Abstract
Objectives
The objective of this study was to determine the relationship between specific genetic alterations and malignant transformation in intraductal papillary mucinous neoplasm (IPMN) of the pancreas.
Methods
Quantitative meta-analysis was conducted of studies through October 2010 that adhered to the 1996 World Health Organization guidelines for distinguishing adenoma and borderline IPMN versus carcinoma in surgically resected specimens using a random-effects model. We developed a 6-point scoring system to assess study quality.
Results
Thirty-nine studies (1235 IPMN samples) satisfied the inclusion criteria, and we conducted pooled analysis of 8 genetic markers: MUC1, MUC2, MUC5AC, kRas, p53, hTERT (human telomerase reverse transcriptase), cyclooxygenase 2, and Shh (Sonic hedgehog). Markers having the strongest association with malignant IPMN were hTERT (odds ratio [OR], 11.4; 95% confidence interval [CI], 3.5–36.7) and Shh (OR, 6.9; 95% CI, 2.4–20.2), whereas MUC5AC (OR, 1.0; 95% CI, 0.1–13.9) and kRas (OR, 2.0; 95% CI, 1.0–4.3) showed weak association with IPMN histologic progression.
Conclusions
Expression of hTERT is strongly associated with malignant transformation in IPMN, consistent with up-regulation of hTERT as a key step in progression of IPMN to cancer. Expression of kRas and MUC5AC is common but not strongly associated with IPMN histologic progression. The quality criteria used here may guide future reporting of genetic markers related to malignant transformation of IPMN.
Keywords: telomerase, mucin, pancreatic cancer, intraductal papillary mucinous neoplasm, IPMN, Sonic hedgehog, cyclooxygenase 2, p53, hTERT, Cox2, kRas, marker
Intraductal papillary mucinous neoplasms (IPMNs) describe papillary proliferations of the exocrine pancreas epithelium that secrete copious amounts of thick mucin causing cystic dilatation of the ducts.1 Since the pathologic entity was first described by Ohashi et al in 1982 and designated intraductal papillary mucinous neoplasm by Sessa et al2 in 1994, it has been characterized radiologically, histologically, and molecularly and is thought to represent a lesion that is distinct from and less clinically aggressive than ductal adenocarcinoma.3–5
Histologic analysis of IPMN samples reveals a spectrum of progressive cytoarchitectural atypia. The spectrum of this histologic progression is reflected in the 1996 World Health Organization (WHO) classification of IPMN into 3 categories based on increasing nuclear atypia and mitotic rate: adenoma (IPM-A), borderline (IPM-B), and carcinoma (IPM-C, both in situ and invasive).6 The histologic progression of IPMN from a benign (IPM-A or IPM-B) into a malignant (IPM-C) lesion has a significant impact on patient survival. In patients who underwent pancreatic resection between 1990 and 2007, the 5-year survival rate of patients with benign IPMN ranged from 89% to 95% versus 63% to 65% for patients with malignant IPMN.7–9 Moreover, malignant transformation is not rare: the frequency of malignancy in IPMNs in the main pancreatic duct ranges from 60% to 92% in various reports.10–15
A longstanding question has been whether the histologic progression of IPMN reflects an accumulation of genetic mutations leading to increasing atypia. Many studies have therefore sought to identify specific genetic mutations associated with malignant transformation in IPMN. The objective of the present study was to enhance our understanding of malignant progression in IPMN based on literature published to date. Specifically, our aim was to perform a quantitative meta-analysis of studies from the past 14 years to determine the relationship between individual genetic alterations and malignant transformation in IPMN.
METHODS
Search Methods and Study Selection Criteria
A computerized literature search was performed independently in the PubMed (National Library of Medicine, Bethesda, Md), Cochrane Library, and EMBASE databases by 2 of the authors (S.N., G.E.I.). To identify studies investigating gene expression in IPMN, we used the following search terms: “intraductal papillary mucinous neoplasm,” “IPMN,” “intraductal papillary mucinous tumor,” “IPMT,” “IPMA,” “IPMB,” “IPMC,” “gene expression,” “molecular marker,” “Ras,” “p53,” “MUC,” “telomerase,” and “mutations.” Bibliographies from relevant publications were further reviewed to identify additional published articles not indexed by the major databases.
Studies published from January 1996 to October 2010 were potentially eligible for inclusion based on publication of WHO guidelines for classification of IPMN in 1996.1 For the present meta-analysis, we included studies that satisfied the following criteria: (1) human subjects, (2) histologic confirmation of IPMN in surgically resected tissue, (3) classification of IPMN in accordance to WHO guidelines, (4) inclusion of data regarding genotype frequency and/or risk estimates, and (5) use of validated molecular methods for genotyping.
Individual case reports, editorials, review articles, and duplicate publications were excluded. We further excluded studies that incorporated patients with chronic pancreatitis, bile duct IPMN, and high-risk familial cohorts.
Data Extraction
Data were extracted independently by each reviewer using a standardized data abstraction form. Any disagreements between the 2 reviewers were examined by all 3 investigators and resolved by consensus.
Data Synthesis
The primary outcome was defined as IPMN with presence of malignant transformation (IPM-C). During the initial phase of data extraction, IPMN grade was extracted as presented in the original publication. For purposes of the present meta-analysis, IPMN grade was further dichotomized to either benign (IPM-A, IPM-B) or malignant (IPM-C) lesions.
Assessment of Study Quality
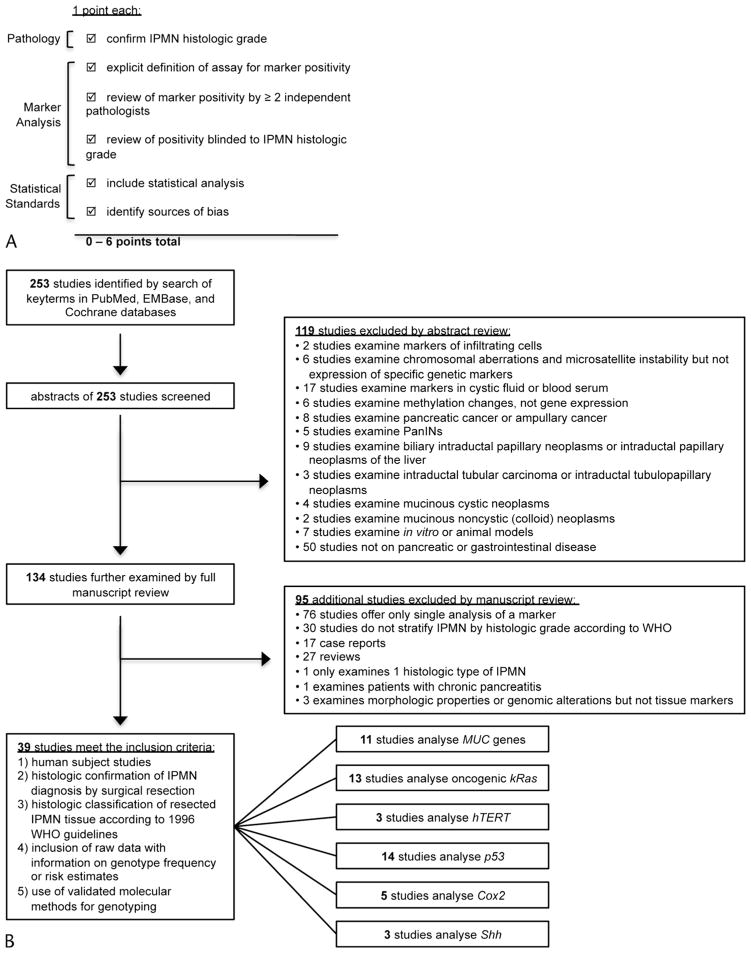
There is currently no validated method to rate the quality of observational studies on IPMN. To provide a means to appraise the methodologic quality of studies included in the present meta-analysis, we formulated a scoring system based on recommendations from the STROBE16 and PRAISE guidelines.17 This checklist consisted of 6 factors summarized in Figure 1A: review of histologic classification of IPMN, in the case of dysplasia or carcinoma review of histology by at least 2 independent pathologists, an explicit definition of genetic assay positivity, blinded genetic assay, description of statistical analysis, and mention of potential sources of bias. One point was assigned for the presence of each of the individual factors.
