Abstract
The complement factor H (FH) mutation R1210C, which was described in association with atypical hemolytic uremic syndrome (aHUS), also confers high risk of age-related macular degeneration (AMD) and associates with C3 glomerulopathy (C3G). To reveal the molecular basis of these associations and to provide insight into what determines the disease phenotype in FH-R1210C carriers, we identified FH-R1210C carriers in our aHUS, C3G, and AMD cohorts. Disease status, determined in patients and relatives, revealed an absence of AMD phenotypes in the aHUS cohort and, vice versa, a lack of renal disease in the AMD cohort. These findings were consistent with differences in the R1210C-independent overall risk for aHUS and AMD between mutation carriers developing one pathology or the other. R1210C is an unusual mutation that generates covalent complexes between FH and HSA. Using purified FH proteins and surface plasmon resonance analyses, we demonstrated that formation of these FH-HSA complexes impairs accessibility to all FH functional domains. These data suggest that R1210C is a unique C-terminal FH mutation that behaves as a partial FH deficiency, predisposing individuals to diverse pathologies with distinct underlying pathogenic mechanisms; the final disease outcome is then determined by R1210C-independent genetic risk factors.
Keywords: complement, hemolytic uremic syndrome, glomerular disease
Complement is a key component of innate immunity that activates in the presence of pathogens, immune complexes, or apoptotic cells. It is able to discriminate between self and foreign/altered-self components and, through a system of molecular labeling, to identify the latter for elimination by opsonophagocytosis or destruction by direct cell lysis. Complement, however, requires strict regulation to focus its action on the surface responsible for its activation and to avoid complete consumption after activation. Loss of complement regulation leads to the generation of proinflammatory components and/or damage to our own tissues, both of which have pathologic consequences.1,2 Factor H (FH), the most important alternative pathway complement regulator, is an abundant plasma glycoprotein composed of 20 repetitive units (short consensus repeats [SCRs]) that regulate complement in the fluid phase and on cellular surfaces. The regulatory activities of the FH molecule depend on three major functional domains: an N-terminal domain (SCR1–SCR4), which sustains the cofactor and decay-accelerating activities, and two domains in SCR6/SCR7 and SCR19/SCR20, which are relevant for ligand binding and cell surface recognition.3 As described, FH is a multifunctional protein and it should be expected that mutations that completely or partially eliminate all or some of these functional sites would cause specific FH dysfunctions with distinct pathologic consequences.
Atypical hemolytic uremic syndrome (aHUS), age-related macular degeneration (AMD), and C3 glomerulopathy (C3G) are examples of diseases in which chronic inflammation and tissue damage play an important role, and these diseases have been associated with mutations and polymorphisms in different genes encoding complement proteins.4–6 Despite sharing complement dysregulation, they are distinct pathologies with different underlying pathogenic mechanisms. These differences are reflected by strong genotype-phenotype correlations in different complement genes. Among these correlations, mutations that alter the C-terminal region of FH are prototypical of aHUS,7–9 whereas the Y402H polymorphism in SCR7 of FH is a unique risk factor for AMD10–13 and complete FH deficiency or homozygous mutations in the N-terminal region of FH associate with C3G.14,15 In this context, the association of the C-terminal FH-R1210C mutation with aHUS,16 AMD,17–19 and C3G20 challenges these genotype-phenotype correlations, suggesting previously unrecognized pathogenic links between these disorders. Here, we performed experiments to reveal the molecular basis of these associations and to identify what determines the disease outcome in FH-R1210C carriers.
