Abstract
The mTORC1 inhibitor everolimus (Afinitor/RAD001) has been approved for multiple cancer indications, including ER+/HER2− metastatic breast cancer. However, the combination of everolimus with the dual PI3K/mTOR inhibitor BEZ235 was shown to be more efficacious than either everolimus or BEZ235 alone in preclinical models. Herein, we describe a male breast cancer (MBC) patient who was diagnosed with hormone receptor-positive (HR+)/HER2− stage IIIA invasive ductal carcinoma and sequentially treated with chemoradiotherapy and hormonal therapy. Upon the development of metastases, the patient began a 200 mg twice-daily BEZ235 and 2.5 mg weekly everolimus combination regimen. The patient sustained a prolonged stable disease of 18 mo while undergoing the therapy, before his tumor progressed again. Therefore, we sought to both better understand MBC and investigate the underlying molecular mechanisms of the patient's sensitivity and subsequent resistance to the BEZ235/everolimus combination therapy. Genomic and immunohistochemical analyses were performed on samples collected from the initial invasive ductal carcinoma pretreatment and a metastasis postprogression on the BEZ235/everolimus combination treatment. Both tumors were relatively quiet genomically with no overlap to recurrent MBC alterations in the literature. Markers of PI3K/mTOR pathway hyperactivation were not identified in the pretreatment sample, which complements previous reports of HR+ female breast cancers being responsive to mTOR inhibition without this activation. The postprogression sample, however, demonstrated greater than fivefold increased estrogen receptor and pathogenesis-related protein expression, which could have constrained the PI3K/mTOR pathway inhibition by BEZ235/everolimus. Overall, these analyses have augmented the limited episteme on MBC genetics and treatment.
Keywords: neoplasm of the breast
INTRODUCTION
The phosphatidylinositol-3-kinase (PI3K)/serine-threonine protein kinase AKT/mammalian target of rapamycin (mTOR) signaling pathway plays a crucial role in several cellular functions, including growth, differentiation, survival, and metabolism. Aberrations of this pathway are key drivers of carcinogenesis in many malignancies, and they are frequently implicated in both primary and acquired resistance to anticancer targeted therapies, as well as to cytotoxic agents and radiation (Burris 2013). In particular, the activation of the PI3K/AKT/mTOR pathway has been indicated as a main mechanism of resistance to endocrine therapy in hormone receptor (HR)-positive breast cancers, and several studies suggest a mutual regulatory effect between PI3K/AKT/mTOR and estrogen receptor (ER) signaling (Ciruelos Gil 2014).
The mTOR inhibitor everolimus (Afinitor/RAD001) was seen to significantly potentiate the activity and efficacy of the steroidal aromatase inhibitor exemestane in patients with HR+/HER2– advanced breast cancers refractory to nonsteroidal aromatase inhibitors (Baselga et al. 2012; Piccart et al. 2014), and, thus, this combination has been approved as first-line treatment for this setting. However, evidence indicates that the inhibition of mTOR is not sufficient for achieving a complete blockade of the PI3K/AKT/mTOR pathway, given the numerous regulatory loops that provide potential escape mechanisms (Efeyan and Sabatini 2010).
BEZ235 inhibits class I PI3K molecules and both mTORC1 and mTORC2 complexes. Preclinical models demonstrated that BEZ235 inhibited downstream molecules of mTOR, classic rapamycin-induced AKT activation, and tumor growth in xenografts (Maira et al. 2008). In dose-escalation trials of BEZ235, 20 of 27 patients undergoing FDG-PET (fluorodeoxyglucose-positron emission tomography) imaging demonstrated a decreased uptake, suggestive of some level of tumor shrinkage (Bendell et al. 2015). Xu et al. (2011) first showed that everolimus and BEZ235 had a synergistic effect in lung cancer cell lines and xenografts. Additional preclinical studies on multiple cancer models, including breast cancer, have confirmed this synergy and that the combination of subtherapeutic doses of everolimus synergized with BEZ235 to increase its potency more than 10-fold (Nyfeler et al. 2012; Thomas et al. 2012; Passacantilli et al. 2014; Leung et al. 2015).
In this report, we describe a male breast cancer patient who was treated with the everolimus/BEZ235 combination therapy as third-line treatment for his metastatic disease and experienced a prolonged stable disease. Therefore, we investigated molecular mechanisms to explain the benefit and subsequent resistance to this combination treatment.
RESULTS
Clinical Presentation
A 66-yr-old Caucasian man underwent a right radical mastectomy and sentinel lymph node biopsy for an infiltrative breast carcinoma of papillary histotype. Previous medical history included hypertension, carotid atherosclerosis, and diabetes mellitus. There was no family history of breast cancer or other cancers. The tumor was pT1c, N0/1, M0, R0, G2, Ki67 15%, ER+, PR+, HER2–. He started adjuvant tamoxifen treatment.