FIGURE 1.
Methodology used in this meta-analysis. A, We developed a 6-point scoring system to assess quality of studies based on recommendations from the STROBE16 and PRAISE17 guidelines. B, Flow diagram of the literature search and study selection.
Statistical Analysis
Effect size was expressed as an odds ratio (OR) with the corresponding 95% confidence interval (CI). Quantitative meta-analysis was performed when 3 or more studies evaluated the same gene candidate. In the meta-analysis, pooled ORs were generated based on the individual studies using a random-effects model (DerSimonian and Laird) chosen based on the ability to incorporate between-study variance potentially related to differences among the populations included among studies. The I2 test was used to evaluate study heterogeneity. For genetic markers with 5 or more studies, we performed further subgroup analyses based on methodologic quality (high quality, ≥3 points). Analyses were performed using Quantitative Meta-analysis version 2.2 (Biostat, Englewood, NJ).
RESULTS
The computerized literature search yielded 253 studies. Abstracts of these studies were reviewed, and 119 studies were excluded for the reasons delineated in Figure 1B. A full manuscript review was performed on the remaining 134 studies, and 95 additional studies were excluded. The most common reasons for exclusion were studies totaling less than 3 for a particular gene marker and studies that did not stratify IPMN samples by histologic grade according to the1996 WHO guidelines.
A total of 39 studies between January 1996 and October 2010 (representing a total of 1235 IPMN samples) met our predetermined inclusion criteria (Table 1). A pooled analysis was performed to determine the risk of malignant transformation for each genetic marker. Results are summarized according to individual gene candidates.
TABLE 1.
Studies Meeting Inclusion Criteria for Meta-Analysis Organized by Genetic Marker
| First Author | Year | Total No. IPMNs | Study Area | Gene | Assay, Definition of Positivity | OR | 95% CI | Study Quality (0–6) |
|---|---|---|---|---|---|---|---|---|
| Luttges et al18 | 2001 | 51 | Germany, Italy, United States | MUC1 | IHC | 22.077 | 1.210–402.833 | 1 |
| Adsay et al19 | 2002 | 74 | United States | MUC1 | >10% Cells positive by IHC | 3.158 | 0.636–15.671 | 2 |
| Terris et al20 | 2002 | 57 | France | MUC1 | IHC | 21.375 | 3.861–118.345 | 1 |
| Ito et al21 | 2005 | 21 | Japan | MUC1 | >5% Cells positive by IHC | 55.8 | 2.246–1386.315 | 5 |
| Moriya et al22 | 2005 | 37 | Japan | MUC1 | >10% Cells positive by IHC | 3.125 | 0.377–25.918 | 3 |
| Ueda et al23 | 2005 | 24 | Japan | MUC1 | IHC | 31.909 | 1.523–668.749 | 1 |
| Jang et al24 | 2009 | 41 | South Korea | MUC1 | >10% Cells positive by IHC | 0.733 | 0.123–4.367 | 3 |
| Cao et al25 | 2010 | 17 | United States | MUC1 | IHC | 0.333 | 0.014–7.875 | 1 |
| Luttges et al18 | 2001 | 51 | Germany, Italy, United States | MUC2 | IHC | 0.316 | 0.016–6.288 | 1 |
| Adsay et al19 | 2002 | 74 | United States | MUC2 | >10% Cells positive by IHC | 2.659 | 0.970–7.286 | 2 |
| Terris et al20 | 2002 | 57 | France | MUC2 | IHC | 0.296 | 0.090–0.975 | 1 |
| Ito et al21 | 2005 | 21 | Japan | MUC2 | >5% Cells positive by IHC | 6.810 | 0.321–144.629 | 5 |
| Moriya et al22 | 2005 | 37 | Japan | MUC2 | >10% Cells positive by IHC | 11.250 | 1.245–101.635 | 3 |
| Ueda et al23 | 2005 | 24 | Japan | MUC2 | IHC | 2.700 | 0.507–14.372 | 1 |
| Jang et al24 | 2009 | 41 | South Korea | MUC2 | >10% Cells positive by IHC | 22.500 | 3.689–137.235 | 3 |
| Cao et al25 | 2010 | 17 | United States | MUC2 | IHC | 7.000 | 0.306–160.322 | 1 |
| Terris et al20 | 2002 | 57 | France | MUC5AC | IHC | 0.096 | 0.017–0.547 | 1 |
| Kanno et al26 | 2006 | 51 | Japan | MUC5AC | IHC | 11.471 | 0.617–213.276 | 4 |
| Jang et al24 | 2009 | 41 | South Korea | MUC5AC | >10% Cells positive by IHC | 1.714 | 0.298–9.869 | 3 |
| Z’graggen et al27 | 1997 | 16 | United States | kRas | Sequence mutation | 6.000 | 0.257–140.045 | 4 |
| Mulligan et al28 | 1999 | 7 | United States | kRas | Sequence mutation | 1.500 | 0.055–40.633 | 3 |
| Paal et al29 | 1999 | 22 | United States | kRas | Immunohistochemistry | 41.000 | 1.115–1507.260 | 2 |
| Yoshizawa et al30 | 2002 | 7 | Japan | kRas | Sequence mutation | 18.000 | 0.812–399.154 | 2 |
| Mueller et al31 | 2003 | 13 | Germany | kRas | Sequence mutation | 1.167 | 0.074–18.346 | 2 |
| Uemura et al32 | 2003 | 15 | Japan | kRas | Sequence mutation | 4.000 | 0.447–35.788 | 2 |
| Kitago et al33 | 2004 | 20 | Japan | kRas | Sequence mutation | 1.364 | 0.112–16.577 | 3 |
| Wada et al34 | 2004 | 23 | Japan | kRas | Sequence mutation | 3.063 | 0.472–19.879 | 2 |
| Kobayashi et al35 | 2008 | 4 | Japan | kRas | Sequence mutation | 0.200 | 0.005–8.825 | 2 |
| Schonleben et al36 | 2008 | 38 | United States | kRas | Sequence mutation | 1.429 | 0.270–7.549 | 2 |
| Schonleben et al37 | 2007 | same data as above | United States | kRas | Sequence mutation | 2 | ||
| Schonleben et al38 | 2008 | same data as above | United States | kRas | Sequence mutation | 1 | ||
| Chadwick et al39 | 2009 | 52 | United States | kRas | Sequence mutation | 0.406 | 0.119–1.381 | 3 |
| Fritz et al40 | 2009 | 20 | United States | kRas | Sequence mutation | 0.333 | 0.047–2.366 | 3 |
