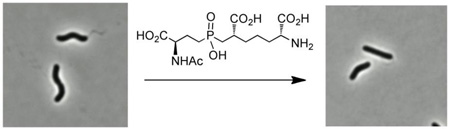
Abstract
Helicobacter pylori and Campylobacter jejuni are human pathogens and causative agents of gastric ulcers/cancer and gastroenteritis, respectively. Recent studies have uncovered a series of proteases that are responsible for maintaining the helical shape of these organisms. The H. pylori metalloprotease Csd4, and its C. jejuni homologue Pgp1, cleave the amide bond between meso-diaminopimelate and iso-d-glutamic acid in truncated peptidoglycan side chains. Deletion of either csd4 or pgp1 results in bacteria with a straight rod phenotype, a reduced ability to move in viscous media, and reduced pathogenicity. In this work, a phosphinic acid-based pseudodipeptide inhibitor was designed to act as a tetrahedral intermediate analog against the Csd4 enzyme. The phosphinic acid was shown to inhibit the cleavage of the alternate substrate, Ac-l-Ala-iso-d-Glu-meso-Dap with a Ki value of 1.5 µM. Structural analysis of the Csd4-inhibitor complex shows that the phosphinic acid displaces the zinc-bound water and chelates the metal in a bidentate fashion. The phosphinate oxygens also interact with the key acid/base residue, Glu222, and the oxyanion-stabilizing residue, Arg86. The results are consistent with the "promoted-water pathway" mechanism for carboxypeptidase A catalysis. Studies on cultured bacteria showed that the inhibitor causes significant cell straightening when incubated with H. pylori at millimolar concentrations. A diminished, yet observable, effect on the morphology of C. jejuni was also apparent. Cell straightening was more pronounced with an acapsular C. jejuni mutant strain compared to the wild type, suggesting that the capsule impaired inhibitor accessibility. These studies demonstrate that a highly polar compound is capable of crossing the outer membrane, inhibiting cell shape determinant proteases and altering cell shape, thereby affecting the pathogenic attributes of these organisms. Peptidoglycan proteases acting as cell shape determinants represent novel targets for the development of antimicrobials against these human pathogens.
Graphical abstract
INTRODUCTION
Peptidoglycan is a key component of the bacterial cell wall that acts as a rigid barrier essential for protecting the bacteria from lysis due to osmotic pressure, and for maintaining bacterial cell shape.1–6 It is composed of glycan polymers of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues that are crosslinked via peptides attached to the lactate side chain of the MurNAc units. The importance of peptidoglycan in bacterial physiology is highlighted by the fact that many clinically used antibiotics target enzymes involved in the biosynthesis of this macromolecule.7
Helicobacter pylori and Campylobacter jejuni are Gram-negative, helical shaped bacteria that colonize the human gastrointestinal tract. H. pylori is the causative agent of gastric ulcers and has been linked to the development of gastric cancer.8 C. jejuni is a leading cause of human gastroenteritis worldwide and has been linked to the development of the debilitating Guillain-Barré syndrome in infected individuals.9 In recent years, a series of genes have been identified that are responsible for maintaining the helical cell shape in these bacteria.5,6 In the case of H. pylori they have been named cell shape-determinant genes or csds.10–13 Most of these genes have been found to encode for either endo- or exo-proteases that act on the peptide chains of peptidoglycan. Similar genes have been identified in C. jejuni and have been named peptidoglycan peptidases or pgps.14,15 Deletion of these genes in both organisms leads to abnormal cell shapes including straight rods, curved rods, "c" shaped cells, and stocky/branched phenotypes.
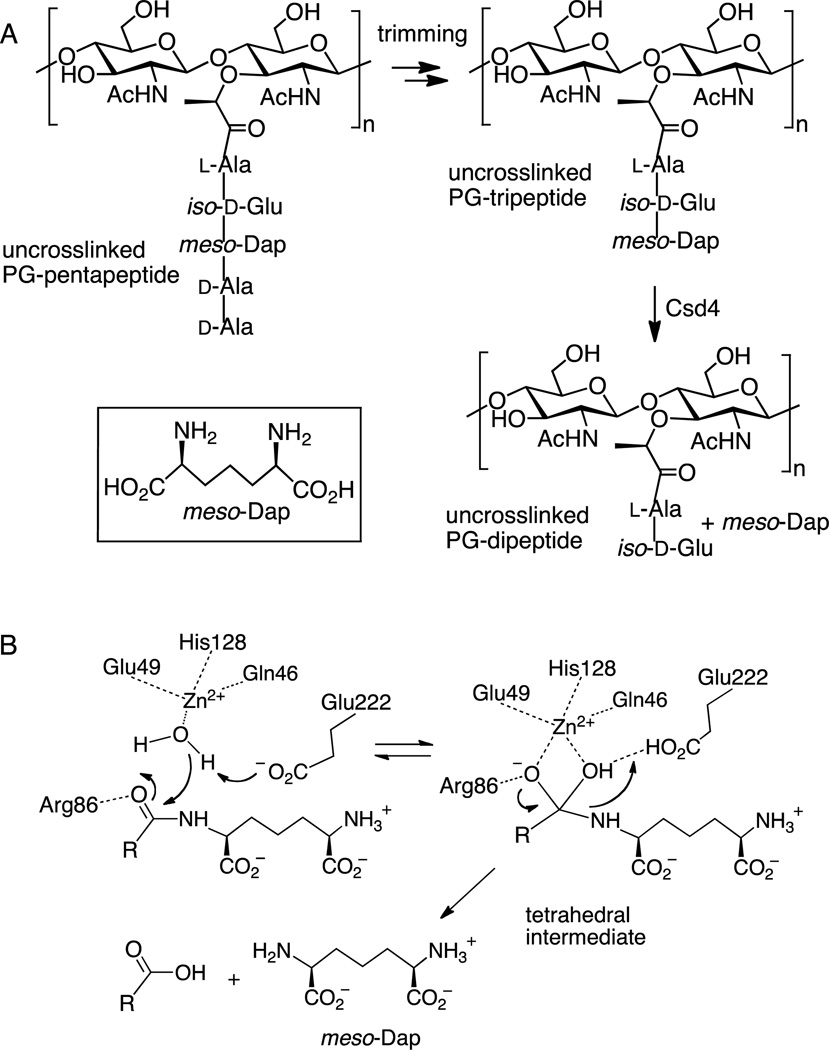
This study focuses on the H. pylori enzyme Csd4 that plays a role in trimming uncrosslinked peptidoglycan peptide chains (Figure 1A).11,16,17 During peptidoglycan biosynthesis, each MurNAc residue bears a pentapeptide chain with the structure l-Ala-iso-d-Glu-meso-Dap-d-Ala-d-Ala. The amino group in the side chain of meso-diaminopimelic acid (meso-Dap) is responsible for attacking a neighboring chain in the transpeptidase-catalyzed crosslinking reaction that is crucial for the structural integrity of the cell wall. In H. pylori, however, many of the pentapeptide chains remain uncrosslinked and can be "trimmed" by a series of proteases that sequentially remove the C-terminal residues. Csd4 has been found to be responsible for hydrolyzing the bond between meso-Dap and iso-d-Glu in the trimming of a peptidoglycan tripeptide to a dipeptide.11,16,17 As the dipeptide no longer bears a meso-Dap residue, it can no longer participate in a crosslinking event. Deletion of csd4 does not impair growth rate, but results in a straight rod phenotype.11 Mutant strains lacking this gene have impaired motility in viscous media and show markedly reduced stomach colonization.11 It has been postulated that the lack of helical structure results in a reduced ability to traverse the gastric mucus, impairing H. pylori’s ability to escape the low pH environment of the stomach and colonize the mucus covered gastric epithelial surface. While the exact link between the activity of Csd4 and the maintenance of helical shape remains unknown, it is thought that localized control of the extent of crosslinking plays a key role.
Figure 1.
A) Peptidoglycan trimming and the reaction catalyzed by Csd4. The inset shows the structure of meso-Dap. B) The proposed catalytic mechanism for the Csd4 reaction.
A Csd4 homolog has also been identified in C. jejuni and has been named Pgp1.15 Deletion of pgp1 also results in a straight rod phenotype and a motility defect, as well as biofilm defects, an impaired ability to colonize chicks, and altered host-pathogen interactions. Activity assays have confirmed that this enzyme also acts as a carboxypeptidase and cleaves the bond between iso-Glu and meso-Dap.