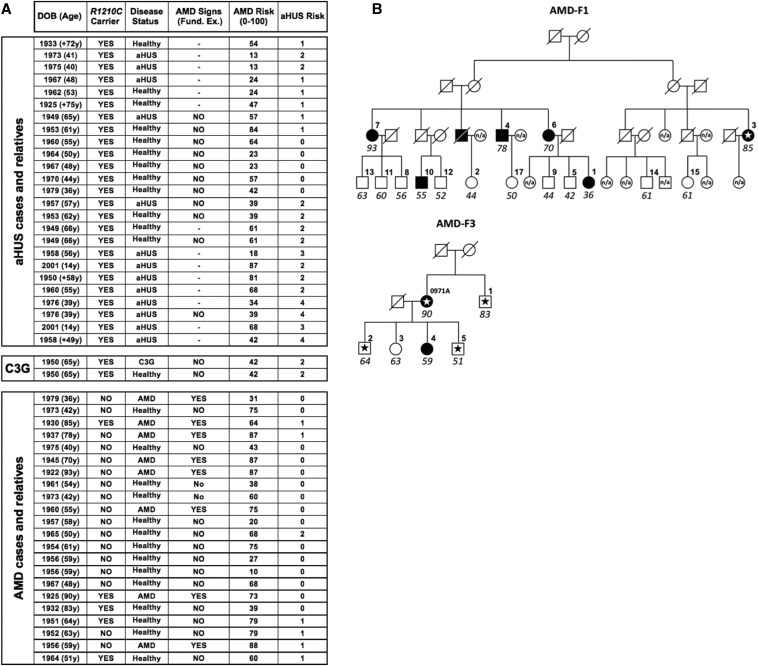
Consistent with previous reports, we identified 25 R1210C carriers in our aHUS cohort (n=1030), 2 in the C3G cohort (n=187), 5 in the AMD cohort (n=259), and none in a control group (n=330). There are both affected and healthy individuals among these carriers (Figure 1A), illustrating the incomplete penetrance of the three diseases among R1210C mutation carriers. We also observed that in the two R1210C AMD pedigrees that we have identified, some individuals developed AMD in the absence of the R1210C mutation, similarly to earlier data17 (Figure 1B). These data are in agreement with FH-R1210C being associated with aHUS, AMD, and C3G. They also indicate that, at least in our R1210C AMD pedigrees, there is an independent and strong genetic predisposition to AMD.
Figure 1.
Patients and relatives carrying the FH-R1210C mutation. (A) Thirteen aHUS patients and 1 C3G patient carrying the R1210C mutation identified in the Spanish and Italian cohorts are listed together with 13 healthy first-degree relatives who also carry the mutation. Also shown are 8 AMD members and 14 healthy members of the two R1210C pedigrees that we identified in the AMD Spanish cohort. Age, disease status, results of the ophthalmologic examination, and the overall genetic risks for aHUS and AMD (see the Concise Methods and Supplemental Figure 1) are provided for all individuals. (B) AMD pedigrees segregating the FH-R1210C mutation. Schematic representation of the two pedigrees identified in the AMD Spanish cohort is provided. Notice that in these pedigrees, in addition to healthy carriers of the FH R1210C mutation, there are also individuals who develop AMD and do not carry the FH-R1210C mutation. Numbers identify the individual in the pedigree and their age (italics). R1210C are identified with a star. Fund. ex., fundus examination.
Notably, within a given pedigree, R1210C carriers only develop one type of disease. Raychaudhuri et al. found no evidence of clinically significant renal dysfunction among 17 unrelated R1210C heterozygotes with advanced AMD.17 Likewise, we found that none of the 27 carriers of the R1210C mutation among our aHUS and C3G patients and their relatives, including 8 individuals aged >55 years, reported AMD-like visual impairment or presented signs of AMD in a fundus examination using standardized protocols (Figure 1A). We previously reported that the presence of additional aHUS genetic risk factors significantly contributes to the manifestation of aHUS among R1210C carriers.16 Therefore, we hypothesized that differences in the R1210C-independent overall genetic risk for AMD and aHUS influence the penetrance of one or other disease among these individuals.