| Jang et al24 | 2009 | 41 | South Korea | kRas | Sequence mutation | 5.867 | 1.395–24.673 | 4 |
| Biankin et al41 | 2002 | 18 | Australia | p53 | >10% Cells positive by IHC | 3.889 | 0.543–27.866 | 4 |
| Jinfeng et al42 | 2002 | 22 | Japan | p53 | >10% Cells positive by IHC | 25.000 | 1.168–535.241 | 2 |
| Mueller et al31 | 2003 | 13 | Germany | p53 | PCR amplification and single-strand conformation polymorphism analysis of exons 5, 6, 7, 8 | 4.500 | 0.190–106.823 | 2 |
| Sasaki et al43 | 2003 | 38 | Japan | p53 | >5% Cells positive by IHC | 33.000 | 1.647–661.051 | 2 |
| Uemura et al32 | 2003 | 15 | Japan | p53 | focal aggregates of >30 positive cells with nucleus stained brown | 5.769 | 0.232–143.371 | 2 |
| Wada et al34 | 2004 | 23 | Japan | p53 | Fluorescence-labeled microsatellite markers to detect loss of heterozygosity of 17p13 | 50.143 | 2.242–1121.352 | 1 |
| Ito et al21 | 2005 | 21 | Japan | p53 | >5% Cells positive by IHC | 3.250 | 0.340–31.074 | 5 |
| Ueda et al23 | 2005 | 24 | Japan | p53 | A few positive cells, scattered staining, diffuse staining | 11.118 | 0.508–243.105 | 1 |
| Nishikawa et al44 | 2006 | 37 | Japan | p53 | >10% Cells positive by IHC | 3.300 | 0.475–22.942 | 4 |
| Abe et al45 | 2007 | 47 | Japan | p53 | >10% Cells positive by IHC | 4.688 | 1.016–21.619 | 4 |
| Miyasaka et al46 | 2007 | 128 | Japan | p53 | >10% Cells positive by IHC | 25.000 | 3.153–198.205 | 4 |
| Chadwick et al39 | 2009 | 52 | United States | p53 | >5% Cells positive by IHC | 0.535 | 0.146–1.962 | 3 |
| Jang et al24 | 2009 | 41 | South Korea | p53 | >10% Cells positive by IHC | 8.767 | 0.458–167.937 | 3 |
| Nodari et al47 | 2003 | 2 | Italy | Telomerase | Reverse transcriptase–PCR | 9.000 | 0.097–831.780 | 0 |
| Hashimoto et al48 | 2008 | 12 | Japan | Telomerase | IHC | 39.667 | 1.279–1229.867 | 4 |
| Hashimoto et al49 | 2008 | 68 | Japan | Telomerase | IHC | 9.750 | 2.676–35.528 | 5 |
| Koshiba et al50 | 1999 | 26 | Japan | Cox2 | IHC | 23.000 | 1.070–494.573 | 2 |
| Kokawa et al51 | 2001 | 29 | Japan | Cox2 | >5% Cells positive by IHC | 1.697 | 0.332–8.666 | 2 |
| Aoki et al52 | 2002 | 30 | Japan | Cox2 | IHC | 5.833 | 1.119–30.403 | 3 |
| Niijima et al53 | 2002 | 32 | Japan | Cox2 | >10% Cells positive by IHC | 1.244 | 0.262–5.915 | 3 |
| Jang et al24 | 2009 | 41 | South Korea | Cox2 | IHC | 2.955 | 0.310–28.140 | 2 |
| Jang et al54 | 2007 | 55 | South Korea | Shh | IHC | 6.691 | 1.946–23.000 | 3 |
| Liu et al55 | 2007 | 18 | Taiwan | Shh | IHC | 10.455 | 0.425–256.957 | 0 |
| Satoh et al56 | 2008 | 63 | Japan | Shh | >20% Cells positive by IHC | 5.940 | 0.322–109.490 | 4 |
MUC Expression
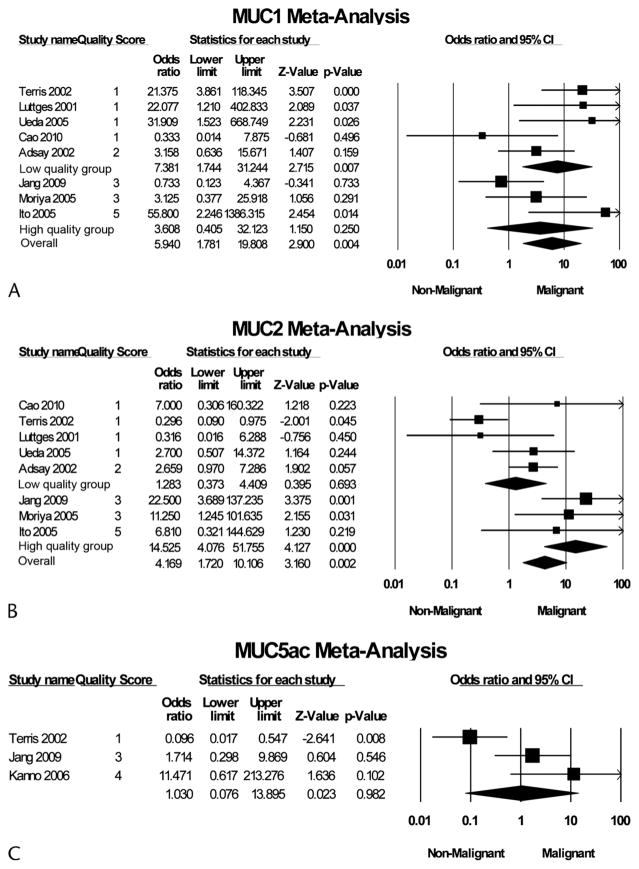
Mucins are large, heavily glycosylated proteins that are differentially expressed in epithelial cells of glandular tissues and various tumor types.19,21,57 Of the 19 mucin genes identified, MUC1, MUC2, and MUC5AC genes have been most frequently characterized in the pancreas. Eleven studies with 417 IPMN samples reported MUC expression.18–21,23–26,58 These studies examined MUC expression at the mRNA level by in situ hybridization20 or at the protein level by immunohistochemistry (IHC).18,19,21,23–26,58 Because the expression of different mucin genes appears to vary with grade of IPMN, we pooled studies according to analysis of the most frequently reported mucin genes: MUC1, MUC2, and MUC5AC. Figure 2 summarizes the individual ORs.
FIGURE 2.
Forest plot of studies examining association of MUC1 (A), MUC2 (B), or MUC5AC (C) expression and malignant transformation of IPMN. Methodologic quality of each study was assigned a score from 1 to 6 based on criteria summarized in Table 1. For each study, the quality score, OR; 95% CI, and relative weight are shown. The size of the data markers (squares) represents the statistical weight that each study contributed to the random-effects summary estimates; horizontal lines represent the 95% CI. The diamonds indicate the summary OR. I2 test P values evaluating study heterogeneity are shown.
Eight studies representing a total of 322 IPMN samples examined the expression of MUC1 by IHC.18–25 MUC1 was expressed in 8.6% (15/174) of IPM-A/B samples and 35.8% (53/148) of IPM-C samples. The pooled OR for MUC1 and malignant transformation was 5.9 (95% CI, 1.8–19.8). Grouped according to study quality, high-quality studies’ pooled OR was 3.6 (95% CI, 0.4–32.1), whereas low-quality studies’ pooled OR was 7.4 (95% CI, 1.7–31.2).
Eight studies representing a total of 322 IPMN sample examined the expression of MUC2 by IHC.18–25 MUC2 was expressed in 51.7% (90/174) of IPM-A/B samples and 68.9% (102/148) of IPM-C samples. The overall pooled OR for MUC2 and malignant transformation was 4.2 (95% CI, 1.7–10.1). Grouped according to study quality, high-quality studies’ pooled OR was 14.5 (95% CI, 4.1–51.8), whereas low-quality studies’ pooled OR was 1.3 (95% CI, 0.4–4.4).
Three studies representing a total of 231 IPMN samples examined the expression of MUC5AC by IHC.20,24,26 Five studies were excluded from pooled analysis because all IPMN samples were positive for MUC5AC.18,21,25,58,59 MUC5AC was expressed in 84.7% (149/176) of IPM-A/B samples and 92.4% (97/105) of IPM-C samples. The pooled OR for MUC5AC and malignant transformation was 1.0 (95% CI, 0.1–13.9).