Two years later, multiple abnormal lymph nodes in the left axilla and a subcentimetric lesion with malignant features in the left mammary gland were detected. The patient underwent left radical mastectomy with axillary lymph node dissection (pretreatment sample). Histology revealed a stage IIIa infiltrative ductal carcinoma, pT1a, N+ 9/11, R0, Ki67 8%, ER+, PR+, HER2–. No distant metastases were detected. The patient was subsequently treated with standard adjuvant chemotherapy, consisting of four cycles of doxorubicin plus paclitaxel every 3 wk; followed by four cycles of cyclophosphamide, methotrexate, and 5-fluorouracil every 4 wk; and 5 wk of radiotherapy. Subsequently, he started adjuvant hormonotherapy with the nonsteroidal aromatase inhibitor letrozole.
The patient developed multiple nodal and bilateral lung metastases 13 mo later. Fine-needle aspiration was performed on the largest left axillary lymph node, confirming the recurrence of ductal breast carcinoma. The patient started 12 courses of first-line chemotherapy with vinorelbine plus capecitabine every 3 wk. Given the achievement of stable disease control by radiological assessments, the patient was shifted to maintenance hormonotherapy with high-dose estrogen receptor antagonist fulvestrant (500 mg i.m. every 4 wk). After 8 mo, the left axillary nodal metastases progressed.
The patient was treated by an investigational combination of BEZ235 (200 mg orally, twice daily) and subtherapeutic everolimus (2.5 mg orally, weekly). The combination therapy was generally well tolerated, except for a G3 skin rash developed likely as a cumulative toxicity after 16 mo of treatment. Standard everolimus dosing is 10 mg daily, whereas 300 mg b.i.d. is the maximum tolerated single-agent dose for BEZ235 (Bendell et al. 2015). The combination treatment enabled sustained disease control for 18 mo.
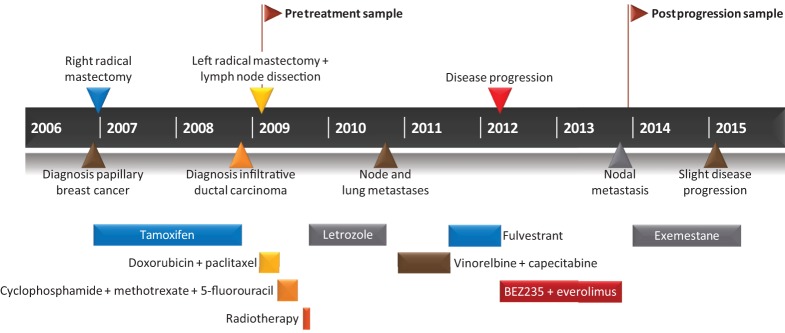
When a new nodal metastasis occurred in the left infraclavicular region during continuous BEZ235/everolimus combination treatment, this treatment was discontinued and the infraclavicular lymph node underwent surgical biopsy (postprogression sample). Immunohistochemistry revealed the tumor as Ki67 10%–12%, ER+, PR+, and HER2–. The patient then started endocrine therapy with the steroidal aromatase inhibitor exemestane. Sixteen months later, a modest increase in the volume of left axillary lymph nodes was reported on computed tomography (CT) scan. The clinical history is depicted in Figure 1.
Figure 1.
Clinical history of the patient. A 66-yr-old Caucasian man was diagnosed with infiltrative papillary breast cancer. While on tamoxifen, he developed a contralateral infiltrative ductal breast carcinoma (pretreatment sample). Following disease progression of his metastasis, he was treated with BEZ235 and everolimus. Stable disease was maintained for 18 mo. Upon nodal metastasis, treatment was discontinued and the postprogression sample collected.
Pharmacokinetics
A full pharmacokinetic (PK) profile of BEZ235 was collected from this patient with Cmax (peak plasma concentration of the drug after administration), AUC (area under the curve), and T1/2 (elimination half-life) of 766 ng/mL, 6308 ng h/mL, and 5.53 h, respectively. The predose trough concentration (Cmin) of BEZ235 in plasma after 200 mg b.i.d. observed over the study period had values of 504, 262, 250, 75.4, and 363 ng/mL, respectively, for Period 1 Day 8 morning dose, Period 1 Day 8 evening dose, Period 2 Day 1 morning dose, Period 5 Day 1 morning dose, and Period 8 Day 1 morning dose. Everolimus Cmin after weekly administration was near or below the limit of detection of the analytical method as expected based on the half-life of the drug of ∼30 hr.
Genomic Analyses
Genomic analysis was undertaken to attempt to understand why this patient sustained prolonged stable disease to the BEZ235/everolimus combination treatment and to better characterize male breast cancer (MBC), a rare disease (1% of all breast cancer cases). Therefore, whole-exome sequencing and analysis was performed on the pretreatment and postprogression samples and compared with a whole-blood normal control.
Overall, the tumors had few somatic alterations. Each tumor encoded 18 shared and nine private somatic short variants (point mutations or indels; Table 1). All but two of the private short variants (RNF212 p.L105S and FUBP1 p.W537*) were determined to be subclonal after adjusting for tumor purity and copy-number alterations. The tumors were also relatively stable at the chromosome level, sharing only a copy-number-neutral loss of heterozygosity in Chromosome 1p, gain of Chromosome 16p, and loss of Chromosome 16q.