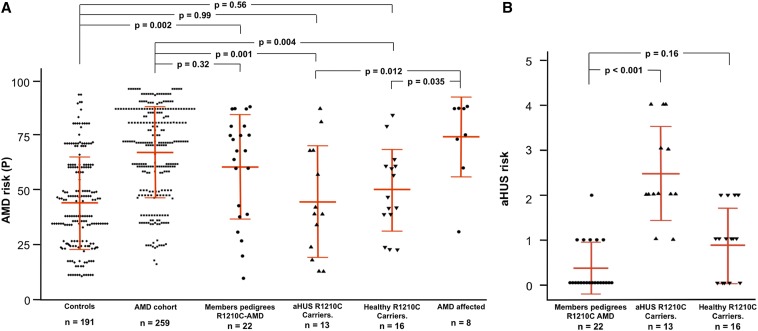
The AMD risk genotypes for previously published AMD cases carrying R1210C have not been reported and we only found two affected R1210C carriers in our AMD cohort. Although this is a very small number, these two AMD cases belong to pedigrees with multiple affected individuals (Figure 1B). As indicated above, these findings suggest that there is an elevated overall AMD risk in these families that is independent of the R1210C mutation. In fact, the average overall AMD genetic risk (see the Concise Methods) for all members in these families (n=22) is 60.6 (Figure 2, Supplemental Figure 1), rising to 74 for affected members only (n=8), which are very high AMD risks considering that the average AMD risk for the whole Spanish AMD cohort (n=259) is 67.3 (Figure 2, Table 1). By contrast, the overall AMD risks among the aHUS (n=13; 44.8) and healthy (n=16; 49.9) R1210C carriers are similar to the control Spanish group (n=191; 44.3) (Figure 2, Table 1). Correspondingly, aHUS risk scores (see the Concise Methods) in the 13 aHUS patients were significantly elevated compared with those in the 16 healthy R1210C mutation carriers (2.46 versus 0.88; p<0.001); aHUS risk scores were consistently significantly decreased among members of the two AMD pedigrees (n=22) compared with the 13 aHUS patients (0.38 versus 2.46; p<0.001) (Figure 2, Table 1). As a whole, these data indicate that additional genetic risk factors independent of the FH-R1210C mutation contribute significantly to the disease outcome in R1210C carriers in our cohorts.
Figure 2.
Graphical representation of the individual AMD and aHUS overall risks. (A and B) Overall AMD (A) and aHUS (B) risk for each individual in the different cohorts included in these studies are shown. Mean values and SDs are indicated with red lines. A one-way ANOVA with Tukey’s post hoc test is used to test differences between the groups. All tests are performed using SPSS statistical software (version 21). A p-value <0.05 is considered statistically significant.
Table 1.
Differences in AMD and aHUS overall risk scores
| Groups Compared | Mean Overall Risk | ||
| AMD risk | |||
| aHUS R1210C carriers (n=13) | 44.8 ± 25.3 | 44.8 ± 25.3 | |
| AMD cohort (n=259) | 67.3 ± 20.8 | ||
| Controls (n=191) | 44.3 ± 21.1 | ||
| p-value | 0.001 | 0.99 | |
| Healthy R1210C carriers (n=16) | 49.9 ± 18.5 | 49.9 ± 18.5 | |
| AMD cohort (n=259) | 67.3 ± 20.8 | ||
| Controls (n=191) | 44.3 ± 21.1 | ||
| p-value | 0.004 | 0.56 | |
| R1210C AMD pedigrees (n=22) | 60.6 ± 23.8 | 60.6 ± 23.8 | |
| AMD cohort (n=259) | 67.3 ± 20.8 | ||
| Controls (n=191) | 44.3 ± 21.1 | ||
| p-value | 0.33 | 0.002 | |
| aHUS risk | |||
| aHUS R1210C carriers (n=13) | 2.46 ± 1.05 | 2.46 ± 1.05 | |
| R1210C AMD pedigrees (n=22) | 0.38 ± 0.59 | ||
| Healthy R1210C carriers (n=13) | 0.88 ± 0.81 | ||
| p-value | <0.001 | <0.001 | |