Oncogenic kRas Expression
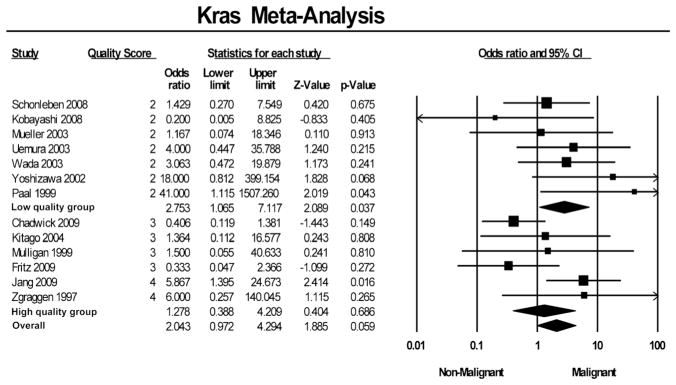
Activating point mutations in the GTP-binding protein kRas have been found in virtually all advanced pancreatic ductal adenocarcinoma and are thought to be an important step in pancreatic oncogenesis.60 Thirteen studies representing a total of 285 samples examined the presence of the kRas oncogenic mutation by polymerase chain reaction (PCR) and sequencing24,27–36,39,40 or IHC29 on resected tissue. Figure 3 summarizes the individual ORs.
FIGURE 3.
Forest plot of studies examining association of kRas expression and malignant transformation of IPMN.
In pooled analysis, the oncogenic mutation in kRas was found in 48.3% (71/147) of IPM-A/B samples and 55.1% (76/ 138) of IPM-C samples. The pooled OR for kRas mutation and malignant transformation was 2.0 (95% CI, 1.0–4.3). Grouped according to study quality, high-quality studies’ pooled OR was 1.3 (95% CI, 0.4–4.2), whereas low-quality studies’ pooled OR was 2.8 (95% CI, 1.1–7.1).
p53 Nuclear Expression
Most studies investigating p53 mutations in IPMN examined nuclear immunostaining of p53. This positivity criterion is based on the observation that wild-type p53 protein is present in the nucleus only at low levels, but mutant forms common to many types of cancer accumulate in the nucleus and can be visualized by IHC. Nuclear localization of p53 has been investigated as a prognostic factor in several kinds of gastrointestinal tumors including pancreatic cancer,61 hepatocellular cancer,62 colorectal cancer,63 and gastric cancer.64
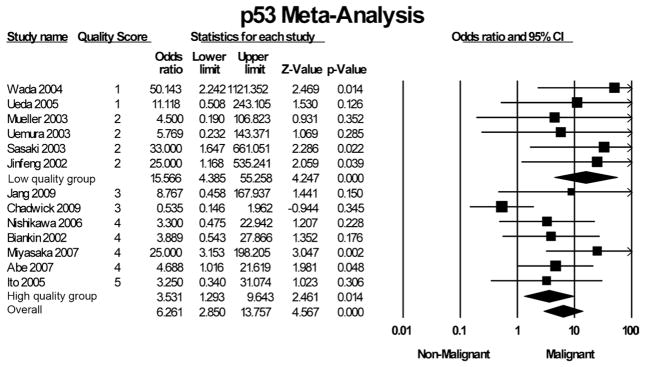
Six studies24,41,42,44–46 measured p53 mutations according to recommendations by Kawai et al,65 in which samples considered positive had greater than 10% of cells with positive nuclear immunostaining of p53. These 6 studies representing a total of 293 IPMNs identified positive p53 staining in 16.9% (31/183) of IPM-A/B samples and 40.9% (45/110) of IPM-C samples.24,41,42,44–46 Three studies used a threshold of 5% positive cells to define positive p53 staining.21,39,43 Together, these 3 studies examined 111 IPMN samples and identified positive staining in 17.1% (13/76) of IPM-A/B samples and 27.5% (11/40) of IPM-C samples.21,39,43 Uemura et al32 examined 15 IPMN samples and, using a criterion of focal aggregates of more than 30 cells with positive nuclear staining, found positive p53 staining in 0% (0/7) of IPM-A/B samples and 25% (2/8) of IPM-C samples. Ueda et al23 examined 24 IPMN samples and found that no IPM-A/B samples had any p53-positive nuclear staining, but 27% (3/11) of IPM-C samples had scattered or diffuse positive cells. Mueller et al31 performed PCR and single-strand conformation polymorphism analysis to find sequence changes in 13 IPMN samples and in this manner identified mutations in 10% (1/10) of IPM-A/B samples and 33.3% (1/3) of IPM-C samples. Wada et al34 used fluorescence-labeled microsatellite markers to detect loss of heterozygosity in the 17p13 chromosomal locus of p53 and found loss of heterozygosity in no IPM-A/B samples but in 66.7% (6/9) of IPM-C samples.
Pooling all the above studies examining p53 mutations by IPMN histologic grade, a total of 478 IPMN samples are examined, of which p53 mutations were identified in 14.9% (45/ 302) of IPM-A/B samples and 38.6% (68/176) of IPM-C samples. The pooled OR for p53 mutations and malignant transformation was 6.3 (95% CI, 2.9–13.8). Grouped according to study quality, high-quality studies’ pooled OR was 3.5 (95% CI, 1.3–9.6), whereas low-quality studies’ pooled OR was 15.6 (95% CI, 4.4–55.3). Figure 4 summarizes the individual ORs.
FIGURE 4.
Forest plot of studies examining association of altered p53 expression and malignant transformation of IPMN.
Telomerase Expression
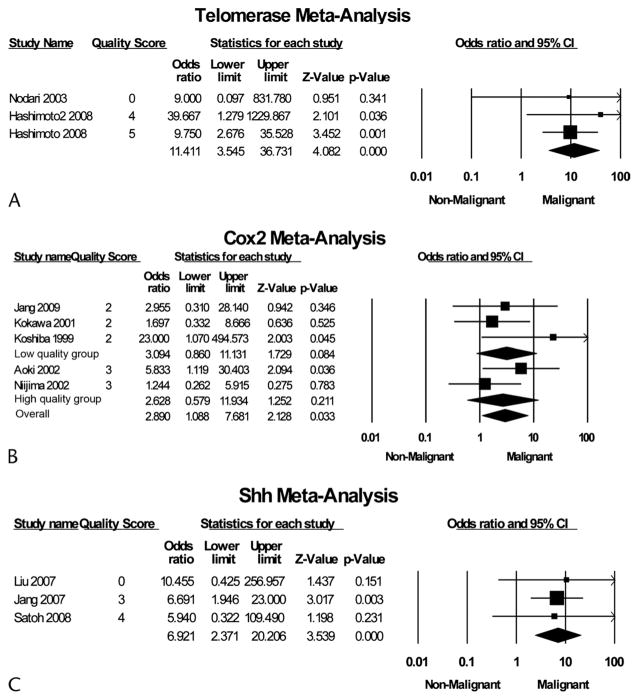
The human telomerase reverse transcriptase (hTERT) gene encodes the catalytic component of telomerase required to overcome telomere shortening and cellular senescence. Therefore, activation of hTERT is considered a hallmark of cancer.66,67 Three studies representing a total of 82 samples examined expression of hTERT by IHC48,49 or reverse transcriptase–PCR47 on resected tissue specimens (Fig. 5). hTERT expression was found in 23.7% (9/38) of IPM-A/B samples and 88.6% (39/44) of IPM-C samples. The pooled OR for hTERT mutation and malignant transformation was 11.4 (95% CI, 3.5–36.7).
FIGURE 5.
Forest plot of studies examining association of hTERT (A), Cox2 (B), or Shh (C) expression and malignant transformation of IPMN.
Cyclooxygenase 2 Expression
The biosynthesis of prostaglandins by the cyclooxygenase 2 (Cox2) enzyme is thought to mediate many properties of carcinogenesis. Expression of Cox2 is upregulated in pancreatic cancer68,69 and is thought to stimulate invasion70 and angiogenesis.71 Five studies representing a total of 158 IPMN samples examined the expression of Cox2 by IHC (Fig. 5).24,50–53 Cox2 was expressed in 53.3% (48/90) of IPM-A/B samples and 75.4% (52/69) of IPM-C samples. The pooled OR for Cox2 and malignant transformation was 2.9 (95% CI, 1.1–7.7). Grouped according to study quality, high-quality studies’ pooled OR was 2.6 (95% CI, 0.6–11.9), whereas low-quality studies’ pooled OR was 3.1 (95% CI, 0.9–11.1).
