Abstract
During the last decade miltefosine (MIL) has been used as first-line treatment for visceral leishmaniasis in endemic areas with antimonial resistance, but a decline in clinical effectiveness is now being reported. While only two MIL-resistant Leishmania infantum strains from HIV co-infected patients have been documented, phenotypic MIL-resistance for L. donovani has not yet been identified in the laboratory. Hence, a better understanding of the factors contributing to increased MIL-treatment failure is necessary. Given the paucity of defined MIL-resistant L. donovani clinical isolates, this study used an experimental amastigote-selected MIL-resistant L. infantum isolate (LEM3323). In-depth exploration of the MIL-resistant phenotype was performed by coupling genomic with phenotypic data to gain insight into gene function and the mutant phenotype. A naturally MIL-resistant L. infantum clinical isolate (LEM5159) was included to compare both datasets. Phenotypically, resistance was evaluated by determining intracellular amastigote susceptibility in vitro and actual MIL-uptake. Genomic analysis provided supportive evidence that the resistance selection model on intracellular amastigotes can be a good proxy for the in vivo field situation since both resistant strains showed mutations in the same inward transporter system responsible for the acquired MIL-resistant phenotype. In line with previous literature findings in promastigotes, our data confirm a defective import machinery through inactivation of the LiMT/LiRos3 protein complex as the main mechanism for MIL-resistance also in intracellular amastigotes. Whole genome sequencing analysis of LEM3323 revealed a 2 base pair deletion in the LiMT gene that led to the formation an early stop codon and a truncation of the LiMT protein. Interestingly, LEM5159 revealed mutations in both the LiMT and LiRos3 genes, resulting in an aberrant expression of the LiMT protein. To verify that these mutations were indeed accountable for the acquired resistance, transfection experiments were performed to re-establish MIL-susceptibility. In LEM3323, susceptibility was restored upon expression of a LiMT wild-type gene, whereas the MIL-susceptibility of LEM5159 could be reversed after expression of the LiRos3 wild-type gene. The aberrant expression profile of the LiMT protein could be restored upon rescue of the LiRos3 gene both in the LEM5159 clinical isolate and a ΔLiRos3 strain, showing that expression of LdMT is dependent on LdRos3 expression. The present findings clearly corroborate the pivotal role of the LiMT/LiRos3 complex in resistance towards MIL.
Introduction
Visceral leishmaniasis (VL) is a tropical protozoan disease caused by Leishmania donovani and L. infantum. More than 500.000 new cases do occur annually [1] and control of this life-threatening condition has long been based on treatment with pentavalent antimonials (SbV) [2]. To tackle the widespread Sb-resistance in the Indian subcontinent, miltefosine (MIL) was introduced in 2005 as first-line treatment for VL as part of the Kala-azar elimination program [3]. Although the phase-III trial that led to clinical approval of MIL in India demonstrated a 6-month cure rate of 94% [4], recent reports now indicate relapse rates of up to 20% [5,6]. At present, MIL is used in combination with paromomycin if a cold chain cannot be guaranteed. In Brazil, MIL-treatment of VL by L. infantum revealed a cure rate of only 43% [7]. To safeguard drug efficacy, the parasite-, host- and drug-related factors that contribute to MIL-treatment failure require further exploration. On the one hand, its pharmacokinetic properties [8] in addition to the long unsupervised treatment regimen [6,9] indeed put MIL at a considerable risk of selecting drug resistant parasites. While in the Indian subcontinent relapse after MIL-treatment could not yet be firmly linked to phenotypic resistance in L. donovani using the standard in vitro susceptibility assays [6,10], a potentially reduced MIL-susceptibility has been demonstrated in Brazilian L. infantum relapse isolates [7]. Rather surprisingly, only two L. infantum strains with definite natural MIL-resistance have been documented [11,12]. Given the overall paucity of MIL-resistant clinical field isolates, laboratory studies must generally rely on experimentally selected strains to explore MIL-resistance mechanisms and dynamics. It is noteworthy that most studies have used exposure of promastigotes to increasing MIL-concentrations, although selection of drug resistance on the more clinically relevant intracellular amastigote stage should be considered [13]. A common feature in MIL-resistant promastigotes is a decreased MIL-accumulation that is caused either by a defect in inward transport of MIL through inactivation of the L. donovani putative MIL-transporter (LdMT) [14] and/or its beta-subunit LdRos3 [15] or by an increased efflux mediated by the overexpression of ABC-transporter proteins [16].
In the present study, the experimentally selected MIL-resistant L. infantum strain LEM3323 [17] was subjected to an in-depth phenotypic and molecular characterization in direct comparison to its drug-susceptible wild-type parent counterpart. To further validate the in vitro intracellular amastigote resistance selection assay [13], the naturally MIL-resistant L. infantum clinical isolate LEM5159 was also investigated [12,18]. Characterisation of phenotypic resistance was based on in vitro amastigote and promastigote susceptibility and actual MIL-uptake, whereas next-generation sequencing explored the genomic basis of the resistant phenotypes in combination with functional validation of the detected mutations to confirm their contribution to the acquisition of resistance. Unravelling the genomic and molecular background of the laboratory experimental selected and clinical MIL-resistant L. infantum strains supports the relevance and validity of the in vitro amastigote model as a close proxy for the study of MIL-resistance in the field.
Materials and Methods
Chemical compounds
Miltefosine (hexadecylphosphocholine, Sigma-Aldrich, Diegem, Belgium) was dissolved in MilliQ water and stored at 4°C. The fluorescent analog of MIL (BODIPY-MIL) was kindly provided by L. Rivas (Madrid, Spain) [19]. [14C]MIL (1.33 MBq/mmol) was synthesized by Amersham Pharmacia Biotech (Buckinghamshire, United Kingdom). All other chemicals were of the highest quality and obtained from commercial vendors.
Leishmania infantum strains
MHOM/FR/96/LEM3323 was obtained from a HIV-positive patient from the Languedoc area in Southern France and provided by CNRL, Montpellier, France. MHOM/FR/95/LEM3049 and MHOM/FR/2005/LEM5159 were isolated from the same patient, but with a ten-year time difference (provided by BRC-Leish, Montpellier, France). This patient had received several successive treatments with liposomal amphotericin B (AmB) [18] and MIL (personal communication Lachaud). Species identification was done by isoenzyme electrophoresis and pteridine-reductase 1 (PTR1) sequencing. Promastigote cultures were maintained at 25°C in haemoflagellate-modified minimal essential medium (HOMEM) (Gibco®, Life technologies, Ghent, Belgium) supplemented with 200 mM L-glutamine, 16.5 mM NaHCO3, 10% heat-inactivated fetal calf serum (iFCS), 40 mg/L adenine, 3 mg/L folic acid, 2 mg/L D-biotin and 2.5 mg/L hemin. Promastigotes of L. infantum ΔLiRos3 [20] and L. donovani ΔLdMT [15] null mutants were maintained at 28°C in RPMI-1640 medium (Gibco®, Life technologies, Ghent, Belgium) supplemented with 10% iFCS. The number of passages was kept as low as possible to maintain parasite virulence.
