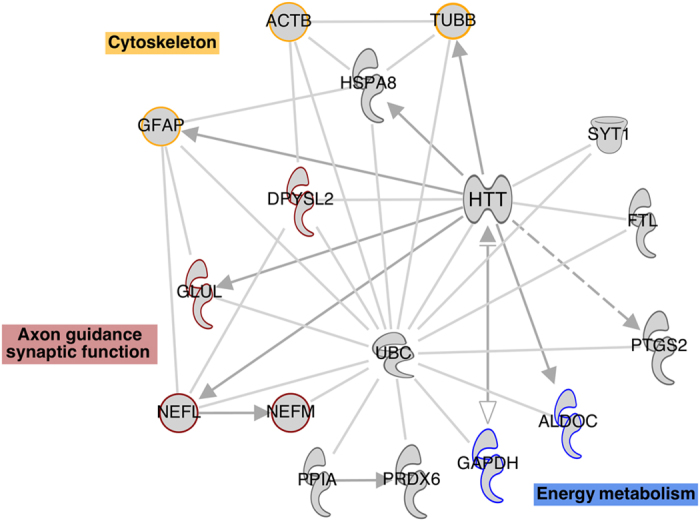
Protein network of regulated proteins in schizophrenia brain, CSF, and blood samples, analyzed by ingenuity pathways knowledge database. ALDOC, aldolase C; CSF, cerebrospinal fluid; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Neuronal transmission and synaptic function
Differentially expressed proteins in schizophrenia proteomic studies have been found to be involved in neuronal transmission, synaptic plasticity, and neurites outgrowth, including several cytoskeletal constituents. Most significant proteomic changes included downregulation of neuroreceptors such as NMDA receptors and alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA), in addition to glutamatergic signaling molecules, such as neurofilaments (NEFL and NEFM), glutamate-ammonia ligase (GLUL), and guanine nucleotide-binding proteins (G proteins) (GNB1), or dihydropyrimidinase-related protein 2 (DPYSL2), which are involved in synaptic function, axon guidance, and signal transduction impairment in schizophrenia.59,113 NEFL, in addition to its role in neuronal morphogenesis, is directly associated with NMDA receptors. NMDAR hypofunction was associated with neurotransmitter dysfunction in NR1 transgenic mice,105 with variations in bioactive peptides and proteins. As GLUL is responsible for removing glutamate from neuronal synapses, it is most likely involved in glutamate imbalance in schizophrenia.2
Other proteins related to NMDA functionality and synaptic plasticity, such as MAPK3, SYNPO, CYFIP2, VDAC, CAMK2B, PRDX1, and ESYT, were also observed differentially expressed in postsynaptic density-enriched samples of postmortem brain tissues.59 Data corresponding to a genomic study of schizophrenia34 found an excess of copy number variants in schizophrenia, confirming several of the proteins differentially regulated with functions in the postsynaptic membrane.
Calcium homeostasis and signaling
Calcium signaling has also been found to be differentially regulated in schizophrenia proteomic studies.54,76,113,116 Calcium is a pivotal metabolite for the dopaminergic hypothesis in schizophrenia, mainly because it has a central role in the function of dopamine receptors D1 and D2.117 Proteins such as calmodulin (CALM1, CALM2), calcium/calmodulin-dependent protein kinase II (CAMK2B, CAMK2D, CAMK2G), voltage-dependent anion channels (VDAC1, VDAC2), and the plasma membrane calcium-transporting ATPase 4 (PMCA-4) are some of the calcium-related proteins found downregulated in the brains of schizophrenia patients.59,116 Some proteins were found differentially expressed in secretion fluids of schizophrenic patients—for example, calmodulin-like proteins and the S100 family of calcium-binding proteins (S100A6, S100A12)—such as in eccrine sweat118 and saliva.76 Complementing these findings, S100B was found downregulated in the nuclear proteome of schizophrenia corpus callosum.119 In addition, calcium activated differential expression of calmodulin-dependent protein kinase II (CAMK2), and calcineurin A in phencyclidine-treated rats.113
Energy metabolism
The brain has a high glucose uptake to supply its major metabolic activity rate. Thus, one of the most consistent dysfunctions underlying the pathophysiology of schizophrenia is in energy metabolism pathways, along with mitochondrial dysfunction and oxidative stress.120,121 Glucose metabolism is confirmed by hyperglycemia, impaired glucose tolerance, and/or insulin resistance in first-onset, antipsychotic, naive schizophrenic patients.110,122 Numerous proteomic studies have identified the glycolysis–gluconeogenesis pathway as being consistently disrupted both in brain and CSF,41,57,58,60,123 and is followed by peripheral tissues.77,90,113,120 The expression of proteins associated with the energy metabolism pathway, such as aldolase C (ALDOC), enolase 1 (ENO1), neuronal enolase 2 (ENO2), lactate dehydrogenase B (LDHB), phosphoglycerate mutase 1 (brain) (PGAM1), phosphoglycerate kinase 1 (PGK1), pyruvate kinase isozyme R/L [PKLR], and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), are often significantly deregulated in schizophrenic patients compared with controls.55,77,120 The most consistent differentially expressed enzyme is aldolase C (ALDOC), which was found altered in several brain samples58,61,66 and also as a marker on blood serum samples.77 Likewise, pyruvate, the final product of glycolysis, and NADPH have been quantified in lower amounts in schizophrenic samples compared with controls, in the thalamus,57 and is replicated in phencyclidine-treated rats, a model of schizophrenia research.113 Whereas schizophrenia seems to be more associated with glycolysis, major depressive disorders are likely to be more associated with oxidative phosphorylation.124
DISC1, a major risk factor of the schizophrenia-susceptibility gene candidate,22 can affect mitochondrial morphology and axonal trafficking.125,126 Alterations of mitochondria morphology were reinforced by the imbalance of the oxidative phosphorilation system, including proteins such as NADH dehydrogenases (i.e., NDUFA1, NDUFV2, NDUFS3, NDUFB5), and ATPases (ATP5B, ATP6V1B2, ATP6V1A1), which have been previously shown altered in animal models of schizoprenia,113,127–129 but also had significant regulation in human brains.57,59 Other molecules such as dopamine have been shown to inhibit electron transport chain complex I (NADH dehydrogenase).130