Table 1.
Genomic variants detected by whole-exome sequencing
| Gene symbol | Chromo-some | HGVS DNA reference | HGVS protein reference | Variant type | dbSNP/COSMIC ID | Comment | Pre-tx depth | Pre-tx allelic ratio | Post-tx depth | Post-tx allelic ratio | Normal depth | Normal allelic ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FUBP1 | Chr1 | g.chr1:78422351C>T | p.W537* | Stop Gained | n/a | Pre-tx only | 107 | 0.26 | 132 | 0 | 61 | 0 |
| RNF212 | Chr4 | g.chr4:1079732A>G | p.L105S | Missense | n/a | Pre-tx only | 99 | 0.32 | 153 | 0 | 94 | 0 |
| RP1 | Chr8 | g.chr8:55541441TC>T | p.S1667fs | Frameshift | n/a | Pre-tx only | 125 | 0.04 | 267 | 0 | 163 | 0 |
| CPXM2 | Chr10 | g.chr10:125622146C>T | p.G166E | Missense | n/a | Pre-tx only | 39 | 0.08 | 53 | 0 | 49 | 0 |
| CACNA1C | Chr12 | g.chr12:2788661C>T | p.R1704W | Missense | rs373996684 | Pre-tx only | 184 | 0.18 | 322 | 0 | 175 | 0 |
| ABCC9 | Chr12 | g.chr12:22005137C>A | p.G888V | Missense | n/a | Pre-tx only | 72 | 0.18 | 182 | 0 | 102 | 0 |
| NTS | Chr12 | g.chr12:86272172G>A | p.C62Y | Missense | n/a | Pre-tx only | 89 | 0.15 | 266 | 0 | 108 | 0 |
| CACNA1H | Chr16 | g.chr16:1252074G>A | p.E542K | Missense | n/a | Pre-tx only | 58 | 0.07 | 158 | 0 | 35 | 0 |
| AMFR | Chr16 | g.chr16:56396967G>A | p.R596C | Missense | n/a | Pre-tx only | 33 | 0.09 | 29 | 0 | 28 | 0 |
| SPEN | Chr1 | g.chr1:16259731CCAGAAAACAACCCGAT>C | p.QKTTRS2333fs | Frameshift | n/a | Both | 138 | 0.35 | 184 | 0.66 | 122 | 0 |
| SYT6 | Chr1 | g.chr1:114640393C>T | p.A406T | Missense | rs142164979 | Both | 61 | 0.23 | 83 | 0.29 | 42 | 0 |
| ZDBF2 | Chr2 | g.chr2:207175047G>A | p.R1932H | Missense | COSM2150473 | Both | 173 | 0.21 | 209 | 0.42 | 145 | 0 |
| YEATS2 | Chr3 | g.chr3:183515744C>T | p.P1044L | Missense | rs376025260 | Both | 70 | 0.21 | 115 | 0.30 | 58 | 0 |
| CCDC71L | Chr7 | g.chr7:106300865T>G | p.S160R | Missense | n/a | Both | 13 | 0.23 | 17 | 0.35 | 15 | 0 |
| KIAA1045 | Chr9 | g.chr9:34971537G>A | p.R81Q | Missense | rs374394658 | Both | 139 | 0.18 | 173 | 0.39 | 97 | 0 |
| CCDC180 | Chr9 | g.chr9:100092618C>G | p.P659A | Missense | n/a | Both | 107 | 0.26 | 148 | 0.38 | 93 | 0 |
| CNNM1 | Chr10 | g.chr10:101090101G>C | p.E319D | Missense | n/a | Both | 158 | 0.15 | 189 | 0.35 | 132 | 0 |
| MICAL2 | Chr11 | g.chr11:12278507A>G | p.D1044G | Missense | n/a | Both | 34 | 0.29 | 50 | 0.36 | 23 | 0 |
| KDM4E | Chr11 | g.chr11:94760214A>G | p.N498S | Missense | n/a | Both | 69 | 0.13 | 63 | 0.33 | 35 | 0 |
| TSPAN8 | Chr12 | g.chr12:71526521A>T | p.D176E | Missense | n/a | Both | 182 | 0.18 | 287 | 0.37 | 180 | 0 |
| CORO6 | Chr17 | g.chr17:27942821C>T | p.V450M | Missense | n/a | Both | 122 | 0.25 | 171 | 0.32 | 92 | 0 |
| AKAP1 | Chr17 | g.chr17:55183750AGCTTGGATAGAAATGAGGAGG>A | p.SLDRNEEG309S | Codon Deletion | COSM1244721 | Both | 151 | 0.11 | 181 | 0.06 | 84 | 0 |
| NOTCH3 | Chr19 | g.chr19:15280935A>T | p.W1721R | Missense | n/a | Both | 12 | 0.50 | 12 | 0.25 | 10 | 0 |
| CPAMD8 | Chr19 | g.chr19:17017774C>T | p.A1386T | Missense | n/a | Both | 148 | 0.23 | 159 | 0.37 | 104 | 0 |
| ZNF420 | Chr19 | g.chr19:37618255A>G | p.N121S | Missense | n/a | Both | 99 | 0.21 | 244 | 0.35 | 134 | 0 |
| ZNF473 | Chr19 | g.chr19:50550059A>T | p.R787* | Stop Gained | n/a | Both | 180 | 0.26 | 205 | 0.36 | 135 | 0 |
| KRTAP10-3 | Chr21 | g.chr21:45978592T>C | p.T3A | Missense | rs452472 | Both | 13 | 0.62 | 7 | 0.43 | 0 | 0 |
| HTRA2 | Chr2 | g.chr2:74757932T>A | p.L232Q | Missense | n/a | Post-tx only | 256 | 0.00 | 225 | 0.23 | 103 | 0 |
| LRAT | Chr4 | g.chr4:155665556T>A | p.S26R | Missense | n/a | Post-tx only | 352 | 0 | 235 | 0.03 | 169 | 0 |
| CREB3 | Chr9 | g.chr9:35736430T>G | p.Y275D | Missense | n/a | Post-tx only | 87 | 0 | 81 | 0.07 | 68 | 0 |
