Abstract
Throughout the past decade, there have been substantial advances in understanding the pathogenesis of idiopathic pulmonary fibrosis (IPF). Recently, several large genome-wide association and linkage studies have identified common genetic variants in more than a dozen loci that appear to contribute to IPF risk. In addition, family-based studies have led to the identification of rare genetic variants in genes related to surfactant function and telomere biology, and mechanistic studies suggest pathophysiologic derangements associated with these rare genetic variants are also found in sporadic cases of IPF. Current evidence suggests that rather than existing as distinct syndromes, sporadic and familial cases of IPF (Familial Interstitial Pneumonia, FIP) likely reflect a continuum of genetic risk. Rapidly evolving bioinformatic and molecular biology techniques, combined with next-generation sequencing technologies, hold great promise for developing a comprehensive, integrated approach to defining the fundamental molecular mechanisms that underlie IPF pathogenesis.
Introduction
Idiopathic Pulmonary Fibrosis (IPF), the most common of the idiopathic interstitial pneumonias (IIPs), is characterized by clinical symptoms of cough and dyspnea, restrictive pulmonary function tests with impaired gas exchange, and progressive lung scarring [1]. Recently, two modestly effective drugs for treating IPF have been identified [2, 3]; however, the prognosis of IPF remains grave, emphasizing a need for a more complete understanding of the mechanisms of disease pathogenesis. Available data indicate that both genetic and environmental factors contribute to risk of IPF and other IIPs [1, 4, 5]. The first insights into IIP genetics came from studies of families with heritable cases of IIP, a syndrome termed Familial Interstitial Pneumonia (FIP). As early as the 1950’s, it was recognized that on occasion, IIP cases clustered in families [6, 7], suggesting a genetic basis to at least a subset of disease. By the 1990’s, it was reported that FIP represented a rare subset of IIP, comprising 3-5% of cases [8]. More recently, estimates from several independent groups have suggested that as many as 20% of IIP cases are familial [9-12]. Studies in families have uncovered rare genetic variants in eight genes that are linked to FIP, including three surfactant-related proteins [surfactant protein C, SFTPC [13-16] and surfactant protein A2, SFTPA2 [17] and ATP-binding cassette member A3 (ABCA3)[18, 19]] as well as five genes linked to telomere function [telomerase reverse transcriptase, TERT [20, 21], human telomerase RNA component, hTR [20, 21], dyskerin, DKC1 [22-24], telomere interacting factor 2, TINF2[25-27] and regulator of telomere elongation helicase, RTEL1[28]]. Rare genetic variants in FIP-associated genes can be found in some cases of sporadic IPF[12], and investigations into the mechanisms through which these mutated genes contribute to disease have uncovered common underlying pathobiological changes that likely contribute to progressive fibrotic remodeling in FIP and sporadic IPF[4, 5].
Common genetic variants in IPF
Several studies since the early 2000’s have investigated the role of functional polymorphisms in a variety of genes in relationship to IPF risk (Table 1). Variants in several genes related to inflammation and immune response, including transforming growth factor beta-1 (TGFB1)[29, 30], interleukin-1 receptor alpha (IL1RN)[31-33], interleukin 8 (IL8)[34], toll-like receptor 3 (TLR3)[35], HLA DRB1*1501[36], as well as cell-cycle progression related genes CDKN1A and TP53 [37], have been nominally associated with IPF risk or progression. However, results from these small studies have not yet been validated in independent cohorts. More recently, several large genome-wide linkage and association studies have been completed and identified numerous additional loci that appear to confer risk for IPF.
Table 1.
Summary of common genetic variants linked to IPF
| Locus | Gene | SNP | IPF Risk |
IPF Survival |
Reference |
|---|---|---|---|---|---|
| 2q14 | IL1RN | rs408392 rs419598 rs2637988 |
Y Y Y |
[31-33] | |
| 3q26 | hTR | rs6793295 | Y | [40] | |
| 4q13 | IL8 | rs4073 rs2227307 |
Y Y |
[34] | |
| 4q22 | FAM13A | rs2609255 | Y | [40] | |
| 4q35 | TLR3 | rs3775291 | Harmful | [35] | |
| 5p15 | TERT | rs2736100 | Y | [40, 112] | |
| 6p21 6p21 |
CDKN1A
HLA- DRB1 |
rs2395655 | Y | Harmful Y |
[37] [36] |
| 6q24 | DSP | rs2076295 | Y | [40] | |
| 7q22 | Intergenic | rs47274443 | Y | [40] | |
| 10q24 | OBFC1 | rs11191865 | Y | [40] | |
| 11p15 |
MUC5B
MUC2 TOLLIP TOLLIP TOLLIP |
rs35705950 rs7934606 rs111521887 rs5743894 rs2743890 |
Y Y Y Y Y |
Protective Protective |
[38-43, 50] [40] [41] [41] [41] |
| 13q34 | ATP11A | rs1278769 | Y | [40] | |
| 14q21 | MDGA2 | rs7144383 | Y | [41] | |
| 15q14- 15 |
Intergenic | rs2034650 | Y | [40] | |
| 17q13 |
TP53
TP53 |
rs12951053 rs12602273 |
N N |
Harmful Harmful |
[37] |
| 17q21 17q21 |
MAPT
SPPL2C |
rs1981997 rs17690703 |
Y Y |
[40] [41] |
|
| 19q13 19q13 |
DPP9
TGFB1 |
rs12610495 rs1800470 |
Y N |
Harmful |
[40] [29, 30] |
MUC5B
In 2011, a genome-wide linkage study identified a locus on chromosome 11 that was significantly associated with IPF risk[38]. Resequencing of this region subsequently identified a common single nucleotide polymorphism (SNP) (rs35705950) in the promoter of the gene encoding for Mucin 5B (Muc5B) that was associated with a 6-8 fold increased risk for IPF. The association of this MUC5B promoter polymorphism and IPF has since been confirmed in several independent cohorts, predominantly in Caucasians [39-44]. Interestingly, it appears the MUC5B SNP has a similar frequency in FIP and sporadic IPF cases[38]. This association, however, may be specific to IIP among interstitial lung diseases (ILD) since reports indicate rs35705950 does not confer increased risk of scleroderma-related ILD or sarcoidosis [39, 42, 45]. This association of rs35705950 with IPF was confirmed in a cohort of Mexican patients [46]; however, rs35705950 was found to be rare in a Korean cohort of IPF patients. Similarly, in a Chinese population, rs35705950 was rare in IPF patients but different MUC5B polymorphisms were associated with disease [47].
While the rs35705950 MUC5B SNP was associated with increased MUC5B mRNA expression in lungs of control subjects, MUC5B expression was uniformly increased in lungs of IPF patients compared to controls regardless of whether the MUC5B SNP was present[38]. Consistent with this observation, increased numbers of MUC5B expressing cells have been detected in the distal airways of IPF patients[48]. MUC5B rs35705950 has also been reported as a risk factor for asymptomatic interstitial lung abnormalities detected on CT scan among subjects over age 50 in the Framingham cohort[49]. Surprisingly, although minor allele carriers of rs35705950 have increased risk of developing disease, IPF patients who carry the minor (risk) allele appear to have improved survival compared to noncarriers[50]. Previous animal studies have suggested that MUC5B regulates airway host defense[51], but the mechanisms by which MUC5B influences fibrotic remodeling are uncertain at present.
Insights from Genome Wide Association Studies
A major advance of the past several years has been the development of large, robust datasets with sufficient statistical power for genome wide association studies (GWAS). Two large independent GWAS of IPF patients have now been conducted and identified numerous genetic loci that confer IPF risk. The first, published in 2013[40], evaluated 1,616 IIP cases (the vast majority of which were IPF) and 4,683 controls subjects with replication in an additional 876 cases and 1,890 controls. In addition to confirming the previously reported association with MUC5B, 9 additional loci were significantly associated with IIP, predominantly IPF (summarized in Table 1), including SNPs near TERT and hTR. Ten SNPs on chromosome 11p15 nominally met genome-wide significance, but after controlling for MUC5B rs35705950 these loci no longer met genome-wide significance, suggesting weak linkage disequilibrium (LD) with the MUC5B promoter polymorphism was largely responsible for this association.
