Cells can maneuver through 3D matrices by using tracks that exist in the matrix. The focal adhesion protein vinculin mediates the unidirectional movement through these tracks. Moreover, vinculin also promotes directional migration in 2D and 3D matrices. Vinculin’s role in migration is mediated by FAK activation.
Abstract
During metastasis, cells can use proteolytic activity to form tube-like “microtracks” within the extracellular matrix (ECM). Using these microtracks, cells can migrate unimpeded through the stroma. To investigate the molecular mechanisms of microtrack migration, we developed an in vitro three-dimensional (3D) micromolded collagen platform. When in microtracks, cells tend to migrate unidirectionally. Because focal adhesions are the primary mechanism by which cells interact with the ECM, we examined the roles of several focal adhesion molecules in driving unidirectional motion. Vinculin knockdown results in the repeated reversal of migration direction compared with control cells. Tracking the position of the Golgi centroid relative to the position of the nucleus centroid reveals that vinculin knockdown disrupts cell polarity in microtracks. Vinculin also directs migration on two-dimensional (2D) substrates and in 3D uniform collagen matrices, as indicated by reduced speed, shorter net displacement, and decreased directionality in vinculin-deficient cells. In addition, vinculin is necessary for focal adhesion kinase (FAK) activation in three dimensions, as vinculin knockdown results in reduced FAK activation in both 3D uniform collagen matrices and microtracks but not on 2D substrates, and, accordingly, FAK inhibition halts cell migration in 3D microtracks. Together these data indicate that vinculin plays a key role in polarization during migration.
INTRODUCTION
Cancer cell migration is a key step in the dissemination of cells from a primary tumor through the collagenous stromal extracellular matrix (ECM) during cancer metastasis. Metastatic cancer cells escape from primary tumors using diverse microenvironment-dependent migration strategies, and cells can migrate through the stroma both individually and as collectives of cells, forming sheets, files, or clusters (Friedl and Wolf, 2003). Critically, proteolytic- and force-mediated matrix remodeling by migrating cells can lead to the formation of cleared pathways or “microtracks” within the ECM (Gaggioli et al., 2007; Friedl and Gilmour, 2009; Giampieri et al., 2009). Of interest, cancer cells can escape from the primary tumor in a proteolysis-independent manner by coopting these microtracks, and cells’ unidirectional migration within microtracks includes elements of both two- and three-dimensional (2D and 3D) cell migration (Kraning-Rush et al., 2013; Carey et al., 2015). Migration through these microtracks reflects 2D migration, in part, because cells do not require proteolytic activity, whereas it also resembles 3D migration because cells can bind to fibrillar collagen in three dimensions during migration. Previously we developed in vitro–micropatterned 3D collagen microtracks using state-of-the- art microfabrication techniques (Kraning-Rush et al., 2013) and showed that mechanical cell–matrix interactions are not necessarily required for migration through 3D collagen microtracks, as matrix-free cleared pathways provide less resistance and eliminate the need for matrix remodeling, traction generation, and extensive cell body deformation (Carey et al., 2015). However, it is not yet clear how cells adhere to and migrate through these physiological collagen microtracks and which molecular mechanisms govern this unique mode of migration.
Various molecular mechanisms, including regulation of cytoskeletal protrusion, cell–matrix adhesion, proteolysis, and actomyosin contractility, are essential for effective cell migration (Lauffenburger and Horwitz, 1996; Friedl and Gilmour, 2009). Many studies have comparatively explored molecular mechanisms of migration in 2D substrates and 3D environments, often finding that 2D migration behavior does not always reflect 3D migration behavior. For example, Meyer et al. (2012) failed to observe a correlation between growth factor–induced cell migration responses on a 2D substrate compared with those within a 3D ECM. Alternatively, they found that 2D protrusions can predict growth factor–induced cell migration in 3D matrices. Zaman et al. (2006) showed that the tumor cell migratory response to matrix stiffness is fundamentally different in 3D matrices than with 2D substrates. In addition, little association has been found between the roles of specific focal adhesion proteins during 2D and 3D migration. Numerous proteins involved in focal adhesion assembly and disassembly in two dimensions play different roles and have differing degrees of importance in regulating 3D cell migration (Fraley et al., 2012; Friedl et al., 2012; Doyle et al., 2013). Because cell–matrix adhesion is a fundamental element of cell migration, it is critical to understand the role of adhesion molecules during 3D microtrack migration separately from their role in 2D substrate and 3D uniform collagen matrix migration.
Vinculin, a prominent and well-characterized focal adhesion protein, has been shown to be critical in regulating cell motility through both structural and signaling means (Mierke et al., 2010; Goldmann et al., 2013). It mediates the connection of the cell cytoskeleton to the extracellular matrix and helps to transmit cellular forces by binding F-actin as well as talin and α-actinin, which bind to integrins (Ziegler et al., 2006). Vinculin is also required in early focal adhesion formation (Galbraith et al., 2002), in which nascent focal adhesions can be highly transient (Tan et al., 2010). Moreover, vinculin knockdown has been correlated with rapid focal adhesion turnover, resulting in the formation of smaller and fewer focal adhesions (Saunders et al., 2006; Srichai and Zent, 2010). In addition to vinculin, zyxin and p130Cas (a scaffolding protein) have been suggested to play a role in mediating migration within 3D collagen matrices (Fraley et al., 2012). Previous studies suggest that both of these proteins are recruited to mature focal adhesions on 2D substrates and may be less common or absent in nascent adhesions (O’Neill et al., 2000; Beningo et al., 2001). However, it is not clear how their role in two dimensions translates into their role in three dimensions or microtrack migration.
To examine whether and how focal adhesions contribute to unidirectional microtrack migration, we used a microfabricated collagen microtrack platform that closely mimics the structural, mechanical, and chemical properties of migration-enabling microtracks within the stromal ECM (Kraning-Rush et al., 2013). Recently we used this system to show that microtracks within 3D ECM eliminate resistance to migration and thus reduce the need for traction force generation and matrix remodeling, whereas 3D collagen matrix migration requires mechanical cell–matrix interaction (Carey et al., 2015). Here our primary goal is to understand the role of focal adhesion molecules in 3D microtrack migration. Of interest, our results indicate that vinculin, an early focal adhesion molecule, plays a unique role in maintaining unidirectional cell migration in microtracks. MDA-MB-231 cells persistently migrate in one direction in microtracks, whereas vinculin-deficient cells show impaired unidirectional migration, with cells repeatedly reversing their direction of movement. Vinculin provides directional cues and mediates polarity during migration. Further investigation demonstrated a 3D-specific role for vinculin in activation and localization of focal adhesion kinase (FAK) at the protruding end of the migrating cells in 3D migration.