The association of the R1210C mutation with AMD and C3G challenges the strong genotype-phenotype correlation that characterizes the FH C-terminal mutations. FH C-terminal mutations are prototypical of aHUS and are the most prevalent genetic alteration among these patients.7,21,22 It is now well established that the FH C-terminal region simultaneously binds C3b (or C3dg) and polyanions, like glycosaminoglycans, sulfated polysaccharides or sialylated glycans, on the host cell surfaces through separate domains in SCR19 and SCR20.23 Compelling evidence has shown that C-terminal mutations in FH associated with aHUS impair the capacity of FH to protect host endothelial cells from complement lysis.21,22 Similarly, the FH-Y402H polymorphism in SCR7 is characteristically associated with AMD.9–12 The mechanism by which the FH-Y402H polymorphism affects AMD risk is as yet unsolved, although it is thought that it alters the ligand specificity of the FH SCR6/7 domain, which may result in failure to recruit FH to sites where complement is activated by the accumulation of endogenous complement-activating compounds in the retina. In this respect, it has been reported that the AMD risk FH-402H variant fails to bind a number of ligands, including C-reactive protein, heparan sulfate, and malondialdehyde, a common lipid peroxidation product found in the aged retina and implicated in AMD.24–26 Integrity of the surface recognition domains in SCR6/SCR7 and SCR19/SCR20 are, therefore, distinctively critical in the pathogenesis of AMD and aHUS, respectively. Then, why does the C-terminal FH-R1210C mutation predispose to AMD and C3G?
Crucially, from a structural point of view, the R1210C mutation is very different from other FH C-terminal mutations in that this mutant forms covalently linked complexes with human serum albumin (HSA) via the mutant cysteine residue.22 An explanation for the association of FH-R1210C with aHUS, AMD, and C3G is that the HSA molecule linked to the C-terminal region of FH not only interferes the interaction of this region with its ligands, impairing functionalities of the C terminus, but it also disturbs the interaction of the SCR6/SCR7 and the N-terminal regions with their ligands on the complement-activating surface.
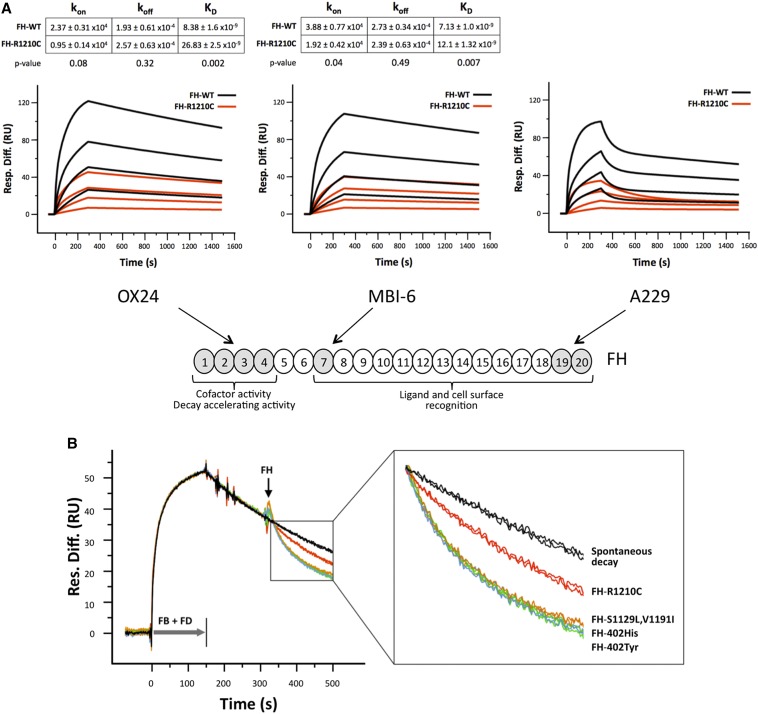
Testing this possibility is complex because we lack ligands uniquely recognized by each of these FH functional domains. To overcome these difficulties we selected three mAbs that specifically impair the function of each of these FH regions, the N-terminal mAb OX24, the anti–FH-402Tyr mAb MBI-6 (or anti–FH-402His MBI-7), and the C-terminal mAb A229, respectively. Using FH purified proteins (Supplemental Figure 2) and surface plasmon resonance analysis, we were able to show that the binding affinities of the three mAbs for FH-R1210C were significantly decreased compared with those for wild-type FH (Figure 3A). Importantly, these differences in all cases resulted from a decreased association constant (Kon), demonstrating that accessibility to all three FH functional regions was impaired in the FH-R1210C mutant (Figure 3A). As an additional control, these analyses included a FH mutant protein carrying two missense mutations (S1129L and V1191I) in the SCR20 of FH, which exclusively associates with aHUS. This additional mutant illustrates that it is the albumin bound to FH and not the C-terminal mutation that impairs accessibility to the FH functional domains in the R1210C mutant (Supplemental Figure 3).
