Live imaging of microtubule dynamics in Caenorhabditis elegans muscle cells reveals a novel microtubule behavior characterized by an abrupt change in growth rate, named “microtubule growth deceleration.” The conserved protein ZYG-9TOGp and two novel ORFs, cylc-1 and cylc-2, are involved in the regulation of this novel microtubule behavior.
Abstract
Microtubules (MTs) are cytoskeletal polymers that participate in diverse cellular functions, including cell division, intracellular trafficking, and templating of cilia and flagella. MTs undergo dynamic instability, alternating between growth and shortening via catastrophe and rescue events. The rates and frequencies of MT dynamic parameters appear to be characteristic for a given cell type. We recently reported that all MT dynamic parameters vary throughout differentiation of a smooth muscle cell type in intact Caenorhabditis elegans. Here we describe local differences in MT dynamics and a novel MT behavior: an abrupt change in growth rate (deceleration) of single MTs occurring in the cell periphery of these cells. MT deceleration occurs where there is a decrease in local soluble tubulin concentration at the cell periphery. This local regulation of tubulin concentration and MT deceleration are dependent on two novel homologues of human cylicin. These novel ORFs, which we name cylc-1 and -2, share sequence homology with stathmins and encode small, very basic proteins containing several KKD/E repeats. The TOG domain–containing protein ZYG-9TOGp is responsible for the faster polymerization rate within the cell body. Thus we have defined two contributors to the molecular regulation for this novel MT behavior.
INTRODUCTION
Microtubule dynamic instability was first described as such in 1984 (Mitchison and Kirschner, 1984) and has since been observed in many different organisms and in vitro reconstitution systems (Desai and Mitchison, 1997). Microtubule (MT) dynamics comprises the rates of assembly and disassembly and the frequencies of transition between growth and shrinking, called catastrophe and rescue, as well as the time spent apparently static (“pause”). Microtubule assembly is regulated in part by the concentration of its tubulin dimer building blocks such that the growth rate increases with increasing tubulin concentration in vitro (Walker et al., 1988). The dependence of the other dynamic parameters on tubulin concentration in vitro is less evident. In vitro experiments revealed that tubulin must be bound to GTP to assemble into MTs (Weisenberg et al., 1976). The presence of a population of GTP-tubulin subunits at the fast- growing end of the MT (the plus end) prevents depolymerization. Within the MT, GTP is hydrolyzed, and the resulting GDP-tubulin heterodimers rapidly dissociate, inducing catastrophe and depolymerization (reviewed in Bowne-Anderson et al., 2013).
In vivo, MT dynamics is further affected by tens to hundreds of tubulin- and MT-associated proteins (MAPs), as well as by the different properties of distinct tubulin isoforms and a variety of posttranslational modifications. Proteins that bind to the MT plus end affect growth rate and catastrophe frequency (Lansbergen and Akhmanova, 2006). Proteins that bind tubulin subunits also affect these parameters, for example, by sequestering the subunits (reviewed in van der Vaart et al., 2009). Because MAPs influence the complexity and range of MT dynamics, their study may ultimately offer insights into molecular and physical mechanisms of MT dynamic instability. We are only beginning to understand how the full ensemble of MAPs combines to control cellular MT dynamics in space and time.
The parameters of MT dynamic instability are significantly different among species and cell types and can vary among different regions of the same cell. For example, astral- and kinetochore-MTs within the same mitotic spindle in the Caenorhabditis elegans embryo display distinct dynamics parameters (Srayko et al., 2005). MTs at the leading edge of motile cells exhibit organization and dynamics distinct from those in the cell body. Specifically, the orientation of noncentrosomal MTs, that is, parallel or perpendicular to the cell edge, correlates with significant differences in dynamic parameters (Waterman-Storer and Salmon, 1997; Wittmann et al., 2003). Growth rate, and to a lesser extent other aspects of MT dynamics, varies between the leading edge and cell body of the same cell (Wadsworth, 1999). Growth rate is reduced and time spent growing is increased at the cell edge during cell migration (Komarova et al., 2002; Mimori-Kiyosue et al., 2005; Kumar et al., 2009; Matov et al., 2010). It is unknown whether regional differences in MT dynamics also occur in nonmotile differentiated cells. Subcellular MT regulation has been reported for cells in two-dimensional culture, as mentioned earlier, but the effect of a three-dimensional environment on spatial differentiation of MT dynamics is not known. Furthermore, whether the dynamic behavior of a MT during a single growth excursion can be spatially regulated remains unknown.
We recently reported that MT dynamics changes over the course of tissue biogenesis and differentiation of a single cell lineage in intact C. elegans (Lacroix and Maddox, 2014). We studied the two precursor cells (the left and right sex myoblasts [SMs]) that divide and differentiate within 27 h from the third larval stage to adulthood to form muscles required for egg laying (the uterine muscle cells [UMCs]). We found that all aspects of MT dynamics differ between the SM and the UMCs. These different dynamics likely result from changes in expression and activity levels for many MAPs, since we showed that distinct complements of MAPs are required for tissue biogenesis and tissue function.
In addition to changes in MAP expression and activity during differentiation, another factor that could affect MT organization and dynamics is cell shape modification. The precursor SM cells are small and round, whereas differentiated UMCs have a complex three-dimensional geometry. We performed live imaging of MTs in the UMCs in intact C. elegans with high spatial and temporal resolution. We then analyzed how dynamic parameters vary within UMCs depending on topological subcellular area. In doing so, we identified an atypical and novel MT behavior that we named “MT growth deceleration.” This phenomenon occurs specifically in differentiated UMCs and is characterized by the abrupt change in assembly rate of a single MT as it enters the dorsal periphery of these cells. Investigating the role of candidate regulators of MT growth deceleration, we identified two novel C. elegans open reading frames (ORFs) that share homology with the protein stathmin. Thus we begin to define the regulation by conserved proteins of a novel MT behavior in differentiated cells.