RESULTS
Cancer cells exhibit unidirectional migration in microtracks
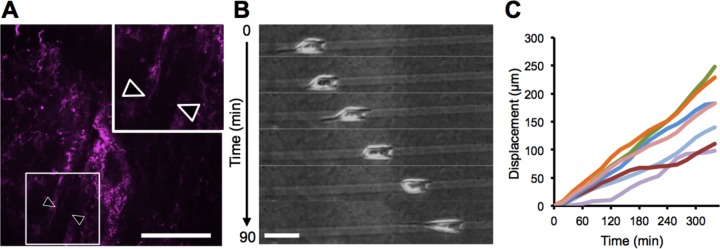
In previous studies, we developed in vitro–microfabricated collagen microtracks that closely mimic tube-like ECM structures found in the stromal matrix in vivo (Kraning-Rush et al., 2013; Carey et al., 2015). These microtrack-like features exist in the stromal ECM surrounding primary tumors in the MMTV-PyMT murine mammary tumor model (Figure 1A). When seeded into our fabricated microtracks, MDA-MB-231 cancer cells generally migrate unidirectionally (Figure 1B). The cells migrate long distances (100–250 μm) in one direction for an average of >300 min in 3D collagen microtracks (Figure 1C) before changing direction.
FIGURE 1:
Unidirectional migration of cancer cells in microtracks. (A) Ex situ confocal reflectance image of an in vivo microtrack in the stromal extracellular matrix (purple) in a murine mammary tumor model; arrowheads denote a microtrack, width 10 μm; 3× inset magnification. (B) Time-lapse phase contrast image of an MDA-MB-231 cell migrating unidirectionally through a microfabricated 3D collagen microtrack in vitro. (C) Quantification of the displacement of MDA-MB-231 cells migrating through in vitro collagen microtracks, seven cells. Scale bars, 50 μm.
Vinculin promotes persistent, unidirectional microtrack migration
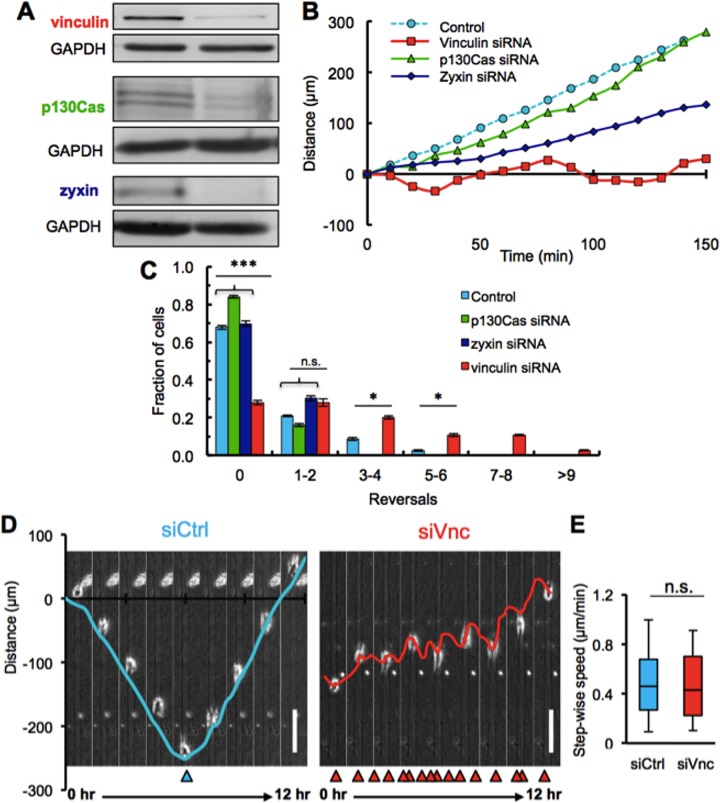
We previously showed that microtrack migration has a unique and diminished requirement for cell–matrix mechanocoupling (Carey et al., 2015). Because focal adhesions are critical to bidirectional cell–matrix signaling (Geiger et al., 2009; Prager-Khoutorsky et al., 2011; Carisey et al., 2013), we sought here to determine the role of focal adhesions in microtrack migration. We targeted the focal adhesion proteins vinculin, p130Cas, and zyxin using small interfering RNA (siRNA) and confirmed the knockdown of these three proteins by Western blotting after siRNA transfection (Figure 2A). To examine the role of focal adhesions in microtrack migration, we seeded MDA-MB-231 cells treated with scrambled (control) or focal adhesion protein–targeting siRNA in 3D microtracks. Whereas p130Cas- and zyxin-knockdown cells were able to migrate unidirectionally for several hours similarly to control cells, vinculin-deficient cells failed to migrate persistently in one direction (Figure 2B). Specifically, vinculin-knockdown cells reversed direction multiple times. Similar to control cells, ∼70% of p130Cas- and zyxin-deficient cells migrated unidirectionally in microtracks, but only ∼30% of the vinculin-knockdown cells were able to maintain persistent directionality (Figure 2C). Whereas control cells eventually reversed direction over longer time periods (typically on reaching the end of the channel), vinculin-deficient cells showed frequent reversals, often in rapid succession (Figure 2D). Of interest, despite their unique migration behaviors, vinculin-knockdown cells exhibited similar migration speeds to control cells treated with scrambled siRNA (Figure 2E).
FIGURE 2:
Vinculin is critical for maintaining directionality within collagen microtracks. (A) Western blotting confirmed knockdown of focal adhesion proteins vinculin, p130Cas, and zyxin. (B) Quantification of MDA-MB-231 cell displacement from the cells’ original location as a function in time after knockdown of p130Cas, zyxin, or vinculin. Note that vinculin siRNA–treated MDA-MB-231 cells migrate back and forth, reversing direction several times. (C) Quantification of the number of reversals in migration direction over a 6-h period of time in micromolded microtracks in control or p130Cas-, zyxin-, or vinculin-knockdown 231 cells; ***p < 0.001, *p < 0.05; >50 cells. (D) Time-lapse phase contrast images and displacement curves of a representative single control (blue) and vinculin (red) siRNA–treated MDA-MB-231 cell migrating through an in vitro collagen microtrack. Whereas control MDA-MB-231 cells migrate in one direction, vinculin siRNA–treated MDA-MB-231 cells reverse directions several times; arrowheads indicate reversals. Scale bars, 100 μm. (E) Quantification of MDA-MB-231 cell migration speed with and without vinculin knockdown; >100 cells. All quantitative data are pooled from three individual equivalent experiments.