Laboratory animals
Laboratory animals were used to provide primary peritoneal macrophages for the in vitro work. Female Swiss mice were obtained from Janvier (Le Genest Saint Isle, France) and kept on a regular rodent diet and given drinking water ad libitum. Twenty-four hours after intraperitoneal stimulation with a 0.2% potato starch solution, animals were euthanized with a CO2 overdose. Primary peritoneal macrophages (PPM) were collected as previously described [21].
Drug susceptibility assays
The in vitro MIL-susceptibility was determined at both promastigote and intracellular amastigote level as previously described [22]. In brief, MIL IC50-values of log-phase promastigotes were assessed by exposing the parasites to serial two-fold MIL-dilutions. After 72h incubation, viability testing was performed by adding resazurin and measuring the fluorescence by spectrophotometry (Tecan®, GENios). Evaluation of the susceptibility of intracellular amastigotes was done after five days of MIL-treatment and microscopic determination of the reduction in amastigote burdens per cell upon Giemsa-staining.
Experimental selection of MIL-resistance
The parent clone of L. infantum (MHOM/FR/96/LEM3323 Cl-4) was subjected to resistance selection on intracellular amastigotes, as previously described [13,17]. The resistance selection cycles were repeated until the arbitrarily set cut-off value of 15 μM for MIL-resistance on amastigote level was achieved [23]. The selected resistant population was cloned again using the micro-drop method and one clone was randomly selected to perform all experiments [13]. A stable MIL-resistant phenotype (LEM3323-MIL) had already been experimentally selected on intracellular amastigotes of the parent LEM3323 strain [17].
Whole-genome sequencing
Next generation sequencing was performed in collaboration with the Center of Medical genetics (CMG, University of Antwerp, Belgium) and the Welcome Trust Sanger Institute (WTSI, Hinxton, United Kingdom). DNA was isolated from a pellet of stationary-phase promastigotes of L. infantum strains LEM3049, LEM5159, LEM3323 and LEM3323-MIL using QIAamp DNA Mini kit (Qiagen, Netherlands). DNA concentration was measured by Qubit® fluorimeter using the Qubit® dsDNA BR Assay Kit (Thermo Scientific, Belgium). Libraries of LEM3323 and LEM3323-MIL were prepared with the Nextera XT sample prep kit (Illumina) and sequenced using the Illumina Miseq at CMG. LEM3049 and LEM5159 were sequenced according to Shaw et al. (in press) [24] at WTSI and deposited in the European Nucleotide Archive with the accession numbers ERS340107 and ERS340108 respectively. Reads (with an average 32X coverage) were aligned to the L. infantum JPCM5 reference genome (TriTrypDB version 8.0) with Bowtie2 [25–27] and variants were called with Samtools Mpileup [28]. Using a Python script, variants were selected with a read coverage of at least 5 for each strain, a variant quality of at least 50 and mapping quality of minimum 30. Alleles that differed between the MIL-susceptible LEM3323 and MIL-resistant strain LEM3323-MIL were retained and manually verified in IGV [29]. Chromosome copy number was determined by measuring the median read depth of each chromosome di, and obtaining the median depth of the 36 chromosomes dm. The somy status of each chromosome was defined by di/dm and the biological ploidy value was defined as 2*di/dm for a strain whose major ploidy status was diploid.
DNA constructions and generation of transfected parasites
Generation of the Leishmania expression vectors containing LiMT and LiRos3 and the LiMT/GFP fusion protein were previously described [14,20]. The LiMTE926QGFP mutant construct with GFP at the C-terminus was developed using the primers P1 (5' GAAGATGCCCTGCTGCAGCGGCCGAAGCTGTAC 3') and P2 (5' GTACAGCTTCGGCCGCTGCAGCAGGGCATCTTC 3') and the QuikChange site-directed mutagenesis kit (Agilent Technologies, Diegem, Belgium) using LiMT-GFP as template. Promastigotes (3 x 107) were transfected by electroporation (450 V, 500 μF) and the LiMT and LiRos3 transfects were selected with 200 μg/ml hygromycin [14]. The transfected ΔLdMT null mutants were selected with 200 μg/ml geneticin. The expression level of GFP-fused proteins in LiMT-GFP and LiMTE926QGFP transfected parasites was determined by flow cytometry in a FACScan flow cytometer (Becton-Dickinson).
Determination of intracellular MIL-accumulation
Preliminary work on differences in MIL-uptake between resistant and susceptible parasites was done using BODIPY-MIL, a highly fluorescent and photostable MIL-analogue that allows visualization of MIL-uptake in both extracellular promastigotes and intracellular amastigotes [19]. The intracellular accumulation of MIL in transfected Leishmania promastigotes was quantitatively evaluated by measuring intracellular [14C]MIL-accumulation [30]. Briefly, 2 x 107 promastigotes were incubated with 0.09 μCi/ml [14C]MIL (2.5 μM) for 60 min at 28°C in culture medium. The parasites were then washed with ice-cold 1% BSA-PBS for removal of the drug fraction bound to the outer plasma membrane, followed by a second wash. Both protein concentration and counts per minute were determined.
Expression level analysis
Promastigotes (3 x 107 cells/ml) were harvested by centrifugation and washed three times in cold PBS. Parasites were suspended in PBS supplemented with protease inhibitor cocktail (Sigma-Aldrich, Diegem, Belgium) and solubilised with lysis buffer containing 50 mM Tris, 150 mM NaCl and 2% dodecyl maltoside (DDM). Protein samples were fractionated by SDS-polyacrylamide gel electrophoresis using standard conditions and electro-transferred onto Immobilon-P membranes (Merck Millipore, Belgium). Immunodetection was performed with 1:300 dilution of rabbit anti-LdMT antibody and 1:1000 dilution of rabbit anti-LdRos3 antibody [31] in PBS containing 0.1% Tween 20 and 0.1% BSA; α-tubulin was detected using a 1:12500 dilution of a mouse monoclonal anti-α-tubulin antibody (Sigma-Aldrich, Diegem, Belgium); the GFP-fused proteins were detected using a 1:5000 dilution of a rabbit anti-GFP antibody (Life technologies, Ghent, Belgium). After washing, membranes were incubated with 1:5000 dilution of horseradish peroxidase-conjugated secondary goat anti-rabbit or anti-mouse immunoglobulin G (Dako, Agilent Technologies, Belgium). Signals were detected by the ECL chemiluminescent substrate (Thermo ScientificTM PierceTM Protein Biology, Life Technologies, Belgium).
Ethics statement
This study using laboratory rodents was carried out in strict accordance to all mandatory guidelines (EU directives, including the Revised Directive 2010/63/EU on the protection of Animals used for Scientific Purposes that came into force on 01/01/2013, and the declaration of Helsinki in its latest version) and was approved by the ethical committee of the University of Antwerp, Belgium (UA-ECD 2010–17).