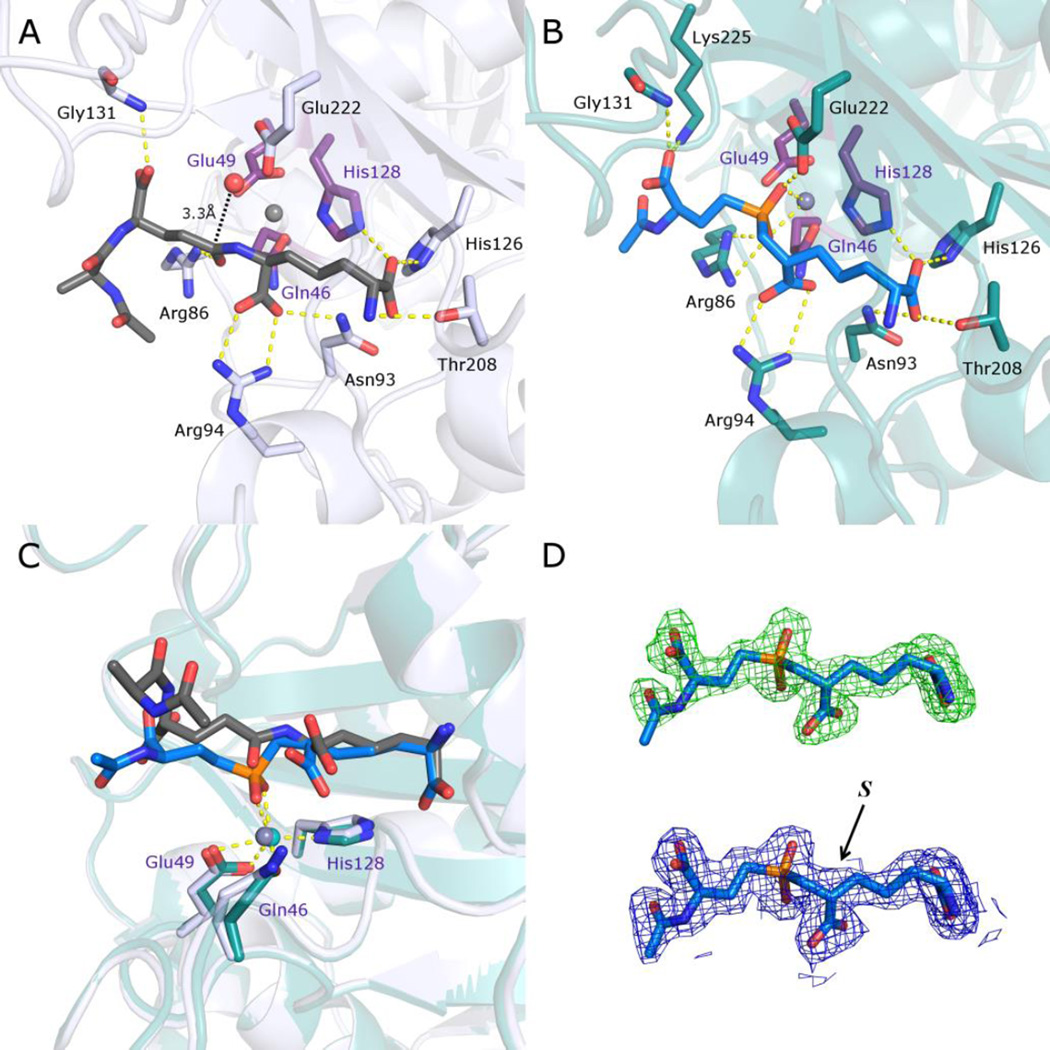
Recent crystallographic studies on Csd4 have confirmed that it is a member of the M14 family of zinc dependent proteases that contain the prototypical member carboxypeptidase A.16,17 The Csd4 enzyme bears an N-terminal domain with a typical carboxypeptidase fold as well as two additional domains of unknown function. The enzyme has been shown to accept peptidoglycan fragments (MurNAc-GlcNAc tripeptide and N-acetylated tripeptide) as alternate substrates (where the tripeptide is l-Ala-iso-d-Glu-meso-Dap). Structures of Csd4 in complex with the MurNAc tripeptide and the acetylated tripeptide are available and provide an informative model of the Michaelis complex. The iso-d-Glu-meso-Dap portion of these substrates is buried within the active site of the carboxypeptidase domain and makes extensive interactions with active site residues (Figure 2A). The MurNAc-l-Ala or Ac-l-Ala portions protrude outside of the active site and make few contacts. The active site zinc is coordinated by a rare His-Glu-Gln motif that differs from the normal His-Glu-His arrangement found in other M14 carboxypeptidases.18 A zinc-bound water is poised appropriately for attack on the iso-Glu γ-carbonyl and is hydrogen bonded to Glu222. This glutamate is thought to serve as the general base that deprotonates the water in the proposed catalytic mechanism (Figure 1B).19,20 Arg86 is hydrogen bonded to the iso-Glu γ-carbonyl and serves to stabilize the anionic tetrahedral intermediate. Breakdown of the tetrahedral intermediate presumably involves protonation of the departing meso-Dap by Glu222. Overall, the active site of Csd4 closely resembles that of other family M14 carboxypeptidases with the exception of the unusual metal ligands.
Figure 2.
Comparison of structures of Csd4 in complex with a N-acetyltripeptide substrate and inhibitor 1. A) The structure of N-acetyltripeptide complexed with Csd4 (produced using PDB ID: 4WCN). The substrate carbon atoms are in dark grey; selected Csd4-substrate interactions are shown as yellow dotted lines; zinc ligands are colored purple; carbon atoms of residues interacting with the tripeptide are colored white; the predicted catalytic water is red; zinc is grey. B) Structure of the Csd4-inhibitor 1 complex. The bound inhibitor (carbon atoms in blue) with key active site residues are highlighted. Carbon atoms of selected amino acid residues that interact with the inhibitor are shown in teal. C) Superposition of the structures of Csd4 with bound substrate and inhibitor 1. The substrate, inhibitor 1 and zinc ligands are shown as in panels A and B. D) Top, the initial omit Fo−Fc map contoured at 3.5 σ prior to modelling of the inhibitor, and below, the final refined 2Fo−Fc map of the inhibitor contoured at 1.5 σ. In both maps, the final refined inhibitor model is included for visualization purposes. The (S)-configuration of the inhibitor stereocenter that is closest to the phosphinate is labeled. In all panels, oxygen, nitrogen and phosphorus atoms are shown in red, blue and orange, respectively.
In this study we report on the synthesis of a phosphinic acid-based inhibitor of Csd4 that mimics the tetrahedral intermediate formed during catalysis. A structure of the resulting Csd4-inhibitor complex is presented and studies with both H. pylori and C. jejuni demonstrate that the inhibitor is effective in promoting cell straightening in living cells.
RESULTS
Inhibitor Design and Synthesis
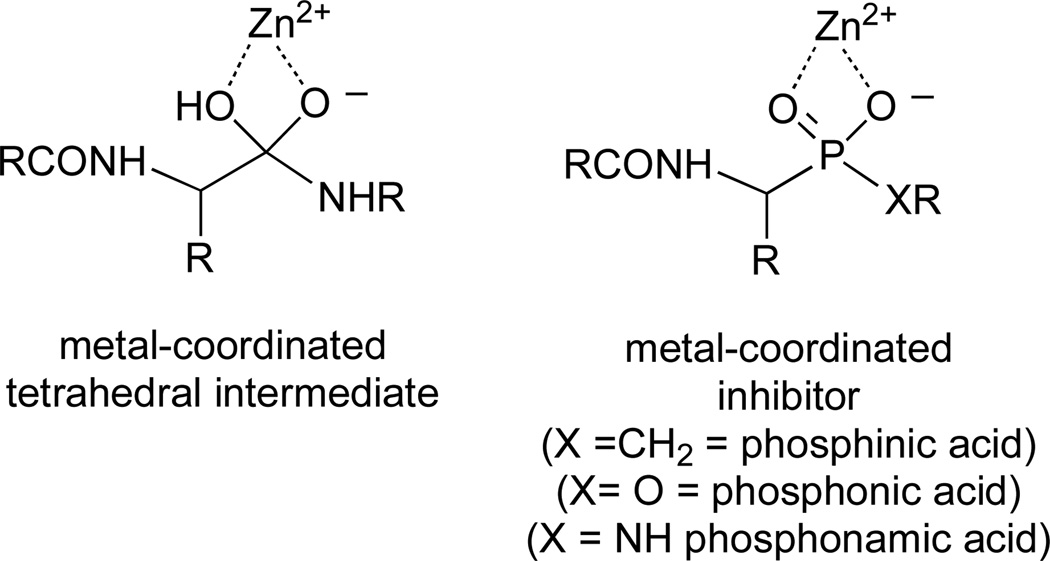
A strategy that has been successfully used in the design of reversible mechanism-based inhibitors of metalloproteases is to prepare phosphorus-containing peptide analogs (Figure 3).21–23 The negative charge and tetrahedral geometry about the phosphorus allow for a bidentate metal-chelation that closely resembles features of the bound transition state or tetrahedral intermediate formed during catalysis. Potent inhibition of metalloproteases has been observed with phosphinic acids (X = CH2), phosphonic acids (X = O) and phosphonamic acids (X = NH). In most cases, inhibition constants are in the low micromolar or nanomolar range; however, in the case of carboxypeptidase A, inhibition constants have been reported to be in the femtomolar range.24
Figure 3.
A metal-coordinated tetrahedral intermediate and the corresponding phosphorous-based inhibitors.