Vinculin regulates cell polarity in 3D microtrack migration
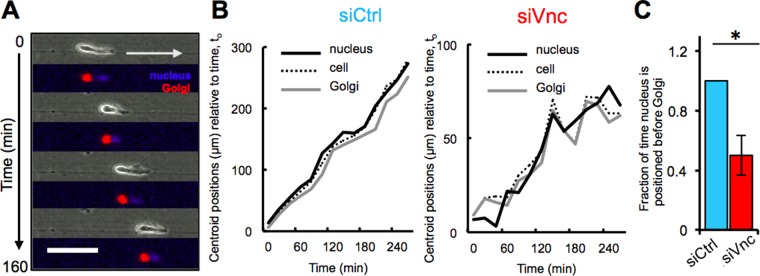
Because cell polarity is essential for unidirectional migration (Etienne-Manneville, 2008) and our data indicate that vinculin mediates unidirectional migration, our next focus was to examine whether cell polarity is perturbed in nonunidirectional vinculin siRNA–treated MDA-MB-231 cells. It was previously established that unidirectional migration requires orientation and maintenance of a front–rear cell polarity axis (Ridley et al., 2003; Etienne-Manneville, 2008). Because the relative localization of organelles such as the Golgi, centrosomes, and nucleus are indicative of cell polarization during cell migration (Iden and Collard, 2008; Hehnly et al., 2010), we examined the relative centroid position of the cell, nucleus, and Golgi in control siRNA–treated and vinculin siRNA–treated cells during collagen microtrack migration (Figure 3A). Of interest, the nucleus was consistently positioned ahead of the cell centroid toward the leading edge of cells and in the direction of movement in control siRNA–treated cells, whereas the nucleus and Golgi repeatedly switched positions relative to the cell centroid in vinculin siRNA–treated cells (Figure 3B). Unidirectional microtrack migration in control cells was accompanied by anterior Golgi localization relative to the cell centroid 100% of the time, whereas vinculin-deficient cells displayed anterior Golgi localization ∼50% of the time (Figure 3C).
FIGURE 3:
Control siRNA cells maintain cell polarity during unidirectional microtrack migration. (A) Representative time-lapse phase contrast and confocal fluorescence images (overlaid) of a control siRNA–treated MDA-MB-231 cell, which maintains cell polarity during unidirectional microtrack migration by positioning the nucleus (blue) at the leading edge, followed by the Golgi apparatus (red). Arrow indicates the direction of movement. Scale bar, 50 μm. (B) Quantification of the position of the cell centroid relative to the centroid of the nucleus and Golgi during migration in a microtrack. Whereas the nucleus is strictly positioned at the leading edge of a control siRNA–treated MDA-MB-231 cell, the nucleus and Golgi positions alternate in an MDA-MB-231 cell treated with vinculin siRNA during microtrack migration. (C) Quantification of the centroid position of the nucleus relative to the Golgi of migrating MDA-MB-231 cells in microtracks. The nucleus is observed ahead of the cell centroid and Golgi relative to the direction of cell motion in control siRNA–treated MDA-MB-231 cells throughout the period of observation and only ∼50% of the time in vinculin siRNA–treated MDA-MB-231 cells; *p < 0.05.
Vinculin is required for directional migration in two and three dimensions
Cell migration behaviors are not always conserved between 2D and 3D environments, and focal adhesions have been shown to have unique mechanistic roles in regulating 2D and 3D migration (Meyer et al., 2012). Because microtrack migration includes elements of both 2D and 3D migration (Kraning-Rush et al., 2013; Carey et al., 2015), we next sought to test whether the unique migration behavior of vinculin-deficient cells observed in 3D microtracks was conserved in 2D substrate and 3D uniform collagen matrix migration systems.
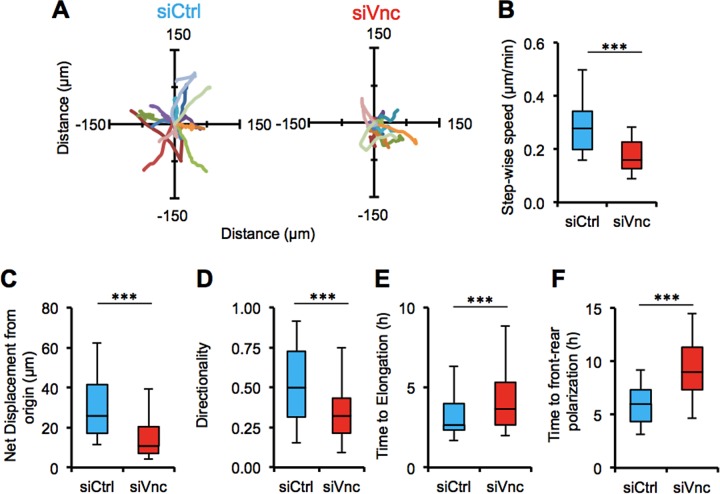
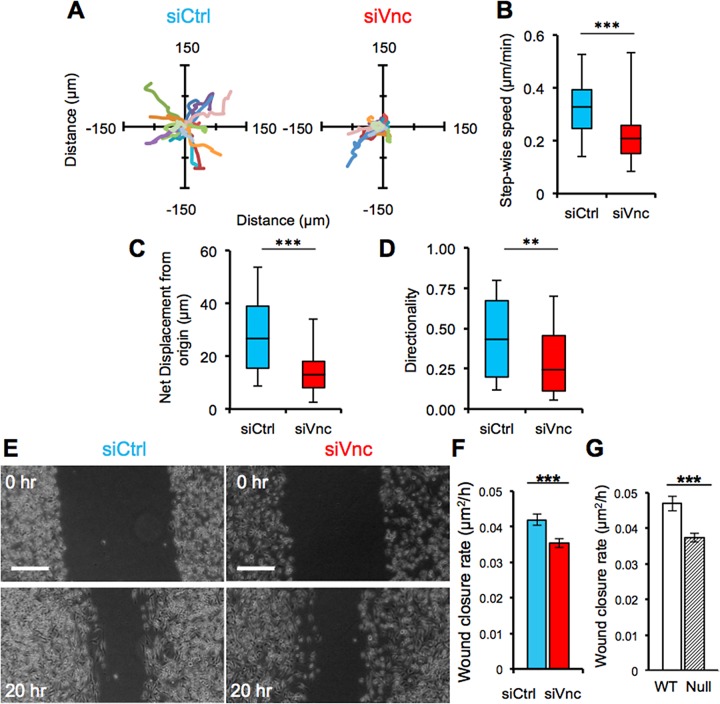
In 3D uniform collagen matrices, cells treated with vinculin-targeting siRNA generally traveled shorter distances than in control siRNA–treated cells (Figure 4A). Vinculin-knockdown cells exhibited significantly reduced stepwise cell migration speed (Figure 4B) and net displacement from their initial position (Figure 4C) compared with control siRNA–treated cells. In addition, control siRNA–treated cells showed higher 3D migration directionality than vinculin-deficient cells (Figure 4D). Analysis of initial cell spreading in 3D uniform collagen matrices indicated that vinculin-knockdown cells spread more slowly and achieved clear front–rear morphologies indicative of mesenchymally migrating cells (Friedl and Gilmour, 2009) compared with control siRNA–treated cells (Figure 4E). Together these data indicate that vinculin plays an important role in cell spreading, migration efficiency, and migration directionality in 3D ECM microenvironments.