Figure 3.
The R1210C mutation impairs all FH functional domains. (A) SPR analysis showing mAb binding to functional domains of FH variants. Panels depict the binding of purified FH-WT (black) and FH-R1210C (red) (both 402Y) to captured Ox24, MBI-6, and A229 mAbs at increasing concentrations (1 μM, 333 nM, 111 nM, and 37 nM). Sensograms for Ox24 and MBI-6 mAbs show the fitted curves and kinetic constants calculated using a one-to-one interaction model. Values are the means of three independent experiments. In contrast with mAbs OX24 and MBI-6, the binding of FH-WT and FH-R1210C to mAb A229 only fits a heterogeneous ligand model, which is defined by multiple kinetic constants. Note that it requires approximately 4–6 times more FH-R1210C than FH-WT protein to achieve similar binding to all three mAbs. (B) SPR analysis of AP C3 convertase decay-accelerating activity of FH variants. In these experiments, we use a very low density C3b-coated Biacore chip (450 RU) to avoid the possibility that the FH N- and C-terminal regions interact simultaneously with two different C3b molecules on the chip surface; AP C3 convertase is formed by flowing FB (1 µM) and FD (40 nM) for 150 seconds. Spontaneous decay (no FH added) is allowed to proceed for another 150 seconds before flowing FH-WT (402Y and 402H), FH-R1210C, and FH-S1129L-V1191I at 70 nM. Changes in RU during FH injection, compared with the spontaneous decay, represent loss of Bb from the C3 convertase due to FH-mediated decay-accelerating activity. The figure depicts two independent assays for each FH variant. An enlarged view of the boxed part of the sensograms is provided to identify the different FH variants. FB, factor B; FD, factor D; Resp. Diff., response difference; RU, resonance units; SPR, surface plasmon resonance; WT, wild-type.
Fifteen years ago we were unable to document differences between wild-type FH and FH-R1210C in N-terminal–dependent functional activities.22 However, consistent with the new data reported here, a reevaluation of these FH activities using surface plasmon resonance demonstrated that the AP C3 convertase decay-accelerating activity of FH-R1210C, which is dependent of the FH N-terminal region, is impaired in the FH-R1210C mutant but not in the aHUS-specific C-terminal FH-S1129L, V1191I mutant (Figure 3B).
As a whole, these data explain that carriers of the FH-R1210C mutation develop aHUS, AMD, or C3G not because these disorders share unrecognized pathogenic links, but because this FH mutant is an exception to the strict genotype-phenotype correlations that characterize these diseases. The R1210C mutation impairs all FH functional domains and in this respect its consequences are equivalent to a partial FH deficiency, which is well documented to be associated with a broad spectrum of pathologies with distinct underlying pathogenic mechanisms. In carriers of partial FH deficiencies, like in FH-R1210C carriers, the final outcome is determined by the individual’s overall genetic risk to each of these diseases. Thus, available evidence suggests that concurrence of a partial FH deficiency with other mutations/polymorphisms decreasing protection to host cells will result in aHUS, whereas the coincidence of partial FH deficiencies with strong fluid phase complement activators, galactose-deficient IgA1-containing immune complexes, or AMD risk polymorphisms will trigger C3G, IgA nephropathy, or AMD,14,27–30 respectively. Additional genetic and/or environmental factors altering host surfaces, depositing activating molecules (i.e., C-reactive protein, malondialdehyde), or removing FH ligands (i.e., heparan sulfate, sialic acid) might also influence the pathologic outcome in these individuals. Our report further illustrates the relevance of understanding the functional consequences of the disease-associated mutations to provide insight into pathogenic mechanisms.