RESULTS
Subcellular compartmentalization of MT dynamics in a differentiated cell in situ
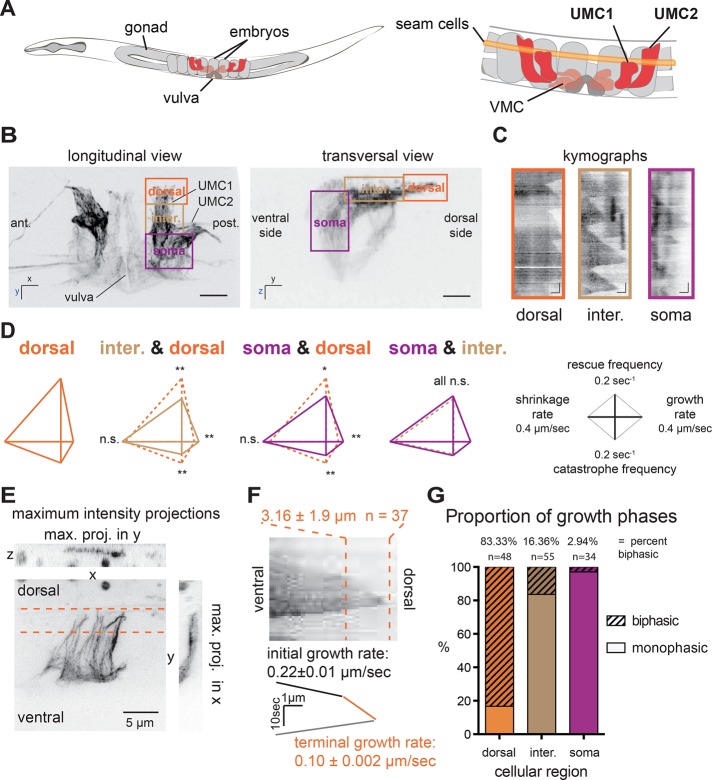
We recently developed live-imaging methods to monitor MT dynamics over the course of differentiation and tissue biogenesis in C. elegans using a strain expressing green fluorescent protein (GFP)–β-tubulin specifically in the SM lineage (Lacroix et al., 2014). This lineage is composed of two precursors called SML and SMR (for left and right, respectively) that divide and differentiate during postembryonic development to give rise to 16 egg-laying muscle cells, including the UMCs (Figure 1A). Given that UMCs are large and polarized, we examined whether MT dynamics are locally regulated within this cell type. The cell bodies of UMCs are located near the vulva on the ventral side on the worm, and their flattened dorsal extensions surround the uterine cavity, terminating at the seam cells just under the birefringent alae (Figure 1, A and B). We compared MT dynamics among three regions of the cell: the ventral cell body (soma), the dorsal extension, and the intermediate zone between these regions (Figure 1, B and C). Several parameters of MT dynamics varied significantly between dorsal cell periphery and the regions deeper within the cell (Figure 1D). As observed in kymographs (Figure 1C) and in dynamics analyses (Figure 1D and Supplemental Table S1), MT growth rate was higher (0.19 μm/s) in the soma than in the periphery (0.11 μm/s; Figure 1, dorsal region). MTs in the periphery also had double the catastrophe than elsewhere in the cell, and rescue frequencies was increase by ∼44% in this region (Supplemental Table S1 and Figure 1D). Thus MT dynamics are regionally distinct in differentiated cells in situ.
FIGURE 1:
Localized differences in MT dynamics in differentiated cells (UMCs) reveal novel MT dynamic behavior: growth deceleration. (A) Schematic of an adult C. elegans, highlighting egg-laying muscles of the SM lineage. VMCs, vulval muscle cells. (B) Maximum intensity x, y- (left) and y, z- (right) projections of a worm expressing GFP-tagged tubulin specifically in the SM lineage. The dorsal cell periphery, an intermediate region, and the cell body are boxed and labeled dorsal, intermediate, and soma, respectively. Scale bar, 10 μm. (C) Kymographs of GFP-tubulin fluorescence depict typical MT dynamics in the different regions. Scale bars, 10 s (vertical), 1 μm (horizontal). (D) Diamond graphs summarize and compare MT dynamic parameters in dorsal, intermediate, and soma regions. Growth and shortening rates are shown on the right and left horizontal axes, respectively; rescue and catastrophe frequencies are shown on the upward and downward horizontal axes, respectively **p < 0.01, *p < 0.05; n.s., not significant, p > 0.05. (E) Maximal intensity projections in x, y, and z of the dorsal extension of UMC1 showing MTs in a thin layer contained within a confocal section. (F) Kymograph reveals growth deceleration of a single MT. Deceleration usually occurs 3.57 ± 1.92 μm before the cell edge. Six worms, 37 MTs. Terminal and initial growth rate are significantly different, p < 0.001, n = 69. (G) Proportion of MTs displaying uniphasic or biphasic growth behavior in dorsal, intermediate, and soma regions; >30 MTs/region.
Identification of microtubule deceleration
Nucleation of MTs was rarely observed in the dorsal extension. Thus most MTs in UMCs emanate from the ventral soma and grow in an almost parallel array toward the dorsal cell extension (Figure 1E). In examining the kymographs of MT behavior in UMCs, we noticed that MT polymerization was relatively regular throughout a growth excursion within most of the cell but that an angle appeared on kymographs tracking polymerizing MTs into UMC dorsal extensions (Figure 1F). This atypical MT behavior, indicating abrupt growth deceleration, occurred more frequently in the UMC1s, the most central UMCs along the worm anteroposterior axis (Figure 1A), and was never observed in SM precursor cells. MT growth deceleration was seldom observed in the soma and primarily occurred in the dorsal extension for 83% of MTs (Figure 1G). MT deceleration occurred 3.6 ± 1.9 μm from the cell periphery (Figure 1, E and F), before force-dependent catastrophe due to interaction with the cell periphery was expected to take place (Brunner and Nurse, 2000; Komarova et al., 2002; Janson et al., 2003; Alieva et al., 2010). On average, MTs grow for 2.2 ± 0.2 μm after deceleration before undergoing catastrophe at or before encountering the cell edge. After catastrophe, MTs often reinitiated growth within this ∼3-μm dorsal zone (Figure 1C, kymograph A). Therefore MTs in the dorsal extension of UMCs undergo biphasic growth excursions composed of a rapid “initial” phase and a slower “terminal” phase near the cell edge in which they exhibit distinct dynamic behavior (Figure 1F and Supplemental Table S2).
The TOG domain protein ZYG-9TOGp promotes the rapid polymerization that precedes growth deceleration
The sharp MT growth deceleration observed in UMCs could be due to either promotion of rapid growth in the cell center or suppression of MT assembly at the cell periphery. We next used RNA interference (RNAi) to test whether MT deceleration requires conserved MAPs implicated in the regulation of MT growth. We tested several conserved positive or negative regulators of MT dynamics that could account for MT growth deceleration (Figure 2A). Among these targets, only depletion of ZYG-9TOGp resulted in a strong reduction in the frequency of biphasic growth (Figure 2A; 10.8% of MTs reaching the cell edge exhibited growth deceleration; control, 73.2%). TOG domain–containing proteins are believed to promote MT growth by stabilizing the MT lattice, facilitating tubulin monomer addition, or both (Tournebize et al., 2000; Al-Bassam and Chang, 2011). In zyg-9TOGp(RNAi) worms, UMCs have a normal morphology, with their dorsal extension reaching to the alae (Supplemental Figure S1A). The rate of MT growth was lower in ZYG-9TOGp(RNAi) than the “initial” (faster) growth rate in control cells and was statistically indistinguishable from the “terminal” slow rate of growth exhibited by MTs in control cells after deceleration (Figure 2, B and C). The Xenopus homologue of ZYG-9, XMAP215, has been found to increase both growth rate and catastrophe frequency (Zanic et al., 2013). Indeed, the catastrophe frequency of MTs in the dorsal extension was strongly reduced in absence of ZYG-9TOGp (Supplemental Table S3). These modifications of MT dynamics in cell soma and in the dorsal extension after depletion of ZYG-9TOGp result in MT dynamic behavior in the dorsal extension that is similar to that in the cell body (Supplemental Table S3). Thus, after depletion of ZYG-9TOGp, MTs emanating from the cell center do not undergo rapid assembly but grow at a significantly reduced rate and do not decelerate when they reach the cell periphery. These results suggest that ZYG-9TOGp promotes rapid MT growth observed in the soma and the intermediate regions of UMCs.