| ALOX5 | Chr10 | g.chr10:45907697T>G | p.L164V | Missense | n/a | Post-tx only | 280 | 0 | 219 | 0.05 | 133 | 0 |
| KNDC1 | Chr10 | g.chr10:135013889G>T | p.E972* | Stop Gained | n/a | Post-tx only | 87 | 0 | 62 | 0.15 | 45 | 0 |
| SLITRK6 | Chr13 | g.chr13:86368728T>TA | p.Y639Lfs | Frameshift | n/a | Post-tx only | 278 | 0 | 762 | 0.14 | 175 | 0 |
| PML | Chr15 | g.chr15:74336964G>C | p.R755P | Missense | n/a | Post-tx only | 471 | 0 | 363 | 0.07 | 235 | 0 |
| ARHGAP35 | Chr19 | g.chr19:47423891GC>AA | p.P654T | Missense | n/a | Post-tx only | 217 | 0 | 179 | 0.13 | 109 | 0 |
| RRP1 | Chr21 | g.chr21:45213265C>T | p.R114C | Missense | n/a | Post-tx only | 196 | 0 | 136 | 0.14 | 83 | 0 |
HGVS, Human Genome Variation Society; dbSNP, Database of Single Nucleotide Polymorphisms; COSMIC, Catalogue of Somatic Mutations in Cancer; Pre-tx, pretreatment sample; Post-tx, postprogression sample; n/a, not applicable.
Although there was no family history of breast cancer, we examined BRCA1 and BRCA2 for genomic alterations given that the patient developed two histologically distinct cases of breast cancer. No somatic alterations were detected in either gene, but three germline missense mutations were identified: BRCA1 p.Q309R, BRCA2 p.V2466R, and BRCA2 p.N372H. None are clear breast cancer susceptibility loci; however, certain contradictory studies have suggested that these BRCA2 polymorphisms may or may not confer increased risk (Cox et al. 2005; Palli et al. 2007; Cecener et al. 2014; Xue et al. 2014).
We assessed the mutations noted in Table 1 in the context of the targeted pathway and previously reported alterations. No somatic mutations or copy-number variants were identified in genes encoding members of the PI3K/mTOR pathway, such as PIK3CA, PTEN, MTOR, TSC1, or TSC2. Gene or chromosomal region alterations previously uncovered by other studies (Dawson et al. 1996; The Cancer Genome Atlas Network 2012; Deb et al. 2013), such as TP53, KRAS, 8q gain, 11q gain, 17q gain, 20q gain, or 9q loss, were absent in this patient. Only two of the identified short variants (point mutations or indels) were previously reported in the COSMIC database, but with not more than two supporting records each. Of the 36 total genes with point mutations or indels, only SLITRK6 and CACNA1C were also mutated in the 10 MBC patients from the Cancer Genome Atlas (TCGA; The Cancer Genome Atlas 2012); however, the impact of these genes on cancer is unclear.
Three of the somatic variants were of greater interest, however. One of the shared short variants was a 16-bp frameshift deletion in the Msx2-binding domain of SPEN, which encodes a potential negative regulator of estrogen receptor (ER) transcriptional activity (Shi et al. 2001). The pretreatment sample had a private nonsense mutation in FUBP1, a gene commonly mutated in lower-grade gliomas (The Cancer Genome Atlas Research Network 2015). The postprogression sample also had a 9-megabase loss in Chromosome 12p (Supplemental Fig. S1), which contains the tumor suppressor p27Kip1 (encoded by CDKN1B).
Protein Expression Analyses
To determine whether important signaling molecules were being regulated at the protein level, immunohistochemistry (IHC) was performed on the pretreatment and postprogression tumor samples (Supplemental Data). The examined signaling pathway biomarkers (HER2, PTEN, pAKT, pS6-235/235, pS6-240/244, pMAPK, pMEK, and pEGFR) presented no or minimal expression changes between the two tumor specimens. These results suggest that these proteins were noncontributory in the tumor's resistance to the BEZ235/everolimus combination therapy. Despite no differences between the time points, overall expression of phosphorylated ribosomal protein S6 at Ser240/244, a downstream target of mTORC1, in both samples was moderate (H-score 140).