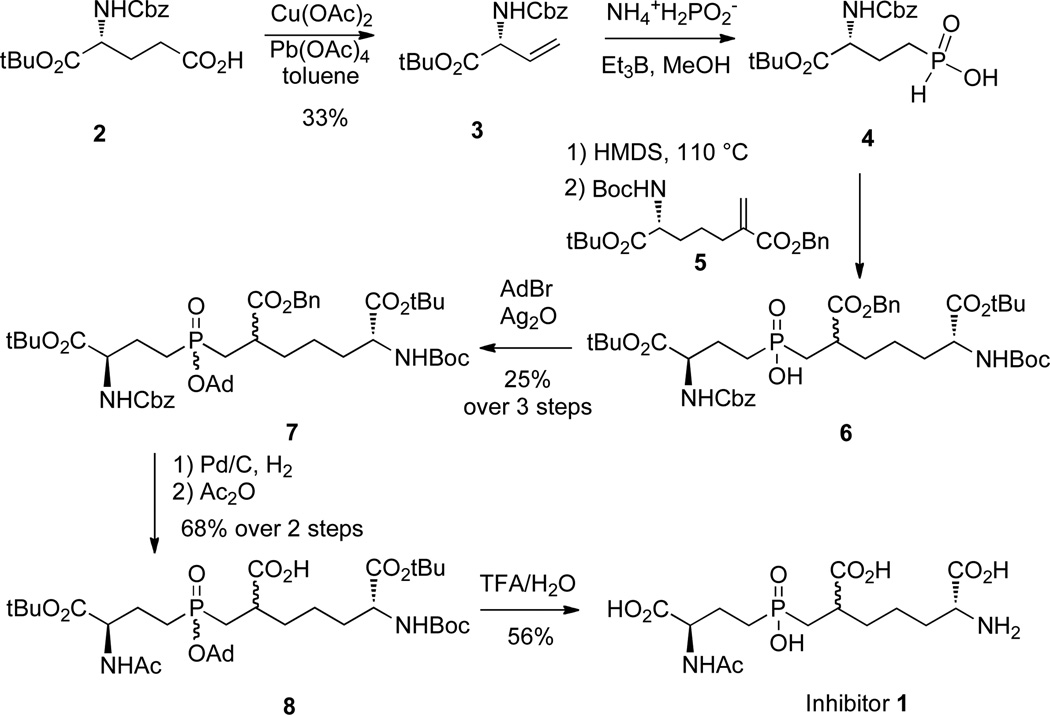
Our initial strategy for the design of a Csd4 inhibitor was to prepare inhibitor 1, which contains a phosphinic acid in place of the amide bond linking iso-d-Glu and meso-Dap (Scheme 1). The choice of a phosphinic acid for the first target was due to the ease of synthesis and the stability of the compound towards hydrolysis. As the structural studies showed that the l-Ala moiety of the N-acetylated tripeptide substrate made few contacts with the active site of Csd4,16,17 this residue was replaced by a simple acetylation of the "d-Glu" α-amino group of the phosphinic acid. A commercially available derivative of d-Glu, compound 2, was oxidatively decarboxylated to give alkene 3 and then converted into the phosphinic acid 4 by treatment with ammonium hypophosphite and triethylborane.25,26 A hexamethyldisilane-mediated conjugate addition of compound 4 to the literature known benzylacrylate 5 provided the phosphinic acid 6,27,28 which was not purified, but was immediately protected with adamantyl bromide to give compound 7 as a mixture of four stereoisomers. Hydrogenolysis removed both the benzyl ester and carboxybenzyl protecting groups and treatment with acetic anhydride gave the N-acetylated compound 8. A final deprotection with trifluoroacetic acid, followed by anion exchange chromatography, provided the target inhibitor 1 as a mixture of two diastereomers (approx. 1:1 ratio). No attempts were made to separate these diastereomers and the mixture was directly used in the inhibition and crystallographic studies.
Scheme 1.
The synthesis of inhibitor 1.
Enzyme Kinetics
A continuous coupled assay for Csd4 activity was developed that was based on a published assay for meso-Dap formed in the reaction catalyzed by diaminopimelate epimerase.16,29 For this assay, His-tagged diaminopimelate dehydrogenase (DAPDH) from Corynebacterium glutamicum was overproduced in Escherichia coli and purified by immobilized metal affinity chromatography for use as a coupling enzyme.16 NADH formed from the oxidation of meso-Dap by DAPDH was monitored by UV spectroscopy. This assay was used to determine the kinetic constants for the N-acetylated tripeptide substrate (Ac- l-Ala-iso-d-Glu-meso-Dap) in the presence of 30 µM zinc (pH 6.5) and the values of kcat = 0.015 ± 0.001 s−1 and KM = 112 ± 5 µM were obtained (Figure S1). The low value of kcat may reflect the fact that the substrate is truncated and is not associated with the peptidoglycan polymer. Additionally, it has been postulated that the two domains of unknown function may bind to other Csd proteins, or to peptidoglycan, and the absence of these partners in an in vitro analysis could affect Csd4 activity.16,17 The value of KM was reasonably low, which is consistent with the observation that contacts within the active site are primarily made with the iso-d-Glu-meso-Dap portion of the substrate and the rest of the polymeric structure protrudes into solution.
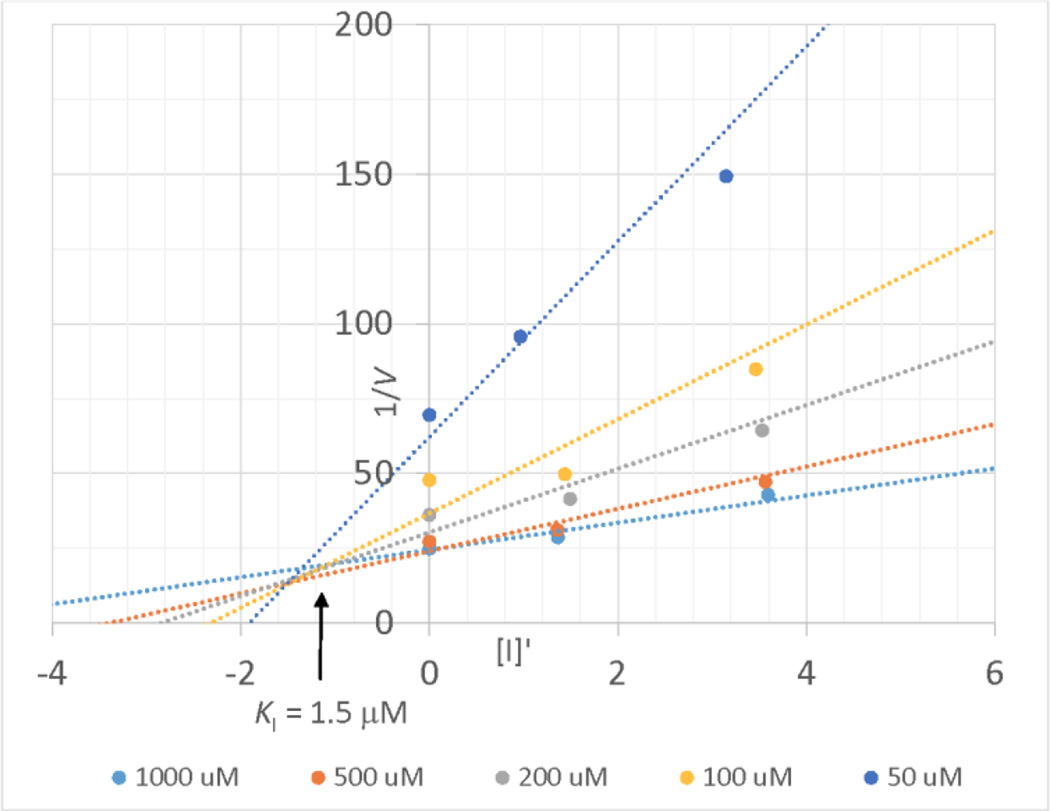
The inhibition of Csd4 by inhibitor 1 was investigated using the same coupled assay. Slow binding inhibition was observed so the reaction mixtures were preincubated for 20 minutes prior to initiation with substrate. Initial studies showed that the inhibitor was active in the low micromolar range. Due to the low activity of Csd4, the enzyme concentration was also in the low micromolar range. In order to account for a scenario where the inhibitor is not in vast excess of the enzyme, the correction factor of Singh et al. was employed that corrects the free inhibitor concentration using the enzyme concentration and the relative reaction rates in the presence and absence of inhibitor.30 Given the precedence for such inhibitors to act as transition state analogs of carboxypeptidase A,23 and the structure of the Csd4-inhibitor complex (vide infra), inhibitor 1 was assumed to act as a competitive inhibitor. Since the use of the correction factor led to varying inhibitor concentrations, a Dixon plot was employed that showed intersecting lines corresponding to a Ki value of 1.5 ± 0.3 µM (Figure 4). This value is considerably lower than the KM value of the substrate (112 µM) as expected for a compound that acts as a tetrahedral intermediate analog.
Figure 4.