FIGURE 2:
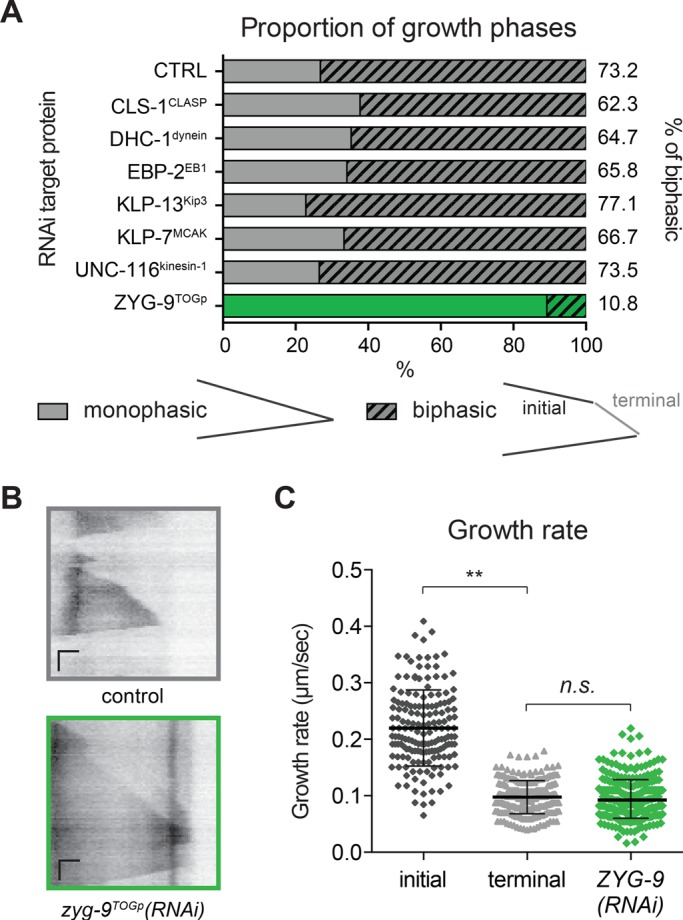
ZYG-9TOGp is required for MT growth deceleration. (A) Proportions of biphasic growth excursions measured in UMC dorsal extension in RNAi-treated worms. CLS-1 (C07H6.3) is one of the three cytoplasmic linker–associated protein (CLASP) homologues in C. elegans and is required for normal SM lineage development (Lacroix et al., 2014). Dynein heavy chain (DHC-1) and kinesin-1 (UNC-116, R05D3.7) are expected to affect retrograde or anterograde MT-based transport (Hirokawa, 1998; Siddiqui, 2002; Koushika et al., 2004). EBP-2 (VW02B12L.3), a homologue of EB1, promotes MT assembly by binding the growing MT plus end (Mimori-Kiyosue et al., 2000; Tirnauer et al., 2002; Sandblad et al., 2006). KLP-13Kip3 (F22F4.3, a kinesin-8 family member) and KLP-7MCAK (K11D9.1, the sole kinesin-13 in C. elegans) are the two C. elegans depolymerizing kinesins, whose homologues can reduce growth rate and induce depolymerization (Gupta et al., 2006; Gardner et al., 2011; Stumpff et al., 2011). Targeting of CLS-1CLASP, DHC-1dynein, EBP-2EB1, KLP-13Kip3, KLP-7MCAK, or UNC-116kinesin-1 did not result in a significant difference in the proportion of MTs exhibiting biphasic growth compared with controls; ZYG-9TOGp depletion did; p > 0.5, chi-square Fisher’s exact test. n(worm) >6, n(MTs) > 60. (B) Representative kymographs for control and ZYG-9TOGp depleted worms. Horizontal scale bar = 1 μm, vertical scale bar = 10 s. (C) Growth rate in ZYG-9TOGp depleted worms was measured in UMCs and compared with initial and terminal growth rate in control UMCs. At least six worms and at least 70 MTs. **p < 0.001, n.s., not significant, p = 0.2408, using unpaired t test.
MT deceleration coincides with a local alteration of tubulin concentration
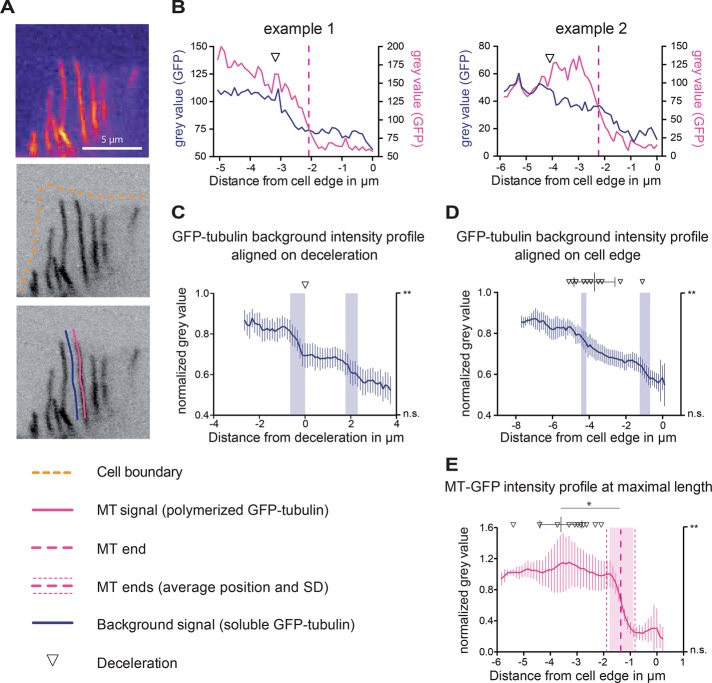
Although ZYG-9TOGp depletion strongly reduced the occurrence of MT deceleration, this result did not explain how deceleration happens at a precise location within control UMCs. Given that tubulin concentration can directly influence MT dynamics (Walker et al., 1988; Erickson and O’Brien, 1992; Gardner et al., 2011), we next examined whether a change in free tubulin availability correlated with the location of MT deceleration. The specific expression of GFP-tubulin expression driven by the unc-62 promoter in the SM lineage allowed us to distinguish soluble GFP-tagged tubulin from the unlabeled cytoplasm of neighboring tissues (Figure 3A). In the ∼5-μm dorsal extension of the UMC, MTs can be imaged in a single confocal section (Figure 1E). From time-lapse image sequences of single confocal planes, we measured the intensity of soluble GFP-tagged tubulin as a proxy for the local concentration of free tubulin in corridors of cytoplasm parallel to MTs in the UMC dorsal extension (Figure 3, A and B). The soluble GFP-tubulin signal (dark blue trace) decays sharply or more gradually (examples 1 and 2, respectively; Figure 3B) within the last 6 μm of UMC dorsal extension. In contrast, GFP-tubulin signal within MTs themselves does not decay. When aligned on the point of MT deceleration and averaged, the soluble GFP-tubulin traces depict a statistically significant decay in intensity in the ∼1 μm before the corresponding deceleration (Figure 3C and Supplemental Figure S2C). The average distance between the deceleration point of a single MT (downward-pointing open triangles) and the drop in intensity of the neighboring background is 0.23 ± 0.16 μm (Figure 3D; approximately our resolution limit), suggesting that the two events are concomitant. Similar to MT deceleration, the drop in intensity occurs at a somewhat variable position with respect to the cell edge (3.8 ± 0.6 μm; Figure 3D). Thus averaging intensity traces aligned at the cell edge reveals a progressive (less sharp) decay in soluble GFP-tubulin signal from the ventral to the dorsal side of UMCs (Figure 3D and Supplemental Figure S2C). In contrast, GFP-tubulin intensity within MTs does not decrease simultaneously with the background or as the MT undergoes deceleration (pink curves, Figure 3, B, individual traces, and E, average), suggesting that the local decrease in tubulin intensity is not due to an overall signal decay in this area of the cell. A drop in soluble GFP-tubulin intensity was also observed in corridors of cytoplasm adjacent to the ∼17% of MTs that do not undergo deceleration (Supplemental Figure S1B), suggesting that global regulation of tubulin exists in the dorsal extension and that a small fraction of MTs remains insensitive to this regulation.