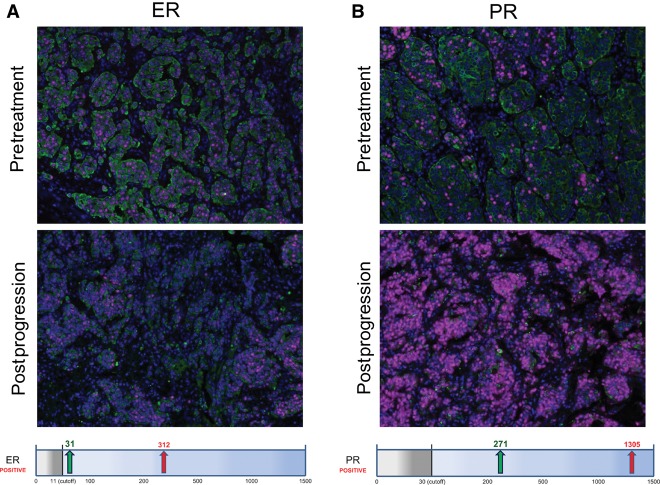
Hormone receptor status was also evaluated for this patient. IHC indicated low ER/PR expression (ER 30% 1+; PR 20% 1+, 10% 3+) in the pretreatment sample, but expression levels of both receptors were increased (60% 1+) in the postprogression sample. Analysis of these samples using the more quantitative Automated Quantitative Analysis (AQUA) IHC technology platform verified a 10-fold score increase of ER from 31 to 312 and an almost fivefold increase of PR from 271 to 1305 (Fig. 2).
Figure 2.
AQUA quantitative immunohistochemistry showed a 10-fold increase in estrogen receptor (ER) (A) and an almost fivefold increase in progesterone receptor (PR) (B) between the pretreatment and postprogression samples. HER2 expression remained negative in both samples. The graphs below the image display the AQUA scores. The gray box indicates the cutoff, the green arrow, the pretreatment score, and the red arrow, the postprogression score. Microscopy images at 40× magnification.
DISCUSSION
MBC is a rare disease, with only 2350 estimated new cases and 440 deaths in the United States in 2015, ∼1% of female breast cancer and <0.5% of male cancer overall (American Cancer Society 2015). Because of its infrequency, the treatment and genetics of MBC is not as well understood as for females.
Here we described a male breast cancer patient who received combination therapy of the dual PI3K/mTOR inhibitor BEZ235 and low-dose everolimus as a third-line treatment for his metastatic ductal carcinoma. This treatment yielded stable disease for 18 mo before progression. Therefore, we chose to pursue genomic and immunohistochemical analyses to understand the molecular mechanism of this sensitivity and subsequent resistance, as well as the underlying genetics of MBC.
Because of the extended stable disease the patient sustained from the PI3K/mTOR inhibitors, we had hypothesized that genomic and/or protein expression alterations in this pathway would suggest a clear biological explanation. The results, though, did not suggest hyperactivation of this pathway in the patient's tumor prior to the BEZ235/everolimus combination therapy. This lack of obvious hyperactivation could potentially be due to the pretreatment sample being from initial diagnosis of the invasive ductal carcinoma, before the development of detectable metastases and the several additional lines of therapy prior to the combination treatment. However, previous research has demonstrated that PI3K pathway alterations are not necessary for response to PI3K or mTOR inhibitors (Juric and Baselga 2012; Hortobagyi et al. 2015), reinforcing that the clinical benefit from PI3K or mTOR inhibition is not dependent on PI3K pathway hyperactivation. Nevertheless, there was moderate expression of the mTORC1-specific phosphorylation of ribosomal protein S6, suggesting that this pathway may be active in the tumors. Additionally, the tumor had a very quiet copy-number profile, which has been associated in general with better prognosis (Ott et al. 2003; Burrell et al. 2010; A'Hern et al. 2013). In the BOLERO-2 clinical trial in HR+/HER2− metastatic female BC patients, patients with lower chromosomal instability had increased progression-free survival gain from everolimus treatment (Hortobagyi et al. 2015). Therefore, although there is no clear genomic or protein biomarker explanation for the patient's sensitivity to the BEZ235/everolimus combination therapy, the patient's HR positivity and chromosomal stability correspond similarly with response to other PI3K or mTOR inhibition in female BC patients.