Statistical analysis
Statistical comparisons between groups were performed using Student’s t-test. Differences were considered significant at a level of p < 0.05.
Results
In vitro MIL-susceptibility
Promastigote and amastigote susceptibilities of the parent and the derived resistant lines are summarized in Table 1. Noting that an infection ratio of only 2 parasites per macrophage was used, both strains showed very high infectivity with an average infection index of 13.4 ± 1.7 intracellular amastigotes/macrophage at 24 hours post-infection for the WT and of 7.3 ± 1.9 for the MIL-R strain. Both promastigotes and amastigotes of LEM5159, isolated from an HIV-infected patient after ten years of therapeutic intervention [18], confirmed a stable MIL-unresponsiveness for at least twenty successive passages without drug pressure [12]. LEM3049 that was collected from the same patient before receiving several MIL-treatment rounds showed full MIL-susceptibility (Table 1).
Table 1. Susceptibility to miltefosine (MIL) of the different L. infantum strains.
| Strain | Intracellular amastigotes IC50 (mean ± SEM) | Promastigotes IC50 (mean ± SEM) |
|---|---|---|
| LEM3323 | 2.3 ± 0.5 | 5.3 ± 0.3 |
| LEM3323-MIL | >20.0 | >40.0 |
| LEM3323-MIL + LiMT | 3.0 ± 1.0 | 2.4 ± 1.2 |
| LEM3049 | 0.8 ± 0.4 | 5.6 ± 1.1 |
| LEM5159 | > 20.0 | > 40.0 |
| LEM5159 + LiRos3 | 0.5 ± 0.3 | 3.8 ± 1.3 |
| LEM5159 + LiMT | > 20.0 | > 40.0 |
| ΔLdMT | > 20.0 | > 40.0 |
| ΔLdMT + LiMTGFP | 3.5 ± 2.3 | 12.8 ± 0.8 |
| ΔLdMT + LiMTE926QGFP | 4.3 ± 1.6 | 16.1 ± 0.8 |
Li: L. infantum; Ld: L. donovani
The average IC50-values (μM) ± the standard error of the mean (SEM) of intracellular amastigotes and promastigotes are shown. LEM3323 was subjected to in vitro MIL-resistance selection, whereas LEM5159 had a natural MIL-resistant phenotype. The data shown are the result of three independent tests run in duplicate.
Whole-genome sequencing
To unravel the underlying mechanisms responsible for the resistant phenotype, whole-genome sequencing was used to compare LEM5159 and LEM3323-MIL with the pre-treatment isolate LEM3049 and the wild-type (WT) LEM3323 (Table 2).
Table 2. Coding sequence mutations within the LiMT and LiRos3 MIL-transporting complex, identified in the L. infantum MIL-resistant strains.
| Strain | LiMT gene (LinJ.13.1590) | LiRos3 gene (LinJ.32.0540) |
|---|---|---|
| LEM3323 | ||
| LEM3323-MIL | INDEL CCACA to CCA (619572) | |
| LEM3049 | ||
| LEM5159 | codon change GAG to CAG (617837) | INDEL TTTTTTA to TTTTTA (189831) |
LEM3323-MIL: from the genome sequence point of view, LEM3323-MIL was rather similar to its WT parent: in total, between the two strains, we found only 40 single nucleotide polymorphisms (SNPs) that passed the quality filters: 7 variants were present within a coding region and passed manual verification in IGV. Only one gene was changed from a homozygous reference sequence to a homozygous variant (homozygous, non-synonymous single nucleotide polymorphism SNP) (indel LinJ.13.1590 or LiMT). The other 6 changes were still heterozygous for the reference allele (and hence should still produce approximately 50% of the functional protein) or comprised synonymous mutations (see S1 Table). The homozygous two base pair (CA) deletion in the LiMT transporter gene on position 1037–1038 of the gene (equivalent to position 619572–619573 within chromosome 13) caused a shift of the reading frame generating a stop codon after 369 of the 1097 amino acids. No variant was found in the LiRos3 gene (LinJ.32.0540) coding sequence or in 5’ or 3’UTRs. From the karyotype point of view, both LEM3323-MIL and the WT strain were aneuploid with a decrease in copy number of four chromosomes (1, 2, 9, 12) in LEM3323-MIL, the maximal being observed for chr1 (-1 copy, S1 Fig, panel A).
LEM5159: The genome sequence of LEM5159 was drastically different from that of the putative pre-treatment isolate LEM3049, with 11,570 SNPs between both lines: possible traces of LEM5159-specific reads were searched for in the sequencing reads of LEM3049, but were not encountered. Considering the average 32x coverage achieved in our sequencing, this indicates that the patient was re-infected with LEM5159 during his long clinical history or that LEM5159 was present at the onset at a proportion lower than about 1/32 (vs LEM3049). Particular attention was given to the two genes incriminated in the experimental MIL-resistance: (i) a single SNP was detected in the aminophospholipid translocase LiMT gene resulting in an E to Q substitution at codon 926 (GAG → CAG) and (ii) a frameshift mutation (deletion of T base at base 103 of the gene) in the LiRos3 gene causing an early stop codon at amino acid 49. Both isolates were aneuploid with more changes than in the experimental pair described above (S1 Fig, panel B): eight chromosomes (2, 8, 10, 20, 22, 23, 29, 35) showed a higher ploidy in the MIL-resistant isolate LEM5159 (vs LEM3049), the largest being for chromosome 23 (+2 copies, S1 Fig, panel B); two chromosomes (12, 31) showed a lower ploidy in LEM5159, the largest (-2 copies, S1 Fig, panel B) in chromosome 12.
Cell transfections and reconstitution of the MIL-susceptible phenotype
To study the role of the mutations found in the LiMT/LiRos3 genes in the loss of translocase activity and the acquisition of the resistant phenotypes, transfections with expression vectors containing the gene of interest were performed, followed by drug susceptibility testing on promastigotes and intracellular amastigotes (Table 1).
LEM3323-MIL: Transfection with a LiMT gene obtained from a wild-type L. infantum strain resulted in rescue of the resistant phenotype in both parasite stages, confirming that the single indel found in the LiMT gene fully accounts for the intrinsic resistance phenotype of the LEM3323-MIL strain.
LEM5159: To verify if the truncation in the LiRos3 gene is accountable for the acquired resistance, transfection with LiRos3 gene obtained from a wild-type L. infantum strain re-established MIL-susceptibility in the transfected strain (LEM5159 + LiRos3) in both parasite stages (Table 1). However, transfection of LEM5159 with a LiMT wild-type gene obtained by PCR from a wild-type L. infantum strain was not able to rescue susceptibility since both proteins are necessary in the MIL- transporter complex [15]. The specific contribution of the LiMT mutation (E926Q) in the acquisition of MIL-resistance was further analysed. A plasmid containing the mutation present in the LiMT gene (LiMTE926QGFP) was introduced in a ΔLdMT strain using the ΔLdMT + LiMT as a control, after determining that the level of wild type LiMTGFP and LiMTE926QGFP were similar across the two transfected strains. MIL-susceptibility was checked in vitro comparing both transfected lines and both parasite stages and no differences were found. Altogether, these results suggest that the point mutation E926Q does not affect the function of LiMT and that the SNP found in the LiMT gene is not involved in the reduction of MIL-susceptibility of LEM5159.