FIGURE 3:
MT deceleration correlates with local soluble tubulin distribution. (A) Single confocal plane of UMCs imaged in MDX12 worms. Signal corresponds to GFP-tubulin expressed in UMCs with a specific promoter (see Materials and Methods). Pseudocolor enhances the visibility of GFP background in a UMC. Pink trace, MT; dark blue trace, background adjacent to MTs. (B) Two examples of the intensity of tubulin in a MT (pink trace) and the intensity of the adjacent background (dark blue trace); graphs depict a representative single track. The deceleration of each MT was determined by kymograph analysis and is noted on each intensity profile (inverted triangle). A drop in background intensity is observed in the vicinity of deceleration. No drop is observed in the intensity of GFP-tubulin along a MT. (C) Sixteen background intensity profiles (blue) aligned relative to the point corresponding to deceleration of the neighboring MT. Vertical shaded blue bars represent the significance of intensity decay measured as described in Materials and Methods. **p ≤ 0.01. (D) The same traces as presented in C aligned relative to the location of the cell edge. Triangles and bars on top represent the position of individual deceleration events and the average position and SD of deceleration position relative to cell edge, respectively. The vertical shaded blue bars represent the significance of intensity decay measured as described in Materials and Methods. **p ≤ 0.01. (E) Average of 16 normalized GFP-tubulin intensities within decelerating MTs. Traces are aligned relative to the cell edge. The vertical pink dashed line represents the average position of MT ends relative to the cell edge (1.4 ± 0.6 μm). The average and SD of the deceleration positions are plotted above the curve. Vertical shaded pink bar indicates significant intensity decay, measured as described Materials and Methods. p value is indicated on the right y-axis. **p ≤ 0.01; n.s., not significant, *p = 0.0009, unpaired t test. (C–E) Individual background trace (dark blue) or MT trace (pink) intensity values were normalized as described in Materials and Methods before being averaged. Error bars indicate 95% confidence interval.
The UMC dorsal extensions where MT growth deceleration occurs are approximately ¼ μm thick, less than our optical resolution in Z. Thus we considered the possibility that the intensity of soluble GFP-tubulin appears to drop off sharply due to an optical artifact generated by the dorsal extension occupying only a fraction of the imaging volume. To test this possibility, we expressed soluble GFP in the UMCs and examined the profile of its intensity in corridors of cytoplasm. We found no comparable drop or decay in GFP intensity as had been observed with GFP-tubulin intensity (Supplemental Figure S2, A–C). These results suggest that the appearance of a lower abundance of soluble GFP-tubulin is not an effect of cell topology but instead might reflect subcellular compartmentalization of soluble tubulin. Thus, in addition to ZYG-9TOGp promoting fast MT polymerization in the cell body, MT growth rate might also be affected by cellular compartmentalization of free tubulin, since growth rate is decreased in regions of lower tubulin concentration in the dorsal extension of UMCs. In contrast, tubulin compartmentalization could be a consequence of MT growth regulation in this cellular area.
Two novel proteins with homology to mammalian cylicin regulate tubulin compartmentalization and MT deceleration
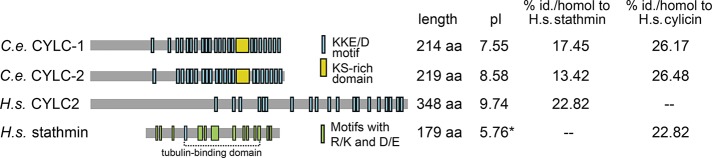
To understand how spatial differences in MT growth rate are regulated and how the compartmentalization of tubulin concentration is achieved, we searched for potential candidate regulators. Stathmin/Op18 is a tubulin-sequestering protein that can reduce the concentration of available tubulin and thus suppress MT assembly (Belmont and Mitchison, 1996; Cassimeris, 2002). Direct comparison of vertebrate and arthropod genomes with C. elegans did not result in the identification of a homologue of stathmin, so we searched for related genes by analyzing the genomes of other nematodes (Supplemental Figure S3). We examined the genome of the nematode Caenorhabditis remanei for a gene product with sequence or domain homology to Xenopus laevis stathmin (Supplemental Figure S3). These searches revealed a C. remanei uncharacterized protein CRE12689 (National Center for Biotechnology Information sequence XP_003108008.1). Alignment of this ORF with the C. elegans genome revealed two uncharacterized ORFs, Y59E9AL.6 and C41G7.6 (Supplemental Figure S3). These genes encode two similar proteins that are small, extremely basic, and contain multiple KKD/E repeats, which are commonly found in chromatin- and MT-interacting proteins (Noble et al., 1989). Comparing these two ORFs with mammalian genomes demonstrated that these genes share more similarity with mammalian cylicin (Figure 4), a putative cytoskeletal protein with unknown function, which is enriched in sperm (Hess et al., 1993; Rousseaux-Prevost et al., 2003). We thus assigned the names CYLC-1 and CYLC-2 to the gene products of ORFs Y59E9AL.6 and C41G7.6, respectively, although these two proteins are distant from the human cylicins.