To explain the tumor's subsequent resistance to the combination PI3K/mTOR inhibition, genomic and histological examination revealed both the loss of a tumor suppressor and increased expression of the hormone receptors as potential contributing factors. The loss of a 9-megabase region of Chromosome 12p in the postprogression tumor included the cell cycle inhibitor p27Kip1, a protein normally inhibited by the PI3K pathway. Therefore, deletion of one copy of the gene may have partially released the inhibition on cell proliferation that occurred because of up-regulation of p27Kip1 from shutting down the PI3K pathway. Supporting this notion, previous research has shown that the level of p27Kip1 expression correlates with response to BEZ235 in mouse embryo fibroblasts (Lee et al. 2011). Additionally, immunohistochemistry detected a five- to 10-fold increase in hormone receptors in the postprogression sample, compared with the pretreatment sample. Resistance associated with an increase in hormone receptor levels has been seen in a number of patients treated with a PI3Kα inhibitor, and in vitro work revealed that suppression of the PI3K pathway increases transcription of ESR1 and expression of ER (Bosch et al. 2015). The patient's tumor's renewed dependence on the hormone receptors is supported by further disease control upon treatment with the aromatase inhibitor exemestane. Therefore, the decrease of cell cycle inhibition combined with an increase in ER and PR levels may have been enough to abrogate the effect of the PI3K/mTOR pathway blockade by BEZ235/everolimus.
To better understand the genomics of sporadic MBC, we profiled the patient's pretreatment and postprogression tumors and found 27 short variants detected in each sample, gain of Chromosome 16p, and loss of Chromosome 16q. No mutations were found in the common breast cancer genes PIK3CA, TP53, CDH1, GATA3, or MAP3K1; no overlap was discovered with common alterations from the 10 reported MBC cases by TCGA; and no high-level amplifications of oncogenes were detected. Although the contribution of most of the identified genomic alterations are unclear, the shared SPEN 16-bp frameshift deletion may be biologically relevant: Previous research suggests that SPEN is a negative regulator of ER transcriptional activity whose expression is increased in the presence of estradiol, creating a potential self-regulatory loop for the estrogen response (Shi et al. 2001). The loss of function of this repressor could therefore potentially allow for increased ER transcriptional activity in this ER-positive tumor. Additionally, the nonsense mutation in FUBP1 in the localized pretreatment tumor is interesting because loss-of-function mutations are common in lower-grade gliomas (The Cancer Genome Atlas Research Network 2015); however, its role in breast cancer is unknown. Overall, however, the relatively quiet genomes suggest that the patient's tumors could be driven by transcriptional, epigenetic, and/or cell signaling mechanisms, particularly those related to hormone receptors.
In conclusion, genomic and histological examination of this MBC patient with prolonged stable disease while under BEZ235/everolimus combination treatment has given us further insight into both MBC and response/resistance to this treatment. Additional studies will be necessary to understand the role of FUBP1 inactivation in MBC and to confirm the hypothesis of decreased p27Kip1 expression and increased hormone receptor expression as a mechanism of resistance to PI3K inhibitors.
METHODS
Clinical Assessment
The patient had tumor assessments by CT scan every 8 wk from the start of BEZ235/everolimus combination treatment until disease progression. The patient had weekly visits for clinical examination during the first 8 wk of treatment; thereafter, the patient returned for visits every 28 d.
Pharmacokinetics
A full PK profile of BEZ235 was collected in this patient at Cycle 1 Day 8. Concentration of BEZ235 in plasma was determined by a LC-MS/MS (liquid chromatography–tandem mass spectrometry) method following protein precipitation. The method has an LLOQ (lower limit of quantification) of 2.0 ng/mL. Everolimus blood concentration in whole blood (K2 EDTA) was determined by the LC-MS/MS method following solid-phase extraction. The method has an LLOQ of 0.300 ng/mL. PK parameters were calculated using noncompartmental methods.
Immunohistochemistry Assays
During treatment, formalin-fixed paraffin-embedded (FFPE) slides were stained and reviewed by a certified pathologist at the University of Verona. Pretreatment and postprogression results were further validated for this study. HER2 negativity was confirmed via IHC by ARUP Laboratories. FFPE slides were stained and expression of Ki-67, pMAPK, pEGFR, pMEK, pAKT, pS6-235/236, pS6-240/244, and PTEN was assessed by a certified pathologist. ER and PR expression was confirmed and quantified using AQUA technology (Genoptix); Fluorescent antibodies were bound, digitized using the slide-scanner system, and measured based on nuclear expression in tumor cells.
Genomic Sequencing and Analysis
Tumor purity of pretreatment and postprogression tumor FFPE sections was pathologically estimated at 85% and 75%, respectively. DNA was extracted from FFPE and whole-blood control samples using the Promega Maxwell DNA extraction kit. Libraries were constructed using Illumina TruSeq, captured with the Agilent SureSelect Whole Exome V4 baits and protocol, and sequenced on an Illumina HiSeq 2500 as 100-bp paired-end reads. Resulting FASTQ files were aligned to the hg19 reference genome using BWA-MEM (Li 2013), duplicates were marked with Picard (http://broadinstitute.github.io/picard/), and local realignment and base quality recalibration were performed with GATK (Genome Analysis Toolkit; McKenna et al. 2010; DePristo et al. 2011). Resulting mean target coverage of the pretreatment and postprogression FFPE tumor specimens were 142× and 179×, respectively (Table 2). The whole-blood control normal control was purposely sequenced to a lower coverage of 82×.
Table 2.