Determination of intracellular MIL-accumulation
Preliminary experiments with BODIPY-MIL in promastigotes (S2 Fig) and intracellular amastigotes (S3 Fig) already indicated marked differences between MIL-sensitive and MIL-resistant parasites in terms of uptake. To confirm these preliminary data, uptake of [14C]MIL was evaluated in the WT parent and MIL-resistant strains and in the transfected strains (Fig 1).
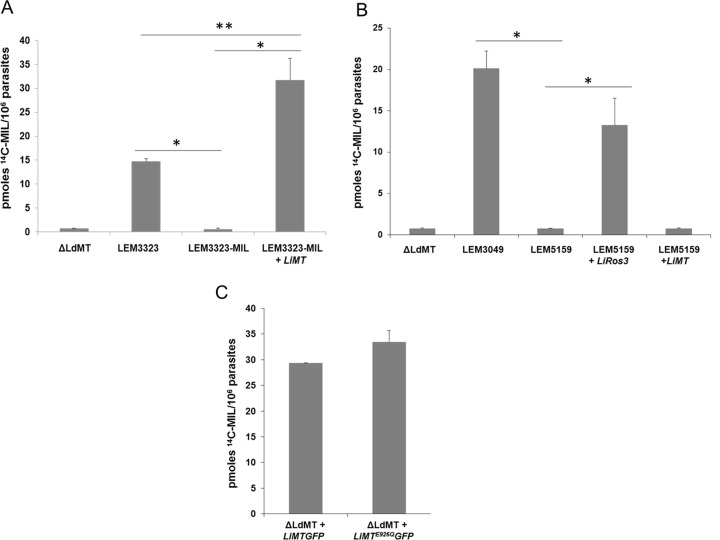
Fig 1. Determination of intracellular MIL-accumulation.
Uptake of [14C]MIL by L. infantum and L. donovani promastigotes was measured after incubation for 60 min at 28◦C (A-C). Results are expressed as the mean ± the standard deviation of three independent experiments in duplicate. (A) L. donovani ΔLdMT line L. infantum LEM3323, LEM3323-MIL (LEM3323MIL), LiMT-transfected LEM3323-MIL (LEM3323-MIL + LiMT), (B) LEM3049, LEM5159, LiRos3- and LiMT-transfected LEM5159 (LEM5159 + LiRos3; LEM5159 + LiMT) and (C) L. donovani ΔLdMT promastigotes transfected with LiMT GFP and with LiMTE926QGFP. Significant differences were determined using the Student's t test (*, p < 0.001, ** p < 0.005).
LEM3323-MIL: Given the presence of the indel in the LiMT gene, internalization of [14C]MIL was evaluated in the transfected strain LEM3323-MIL + LiMT to study whether the MIL-internalization was restored within 60 min incubation. While LEM3323-MIL showed a strong reduction in [14C]MIL accumulation compared to the MIL-susceptible parent strain, recovery of MIL-uptake in the transfected strain was complete (Fig 1A) and identifies the involvement of the major truncation in LiMT gene in the acquisition of resistance in LEM3323-MIL.
LEM5159: The same experiment was performed on LEM3049, LEM5159 and the transfected strains LEM5159 + LiRos3 and LEM5159 + LiMT (Fig 1B). Since LEM3049 was isolated from the patient before the start of MIL-treatment, its extensive MIL-accumulation confirms the presence of an intact LiMT/LiRos3 transporter complex. In contrast, LEM5159 showed almost no MIL-uptake while transfection with LiRos3 was able to rescue the defect in MIL-internalization (Fig 1B) while LiMT-transfected LEM5159 parasites were still defective in MIL-uptake (Fig 1B). To examine the contribution of the mutation LiMTE926QGFP in the decreased MIL-susceptibility profile of LEM5159, MIL-internalization was also measured in the LiMTE926QGFP transfected ΔLdMT line (Fig 1C). The ΔLdMT + LiMTE926QGFP line showed a high ability to take up MIL, with no significant difference to the control line ΔLdMT + LiMTGFP (Fig 1C), endorsing that the E926Q mutation in the LiMT gene is not responsible for the resistant phenotype of LEM5159.
Expression level analysis
LEM3323-MIL: To validate the impact of the truncation present in the LiMT gene on LiMT and LiRos3 protein expression levels, western blot experiments were performed using anti-LdMT and anti-LdRos3 antibodies (Fig 2). Expression levels of α-tubulin was used as a probe for protein loading control. LEM3323-MIL showed similar expression levels of LiRos3 as LEM3323 but no expression of the LiMT protein, clearly demonstrating that the absence of the LiMT protein renders the wild-type LEM3323 refractory to MIL (Fig 2A).
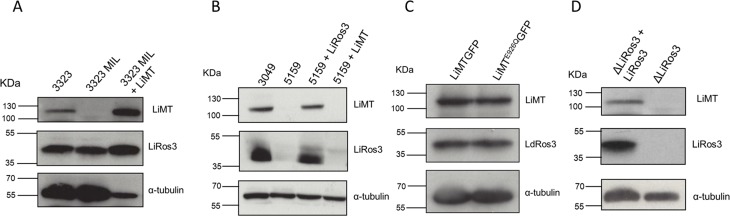
Fig 2. Analysis of the expression levels of the MIL-translocation machinery in different Leishmania strains.
Extracts from (A) L. infantum LEM3323 (3323), LEM3323-MIL (3323MIL), LiMT-transfected LEM3323-MIL (LEM3323-MIL + LiMT), (B) LEM3049, LEM5159, LiRos3- and LiMT-transfected LEM5159 (LEM5159 + LiRos3; 5159 + LiMT), (C) L. donovani ΔLdMT promastigotes transfected with LiMT GFP and with LiMTE926QGFP and (D) ΔLiRos3 and ΔLiRos3 + LiRos3 lines were subjected to SDS/PAGE and immunoblotted with the rabbit polyclonal anti-LdMT and anti-LdRos3 antibodies. Anti-α-tubulin monoclonal antibody was used as a probe for a protein loading control.