Oxidative stress
This overall imbalance of mitochondrial energy metabolism, associated with elevated calcium, leads to hazardous ROS concentrations and oxidative stress events in brain cells.131 The resultant ROS may cause oxidative damage in cellular DNA, RNA, proteins, and membrane lipids. Proteomics of the brain have shown several enzymes involved in redox activities (responsible for removing ROS and protecting cells against oxidative injury) to be differentially expressed in schizophrenia brain tissues. Proteins such as superoxide dismutase (which catalyze the dismutation of superoxide (O2 −) into oxygen and hydrogen peroxide), peroxiredoxins (PRDX1, PRDX2, and PRDX3) (which are responsible for reducing hydrogen peroxide), glutathione S-transferases (i.e., GSTM3, GSTTLp28, and GSTP1) (which are a family of multifunctional enzymes involved in cellular detoxification, glutathione reduction, and neutralization of ROS), and NADPH-dependent oxidoreductases such as carbonyl reductases (CBR1 and CBR2) and quinoid dihydropteridine reductase (QDPR) (which might be involved in the NADP/NADPH imbalance observed in the thalamus) were often found regulated in brain tissue,41,57–59,61,113 but could also be detected to be differentially regulated in peripheral tissues such as blood and fibroblast samples .70,73,75,132 Proteomics and combined metabolomics support evidence that slight imbalance in energy glucose metabolism, disrupting mitochondria and the oxidative phosphorylation system, results in compromised ATP production and oxidative stress, which is central in the pathophysiology of schizophrenia.41,113,133
Cytoskeleton
Cytoskeleton constituents are proteins that have shown broad differential expressions in schizophrenia—namely, microtubules such as tubulins (TBA1B, TUBB2A), microfilaments such as actins (ACTG1, ACTB) and actin-binding proteins such as tropomyosins (TPM1, TPM2, TPM3, TPM4), and intermediate filaments (i.e., GFAP, vimetin) and endocytosis proteins, such as dynamin (DNM1), a protein involved in clathrin-mediated endocytosis and other vesicular trafficking processes.55,58,59,113,134,135 Such modifications impact the cellular structure, axonal function, and neurite outgrowth, influencing synaptic plasticity and metabolism, all significantly influencing disturbed cytoskeleton arrangement in schizophrenia.44 Protein components of the cytoskeleton, such as the above-mentioned neurofilaments M and L (NEFL, NEFM) and DPYSL2, a regulator of cytoskeleton remodeling, have a role in axon guidance, neuronal growth, and cell migration. Glial fibrillary acidic protein (GFAP), the major intermediate filament of astrocytes, was found to be strongly regulated in brain tissues, both up- and downexpressed, indicating a precise protein expression across the brain.55,59,61,135,136 In addition, actin was often found downregulated in brain tissues,41,61,62,75 but was upregulated in fibroblasts75 or liver74 of schizophrenic patients.
Immune system and inflammation
Several abnormalities were found in schizophrenia proteomics, including changes in immune- and inflammation-related pathways in first-onset schizophrenic patients compared with controls. Molecules such as α-defensins (DEFA1, DEFA2, DEFA3, DEFA4), migration inhibitory factor, and several interleukins (IL-1ra, IL-8, IL-10, IL-15, IL-16, IL-17, and IL-18), including growth factors such as brain-derived neurotrophic factor, have been differentially regulated in blood samples from schizophrenic patients compared with controls.70,73,76,109,115,132 In addition, extracellular calcium-binding S100A12 exhibits cytokine-like characteristics, recruiting inflammatory cells to the sites of tissue damage. Indeed, anti-inflammatory treatment with cyclooxygenase-2 (COX-2) inhibitors has shown diminished schizophrenic behavior by blocking the synthesis of proinflammatory prostaglandins.137 Multiplex immunoassay profiling studies of blood serum have found numerous components of inflammation signaling pathways.109,111,115 Levels of anti-inflammatory cytokines IL-1ra and IL-10 were decreased after treatment with atypical antipsychotics, which correlated with symptom improvement.109 In addition, profiling studies using a subset of cytokines found increased levels of interleukins (i.e., IL-1β) in the cerebrospinal fluid of first-episode schizophrenic patients, indicative of immune system activation in the brain of some patients.138 Therefore, a proper subset of those altered molecular inflammatory molecules could be included in a sensitive and specific biomarker panel, both for diagnosis and treatment follow-up response.
An overview
Diverse proteomic techniques provided non-biased screening analyses of postmortem brain tissue from schizophrenic patients, and insights into pathways affected in the disease.10,57 In addition, more accessible tissues, such as cerebrospinal fluid, blood serum and plasma, and others such as fibroblasts, liver, and urine,57,66,70,74,75,139 have complemented those findings, suggesting several proteins that could be used as biomarkers to improve diagnosis. We have not gone through the details of the role of oligodendrocytes in schizophrenia, as these were recently tackled somewhere else,140–142 although these are as important as all that are listed here.