Sequencing metrics
| Sample | Total reads | Unique mapped reads | Mean target coverage |
|---|---|---|---|
| Pretreatment | 118,318,058 | 92,907,513 | 142 |
| Postprogression | 137,438,824 | 120,704,547 | 179 |
| Normal | 59,691,734 | 56,623,598 | 82 |
Single-nucleotide variants were called in paired normal mode using MuTect (Cibulskis et al. 2013) and phased with GATK, whereas indels were called with Pindel (Ye et al. 2009) and filtered against a pool of normal samples. Annotation was performed with SnpEff (Cingolani et al. 2012), using dbSNP v141 (http://www.ncbi.nlm.nih.gov/SNP/), COSMIC v70 (Forbes et al. 2015), and dbNSFP v2.4 (Liu et al. 2013). Nonsynonymous protein-coding SNVs or indels with coverage of at least 10× or 30×, respectively, in at least one tumor sample were retained. All calls were manually reviewed in IGV (Integrative Genomics Viewer; Robinson et al. 2011). Copy number, tumor purity, loss of heterozygosity, and subclonality were performed using an in-house implementation, the ABSOLUTE algorithm (Carter et al. 2012), after the GC normalization step from Control-FREEC (Janevski et al. 2012). Tumor purity was calculated to be 47% in the pretreatment sample and 72% in the posttreatment sample. Common alterations in the 10 MBC patients from the BRCA TCGA study were accessed via the Breast Invasive Carcinoma (TCGA, Provisional) data set at http://cBioPortal.org (Cerami et al. 2012; Gao et al. 2013).
ADDITIONAL INFORMATION
Ethics Statement
The treatment of this patient in the clinical trial was approved by the University of Verona's Institutional Review Board and conducted in accordance with Good Clinical Practice, the Declaration of Helsinki, and other applicable local regulations. The patient provided written informed consent for sequence analysis.
Database Deposition and Access
The somatic short variants (point mutations and indels), BRCA germline variants, copy number alterations, and LOH regions for both the pretreatment and postprogression samples (Supplemental Table 2) identified in this study have been deposited in ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) under accession numbers SCV000258645–SCV000258901. Permission has not been granted for deposition of sequencing data.
Acknowledgments
We extend our gratitude to the patient who consented for this investigational combination therapy and exploratory biomarker analysis and provided tumor and blood samples.
Author Contributions
A.R.B., M.F., M.R., P.P., M.M., D.C., and D.M. conceived/designed the study. M.F., C.C., S.C., P.P., G.T., and D.M. contributed to patient material collection. M.F. and D.M. provided patient history information. A.R.B., J.H., W.C., M.R., J.G., G.T., M.M., and D.C. contributed to analysis and interpretation of genomic and histological results. All authors contributed to manuscript writing, editing, and approval.
Funding
Research in the laboratory of D.M. is supported in part by the Associazione Italiana per la Ricerca sul Cancro (AIRC) Start-Up no. 10129, and 5 per mille no. 10016 grants. Part of the work was performed at the Laboratorio Universitario di Ricerca Medica (LURM) Research Center, University of Verona. Novartis provided support, including the generation and analysis of sequence data for this study.
Competing Interest Statement
A. Rose Brannon, David Chen, Jennifer Hummel, Jorge Gallo, Markus Riester, Parul Patel, Wing Cheung, and Michael Morrissey work for Novartis.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- A'Hern RP, Jamal-Hanjani M, Szasz AM, Johnston SR, Reis-Filho JS, Roylance R, Swanton C. 2013. Taxane benefit in breast cancer—a role for grade and chromosomal stability. Nat Rev Clin Oncol 10: 357–364. [DOI] [PubMed] [Google Scholar]
- American Cancer Society. 2015. Cancer facts & figures 2015. American Cancer Society, Atlanta. [Google Scholar]
- Baselga J, Campone M, Piccart M, Burris HA III, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, et al. 2012. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 366: 520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendell JC, Kurkjian C, Infante JR, Bauer TM, Burris HA III, Greco FA, Shih KC, Thompson DS, Lane CM, Finney LH, et al. 2015. A phase 1 study of the sachet formulation of the oral dual PI3K/mTOR inhibitor BEZ235 given twice daily (BID) in patients with advanced solid tumors. Invest New Drugs 33: 463–471. [DOI] [PubMed] [Google Scholar]
- Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, Tao JJ, Spratt DE, Viola-Villegas NT, Castel P, et al. 2015. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med 7: 283ra251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, Juul N, Johnston SR, Reis-Filho JS, Szallasi Z, Swanton C. 2010. Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J Cell Biochem 111: 782–790. [DOI] [PubMed] [Google Scholar]
- Burris HA III. 2013. Overcoming acquired resistance to anticancer therapy: focus on the PI3K/AKT/mTOR pathway. Cancer Chemother Pharmacol 71: 829–842. [DOI] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. 2012. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecener G, Egeli U, Tunca B, Erturk E, Ak S, Gokgoz S, Tasdelen I, Tezcan G, Demirdogen E, Bayram N, et al. 2014. BRCA1/2 germline mutations and their clinical importance in Turkish breast cancer patients. Cancer Invest 32: 375–387. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang Le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruelos Gil EM. 2014. Targeting the PI3K/AKT/mTOR pathway in estrogen receptor–positive breast cancer. Cancer Treat Rev 40: 862–871. [DOI] [PubMed] [Google Scholar]