Results of a second GWAS again implicated a locus on chromosome 11p15 as significantly associated with IPF, but did not replicate other risk loci identified by Fingerlin and colleagues. This GWAS [41] performed a three-stage analysis, including a discovery and two replication cohorts, comprising in total 1410 IPF cases and 2934 control individuals. Five loci achieved genome-wide significance, including 4 SNPs on chromosome 11p15 and one on 17q21. Among the 11p15 SNPs were MUC5B rs35705950 and 3 SNPs within the Toll-interacting protein (TOLLIP) locus. LD was reported to be low with rs35705950, suggesting TOLLIP may represent an independent risk locus. Similar to MUC5B rs35705950, IPF cases with the TOLLIP risk allele (the major allele) had decreased mortality compared to minor allele carriers.
Deciphering the biological effects of common genetic variants identified by GWAS has proven challenging so far. It is possible that the relevant biological effect of most individual SNPs is subtle or manifests only in the context of unique additional genetic or environmental factors to confer disease risk. Despite challenges, future studies are needed to clarify the biological role of disease-associated common genetic variants.
Rare genetic variants in FIP and IPF
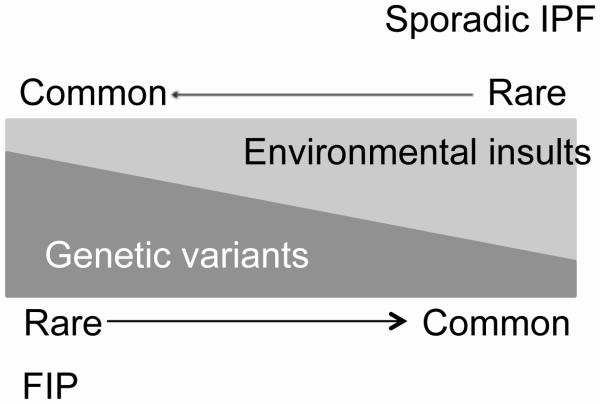
FIP and sporadic IPF share many clinical and histopathologic features[52], which has led to the hypothesis that similar mechanisms underlie the pathogenesis of sporadic and familial disease. Additionally, Scholand et al. employed an extensive genealogical database and found unexpected relatedness among patients who died of what was believed to be sporadic IPF[53], further supporting the idea that the genetic landscapes of sporadic IPF and FIP overlap considerably. Together, studies to date suggest that sporadic and familial disease reflect of spectrum of genetic risk for pulmonary fibrosis (Figure 1). In this model, genetic risk factors of small and large effects interact with rare and common environmental stimuli to produce the phenotype of pulmonary fibrosis. Most FIP kindreds appear to have an autosomal dominant inheritance pattern with incomplete penetrance, suggesting an important role for genetic rare variants (RVs) of large effect. In contrast, sporadic IPF may occur more often in the setting of de-novo or low penetrance RVs, or a combination of more common, less severe genetic risk alleles. As described below, in some cases genetic RVs of large effect may be found in genes that lie within loci also containing common variants associated with IPF risk. Focusing on familial disease offers the ability to use Mendelian approaches to identify disease-associated RVs. This represents a promising approach to enhance mechanistic understanding of the impact of genetic risk factors on development of FIP and potentially sporadic IPF.
Figure 1.
Proposed model of gene × environment interactions in the pathogenesis of pulmonary fibrosis.
Telomerase and short telomeres
Pulmonary fibrosis occurs in approximately 20% of patients with dyskeratosis congenita (DC)[54], a rare inherited genetic disorder characterized by leukoplakia, bone marrow failure and dystrophic nails that typically affects young males. RVs in genes related to telomere biology have been implicated in DC[55]. In 2007, using candidate gene approaches, two groups identified heterozygous loss-of-function RVs in telomere-related genes in 7-15% of FIP families who did not have a history of DC[20, 21]. These variants in TERT and hTR lead to short telomeres in peripheral blood and in the lung [20, 21, 56, 57]. To date, TERT RVs are the most commonly identified mutations linked to FIP; however, TERT RVs are rarely identified in sporadic cases of IPF[56]. In addition to variants in TERT and hTR, two recent reports identified FIP patients with RVs in the gene encoding for dyskerin (DKC1)[22, 23], another component of the telomerase complex. Several reports have also identified pulmonary fibrosis in families with DC associated with RVs in TINF2 [26, 27]. In one of these families, there was evidence of somatic or acquired mosaicism for a deletion which abolished expression of the missense variant [25]. This interesting observation suggests acquired genetic variation may represent one mechanism regulating the clinical spectrum of disease linked to telomere pathway RVs.
Using whole-exome sequencing in a cohort of >180 FIP kindreds, our group has identified heterozygous loss-of-function RVs in another telomere related gene, regulator of telomere elongation helicase (RTEL1) in 9 families with IPF[28]. Similar to TERT, hTR and DKC1 RVs, these RTEL1 RVs are associated with short telomeres in peripheral blood. In addition to a role in telomere maintenance, RTEL1 appears to play a more general role in genome stability, DNA-repair, and replication[58-60], suggesting it may confer disease risk through additional mechanisms. As TERT deficiency has also been associated with abnormal DNA-repair[61], it is possible that this mechanism, rather than direct effects on telomere length, could be an important in mediating disease risk associated with telomerase pathway RVs. Cumulatively, RVs in these five telomere-related genes are reported in approximately 15-20% of FIP families.
Notably, the short telomere phenotype in peripheral blood mononuclear cells (PBMCs) is not limited to patients with loss of function RVs in telomerase complex genes. Approximately 1/3 of sporadic IPF and FIP patients have short telomeres (<10th percentile for age) in PBMCs [56, 57]. In addition, it appears that the majority of IPF patients have short telomeres in alveolar epithelial cells[23, 56], suggesting that additional factors (besides genetic risk) contribute to telomere shortening in lungs of patients with IPF and FIP. Interestingly, asymptomatic first-degree relatives of FIP patients have decreased alveolar epithelial cell telomere length compared to controls, and alveolar epithelial cell telomere length is significantly associated with the presence of interstitial changes on high-resolution chest CT [62]. In addition, it appears that PBMC telomere length within families with TERT RVs can be inherited at least in part independent of a known RV, producing a unique scenario of inherited genetic risk without the risk allele[21, 56, 63]. PBMC telomere length appears to be predictive of survival among patients with IPF, wherein IPF patients with short telomeres have reduced survival compared to those with “normal” length PBMC telomeres [64]. In addition to rare genetic variants in telomere related genes, common genetic variants in loci near TERT, hTR and telomere gene OBFC1 have been linked to sporadic IPF by GWAS [40] and may be an important factor in determining telomere length. Environmental factors, including cigarette smoke exposure, may also play a role in telomere shortening in FIP and sporadic IPF[65].
Genetic and clinical evidence provide a compelling association between lung fibrosis and telomere biology. Although the mechanisms through which telomerase pathway RVs lead to lung fibrosis are uncertain, it has been suggested that these loss of function variants disrupt lung epithelial repair mechanisms[66]. Murine models of telomerase dysfunction have been developed but present a number of challenges that limit their utility for mechanistic studies [4]. In spite of these limitations, several studies have reported attempts to model telomerase deficiency in the lung. Tert null mice have decreased numbers of alveolar epithelial cells and modest architectural changes in the lung[67]. These mice have increased susceptibility to cigarette smoke induced-emphysema[68], but do not develop lung fibrosis. Studies using pro-fibrotic stimuli such as bleomycin to investigate fibrotic susceptibility in Tert and Terc null mice have yielded conflicting results[69, 70]. Together, it appears that recapitulating the biology of telomere dysfunction in humans using mouse models is problematic. Therefore, new approaches are needed in this area.
Surfactant Protein-Related Genes
Nogee and colleagues first described a heterozygous mutation in the gene encoding surfactant protein C (SFTPC) in a young woman and her child with IIP in 2001[13]. Soon after, we identified the first association between SFTPC and FIP in a large family with 11 affected individuals[14]. Subsequently, other groups have reported heterozygous RVs in SFTPC in 1-2% of FIP [9, 12, 14-16, 71]. Although one group reported SFTPC RVs in 25% of FIP kindreds [15], this high frequency was likely related to founder effects. The mechanisms through which SFTPC RVs contribute to disease pathogenesis were recently reviewed elsewhere[4, 72]. In brief, it appears that C-teminal BRICHOS domain mutants result in defects in folding of the propeptide within the endoplasmic reticulum (ER), leading to ER stress and activation of the unfolded protein response[73-77]. Linker domain mutants (such as the I73T RV) appear to alter trafficking of the pro-peptide[78] and lead to dysregulated proteastasis.[79]. Animal modeling suggests induction of ER stress in alveolar epithelial cells is not sufficient to induce spontaneous fibrosis but results in an exaggerated fibrotic response following low-dose bleomycin challenge[80]. Induction of ER stress in alveolar epithelial cells increases susceptibility to apoptotic stimuli[80, 81], increases expression of mesenchymal markers, and enhances production of profibrotic mediators [82, 83]. In addition, RVs in another surfactant protein (SFTPA2)[17] has been linked to FIP. SFTPA2 RVs also result in ER stress and may increase latent TGFβ activation[84, 85].