LEM5159: The expression levels of LiMT and LiRos3 protein of LEM3049, LEM5159 and the LiMT- and LiRos3-transfected lines were analysed by western blotting (Fig 2B). The MIL-susceptible LEM3049 displayed a clear band for both LiMT and LiRos3 proteins, indicating the presence of a fully functional inward transporter complex. LEM5159 was defective in the expression of both LiMT and LiRos3 protein. Surprisingly, protein expression of LiMT and LiRos3 was restored after transfection with LiRos3 gene implying that the activity and location of both proteins depend upon one another (Fig 2B). Since LiRos3 protein was seriously affected by the frameshift mutation, expression of LiMT did not occur, as illustrated in the LiMT-transfected LEM5159 line (Fig 2B). Hence, LiRos3 protein is necessary to maintain correct levels of LiMT protein, as is demonstrated in the ΔLiRos3 strain which revealed no expression of both proteins (Fig 2D). Demonstrating that ΔLdMT + LiMTE926QGFP and ΔLdMT + LiMTGFP lines had similar levels of both proteins (Fig 2C), MIL-susceptibility and internalization assays were performed to fully corroborate that E926Q substitution in LiMT does not affect the MIL uptake (Fig 2C).
Discussion
The current increasing trend for MIL-treatment relapses of L. donovani in endemic areas in the Indian subcontinent [5,6] in combination with the fact that intrinsic phenotypic resistance in isolates from relapse patients has not unequivocally been demonstrated using the standard in vitro susceptibility laboratory assays [6,10,32] triggers the need for in-depth exploration of the phenotypic and genotypic characteristics of MIL-resistant Leishmania species/strains. Facing the fact that MIL-resistant L. donovani patient isolates are not yet available while two MIL-resistant L. infantum strains have already been documented in HIV co-infected patients [11,12] directed our focus towards L. infantum. While resistance has mostly been studied in laboratory-selected promastigotes, it should be recognized that focusing such research on the intracellular amastigote stage is definitely more relevant. After having developed such an alternative resistance selection assay on intracellular amastigotes [13], our research succeeded in experimentally selecting full MIL-resistance in an L. infantum field isolate [17,33], adding the particular advantage that the parent drug-susceptible LEM3323 and the derived MIL-resistant mutant LEM3323-MIL could be directly compared for phenotypic and genotypic analysis. Although the LEM3323-MIL strain achieved resistance within five successive cycles of drug pressure, in vitro generation of additional MIL-resistant strains of L. donovani and L. infantum at amastigote level has proven to be quite challenging [12,17] and suggests that MIL-resistance may not that easily be selected in the field as originally anticipated based on pharmacokinetic [8] and treatment compliance [6,9] considerations. The MIL-resistant clinical field isolate LEM5159 was included as a crosscheck for the validity of the in vitro amastigote resistance selection model. LEM5159 was isolated from a HIV-positive patient who relapsed 8 times and received successive cycles of MIL during a five-year period [34]. It is well known that immunocompromised patients develop a chronic infection with poor responsiveness to repetitive MIL-treatments [35,36], also due to the absence of an adequate host response [37]. It is still an open question whether such conditions may also lead to full MIL-resistance in L. donovani or L. infantum in an immune-competent patient.
Unresponsiveness of LEM3323-MIL and LEM5159 was established by in vitro susceptibility assays on promastigotes and intracellular amastigotes (Table 1). Research on MIL-resistant promastigotes already suggested that decreased MIL-accumulation is a plausible resistance mechanism that is achieved by a defect in MIL uptake through inactivation of the LdMT transporter [30] and/or its beta-subunit LdRos3 [15]) or by an increased efflux mediated by the overexpression of ABC-transporter proteins [16]. To check if the observed decreased MIL-accumulation acquired at intracellular amastigote level had indeed the same mechanistic basis as described for promastigotes, whole genome sequencing was conducted to identify mutations involved in MIL-resistance.
Previously, genetic analysis of a natural MIL-resistant L. infantum strain identified a SNP in the LiMT gene, pointing to a correlation between the mutation and the reduced MIL-susceptibility [11]. Unfortunately, no functional experiments on this strain were performed to clarify the exact mechanism of resistance, highlighting the importance of the present study that included a natural (LEM5159) and an experimental (LEM3323-MIL) MIL-resistant strain (Table 1). Whole genome sequencing explored the genomic basis of MIL resistance of both strains and revealed the presence of a 2 bp-deletion in the LiMT gene of LEM3323-MIL leading to an early stop codon and inactivation of the LiMT protein. No mutations in the LiRos3 gene were found. On the other hand, sequencing of the MIL-resistant LEM5159 clinical isolate revealed mutations in both LiMT/LiRos3 transporter genes (Table 2). To assess the individual functional role of these mutations, cell transfection experiments were performed whereby transfection of LEM3323-MIL with wild-type LiMT resulted in the expression of the LiMT protein and full recovery of MIL-susceptibility (Table 1, Figs 1 and 2). These findings clearly indicate that the mutation in the LiMT gene was likely the sole determinant for the acquired resistance. In the natural resistant LEM5159 with both transporter genes showing mutations, re-establishment of full MIL-susceptibility was achieved after transfection with LiRos3 (Table 1) leading to accumulation of [14C]MIL (Fig 1) and expression of both LiRos3 and LiMT proteins (Fig 2). These findings indicate that a major defect in the LiRos3 gene results in loss of functionality of the inward-directed transporter. The contribution of the E926Q substitution in the LiMT gene to the level of resistance was negligible since transfection of LEM5159 with an LiMT plasmid failed to restore MIL-susceptibility (Table 1). Surprisingly, no expression of the LiMT protein could be detected in both the parental and LiMT transfected LEM5159 strain (Fig 2). Moreover, introduction of an LiMTE926QGFP plasmid containing the LEM5159 characteristic LiMT mutation into a ΔLdMT strain was equally efficient as an LiMTGFP plasmid to reconstitute MIL-susceptibility, hereby reinforcing that the non-synonymous mutation in LiMT does not add to the overall resistance level of LEM5159 (Table 1, Fig 2). This fully supports the functional characterization of the translocation machinery for which it was shown that the LdMT and LdRos3 are part of the same inward phospholipid transporter [15]. LdMT, a member of the P4 subfamily of P-type ATPases, is involved in phospholipid translocation across the plasma membrane (PM) of Leishmania parasites together with its β-subunit LdRos3, a member of the Lem3/CDC50 family [14,38]. LdMT and LdRos3 are required for the translocation activity and normally become localized in the PM, but are retained inside the endoplasmic reticulum in the absence of the other protein or when inactivating point mutations are introduced in LdMT [38]. The presence of a functional LiRos3 in the LiMT-transfected LEM3323-MIL and the LiRos3-transfected LEM5159 allowed the functional expression of LiMT, while the LiMT-transfected LEM5159 showed no expression of LiMT due to lack of a functional LiRos3 protein (Fig 2). However, the absence of LiMT protein expression could be restored upon the introduction of the LiRos3 gene in both the LEM5159 strain and the ΔLiRos3 strain, endorsing the pivotal role for the Ros3 subunit for LiMT protein expression. However, systematic sequencing of clinical L. donovani isolates from patients showing a relapse after MIL treatment did not (yet) reveal any mutations in LdMT and LdRos3 (Imamura, unpublished results). Extrapolation of genetic markers identified in experimentally-induced parasites to the field may not be straightforward due to changing environments and host immunity [39]. In addition to the findings of Cojean et al, 2012, our results demonstrate that different genes can be involved in the dysfunctionality of LiRos/LiMT of which both can be affected by deletions, point mutations or frame shifts. Hence, defining suitable markers for MIL-resistance may still prove to be very difficult and would imply that a genetic diagnostic test would have to rely on full length sequencing of both LiMT and LiRos, which appears practically unfeasible at large-scale. Efflux rates were not measured as there were no indications that efflux is involved in the strains characterized in this study, particularly given the fact that reconstitution of resistant strains with the inward transporter is sufficient to fully restore susceptibility. Literature data suggest that drug efflux may only be relevant in conditions of high MIL-uptake, for example in cancer cells [40].