FIGURE 4:
C. elegans cylicin homologues CYLC-1 and CYLC-2 share similarities with human stathmin. Sequence comparison of C. elegans CYLC-1 and CYLC-2 and human cylicin and stathmin. C.e. CYLC-1, Y59E9AL.6; C.e. CYLC-2, C41G7.6; H.s. stathmin (NP_005554); and H.s. CYLC2 (NP_001331). Charged motifs are highlighted as possible areas for tubulin binding. KKE/D motifs are highlighted in blue in all proteins. C.e. CYLC-1 and -2 both contain a KS-rich domain (∼18 amino acids long), highlighted in yellow, between two stretches of KKE/d-rich domains. Because H.s. stathmin contains only one KKD domain, short motifs containing both positively and negatively charged amino acids are highlighted in green. *The pI value of the tubulin-binding domain of H.s stathmin is 7.06.
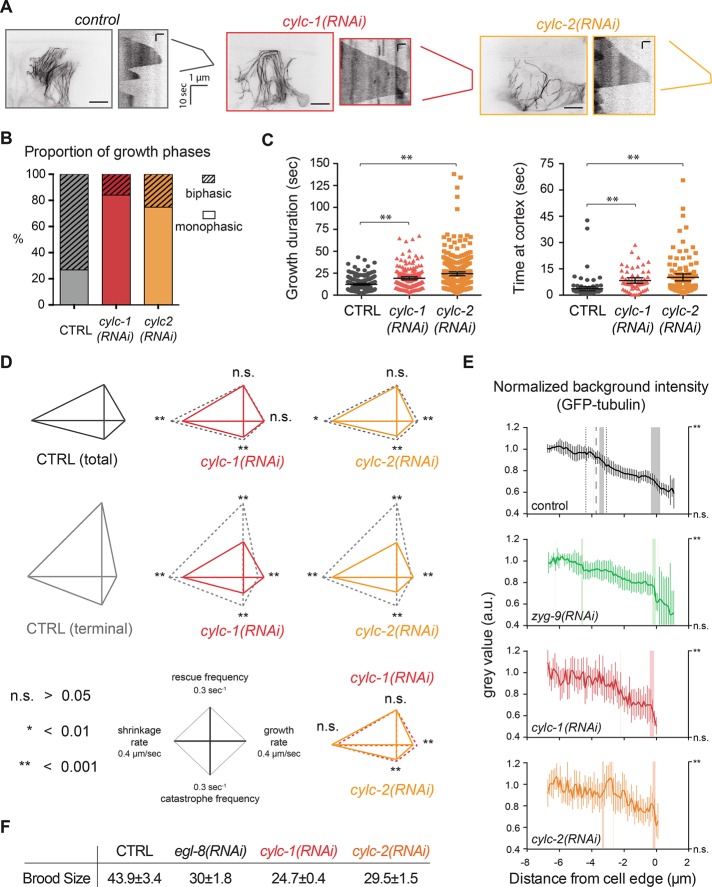
To test the function of CYLC-1 and -2, we targeted these proteins by RNAi and monitored MT organization and dynamics. In CYLC-1– or -2–depleted worms, MT organization in the UMCs was strongly altered compared with control worms (Figure 5A). The incidence of biphasic growth was strongly reduced by depletion of CYLC-1 or -2 (Figure 5, A and B). Unlike ZYG-9TOGp depletion, which eliminates deceleration by reducing growth speed throughout the cell to the slow speed normally occurring in the dorsal extension (Figure 2C), CYLC-1 or -2 depletion allowed persistent growth (long growth excursions; Figure 5C, left) into the cell periphery at speeds significantly higher (0.18 and 0.13 μm/s, respectively) than that normally observed in the periphery (0.11 μm/s; Figure 5, A and D, and Supplemental Table S3). MTs in CYLC-1– or -2–depleted UMCs exhibited a substantial (8.3 ± 0.8 and 10.1 ± 1 s, respectively) pause upon reaching the cell periphery, unlike in control cells, in which catastrophe occurred within 3.6 ± 0.6 s of a MT reaching the periphery (Figure 5C, right). In addition, both depletions significantly reduced catastrophe frequency and shrinkage rate (Figure 5D and Supplemental Table S3).
FIGURE 5:
CYLC-1 and CYLC-2 abrogate MT deceleration and soluble tubulin compartmentalization in UMCs. (A) Maximal intensity projection of UMCs in adult worms after CYLC-1 or -2 depletion. Scale bar, 10 μm. Kymographs of single MTs in these cells show suppression of deceleration in CYLC-1– or -2–depleted worms. Scale bar, 10 s (vertical), 1 μm (horizontal). (B) Proportion of MTs exhibiting monophasic or biphasic growth excursions in the terminal area of UMCs in CYLC-1– or CYLC-2–depleted worms. At least six worms and at least 69 MTs. (C) Left, the time of MT growing excursion without catastrophe is increased in CYLC-1– and CYLC-2–depleted worms. Right, time at cortex represents the time spent by MTs at the cell periphery before undergoing catastrophe. **p < 0.001, unpaired t test, compared with total control MT population. (D) Diamond graphs representing dynamic parameters in control and CYLC-1– or -2–depleted worms. n.s., not significant; *p < 0.01, **p < 0.001, unpaired t test. More than 12 worms and >160 MTs. (E) As in Figure 3, background intensity was measured next to MT tracks and normalized in each condition. Vertical lines at 3.8 μm in control (black) represent average location and SD (± 0.6 μm) of the intensity drop in control cells. The vertical shaded bar indicates significant intensity increase or decrease as described in Materials and Methods. The p value is indicated on the right y-axis. **p ≤ 0.01. At least 12 worms. (F) CYLC-1 or CYLC-2 depletion affects egg-laying function, as evidenced by reduced brood size. Number of eggs laid per day and per worm in each condition correspond to the average brood size of five worms in three different experiments. EGL-8 is a phospholipase Cβ isoform involved in egg laying (Lackner et al., 1999) and served as a positive control for the experiment.
To test whether CYLC-1 and -2 promote MT deceleration by altering the cytoplasmic compartmentalization of soluble tubulin, we measured the background GFP-tubulin intensity in the dorsal extension of UMCs in CYLC-1– or -2–depleted worms. We did not detect a reduction of GFP-tubulin intensity in the dorsal extension of CYLC-1– or -2–depleted cells as in controls (Figure 5E, red and orange curves, respectively). After ZYG-9TOGp depletion, a significant decrease in tubulin fluorescence occurred ∼4 μm before the cell edge, and the intensity profile was similar to control (Figure 5E, green and black curves, respectively). For reasons that we do not understand, the intensity of soluble GFP-tubulin signal was more variable in cells depleted of CYLC-1 or -2 (Figure 5E). Together these results suggest that the effect of CYLC-1 and -2 on MT deceleration is due to a role in tubulin compartmentalization or stabilization or, alternatively, that CYLCs affect MT dynamics in a way that alters the equilibrium and/or the distribution of soluble tubulin in this subcellular compartment.