While the frequency of surfactant protein RVs in sporadic IPF appears to be low[86, 87], several groups have reported that ER stress and UPR activation is a common feature of FIP and sporadic IPF [74, 88], suggesting that environmental factors (such as herpesviruses [74, 89] and tobacco smoke[90-92]) may contribute to this phenotype. Promisingly, it has recently been shown that pharmacologic chaperones might improve processing of mutant surfactant proteins in alveolar epithelial cell lines[93]. These exciting developments raise the possibility of targeted therapies for at least a subset of patients with pulmonary fibrosis.
In addition, RVs in another gene involved in surfactant processing, ATP-binding cassette-type 3 (ABCA3) (previously linked to pediatric interstitial lung disease) have been reported in several FIP families[18, 19, 94], as well as in sporadic cases of IPF[12]. In one consanguineous family[18], homozygous RVs in ABCA3 were identified. A heterozygous RV in ABCA3 was also reported in a patient with “combined pulmonary fibrosis and emphysema”[19]. In another family carrying the I73T SFTPC RV, a second heterozygous RV in ABCA3 modified disease penetrance [94]. The exact mechanisms by which ABCA3 variants confer FIP risk are unclear at present, but presumably relate to epithelial cell dysfunction.
ELMOD2
A report using linkage in a cohort of Finnish FIP families suggested ELMOD2 as a candidate FIP gene[95] and in vitro studies suggest ELMOD2 may play a role in anti-viral responses[96]. Whether ELMOD2 plays an important role in lung fibrosis has not yet been elucidated.
Missing heritability and future genetic discovery
Cumulatively, available literature suggests that rare-variants in FIP genes SFTPC, SFTPA2, ABCA3, TERT, hTR, DKC1, TINF2 and RTEL1 comprise 15-20% of FIP cases (Table 2). However, it is possible that selection of families for these candidate-based genetic studies, as well as founder effects in certain populations, may have overestimated the frequency of some variants among all FIP families. While common genetic variants also confer FIP risk and may explain as much as 30% of FIP risk [40], there remains substantial “missing heritability.” In an effort to identify novel FIP genes, recently, our group has performed extensive whole-exome sequencing of subjects from FIP kindreds. While this approach has implicated RTEL1 as an FIP gene, ongoing analysis suggests that RVs in a single gene (or small group of related genes) do not account for disease in a majority of families [28]. Compared to other genetic lung diseases such as cystic fibrosis and familial pulmonary arterial hypertension, it appears the genetic basis of FIP is substantially more heterogenous. Given the lack of a dominant gene in FIP, the principal challenge is one of power, both within families and across subjects. Linkage or co-segregation based approaches rely upon the power of large, multigenerational pedigrees with numbers of affected individuals, a well-established inheritance model, and ability to clearly ascertain affection status. Considering that each individual typically harbors 200 rare genetic variants in their exomes[97] and most FIP kindreds are small, dozens of RVs will be shared among affected individuals making it difficult to identify the culprit RV by this approach. In light of what appears to be substantial allelic heterogeneity, similar to what has been observed for other familial disorders including hypercholesterolemia [98], neurodegenerative diseases [99] and cardiomyopathies[100], functional testing and validation in cell and/or animal models will be critical to determine the pathogenicity of specific variants and genes tentatively linked to disease.
Table 2.
Rare genetic variants linked to FIP
These challenges suggest that novel and creative approaches will be necessary to further genetic discovery in FIP and IPF. As elaborated further below, we anticipate that evolving bioinformatic approaches to variant prioritization [101, 102], coupled with network and pathway based analysis [103] hold promise for identifying, validating, and integrating disease-associated variants. In addition to rare coding variants, future studies investigating intronic variants or variants in more distant cis- and trans- regulatory regions should further inform understanding of the spectrum of genetic risk for FIP.
Implications for other IIPs
The evolving understanding of the genetic landscape of FIP and sporadic IPF leads to the question of whether the same genes and genetic variants confer risk for other ILDs, including other forms of IIP. Genetic predisposition for IIPs other than IPF has been poorly characterized; however, several clues suggest there are both conserved and distinct genetic risk factors. Within FIP pedigrees, it has been well recognized that individuals may harbor the same disease-associated RV yet present with different histopathologies [13, 52]. This finding indicates that differential environmental exposures overlaid on a common set of genetic risk factors may play a role in determining IIP phenotype. No study to date has been adequately powered to assess whether common genetic variants linked to IPF also confer risk to other IIPs. Future studies are required to address this important issue.
Implications for clinical management
Impact on clinical trial design and treatment
Although no clinical trials to date have stratified patients based on genetic risk, this strategy could prove useful in light of the now recognized associations of the 11p15 risk variants (MUC5B and TOLLIP) with reduced disease progression and improved survival. As additional genes and risk alleles are identified, more complex stratification schemes may be developed to enhance study design. With the recent publication of randomized controlled trials of two pharmacologic agents, pirfenidone [2] and nintedanib[3], that reduce lung function decline among IPF patients, a reasonable question for future study is whether genetic factors influence response to treatment with one or both of these medications.
Whether genetic factors influence outcomes after lung transplant remains an underexplored question. One small series described successful lung transplant in 8 patients with TERT RVs, however a high rate of hematologic and renal toxicities were noted[104]. The role of other rare or common genetic variants on outcome after lung transplant is not known.
Genetic testing
As our understanding of the influence of genetic factors on risk of IPF and its natural history, a potential role for clinical genetic testing emerges. At present, we suggest that evidence is not sufficient to recommend routine genetic testing for rare or common genetic variants for patients with sporadic IPF. In families with FIP, clinical testing for variants in SFTPC, SFTPA2, and telomerase-related genes including TERT, hTR, DKC1 and RTEL1 is available from commercial and academic sources. Our practice is to offer genetic counseling and consideration of genetic testing to patients with FIP and a family history suggestive of a telomerase dysfunction syndrome (including diagnoses of aplastic anemia, cryptogenic cirrhosis, premature graying). Decisions to undergo genetic testing are complex and highly individual, and no studies have evaluated the impact of genetic testing on patients/families in the context of FIP. Extrapolating from our experience and literature from other disorders [105-107], continued close follow-up of patients who undergo genetic testing appears essential regardless of whether they are found to carry a disease-associated RV. In light of the incomplete penetrance and variable expressivity of FIP associated RVs, we recommend patients and their families confer with genetic counselors before consideration of genetic tests that are currently available or may become available in the future. Over time, we anticipate there may be a role for broader screening of common and rare genetic variants associated with IPF.
Overcoming challenges and future directions
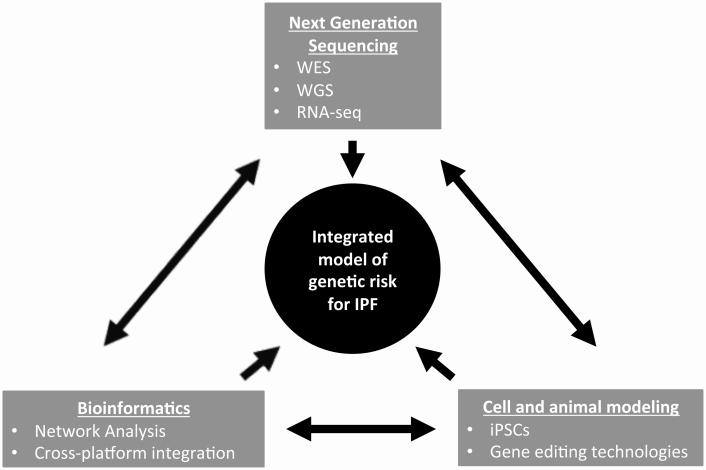
To date, investigations of the genetic basis of FIP and sporadic IPF have provided crucial insights into underlying mechanisms of progressive pulmonary fibrosis. We suggest that there is likely not a single “road to IPF” but rather there are “multiple paths” that converge to a common phenotype. The development of exciting next-generation sequencing capabilities, along with continuously evolving bioinformatic approaches to analysis of large data sets and rapidly improving molecular biological techniques, offer unique possibilities to identify additional novel genes and pathways that contribute to the pathogenesis of progressive pulmonary fibrosis (Figure 2). Whole-genome sequencing is rapidly becoming feasible, and although it presents new and greater bioinformatic challenges, the possibility of analyzing non-coding variants, as well as interactions among variants holds promise in identifying as-of-yet unexplained heritability of FIP and sporadic IPF. In addition, the rapidly evolving field of stem-cell biology offers the possibility of studying the effects of genetic variants in primary human cell-types of interest using inducible pluripotent stem cell (iPSC) differentiation strategies[108, 109]. The development of improved gene-editing technologies including the CRISPR-Cas9-based system should facilitate enhanced ability to characterize genetic variants with functional testing in vitro and in vivo[110, 111].