FIGURE 4:
3D collagen matrix migration is regulated by vinculin. (A) Rose plots show trajectories of control siRNA– and vinculin siRNA–treated MDA-MB-231 cells in 3D collagen matrices over several hours. Vinculin siRNA cells (B) show slower migration speed (μm/min), (C) travel less far over the same observation window (μm), (D) lose persistent directionality, and take more time to (E) elongate and (F) establish front–rear polarization compared with control siRNA cells. ***p < 0.001; 45 cells. All quantitative data are pooled from three individual equivalent experiments.
As in 3D uniform collagen matrix migration, cells treated with vinculin siRNA and seeded on 2D substrates traveled shorter distances (Figure 5A), exhibited reduced stepwise cell migration speeds (Figure 5B), and showed reduced net displacement compared with cells treated with nontargeting control siRNA (Figure 5C). In addition, vinculin knockdown induced a significant decrease in migration directionality (Figure 5D). Despite the significant differences in 2D migratory behavior induced by vinculin siRNA treatment, cell migration on planar substrates is unconstrained and is therefore largely random (Wu et al., 2014). Thus, we further examined the role of vinculin in a wound-healing model, in which directional 2D migration can be more effectively measured. Vinculin knockdown significantly reduced closure rate (Figure 5, E and F). To determine whether vinculin regulates directional motility in cell types other than MDA-MB-231s, we studied the motility of vinculin-null and wild-type vinculin-expressing fibroblasts (Dumbauld et al., 2013). Similar to siRNA-treated MDA-MB-231 cells, vinculin-null mouse embryonic fibroblasts (MEFs) displayed a significantly slower wound closure rate than did wild-type vinculin-expressing MEFs (Figure 5G). Taken together, these data show that vinculin is important for maintaining efficient and directional migration in 2D microenvironments.
FIGURE 5:
2D migration is regulated by vinculin. (A) Rose plots of the trajectories of control siRNA– and siRNA vinculin–treated MDA-MB-231 cells on a 2D plastic surface over several hours. (B) Migration speed of control and siRNA vinculin–treated MDA-MB-231 cells (***p < 0.001; 45 cells). (C) Net displacement of control and siRNA vinculin–treated MDA-MB-231 cells in 2D (***p < 0.001; 45 cells). (D) Directionality of control and siRNA vinculin–treated MDA-MB-231 cells, **p < 0.01; 45 cells. (E) Time-lapse phase contrast images of control and vinculin siRNA–treated MDA-MB-231 cells during wound healing; scale bar, 100 μm. (F) Wound closure rate for control and siRNA vinculin–treated MDA-MB-231 cells; ***p < 0.001. (G) Quantification of the wound closure rate for wild-type vinculin MEFs compared with vinculin-null MEFs; ***p < 0.001. All quantitative data are pooled from three individual equivalent experiments.
Vinculin regulates traction force generation
Because force generation is a fundamental component of cell motility (Pelham and Wang, 1998; Dembo and Wang, 1999) and is mediated by focal adhesions (Beningo et al., 2001; Oakes et al., 2012; Sim et al., 2015), we investigated whether vinculin regulates traction force generation. We used traction force microscopy on control and vinculin-knockdown MDA-MB-231 cells to measure the effect of vinculin knockdown on cell contractility. Vinculin-deficient cells exhibited different contractility profiles (Figure 6A), generating significantly reduced traction forces compared with control siRNA–treated cells (Figure 6B). Of note, control and vinculin siRNA–treated cells also exhibited differences in cell area. Vinculin-knockdown cells showed reduced cell area and spread less compared with control siRNA–treated cells (Figure 6C). Therefore, we measured normalized force (per area) and did not observe a significant difference between control and vinculin- knockdown cells (Figure 6D). Of interest, we observed an increase in traction force generation as an increasing function of cell area when we plotted force as a function of cell area (Figure 6E). The plotted data distinguish between the control siRNA– and vinculin siRNA–treated cells; the control siRNA–treated cells generated more force with greater area change than vinculin siRNA– treated cells (Figure 6E).
FIGURE 6:
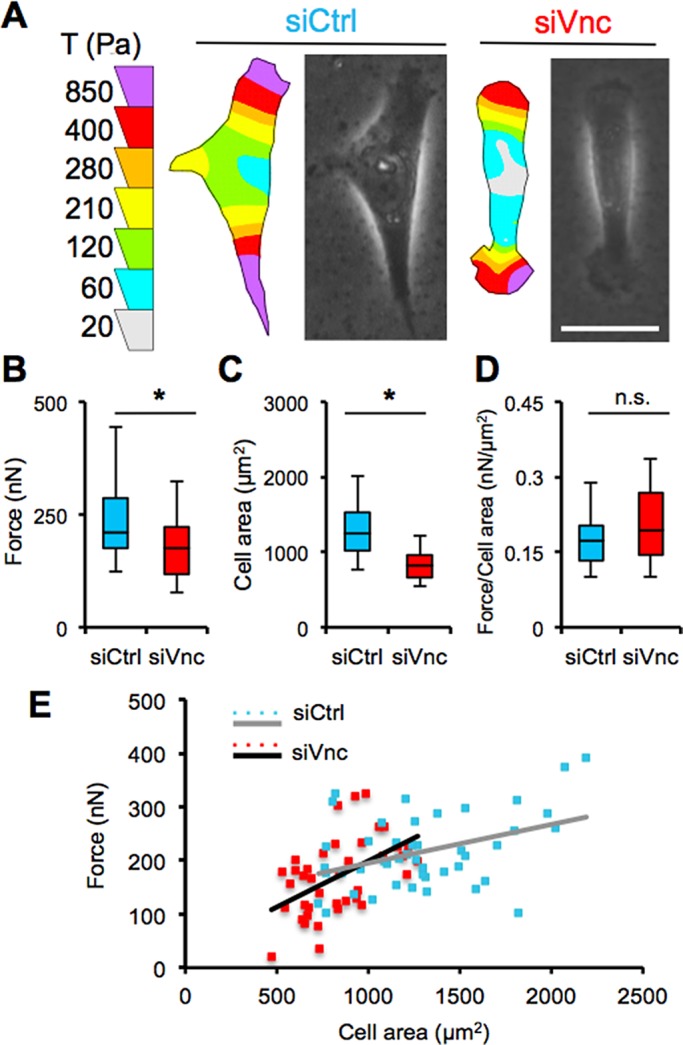
Vinculin siRNA cells generate reduced traction force. (A) Traction force color contour maps and phase contrast images of a control and vinculin siRNA–treated MDA-MB-231 cells; scale bar, 30 μm. (B) Quantification of the total force, |F|, generated (nN), (C) spread area (μm2), and (D) normalized force per cell area (nN/μm2) in control and siRNA vinculin–treated MDA-MB-231 cells. (E) Corresponding scatter plot of traction force as a function of cell area with linear regression lines for control and vinculin knockdown–treated MDA-MB-231 cells. *p < 0.05; >40 cells. All quantitative data are pooled from three individual equivalent experiments.