Shh Expression
The secreted factor Sonic hedgehog (Shh) has an important role in regulating normal pancreas development and has been implicated in tumorigenesis in pancreatic ductal adenocarcinoma.72 Three studies representing a total of 136 IPMN samples examined the expression of Shh by IHC (Fig. 5).54–56 Shh was expressed in 68.7% (57/83) of IPM-A/B samples and 90.6% (48/53) of IPM-C samples. The pooled OR for Shh expression and malignant transformation was 6.9 (95% CI, 2.4–20.2).
Heterogeneity and Impact of Quality
The estimated I2 was low (<25%) for studies evaluating telomerase, Cox2, and Shh. Estimated I2 for studies on kRas and p53 was moderate, 32% and 36%, respectively. Estimated I2 exceeded 50% for each of the MUC genes analyzed (MUC1, 54%; MUC2, 68%; and MUC5AC, 79%). Heterogeneity was significantly lower among high-quality studies of MUC2 (I2, 0%) and MUC5AC (I2, 16%). However, this was not the case for MUC1 (I2 among high-quality studies, 63%).
DISCUSSION
We have performed a quantitative meta-analysis of genetic markers associated with histologic progression of IPMN. Through a computerized literature search of online databases using predetermined inclusion criteria, we identified 39 studies between January 1996 and October 2010 (representing a total of 1235 IPMN samples) that examined the expression of 8 different genetic markers in benign versus malignant IPMN: MUC1, MUC2, MUC5AC, kRas, p53, hTERT, Cox2, and Shh. Pooled analysis of these studies revealed expression of hTERT, Shh, and MUC1 to have the strongest association with malignant progression of IPMN and expression of MUC5AC to have the weakest association with malignant progression of IPMN.
Intraductal papillary mucinous neoplasm of the pancreas has presented challenges in terms of pathophysiology and clinical management. Although the pathologic entity has been recognized for decades, the mechanisms of malignant transformation remain poorly understood. A key hypothesis has been that IPMN is fundamentally a genetic lesion and that an accumulation of somatic mutations drives the histologic progression, ultimately leading to malignant transformation. This hypothesis is a basis for pursuit of genetic markers that can be utilized to improve diagnosis, guide optimal management, and potentially design new therapeutic targets. In this meta-analysis, we investigated the hypothesis that specific genetic alterations are associated with the histologic progression of IPMN.
The risk of cancer associated with various gene mutations ranged from 1.0 to 11.41. The marker found to have the strongest association with malignant IPMN was hTERT (OR, 11.4; 95% CI, 3.5–36.7). One interpretation of this finding is that the genetic alterations leading to abnormal expression of hTERT occur later in the histologic progression of IPMN toward cancer. This is consistent with the hypothesis of “telomere crisis” in carcinogenesis. This model postulates that telomeres are progressively shortened with cell proliferation until cells reach “crisis” at which point most cells will die; the ability of rare cells to overcome crisis by upregulating hTERT is a critical step in carcinogenesis.66 Shortening of telomeres has been reported in some IPM-A but has been noted to progressively worsen with histologic progression.49 Therefore, the dramatic upregulation of hTERT observed in malignant compared with benign IPMN supports the notion of a crisis point in the development of malignancy.49
In the pooled analysis, we also identified markers that had little or no association with malignant progression of IPMN. Although fairly common, MUC5AC and kRas were not strongly associated with malignancy among IPMN lesions. A weaker association with malignant progression may suggest that alterations leading to expression of these markers occur early in the histologic progression of IPMN toward cancer. Activating mutations of kRas are recognized to be among the earliest mutations leading to pancreatic cancer, detected in more than 40% of early pancreatic intraepithelial neoplasia lesions.60,73,74 Alternatively, markers with no association with malignant IPMN may have no role in the pathobiology of IPMN progression. This could be the case with MUC5AC, which had no association (OR, 1.0; 95% CI, 0.1–13.9) as opposed to MUC1, which had a strong association (OR, 5.9; 95% CI, 1.8–19.8) with malignant transformation. MUC5AC is a secreted mucin speculated to form a protective gel around tumors,75 a property that may be advantageous for all neoplasms, benign or malignant. In contrast, MUC1 is a membrane-associated mucin that has been demonstrated to bind and signal through β-catenin and mitogen-activated protein kinase pathways critical in cell proliferation, and thus upregulation of MUC1 may be necessary for the progression of IPMN from benign to malignant lesions.76–78 Interestingly, a knockout of Muc1 in mice significantly slowed the growth rate of oncogene-induced breast tumors and decreased the rates of tumor metastasis.79
A proposed model for genetic alterations associated with malignant progression of IPMN based on the present meta-analysis is presented in Figure 6. How well these ORs in fact correspond to the pathobiology of IPMN progression remains to be validated in future functional studies. A key question is whether the genetic markers are so-called “passenger” or “driver” alterations: do these genetic alterations cause IPMN lesions to acquire malignant behavior, or are they simply a consequence of other genetic alterations that in fact drive IPMN progression?
FIGURE 6.
A model for genetic alterations associated with malignant progression of IPMN based on the findings of this meta-analysis.
The strengths of the approach taken in this meta-analysis include a comprehensive and unbiased search of IPMN literature, the use of standardized systematic review and meta-analysis techniques using predefined inclusion and exclusion criteria, and elaboration of a scoring system to assess quality of included studies.
There were several limitations to the present study. In this meta-analysis, IPMN lesions were stratified according to the WHO IPM-A/B versus IPM-C grades, but insufficient data were available to stratify according to histologic cell type (eg, gastric, intestinal, pancreatobiliary, oncocytic). There are also potential biases derived from limiting our search to published studies. Unpublished data may have an increased proportion of null results in which no association is detected between a genetic marker and IPMN progression. The genetic markers eligible for pooled analysis were limited by the number of studies that met the inclusion criteria. A number of markers such as p1623,43 and Smad480,81 have been frequently investigated but have not been used as consistent criteria for measuring genetic alterations. Additional candidate genetic markers that have been examined in IPMN tissue were unable to be included in the quantitative meta-analysis because of availability of fewer than 3 published reports in the medical literature. Finally, as it is derived from published reports, this meta-analysis is limited to previously characterized markers. Unbiased approaches such as RNASeq will be critical to identify novel genetic alterations that play a role in malignant progression of IPMN.
We developed a 6-point scoring system based on the STROBE16 and PRAISE reporting guidelines17 for observational studies. We found considerable variability in quality among the studies included in the present meta-analysis. In particular, many studies failed to confirm the histologic diagnosis of malignancy by 2 independent pathologists. In our analysis, we found that the risk of malignant transformation associated with a number of gene mutations varied according to study quality. Therefore, we advocate establishment of standardized quality reporting criteria to reduce heterogeneity and enhance accuracy of future studies evaluating genetic risk factors for malignant transformation of IPMN.
Ultimately, the motivation for this and future studies is to elucidate genetic markers that can improve diagnosis, guide optimal management, and offer new therapeutic targets. In addition, elucidating the genetic alterations underlying malignant transformation of IPMN may have relevance to pancreatic cancer. A recent meta-analysis identified Cox2 as a marker associated with poor survival outcome in pancreatic cancer.82 Here, we similarly find that Cox2 expression is associated with malignant IPMN (OR, 2.9), highlighting potential parallels in the pathogenesis of malignant IPMN and pancreatic cancer.