Figure 2. Paradigm to develop an integrated model of genetic risk for IPF.
Future studies identifying and characterizing the role of genetic variants in IPF will require integration of next-generation sequencing technologies, bioinformatics, and use of state-of-the-art molecular biology in cell and animal models. Identification of variants by whole exome sequencing (WES) or whole-genome-sequencing (WGS) will require strategic bioinformatics approaches. These genetic variants will require functional validation in cell and animal models; characterizing the effects of these genetic variants on gene expression profiles will require integration of sequencing and bioinformatics technologies.
We anticipate that by understanding the biological mechanisms through which individual genetic variants contribute to disease pathogenesis, key pathways will be identified that will clarify the crucial molecular mediators of IPF pathogenesis. We anticipate that a role for molecular genetics in the classification of IIPs will emerge. The ultimate challenge that lies ahead is to develop an integrated understanding of the role of genetic variants (rare and common) and to identify how these variants interact with each other and with environmental factors to produce the epigenetic, transcriptomic, proteomic, histopathologic, and clinical features of IPF. With increased understanding of the fundamental mechanisms of disease, the future is promising for development of new, targeted therapies to further improve treatment of IPF.
Take home idea.
Emerging genetic studies offer new insights into the fundamental mechanisms of pulmonary fibrosis.
Novel ideas.
Through the past decade, rapid advances in genetic and genomic technologies have begun to reshape our understanding of the “idiopathic” interstitial pneumonias. Genome-wide association studies have identified more than a dozen common genetic variants associated with IPF risk, and may be linked to altered disease progression and survival. Rare genetic variants in 8 genes have been implicated in familial interstitial pneumonia (FIP), the familial form of IPF, which broadly fall into two categories: genes related to surfactant protein processing and trafficking and those linked to telomere biology. In addition to genetic links, unique disease phenotypes based on transcriptomic changes have been identified. As we go forward, we anticipate that advances in these genetic and genomic technologies will result in a re-organization of the way we define and classify interstitial lung disease based on molecular characterization. As we evolve from a system of diagnosis based on histopathology to one based on a specific genetic/genomic signature reflecting the fundamental biology of the disease, there will be unique opportunities to develop and test therapies in specific patient populations based on the molecular profiles. Coupled with advances in detection of early disease, the coming decade offers an unprecedented opportunity to dramatically change the lives of patients with idiopathic pulmonary fibrosis.
References
- 1.King TE, Jr., Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378(9807):1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 2.King TE, Jr., Bradford WZ, Castro-Bernardini S, Fagan EA, Glaspole I, Glassberg MK, Gorina E, Hopkins PM, Kardatzke D, Lancaster L, Lederer DJ, Nathan SD, Pereira CA, Sahn SA, Sussman R, Swigris JJ, Noble PW. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 3.Richeldi L, du Bois RM, Raghu G, Azuma A, Brown KK, Costabel U, Cottin V, Flaherty KR, Hansell DM, Inoue Y, Kim DS, Kolb M, Nicholson AG, Noble PW, Selman M, Taniguchi H, Brun M, Le Maulf F, Girard M, Stowasser S, Schlenker-Herceg R, Disse B, Collard HR. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 4.Kropski JA, Lawson WE, Young LR, Blackwell TS. Genetic studies provide clues on the pathogenesis of idiopathic pulmonary fibrosis. Dis Model Mech. 2013;6(1):9–17. doi: 10.1242/dmm.010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spagnolo P, Grunewald J, du Bois RM. Genetic determinants of pulmonary fibrosis: evolving concepts. Lancet Respir Med. 2014;2(5):416–428. doi: 10.1016/S2213-2600(14)70047-5. [DOI] [PubMed] [Google Scholar]
- 6.Bonanni PP, Frymoyer JW, Jacox RF. A Family Study of Idiopathic Pulmonary Fibrosis. A Possible Dysproteinemic and Genetically Determined Disease. Am J Med. 1965;39:411–421. doi: 10.1016/0002-9343(65)90208-1. [DOI] [PubMed] [Google Scholar]
- 7.Peabody JW, Peabody JW, Jr., Hayes EW, Hayes EW., Jr Idiopathic pulmonary fibrosis; its occurrence in identical twin sisters. Dis Chest. 1950;18(4):330–344. doi: 10.1016/s0096-0217(15)34710-5. [DOI] [PubMed] [Google Scholar]
- 8.Hodgson U, Laitinen T, Tukiainen P. Nationwide prevalence of sporadic and familial idiopathic pulmonary fibrosis: evidence of founder effect among multiplex families in Finland. Thorax. 2002;57(4):338–342. doi: 10.1136/thorax.57.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez BA, Fox G, Bhatia R, Sala E, Noble B, Denic N, Fernandez D, Duguid N, Dohey A, Kamel F, Edwards L, Mahoney K, Stuckless S, Parfrey PS, Woods MO. A Newfoundland cohort of familial and sporadic idiopathic pulmonary fibrosis patients: clinical and genetic features. Respir Res. 2012;13:64. doi: 10.1186/1465-9921-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Sancho C, Buendia-Roldan I, Fernandez-Plata MR, Navarro C, Perez-Padilla R, Vargas MH, Loyd JE, Selman M. Familial pulmonary fibrosis is the strongest risk factor for idiopathic pulmonary fibrosis. Respir Med. 2011;105(12):1902–1907. doi: 10.1016/j.rmed.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Loyd JE. Pulmonary fibrosis in families. Am J Respir Cell Mol Biol. 2003;29(3 Suppl):S47–50. [PubMed] [Google Scholar]
- 12.Coghlan MA, Shifren A, Huang HJ, Russell TD, Mitra RD, Zhang Q, Wegner DJ, Cole FS, Hamvas A. Sequencing of idiopathic pulmonary fibrosis-related genes reveals independent single gene associations. BMJ open respiratory research. 2014;1(1):e000057. doi: 10.1136/bmjresp-2014-000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nogee LM, Dunbar AE, 3rd, Wert SE, Askin F, Hamvas A, Whitsett JA. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med. 2001;344(8):573–579. doi: 10.1056/NEJM200102223440805. [DOI] [PubMed] [Google Scholar]
- 14.Thomas AQ, Lane K, Phillips J, 3rd, Prince M, Markin C, Speer M, Schwartz DA, Gaddipati R, Marney A, Johnson J, Roberts R, Haines J, Stahlman M, Loyd JE. Heterozygosity for a surfactant protein C gene mutation associated with usual interstitial pneumonitis and cellular nonspecific interstitial pneumonitis in one kindred. Am J Respir Crit Care Med. 2002;165(9):1322–1328. doi: 10.1164/rccm.200112-123OC. [DOI] [PubMed] [Google Scholar]
- 15.van Moorsel CH, van Oosterhout MF, Barlo NP, de Jong PA, van der Vis JJ, Ruven HJ, van Es HW, van den Bosch JM, Grutters JC. Surfactant protein C mutations are the basis of a significant portion of adult familial pulmonary fibrosis in a dutch cohort. Am J Respir Crit Care Med. 2010;182(11):1419–1425. doi: 10.1164/rccm.200906-0953OC. [DOI] [PubMed] [Google Scholar]