A Dixon plot showing the inhibition of Csd4 by inhibitor 1. The value of [I'] was adjusted to account for enzyme-bound inhibitor.30
Structural Analysis of Csd4-Inhibitor Complex
Crystals of Csd4 were grown in Tris buffer (pH 8) containing sodium iodide and 13–18% PEG 3350, as described previously.16 These crystals were then sequentially soaked with solutions of ZnCl2, inhibitor 1, and cryoprotectant. A dataset to 1.9 Å resolution was collected and the structure factors were directly refined against the apo-Csd4 crystal structure. The overall structure of the Csd4-inhibitor complex was very similar to that of the Csd4-tripeptide complex, with the only notable differences occurring in the active site of the carboxypeptidase domain (Figure 2B). Inspection of the bound inhibitor indicated that it had an (S)-configuration at the stereocenter closest to the phosphinic acid (Figure 2D). This mirrors the (S)-configuration of the Dap stereocenter that is attached to iso-Glu in the normal substrate, and indicates that the enzyme has selected the appropriate stereoisomer from solution. Both oxygen atoms of the phosphinic acid are coordinated with the zinc atom (Zn-O distances = 2.4 and 2.3 Å) and the zinc bound water molecule has been displaced. This is as expected for an inhibitor that mimics the transition state or tetrahedral intermediate formed during catalysis.23 When compared to the structure of the bound tripeptide substrate, a twisting of the peptide backbone has occurred that allows for this coordination to take place (Fig. 2C). It should be noted that in the structure of the bound tripeptide substrate, the carbonyl of the iso-Glu side chain amide (the site of hydrolysis) is not coordinated to the zinc atom (Zn-O distance = 3.8 Å, Figure 2A).16,17 This implies that the mechanism proposed by Christianson and Lipscomb is operative, in which the metal acts to acidify the nucleophilic water and stabilize the tetrahedral intermediate, but does not directly activate the carbonyl of the substrate by electrophilic catalysis (Figure 1B).19,20 As expected, the key acid/base catalyst Glu222 that serves to deprotonate the nucleophilic water molecule is hydrogen bonded to a phosphinic acid oxygen of inhibitor 1. Similarly, Arg86 that stabilizes the oxyanion of the tetrahedral intermediate interacts with the other phosphinic acid oxygen. Most other interactions are similar to those seen with the tripeptide substrate with the exception of Lys225 that has moved to form a hydrogen bond with the carboxylate of the "iso-Glu portion" of the inhibitor (N-O distance = 2.8 Å; as compared to 3.4 Å in the tripeptide bound structure) and contributes additional stabilization of the tetrahedral intermediate. Overall, the structure provides an excellent model for the tetrahedral intermediate complex and strongly supports the mechanism outlined in Figure 1B.
Testing of Inhibitor 1 with Cultured Bacteria
Given the phenotypic change in cell shape observed upon disruption of the Csd4 gene in H. pylori,11 it was anticipated that inhibitor 1 may cause similar cell straightening provided it could cross the outer membrane to target Csd4, which is localized in the periplasm. In addition, it was anticipated that inhibitor 1 would also be active against Pgp1 of C. jejuni and cause cell straightening in this organism.
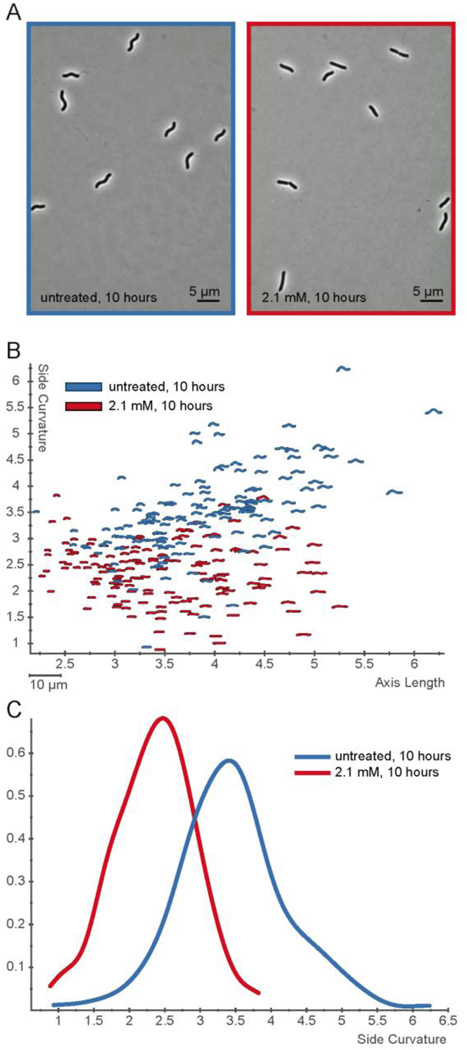
We first tested H. pylori strain J99 with exposure to varying concentration of inhibitor 1 for 24 hours. At lower concentrations (2.5 mM) we observed cell straightening, and at higher concentrations (>4.5 mM) cell growth was inhibited. To better quantify subtle differences in shape, we used H. pylori strain KBH19 (a derivative of the human isolate G27),46 which has higher intrinsic cell curvature than J99. We performed a time course of exposure at 2.1 mM inhibitor 1 using phase contrast microscopy coupled with quantitative image analysis of 100–200 cells at each time point using CellTool. By 10 hours (four doublings) the entire culture showed nearly straight morphology in the presence of inhibitor 1 (Figure 5A–B). A histogram showing the fractional cell population as a function of side curvature shows the loss of curvature after 10 hours of treatment with 2.1 mm inhibitor 1 in comparison to untreated cells (Figure 5C, p<0.00001). A histogram of a time course of the treatment shows that the cell morphology of the untreated population shifts toward lower curvature as the bacteria approach stationary phase (Figure S10A), but even after 2.5 hours (approximately one doubling time) inhibitor 1 treated cells show a significantly higher proportion of cells with lower curvature than untreated cells. This effect becomes more pronounced at subsequent time points (Figure S10B, K-S statistics for comparison of untreated to treated samples at 2.5 hours, p = 0.044; 5, 7.5, and 10 hours, p<0.00001). The observation of cell straightening indicates that the polar inhibitor is able to cross the outer membrane and reach the peptidoglycan layer in the periplasm where it acts to inhibit Csd4. The cause of the growth inhibition due to inhibitor 1 is not clear, as deletion of Csd4 is not lethal to H. pylori.11 It is possible that at these concentrations, it can cross the inner membrane and inhibit the cytosolic meso-Dap adding ligase (MurE) that is required for peptidoglycan biosynthesis.31
Figure 5.
Inhibitor 1 alters H. pylori cell shape. A) Phase contrast images of H. pylori (KBH19) treated for 10 hours without (blue, left) or with 2.1 mM inhibitor 1 (red, right). B) Scatterplot of 100–200 cell contours per condition from phase contrast images of cells grown for 10 hours with 2.1 mM inhibitor 1 (red) or without (blue). Axis length is plotted on the x-axis and side curvature is plotted on the y-axis. C) Smooth histogram of population side curvature values for cells treated for 10 hours without (blue) and with 2.1 mM inhibitor 1 (red). Treated cells have significantly lower side curvatures than u––––ntreated cells (p<0.00001).
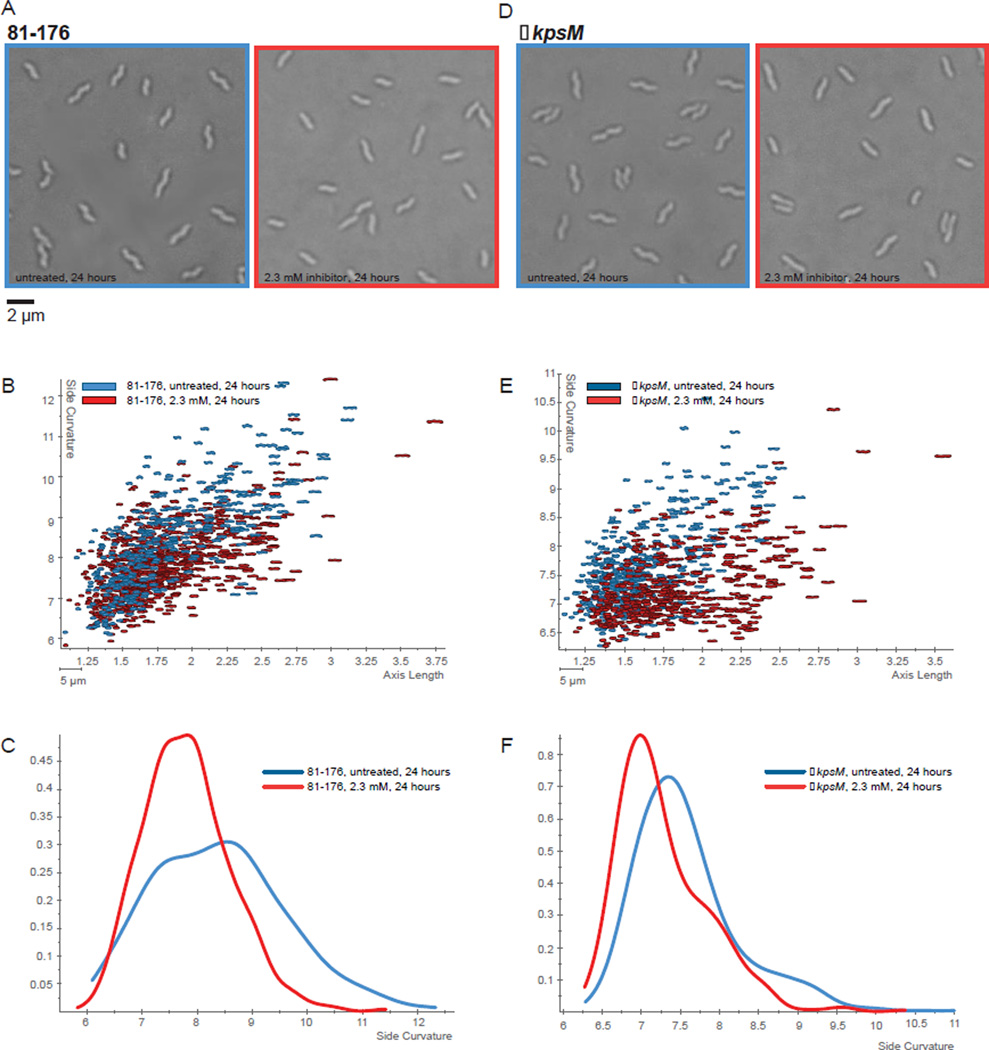
A similar study was performed with C. jejuni 81–176 cells. Unlike H. pylori, which is not encapsulated, wild type C. jejuni is coated with a carbohydrate capsule that provides an additional barrier to polar compounds. For this reason inhibitor 1 was tested against both the wild type and the acapsular mutant strain C. jejuni ΔkpsM. The kpsM gene encodes an ATP-binding cassette transporter protein required for the transport of capsular polysaccharide across the inner membrane. C. jejuni cells were grown in both the presence and the absence of varying concentrations of inhibitor 1 and visualized using DIC microscopy. Similar to the observations with H. pylori, cell straightening was observed after 24 hours of incubation with a maximal effect at a concentration of 2.3 mM (Figure 6A, 81–176 and 7D, ΔkpsM). At higher concentrations of inhibitor 1 (>4.5 mM), growth inhibition was observed. Cell straightening was more pronounced with the C. jejuni ΔkpsM strain than with the wild type, indicating that the presence of a capsule hindered the ability of the inhibitor to cross the outer membrane. Scatter plots of side curvature vs. axis length (Figures 6B, 81–176 and 6E, ΔkpsM) as well as histograms of fractional cell population as a function of side curvature (Figure 6C, 81–176 and 6F, ΔkpsM) confirm that the cell straightening was statistically significant. Similar effects were also seen with the C. jejuni strains 11168-O and 81116 (data not shown). The effects were less pronounced in C. jejuni as compared to H. pylori, particularly in the case of the wild type strain. This is evident from the smaller magnitudes of the shift in the maximal fractional cell populations in the histograms. Nevertheless, it is clear that inhibitor 1 is promoting cell straightening in these bacteria, indicating that this compound is also active against the Pgp1 carboxypeptidase.15 The reduced magnitude of the effect between the two species may be due either to a lower affinity to Pgp1 as compared to Csd4, or to difficulties in crossing the outer membrane in C. jejuni strains, which have a different LPS structure.