In summary, numerous studies have evaluated candidate gene mutations associated with malignant transformation in IPMN. Using quantitative meta-analysis, we identified a strong association of hTERT expression with malignant transformation of IPMN, consistent with up-regulation of hTERT as a key step in progression of IPMN to cancer. Although kRas was a commonly detected mutation, its presence was not strongly associated with histologic progression of IPMN. In the context of this meta-analysis, we have also proposed a set of quality criteria for reporting of genetic studies related to malignant transformation of IPMN. We believe the present findings can be used as a framework to help guide further research aimed at elucidating the genetic basis of malignant progression in IPMN of the pancreas.
Abbreviations
- CI
confidence interval
- Cox2
cyclooxygenase 2
- OR
odds ratio
- IPMN
intraductal mucinous neoplasm
- hTERT
human telomerase reverse transcriptase
- Shh
Sonic hedgehog
- IHC
immunohistochemistry
Footnotes
Guarantor of the article: Dr Wu. Author contributions: Dr Nissim: drafting of manuscript, study concept and design, and analysis and interpretation of data. Dr Idos: study concept and design, analysis and interpretation of data, and critical revision of the manuscript. Dr Wu: study concept and design, analysis and interpretation of data, critical revision of the manuscript, statistical analysis, and study supervision.
The authors declare no conflict of interest.
No financial support was received for this study.
References
- 1.Hruban RH, Takaori K, Klimstra DS, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28(8):977–987. doi: 10.1097/01.pas.0000126675.59108.80. [DOI] [PubMed] [Google Scholar]
- 2.Sessa F, Solcia E, Capella C, et al. Intraductal papillary-mucinous tumours represent a distinct group of pancreatic neoplasms: an investigation of tumour cell differentiation and K-ras, p53 and c-erbB-2 abnormalities in 26 patients. Virchows Arch. 1994;425(4):357–367. doi: 10.1007/BF00189573. [DOI] [PubMed] [Google Scholar]
- 3.Shimada K, Sakamoto Y, Sano T, et al. Invasive carcinoma originating in an intraductal papillary mucinous neoplasm of the pancreas: a clinicopathologic comparison with a common type of invasive ductal carcinoma. Pancreas. 2006;32:281–287. doi: 10.1097/01.mpa.0000202955.33483.e2. [DOI] [PubMed] [Google Scholar]
- 4.Maire F, Hammel P, Terris B, et al. Prognosis of malignant intraductal papillary mucinous tumours of the pancreas after surgical resection. Comparison with pancreatic ductal adenocarcinoma. Gut. 2002;51(5):717–722. doi: 10.1136/gut.51.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sohn TA, Yeo CJ, Cameron JL, et al. Intraductal papillary mucinous neoplasms of the pancreas: an increasingly recognized clinicopathologic entity. Ann Surg. 2001;234(3):313–321. doi: 10.1097/00000658-200109000-00005. discussion 312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kloppel G, Solcia E, Longnecker D, et al. Histological Typing of Tumours of the Exocrine Pancreas. World Health Organization International Histological Classification of Tumours. 2. Berlin: Springer-Verlag; 1996. [Google Scholar]
- 7.Jang JY, Kim SW, Ahn YJ, et al. Multicenter analysis of clinicopathologic features of intraductal papillary mucinous tumor of the pancreas: is it possible to predict the malignancy before surgery? Ann Surg Oncol. 2005;12(2):124–132. doi: 10.1245/ASO.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 8.Murakami Y, Uemura K, Hayashidani Y, et al. Predictive factors of malignant or invasive intraductal papillary-mucinous neoplasms of the pancreas. J Gastrointest Surg. 2007;11(3):338–344. doi: 10.1007/s11605-006-0069-8. [DOI] [PubMed] [Google Scholar]
- 9.Shin SH, Han DJ, Park KT, et al. Validating a simple scoring system to predict malignancy and invasiveness of intraductal papillary mucinous neoplasms of the pancreas. World J Surg. 2010;34(4):776–783. doi: 10.1007/s00268-010-0416-5. [DOI] [PubMed] [Google Scholar]
- 10.Kobari M, Egawa S, Shibuya K, et al. Intraductal papillary mucinous tumors of the pancreas comprise 2 clinical subtypes: differences in clinical characteristics and surgical management. Arch Surg. 1999;134(10):1131–1136. doi: 10.1001/archsurg.134.10.1131. [DOI] [PubMed] [Google Scholar]
- 11.Doi R, Fujimoto K, Wada M, et al. Surgical management of intraductal papillary mucinous tumor of the pancreas. Surgery. 2002;132(1):80–85. doi: 10.1067/msy.2002.125386. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa Y, Unger TA, Taylor S, et al. Mucus is a predictor of better prognosis and survival in patients with intraductal papillary mucinous tumor of the pancreas. J Gastrointest Surg. 2003;7:12–18. doi: 10.1016/S1091-255X(02)00152-X. discussion 18–19. [DOI] [PubMed] [Google Scholar]
- 13.Sugiyama M, Izumisato Y, Abe N, et al. Predictive factors for malignancy in intraductal papillary-mucinous tumours of the pancreas. Br J Surg. 2003;90(10):1244–1249. doi: 10.1002/bjs.4265. [DOI] [PubMed] [Google Scholar]
- 14.Salvia R, Fernández-del Castillo C, Bassi C, et al. Main-duct intraductal papillary mucinous neoplasms of the pancreas: clinical predictors of malignancy and long-term survival following resection. Ann Surg. 2004;239(5):678–685. doi: 10.1097/01.sla.0000124386.54496.15. discussion 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takuma K, Kamisawa T, Anjiki H, et al. Predictors of malignancy and natural history of main-duct intraductal papillary mucinous neoplasms of the pancreas. Pancreas. 2011;40(3):371–375. doi: 10.1097/MPA.0b013e3182056a83. [DOI] [PubMed] [Google Scholar]
- 16.Sanderson S, Tatt ID, Higgins JP. Tools for assessing quality and susceptibility to bias in observational studies in epidemiology: a systematic review and annotated bibliography. Int J Epidemiol. 2007;36(3):666–676. doi: 10.1093/ije/dym018. [DOI] [PubMed] [Google Scholar]
- 17.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269. W264. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 18.Luttges J, Zamboni G, Longnecker D, et al. The immunohistochemical mucin expression pattern distinguishes different types of intraductal papillary mucinous neoplasms of the pancreas and determines their relationship to mucinous noncystic carcinoma and ductal adenocarcinoma. Am J Surg Pathol. 2001;25(7):942–948. doi: 10.1097/00000478-200107000-00014. [DOI] [PubMed] [Google Scholar]
- 19.Adsay NV, Merati K, Andea A, et al. The dichotomy in the preinvasive neoplasia to invasive carcinoma sequence in the pancreas: differential expression of MUC1 and MUC2 supports the existence of two separate pathways of carcinogenesis. Mod Pathol. 2002;15(10):1087–1095. doi: 10.1097/01.MP.0000028647.98725.8B. [DOI] [PubMed] [Google Scholar]
- 20.Terris B, Dubois S, Buisine MP, et al. Mucin gene expression in intraductal papillary-mucinous pancreatic tumours and related lesions. J Pathol. 2002;197(5):632–637. doi: 10.1002/path.1146. [DOI] [PubMed] [Google Scholar]
- 21.Ito H, Endo T, Oka T, et al. Mucin expression profile is related to biological and clinical characteristics of intraductal papillary-mucinous tumors of the pancreas. Pancreas. 2005;30:e96–e102. doi: 10.1097/01.mpa.0000163358.90111.ab. [DOI] [PubMed] [Google Scholar]