- 16.Ono S, Tanaka T, Ishida M, Kinoshita A, Fukuoka J, Takaki M, Sakamoto N, Ishimatsu Y, Kohno S, Hayashi T, Senba M, Yasunami M, Kubo Y, Yoshida LM, Kubo H, Ariyoshi K, Yoshiura K, Morimoto K. Surfactant protein C G100S mutation causes familial pulmonary fibrosis in Japanese kindred. Eur Respir J. 2011;38(4):861–869. doi: 10.1183/09031936.00143610. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Kuan PJ, Xing C, Cronkhite JT, Torres F, Rosenblatt RL, DiMaio JM, Kinch LN, Grishin NV, Garcia CK. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet. 2009;84(1):52–59. doi: 10.1016/j.ajhg.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campo I, Zorzetto M, Mariani F, Kadija Z, Morbini P, Dore R, Kaltenborn E, Frixel S, Zarbock R, Liebisch G, Hegermann J, Wrede C, Griese M, Luisetti M. A large kindred of pulmonary fibrosis associated with a novel ABCA3 gene variant. Respir Res. 2014;15:43. doi: 10.1186/1465-9921-15-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epaud R, Delestrain C, Louha M, Simon S, Fanen P, Tazi A. Combined pulmonary fibrosis and emphysema syndrome associated with ABCA3 mutations. Eur Respir J. 2014;43(2):638–641. doi: 10.1183/09031936.00145213. [DOI] [PubMed] [Google Scholar]
- 20.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA, 3rd, Lansdorp PM, Greider CW, Loyd JE. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356(13):1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 21.Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci U S A. 2007;104(18):7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hisata S, Sakaguchi H, Kanegane H, Hidaka T, Ichinose M, Kojima S, Nukiwa T, Ebina M. A Novel Missense Mutation of DKC1 In Dyskeratosis Congenita With Pulmonary Fibrosis. Sarcoidosis Vasc Diffuse Lung Dis. 2013;30(3):221–225. [PubMed] [Google Scholar]
- 23.Kropski JA, Mitchell DB, Markin C, Polosukhin VV, Choi LA, Johnson JE, Lawson WE, Phillips JA, 3rd, Cogan JD, Blackwell TS, Loyd JE. A novel dyskerin (DKC1) mutation is associated with Familial Interstitial Pneumonia. Chest. 2014 doi: 10.1378/chest.13-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alder JK, Parry EM, Yegnasubramanian S, Wagner CL, Lieblich LM, Auerbach R, Auerbach AD, Wheelan SJ, Armanios M. Penetrant telomere phenotypes in females with heterozygous mutations in the dyskeratosis congenita 1 (DKC1) gene. Hum Mutat. 2013 doi: 10.1002/humu.22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alder JK, Stanley SE, Wagner CL, Hamilton M, Hanumanthu VS, Armanios M. Exome sequencing identifies mutant TINF2 in a family with pulmonary fibrosis. Chest. 2014 doi: 10.1378/chest.14-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukuhara A, Tanino Y, Ishii T, Inokoshi Y, Saito K, Fukuhara N, Sato S, Saito J, Ishida T, Yamaguchi H, Munakata M. Pulmonary fibrosis in dyskeratosis congenita with TINF2 gene mutation. Eur Respir J. 2013;42(6):1757–1759. doi: 10.1183/09031936.00149113. [DOI] [PubMed] [Google Scholar]
- 27.Kannengiesser C, Borie R, Revy P. Pulmonary fibrosis associated with TINF2 gene mutation: is somatic reversion required? Eur Respir J. 2014;44(1):269–270. doi: 10.1183/09031936.00038714. [DOI] [PubMed] [Google Scholar]
- 28.Cogan JD, Kropski JA, Zhao M, Mitchell DB, Rives L, Markin C, Garnett ET, Montgomery KH, Mason WR, McKean DF, Powers J, Murphy E, Olson LM, Choi L, Cheng DS, Blue EM, Young LR, Lancaster LH, Steele MP, Brown KK, Schwarz MI, Fingerlin TE, Schwartz DA, Lawson WE, Loyd JE, Zhao Z, Phillips JA, Iii, Blackwell TS, Genomics UoWCfM Rare Variants in RTEL1 are Associated with Familial Interstitial Pneumonia. Am J Respir Crit Care Med. 2015 doi: 10.1164/rccm.201408-1510OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Son JY, Kim SY, Cho SH, Shim HS, Jung JY, Kim EY, Lim JE, Park BH, Kang YA, Kim YS, Kim SK, Chang J, Park MS. TGF-beta1 T869C polymorphism may affect susceptibility to idiopathic pulmonary fibrosis and disease severity. Lung. 2013;191(2):199–205. doi: 10.1007/s00408-012-9447-z. [DOI] [PubMed] [Google Scholar]
- 30.Xaubet A, Marin-Arguedas A, Lario S, Ancochea J, Morell F, Ruiz-Manzano J, Rodriguez-Becerra E, Rodriguez-Arias JM, Inigo P, Sanz S, Campistol JM, Mullol J, Picado C. Transforming growth factor-beta1 gene polymorphisms are associated with disease progression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2003;168(4):431–435. doi: 10.1164/rccm.200210-1165OC. [DOI] [PubMed] [Google Scholar]
- 31.Whyte M, Hubbard R, Meliconi R, Whidborne M, Eaton V, Bingle C, Timms J, Duff G, Facchini A, Pacilli A, Fabbri M, Hall I, Britton J, Johnston I, Di Giovine F. Increased risk of fibrosing alveolitis associated with interleukin-1 receptor antagonist and tumor necrosis factor-alpha gene polymorphisms. Am J Respir Crit Care Med. 2000;162(2 Pt 1):755–758. doi: 10.1164/ajrccm.162.2.9909053. [DOI] [PubMed] [Google Scholar]
- 32.Barlo NP, van Moorsel CH, Korthagen NM, Heron M, Rijkers GT, Ruven HJ, van den Bosch JM, Grutters JC. Genetic variability in the IL1RN gene and the balance between interleukin (IL)-1 receptor agonist and IL-1beta in idiopathic pulmonary fibrosis. Clin Exp Immunol. 2011;166(3):346–351. doi: 10.1111/j.1365-2249.2011.04468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korthagen NM, van Moorsel CH, Kazemier KM, Ruven HJ, Grutters JC. IL1RN genetic variations and risk of IPF: a meta-analysis and mRNA expression study. Immunogenetics. 2012;64(5):371–377. doi: 10.1007/s00251-012-0604-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn MH, Park BL, Lee SH, Park SW, Park JS, Kim DJ, Jang AS, Shin HK, Uh ST, Kim YK, Kim YW, Han SK, Jung KS, Lee KY, Jeong SH, Park JW, Choi BW, Park IW, Chung MP, Shin HD, Song JW, Kim DS, Park CS, Shim YS. A promoter SNP rs4073T>A in the common allele of the interleukin 8 gene is associated with the development of idiopathic pulmonary fibrosis via the IL-8 protein enhancing mode. Respir Res. 2011;12:73. doi: 10.1186/1465-9921-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Dwyer DN, Armstrong ME, Trujillo G, Cooke G, Keane MP, Fallon PG, Simpson AJ, Millar AB, McGrath EE, Whyte MK, Hirani N, Hogaboam CM, Donnelly SC. The Toll-like receptor 3 L412F polymorphism and disease progression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2013;188(12):1442–1450. doi: 10.1164/rccm.201304-0760OC. [DOI] [PubMed] [Google Scholar]
- 36.Xue J, Gochuico BR, Alawad AS, Feghali-Bostwick CA, Noth I, Nathan SD, Rosen GD, Rosas IO, Dacic S, Ocak I, Fuhrman CR, Cuenco KT, Smith MA, Jacobs SS, Zeevi A, Morel PA, Pilewski JM, Valentine VG, Gibson KF, Kaminski N, Sciurba FC, Zhang Y, Duncan SR. The HLA class II Allele DRB1*1501 is over-represented in patients with idiopathic pulmonary fibrosis. PLoS One. 2011;6(2):e14715. doi: 10.1371/journal.pone.0014715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korthagen NM, van Moorsel CH, Barlo NP, Kazemier KM, Ruven HJ, Grutters JC. Association between variations in cell cycle genes and idiopathic pulmonary fibrosis. PLoS One. 2012;7(1):e30442. doi: 10.1371/journal.pone.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, Fingerlin TE, Zhang W, Gudmundsson G, Groshong SD, Evans CM, Garantziotis S, Adler KB, Dickey BF, du Bois RM, Yang IV, Herron A, Kervitsky D, Talbert JL, Markin C, Park J, Crews AL, Slifer SH, Auerbach S, Roy MG, Lin J, Hennessy CE, Schwarz MI, Schwartz DA. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364(16):1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borie R, Crestani B, Dieude P, Nunes H, Allanore Y, Kannengiesser C, Airo P, Matucci-Cerinic M, Wallaert B, Israel-Biet D, Cadranel J, Cottin V, Gazal S, Peljto AL, Varga J, Schwartz DA, Valeyre D, Grandchamp B. The MUC5B variant is associated with idiopathic pulmonary fibrosis but not with systemic sclerosis interstitial lung disease in the European Caucasian population. PLoS One. 2013;8(8):e70621. doi: 10.1371/journal.pone.0070621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, Loyd JE, Cosgrove GP, Lynch D, Groshong S, Collard HR, Wolters PJ, Bradford WZ, Kossen K, Seiwert SD, du Bois RM, Garcia CK, Devine MS, Gudmundsson G, Isaksson HJ, Kaminski N, Zhang Y, Gibson KF, Lancaster LH, Cogan JD, Mason WR, Maher TM, Molyneaux PL, Wells AU, Moffatt MF, Selman M, Pardo A, Kim DS, Crapo JD, Make BJ, Regan EA, Walek DS, Daniel JJ, Kamatani Y, Zelenika D, Smith K, McKean D, Pedersen BS, Talbert J, Kidd RN, Markin CR, Beckman KB, Lathrop M, Schwarz MI, Schwartz DA. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45(6):613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, Broderick SM, Wade MS, Hysi P, Scuirba J, Richards TJ, Juan-Guardela BM, Vij R, Han MK, Martinez FJ, Kossen K, Seiwert SD, Christie JD, Nicolae D, Kaminski N, Garcia JG. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1(4):309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peljto AL, Steele MP, Fingerlin TE, Hinchcliff ME, Murphy E, Podlusky S, Carns M, Schwarz M, Varga J, Schwartz DA. The Pulmonary Fibrosis-Associated MUC5B Promoter Polymorphism does not Influence the Development of Interstitial Pneumonia in Systemic Sclerosis. Chest. 2012 doi: 10.1378/chest.12-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Noth I, Garcia JG, Kaminski N. A variant in the promoter of MUC5B and idiopathic pulmonary fibrosis. N Engl J Med. 2011;364(16):1576–1577. doi: 10.1056/NEJMc1013504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horimasu Y, Ohshimo S, Bonella F, Tanaka S, Ishikawa N, Hattori N, Kohno N, Guzman J, Costabel U. MUC5B promoter polymorphism in Japanese patients with idiopathic pulmonary fibrosis. Respirology. 2015 doi: 10.1111/resp.12466. [DOI] [PubMed] [Google Scholar]
- 45.Stock CJ, Sato H, Fonseca C, Banya WA, Molyneaux PL, Adamali H, Russell AM, Denton CP, Abraham DJ, Hansell DM, Nicholson AG, Maher TM, Wells AU, Lindahl GE, Renzoni EA. Mucin 5B promoter polymorphism is associated with idiopathic pulmonary fibrosis but not with development of lung fibrosis in systemic sclerosis or sarcoidosis. Thorax. 2013;68(5):436–441. doi: 10.1136/thoraxjnl-2012-201786. [DOI] [PubMed] [Google Scholar]
- 46.Peljto AL, Selman M, Kim DS, Murphy E, Tucker L, Pardo A, Lee JS, Ji W, Schwarz MI, Yang IV, Schwartz DA, Fingerlin TE. The MUC5B promoter polymorphism is associated with IPF in a Mexican cohort but is rare among Asian ancestries. Chest. 2014 doi: 10.1378/chest.14-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei R, Li C, Zhang M, Jones-Hall YL, Myers JL, Noth I, Liu W. Association between MUC5B and TERT polymorphisms and different interstitial lung disease phenotypes. Transl Res. 2014;163(5):494–502. doi: 10.1016/j.trsl.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seibold MA, Smith RW, Urbanek C, Groshong SD, Cosgrove GP, Brown KK, Schwarz MI, Schwartz DA, Reynolds SD. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS One. 2013;8(3):e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunninghake GM, Hatabu H, Okajima Y, Gao W, Dupuis J, Latourelle JC, Nishino M, Araki T, Zazueta OE, Kurugol S, Ross JC, San Jose Estepar R, Murphy E, Steele MP, Loyd JE, Schwarz MI, Fingerlin TE, Rosas IO, Washko GR, O'Connor GT, Schwartz DA. MUC5B promoter polymorphism and interstitial lung abnormalities. N Engl J Med. 2013;368(23):2192–2200. doi: 10.1056/NEJMoa1216076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peljto AL, Zhang Y, Fingerlin TE, Ma SF, Garcia JG, Richards TJ, Silveira LJ, Lindell KO, Steele MP, Loyd JE, Gibson KF, Seibold MA, Brown KK, Talbert JL, Markin C, Kossen K, Seiwert SD, Murphy E, Noth I, Schwarz MI, Kaminski N, Schwartz DA. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. JAMA. 2013;309(21):2232–2239. doi: 10.1001/jama.2013.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, Alexander SN, Bellinghausen LK, Song AS, Petrova YM, Tuvim MJ, Adachi R, Romo I, Bordt AS, Bowden MG, Sisson JH, Woodruff PG, Thornton DJ, Rousseau K, De la Garza MM, Moghaddam SJ, Karmouty-Quintana H, Blackburn MR, Drouin SM, Davis CW, Terrell KA, Grubb BR, O'Neal WK, Flores SC, Cota-Gomez A, Lozupone CA, Donnelly JM, Watson AM, Hennessy CE, Keith RC, Yang IV, Barthel L, Henson PM, Janssen WJ, Schwartz DA, Boucher RC, Dickey BF, Evans CM. Muc5b is required for airway defence. Nature. 2014;505(7483):412–416. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steele MP, Speer MC, Loyd JE, Brown KK, Herron A, Slifer SH, Burch LH, Wahidi MM, Phillips JA, 3rd, Sporn TA, McAdams HP, Schwarz MI, Schwartz DA. Clinical and pathologic features of familial interstitial pneumonia. Am J Respir Crit Care Med. 2005;172(9):1146–1152. doi: 10.1164/rccm.200408-1104OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scholand MB, Coon H, Wolff R, Cannon-Albright L. Use of a genealogical database demonstrates heritability of pulmonary fibrosis. Lung. 2013;191(5):475–481. doi: 10.1007/s00408-013-9484-2. [DOI] [PubMed] [Google Scholar]
- 54.Armanios M. Telomerase and idiopathic pulmonary fibrosis. Mutat Res. 2012;730(1-2):52–58. doi: 10.1016/j.mrfmmm.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calado RT, Young NS. Telomere diseases. N Engl J Med. 2009;361(24):2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alder JK, Chen JJ, Lancaster L, Danoff S, Su SC, Cogan JD, Vulto I, Xie M, Qi X, Tuder RM, Phillips JA, 3rd, Lansdorp PM, Loyd JE, Armanios MY. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci U S A. 2008;105(35):13051–13056. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cronkhite JT, Xing C, Raghu G, Chin KM, Torres F, Rosenblatt RL, Garcia CK. Telomere shortening in familial and sporadic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178(7):729–737. doi: 10.1164/rccm.200804-550OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uringa EJ, Lisaingo K, Pickett HA, Brind'Amour J, Rohde JH, Zelensky A, Essers J, Lansdorp PM. RTEL1 contributes to DNA replication and repair and telomere maintenance. Mol Biol Cell. 2012;23(14):2782–2792. doi: 10.1091/mbc.E12-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, Boulton SJ. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342(6155):239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 60.Vannier JB, Sarek G, Boulton SJ. RTEL1: functions of a disease-associated helicase. Trends in cell biology. 2014;24(7):416–425. doi: 10.1016/j.tcb.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Masutomi K, Possemato R, Wong JM, Currier JL, Tothova Z, Manola JB, Ganesan S, Lansdorp PM, Collins K, Hahn WC. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc Natl Acad Sci U S A. 2005;102(23):8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kropski JA, Pritchett JM, Zoz DF, Crossno PF, Markin C, Garnett ET, Degryse AL, Mitchell DB, Polosukhin VV, Rickman OB, Choi L, Cheng DS, McConaha ME, Jones BR, Gleaves LA, McMahon FB, Worrell JA, Solus JF, Ware LB, Lee JW, Massion PP, Zaynagetdinov R, White ES, Kurtis JD, Johnson JE, Groshong SD, Lancaster LH, Young LR, Steele MP, Phillips JA, Iii, Cogan JD, Loyd JE, Lawson WE, Blackwell TS. Extensive Phenotyping of Individuals At-risk for Familial Interstitial Pneumonia Reveals Clues to the Pathogenesis of Interstitial Lung Disease. Am J Respir Crit Care Med. 2014 doi: 10.1164/rccm.201406-1162OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diaz de Leon A, Cronkhite JT, Katzenstein AL, Godwin JD, Raghu G, Glazer CS, Rosenblatt RL, Girod CE, Garrity ER, Xing C, Garcia CK. Telomere lengths, pulmonary fibrosis and telomerase (TERT) mutations. PLoS One. 2010;5(5):e10680. doi: 10.1371/journal.pone.0010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stuart BD, Lee JS, Kozlitina J, Noth I, Devine MS, Glazer CS, Torres F, Kaza V, Girod CE, Jones KD, Elicker BM, Ma SF, Vij R, Collard HR, Wolters PJ, Garcia CK. Effect of telomere length on survival in patients with idiopathic pulmonary fibrosis: an observational cohort study with independent validation. Lancet Respir Med. 2014;2(7):557–565. doi: 10.1016/S2213-2600(14)70124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theall KP, McKasson S, Mabile E, Dunaway LF, Drury SS. Early hits and long-term consequences: tracking the lasting impact of prenatal smoke exposure on telomere length in children. Am J Public Health. 2013;103(Suppl 1):S133–135. doi: 10.2105/AJPH.2012.301208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Driscoll B, Buckley S, Bui KC, Anderson KD, Warburton D. Telomerase in alveolar epithelial development and repair. Am J Physiol Lung Cell Mol Physiol. 2000;279(6):L1191–1198. doi: 10.1152/ajplung.2000.279.6.L1191. [DOI] [PubMed] [Google Scholar]
- 67.Lee J, Reddy R, Barsky L, Scholes J, Chen H, Shi W, Driscoll B. Lung alveolar integrity is compromised by telomere shortening in telomerase-null mice. Am J Physiol Lung Cell Mol Physiol. 2009;296(1):L57–70. doi: 10.1152/ajplung.90411.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alder JK, Guo N, Kembou F, Parry EM, Anderson CJ, Gorgy AI, Walsh MF, Sussan T, Biswal S, Mitzner W, Tuder RM, Armanios M. Telomere length is a determinant of emphysema susceptibility. Am J Respir Crit Care Med. 2011;184(8):904–912. doi: 10.1164/rccm.201103-0520OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Degryse AL, Xu XC, Newman JL, Mitchell DB, Tanjore H, Polosukhin VV, Jones BR, McMahon FB, Gleaves LA, Phillips JA, 3rd, Cogan JD, Blackwell TS, Lawson WE. Telomerase deficiency does not alter bleomycin-induced fibrosis in mice. Exp Lung Res. 2012;38(3):124–134. doi: 10.3109/01902148.2012.658148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu T, Chung MJ, Ullenbruch M, Yu H, Jin H, Hu B, Choi YY, Ishikawa F, Phan SH. Telomerase activity is required for bleomycin-induced pulmonary fibrosis in mice. J Clin Invest. 2007;117(12):3800–3809. doi: 10.1172/JCI32369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cottin V, Reix P, Khouatra C, Thivolet-Bejui F, Feldmann D, Cordier JF. Combined pulmonary fibrosis and emphysema syndrome associated with familial SFTPC mutation. Thorax. 2011;66(10):918–919. doi: 10.1136/thx.2010.151407. [DOI] [PubMed] [Google Scholar]
- 72.Tanjore H, Blackwell TS, Lawson WE. Emerging evidence for endoplasmic reticulum stress in the pathogenesis of idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;302(8):L721–729. doi: 10.1152/ajplung.00410.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bridges JP, Wert SE, Nogee LM, Weaver TE. Expression of a human surfactant protein C mutation associated with interstitial lung disease disrupts lung development in transgenic mice. J Biol Chem. 2003;278(52):52739–52746. doi: 10.1074/jbc.M309599200. [DOI] [PubMed] [Google Scholar]
- 74.Lawson WE, Crossno PF, Polosukhin VV, Roldan J, Cheng DS, Lane KB, Blackwell TR, Xu C, Markin C, Ware LB, Miller GG, Loyd JE, Blackwell TS. Endoplasmic reticulum stress in alveolar epithelial cells is prominent in IPF: association with altered surfactant protein processing and herpesvirus infection. Am J Physiol Lung Cell Mol Physiol. 2008;294(6):L1119–1126. doi: 10.1152/ajplung.00382.2007. [DOI] [PubMed] [Google Scholar]
- 75.Mulugeta S, Maguire JA, Newitt JL, Russo SJ, Kotorashvili A, Beers MF. Misfolded BRICHOS SP-C mutant proteins induce apoptosis via caspase-4- and cytochrome c-related mechanisms. Am J Physiol Lung Cell Mol Physiol. 2007;293(3):L720–729. doi: 10.1152/ajplung.00025.2007. [DOI] [PubMed] [Google Scholar]
- 76.Mulugeta S, Nguyen V, Russo SJ, Muniswamy M, Beers MF. A surfactant protein C precursor protein BRICHOS domain mutation causes endoplasmic reticulum stress, proteasome dysfunction, and caspase 3 activation. Am J Respir Cell Mol Biol. 2005;32(6):521–530. doi: 10.1165/rcmb.2005-0009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang WJ, Mulugeta S, Russo SJ, Beers MF. Deletion of exon 4 from human surfactant protein C results in aggresome formation and generation of a dominant negative. J Cell Sci. 2003;116(Pt 4):683–692. doi: 10.1242/jcs.00267. [DOI] [PubMed] [Google Scholar]
- 78.Beers MF, Hawkins A, Maguire JA, Kotorashvili A, Zhao M, Newitt JL, Ding W, Russo S, Guttentag S, Gonzales L, Mulugeta S. A nonaggregating surfactant protein C mutant is misdirected to early endosomes and disrupts phospholipid recycling. Traffic. 2011;12(9):1196–1210. doi: 10.1111/j.1600-0854.2011.01223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hawkins A, Guttentag SH, Deterding R, Funkhouser WK, Goralski JL, Chatterjee S, Mulugeta S, Beers MF. A non-BRICHOS SFTPC mutant (SP-CI73T) linked to interstitial lung disease promotes a late block in macroautophagy disrupting cellular proteostasis and mitophagy. Am J Physiol Lung Cell Mol Physiol. 2015;308(1):L33–47. doi: 10.1152/ajplung.00217.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lawson WE, Cheng DS, Degryse AL, Tanjore H, Polosukhin VV, Xu XC, Newcomb DC, Jones BR, Roldan J, Lane KB, Morrisey EE, Beers MF, Yull FE, Blackwell TS. Endoplasmic reticulum stress enhances fibrotic remodeling in the lungs. Proc Natl Acad Sci U S A. 2011;108(26):10562–10567. doi: 10.1073/pnas.1107559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maguire JA, Mulugeta S, Beers MF. Multiple ways to die: delineation of the unfolded protein response and apoptosis induced by Surfactant Protein C BRICHOS mutants. Int J Biochem Cell Biol. 2012;44(1):101–112. doi: 10.1016/j.biocel.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maguire JA, Mulugeta S, Beers MF. Endoplasmic reticulum stress induced by surfactant protein C BRICHOS mutants promotes proinflammatory signaling by epithelial cells. Am J Respir Cell Mol Biol. 2011;44(3):404–414. doi: 10.1165/rcmb.2009-0382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanjore H, Cheng DS, Degryse AL, Zoz DF, Abdolrasulnia R, Lawson WE, Blackwell TS. Alveolar epithelial cells undergo epithelial-to-mesenchymal transition in response to endoplasmic reticulum stress. J Biol Chem. 2011;286(35):30972–30980. doi: 10.1074/jbc.M110.181164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maitra M, Dey M, Yuan WC, Nathanielsz PW, Garcia CK. Lung fibrosis-associated surfactant protein A1 and C variants induce latent transforming growth factor beta1 secretion in lung epithelial cells. J Biol Chem. 2013;288(38):27159–27171. doi: 10.1074/jbc.M113.475335. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Maitra M, Wang Y, Gerard RD, Mendelson CR, Garcia CK. Surfactant protein A2 mutations associated with pulmonary fibrosis lead to protein instability and endoplasmic reticulum stress. J Biol Chem. 2010;285(29):22103–22113. doi: 10.1074/jbc.M110.121467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawson WE, Grant SW, Ambrosini V, Womble KE, Dawson EP, Lane KB, Markin C, Renzoni E, Lympany P, Thomas AQ, Roldan J, Scott TA, Blackwell TS, Phillips JA, 3rd, Loyd JE, du Bois RM. Genetic mutations in surfactant protein C are a rare cause of sporadic cases of IPF. Thorax. 2004;59(11):977–980. doi: 10.1136/thx.2004.026336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Markart P, Ruppert C, Wygrecka M, Schmidt R, Korfei M, Harbach H, Theruvath I, Pison U, Seeger W, Guenther A, Witt H. Surfactant protein C mutations in sporadic forms of idiopathic interstitial pneumonias. Eur Respir J. 2007;29(1):134–137. doi: 10.1183/09031936.00034406. [DOI] [PubMed] [Google Scholar]