Globally, CYLC-1 or -2 depletion reduced MT dynamics in UMCs. We previously demonstrated that general reduction of MT dynamics correlates with loss of UMC muscle function (Lacroix et al., 2014). Indeed, CYLC-1 or -2 depletion abrogated egg laying (Figure 5F). However, depletion of targets that we previously showed to perturb egg laying, including KLP-7MCAK and KLP-13Kip3 (Lacroix et al., 2014), did not affect MT deceleration (Figure 2A), suggesting that the loss of deceleration observed after CYLC-1 or -2 depletion is not a consequence of the egg-laying defect exhibited by these worms. In sum, these results demonstrate that MT growth deceleration is regulated by both established and novel factors.
DISCUSSION
MT dynamic instability is a fascinating intrinsic property of tubulin that allows MTs to exhibit a variety of behaviors. Both in vivo and in vitro, single MTs display a range of growth rates across different growth excursions (Gildersleeve et al., 1992; Shelden and Wadsworth, 1993; Odde et al., 1996; Pedigo and Williams, 2002; discussed in Howard and Hyman, 2009). Fluctuations in the rate of MT assembly in vitro have been attributed to differences in the assembly of individual protofilaments at MT ends (Kerssemakers et al., 2006; Schek et al., 2007) or variations in the size of the GTP cap (VanBuren et al., 2005). In vivo, the cellular environment and MAPs are likely to further influence these behaviors. Careful observation and description of the intricate variations in MT behavior that occur in vivo could help to refine and inform theoretical models describing the underlying principles of MT polymerization.
Here we show how such quantitative observation can reveal novel aspects of MT dynamics. A single MT can decelerate abruptly while entering a specific subcellular compartment. Thus the polymerization of a given MT plus end can be differentially regulated in space and/or time. This abrupt change in the behavior of the plus end could be due to a change in the composition or regulatory state of the plus tip–binding complex that includes ZYG-9TOGp, the change in the availability of free tubulin, the structure of the plus tip, a combination of these factors, or as-yet-undescribed contributors. Of interest, earlier in worm development in embryos or in the precursor cells that give rise to the UMCs, biphasic MT growth occurs only rarely (Lacroix et al., 2014), indicating that this MT behavior is induced by molecular or morphological changes occurring during cell differentiation.
Our work implicates compartmentalization of soluble tubulin in the regulation of MT growth rate. Our findings that soluble GFP alone is not segregated and that depletion of CYLC-1 or CYLC-2 does not affect soluble GFP distribution (Supplemental Figure S2; unpublished data) suggest that CYLC-1 and -2 act on tubulin or associated proteins. Although it is difficult to determine the protein content in a cell-specific manner in C. elegans, we observed an increase in local variability of the tubulin signal throughout the dorsal region of CYLC-1– or -2–depleted UMCs (Figure 5E). We described MT growth deceleration as concomitant with a decrease in local GFP-tubulin concentration, but the causality between these two observations remains to be determined. The compartmentalization of tubulin observed in control UMCs could be a consequence of the change in dynamics, as proposed by a theoretical model (Janulevicius et al., 2006). Furthermore, it is possible that our failure to observe tubulin compartmentalization after depletion of CYLC-1 and -2 is due to the high heterogeneity of the tubulin signal in these cells.
After CYLC-1 or -2 depletion, MT growth rate in the dorsal extension remains high (0.18 and 0.13 μm/s, respectively) even in the dorsal extension, whereas it is reduced from 0.18 to 0.09 μm/s in control UMCs. The abnormally high growth rate in the dorsal extension could result from abnormal ZYG-9TOGp activity in this region after CYLC-1 or -2 depletion, since depletion of ZYG-9TOGp reduces the growth rate to a level comparable to that normally observed in the dorsal extension (Figure 2C). Thus we speculate that CYLCs affect not only tubulin distribution but also localization of ZYG-9TOGp or another MAP that directly or indirectly regulates MT growth rate. Further studies of protein localization are needed to address this question.
Cylicins may contribute to regulation of cellular tubulin levels by stabilizing dimers. Of interest, muscle cells in Drosophila mutants with stathmin loss of function have decreased tubulin content and a MT network with altered morphology (Duncan et al., 2013). The way in which CYLC-1 and -2 affect tubulin levels or MT behavior in general and whether C. elegans cylicins function similarly to their stathmin orthologues remain to be discovered. Further investigation will be required to determine whether CYLC-1 and -2 directly regulate tubulin levels and whether they directly interact with tubulin or MTs to mediate subcellular tubulin compartmentalization and deceleration. Alternatively, their effects on localized MT behavior may stem from their local regulation by posttranslational modification, as was shown for Stathmin/Op18 (Wittmann et al., 2003; Watabe-Uchida et al., 2006).
We observed an increase in rescue frequency in the dorsal extension, which was not predicted, since the reduction of tubulin concentration is expected to have the opposite effect on this aspect of MT dynamics (Walker et al., 1988; O’Brien et al., 1990). This local regulation of rescue could be induced by a modification of the composition or structure of the MT lattice, for example, retention of GTP within the lattice (Dimitrov et al., 2008; Gardner et al., 2013). The idea that local reduction of free tubulin influences the GTP hydrolysis regime of the MT is supported by the observation that GTPase activity was affected by tubulin concentration and interaction of soluble subunits with the MT lattice (Caplow and Shanks, 1990).
The four major MT dynamics parameters—the rates of growth and shrinkage and the frequencies of catastrophe and rescue—can be used to determine the growth velocity of a population of MTs (the rate of the accumulation of polymer; Verde et al., 1992). Verde et al. (1992) devised an equation, the solution to which predicts whether MT length on average remains constant (“bounded”) or MTs continue to elongate with time (“unbounded,” assuming unlimited tubulin and no spatial confinement). Applying our measured parameters to this equation, we found that MTs are in the unbounded state in the cell soma and intermediate area but in the bounded state in the dorsal region (Supplemental Figure S4). Thus a given MT can adopt distinct states depending on its location in the cell, and cellular compartmentalization of MT regulators locally influences whether the local network is bounded or unbounded. These results further suggest that in the soma, a slight perturbation of tubulin availability could limit MT elongation. One the other hand, in the dorsal extension, the model predicts that MT length will remain roughly constant even with an unlimited supply of tubulin subunits. These predictions agree with our observations of MT organization and behavior in UMCs but also suggest that the local modification of MT dynamics in the dorsal region is not necessarily or directly linked to tubulin compartmentalization but perhaps is to the regulation of one or several MAPs (Supplemental Figure S5).
We observed that cylicin depletions affect UMC egg-laying function (Figure 5F). How does MT growth deceleration or the cylicins affect UMC function? MT dynamics, including the duration of pause at the cell edge, could be important for cell–cell communication and contraction, as reported for neurons in which synaptic protein and neurotransmitter deposition can be altered by affecting the MT network (Butler et al., 2007). Alternatively, MT dynamics and organization changes deep in the cell could indirectly affect viscoelastic properties of smooth muscles and their ability to contract correctly (Yamamoto et al., 1998; Zhang et al., 2000).