- Cox DG, Hankinson SE, Hunter DJ. 2005. No association between BRCA2 N372H and breast cancer risk. Cancer Epidemiol Biomarkers Prev 14: 1353–1354. [DOI] [PubMed] [Google Scholar]
- Dawson PJ, Schroer KR, Wolman SR. 1996. ras and p53 genes in male breast cancer. Mod Pathol 9: 367–370. [PubMed] [Google Scholar]
- Deb S, Do H, Byrne D, Jene N, kConFab I, Dobrovic A, Fox SB. 2013. PIK3CA mutations are frequently observed in BRCAX but not BRCA2-associated male breast cancer. Breast Cancer Res 15: R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Sabatini DM. 2010. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol 22: 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. 2015. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 43(Database issue): D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortobagyi GN, Chen D, Piccart M, Rugo HS, Burris HA III, Pritchard KI, Campone M, Noguchi S, Perez A, Deleu I, et al. 2015. Correlative analysis of genetic alterations and everolimus benefit in hormone receptor–positive, human epidermal growth factor receptor 2–negative advanced breast cancer: results from BOLERO-2. J Clin Oncol 10.1200/JCO.2014.60.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janevski A, Varadan V, Kamalakaran S, Banerjee N, Dimitrova N. 2012. Effective normalization for copy number variation detection from whole genome sequencing. BMC Genomics 13Suppl 6: S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D, Baselga J. 2012. Tumor genetic testing for patient selection in phase I clinical trials: the case of PI3K inhibitors. J Clin Oncol 30: 765–766. [DOI] [PubMed] [Google Scholar]
- Lee M, Theodoropoulou M, Graw J, Roncaroli F, Zatelli MC, Pellegata NS. 2011. Levels of p27 sensitize to dual PI3K/mTOR inhibition. Mol Cancer Ther 10: 1450–1459. [DOI] [PubMed] [Google Scholar]
- Leung EY, Askarian-Amiri M, Finlay GJ, Rewcastle GW, Baguley BC. 2015. Potentiation of growth inhibitory responses of the mTOR inhibitor everolimus by dual mTORC1/2 inhibitors in cultured breast cancer cell lines. PLoS One 10: e0131400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. http://arxivorg/abs/13033997.
- Liu X, Jian X, Boerwinkle E. 2013. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat 34: E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chene P, De Pover A, Schoemaker K, et al. 2008. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther 7: 1851–1863. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler B, Chen Y, Li X, Pinzon-Ortiz M, Wang Z, Reddy A, Pradhan E, Das R, Lehar J, Schlegel R, et al. 2012. RAD001 enhances the potency of BEZ235 to inhibit mTOR signaling and tumor growth. PLoS One 7: e48548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott K, Vogelsang H, Mueller J, Becker K, Muller M, Fink U, Siewert JR, Hofler H, Keller G. 2003. Chromosomal instability rather than p53 mutation is associated with response to neoadjuvant cisplatin-based chemotherapy in gastric carcinoma. Clin Cancer Res 9: 2307–2315. [PubMed] [Google Scholar]
- Palli D, Falchetti M, Masala G, Lupi R, Sera F, Saieva C, D'Amico C, Ceroti M, Rizzolo P, Caligo MA, et al. 2007. Association between the BRCA2 N372H variant and male breast cancer risk: a population-based case–control study in Tuscany, Central Italy. BMC Cancer 7: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passacantilli I, Capurso G, Archibugi L, Calabretta S, Caldarola S, Loreni F, Delle Fave G, Sette C. 2014. Combined therapy with RAD001 e BEZ235 overcomes resistance of PET immortalized cell lines to mTOR inhibition. Oncotarget 5: 5381–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccart M, Hortobagyi GN, Campone M, Pritchard KI, Lebrun F, Ito Y, Noguchi S, Perez A, Rugo HS, Deleu I, et al. 2014. Everolimus plus exemestane for hormone-receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: overall survival results from BOLERO-2. Ann Oncol 25: 2357–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev 15: 1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2015. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas HE, Mercer CA, Carnevalli LS, Park J, Andersen JB, Conner EA, Tanaka K, Matsutani T, Iwanami A, Aronow BJ, et al. 2012. mTOR inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Sci Transl Med 4: 139ra184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CX, Li Y, Yue P, Owonikoko TK, Ramalingam SS, Khuri FR, Sun SY. 2011. The combination of RAD001 and NVP-BEZ235 exerts synergistic anticancer activity against non-small cell lung cancer in vitro and in vivo. PLoS One 6: e20899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue WQ, He YQ, Zhu JH, Ma JQ, He J, Jia WH. 2014. Association of BRCA2 N372H polymorphism with cancer susceptibility: a comprehensive review and meta-analysis. Sci Rep 4: 6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.