Interestingly, our data showed that the experimental amastigote model correlated fairly well with the in vivo situation, hereby supporting the use of LiMT and LiRos3 transporter genes as relevant molecular markers of MIL-resistance in the field. Whole genome sequencing of an extended selection of clinical isolates derived from MIL-treatment failures should give more valuable information about the appearance of particular gene mutations in relation to the occurrence of treatment relapses. On the other hand, treatment failure has a more multifactorial origin and should not solely be linked to drug resistance [41,42]. For example, L. donovani isolates from cured and relapsed patients in the Indian subcontinent showed similar MIL-susceptibility [6] while those of post-kala-azar dermal leishmaniasis (PKDL) patients showed reduced in vitro susceptibility [10,43] suggesting the involvement of different mechanisms in treatment failure. Rai et al. reported an association between the increased infectivity of L. donovani parasites with MIL-relapse of VL patients, indicating the importance to assess other phenotypes than drug susceptibility when characterizing parasites from relapse patients [44]. Moreover, it is recommended to combine the genomic screening of clinical isolates with functional studies to validate the exact contribution of identified mutations in the acquisition of resistance. Metabolomic studies may further shed light on the very complex nature of MIL-resistance, treatment failure and relapse [45–46].
In conclusion, this study is the first to explore the genomic and functional molecular basis of MIL-resistance in two L. infantum strains that definitively developed resistance in the intracellular amastigote stage. The natural resistant patient isolate LEM5159 and the experimentally in vitro selected LEM3323-MIL both showed changes in the MIL-translocation machinery leading to the acquisition of the full blown MIL-resistance phenotype, providing compelling evidence that the in vitro amastigote resistance selection model could be a good proxy of what may happen in the in vivo field situation. A defect in the inward translocation machinery through inactivation of the LiMT/LiRos3 proteins remains the main mechanism of MIL-resistance [47] and supports current literature findings obtained for promastigotes. It is evident that similar work needs to be carried out with L. donovani strains, but ongoing in vitro and in vivo laboratory work already indicates that MIL-resistance selection in L. donovani appears much more slowly requiring a greater number of passages [12,17] and that the availability of MIL-resistant clinical isolates certainly remains a critical issue.
Supporting Information
Somy of the 36 chromosomes of L. infantum, inferred by whole genome sequencing: (A) Comparison between the parent LEM3323 and the experimentally derived MIL-resistant LEM3323-MIL; (B) Comparison between LEM3049 and the natural MIL-resistant isolate LEM5159. The error bars indicate the ploidy standard deviation within individual chromosomes.
(TIF)
Parasites were incubated with 2 µM BODIPY-MIL for 1h. (a) MIL-susceptible LEM3323; (b) MIL-resistant strains LEM3323-MIL and (c) LEM5159. Excitation/emission wavelengths were 529/536 nm for BODIPY-labelled MIL.
(TIF)
(a) MIL-susceptible LEM3323, (b) in vitro MIL-resistant LEM3323-MIL and (c) MIL-resistant clinical isolate LEM5159. The intracellular amastigotes appear as small blue spots while the PMM nucleus is a big blue spot. The wild-type strain shows a clear association between the DAPI spot and the green fluorescence. Excitation/emission wavelengths were 529/536 nm for BODIPY-labelled MIL and 365/445 nm for DAPI.
(TIF)
List of SNPs or indels in coding regions of LEM3323 and LEM3323-MIL that differed between these two isolates; 0/0, homozygous reference; 0/1, heterozygous altered, 1/1, homozygous altered. The variants were called against the L. infantum JPCM5 reference genome.
(DOCX)
Acknowledgments
The authors thank Stephen M. Beverley (Washington University, School of Medicine, USA) for providing the vectors pXG and pXG-'GFP+ used throughout this research work. We also acknowledge the Center of Medical Genetics (Edegem, Belgium) and the Hercules Foundation for providing and managing the sequencing infrastructure. The authors thank Dr. Luis Rivas (Centro de Investigaciones Biológicas, CSIC, Madrid, Spain) for providing the BODIPY-labelled MIL; An Matheeussen, Margot Desmet, Pim-Bart Feijens, Mandy Vermont and members of the Wellcome Trust Sanger Institute sequencing pipeline for their technical assistance and Dr Matthew Berriman for supporting this project. LMPH is a partner of the Antwerp Drug Discovery Network (ADDN, www.addn.be).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the European Commission’s 7th Framework Programme (Kaladrug-R project, Grant 222895) (to LM and JCD), the Flanders Research Fund (FWO) (PhD grant 11V4315N) (to EE), the Agency for Innovation by Science and Technology in Flanders (IWT) (PhD grant 121474 (to AM) and project grants G051812N and G058916N) (to LM), the Spanish Grants SAF2011-28102 (to SC), SAF2012-34267 (to FG) and by the Proyecto de Excelencia, Junta de Andalucia, Ref. CTS-7282 (to FG). MJS and JAC are supported by the Wellcome Trust via core funding of the Wellcome Trust Sanger Institute (grant 098051). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Matlashewski G, Arana B, Kroeger A, Battacharya S, Sundar S, Das P, et al. Visceral leishmaniasis: elimination with existing interventions. Lancet Infect Dis. 2011;11(4): 322–325. 10.1016/S1473-3099(10)70320-0 [DOI] [PubMed] [Google Scholar]
- 2.Sundar S, Chakravarty J. An update on pharmacotherapy for leishmaniasis. Expert Opin Pharmacother. 2015;16(2): 237–252. 10.1517/14656566.2015.973850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhillon GP, Sharma SN, Nair B. Kala-azar elimination programme in India. J Indian Med Assoc 2008;106: 664, 666–668. [PubMed] [Google Scholar]
- 4.Sundar S, Jha TK, Thakur CP, Engel J, Sindermann H, Fischer C, et al. Oral miltefosine for Indian visceral leishmaniasis. N Engl J Med. 2002;347(22): 1739–1746. [DOI] [PubMed] [Google Scholar]