Concise Methods
Further information is available in Supplemental Methods.
Overall Genetic Risks for aHUS and AMD and Statistical Analyses
To evaluate the AMD risk scores, we used a model developed in our laboratory that includes the most relevant polymorphisms associated with AMD.31 Briefly, this model considers the genotypes at the triallelic locus CFHR1 in addition to the genotypes of the CFH Tyr402His, CFB Leu9His, CFB Arg32Gln/Trp, and ARMS2 Ala69Ser polymorphisms. Regression coefficients (risk scores) were determined for all of the genotypes using multivariate logistic regression analysis as previously described.31 Cumulative risk scores (Z) were determined by the equation Z = α + ΣβiXi, where the regression coefficients α (constant) and βi (risk score specific for each genotype X) are taken directly from the multivariate logistic regression analysis. In this work, we are using probabilities of developing AMD (P values). P values were calculated as P = eZ/(1 + eZ). As an example, for a population like Spain with a prevalence of AMD (grades 2, 3, and 4) of 16% over 65 years of age, 19% of individuals with a P value of 60% will present AMD, whereas this percentage decreases to 9.3% for those with a P value of 40%.31 Because we have no similar models for the calculation of the overall risk for aHUS, we have considered in this estimation the presence of mutations in additional aHUS candidate genes and the genotypes for the CFH-H3 and MCPggaac aHUS risk haplotypes.32 We therefore gave 1 point for each additional mutation or risk polymorphism, and the sum of the codes was used as a continuous variable. In contrast with the overall AMD risk (P values), which range from 0% to 100%, the overall risks for aHUS are increasing numerals (i.e., 0, 1, 2, 3, etc.). One-way ANOVA with Tukey’s post hoc test was used to test differences between the groups. All tests were performed using SPSS statistical software (version 21; SPSS Inc., Chicago, IL). A p-value < 0.05 was considered statistically significant.
Study Approval
Samples from all individuals included in these studies were obtained with informed consent in accordance with the Declaration of Helsinki and our institutional review boards.
Disclosures
S.R.d.C. and M.N. have received honoraria from Alexion Pharmaceuticals for giving lectures and participating in advisory boards. None of these activities has had any influence on the results or interpretation in this article.
Supplementary Material
Acknowledgments
The authors thank all of the patients and relatives participating in this study. They also thank Dr. Claire Harris for advice in the surface plasmon resonance experiments.
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (SAF2011-26583, PI11/00898, and RETICS RD12/0034), the Iñigo Alvarez de Toledo Renal Foundation, the European Union Seventh Framework Programme EURenOmics Project (305608), and the Autonomous Region of Madrid (S2010/BMD-2316). R.M. is recipient of a fellowship from the Aid Foundation for Research on Rare Diseases.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2015050580/-/DCSupplemental.