Figure 6.
C. jejuni wild type 81–176 and ΔkpsM show cell straightening in the presence of inhibitor 1. DIC microscopy images of C. jejuni 81–176 (A) and the acapsular ΔkpsM (D) without inhibitor (left) and treated with 2.3 mM inhibitor for 24h (right). Scatter plots arraying cell length (x-axis, µm) and side curvature (y-axis, arbitrary units) for 81–176 (B) and ΔkpsM (E) grown for 24 h with and without inhibitor 1. Each contour represents the morphology of a single cell from a 1000× DIC image as determined using CellTool software. Smooth histograms displaying population side curvature (x-axis, arbitrary units) as a density function (y-axis) for 81–176 (C) and ΔkpsM (F) grown for 24 h with and without inhibitor 1. Kolmogorov–Smirnov statistical comparisons of population cell side curvature distributions indicate that treated cells have significantly lower side curvatures than untreated cells (81–176, p<0.00001; ΔkpsM, p<0.00001, with p<0.05 indicating significance).
DISCUSSION
This study describes the design and synthesis of a phosphinic acid-based inhibitor against the cell shape determinant enzyme Csd4 from H. pylori. The inhibitor is an acetylated iso-d-Glu-meso-Dap pseudopeptide in which a phosphinic acid replaces the peptide bond in order to mimic the tetrahedral oxyanion intermediate. The inhibitor was found to bind with a Ki value of 1.5 µM, which is markedly lower than the KM value of 112 µM for the tripeptide substrate. Crystallographic analysis of the enzyme-inhibitor complex showed that the inhibitor bound in the carboxypeptidase active site, as one would expect for a competitive inhibitor.
Structures of the tripeptide substrate complex and the inhibitor 1 complex with Csd4 provide snapshots of the Michaelis complex and the tetrahedral intermediate complex, respectively. Together these structures provide strong support for the "promoted-water pathway" mechanism proposed by Christianson and Lipscomb for carboxypeptidase A.19,20 In this mechanism, the zinc ion serves to acidify a bound water molecule but does not coordinate to the substrate amide (Figure 1B). Instead, Arg86 serves to activate the carbonyl for attack. Glu222 deprotonates the water and the resulting hydroxide attacks the scissile amide bond to form the tetrahedral intermediate. As the tetrahedral center is formed, the incipient oxyanion rotates into coordination with the metal so that bidentate coordination results. Such a rotation could help to explain why slow binding kinetics were observed with inhibitor 1, as the free enzyme adopts a slightly different conformation than the complex with the tetrahedral intermediate. Arg86 and Glu222 provide further stabilization of the tetrahedral intermediate. Finally, collapse of the tetrahedral intermediate is facilitated by protonation of the departing meso-Dap amine by Glu222.
Gene deletion and active site disruption mutants have demonstrated that Csd4 activity is responsible for maintaining the helical shape of H. pylori that is required for motility and colonization.11,16 This suggests that a Csd4 inhibitor could induce cultures of H. pylori to adopt a straight rod phenotype and thereby dramatically reduce pathogenicity. Our results have shown that inhibitor 1 has a pronounced effect on the shape of cultured H. pylori and results in significant cell straightening when present in millimolar concentrations. The requirement of a concentration that is higher than the Ki value likely reflects the need of the highly polar compound to cross the outer membrane. Straightening was also observed with C. jejuni cells at similar inhibitor concentrations, and the effects were more pronounced with an acapsular strain. This indicates that inhibitor 1 also targets Pgp1 and that polysaccharide capsules retard the ability of the compound to enter the cell.
This study represents the first usage of an inhibitor to directly target cell shape determining enzymes in helical bacteria. Perhaps the most closely related work involves the development of inhibitors against the bacterial cytoskeleton analogs MreB and FtsZ. MreB is the prokaryotic version of actin, and inhibitors that interfere with the controlled polymerization of this protein can cause normally rod-shaped bacteria to assume a spherical shape.32–38 FtsZ is the prokaryotic version of tubulin, and inhibitors that interfere with its ability to self-assemble halt bacterial cell division and result in filamentous undivided cells.39–45 It has also been reported that inhibitors of peptidoglycan biosynthesis/degradation such β-lactams and lytic transglycosylase inhibitors can result in spheroplast formation and ultimately cell death.46–48
The demonstration that hydrophilic small molecules can cross the outer membrane and cause cell straightening in pathogenic helical bacteria suggests that Csd4 is a novel target for antibiotic development. Further work will involve the synthesis of more potent versions of Csd4 inhibitors, such as phosphonic acids and phosphonamic acids that often bind more tightly than the corresponding phosphinic acids, presumably due to the presence of an additional hydrogen bond.23,24,49 Such compounds are predicted to demonstrate activities at more physiologically relevant concentrations.
MATERIALS AND METHODS
General
Chemicals were purchased from Aldrich Chemical Co., Alfa Aesar Co. or Fisher Scientific and used without further purification unless otherwise noted. Ion exchange resin AG-1X8 was purchased from Bio-Rad Laboratories and Amberlite IR-120H resin was purchased from Aldrich Co. CH2Cl2, MeOH and TEA were distilled under Ar from CaH2. 1H NMR and proton-decoupled 31P NMR were recorded on a Bruker AV400dir spectrometer or a Bruker AV400inv spectrometer at field strengths of 400 MHz and 162 MHz, respectively. Mass spectrometry was performed by electrospray ionization (ESI-MS) using an Esquire LC mass spectrometer. Neutral compounds were detected as positive ions and negatively charged compounds were detected as negative ions. Recombinant H. pylori Csd4 and Corynebacterium glutamicum DAPDH were overproduced in Escherichia coli and purified as described previously.16
Synthesis of Inhibitor 1
Compound 3
Compound 2 (1.54 g, 4.6 mmol) was added to 20 mL anhydrous benzene in a round bottom flask. To the resulting suspension, cupric acetate monohydrate (208 mg, 1.2 mmol) was added. The mixture was stirred for 1 h at rt under argon. Fresh lead tetraacetate (4.1 g, 9.2 mmol) was then added and the reaction was allowed to stir for another 1 h at rt. The reaction was heated to reflux overnight under argon. After filtering through celite, the filtrate was diluted with EtOAc and washed three times with water and one time with brine. Then the organic layer was dried with MgSO4. After filtration, the filtrate was concentrated under reduced pressure. Further purification with flash chromatography (silica, 10:1 Hexane:EtOAc) gave 450 mg (33 %) compound 3 as a colorless oil. 1H NMR (400 MHz, CDCl3) δ 7.49 − 7.29 (m, 5H), 5.90 (m, 1H), 5.35 (d, J = 17.0 Hz, 1H), 5.25 (d, J = 10.4 Hz, 1H), 5.13 (s, 2H), 3.45 (m, 1H), 1.47 (s, 9H). MS (ESI) (m/z) 314.3 [M+Na]+.
Compound 7
Ammonium hypophosphite (350 mg, 4.2 mmol) and triethylborane (1.0 M in THF, 2.8 mL) was added to compound 3 (400 mg, 1.4 mmol) in MeOH (20 mL). The solution was stirred for 3h, then evaporated to dryness under reduced pressure. The resulting residue was dissolved in aqueous KHSO4 (1.0 M, 10 mL) and was extracted with EtOAc (3 × 50 mL). The combined organic phases were dried over anhydrous Na2SO4. Removal of the solvent in vacuo gave compound 4 as a colorless oil, which was used in the next step without further purification.