- 5.Sundar S, Singh A, Rai M, Prajapati VK, Singh AK, Ostyn B, et al. Efficacy of miltefosine in the treatment of visceral leishmaniasis in India after a decade of use. Clin Infect Dis 2012;55: 543–550. 10.1093/cid/cis474 [DOI] [PubMed] [Google Scholar]
- 6.Rijal S, Ostyn B, Uranw S, Rai K, Bhattarai NR, Dorlo TP, et al. Increasing failure of miltefosine in the treatment of Kala-azar in Nepal and the potential role of parasite drug resistance, reinfection, or noncompliance. Clin Infect Dis. 2013;56(11): 1530–1538. 10.1093/cid/cit102 [DOI] [PubMed] [Google Scholar]
- 7.Carnielli JB, de Andrade HM, Pires SF, Chapeaurouge AD, Perales J, Monti-Rocha R, et al. Proteomic analysis of the soluble proteomes of miltefosine-sensitive and -resistant Leishmania infantum chagasi isolates obtained from Brazilian patients with different treatment outcomes. J Proteomics 2014;108: 198–208. 10.1016/j.jprot.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 8.Dorlo TP, Balasegaram M, Beijnen JH, de Vries PJ. Miltefosine: a review of its pharmacology and therapeutic efficacy in the treatment of leishmaniasis. J Antimicrob Chemother. 2012;67(11): 2576–2597. 10.1093/jac/dks275 [DOI] [PubMed] [Google Scholar]
- 9.Dorlo TP, Rijal S, Ostyn B, de Vries PJ, Singh R, Bhattarai N, et al. Failure of miltefosine in visceral leishmaniasis is associated with low drug exposure. J Infect Dis. 2014;210(1): 146–153. [DOI] [PubMed] [Google Scholar]
- 10.Bhandari V, Kulshrestha A, Deep DK, Stark O, Prajapati VK, Ramesh V, et al. Drug susceptibility in Leishmania isolates following miltefosine treatment in cases of visceral leishmaniasis and post kala-azar dermal leishmaniasis. PLoS Negl Trop Dis 2012;6: e1657 10.1371/journal.pntd.0001657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cojean S, Houze S, Haouchine D, Huteau F, Lariven S, Hubert V, et al. Leishmania resistance to miltefosine associated with genetic marker. Emerg Infect Dis 2012;18: 704–706 10.3201/eid1804.110841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrickx S, Boulet G, Mondelaers A, Dujardin JC, Rijal S, Lachaud L, et al. Experimental selection of paromomycin and miltefosine resistance in intracellular amastigotes of Leishmania donovani and L. infantum. Parasitol Res. 2014;113(5):1875–1881. 10.1007/s00436-014-3835-7 [DOI] [PubMed] [Google Scholar]
- 13.Hendrickx S, Inocêncio da Luz RA, Bhandari V, Kuypers K, Shaw CD, Lonchamp J, et al. Experimental induction of paromomycin resistance in antimony-resistant strains of L. donovani: outcome dependent on in vitro selection protocol. PLoS Negl Trop Dis. 2012;6(5): e1664 10.1371/journal.pntd.0001664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pérez-Victoria FJ, Gamarro F, Ouellette M, Castanys S. Functional cloning of the miltefosine transporter. A novel P-type phospholipid translocase from Leishmania involved in drug resistance. J Biol Chem. 2003b;278(50): 49965–49971. [DOI] [PubMed] [Google Scholar]
- 15.Pérez-Victoria FJ, Sánchez-Cañete MP, Castanys S, Gamarro F. Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J Biol Chem. 2006a;281(33): 23766–23775. [DOI] [PubMed] [Google Scholar]
- 16.Castanys-Muñoz E, Pérez-Victoria JM, Gamarro F, Castanys S. Characterization of an ABCG-like transporter from the protozoan parasite Leishmania with a role in drug resistance and transbilayer lipid movement. Antimicrob Agents Chemother. 2008;52(10): 3573–3579. 10.1128/AAC.00587-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickx S, Mondelaers A, Eberhardt E, Lachaud L, Delputte P, Cos P, et al. Intracellular amastigote replication may not be required for successful in vitro selection of miltefosine resistance in Leishmania infantum. Parasitol Res. 2015b;114(7): 2561–2565. 10.1007/s00436-015-4460-9 [DOI] [PubMed] [Google Scholar]
- 18.Lachaud L, Bourgeois N, Plourde M, Leprohon P, Bastien P, Ouellette M. Parasite susceptibility to amphotericin B in failures of treatment for visceral leishmaniasis in patients coinfected with HIV type 1 and Leishmania infantum. Clin Infect Dis. 2009;48(2): e16–22. 10.1086/595710 [DOI] [PubMed] [Google Scholar]
- 19.Hornillos V, Carrillo E, Rivas L, Amat-Guerri F, Acuña AU. Synthesis of BODIPY-labeled alkylphosphocholines with leishmanicidal activity, as fluorescent analogues of miltefosine. Bioorg Med Chem Lett. 2008;18(24): 6336–6339. 10.1016/j.bmcl.2008.10.089 [DOI] [PubMed] [Google Scholar]
- 20.García-Sánchez S, Sánchez-Cañete MP, Gamarro F, Castanys S. Functional role of evolutionarily highly conserved residues, N-glycosylation level and domains of the Leishmania miltefosine transporter-Cdc50 subunit. Biochem J. 2014;459(1): 83–94. 10.1042/BJ20131318 [DOI] [PubMed] [Google Scholar]
- 21.Inocêncio da Luz R, Vermeersch M, Dujardin JC, Cos P, Maes L. In vitro sensitivity testing of Leishmania clinical field isolates: preconditioning of promastigotes enhances infectivity for macrophage host cells. Antimicrob Agents Chemother. 2009;53(12): 5197–5203. 10.1128/AAC.00866-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermeersch M, da Luz RI, Toté K, Timmermans JP, Cos P, Maes L. In vitro susceptibilities of Leishmania donovani promastigote and amastigote stages to antileishmanial reference drugs: practical relevance of stage-specific differences. Antimicrob Agents Chemother. 2009;53(9): 3855–3859. 10.1128/AAC.00548-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maes L, Cos P, Croft S. The relevance of susceptibility tests, breakpoints and markers In: Ponte-Sucre A, Diaz E., Padrón-Nieves M., eds. Drug Resistance in Leishmania Parasites: Springer Vienna, 2013;407–429. [Google Scholar]