- 22.Moriya T, Kimura W, Semba S, et al. Biological similarities and differences between pancreatic intraepithelial neoplasias and intraductal papillary mucinous neoplasms. Int J Gastrointest Cancer. 2005;35(2):111–119. doi: 10.1385/IJGC:35:2:111. [DOI] [PubMed] [Google Scholar]
- 23.Ueda M, Miura Y, Kunihiro O, et al. MUC1 overexpression is the most reliable marker of invasive carcinoma in intraductal papillary-mucinous tumor (IPMT) Hepatogastroenterology. 2005;52(62):398–403. [PubMed] [Google Scholar]
- 24.Jang JY, Park YC, Song YS, et al. Increased K-ras mutation and expression of S100A4 and MUC2 protein in the malignant intraductal papillary mucinous tumor of the pancreas. J Hepatobiliary Pancreat Surg. 2009;16(5):668–674. doi: 10.1007/s00534-009-0105-7. [DOI] [PubMed] [Google Scholar]
- 25.Cao W, Adley BP, Liao J, et al. Mucinous nonneoplastic cyst of the pancreas: apomucin phenotype distinguishes this entity from intraductal papillary mucinous neoplasm. Hum Pathol. 2010;41:513–521. doi: 10.1016/j.humpath.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanno A, Satoh K, Kimura K, et al. The expression of MUC4 and MUC5AC is related to the biologic malignancy of intraductal papillary mucinous neoplasms of the pancreas. Pancreas. 2006;33:391–396. doi: 10.1097/01.mpa.0000236742.92606.c1. [DOI] [PubMed] [Google Scholar]
- 27.Z’graggen K, Rivera JA, Compton CC, et al. Prevalence of activating K-ras mutations in the evolutionary stages of neoplasia in intraductal papillary mucinous tumors of the pancreas. Ann Surg. 1997;226(4):491–498. doi: 10.1097/00000658-199710000-00010. discussion 498–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulligan NJ, Yang S, Andry C, et al. The role of p21ras in pancreatic neoplasia and chronic pancreatitis. Hum Pathol. 1999;30(6):602–610. doi: 10.1016/s0046-8177(99)90082-5. [DOI] [PubMed] [Google Scholar]
- 29.Paal E, Thompson LD, Przygodzki RM, et al. A clinicopathologic and immunohistochemical study of 22 intraductal papillary mucinous neoplasms of the pancreas, with a review of the literature. Mod Pathol. 1999;12(5):518–528. [PubMed] [Google Scholar]
- 30.Yoshizawa K, Nagai H, Sakurai S, et al. Clonality and K-ras mutation analyses of epithelia in intraductal papillary mucinous tumor and mucinous cystic tumor of the pancreas. Virchows Arch. 2002;441(5):437–443. doi: 10.1007/s00428-002-0645-6. [DOI] [PubMed] [Google Scholar]
- 31.Mueller J, Gansauge S, Mattfeldt T. P53 mutation but not p16/MTS1 mutation occurs in intraductal papillary mucinous tumors of the pancreas. Hepatogastroenterology. 2003;50(50):541–544. [PubMed] [Google Scholar]
- 32.Uemura K, Hiyama E, Murakami Y, et al. Comparative analysis of K-ras point mutation, telomerase activity, and p53 overexpression in pancreatic tumours. Oncol Rep. 2003;10(2):277–283. [PubMed] [Google Scholar]
- 33.Kitago M, Ueda M, Aiura K, et al. Comparison of K-ras point mutation distributions in intraductal papillary-mucinous tumors and ductal adenocarcinoma of the pancreas. Int J Cancer. 2004;110(2):177–182. doi: 10.1002/ijc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Wada K, Takada T, Yasuda H, et al. Does “clonal progression” relate to the development of intraductal papillary mucinous tumors of the pancreas? J Gastrointest Surg. 2004;8:289–296. doi: 10.1016/j.gassur.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi N, Inamori M, Fujita K, et al. Characterization of K-ras gene mutations in association with mucinous hypersecretion in intraductal papillary-mucinous neoplasms. J Hepatobiliary Pancreat Surg. 2008;15(2):169–177. doi: 10.1007/s00534-007-1223-8. [DOI] [PubMed] [Google Scholar]
- 36.Schonleben F, Allendorf JD, Qiu W, et al. Mutational analyses of multiple oncogenic pathways in intraductal papillary mucinous neoplasms of the pancreas. Pancreas. 2008;36:168–172. doi: 10.1097/MPA.0b013e318158a4d2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schonleben F, Qiu W, Bruckman KC, et al. BRAF and KRAS gene mutations in intraductal papillary mucinous neoplasm/carcinoma (IPMN/IPMC) of the pancreas. Cancer Lett. 2007;249:242–248. doi: 10.1016/j.canlet.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schonleben F, Qiu W, Remotti HE, et al. PIK3CA, KRAS, and BRAF mutations in intraductal papillary mucinous neoplasm/carcinoma (IPMN/C) of the pancreas. Langenbecks Arch Surg. 2008;393(3):289–296. doi: 10.1007/s00423-008-0285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chadwick B, Willmore-Payne C, Tripp S, et al. Histologic, immunohistochemical, and molecular classification of 52 IPMNs of the pancreas. Appl Immunohistochem Mol Morphol. 2009;17(1):31–39. doi: 10.1097/PAI.0b013e31817c02c6. [DOI] [PubMed] [Google Scholar]
- 40.Fritz S, Fernandez-del Castillo C, Mino-Kenudson M, et al. Global genomic analysis of intraductal papillary mucinous neoplasms of the pancreas reveals significant molecular differences compared to ductal adenocarcinoma. Ann Surg. 2009;249(3):440–447. doi: 10.1097/SLA.0b013e31819a6e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biankin AV, Biankin SA, Kench JG, et al. Aberrant p16(INK4A) and DPC4/Smad4 expression in intraductal papillary mucinous tumours of the pancreas is associated with invasive ductal adenocarcinoma. Gut. 2002;50(6):861–868. doi: 10.1136/gut.50.6.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jinfeng M, Kimura W, Sakurai F, et al. Histopathological study of intraductal papillary mucinous tumor of the pancreas: special reference to the roles of Survivin and p53 in tumorigenesis of IPMT. Int J Gastrointest Cancer. 2002;32:73–81. doi: 10.1385/IJGC:32:2-3:73. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki S, Yamamoto H, Kaneto H, et al. Differential roles of alterations of p53, p16, and SMAD4 expression in the progression of intraductal papillary-mucinous tumors of the pancreas. Oncol Rep. 2003;10(1):21–25. [PubMed] [Google Scholar]
- 44.Nishikawa N, Kimura Y, Okita K, et al. Intraductal papillary mucinous neoplasms of the pancreas: an analysis of protein expression and clinical features. J Hepatobiliary Pancreat Surg. 2006;13(4):327–335. doi: 10.1007/s00534-005-1073-1. [DOI] [PubMed] [Google Scholar]
- 45.Abe K, Suda K, Arakawa A, et al. Different patterns of p16INK4A and p53 protein expressions in intraductal papillary-mucinous neoplasms and pancreatic intraepithelial neoplasia. Pancreas. 2007;34:85–91. doi: 10.1097/01.mpa.0000240608.56806.0a. [DOI] [PubMed] [Google Scholar]
- 46.Miyasaka Y, Nagai E, Yamaguchi H, et al. The role of the DNA damage checkpoint pathway in intraductal papillary mucinous neoplasms of the pancreas. Clin Cancer Res. 2007;13:4371–4377. doi: 10.1158/1078-0432.CCR-07-0032. [DOI] [PubMed] [Google Scholar]
- 47.Nodari F, Baiocchi GL, Salvi A, et al. Telomerase gene expression in intraductal papillary-mucinous tumors (IPMT): preliminary findings. Acta Biomed. 2003;74(suppl 2):59–64. [PubMed] [Google Scholar]
- 48.Hashimoto Y, Murakami Y, Uemura K, et al. Detection of human telomerase reverse transcriptase (hTERT) expression in tissue and pancreatic juice from pancreatic cancer. Surgery. 2008;143:113–125. doi: 10.1016/j.surg.2007.07.042. [DOI] [PubMed] [Google Scholar]