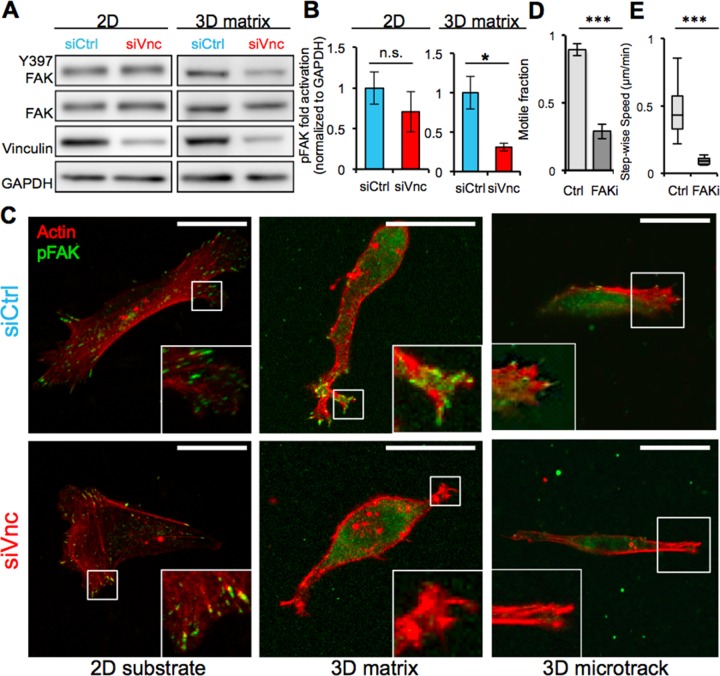
Vinculin coregulates FAK in three but not two dimensions
Like other focal adhesion proteins, a critical function of vinculin is to establish both physical and biochemical connections between the ECM and intracellular domain of the cell by binding to the cytoplasmic actin cytoskeleton and transmembrane integrins (Wozniak et al., 2004). As integrins associate with the actin cytoskeleton through vinculin, other focal adhesion proteins are recruited to the complex, including various scaffolding proteins and adaptor proteins, which can further associate with additional signaling molecules and activate various pathways (Amano et al., 2010). One such protein is FAK, which modulates focal adhesive strength by reducing vinculin localization to the focal adhesion complex (Dumbauld et al., 2010). Of interest, whereas vinculin knockdown in MDA-MB-231 cells induced a modest but statistically insignificant reduction in FAK activation on 2D substrates, vinculin knockdown dramatically and significantly reduced FAK activation in 3D collagen matrices (Figure 7, A and B). Similarly, whereas vinculin knockdown had no effect on pFAK accumulation at focal adhesions on 2D substrates (Figure 7C, left), vinculin-deficient cells were unable to localize pFAK to cell protrusions in 3D uniform collagen matrices (Figure 7C, middle). Moreover, vinculin-deficient cells also failed to localize pFAK at the protruding end in 3D collagen microtracks (Figure 7C, right). To test directly whether FAK activity regulates cell migration in 3D collagen microtracks, we treated MDA-MB-231 cells with the FAK inhibitor PF573228 and found that FAK inhibition significantly reduced the fraction of motile cells (Figure 7D) as well as cell migration speed (Figure 7E) in microtracks. Taken together, these results demonstrate that vinculin uniquely contributes to FAK activation and localization in 3D collagen microenvironments but not on 2D substrates.
FIGURE 7:
Vinculin regulates FAK activity in 3D migration. (A) Western blotting of MDA-MB-231 cells in 2D and 3D environments with and without knockdown of vinculin. Vinculin knockdown reduces FAK phosphorylation of MDA-MB-231 cells in 3D matrices but not 2D substrates. (B) Quantification of pFAK activation in control and vinculin siRNA–treated MDA-MB-231 cells during 2D (p = 0.475) and 3D matrix migration; *p < 0.05. (C) Confocal fluorescence images of control and vinculin siRNA cells on 2D substrate, in 3D uniform collagen matrices, and in 3D collagen microtracks; scale bars, 25 μm. (D) Fraction of motile cells in microtracks with and without FAK inhibition using PF573228; ***p < 0.001; >60 cells. (E) Migration speed of control MDA-MB-231 cells and MDA-MB-231 cells treated with the FAK inhibitor in microtracks; ***p < 0.001; >60 cells. All quantitative data are pooled from three individual equivalent experiments.
DISCUSSION
In this work, we demonstrated that the focal adhesion protein vinculin helps to maintain cell polarity and unidirectional migration in 3D in vitro collagen microtracks. Similarly, vinculin expression is also required for efficient, directional migration in isotropic 2D substrates and 3D uniform collagen matrices. Specifically, we found that vinculin-deficient cells displace significantly shorter distances, migrate with lower stepwise speeds, and exhibit reduced migration directionality in 2D substrates and 3D collagen matrices. In addition, vinculin-deficient cells generate less traction force, and both siRNA-treated MDA-MB-231 cells and vinculin-depleted fibroblasts show significantly reduced directional migration in a wound- healing migration model. Of interest, we found that vinculin plays a microenvironment-specific role in the regulation of FAK by contributing to its activation and localization in 3D microtracks and 3D uniform collagen matrices but not on 2D substrates.
The ability of adherent cells to migrate and invade depends on the dynamic regulation of cell–matrix linkages at focal adhesions (Parsons et al., 2010). Moreover, directional migration of the cell requires continuous, rapid, and coordinated formation and turnover of focal adhesions at the leading edge of the cell body (Webb et al., 2002; Nagano et al., 2012). The focal adhesion protein vinculin is required for early focal adhesion formation, with some evidence indicating its presence in nascent focal adhesions, which are highly transient (Tan et al., 2010). In addition, vinculin knockdown has been shown to correlate with smaller and fewer focal adhesions, which turn over rapidly (Saunders et al., 2006; Srichai and Zent, 2010). Recent findings show that vinculin is up-regulated in highly migratory cells, and vinculin expression at focal adhesion complexes has been associated with increased cell migration, as well as metastasis formation in vivo (Mierke, 2009). In this study, we found that vinculin regulates directional cell migration on 2D substrates, in 3D uniform collagen matrices, and in 3D collagen microtracks, where cells typically migrate with high unidirectional persistence.
Previous work demonstrated that vinculin-deficient cells are significantly less invasive and show decreased directional persistence in 3D matrices (Mierke et al., 2010). Our finding that vinculin enables efficient and directional migration on 2D substrates and in 3D uniform collagen matrices is consistent with these results. Furthermore, the loss of unidirectional microtrack migration in vinculin-deficient cells provides additional insight into microtrack-specific migration mechanisms (Kraning-Rush et al., 2013; Carey et al., 2015). Persistent, directional migration is enabled by front–rear cell polarization, which can include anisotropic organization of cell organelles, intracellular signaling networks, and the cytoskeleton (Pegtel et al., 2007; Iden and Collard, 2008; Hehnly et al., 2010). Given that nucleus–Golgi organization, front–rear cell polarity, and FAK activation in 3D microenvironments were perturbed by vinculin knockdown and that direct FAK inhibition significantly reduced microtrack motility, our results suggest that vinculin contributes to cell polarity during migration and may do so by regulating signaling through FAK. Future work will examine the role of vinculin–FAK signaling in establishing and maintaining cell polarity during migration.