- 88.Korfei M, Ruppert C, Mahavadi P, Henneke I, Markart P, Koch M, Lang G, Fink L, Bohle RM, Seeger W, Weaver TE, Guenther A. Epithelial endoplasmic reticulum stress and apoptosis in sporadic idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178(8):838–846. doi: 10.1164/rccm.200802-313OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Isler JA, Skalet AH, Alwine JC. Human cytomegalovirus infection activates and regulates the unfolded protein response. J Virol. 2005;79(11):6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geraghty P, Wallace A, D'Armiento JM. Induction of the unfolded protein response by cigarette smoke is primarily an activating transcription factor 4-C/EBP homologous protein mediated process. Int J Chron Obstruct Pulmon Dis. 2011;6:309–319. doi: 10.2147/COPD.S19599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hengstermann A, Muller T. Endoplasmic reticulum stress induced by aqueous extracts of cigarette smoke in 3T3 cells activates the unfolded-protein-response-dependent PERK pathway of cell survival. Free Radic Biol Med. 2008;44(6):1097–1107. doi: 10.1016/j.freeradbiomed.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 92.Zhao H, Yang J, Shan L, Jorgensen ED. Measuring the impact of cigarette smoke on the UPR. Methods Enzymol. 2011;489:147–164. doi: 10.1016/B978-0-12-385116-1.00009-1. [DOI] [PubMed] [Google Scholar]
- 93.Stewart GA, Ridsdale R, Martin EP, Na CL, Xu Y, Mandapaka K, Weaver TE. 4-Phenylbutyric acid treatment rescues trafficking and processing of a mutant surfactant protein-C. Am J Respir Cell Mol Biol. 2012;47(3):324–331. doi: 10.1165/rcmb.2012-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Crossno PF, Polosukhin VV, Blackwell TS, Johnson JE, Markin C, Moore PE, Worrell JA, Stahlman MT, Phillips JA, 3rd, Loyd JE, Cogan JD, Lawson WE. Identification of early interstitial lung disease in an individual with genetic variations in ABCA3 and SFTPC. Chest. 2010;137(4):969–973. doi: 10.1378/chest.09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hodgson U, Pulkkinen V, Dixon M, Peyrard-Janvid M, Rehn M, Lahermo P, Ollikainen V, Salmenkivi K, Kinnula V, Kere J, Tukiainen P, Laitinen T. ELMOD2 is a candidate gene for familial idiopathic pulmonary fibrosis. Am J Hum Genet. 2006;79(1):149–154. doi: 10.1086/504639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pulkkinen V, Bruce S, Rintahaka J, Hodgson U, Laitinen T, Alenius H, Kinnula VL, Myllarniemi M, Matikainen S, Kere J. ELMOD2, a candidate gene for idiopathic pulmonary fibrosis, regulates antiviral responses. FASEB J. 2010;24(4):1167–1177. doi: 10.1096/fj.09-138545. [DOI] [PubMed] [Google Scholar]
- 97.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 98.Futema M, Plagnol V, Li K, Whittall RA, Neil HA, Seed M, Simon Broome C, Bertolini S, Calandra S, Descamps OS, Graham CA, Hegele RA, Karpe F, Durst R, Leitersdorf E, Lench N, Nair DR, Soran H, Van Bockxmeer FM, Consortium UK, Humphries SE. Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J Med Genet. 2014;51(8):537–544. doi: 10.1136/jmedgenet-2014-102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nature reviews Cardiology. 2013;10(9):531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 101.Esteban-Jurado C, Vila-Casadesus M, Garre P, Lozano JJ, Pristoupilova A, Beltran S, Munoz J, Ocana T, Balaguer F, Lopez-Ceron M, Cuatrecasas M, Franch-Exposito S, Pique JM, Castells A, Carracedo A, Ruiz-Ponte C, Abuli A, Bessa X, Andreu M, Bujanda L, Caldes T, Castellvi-Bel S. Whole-exome sequencing identifies rare pathogenic variants in new predisposition genes for familial colorectal cancer. Genetics in medicine : official journal of the American College of Medical Genetics. 2014 doi: 10.1038/gim.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu H, Roach JC, Coon H, Guthery SL, Voelkerding KV, Margraf RL, Durtschi JD, Tavtigian SV, Shankaracharya, Wu W, Scheet P, Wang S, Xing J, Glusman G, Hubley R, Li H, Garg V, Moore B, Hood L, Galas DJ, Srivastava D, Reese MG, Jorde LB, Yandell M, Huff CD. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat Biotechnol. 2014;32(7):663–669. doi: 10.1038/nbt.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, Masri A, Kayserili H, Al-Aama JY, Abdel-Salam GM, Karminejad A, Kara M, Kara B, Bozorgmehri B, Ben-Omran T, Mojahedi F, Mahmoud IG, Bouslam N, Bouhouche A, Benomar A, Hanein S, Raymond L, Forlani S, Mascaro M, Selim L, Shehata N, Al-Allawi N, Bindu PS, Azam M, Gunel M, Caglayan A, Bilguvar K, Tolun A, Issa MY, Schroth J, Spencer EG, Rosti RO, Akizu N, Vaux KK, Johansen A, Koh AA, Megahed H, Durr A, Brice A, Stevanin G, Gabriel SB, Ideker T, Gleeson JG. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343(6170):506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Silhan LL, Shah PD, Chambers DC, Snyder LD, Riise GC, Wagner CL, Hellstrom-Lindberg E, Orens JB, Mewton JF, Danoff SK, Arcasoy MO, Armanios M. Lung transplantation in telomerase mutation carriers with pulmonary fibrosis. Eur Respir J. 2014;44(1):178–187. doi: 10.1183/09031936.00060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Etchegary H, Pullman D, Simmonds C, Young TL, Hodgkinson K. 'It had to be done': genetic testing decisions for arrhythmogenic right ventricular cardiomyopathy. Clin Genet. 2014 doi: 10.1111/cge.12513. [DOI] [PubMed] [Google Scholar]
- 106.Schuurman AG, van der Kolk DM, Verkerk MA, Birnie E, Ranchor AV, Plantinga M, van Langen IM. Maximising the efficiency of clinical screening programmes: balancing predictive genetic testing with a right not to know. European journal of human genetics : EJHG. 2015 doi: 10.1038/ejhg.2014.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chio A, Battistini S, Calvo A, Caponnetto C, Conforti FL, Corbo M, Giannini F, Mandrioli J, Mora G, Sabatelli M, Consortium I, Ajmone C, Mastro E, Pain D, Mandich P, Penco S, Restagno G, Zollino M, Surbone A. Genetic counselling in ALS: facts, uncertainties and clinical suggestions. Journal of neurology, neurosurgery, and psychiatry. 2014;85(5):478–485. doi: 10.1136/jnnp-2013-305546. [DOI] [PubMed] [Google Scholar]
- 108.Ghaedi M, Calle EA, Mendez JJ, Gard AL, Balestrini J, Booth A, Bove PF, Gui L, White ES, Niklason LE. Human iPS cell-derived alveolar epithelium repopulates lung extracellular matrix. J Clin Invest. 2013;123(11):4950–4962. doi: 10.1172/JCI68793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yan Q, Quan Y, Sun H, Peng X, Zou Z, Alcorn JL, Wetsel RA, Wang D. A site-specific genetic modification for induction of pluripotency and subsequent isolation of derived lung alveolar epithelial type II cells. Stem Cells. 2014;32(2):402–413. doi: 10.1002/stem.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 111.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mushiroda T, Wattanapokayakit S, Takahashi A, Nukiwa T, Kudoh S, Ogura T, Taniguchi H, Kubo M, Kamatani N, Nakamura Y. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. J Med Genet. 2008;45(10):654–656. doi: 10.1136/jmg.2008.057356. [DOI] [PubMed] [Google Scholar]