Proteomic insights from naive first-onset patients’ impaired protein pathways confirm patterns of disease onset, which along with genetic predisposition could be used as biomarkers for stratification of patients, improving the diagnosis and treatment classification. Also, this valuable information can lead to a more individualized medication, selected according to specific molecular dysfunctions and phenotype observed in schizophrenic individuals. Understanding the pathways affected by medication might also lead to reliable analytical platforms to evaluate individual response to treatment in a personalized-medicine mode. Moreover, the ability to monitor levels of molecules in noninvasive body fluids, such as saliva, urine, or blood serum or plasma, is a great advance. In addition, knowledge of gene–protein pathway networks affected and impaired by the disease can give clues for the development of new and more efficient targeted drugs to those relevant pathways.51,121
Perspectives
Psychiatric disorders are one of the biggest burdens to society,3 and consequently one of the most challenging fields of medical research, with complex and multifactorial characteristics, along with genetic, neurodevelopmental, environmental, and molecular components. Hence, proteomics can add valuable insights into revealing psychiatric disorder connections, as it is closely linked to phenotype, and, by definition, proteomics constitute one of the most suitable approaches for this purpose.143
In 2010, the Human Proteome Organization (HUPO) started a project aiming to map the entire human proteome, the Human Proteome Project (HPP) initiative, with joint initiatives such as the Chromosome-centric Human Proteome Project.144–146 Thus, at the beginning of 2014, two extensive drafts of this map were released,79,80 showing progress in the identification of proteins from high-quality proteomic data to complement genomic annotation. The Human Brain Proteome Project (HBPP) initiative, specifically addressing the proteomic landscape of the human brain, aims to study individuals affected by neurodegenerative diseases, understanding its many different cell types and their particular structure at the cellular and tissue level.147,148 Another main focus is to untangle the human plasma proteome149 on health and disease, to support biomarker validation and development of new tools for diagnosis, disease progression, and medication efficiency, considering the confounding factors present in those body fluids.
From the schizophrenia research point of view, this are exciting news, because of the potential of information that can be extracted, as, regardless of efforts in the search for biomarkers, by investigating the transcriptome and proteome in the post-genomic era, schizophrenia is one more psychiatric disorder without a reliable marker. Those recent advances in ‘omics’ technologies, such as genomics, transcriptomics, proteomics, and metabolomics, which are not only expanding coverage and resolution but also becoming cheaper and more accessible, present new prospects for a global comprehension of biological characteristics of disease mechanisms.150
While genomic and transcriptomic technologies have achieved single-nucleotide resolution, the protein coverage of the amino-acid sequence is still restricted. State-of-the-art shotgun mass spectrometry has improved immensely, such as targeted proteomic measurements, and is useful for biomarker identification. Although the detection of some protein variants, such as differential splice products and posttranslational modifications, remains a challenge for proteomics to get a more comprehensive picture of the whole proteome using a systematic approach. This high-throughput investigation of nucleic acids, proteins, and metabolites from particular tissues and cells provides essential data, which is basic to system biology studies, in order to create integral models of cellular processes.151 Therefore, integrating biological data from omics studies to the expertise of complementary disciplines such as mathematics, physics, and computational sciences, toward better conceptual analysis and predictive models, provides new tools for understanding biological systems at different levels. Hence, we can analyze the cellular space-time and hierarchical organization,152 aiming for complete understanding of psychiatric diseases and identifying candidate biomarkers, especially before and after the onset of clinical manifestations, as well as target metabolic pathways impaired and/or affected by antipsychotics, in order to distinguish subgroups of patients who respond to medication on the basis of their molecular profiles.51
Furthermore, promising investigative tools such as stem cell technologies, which made it possible to generate functional live human neurons in vitro, have emerged in molecular and cellular analyses of complex diseases such as schizophrenia.153,154 Reprogrammed (induced) pluripotent stem cells from the somatic cells of patients offer a novel model for investigating mechanisms underlying complex human diseases.155 The brain remains one of the least understood tissues in our body, partly because of its complexity and also because of limitations associated with in vivo studies. Research on neurodevelopment and the biological basis of schizophrenia and other psychiatric disorders has been hindered by the unavailability of live human brain cells. Pluripotent stem cells carry the genetic diversity of donors, and have been a major focus of brain research, allowing the study of individual molecular pathways, and are evidently interesting as a tool to develop in vitro cell models for drug discovery, disease mechanisms, biomarker validation, and cell therapy.9
Currently, a challenge is to develop differentiation protocols to produce disease-relevant cells, such as subtypes of neurons,156 in addition to more complex structures such as cerebral organoid cultures,157 which are expected to have spontaneous differentiation of neural as well as non-neural cell types, which are important for characterizing behavioral and cell communication aspects in schizophrenia. In vitro models of differentiated cells and organoids are still a simplification of the schizophrenia brain; nevertheless, those models recapitulate distinguishing features of the human brain complexity and are valuable tools to understanding the mechanisms and pathways underlying the pathology of schizophrenia.24 Therefore, proteomics consitute a fundamental tool for studying schizophrenia pluripotent stem cell differentiation, demonstrating individual genetic functionality by analyzing proteins in specific brain cell types. Proteomics can validate this study model, still in development, toward a more complex cell and molecular analysis of neuronal and non-neuronal cell type function and development. Global expression analysis of proteins in different states of the disease provides a powerful tool by direct comparison of signaling pathways and impaired features of individual cells. System-level analysis changes the focus from a single particle, complex, or pathway to a more holistic view including all cellular components and their dynamic cross talk.152 Therefore, association of proteomics of pluripotent stem cell technologies applied to schizophrenia research is also an important tool for drug discovery and biomarker validation, supported by proteomics and system biology analysis to unravel critical differences.150
Acknowledgments
We thank Professor Stevens K. Rehen for his support. We also thank the São Paulo Research Foundation (FAPESP) and National Council for Scientific and Technological Development (CNPq) for financial support.
Funding
This work is supported by São Paulo Research Foundation (FAPESP) grants 13/08711-3 and 14/10068-4.
The authors declare no conflict of interest.