Of note, vinculin plays a key role in the transmission of extracellular mechanical stimuli leading to the reorganization of cell polarity (Carisey et al., 2013) and also contributes to traction force strength (Dumbauld et al., 2013; Dumbauld and García, 2014). Mierke et al. (2010) demonstrated that vinculin-expressing cells generated enhanced traction forces that enable them to overcome the restrictive environment of a dense 3D matrix more effectively than their low vinculin–expressing counterparts. Here we find that, in addition to generating reduced traction forces, vinculin-knockdown cells show a phenotypic change accordingly by exhibiting reduced cell area compared with controls. This phenotypic change could also potentially explain cells’ impaired ability to move efficiently within permissive collagen microtracks that provide little resistance to migration and reduce the mechanistic burden of movement (Kraning-Rush et al., 2013; Carey et al., 2015).
The absence of vinculin may trigger other subsequent signaling in 3D and 2D migration through different mechanisms (Wozniak et al., 2004). Our results show that vinculin deficiency results in lower speed on 2D substrates and in 3D collagen matrices, but there was no significant difference in cell speed in 3D microtrack migration. Recent studies also showed that vinculin–FAK coregulation in 2D migration systems can be quite different from the results obtained with a 3D migration system (Mierke, 2013). Whereas cytoskeletal dynamics, adhesion turnover, and the transmission and generation of traction forces play essential roles in 2D motility, cell motility in 3D systems is further regulated by cellular stiffness, cell deformability, and matrix remodeling (Mierke, 2009; Carey et al., 2012). Previous work demonstrated increased activation of adhesion adapter proteins like FAK in vinculin-knockout MEFs on 2D culture systems (Saunders et al., 2006; Mierke, 2009). Further, vinculin recruitment and localization to focal adhesions is regulated by FAK on micropatterned substrates (Dumbauld et al., 2010). Thus our finding that vinculin knockdown reduces FAK activation uniquely in both 3D collagen matrices and 3D microtracks but not on 2D substrates is not inconsistent with these data, and in fact provides further insight into the microenvironment-specific regulation of cell–matrix adhesion signaling networks. Because of the differences in adhesion substrate composition, conformation, and mechanics between 2D and 3D ECM (Cukierman et al., 2001), it is plausible that cues in the microenvironment mediate both upstream and downstream elements of adhesion signaling. Future work should continue to address extracellular control of cell–matrix adhesion composition and signaling, as well as downstream outcomes including, but not limited to, cell polarity and migration.
MATERIALS AND METHODS
Cell culture and reagents
Highly metastatic MDA-MB-231 breast adenocarcinoma cells (HTB-26; American Type Culture Collection, Manassas, VA) were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (Life Technologies, Grand Island, NY). Cells were maintained in culture at 37°C and 5% CO2. Wild-type vinculin and vinculin-null MEFs were maintained as previously described (Dumbauld et al., 2013).
Preparation of murine tumor model and imaging
MMTV-PyMT transgenic mice on the FVB strain background were obtained from the Jackson Laboratory (Bar Harbor, ME). All mice were maintained following a protocol approved by the Cornell University Institutional Animal Care and Use Committee. Mice were killed at 8 wk of age by CO2 asphyxiation, and mammary tumors were collected and snap frozen. Confocal reflectance imaging of freshly thawed tumor stroma was performed ex situ as described previously (Carey et al., 2015).
RNA interference
MDA-MB-231 cells were transfected with 2 nM nontargeting scrambled (control) siRNA or siRNA targeting vinculin, p130Cas, or zyxin using Lipofectamine 2000 (2 μg/ml; Invitrogen, Carlsbad, CA). The nontargeting sequence was 5′-UUCCUCUCCACGCGCAGUACAUUUA-3′. Targeting siRNA accession numbers and sequences were, respectively, for vinculin, NM_001191370.1 and 5′-UCCUGGAAAUCAAGCUGCUUAUGAA-3′; for p130Cas, NM_001170715.1 and 5′-GCCUCAAGAUCUUGGUGGGCAUGUA-3′, and for zyxin, NM_003461.4 and 5′-CAUGACCAAGAAUGAUCCUUUCAAA-3′. All siRNAs were custom synthesized by Life Technologies, and RNA interference–mediated knockdown was confirmed using Western blot.
Western blotting
MDA-MB-231 cells were rinsed with ice-cold phosphate-buffered saline and lysed using preheated (90°C) 2× Laemmli sample buffer as described previously (Huynh et al., 2013). For the study of FAK-inhibited cells embedded within 1.5 mg/ml collagen gels, cultures were subjected to a quick spin to remove excess medium before lysing and snap freezing. Afterward, all frozen collagen samples were ground using a liquid nitrogen–cooled mortar and pestle and lysed. All cell lysates were cleared by centrifugation at 14,000 × g, and the supernatant was snap frozen and stored at −80°C until analysis. All samples were subjected to SDS–PAGE with a Mini-PROTEAN Tetra System (Bio-Rad, Hercules, CA) and electrotransferred onto a polyvinylidene difluoride membrane. Blots were probed using antibodies against vinculin (V284; Millipore, Billerica, MA), zyxin (Z4751; Sigma-Aldrich, St. Louis, MO), p130cas (sc-860; Santa Cruz Biotechnology, Santa Cruz, CA), phosphorylated FAK at tyrosyne-397 (pMLC; 3283; Cell Signaling Technology, Beverly, MA), total FAK (3285; Cell Signaling Technology), and glyceraldehyde-3-phosphate dehydrogenase (MAB374; Millipore). Anti-rabbit and anti-mouse horseradish peroxidase–conjugated secondary antibodies were obtained from Rockland (Limerick, PA). After incubation with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL), blots were exposed and imaged using an ImageQuant LAS-4000 (Fujifilm, Tokyo, Japan). Protein densitometry of blots was determined with ImageJ software (version 1.47k; National Institutes of Health, Bethesda, MD).