A mixture of crude compound 4 (330 mg) and hexamethyldisilazane (640 mg, 4.0 mmol) was heated for 1 h at 110 °C under argon. Compound 5 (300 mg, 0.70 mmol) was then added. The resulting mixture was heated for 3 h at 90 °C. After cooling to 70 °C, the reaction was quenched with MeOH (10 mL). The resulting solution was evaporated to dryness in vacuo, then extracted with a NaHCO3 solution (10%, 15 mL) and diethyl ether (10 mL). The aqueous phase was separated, and extracted with EtOAc (3 × 50 mL). The combined organic phases were dried over anhydrous Na2SO4, filtered, and the solvent was removed under reduced pressure to give crude compound 6 as a yellow oil (215 mg). This oil was used in the next step without further purification.
Crude compound 6 (400 mg) and 1-adamantylbromide (235 mg, 1.2 mmol) were dissolved in chloroform (20 mL). This solution was heated to reflux. Silver oxide (4 × 140 mg, 2.4 mmol) was added in a quarter portion each time, over 40 min. This solution was refluxed for an additional hour. The solvent was removed in vacuo and the residue was suspended in diethylether and filtered through celite. The filtrates were concentrated. The residue was purified by column chromatography using chloroform/methanol (9.6:0.4) as eluent. Compound 7 was obtained as a colorless foamed gum (mixture of four diastereomers, 265 mg, 0.286 mmol, 25%). 1H NMR (400 MHz, CDCl3) δ 7.52 − 7.28 (m, 10H), 5.10 (s, 4H), 4.40 − 4.05 (m, 2H), 2.26 − 2.02 (m, 15H), 1.75 − 1.40 (m, 35H), 1.34 − 1.24 (m, 3H), 1.05 − 0.81 (m, 2H). 31P NMR (162 MHz, CDCl3) δ 50.76, 50.32, 50.19. MS (ESI) (m/z) 947.8 [M+Na]+.
Compound 8
To a solution of compound 7 (260 mg, 0.28 mmol) in MeOH (20 mL) was added Pd/C (10%, 50 mg). The resulting mixture was stirred under hydrogen gas (1 atm) for 5 h, and then filtered through celite. The filtrate was evaporated in vacuo and dried under reduced pressure to give a colorless oil (210 mg).
The oil was dissolved in an aqueous solution of acetic acid (0.1 M, 20 mL). Acetic anhydride (285 mg, 2.8 mmol) was added dropwise and the pH was adjusted to 6 by the addition of triethylamine. The resulting solution was stirred for 10 min and was acidified to pH 2 by the addition of Amberlite IR-120H ion exchange resin. Filtration and removal of the solvent in vacuo gave a colorless oil as crude. The oil was purified by silica gel chromatography (5% MeOH in chloroform) and gave compound 8 as a colorless oil (mixture of four diastereomers, 114 mg, 0.19 mmol, 68%). 1H NMR (400 MHz, CDCl3) δ 6.60 − 6.52 (m, 1H), 4.58 − 4.41 (m, 2H), 2.20 − 2.06 (m, 21H), 2.05 − 2.01 (s, 3H), 2.02 (s, 3H), 1.87 − 1.37 (m, 35H). 31P NMR (162 MHz, CDCl3) δ 54.30, 52.82. MS (ESI) (m/z) 741.6 [M−H]−.
Inhibitor 1
Compound 8 (141 mg, 0.19 mmol) was dissolved in TFA (9.5 ml)/H2O (0.5 ml). The resulting solution was stirred for 3 h at rt, and then the solvent was evaporated in vacuo. The residue was redissolved in H2O (2.0 ml) and the pH of the solution was adjusted to 8 by adding NaHCO3 (0.5 M). This was loaded onto a column of AG 1-X8 resin (formate form, 100–200 mesh, 5 ml). The column was washed with water (50 ml) and formic acid (0.1 M, 50 ml; 0.5 M, 50 mL) and then was eluted by formic acid (4.0 M, 100 ml). The fractions containing the compound were combined and evaporated to dryness in vacuo to give inhibitor 1 as a colorless oil (mixture of two diastereomers in a 54:46 ratio, 42 mg, 0.106 mol, 56%). 1H NMR (400 MHz, MeOD) δ 4.44 (s, 2H), 2.22 − 2.08 (m, 3H), 2.03 − 2.00 (m, 2H), 2.00 (s, 3H), 2.00 − 1.91 (m, 4H), 1.82 − 1.70 (m, 4H). 31P NMR (162 MHz, MeOD) δ 29.50, 28.24. MS (ESI) (m/z) 395.3 [M−H]−.
Enzyme Kinetics
Csd4 activity with the tripeptide substrate was monitored using a continuous coupled assay that involves m-Dap dehydrogenase (DAPDH) and NADP. The production of NADPH occurred during the oxidation of m-Dap by DAPDH, and the rate of tripeptide substrate hydrolysis was followed by monitoring the increase in absorbance at 340 nm using a Cary300 UV-Vis spectrophotometer. All kinetic assays were performed at 30 °C in 100 mM phosphate buffer (pH 6.5) in the presence of 500 mM NaCl and 2.5 mM NADP. The concentration of Csd4 and DAPDH were set to 3.0 µM and 10.0 µM, respectively. The concentration of ZnCl2 was fixed to 30 µM. Assay mixtures were pre-incubated at 30 °C for 10 – 15 min, and then initiated by addition of the substrate at different concentrations. A non-linear lag associated with the coupled assay was observed in the first 5 min, therefore the initial velocity data was measured based on the slope of the linear plot after 5 min (6 – 20 min) using Microsoft Office Excel 2013. The initial rates measured in this manner were found to be linearly dependent on the concentration of Csd4, indicating the assay was efficiently coupled. Each data point in the Csd4 substrate concentration vs. initial velocity plot was calculated from the average of triplicate assays. The data were fitted by GraphPad® software using a non-linear fit program, and the kinetic parameters were determined based on this fit.
In the inhibition assay, the initial velocity was monitored in the presence of inhibitor 1 at several concentration levels. The concentration of all the components is consistent with the activity assays. The pH and temperature of the solution were also kept identical. Initial assay attempts run without preincubation of enzyme and inhibitor showed non-linear initial velocities that decreased with time, as expected for a slow binding inhibitor. A 20 minute preincubation of enzyme and inhibitor was sufficient to produce linear initial velocities, indicating that binding equilibrium had been reached. The inhibitor was therefore added to the mixture (without the tripeptide substrate), and pre-incubated for 20 min prior to initiating the reaction by the addition of the tripeptide substrate. The initial velocity values were calculated from the linear slope between 5 – 20 min after initiation using Microsoft Office Excel 2013. The data were collected at inhibitor concentrations of 0 µM, 2.0 µM, 5.0 µM and 10.0 µM in triplicate. For each data point the concentration of free inhibitor (I') was corrected using equation 1 as described by Singh et al. for situations in which enzyme and inhibitor concentrations are similar.30 The data were globally fitted to a Dixon equation for competitive inhibition and the error was calculated used a Maximum Likelihood Estimation (MLE) analysis.50
| Eq. 1 |
Crystallization and Structure Determination
Csd4 was crystallized in the P212121 space group with a reservoir solution containing 13–18% PEG 3350, 15 µM Tris pH 8 and 0.4 M sodium iodide by hanging drop vapor diffusion. Crystals were grown for approximately two weeks to reach a sufficient size. The inhibitor-bound structure was then obtained by sequentially soaking crystals of Csd4 in freshly prepared well solution with 1.6 mM ZnCl2 and PEG 3350 in increments of 3% up to 28% (~5 min per step) followed by the addition of an equal volume of 28% PEG 3350 soaking solution supplemented with 2 mM inhibitor for 80 min. These crystals were then briefly soaked in cryoprotectant consisting of 25% (v/v) ethylene glycol prepared with soaking solution and flash frozen. A native 0.9796 Å wavelength dataset to 1.9 Å resolution was collected at 100 K at the Canadian Light Source CMCF-BM Beamline and processed using XDS.51 The structure factors were directly refined against our apo-Csd4 crystal structure (PDB ID: 4WCK) using PHENIX and manually completed using Coot (Table S1).52,53 The structure has excellent stereochemistry with 97.1% of residues in the favored region of the Ramachandran plot and no outliers. The atomic coordinates for the inhibitor-bound crystal structure Csd4 are available in the Research Collaboratory for Structural Bioinformatics Protein Databank under PDB ID: 5D2R.
Inhibitor testing with H. pylori
H. pylori strains J99 and KBH19 were used in this study. J99 was isolated from a patient in the USA with a duodenal ulcer.54 KBH19 is a derivative of human clinical isolate G27 with three tandem copies of the FLAG tag at the C-terminus of Csd4.16,55
KBH19 was grown at 37 °C in a Sanyo tri-gas incubator (10% CO2, 10% O2 from air, balanced with N2). KBH19 was first grown up on horse blood agar plates supplemented with vancomycin (10 µg/mL), polymyxin B (2.5 U/mL), and amphotericin B (8 µg/mL). This culture was used to inoculate liquid Brucella broth (BD Biosciences) with 10% fetal bovine serum (FBS; Gibco) with no antibiotics (BB10). This culture was back-diluted after 5–7 hours and grown overnight to reach an OD600 of 0.4.