References
- Lewis DA, Lieberman JA. Catching up on schizophrenia: natural history and neurobiology. Neuron 2000; 28: 325–334. [DOI] [PubMed] [Google Scholar]
- Freedman R. Schizophrenia. N Engl J Med 2003; 349: 1738–1749. [DOI] [PubMed] [Google Scholar]
- World Health Organization. The Global Burden of Disease. World Health Organization, Geneva, Switzerland, 2008. [Google Scholar]
- Rodriguez-Murillo L, Gogos JA, Karayiorgou M. The genetic architecture of schizophrenia: new mutations and emerging paradigms. Annu Rev Med 2012; 63: 63–68. [DOI] [PubMed] [Google Scholar]
- Balter M. Talking back to madness. Science 2014; 343: 1190–1193. [DOI] [PubMed] [Google Scholar]
- Chakos M, Lieberman J, Hoffman E, Bradford D, Sheitman B. Effectiveness of second-generation antipsychotics in patients with treatment-resistant schizophrenia: a review and meta-analysis of randomized trials. Am J Psychiatry 2001; 158: 518–526. [DOI] [PubMed] [Google Scholar]
- Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO et al. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med 2005; 353: 1209–1223. [DOI] [PubMed] [Google Scholar]
- Tandon R. Antipsychotics in the treatment of schizophrenia: an overview. J Clin Psychiatry 2011; 72 (Suppl 1): 4–8. [DOI] [PubMed] [Google Scholar]
- Hyman SE. Perspective: revealing molecular secrets. Nature 2014; 508: S20–S20. [DOI] [PubMed] [Google Scholar]
- Guest PC, Martins-de-Souza D, Schwarz E, Rahmoune H, Alsaif M, Tomasik J et al. Proteomic profiling in schizophrenia: enabling stratification for more effective treatment. Genome Med 2013; 5: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard TM, Sikich L, Lieberman JA, LaMantia AS. Neural development, cell-cell signaling, and the “two-hit” hypothesis of schizophrenia. Schizophr Bull 2001; 27: 457–476. [DOI] [PubMed] [Google Scholar]
- Jaaro-Peled H, Ayhan Y, Pletnikov MV, Sawa A. Review of pathological hallmarks of schizophrenia: comparison of genetic models with patients and nongenetic models. Schizophr Bull 2010; 36: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocklington AJ, O'Donovan M, Owen MJ. The synapse in schizophrenia. Eur J Neurosci 2014; 39: 1059–1067. [DOI] [PubMed] [Google Scholar]
- Do KQ, Cabungcal JH, Frank A, Steullet P, Cuenod M. Redox dysregulation, neurodevelopment, and schizophrenia. Curr Opin Neurobiol 2009; 19: 220–230. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Gattaz WF, Schmitt A, Maccarrone G, Hunyadi-Gulyás E, Eberlin MN et al. Proteomic analysis of dorsolateral prefrontal cortex indicates the involvement of cytoskeleton, oligodendrocyte, energy metabolism and new potential markers in schizophrenia. J Psychiatr Res 2009; 43: 978–986. [DOI] [PubMed] [Google Scholar]
- Fournier M, Ferrari C, Baumann PS, Polari A, Monin A, Bellier-Teichmann T et al. Impaired metabolic reactivity to oxidative stress in early psychosis patients. Schizophr Bull 2014; 40: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium SWGOTPG. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG. Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to Star Wars Mx and functional genomics. Am J Med Genet 2001; 97: 12–17. [PubMed] [Google Scholar]
- Craddock N. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet 2005; 42: 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014; 506: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012; 13: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson PA, Malavasi ELV, Grünewald E, Soares DC, Borkowska M, Millar JK. DISC1 genetics, biology and psychiatric illness. Front Biol (Beijing) 2013; 8: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y et al. Disrupted-in-schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 2007; 130: 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 2014; 515: 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gejman PV, Sanders AR, Duan J. The role of genetics in the etiology of schizophrenia. Psychiatr Clin North Am 2010; 33: 35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, McEvoy JP, Gennarelli M, Heinzen EL, Ge D, Maia JM et al. Exome sequencing followed by large-scale genotyping suggests a limited role for moderately rare risk factors of strong effect in schizophrenia. Am J Hum Genet 2012; 91: 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M, Dorschner M, Tsuang D. Next-generation sequencing in schizophrenia and other neuropsychiatric disorders. Am J Med Genet B Neuropsychiatr Genet 2013; 162B: 671–678. [DOI] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014; 506: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee TPGCS. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry 2008; 14: 10–17. [DOI] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DHR, Penninx BWJH, Gordon SD, Nyholt DR et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry 2010; 17: 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D et al. Common variants conferring risk of schizophrenia. Nature 2009; 460: 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 2012; 17: 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon ES, Alliey-Rodriguez N, Liu C. After GWAS: searching for genetic risk for schizophrenia and bipolar disorder. Am J Psychiatry 2011; 168: 253–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Feng G, Hyman SE. Genome-scale neurogenetics: methodology and meaning. Nat Neurosci 2014; 17: 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TSPG-WASG. Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43: 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry 2006; 60: 163–176. [DOI] [PubMed] [Google Scholar]
- Horváth S, Janka Z, Mirnics K. Analyzing schizophrenia by DNA microarrays. Biol Psychiatry 2011; 69: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazisar M, Cammaerts S, van der Ven K, Forero DA, Lenaerts A-S, Nordin A et al. MIR137 variants identified in psychiatric patients affect synaptogenesis and neuronal transmission gene sets. Mol Psychiatry 2014, e-pub ahead of print 10.1038/mp.2014.53. [DOI] [PubMed]
- Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT-J, Griffin JL et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry 2004; 9: 684–97–643. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron 2000; 28: 53–67. [DOI] [PubMed] [Google Scholar]
- Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci 2002; 22: 2718–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu S-A et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry 2014, epub ahead of print 10.1038/mp.2014.82. [DOI] [PMC free article] [PubMed]
- Xu J, Sun J, Chen J, Wang L, Li A, Helm M et al. RNA-Seq analysis implicates dysregulation of the immune system in schizophrenia. BMC Genomics 2012; 13: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Kim J, Shin J-Y, Kim J-I, Seo J-S, Webster MJ et al. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl Psychiatry 2013; 3: e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry 2007; 62: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA 2001; 98: 4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloyianni N, Tsangaris GT. The audacity of proteomics: a chance to overcome current challenges in schizophrenia research. Expert Rev Proteomics 2009; 6: 661–674. [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012; 13: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest PC, Chan MK, Gottschalk MG, Bahn S. The use of proteomic biomarkers for improved diagnosis and stratification of schizophrenia patients. Biomarkers Med 2014; 8: 15–27. [DOI] [PubMed] [Google Scholar]
- Vargas G. Biomarkers in schizophrenia. Biomarkers Med 2014; 8: 1–3. [DOI] [PubMed] [Google Scholar]
- English JA, Pennington K, Dunn MJ, Cotter DR. The neuroproteomics of schizophrenia. Biol Psychiatry 2011; 69: 163–172. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Gattaz WF, Schmitt A, Rewerts C, Maccarrone G, Dias-Neto E et al. Prefrontal cortex shotgun proteome analysis reveals altered calcium homeostasis and immune system imbalance in schizophrenia. Eur Arch Psychiatry Clin Neurosci 2009; 259: 151–163. [DOI] [PubMed] [Google Scholar]
- English JA, Dicker P, Föcking M, Dunn MJ, Cotter DR. 2-D DIGE analysis implicates cytoskeletal abnormalities in psychiatric disease. Proteomics 2009; 9: 3368–3382. [DOI] [PubMed] [Google Scholar]
- Johnston-Wilson NL, Sims CD, Hofmann JP, Anderson L, Shore AD, Torrey EF et al. Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. The Stanley Neuropathology Consortium. Mol Psychiatry 2000; 5: 142–149. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Maccarrone G, Wobrock T, Zerr I, Gormanns P, Reckow S et al. Proteome analysis of the thalamus and cerebrospinal fluid reveals glycolysis dysfunction and potential biomarkers candidates for schizophrenia. J Psychiatr Res 2010; 44: 1176–1189. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Schmitt A, Röder R, Lebar M, Schneider-Axmann T, Falkai P et al. Sex-specific proteome differences in the anterior cingulate cortex of schizophrenia. J Psychiatr Res 2010; 44: 989–991. [DOI] [PubMed] [Google Scholar]
- Föcking M, Lopez LM, English JA, Dicker P, Wolff A, Brindley E et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol Psychiatry; advance online publication, 22 July 2014; doi:10.1038/mp.2014.63 (e-pub ahead of print). [DOI] [PubMed]
- Clark D, Dedova I, Cordwell S, Matsumoto I. A proteome analysis of the anterior cingulate cortex gray matter in schizophrenia. Mol Psychiatry 2006; 11: 459–470, 423. [DOI] [PubMed] [Google Scholar]
- Föcking M, Dicker P, English JA, Schubert KO, Dunn MJ, Cotter DR. Common proteomic changes in the hippocampus in schizophrenia and bipolar disorder and particular evidence for involvement of cornu ammonis regions 2 and 3. Arch Gen Psychiatry 2011; 68: 477–488. [DOI] [PubMed] [Google Scholar]
- Nesvaderani M, Matsumoto I, Sivagnanasundaram S. Anterior hippocampus in schizophrenia pathogenesis: molecular evidence from a proteome study. Aust N Z J Psychiatry 2009; 43: 310–322. [DOI] [PubMed] [Google Scholar]
- Sivagnanasundaram S, Crossett B, Dedova I, Cordwell S, Matsumoto I. Abnormal pathways in the genu of the corpus callosum in schizophrenia pathogenesis: a proteome study. Proteomics Clin Appl 2007; 1: 1291–1305. [DOI] [PubMed] [Google Scholar]
- Pennington K, Beasley CL, Dicker P, Fagan A, English J, Pariante CM et al. Prominent synaptic and metabolic abnormalities revealed by proteomic analysis of the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder. Mol Psychiatry 2007; 13: 1102–1117. [DOI] [PubMed] [Google Scholar]
- Albertini V, Benussi L, Paterlini A, Glionna M, Prestia A, Bocchio-Chiavetto L et al. Distinct cerebrospinal fluid amyloid-beta peptide signatures in cognitive decline associated with Alzheimer’s disease and schizophrenia. Electrophoresis 2012; 33: 3738–3744. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Wobrock T, Zerr I, Schmitt A, Gawinecka J, Schneider-Axmann T et al. Different apolipoprotein E, apolipoprotein A1 and prostaglandin-H2 D-isomerase levels in cerebrospinal fluid of schizophrenia patients and healthy controls. World J Biol Psychiatry 2010; 11: 719–728. [DOI] [PubMed] [Google Scholar]
- Huang JTJ, Leweke FM, Oxley D, Wang L, Harris N, Koethe D et al. Disease biomarkers in cerebrospinal fluid of patients with first-onset psychosis. PLoS Med 2006; 3: e428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JT-J Wang L, Prabakaran S, Wengenroth M, Lockstone HE, Koethe D et al. Independent protein-profiling studies show a decrease in apolipoprotein A1 levels in schizophrenia CSF, brain and peripheral tissues. Mol Psychiatry 2007; 13: 1118–1128. [DOI] [PubMed] [Google Scholar]
- Schwarz E, Izmailov R, Spain M, Barnes A, Mapes JP, Guest PC et al. Validation of a blood-based laboratory test to aid in the confirmation of a diagnosis of schizophrenia. BMI 2010; 2010: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Guest PC, Rahmoune H, Harris LW, Wang L, Leweke FM et al. Identification of a biological signature for schizophrenia in serum. Mol Psychiatry 2011; 17: 494–502. [DOI] [PubMed] [Google Scholar]