- 24.Shaw CD, Lonchamp J, Downing T, Imamura H, Freeman TM, Cotton JA, et al. In vitro selection of miltefosine resistance in promastigotes of Leishmania donovani from Nepal: genomic and metabolomics characterization. Mol Microbiol. 2015. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nature genetics 2007;39(7): 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic acids research 2010;38: D457–D462. 10.1093/nar/gkp851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25(16): 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29(1): 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Victoria FJ, Castanys S, Gamarro F. Resistance to miltefosine in Leishmania donovani involves a defective inward translocation of the drug. Antimicrob Agents Chemother. 2003a;47(8): 2397–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Cañete MP, Carvalho L, Perez-Victoria FJ, Gamarro F, Castanys S. The low plasma membrane expression of the miltefosine transport complex renders Leishmania braziliensis refractory to the drug. Antimicrobial Agents Chemother. 2009; 53:1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prajapati VK, Sharma S, Rai M, Ostyn B, Salotra P, Vanaerschot M, et al. In vitro susceptibility of Leishmania donovani to miltefosine in Indian visceral leishmaniasis. Am J Trop Med Hyg. 2013;89(4):750–754. 10.4269/ajtmh.13-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendrickx S, Mondelaers A, Eberhardt E, Delputte P, Cos P, Maes L. In Vivo Selection of Paromomycin and Miltefosine Resistance in Leishmania donovani and L. infantum in a Syrian Hamster Model. Antimicrob Agents Chemother. 2015a;59(8): 4714–4718. 10.1128/AAC.00707-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourgeois N, Lachaud L, Reynes J, Rouanet I, Mahamat A, Bastien P. Long-term monitoring of visceral leishmaniasis in patients with AIDS: relapse risk factors, value of polymerase chain reaction, and potential impact on secondary prophylaxis. J Acquir Immune Defic Syndr. 2008;48(1): 13–19. 10.1097/QAI.0b013e318166af5d [DOI] [PubMed] [Google Scholar]
- 35.Ter Horst R, Collin SM, Ritmeijer K, Bogale A, Davidson RN. Concordant HIV infection and visceral leishmaniasis in Ethiopia: the influence of antiretroviral treatment and other factors on outcome. Clin Infect Dis. 2008;46(11): 1702–1709. 10.1086/587899 [DOI] [PubMed] [Google Scholar]
- 36.Troya J, Casquero A, Refoyo E, Fernández-Guerrero ML, Górgolas M. Long term failure of miltefosine in the treatment of refractory visceral leishmaniasis in AIDS patients. Scand J Infect Dis. 2008;40(1): 78–80. [DOI] [PubMed] [Google Scholar]
- 37.Das M, Saudagar P, Sundar S, Dubey VK. Miltefosine-unresponsive Leishmania donovani has a greater ability than miltefosine-responsive L. donovani to resist reactive oxygen species. FEBS J. 2013;280(19): 4807–4815. 10.1111/febs.12449 [DOI] [PubMed] [Google Scholar]
- 38.Pérez-Victoria FJ, Sánchez-Cañete MP, Seifert K, Croft SL, Sundar S, Castanys S, et al. Mechanisms of experimental resistance of Leishmania to miltefosine: Implications for clinical use. Drug Resist Updat. 2006b;9(1–2): 26–39. [DOI] [PubMed] [Google Scholar]
- 39.Berg M, Mannaert A, Vanaerschot M, Van Der Auwera G, Dujardin JC. (Post-) Genomic approaches to tackle drug resistance in Leishmania. Parasitology. 2013;140(12): 1492–505. 10.1017/S0031182013000140 [DOI] [PubMed] [Google Scholar]
- 40.Rybczynska M, Liu R, Lu P, Sharom FJ, Steinfels E, Pietro AD, et al. MDR1 causes resistance to the antitumour drug miltefosine. Br J Cancer. 2001. May 18;84(10):1405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulshrestha A, Sharma V, Singh R, Salotra P. Comparative transcript expression analysis of miltefosine-sensitive and miltefosine-resistant Leishmania donovani. Parasitol Res. 2014;113(3): 1171–1184. 10.1007/s00436-014-3755-6 [DOI] [PubMed] [Google Scholar]
- 42.Vanaerschot M, Dumetz F, Roy S, Ponte-Sucre A, Arevalo J, Dujardin JC. Treatment failure in leishmaniasis: drug-resistance or another (epi-) phenotype? Expert Rev Anti Infect Ther. 2014;12(8):937–946. 10.1586/14787210.2014.916614 [DOI] [PubMed] [Google Scholar]
- 43.Mishra J, Madhubala R, Singh S. Visceral and post-Kala-Azar dermal leishmaniasis isolates show significant difference in their in vitro drug susceptibility pattern. Parasitol Res. 2013;112(3):1001–1009. 10.1007/s00436-012-3222-1 [DOI] [PubMed] [Google Scholar]
- 44.Rai K, Cuypers B, Bhattarai NR, Uranw S, Berg M, Ostyn B, et al. Relapse after Treatment with Miltefosine for Visceral Leishmaniasis Is Associated with Increased Infectivity of the Infecting Leishmania donovani Strain. mBio. 2013;4(5): e00611–13. 10.1128/mBio.00611-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canuto GA, Castilho-Martins EA, Tavares MF, Rivas L, Barbas C, López-Gonzálvez Á. Multi-analytical platform metabolomic approach to study miltefosine mechanism of action and resistance in Leishmania. Anal Bioanal Chem. 2014;406(14): 3459–3476. 10.1007/s00216-014-7772-1 [DOI] [PubMed] [Google Scholar]
- 46.Berg M, García-Hernández R, Cuypers B, Vanaerschot M, Manzano JI, Poveda JA, et al. Experimental resistance to drug combinations in Leishmania donovani: metabolic and phenotypic adaptations. Antimicrob Agents Chemother. 2015;59(4): 2242–55. 10.1128/AAC.04231-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seifert K, Pérez-Victoria FJ, Stettler M, Sánchez-Cañete MP, Castanys S, Gamarro F, et al. Inactivation of the miltefosine transporter, LdMT, causes miltefosine resistance that is conferred to the amastigote stage of Leishmania donovani and persists in vivo. Int J Antimicrob Agents. 2007;30(3): 229–235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Somy of the 36 chromosomes of L. infantum, inferred by whole genome sequencing: (A) Comparison between the parent LEM3323 and the experimentally derived MIL-resistant LEM3323-MIL; (B) Comparison between LEM3049 and the natural MIL-resistant isolate LEM5159. The error bars indicate the ploidy standard deviation within individual chromosomes.
(TIF)
Parasites were incubated with 2 µM BODIPY-MIL for 1h. (a) MIL-susceptible LEM3323; (b) MIL-resistant strains LEM3323-MIL and (c) LEM5159. Excitation/emission wavelengths were 529/536 nm for BODIPY-labelled MIL.
(TIF)
(a) MIL-susceptible LEM3323, (b) in vitro MIL-resistant LEM3323-MIL and (c) MIL-resistant clinical isolate LEM5159. The intracellular amastigotes appear as small blue spots while the PMM nucleus is a big blue spot. The wild-type strain shows a clear association between the DAPI spot and the green fluorescence. Excitation/emission wavelengths were 529/536 nm for BODIPY-labelled MIL and 365/445 nm for DAPI.
(TIF)
List of SNPs or indels in coding regions of LEM3323 and LEM3323-MIL that differed between these two isolates; 0/0, homozygous reference; 0/1, heterozygous altered, 1/1, homozygous altered. The variants were called against the L. infantum JPCM5 reference genome.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.