References
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD: Complement: A key system for immune surveillance and homeostasis. Nat Immunol 11: 785–797, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holers VM: The spectrum of complement alternative pathway-mediated diseases. Immunol Rev 223: 300–316, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Rodríguez de Córdoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sánchez-Corral P: The human complement factor H: Functional roles, genetic variations and disease associations. Mol Immunol 41: 355–367, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Noris M, Remuzzi G: Atypical hemolytic-uremic syndrome. N Engl J Med 361: 1676–1687, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Xiao X, Pickering MC, Smith RJ: C3 glomerulopathy: The genetic and clinical findings in dense deposit disease and C3 glomerulonephritis. Semin Thromb Hemost 40: 465–471, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Gorin MB: Genetic insights into age-related macular degeneration: Controversies addressing risk, causality, and therapeutics. Mol Aspects Med 33: 467–486, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-Caballero D, González-Rubio C, Gallardo ME, Vera M, López-Trascasa M, Rodríguez de Córdoba S, Sánchez-Corral P: Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am J Hum Genet 68: 478–484, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caprioli J, Bettinaglio P, Zipfel PF, Amadei B, Daina E, Gamba S, Skerka C, Marziliano N, Remuzzi G, Noris M; Italian Registry of Familial and Recurrent HUS/TTP : The molecular basis of familial hemolytic uremic syndrome: Mutation analysis of factor H gene reveals a hot spot in short consensus repeat 20. J Am Soc Nephrol 12: 297–307, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Richards A, Buddles MR, Donne RL, Kaplan BS, Kirk E, Venning MC, Tielemans CL, Goodship JA, Goodship TH: Factor H mutations in hemolytic uremic syndrome cluster in exons 18-20, a domain important for host cell recognition. Am J Hum Genet 68: 485–490, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards AO, Ritter R 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA: Complement factor H polymorphism and age-related macular degeneration. Science 308: 421–424, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R: A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A 102: 7227–7232, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA: Complement factor H variant increases the risk of age-related macular degeneration. Science 308: 419–421, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J: Complement factor H polymorphism in age-related macular degeneration. Science 308: 385–389, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Licht C, Heinen S, Józsi M, Löschmann I, Saunders RE, Perkins SJ, Waldherr R, Skerka C, Kirschfink M, Hoppe B, Zipfel PF: Deletion of Lys224 in regulatory domain 4 of Factor H reveals a novel pathomechanism for dense deposit disease (MPGN II). Kidney Int 70: 42–50, 2006 [DOI] [PubMed] [Google Scholar]
- 15.de Córdoba SR, de Jorge EG: Translational mini-review series on complement factor H: Genetics and disease associations of human complement factor H. Clin Exp Immunol 151: 1–13, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Barricarte R, Pianetti G, Gautard R, Misselwitz J, Strain L, Fremeaux-Bacchi V, Skerka C, Zipfel PF, Goodship T, Noris M, Remuzzi G, de Cordoba SR; European Working Party on the Genetics of HUS : The complement factor H R1210C mutation is associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol 19: 639–646, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychaudhuri S, Iartchouk O, Chin K, Tan PL, Tai AK, Ripke S, Gowrisankar S, Vemuri S, Montgomery K, Yu Y, Reynolds R, Zack DJ, Campochiaro B, Campochiaro P, Katsanis N, Daly MJ, Seddon JM: A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet 43: 1232–1236, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhan X, Larson DE, Wang C, Koboldt DC, Sergeev YV, Fulton RS, Fulton LL, Fronick CC, Branham KE, Bragg-Gresham J, Jun G, Hu Y, Kang HM, Liu D, Othman M, Brooks M, Ratnapriya R, Boleda A, Grassmann F, von Strachwitz C, Olson LM, Buitendijk GH, Hofman A, van Duijn CM, Cipriani V, Moore AT, Shahid H, Jiang Y, Conley YP, Morgan DJ, Kim IK, Johnson MP, Cantsilieris S, Richardson AJ, Guymer RH, Luo H, Ouyang H, Licht C, Pluthero FG, Zhang MM, Zhang K, Baird PN, Blangero J, Klein ML, Farrer LA, DeAngelis