- Levin Y, Wang L, Schwarz E, Koethe D, Leweke FM, Bahn S. Global proteomic profiling reveals altered proteomic signature in schizophrenia serum. Mol Psychiatry 2009; 15: 1088–1100. [DOI] [PubMed] [Google Scholar]
- Jaros JAJ, Martins-de-Souza D, Rahmoune H, Rothermundt M, Leweke FM, Guest PC et al. Protein phosphorylation patterns in serum from schizophrenia patients and healthy controls. J Proteomics 2012; 76: 43–55. [DOI] [PubMed] [Google Scholar]
- Domenici E, Willé DR, Tozzi F, Prokopenko I, Miller S, McKeown A et al. Plasma protein biomarkers for depression and schizophrenia by multi analyte profiling of case-control collections. PLoS One 2010; 5: e9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran S, Wengenroth M, Lockstone HE, Lilley K, Leweke FM, Bahn S. 2-D DIGE analysis of liver and red blood cells provides further evidence for oxidative stress in schizophrenia. J Proteome Res 2007; 6: 141–149. [DOI] [PubMed] [Google Scholar]
- Wang L, Lockstone HE, Guest PC, Levin Y, Palotás A, Pietsch S et al. Expression profiling of fibroblasts identifies cell cycle abnormalities in schizophrenia. J Proteome Res 2010; 9: 521–527. [DOI] [PubMed] [Google Scholar]
- Iavarone F, Melis M, Platania G, Cabras T, Manconi B, Petruzzelli R et al. Characterization of salivary proteins of schizophrenic and bipolar disorder patients by top-down proteomics. J Proteomics 2014; 103: 15–22. [DOI] [PubMed] [Google Scholar]
- Herberth M, Koethe D, Cheng TMK, Krzyszton ND, Schoeffmann S, Guest PC et al. Impaired glycolytic response in peripheral blood mononuclear cells of first-onset antipsychotic-naive schizophrenia patients. Mol Psychiatry 2010; 16: 848–859. [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Sanchez J-C, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF et al. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev 1996; 13: 19–50. [DOI] [PubMed] [Google Scholar]
- Kim M-S, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R et al. A draft map of the human proteome. Nature 2014; 509: 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM et al. Mass-spectrometry-based draft of the human proteome. Nature 2014; 509: 582–587. [DOI] [PubMed] [Google Scholar]
- Oliveira BM, Coorssen JR, Martins-de-Souza D. 2DE: the phoenix of proteomics. J Proteomics 2014; 104: 140–150. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 1999; 17: 676–682. [DOI] [PubMed] [Google Scholar]
- Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol 2006; 7: 391–403. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem 1975; 250: 4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Görg A, Postel W, Günther S, Weser J, Strahler JR, Hanash SM et al. Approach to stationary two-dimensional pattern: influence of focusing time and immobiline/carrier ampholytes concentrations. Electrophoresis 1988; 9: 37–46. [DOI] [PubMed] [Google Scholar]
- Bjellqvist B, Ek K, Righetti PG, Gianazza E, Görg A, Westermeier R et al. Isoelectric focusing in immobilized pH gradients: principle, methodology and some applications. J Biochem Biophys Methods 1982; 6: 317–339. [DOI] [PubMed] [Google Scholar]
- Unlü M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 1997; 18: 2071–2077. [DOI] [PubMed] [Google Scholar]
- Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem Rev 2001; 101: 269–296. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Guest PC, Vanattou-Saifoudine N, Harris LW, Bahn S. Proteomic technologies for biomarker studies in psychiatry: advances and needs. In: PCGAS Bahn ed. Biomarkers of Neurological and Psychiatric Disease. International Review of Neurobiology 2011, 65–94. [DOI] [PubMed]
- Guest PC, Schwarz E, Krishnamurthy D, Harris LW, Leweke FM, Rothermundt M et al. Altered levels of circulating insulin and other neuroendocrine hormones associated with the onset of schizophrenia. Psychoneuroendocrinology 2011; 36: 1092–1096. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Guest PC, Vanattou-Saifoudine N, Rahmoune H, Bahn S. Phosphoproteomic differences in major depressive disorder postmortem brains indicate effects on synaptic function. Eur Arch Psychiatry Clin Neurosci 2012; 262: 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou K, Zhang Z, Sun L, Yang J, Zhang M et al. Label-free quantitative proteomic analysis reveals dysfunction of complement pathway in peripheral blood of schizophrenia patients: evidence for the immune hypothesis of schizophrenia. Mol Biosyst 2012; 8: 2664–2671. [DOI] [PubMed] [Google Scholar]
- Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J Proteome Res 2002; 1: 317–323. [DOI] [PubMed] [Google Scholar]
- Levin Y, Bahn S. Quantification of proteins by label-free LC-MS/MS. In: Cutillas PR, Timms JF eds Methods in Molecular Biology. Humana Press: Totowa, NJ, USA, 2010, 217–231. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 2004; 3: 1154–1169. [DOI] [PubMed] [Google Scholar]
- Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002; 1: 376–386. [DOI] [PubMed] [Google Scholar]
- McClatchy DB, Liao L, Park SK, Xu T, Lu B, Yates Iii JR. Differential proteomic analysis of mammalian tissues using SILAM. PLoS One 2011; 6: e16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert S, Jesse S, Rist W, Steinacker P, Soininen H, Herukka S-K et al. iTRAQ and multiple reaction monitoring as proteomic tools for biomarker search in cerebrospinal fluid of patients with Parkinson’s disease dementia. Exp Neurol 2012; 234: 499–505. [DOI] [PubMed] [Google Scholar]
- Gokhale A, Larimore J, Werner E, So L, Moreno-De-Luca A, Lese-Martin C et al. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J Neurosci 32: 3697–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ML, Ciccimaro E, Prakash A, Banerjee A, Seeholzer SH, Blair IA et al. Biochemical fractionation and stable isotope dilution liquid chromatography-mass spectrometry for targeted and microdomain-specific protein quantification in human postmortem brain tissue. Mol Cell Proteomics 2012; 11: 1670–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods 2012; 9: 555–566. [DOI] [PubMed] [Google Scholar]