It will be interesting and challenging to understand how cylicins elicit MT deceleration and determine whether UMC dysfunction induced by cylicin depletions is directly related to the loss of deceleration or to the global alteration of MT organization and dynamics. Although it is beyond the scope of this work to dissect this mechanism, our work underscores the power of using an intact animal to study the behavioral consequences of cell- and molecule-scale events.
Taken together, our results illustrate the complexity of intracellular influences on MT dynamics and open new avenues of investigation.
Materials and Methods
C. elegans strains and maintenance
MDX12 was generated by crossing JJ1753 and NK682 (Lacroix et al., 2014). JJ1753: unc-119(ed3) III; zuIs151 [pJN326: nmy-2::mRFP; unc-119(+)] was kindly provided by Jeremy Nance (New York University School of Medicine, New York, NY). NK682 contains an integrated transgene qyIs119 generated by amplifying the sequence of GFP–β-tubulin from pJH4.66 (pie-1p::GFP::β-tubulin) and placing it after the unc-62 promoter region, which was amplified from the fosmid WRM0629aH06. Similarly, in NK607, GFP is expressed under the control of the unc-62 promoter (unc-62-::GFP). This strain was kindly provided by David Sherwood and Shinji Ihara (Duke University, Durham, NC).
Sequence analyses
Amino acid sequences were obtained from the Uniprot database, and multiple sequence alignments were performed using the ClustalW algorithm (EMBL-EBI, Cambridge, UK) and rendered in BioEdit Sequence Alignment Editor (Ibis Biosciences, Carlsbad, CA) for visualization. pI values were predicted using the Compute pI/Mw tool (ExPASy; Swiss Institute of Bioinformatics, Lausanne, Switzerland). Percentage identity was determined using the ClustalW web tool (EMBL-EBI).
Worm strain, worm culture, and RNA interference
Worm strains were maintained at 25°C using standard procedures (Brenner, 1974). For in vivo imaging and feeding experiments, worms were synchronized using the alkaline bleach method (Stiernagle, 2006). For protein depletion experiments, 20–30 worms were placed on a plate seeded with HT115 bacterial strain containing the L4440 vector for isopropylthiogalactoside-mediated induction of double-strand RNA expression as described previously (Kamath et al., 2001). Individual bacterial clones come from the Arhinger library and were kindly provided by Jean-Claude Labbé (Institute for Research in Immunology and Cancer, Université de Montréal, Montréal, Canada) and were verified by sequencing. Depletions were all made by L4-feeding experiments: L1-stage synchronized worms were grown on regular nematode growth medium plates for 44 h and then transferred to RNA feeding plates for at least 24 h. Egg-laying function was evaluated by measuring brood size as follows: five hermaphrodites after 24 h of feeding were transferred to a new feeding plate, and eggs laid in 24 h were counted.
Worm mounting and live-imaging conditions
Live imaging was performed as in Lacroix et al. (2014). Briefly, worms were anesthetized in 0.01% tetramisole in M9 buffer for 10 min before being transferred onto a 5% agarose pad. Worms were then covered with a poly-l-lysine–coated coverslip. Coverslip was sealed with a mixture of Vaseline, paraffin wax, and lanolin (1:1:1), and the imaging chamber was filled with M9 to prevent drying. To minimize out-of-focus light and maximize acquisition time, we used either a real-time swept-field confocal (SFC; Nikon Canada, Mississauga, Canada, and Prairie Technologies, Madison, WI) or spinning-disk confocal microscope (Roper Scientific, Evry, France). All SFC acquisitions and additional components, including laser exposure setting, were controlled by Elements software (Nikon Canada), and spinning-disk acquisition was controlled using MetaMorph 7 software (Molecular Devices, Sunnyvale, CA). For Z-series and volume view, 60× or 100×/1.4 numerical aperture (NA) Plan-Apochromat objectives were used to acquire confocal Z-sections with 500- or 600-μm steps. Acquisition of time-lapse images for MT dynamics was performed with the 100×/1.4 NA Plan-Apochromat objective with the 1.5× Optivar for 2 min with a time interval of 1 s and <200-ms exposure time. A maximum of five movies (10 min of acquisition) were made on a single worm to avoid phototoxicity. At least six worms were imaged per condition.
Image processing and analysis of MT dynamics
Three-dimensional views of Z-stacks were generated using NIS-Elements software (Nikon Canada). Images, Z-stacks, and movies were converted into noncompressed .avi files and processed in ImageJ software (32 bits; National Institutes of Health, Bethesda, MD). For Figures 3 and 5 and Supplemental Figure S2, GFP-tubulin fluorescence intensity was normalized to the average of the first 7 pixels of each trace. Image intensity was scaled only for figure construction. In Figure 3A, the pseudocolor scheme Fire was used to highlight signal variations. The ImageJ plug-in Multiple kymograph (www.embl.de/eamnet/html/kymograph.html) was used to extract kymographs. Dynamic parameters were extracted from these kymographs using ImageJ and compiled in Excel (Microsoft, Redmond, WA). Scatter plots, histograms, and statistical analyses were generated and performed using Prism (GraphPad Software, La Jolla, CA). Means were compared using the unpaired Welch t test. Diamond graphs were created using custom MATLAB-based software (MathWorks, Natick, MA). Rates and frequencies were respectively jointly normalized. The characterization of microtubule dynamics primarily refers to the measurement of growth and shrinking rates, as well as to the frequency of transition between events (Desai and Mitchison, 1997). The term catastrophe is defined as the transition between polymerization and depolymerization, and rescue describes the reverse transition. We calculated dynamic parameters by defining catastrophe frequency per unit time or per unit distance as the inverse of the average growth time or the average growth distance, respectively (Walker et al., 1988; Desai and Mitchison, 1997; Komarova et al., 2002). Similarly, rescue frequencies per unit time and per unit distance are the inverses of average shrinkage time and average shrinkage distance, respectively.
Statistical analysis
All p values were determined in Prism 6 (GraphPad) using an unpaired t test, except for Figure 3C (chi-square Fisher’s exact test). Because the dynamic parameters—growth rate, shrinkage rate, and catastrophe and rescue frequency—display different variances for UMCs, for these four parameters we used the unequal-variance unpaired t test with Welch’s correction. This test compares normally distributed populations without assuming equal SD. To identify significant changes in intensity (Figures 3, C–E, and 5E and Supplemental Figure S2C), we used unpaired two-tailed t tests to compare the average intensities of four consecutive 0.11 μm–pixel bins (i.e., positions 1–4 and 5–8, two neighboring 0.44-μm stretches). On the basis of the local signal variation in negative control traces (GFP alone and GFP-tubulin in MTs), we determined that p ≤ 0.01 reflected significant local difference in intensity.