MM, Weeks DE, Gorin MB, Yates JR, Klaver CC, Pericak-Vance MA, Haines JL, Weber BH, Wilson RK, Heckenlively JR, Chew EY, Stambolian D, Mardis ER, Swaroop A, Abecasis GR: Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nat Genet 45: 1375–1379, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seddon JM, Reynolds R, Yu Y, Rosner B: Three new genetic loci (R1210C in CFH, variants in COL8A1 and RAD51B) are independently related to progression to advanced macular degeneration. PLoS One 9: e87047, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Servais A, Frémeaux-Bacchi V, Lequintrec M, Salomon R, Blouin J, Knebelmann B, Grünfeld JP, Lesavre P, Noël LH, Fakhouri F: Primary glomerulonephritis with isolated C3 deposits: A new entity which shares common genetic risk factors with haemolytic uraemic syndrome. J Med Genet 44: 193–199, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manuelian T, Hellwage J, Meri S, Caprioli J, Noris M, Heinen S, Jozsi M, Neumann HP, Remuzzi G, Zipfel PF: Mutations in factor H reduce binding affinity to C3b and heparin and surface attachment to endothelial cells in hemolytic uremic syndrome. J Clin Invest 111: 1181–1190, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sánchez-Corral P, Pérez-Caballero D, Huarte O, Simckes AM, Goicoechea E, López-Trascasa M, de Córdoba SR: Structural and functional characterization of factor H mutations associated with atypical hemolytic uremic syndrome. Am J Hum Genet 71: 1285–1295, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaum BS, Hannan JP, Herbert AP, Kavanagh D, Uhrín D, Stehle T: Structural basis for sialic acid-mediated self-recognition by complement factor H. Nat Chem Biol 11: 77–82, 2015 [DOI] [PubMed] [Google Scholar]
- 24.Clark SJ, Perveen R, Hakobyan S, Morgan BP, Sim RB, Bishop PN, Day AJ: Impaired binding of the age-related macular degeneration-associated complement factor H 402H allotype to Bruch’s membrane in human retina. J Biol Chem 285: 30192–30202, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prosser BE, Johnson S, Roversi P, Herbert AP, Blaum BS, Tyrrell J, Jowitt TA, Clark SJ, Tarelli E, Uhrín D, Barlow PN, Sim RB, Day AJ, Lea SM: Structural basis for complement factor H linked age-related macular degeneration. J Exp Med 204: 2277–2283, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weismann D, Hartvigsen K, Lauer N, Bennett KL, Scholl HP, Charbel Issa P, Cano M, Brandstätter H, Tsimikas S, Skerka C, Superti-Furga G, Handa JT, Zipfel PF, Witztum JL, Binder CJ: Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature 478: 76–81, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dragon-Durey MA, Frémeaux-Bacchi V, Loirat C, Blouin J, Niaudet P, Deschenes G, Coppo P, Herman Fridman W, Weiss L: Heterozygous and homozygous factor h deficiencies associated with hemolytic uremic syndrome or membranoproliferative glomerulonephritis: Report and genetic analysis of 16 cases. J Am Soc Nephrol 15: 787–795, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Hakobyan S, Tortajada A, Harris CL, de Córdoba SR, Morgan BP: Variant-specific quantification of factor H in plasma identifies null alleles associated with atypical hemolytic uremic syndrome. Kidney Int 78: 782–788, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montes T, Goicoechea de Jorge E, Ramos R, Gomà M, Pujol O, Sánchez-Corral P, Rodríguez de Córdoba S: Genetic deficiency of complement factor H in a patient with age-related macular degeneration and membranoproliferative glomerulonephritis. Mol Immunol 45: 2897–2904, 2008 [DOI] [PubMed] [Google Scholar]
- 30.Rosenblad T, Rebetz J, Johansson M, Békássy Z, Sartz L, Karpman D: Eculizumab treatment for rescue of renal function in IgA nephropathy. Pediatr Nephrol 29: 2225–2228, 2014 [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Barricarte R, Recalde S, Fernández-Robredo P, Millán I, Olavarrieta L, Viñuela A, Pérez-Pérez J, García-Layana A, Rodríguez de Córdoba S; Spanish Multicenter Group on AMD : Relevance of complement factor H-related 1 (CFHR1) genotypes in age-related macular degeneration. Invest Ophthalmol Vis Sci 53: 1087–1094, 2012 [DOI] [PubMed] [Google Scholar]
- 32.Sánchez Chinchilla D, Pinto S, Hoppe B, Adragna M, Lopez L, Justa Roldan ML, Peña A, Lopez Trascasa M, Sánchez-Corral P, Rodríguez de Córdoba S: Complement mutations in diacylglycerol kinase-ε-associated atypical hemolytic uremic syndrome. Clin J Am Soc Nephrol 9: 1611–1619, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.