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics 2006; 5: 573–588. [DOI] [PubMed] [Google Scholar]
- Marx V. Targeted proteomics. Nat Methods 2012; 10: 19–22. [DOI] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 2008; 4: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling H, Guest PC, Lee C-M, Wong EH, Rahmoune H, Bahn S. Integrative proteomic analysis of the NMDA NR1 knockdown mouse model reveals effects on central and peripheral pathways associated with schizophrenia and autism spectrum disorders. Mol Autism 2014; 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-de-Souza D, Alsaif M, Ernst A, Harris LW, Aerts N, Lenaerts I et al. The application of selective reaction monitoring confirms dysregulation of glycolysis in a preclinical model of schizophrenia. BMC Res Notes 2012; 5: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy D, Harris LW, Levin Y, Koutroukides TA, Rahmoune H, Pietsch S et al. Targeted proteomics for validation of biomarkers in early psychosis. Biol Psychiatry 2014; 76: e7–e9. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Guest PC, Mann DM, Roeber S, Rahmoune H, Bauder C et al. Proteomic analysis identifies dysfunction in cellular transport, energy, and protein metabolism in different brain regions of atypical frontotemporal lobar degeneration. J Proteome Res 2012; 11: 2533–2543. [DOI] [PubMed] [Google Scholar]
- de Witte L, Tomasik J, Schwarz E, Guest PC, Rahmoune H, Kahn RS et al. Schizophrenia research. Schizophr Res 2014; 154: 23–29. [DOI] [PubMed] [Google Scholar]
- Guest PC, Wang L, Harris LW, Burling K, Levin Y, Ernst A et al. Increased levels of circulating insulin-related peptides in first-onset, antipsychotic naïve schizophrenia patients. Mol Psychiatry 2010; 15: 118–119. [DOI] [PubMed] [Google Scholar]
- Schwarz E, Guest PC, Steiner J, Bogerts B, Bahn S. Identification of blood-based molecular signatures for prediction of response and relapse in schizophrenia patients. Transl Psychiatry 2012; 2: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón-Pérez JM, Lu SC, Mato JM. Sub-proteome approach to the knowledge of liver. Prot Clin Appl 2010; 4: 407–415. [DOI] [PubMed] [Google Scholar]
- Wesseling H, Chan MK, Tsang TM, Ernst A, Peters F, Guest PC et al. A combined metabonomic and proteomic approach identifies frontal cortex changes in a chronic phencyclidine rat model in relation to human schizophrenia brain pathology. Neuropsychopharmacology 2013; 38: 2532–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ. Using our brains: the findings, flaws, and future of postmortem studies of psychiatric disorders. Biol Psychiatry 2011; 69: 102–103. [DOI] [PubMed] [Google Scholar]
- Schwarz E, van Beveren NJM, Ramsey J, Leweke FM, Rothermundt M, Bogerts B et al. Identification of subgroups of schizophrenia patients with changes in either immune or growth factor and hormonal pathways. Schizophr Bull 2013; 40: 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-de-Souza D, Gattaz WF, Schmitt A, Rewerts C, Marangoni S, Novello JC et al. Alterations in oligodendrocyte proteins, calcium homeostasis and new potential markers in schizophrenia anterior temporal lobe are revealed by shotgun proteome analysis. J Neural Transm 2008; 116: 275–289. [DOI] [PubMed] [Google Scholar]
- Bergson C, Levenson R, Goldman-Rakic PS, Lidow MS. Dopamine receptor-interacting proteins: the Ca(2+) connection in dopamine signaling. Trends Pharmacol Sci 2003; 24: 486–492. [DOI] [PubMed] [Google Scholar]
- Raiszadeh MM, Ross MM, Russo PS, Schaepper MA, Zhou W, Deng J et al. Proteomic analysis of eccrine sweat: implications for the discovery of schizophrenia biomarker proteins. J Proteome Res 2012; 11: 2127–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner J, Schmitt A, Schroeter ML, Bogerts B, Falkai P, Turck CW et al. S100B is downregulated in the nuclear proteome of schizophrenia corpus callosum. Eur Arch Psychiatry Clin Neurosci 2014; 264: 311–316. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Harris LW, Guest PC, Bahn S. The role of energy metabolism dysfunction and oxidative stress in schizophrenia revealed by proteomics. Antioxid Redox Signal 2011; 15: 2067–2079. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Guest PC, Rahmoune H, Bahn S. Proteomic approaches to unravel the complexity of schizophrenia. Expert Rev Proteomics 2012; 9: 97–108. [DOI] [PubMed] [Google Scholar]
- Spelman LM, Walsh PI, Sharifi N, Collins P, Thakore JH. Impaired glucose tolerance in first-episode drug-naïve patients with schizophrenia. Diabetic Med 2007; 24: 481–485. [DOI] [PubMed] [Google Scholar]
- Behan ÁT, Byrne C, Dunn MJ, Cagney G, Cotter DR. Proteomic analysis of membrane microdomain-associated proteins in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder reveals alterations in LAMP, STXBP1 and BASP1 protein expression. Mol Psychiatry 2008; 14: 601–613. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Guest PC, Harris LW, Vanattou-Saifoudine N, Webster MJ, Rahmoune H et al. Identification of proteomic signatures associated with depression and psychotic depression in post-mortem brains from major depression patients. Transl Psychiatry 2012; 2: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa F, Malavasi ELV, Crummie DK, Eykelenboom JE, Soares DC, Mackie S et al. DISC1 complexes with TRAK1 and Miro1 to modulate anterograde axonal mitochondrial trafficking. Hum Mol Genet 2014; 23: 906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y-U, Jeong J, Lee H, Mun JY, Kim J-H, Lee JS et al. Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc Natl Acad Sci USA 2010; 107: 17785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji B, La Y, Gao L, Zhu H, Tian N, Zhang M et al. A comparative proteomics analysis of rat mitochondria from the cerebral cortex and hippocampus in response to antipsychotic medications. J Proteome Res 2009; 8: 3633–3641. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M, Brenner-Lavie H, SG-B Ari, Kavushansky A, Ben-Shachar D. Perturbation in mitochondrial network dynamics and in complex I dependent cellular respiration in schizophrenia. Biol Psychiatry 2011; 69: 980–988. [DOI] [PubMed] [Google Scholar]