For inhibitor testing, the overnight KBH19 culture was diluted in BB10 to OD600 reading of 0.1 and 120 µL was added per condition to a 96 well plate. 50 mM inhibitor 1 stock was added to yield final concentrations of 2.1 mM, 4.2 mM, and 8.3 mM. A 120 µL well was left untreated as a control. KBH19 growth was inhibited at 4.2 mM and 8.3 mM inhibitor 1. Cultures were left shaking for 10 hours and 2 µL from each condition was imaged on agarose pads every 2.5 hours. 2 mm-thick agarose pads were made with 2% low melting point agarose (Invitrogen) dissolved in either Dulbecco’s phosphate buffered saline (Gibco) or in water with 10% FBS and 2.8% (w/v) Brucella broth powder to mimic BB10. Cells were spotted on the agarose pad, allowed to sit briefly, then covered with a coverslip and sealed with VALP (1:1:1 Vasoline:lanolin:paraffin). Cells were imaged by phase contrast microscopy with a 100× oil immersion objective using a Nikon TE200 microscope, a Nikon CoolSnap HQ CCD camera, and MetaMorph software (MDS Analytical Technologies).
Phase contrast images for cells grown at 2.1 mM and the untreated control were thresholded using ImageJ and debris was removed manually. These thresholded images were quantitatively analyzed using Celltool (as previously described).12,56 Side curvature and axis length was measured for 100–200 cells. Komolgorov-Smirnov statistics were calculated to ascertain whether the side curvatures of the treated and untreated cells at each time point were significantly different.11 P-values less than or equal to 0.05 were considered to be significant.
Inhibitor testing with C. jejuni
The wild type C. jejuni 81–176 strain isolated from a diarrheic patient was used in this study,57 as well as an acapsular ΔkpsM mutant strain constructed in the 81–176 background strain.58 C. jejuni strains were grown at 38 °C in Mueller-Hinton (MH; Oxoid) broth or on 8.5% (w/v) agar plates supplemented with vancomycin (V, 10 µg/mL) and trimethoprim (T, 5 µg/mL) under microaerobic conditions (6% O2, 12% CO2) in a Sanyo tri-gas incubator for plates or using the Oxoid CampyGen system for broth cultures. Growth media was supplemented with kanamycin (50 µg/mL) for the ΔkpsM mutant strain, where appropriate.
For inhibitor analysis, C. jejuni strains were streaked from fresh plate cultures and grown for 5–7 h. They were harvested in MH-TV broth and inoculated at an OD600 of 0.002 into MH-TV broth and grown shaking for 18 h. Strains were subcultured to an OD600 of 0.0002 to give approximately 1×106 CFU/mL and 50 µL was added to a microtitre plate containing 5 µL of doubling dilutions of inhibitor (final inhibitor concentrations of 4.5 mM to 0.07 mM) and 5 µL of water was used as the control. Cell morphology was monitored by DIC microscopy (described below) over 24 h. At 24 h, changes in cell morphology were quantified by CellTool analysis on images from DIC microscopy (described below). Experiments were carried out in triplicate with three biological replicates.
Microscopy and Shape Analysis
DIC microscopy was carried out on 1 µL of a broth immobilized on a thin 1% agar (w/v in H2O) slab and overlaid with a cover slip. Images were captured with a Nikon Eclipse TE2000-U microscope and a Hamamatsu Orca camera system. For shape analysis, several images were taken for each sample (with a sample size of 350–900 cells) and were thresholded using Phototshop with debris being removed manually. The CellTool software package was used to compare side curvature and central axis length of bacteria in the images as described.12 Komolgorov-Smirnov statistical comparisons were used to determine whether there was a significant difference in side curvature and axis length between the strains.11
Supplementary Material
Acknowledgments
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grants to M.E.P.M and M.E.T., and a Canadian Institutes of Health Research (CIHR) operating grant (MOP-68981) to E.C.G. It was also supported by the US National Institutes of Health under award numbers RO1A1094839 to N.R.S. and T32CA009657 to K.M.B. This material is partially based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant DGE-0718124 (to J.A.T.) and DGE-1256082 (to J.A.T. and K.M.B.) and by the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) Program (to J.M.T.). Support for infrastructure for structural biology was provided by the Canadian Foundation for Innovation to M.E.P.M. Additionally, research described in this paper was performed using beamline 08B1-1 at the Canadian Light Source, which is supported by NSERC, CIHR, the National Research Council Canada, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Data collection and refinement statistics for Csd4-inhibitor structure, plot of initial velocity versus substrate concentration for Csd4 catalysis with N-acetyltripeptide, and NMR spectra of all newly synthesized compounds.
REFERENCES
- 1.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEBS Microbiol. Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 2.Park JT. The murein sacculus. In: Neidhardt FC, Curtis R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Resnikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Vol. 2. Washington, DC: American Society for Microbiology; 1996. pp. 48–57. [Google Scholar]
- 3.Turner RD, Vollmer W, Foster SJ. Different walls for rods and balls: the diversity of peptidoglycan. Mol. Microbiol. 2014;91:862–874. doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cava F, de Pedro MA. Peptidoglycan plasticity in bacteria: emerging variability of the murein sacculus and their associated biological functions. Curr. Opin. Microbiol. 2014;18:46–53. doi: 10.1016/j.mib.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Frirdich E, Gaynor EC. Peptidoglycan hydrolases, bacterial shape, and pathogenesis. Curr. Opin. Microbiol. 2013;16:767–778. doi: 10.1016/j.mib.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Wyckoff TJ, Taylor JA, Salama NR. Beyond growth: novel functions for bacterial cell wall hydrolases. Trends Microbiol. 2012;20:540–547. doi: 10.1016/j.tim.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautam A, Yyas R, Tewari R. Peptidoglycan biosynthesis machinery: a rich source of drug targets. Crit. Rev. Biotech. 2011;31:295–336. doi: 10.3109/07388551.2010.525498. [DOI] [PubMed] [Google Scholar]
- 8.van Amsterdam K, van Vliet AHM, Kusters JG, van der Ende A. Of microbe and man: determinants of Helicobacter pylori-related diseases. FEMS Microbiol. Rev. 2006;30:131–156. doi: 10.1111/j.1574-6976.2005.00006.x. [DOI] [PubMed] [Google Scholar]
- 9.Butzler J-P. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 2004;10:868–876. doi: 10.1111/j.1469-0691.2004.00983.x. [DOI] [PubMed] [Google Scholar]
- 10.Sycuro LK, Rule CS, Petersen TW, Wyckoff TJ, Sessler T, Nagarkar DB, Khalid F, Pincus Z, Biboy J, Vollmer W, Salama NR. Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol. Microbiol. 2013;90:869–883. doi: 10.1111/mmi.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. Multiple peptidoglycan modification networks modulate Helicobacter pylori's cell shape, motility, and colonization potential. PLoS Pathogens. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonis M, Ecobichon C, Guadagnini S, Prevost M-C, Boneca IG. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol. Microbiol. 2010;78:809–819. doi: 10.1111/j.1365-2958.2010.07383.x. [DOI] [PubMed] [Google Scholar]
- 14.Frirdich E, Vermeulen J, Biboy J, Soares F, Taveirne ME, Johnson JG, DiRita VJ, Girardin SE, Vollmer W, Gaynor EC. Peptidoglycan ld-carboxypeptidase Pgp2 influences Campylobacter jejuni helical cell shape and pathogenic properties and provides the substrate for the dl-carboxypeptidase Pgp1. J. Biol. Chem. 2014;289:8007–8018. doi: 10.1074/jbc.M113.491829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frirdich E, Biboy J, Adams C, Lee J, Ellermeier J, Davis Gielda L, DiRita VJ, Girardin SE, Vollmer W, Gaynor EC. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathogens. 2012;8:e1002602. doi: 10.1371/journal.ppat.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan ACK, Blair KM, Liu Y, Frirdich E, Gaynor EC, Tanner ME, Salama NR, Murphy MEP. Helical shape of Helicobacter pylori requires an atypical glutamine as a zinc ligand in the carboxypeptidase Csd4. J. Biol. Chem. 2015;290:3622–3638. doi: 10.1074/jbc.M114.624734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HS, KIm J, Im HN, An DR, Lee M, Hesek D, Mobashery S, Kim JY, Cho K, Yoon HJ, Han BW, Lee BL, Suh SW. Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a d,l-carboxypeptidase controlling the helical cell shape. Acta Crystallogr. D Biol. Crystallogr. 2014;70:2800–2812. doi: 10.1107/S1399004714018732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipscomb WN, Strater N. Recent advances in zinc enzymology. Chem. Rev. 1996;96:2375–2433. doi: 10.1021/cr950042j. [DOI] [PubMed] [Google Scholar]