Phase contrast and confocal microscopy
Phase contrast and confocal fluorescence images were obtained as previously described in custom temperature-, humidity-, and CO2-controlled incubation chambers (Kraning-Rush et al., 2013; Carey et al., 2015). Phase contrast images were obtained using a Zeiss observer Z1m inverted microscope equipped with a Hamamatsu (Hamamatsu, Japan) ORCA-ER camera and operated by AxioVision software (version 4.8.1.0; Carl Zeiss Microscopy, Thornwood, NY). Confocal fluorescence imaging was performed with a Zeiss LSM700 laser scanning confocal microscope on a Zeiss AxioObserver Z1 inverted stand using a long–working distance water immersion C-Apochromat 40×/1.1 numerical aperture objective and operated by Zen software (version 2010). For activated FAK and filamentous actin staining, cells were allowed to spread overnight and stained with anti-pFAKY397 antibody (Cell Signaling Technology) and Alexa Fluor 568–conjugated phalloidin (Life Technologies) as previously described (Kraning-Rush et al., 2013). For nuclei staining, samples were incubated for 5 min with a 1:1000 dilution of Hoechst 33342 and trihydrochloride trihydrate (Invitrogen) in complete medium. Samples were then incubated overnight with Golgi-RFP (Cell Light, BacMam 2.0; Life Technologies) for live-cell fluorescence imaging of the nucleus and Golgi apparatus.
3D and 2D cell migration studies
For 3D migration studies, 3D collagen microtracks and matrices were prepared using type I collagen extracted from rat-tail tendons as previously described (Kraning-Rush et al., 2013; Carey et al., 2015). Briefly, 3 mg/ml (for 3D microtracks) and 1.5 mg/ml (for 3D matrix) collagen solutions were prepared from a 10 mg/ml collagen stock solution using ice-cold complete medium and 1 N NaOH (to neutralize at pH 7.0). MDA-MB-231 cells were seeded within collagen scaffolds at low cell density to obtain isolated cells for single-cell migration studies. Where indicated, siRNA-treated cells were used or complete medium was supplemented with the FAK inhibitor PF573228 (5 μM; Santa Cruz Biotechnology).
For 2D single-cell migration experiments, siRNA-treated MDA-MB-231 cells were seeded on culture plastic. For 2D wound-healing experiments, MDA-MB-231 cells were seeded on 4-cm2 plastic culture wells in complete medium at 50–60% confluence for 12–18 h before transfection. Samples were transfected as described and incubated at 37°C until cells reached 100% confluence to form a monolayer. To create a wound, the cell monolayer was lightly scratched in a straight line with a p200 pipette tip. For the MEF wound-healing assay, plastic culture wells were coated with fibronectin (F1141; Sigma-Aldrich) before seeding cells and scratching.
3D and 2D cell migration analysis
All cell migration imaging was performed using time-lapse phase contrast and confocal microscopy. Image analysis was performed using ImageJ.
For 3D microtrack migration studies, cells were seeded in microtracks and allowed to adhere for 6 h. Time-lapse imaging was performed every 20 min. Cell migration distance was quantified by determining the displacement of the cell from its initial position, and the relative positions of the nucleus and Golgi apparatus were determined by outlining the cell, cell nucleus, and Golgi apparatus and measuring centroid position for each using ImageJ software. Stepwise cell speed was calculated by averaging the distance traveled between each frame and dividing by the time step. Measurements were taken over a minimum of 6 h. The average stepwise speed for all single cells was represented in boxplots (Kraning-Rush et al., 2013; Carey et al., 2015). All data are presented for three experiments per treatment.
For both the 3D and 2D migration studies, time-lapse phase contrast imaging was performed at 10× magnification every 20 min for 24 h. All imaged cells were analyzed between 12 and 18 h after seeding. Cell trajectories, stepwise net displacement, and stepwise cell speed were calculated by measuring cell centroid position based on cell outlines as previously described (Kraning-Rush et al., 2013). Directionality of migration was determined by measuring the ratio of the shortest, linear distance from the starting point of a time-lapse recording to the end point compared with the total net displacement of the cell (Pankov et al., 2005). For 3D migration studies, images were acquired >200 μm above the bottom surface; time to cell elongation and front–rear cell polarity axis were measured, respectively, by recording the time when cells were clearly elongated (aspect ratio >1.75) and the time when a head and tail of cells were clearly distinguished and had initiated migration, not just protrusions. All data are represented for >45 cells and obtained from three independent experiments. For 2D wound- healing migration, time-lapse phase contrast imaging was performed at 5× magnification every hour for 20 h. The wound closure rate (per hour) was determined by outlining and calculating the remaining wound area using ImageJ.
Polyacrylamide gel synthesis and traction force microscopy
Polyacrylamide gels embedded with 0.5-μm-diameter fluorescent microsphere beads (Invitrogen) were synthesized with a bisacrylamide:acrylamide (Bio-Rad) ratio of 7.5:0.175 to achieve a Young's modulus of 5 kPa and were coated with 0.1 mg/ml rat-tail type I collagen (BD Biosciences, Franklin Lakes, NJ) as described previously (Califano and Reinhart-King, 2008). Control and vinculin siRNA–treated MDA-MB-231 cells were seeded on the gel surface and allowed to adhere overnight. Cellular traction vectors and the total magnitudes of force were determined using the LIBTRC analysis library developed by Micah Dembo (Boston University, Boston, MA). The overall force exerted, |F|, was calculated as the integral of the traction vector magnitudes over cell area (Reinhart-King et al., 2005) and is presented for three experiments and >40 cells per treatment.
Statistical analysis
Data are presented as mean ± SEM or box-and-whisker plot or scatter plot. The nonparametric Wilcoxon rank sum test statistical analysis was conducted in JMP (version 10; SAS, Cary, NC). Excel (Microsoft, Redmond, WA) and Matlab (R2010a; MathWorks, Natick, MA) were used to compare data for all cell migration and traction force analysis. p < 0.05 and <0.001 are considered statistically significant.
Acknowledgments
We acknowledge the use of equipment and resources at the Cornell NanoScale Science and Technology Facility. This work was supported by the Cornell Center of the Microenvironment and Metastasis through Award U54CA143876 from the National Cancer Institute and by National Science Foundation–National Institutes of Health Physical and Engineering Sciences in Oncology Award 1233827 to C.A.R. This was work is also supported by National Science Foundation Graduate Fellowships to A.R., S.P.C., and M.C.L.
Abbreviations used:
- 2D
two dimensional
- 3D
three dimensional
- ECM
extracellular matrix
- FAK
focal adhesion kinase
- MEFs
mouse embryonic fibroblasts
- siRNA
small interfering RNA.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-06-0432) on March 9, 2016.