- Karry R, Klein E, Ben-Shachar D. Mitochondrial complex I subunits expression is altered in schizophrenia: a postmortem study. Biol Psychiatry 2004; 55: 676–684. [DOI] [PubMed] [Google Scholar]
- Brenner-Lavie H, Klein E, Ben-Shachar D. Mitochondrial complex I as a novel target for intraneuronal DA: modulation of respiration in intact cells. Biochem Pharmacol 2009; 78: 85–95. [DOI] [PubMed] [Google Scholar]
- Bitanihirwe BKY, Woo T-UW. Oxidative stress in schizophrenia: an integrated approach. Neurosci Biobehav Rev 2011; 35: 878–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Guest PC, Rahmoune H, Martins-de-Souza D, Niebuhr DW, Weber NS et al. Identification of a blood-based biological signature in subjects with psychiatric disorders prior to clinical manifestation. World J Biol Psychiatry 2012; 13: 627–632. [DOI] [PubMed] [Google Scholar]
- Awam Al K, Haußleiter IS, Dudley E, Donev R, Brüne M, Juckel G et al. Multiplatform metabolome and proteome profiling identifies serum metabolite and protein signatures as prospective biomarkers for schizophrenia. J Neural Transm 2014;:1–12. [DOI] [PubMed]
- Baumgartner R, Umlauf E, Veitinger M, Guterres S, Rappold E, Babeluk R et al. Identification and validation of platelet low biological variation proteins, superior to GAPDH, actin and tubulin, as tools in clinical proteomics. J Proteomics 2013; 94: 540–551. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Dias-Neto E, Schmitt A, Falkai P, Gormanns P, Maccarrone G et al. Proteome analysis of schizophrenia brain tissue. World J Biol Psychiatry 2010; 11: 110–120. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Gattaz WF, Schmitt A, Novello JC, Marangoni S, Turck CW et al. Proteome analysis of schizophrenia patients Wernicke’s area reveals an energy metabolism dysregulation. BMC Psychiatry 2009; 9: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N. The role of anti-inflammatory treatment in psychiatric disorders. Psychiatr Danub 2013; 25: 292–298. [PubMed] [Google Scholar]
- Söderlund J, Schröder J, Nordin C, Samuelsson M, Walther-Jallow L, Karlsson H et al. Activation of brain interleukin-1β in schizophrenia. Mol Psychiatry 2009; 14: 1069–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JTJ, Leweke FM, Tsang TM, Koethe D, Kranaster L, Gerth CW et al. CSF metabolic and proteomic profiles in patients prodromal for psychosis. PLoS One 2007; 2: e756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroutunian V, Katsel P, Roussos P, Davis KL, Altshuler LL, Bartzokis G. Myelination, oligodendrocytes, and serious mental illness. Glia 2014; 62: 1856–1877. [DOI] [PubMed] [Google Scholar]
- Nave K-A, Ehrenreich H. Myelination and oligodendrocyte functions in psychiatric diseases. JAMA Psychiatry 2014; 71: 582–584. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D. Proteome and transcriptome analysis suggests oligodendrocyte dysfunction in schizophrenia. J Psychiatr Res 2010; 44: 149–156. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D. Proteomics tackling schizophrenia as a pathway disorder. Schizophr Bull 2012; 38: 1107–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Aebersold R, Archakov A, Bairoch A, Bala K, Beretta L et al. The Human Proteome Project: current state and future direction. Mol Cell Proteomics 2011; 10: M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko-Varga G, Omenn GS, Paik Y-K, Hancock WS. A first step toward completion of a genome-wide characterization of the human proteome. J Proteome Res 2013; 12: 1–5. [DOI] [PubMed] [Google Scholar]
- Paik Y-K, Jeong S-K, Omenn GS, Uhlén M, Hanash S, Cho SY et al. The Chromosome-Centric Human Proteome Project for cataloging proteins encoded in the genome. Nat Biotechnol 2012; 30: 221–223. [DOI] [PubMed] [Google Scholar]
- Meyer HE, Hamacher M. Quintessence from proteomics networks—the HUPO Brain Proteome Project Pilot Studies. Proteomics 2006; 6: 4887–4889. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Carvalho PC, Schmitt A, Junqueira M, Nogueira FCS, Turck CW et al. Deciphering the human brain proteome: characterization of the anterior temporal lobe and corpus callosum as part of the Chromosome 15-centric Human Proteome Project. J Proteome Res 2014; 13: 147–157. [DOI] [PubMed] [Google Scholar]
- Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics 2011; 10: M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J, Krijgsveld J. Proteomic analysis of cell fate decision. Curr Opin Genet Dev 2013; 23: 540–547. [DOI] [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV. The nature of systems biology. Trends Microbiol 2007; 15: 45–50. [DOI] [PubMed] [Google Scholar]
- Breker M, Schuldiner M. The emergence of proteome-wide technologies: systematic analysis of proteins comes of age. Nat Rev Mol Cell Biol 2014; 15: 453–464. [DOI] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011; 473: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen BDS, Maciel R de M, Galina A, da Silveira MS, Souza CDS, Drummond H et al. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant 2012; 21: 1547–1559. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–872. [DOI] [PubMed] [Google Scholar]
- Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports 2014; 2: 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME et al. Cerebral organoids model human brain development and microcephaly. Nature 2013; 501: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy D, Harris LW, Levin Y, Koutroukides TA, Rahmoune H, Pietsch S et al. Metabolic, hormonal and stress-related molecular changes in post-mortem pituitary glands from schizophrenia subjects. World J Biol Psychiatry 2013; 14: 478–489. [DOI] [PubMed] [Google Scholar]