- 19.Christianson DW, Lipscomb WN. Carboxypeptidase A. Acc. Chem. Res. 1989;22:62–69. [Google Scholar]
- 20.Christianson DW, David PR, Lipscomb WN. Mechanism of carboxypeptidase A: hydration of a ketonic substrate analogue. Proc. Natl. Acad. Sci. USA. 1987;84:1512–1515. doi: 10.1073/pnas.84.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dive V, Georgiadis D, Matziari M, Makaritis A, Beau F, Cuniasse P, Yiotakis A. Phosphinic peptides as zinc metalloproteinase inhibitors. Cell. Mol. Life Sci. 2004;61:2010–2019. doi: 10.1007/s00018-004-4050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiratake J, Oda J. Aminophosphonic and aminoboronic acids as key elements of a transition state analogue inhibitor of enzymes. Biosci. Biotech. Biochem. 1997;61:211–218. [Google Scholar]
- 23.Mader MM, Bartlett PA. Binding energy and catalysis: the implications for transition-state analogs and catalytic antibodies. Chem. Rev. 1997;97:1281–1301. doi: 10.1021/cr960435y. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan AP, Bartlett PA. Synthesis and evaluation of an inhibitor of carboxypeptidase-A with a Ki value in the femtomolar range. Biochemistry. 1991;30:8165–8170. doi: 10.1021/bi00247a011. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y, Coward JK. Prodrug forms of N-[(4-deoxy-4-amino-10-methyl)pteroyl]glutamate-γ-[d-P(O)(OH)]-glutarate, a potent inhibitor of folylpoly-γ-glutamate synthetase: synthesis and hydrolytic stability. J. Med. Chem. 2006;49:770–788. doi: 10.1021/jm050871p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Ben RN. C-Linked galactosyl serine AFGP analogues as potent recrystallization inhibitors. Org. Lett. 2005;7:2385–2388. doi: 10.1021/ol050677x. [DOI] [PubMed] [Google Scholar]
- 27.Hin B, Majer P, Tsukamoto T. Facile synthesis of α-substituted acrylate esters. J. Org. Chem. 2002;67:7365–7368. doi: 10.1021/jo026101s. [DOI] [PubMed] [Google Scholar]
- 28.Georgiadis D, Matziari M, Vassilou S, Dive V, Yiotakis A. A convenient method to synthesize phosphinic peptides containing an aspartyl or glutamyl aminophosphinic acid. Use of the phenyl group as the carbonyl synthon. Tetrahedron. 1999;55:14635–14648. [Google Scholar]
- 29.Lam LKP, Arnold LD, Kalantar TH, Kelland JG, Lane-Bell PM, Palcic MM, Pickard MA, Vederas JC. Analogs of diaminopimelic acid as inhibitors of meso-diaminopimelate dehydrogenase and l,l-diaminopimelate epimerase. J. Biol. Chem. 1988;263:11814–11819. [PubMed] [Google Scholar]
- 30.Singh V, Evans GB, Lenz DH, Mason JM, Clinch K, Mee S, Painter GF, Tyler PC, Furneaux RH, Lee JE, Howell PL, Schramm VL. Femtomolar transition state analogue inhibitors of 5-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli. J. Biol. Chem. 2005;280:18265–18273. doi: 10.1074/jbc.M414472200. [DOI] [PubMed] [Google Scholar]
- 31.Zeng B, Wong KK, Pompliano DL, Reddy S, Tanner ME. A phosphinate inhibitor of the meso-diaminopimelic acid-adding enzyme (MurE) of peptidoglycan biosynthesis. J. Org. Chem. 1988;63:10081–10086. [Google Scholar]
- 32.van den Ent F, Izore T, Bharat TAM, Johnson CM, Loewe J. Bacterial actin forms antiparallel double filaments. eLife. 2014;3:eo5394. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker CA, Allison SE, Zlitni S, Nguyen ND, Das R, Melacini G, Capretta AA, Brown ED. Degradation of MAC13243 and studies of the interaction of resulting thiourea compounds with the lipoprotein targeting chaperone LolA. Bioorg. Med. Chem. Lett. 2013;23:2426–2431. doi: 10.1016/j.bmcl.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Taylor PT, Rossi L, De Pascale G, Wright GD. A forward chemical screen identifies antibiotic adjuvants in Escherichia coli. ACS Chem. Biol. 2012;7:1547–1555. doi: 10.1021/cb300269g. [DOI] [PubMed] [Google Scholar]
- 35.Yamachika S, Sugihara C, Tsuji H, Muramatsu Y, Kamai Y, Yamashita M. Anti-Pseudamonas aeruginosa compound 1,2,3,4-tetrahydro-1,3,5-triazine derivative, exerts its action by primarily targeting MreB. Biol. Pharm. Bull. 2012;35:1740–1744. doi: 10.1248/bpb.b12-00397. [DOI] [PubMed] [Google Scholar]
- 36.Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. A22 disrupts the bacterial actin cystoskeleton by directly binding and inducing a low affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gatai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Iwai N, Nagai K, Wachi M. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci. Biotechnol. Biochem. 2002;66:2658–2662. doi: 10.1271/bbb.66.2658. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Ma S. Advances in the discovery of novel antimicrobials targeting the assembly of bacterial cell division protein FtsZ. Eur. J. Med. Chem. 2015;95:1–15. doi: 10.1016/j.ejmech.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 40.Chan F-Y, Sun N, Leung Y-C, Wong K-Y. Antimicrobial activity of a quinuclidine-based FtsZ inhibitor and its synergistic potential with β-lactam antibiotics. J. Antibiotics. 2015;68:253–258. doi: 10.1038/ja.2014.140. [DOI] [PubMed] [Google Scholar]
- 41.Artola M, Ruiz-Avila LB, Vergonos A, Huecas S, Araujo-Bazan L, Martin-Fontecha M, Vasquez-Villa H, Turrado C, Ramirez-Aportela E, Hoegl A, Nodwell M, Barasoain I, Chacon P, Sieber SA, Andreu JM, Lopez-Rodriguez ML. Effective GTP-replacing FtsZ inhibitors and antibacterial mechanism of action. ACS Chem. Biol. 2015;10:834–843. doi: 10.1021/cb500974d. [DOI] [PubMed] [Google Scholar]
- 42.Arjes HA, Heidi A, Kriel A, Sorto NA, Shaw JT, Wang JD, Levin PA. Failsafe mechanisms couple division and DNA replication in bacteria. Curr. Biol. 2014;24:2149–2155. doi: 10.1016/j.cub.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan CM, Therien AG, Lu J, Lee SH, Caron A, Gill CJ, Lebeau-Jacob C, Benton-Perdomo L, Monteiro JM, Pereira PM, Elsen NL, Wu J, Deschamps K, Petcu M, Wong S, Daigneault E, Kramer S, Liang LZ, Maxwell E, Claveau D, Vaillancourt J, Skorey, K,, Tam J, Wang H, Meredith TC, Sillaots S, Wang-Jarantow L, Ramtohul Y, Langlois E, Landry F, Reid JC, Parthasarathy G, Sharma S, Baryshnikova A, Lumb KJ, Pinho MG, Soisson SM, Roemer T. Restoring methicillin-resistant Staphylococcus aureus susceptibility to beta-lactam antibiotics. Sci. Transl. Med. 2012;4:126ra35. doi: 10.1126/scitranslmed.3003592. [DOI] [PubMed] [Google Scholar]
- 44.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, Baker PJ, Barynin VV, Rice DW, Sedelnikova SE, Heal JR, Sheridan JM, Aiwale ST, Chauhan PK, Srivistava A, Taneja A, Collins I, Errington J, Czaplewski LG. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 45.Ohashi Y, Chijiiwa Y, Suzuki K, Takahashi K, Nanamiya H, Sato T, Hosoya Y, Ochi K, Kawamura F. The lethal effect of a benzamide derivative, 3-methoxybenzamide, can be suppressed by mutations within a cell division gene, ftsZ, in Bacillus subtilis. J. Bacteriol. 1999;181:1348–1351. doi: 10.1128/jb.181.4.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reid CW, Blackburn NT, Clarke AJ. The effect of NAG-thiazoline on morphology and surface hydrophobicity of Escherichia coli. FEMS Microbiol. Lett. 2004;234:343–348. doi: 10.1016/j.femsle.2004.03.047. [DOI] [PubMed] [Google Scholar]
- 47.Dalhoff A, Nasu T, Okamoto K. Target affinities of faropenem to and its impact on the morphology of gram-positive and gram-negative bacteria. Chemotherapy. 2003;49:172–183. doi: 10.1159/000071141. [DOI] [PubMed] [Google Scholar]
- 48.Horii T, Mase K, Suzuki Y, Kimura T, Ohta M, Maekawa M, Kanno T, Kobayashi M. Antibacterial activities of beta-lactamase inhibitors associated with morphological changes of cell wall in Helicobacter pylori. Helicobacter. 2002;7:39–45. doi: 10.1046/j.1523-5378.2002.00054.x. [DOI] [PubMed] [Google Scholar]
- 49.Bartlett PA, Marlowe CK. Evaluation of intrinsic binding-energy from a hydrogen bonding group in an enzyme-inhibitor. Science. 1987;235:569–571. doi: 10.1126/science.3810155. [DOI] [PubMed] [Google Scholar]
- 50.Johansen S, Juselius Maximum likelihood estimation and inference on cointegration - with applications to the demand for money. Oxford Bull. Econ. Stat. 1990;52:169–210. [Google Scholar]
- 51.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McMcoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alm RA, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 55.Baltrus DA, et al. The complete genome sequence of Helicobacter pylori strain G27. J. Bacteriol. 2009;191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pincus Z, Theriot JA. Comparison of quantitative methods for cell-shape analysis. J. Microsc. 2007;227:140–156. doi: 10.1111/j.1365-2818.2007.01799.x. [DOI] [PubMed] [Google Scholar]
- 57.Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 1985;152:592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- 58.Stahl MA, et al. Novel Mouse Model of Campylobacter jejuni Gastroenteritis Reveals Key Pro-inflammatory and Tissue Protective Roles for Toll-like Receptor Signaling during Infection. PloS Pathogens. 2014;10:e1004264. doi: 10.1371/journal.ppat.1004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.