- 49.Hashimoto Y, Murakami Y, Uemura K, et al. Telomere shortening and telomerase expression during multistage carcinogenesis of intraductal papillary mucinous neoplasms of the pancreas. J Gastrointest Surg. 2008;12(1):17–28. doi: 10.1007/s11605-007-0383-9. discussion 19–28. [DOI] [PubMed] [Google Scholar]
- 50.Koshiba T, Hosotani R, Miyamoto Y, et al. Immunohistochemical analysis of cyclooxygenase-2 expression in pancreatic tumors. Int J Pancreatol. 1999;26(2):69–76. doi: 10.1007/BF02781733. [DOI] [PubMed] [Google Scholar]
- 51.Kokawa A, Kondo H, Gotoda T, et al. Increased expression of cyclooxygenase-2 in human pancreatic neoplasms and potential for chemoprevention by cyclooxygenase inhibitors. Cancer. 2001;91:333–338. doi: 10.1002/1097-0142(20010115)91:2<333::aid-cncr1006>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 52.Aoki T, Nagakawa Y, Tsuchida A, et al. Expression of cyclooxygenase-2 and vascular endothelial growth factor in pancreatic tumors. Oncol Rep. 2002;9(4):761–765. [PubMed] [Google Scholar]
- 53.Niijima M, Yamaguchi T, Ishihara T, et al. Immunohistochemical analysis and in situ hybridization of cyclooxygenase-2 expression in intraductal papillary-mucinous tumors of the pancreas. Cancer. 2002;94:1565–1573. doi: 10.1002/cncr.10358. [DOI] [PubMed] [Google Scholar]
- 54.Jang KT, Lee KT, Lee JG, et al. Immunohistochemical expression of Sonic hedgehog in intraductal papillary mucinous tumor of the pancreas. Appl Immunohistochem Mol Morphol. 2007;15:294–298. doi: 10.1097/01.pai.0000213132.71041.da. [DOI] [PubMed] [Google Scholar]
- 55.Liu MS, Yang PY, Yeh TS. Sonic hedgehog signaling pathway in pancreatic cystic neoplasms and ductal adenocarcinoma. Pancreas. 2007;34:340–346. doi: 10.1097/mpa.0b013e3180333ab5. [DOI] [PubMed] [Google Scholar]
- 56.Satoh K, Kanno A, Hamada S, et al. Expression of Sonic hedgehog signaling pathway correlates with the tumorigenesis of intraductal papillary mucinous neoplasm of the pancreas. Oncol Rep. 2008;19(5):1185–1190. [PubMed] [Google Scholar]
- 57.Kim YS, Gum JR., Jr Diversity of mucin genes, structure, function, and expression. Gastroenterology. 1995;109(3):999–1001. doi: 10.1016/0016-5085(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 58.Ji Y, Lou WH, Jin DY, et al. A series of 64 cases of pancreatic cystic neoplasia from an institutional study of China. World J Gastroenterol. 2006;12(45):7380–7387. doi: 10.3748/wjg.v12.i45.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Handra-Luca A, Flejou JF, Rufat P, et al. Human pancreatic mucinous cystadenoma is characterized by distinct mucin, cytokeratin and CD10 expression compared with intraductal papillary-mucinous adenoma. Histopathology. 2006;48:813–821. doi: 10.1111/j.1365-2559.2006.02444.x. [DOI] [PubMed] [Google Scholar]
- 60.Hezel AF, Kimmelman AC, Stanger BZ, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20(10):1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 61.Harada N, Gansauge S, Gansauge F, et al. Nuclear accumulation of p53 correlates significantly with clinical features and inversely with the expression of the cyclin-dependent kinase inhibitor p21(WAF1/CIP1) in pancreatic cancer. Br J Cancer. 1997;76(3):299–305. doi: 10.1038/bjc.1997.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bressac B, Galvin KM, Liang TJ, et al. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990;87(5):1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodrigues NR, Rowan A, Smith ME, et al. p53 mutations in colorectal cancer. Proc Natl Acad Sci U S A. 1990;87(19):7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uchino S, Noguchi M, Hirota T, et al. High incidence of nuclear accumulation of p53 protein in gastric cancer. Jpn J Clin Oncol. 1992;22(4):225–231. [PubMed] [Google Scholar]
- 65.Kawai A, Noguchi M, Beppu Y, et al. Nuclear immunoreaction of p53 protein in soft tissue sarcomas. A possible prognostic factor. Cancer. 1994;73(10):2499–2505. doi: 10.1002/1097-0142(19940515)73:10<2499::aid-cncr2820731008>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 66.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297(5581):565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 67.O’Hagan RC, Chang S, Maser RS, et al. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2(2):149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- 68.Okami J, Yamamoto H, Fujiwara Y, et al. Overexpression of cyclooxygenase-2 in carcinoma of the pancreas. Clin Cancer Res. 1999;5(8):2018–2024. [PubMed] [Google Scholar]
- 69.Yip-Schneider MT, Barnard DS, Billings SD, et al. Cyclooxygenase-2 expression in human pancreatic adenocarcinomas. Carcinogenesis. 2000;21(2):139–146. doi: 10.1093/carcin/21.2.139. [DOI] [PubMed] [Google Scholar]
- 70.Ito H, Duxbury M, Benoit E, et al. Prostaglandin E2 enhances pancreatic cancer invasiveness through an Ets-1–dependent induction of matrix metalloproteinase-2. Cancer Res. 2004;64(20):7439–7446. doi: 10.1158/0008-5472.CAN-04-1177. [DOI] [PubMed] [Google Scholar]
- 71.Chu J, Lloyd FL, Trifan OC, et al. Potential involvement of the cyclooxygenase-2 pathway in the regulation of tumor-associated angiogenesis and growth in pancreatic cancer. Mol Cancer Ther. 2003;2(1):1–7. [PubMed] [Google Scholar]
- 72.Morris JPt, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10(10):683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klimstra DS, Longnecker DS. K-ras mutations in pancreatic ductal proliferative lesions. Am J Pathol. 1994;145(6):1547–1550. [PMC free article] [PubMed] [Google Scholar]
- 74.Hruban RH, van Mansfeld AD, Offerhaus GJ, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143(2):545–554. [PMC free article] [PubMed] [Google Scholar]
- 75.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4(1):45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 76.Li Y, Bharti A, Chen D, et al. Interaction of glycogen synthase kinase 3beta with the DF3/MUC1 carcinoma-associated antigen and beta-catenin. Mol Cell Biol. 1998;18(12):7216–7224. doi: 10.1128/mcb.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schroeder JA, Thompson MC, Gardner MM, et al. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. J Biol Chem. 2001;276(16):13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- 78.Yamamoto M, Bharti A, Li Y, et al. Interaction of the DF3/MUC1 breast carcinoma–associated antigen and beta-catenin in cell adhesion. J Biol Chem. 1997;272(19):12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 79.Spicer AP, Rowse GJ, Lidner TK, et al. Delayed mammary tumor progression in Muc-1 null mice. J Biol Chem. 1995;270(50):30093–30101. doi: 10.1074/jbc.270.50.30093. [DOI] [PubMed] [Google Scholar]
- 80.Sunamura M, Lefter LP, Duda DG, et al. The role of chromosome 18 abnormalities in the progression of pancreatic adenocarcinoma. Pancreas. 2004;28:311–316. doi: 10.1097/00006676-200404000-00019. [DOI] [PubMed] [Google Scholar]
- 81.Inoue H, Furukawa T, Sunamura M, et al. Exclusion of SMAD4 mutation as an early genetic change in human pancreatic ductal tumorigenesis. Genes Chromosomes Cancer. 2001;31(3):295–299. doi: 10.1002/gcc.1147. [DOI] [PubMed] [Google Scholar]
- 82.Jamieson NB, Carter CR, McKay CJ, et al. Tissue biomarkers for prognosis in pancreatic ductal adenocarcinoma: a systematic review and meta-analysis. Clin Cancer Res. 2011;17(10):3316–3331. doi: 10.1158/1078-0432.CCR-10-3284. [DOI] [PubMed] [Google Scholar]