REFERENCES
- Amano M, Nakayama M, Kaibuchi K. Rho-kinase/ROCK: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton. 2010;67:545–554. doi: 10.1002/cm.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J Cell Biol. 2001;153:881–887. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano JP, Reinhart-King CA. A balance of substrate mechanics and matrix chemistry regulates endothelial cell network assembly. Cell Mol Bioeng. 2008;1:122–132. [Google Scholar]
- Carey SP, D’Alfonso TM, Shin SJ, Reinhart-King CA. Mechanobiology of tumor invasion: Engineering meets oncology. Crit Rev Oncol Hematol. 2012;83:170–183. doi: 10.1016/j.critrevonc.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey SP, Rahman A, Kraning-Rush CM, Romero B, Somasegar S, Torre OM, Williams RM, Reinhart-King CA. Comparative mechanisms of cancer cell migration through 3D matrix and physiological microtracks. Am J Physiol Cell Physiol. 2015;308:436–447. doi: 10.1152/ajpcell.00225.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carisey A, Tsang R, Greiner AM, Nijenhuis N, Heath N, Nazgiewicz A, Kemkemer R, Derby B, Spatz J, Ballestrem C. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr Biol. 2013;23:271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Petrie RJ, Kutys ML, Yamada KM. Dimensions in cell migration. Curr Opin Cell Biol. 2013;25:642–649. doi: 10.1016/j.ceb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbauld DW, García AJ. A helping hand: how vinculin contributes to cell-matrix and cell-cell force transfer. Cell Adh Migr. 2014;8:550–557. doi: 10.4161/cam.29139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbauld DW, Lee TT, Singh A, Scrimgeour J, Gersbach CA, Zamir EA, Fu J, Chen CS, Curtis JE, Craig SW, García AJ. How vinculin regulates force transmission. Proc Natl Acad Sci USA. 2013;110:9788–9793. doi: 10.1073/pnas.1216209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbauld DW, Michael KE, Hanks SK, García AJ. Focal adhesion kinase-dependent regulation of adhesive forces involves vinculin recruitment to focal adhesions. Biol Cell. 2010;102:203–213. doi: 10.1042/BC20090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27:6970–6980. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- Fraley SI, Feng Y, Giri A, Longmore GD, Wirtz D. Dimensional and temporal controls of three-dimensional cell migration by zyxin and binding partners. Nat Commun. 2012;3:719. doi: 10.1038/ncomms1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley SI, Feng Y, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, Wirtz D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- Friedl P, Sahai E, Weiss S, Yamada KM. New dimensions in cell migration. Nat Rev Mol Cell Biol. 2012;13:743–747. doi: 10.1038/nrm3459. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann WH, Auernheimer V, Thievessen I, Fabry B. Vinculin, cell mechanics and tumour cell invasion. Cell Biol Int. 2013;37:397–405. doi: 10.1002/cbin.10064. [DOI] [PubMed] [Google Scholar]
- Hehnly H, Xu W, Chen JL, Stamnes M. Cdc42 regulates microtubule-dependent golgi positioning. Traffic. 2010;11:1067–1078. doi: 10.1111/j.1600-0854.2010.01082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J, Bordeleau F, Kraning-Rush CM, Reinhart-King CA. Substrate stiffness regulates PDGF-induced circular dorsal ruffle formation through MLCK. Cell Mol Bioeng. 2013;6:138–147. doi: 10.1007/s12195-013-0278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- Kraning-Rush CM, Carey SP, Lampi MC, Reinhart-King CA. Microfabricated collagen tracks facilitate single cell metastatic invasion in 3D. Integr Biol. 2013;5:606–616. doi: 10.1039/c3ib20196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Meyer AS, Hughes-Alford SK, Kay JE, Castillo A, Wells A, Gertler FB, Lauffenburger DA. 2D protrusion but not motility predicts growth factor-induced cancer cell migration in 3D collagen. J Cell Biol. 2012;197:721–729. doi: 10.1083/jcb.201201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke CT. The role of vinculin in the regulation of the mechanical properties of cells. Cell Biochem Biophys. 2009;53:115–126. doi: 10.1007/s12013-009-9047-6. [DOI] [PubMed] [Google Scholar]
- Mierke CT. The role of focal adhesion kinase in the regulation of cellular mechanical properties. Phys Biol. 2013;10:065005. doi: 10.1088/1478-3975/10/6/065005. [DOI] [PubMed] [Google Scholar]
- Mierke CT, Kollmannsberger P, Zitterbart DP, Diez G, Koch TM, Marg S, Ziegler WH, Goldmann WH, Fabry B. Vinculin facilitates cell invasion into three-dimensional collagen matrices. J Biol Chem. 2010;285:13121–13130. doi: 10.1074/jbc.M109.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Hoshino D, Koshikawa N, Akizawa T, Seiki M. Turnover of focal adhesions and cancer cell migration. Int J Cell Biol. 2012;2012:310616. doi: 10.1155/2012/310616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes PW, Beckham Y, Stricker J, Gardel ML. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J Cell Biol. 2012;196:363–374. doi: 10.1083/jcb.201107042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill GM, Fashena SJ, Golemis EA. Integrin signalling: a new Cas(t) of characters enters the stage. Trends Cell Biol. 2000;10:111–119. doi: 10.1016/s0962-8924(99)01714-6. [DOI] [PubMed] [Google Scholar]
- Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Ellenbroek SIJ, Mertens AEE, van der Kammen RA, de Rooij J, Collard JG. The Par-Tiam1 complex controls persistent migration by stabilizing microtubule-dependent front-rear polarity. Curr Biol. 2007;17:1623–1634. doi: 10.1016/j.cub.2007.08.035. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Wang YL. Cell locomotion and focal adhesions are regulated by the mechanical properties of the substrate. Biol Bull. 1998:348–350. doi: 10.2307/1543109. [DOI] [PubMed] [Google Scholar]
- Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, Geiger B, Bershadsky AD. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- Reinhart-King CA, Dembo M, Hammer DA. The dynamics and mechanics of endothelial cell spreading. Biophys J. 2005;89:676–689. doi: 10.1529/biophysj.104.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Saunders RM, Holt MR, Jennings L, Sutton DH, Barsukov IL, Bobkov A, Liddington RC, Adamson EA, Dunn GA, Critchley DR. Role of vinculin in regulating focal adhesion turnover. Eur J Cell Biol. 2006;85:487–500. doi: 10.1016/j.ejcb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Sim JY, Moeller J, Hart KC, Ramallo D, Vogel V, Dunn AR, Nelson WJ, Pruitt BL. Spatial distribution of cell–cell and cell–ECM adhesions regulates force balance while maintaining E-cadherin molecular tension in cell pairs. Mol Biol Cell. 2015;26:2456–2465. doi: 10.1091/mbc.E14-12-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srichai MB, Zent R. Integrin structure and function. In: Zent R, Pozzi A, editors. Cell-Extracellular Matrix Interactions in Cancer. New York: Springer; 2010. pp. 19–41. [Google Scholar]
- Tan L, Meyer T, Pfau B, Hofmann T, Tan TW, Jones D. Rapid vinculin exchange dynamics at focal adhesions in primary osteoblasts following shear flow stimulation. J Musculoskelet Neuronal Interact. 2010;10:92–99. [PubMed] [Google Scholar]
- Webb DJ, Parsons J T, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells—over and over and over again. Nat Cell Biol. 2002;4:E97–E100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Wu PH, Giri A, Sun SX, Wirtz D. Three-dimensional cell migration does not follow a random walk. Proc Natl Acad Sci U SA. 2014;111:3949–3954. doi: 10.1073/pnas.1318967111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman MH, Trapani LM, Sieminski AL, Mackellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]