Supplementary Material
Acknowledgments
We are grateful to Jean-Claude Labbé (Institute for Research in Immunology and Cancer) for providing essential reagents and Kevin Slep and Lionel Pintard for critical reading of the manuscript. We thank David Sherwood, Shinji Ihara, and Jeremy Nance for providing essential reagents. Tim Mitchison provided thoughtful insights into the bounded/unbounded regimes of MTs. This work was supported by operating funds from the U.S. National Institutes of Health (GM102390) to A.S.M. and by grants from the Fondation pour la Recherche Médicale (AJE201112) and the Mairie de Paris (Emergence) to J.D. B.L. was supported by Cancer Research Society of Canada Postdoctoral Fellowship F2011-16307 and FRM ARF20140129055. J.R. was supported by Fonds de recherche du Québec–Santé. This work is dedicated to the victims and first responders of the acts of terrorism in Paris on November 13, 2015.
Abbreviations used:
- GFP
green fluorescent protein
- homol
sequence homology
- MT
microtubule
- Op
oncoprotein
- ORF
open reading frame
- SM
sex myoblast
- TOG
tumor overexpressed gene
- UMC
uterine muscle cell
- Unc
uncoordinated movement C. elegans mutant
- XMAP
Xenopus MT-associated protein
- Zyg
zygotic lethal C. elegans mutant.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0056) on March 16, 2016.
REFERENCES
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alieva IB, Zemskov EA, Kireev II, Gorshkov BA, Wiseman DA, Black SM, Verin AD. Microtubules growth rate alteration in human endothelial cells. J Biomed Biotechnol. 2010;2010:671536. doi: 10.1155/2010/671536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Bowne-Anderson H, Zanic M, Kauer M, Howard J. Microtubule dynamic instability: a new model with coupled GTP hydrolysis and multistep catastrophe. Bioessays. 2013;35:452–461. doi: 10.1002/bies.201200131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- Butler D, Bendiske J, Michaelis ML, Karanian DA, Bahr BA. Microtubule-stabilizing agent prevents protein accumulation-induced loss of synaptic markers. Eur J Pharmacol. 2007;562:20–27. doi: 10.1016/j.ejphar.2007.01.053. [DOI] [PubMed] [Google Scholar]
- Caplow M, Shanks J. Mechanism of the microtubule GTPase reaction. J Biol Chem. 1990;265:8935–8941. [PubMed] [Google Scholar]
- Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Dimitrov A, Quesnoit M, Moutel S, Cantaloube I, Pous C, Perez F. Detection of GTP-tubulin conformation in vivo reveals a role for GTP remnants in microtubule rescues. Science. 2008;322:1353–1356. doi: 10.1126/science.1165401. [DOI] [PubMed] [Google Scholar]
- Duncan JE, Lytle NK, Zuniga A, Goldstein LS. The microtubule regulatory protein stathmin is required to maintain the integrity of axonal microtubules in Drosophila. PLoS One. 2013;8:e68324. doi: 10.1371/journal.pone.0068324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, O’Brien ET. Microtubule dynamic instability and GTP hydrolysis. Annu Rev Biophys Biomol Struct. 1992;21:145–166. doi: 10.1146/annurev.bb.21.060192.001045. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Howard J. Microtubule catastrophe and rescue. Curr Opin Cell Biol. 2013;25:14–22. doi: 10.1016/j.ceb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildersleeve RF, Cross AR, Cullen KE, Fagen AP, Williams RC., Jr. Microtubules grow and shorten at intrinsically variable rates. J Biol Chem. 1992;267:7995–8006. [PubMed] [Google Scholar]
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hess H, Heid H, Franke WW. Molecular characterization of mammalian cylicin, a basic protein of the sperm head cytoskeleton. J Cell Biol. 1993;122:1043–1052. doi: 10.1083/jcb.122.5.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- Janson ME, de Dood ME, Dogterom M. Dynamic instability of microtubules is regulated by force. J Cell Biol. 2003;161:1029–1034. doi: 10.1083/jcb.200301147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulevicius A, van Pelt J, van Ooyen A. Compartment volume influences microtubule dynamic instability: a model study. Biophys J. 2006;90:788–798. doi: 10.1529/biophysj.105.059410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerssemakers JW, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- Komarova YA, Vorobjev IA, Borisy GG. Life cycle of MTs: persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J Cell Sci. 2002;115:3527–3539. doi: 10.1242/jcs.115.17.3527. [DOI] [PubMed] [Google Scholar]
- Koushika SP, Schaefer AM, Vincent R, Willis JH, Bowerman B, Nonet ML. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J Neurosci. 2004;24:3907–3916. doi: 10.1523/JNEUROSCI.5039-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Lyle KS, Gierke S, Matov A, Danuser G, Wittmann T. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J Cell Biol. 2009;184:895–908. doi: 10.1083/jcb.200901042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner MR, Nurrish SJ, Kaplan JM. Facilitation of synaptic transmission by EGL-30 Gqalpha and EGL-8 PLCbeta: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron. 1999;24:335–346. doi: 10.1016/s0896-6273(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Bourdages KG, Dorn JF, Ihara S, Sherwood DR, Maddox PS, Maddox AS. In situ imaging in C. elegans reveals developmental regulation of microtubule dynamics. Dev Cell. 2014;29:203–216. doi: 10.1016/j.devcel.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Maddox AS. Microtubule dynamics followed through cell differentiation and tissue biogenesis in elegans C. Worm. 2014;3:e967611. doi: 10.4161/21624046.2014.967611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G, Akhmanova A. Microtubule plus end: a hub of cellular activities. Traffic. 2006;7:499–507. doi: 10.1111/j.1600-0854.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- Matov A, Applegate K, Kumar P, Thoma C, Krek W, Danuser G, Wittmann T. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat Methods. 2010;7:761–768. doi: 10.1038/nmeth.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Noble M, Lewis SA, Cowan NJ. The microtubule binding domain of microtubule-associated protein MAP1B contains a repeated sequence motif unrelated to that of MAP2 and tau. J Cell Biol. 1989;109:3367–3376. doi: 10.1083/jcb.109.6.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien ET, Salmon ED, Walker RA, Erickson HP. Effects of magnesium on the dynamic instability of individual microtubules. Biochemistry. 1990;29:6648–6656. doi: 10.1021/bi00480a014. [DOI] [PubMed] [Google Scholar]
- Odde DJ, Cassimeris L, Buettner HM. Spectral analysis of microtubule assembly dynamics. AIChE J. 1996;42:1434–1442. [Google Scholar]
- Pedigo S, Williams RC., Jr Concentration dependence of variability in growth rates of microtubules. Biophys J. 2002;83:1809–1819. doi: 10.1016/S0006-3495(02)73946-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux-Prevost R, Lecuyer C, Drobecq H, Sergheraert C, Dacheux JL, Rousseaux J. Characterization of boar sperm cytoskeletal cylicin II as an actin-binding protein. Biochem Biophys Res Commun. 2003;303:182–189. doi: 10.1016/s0006-291x(03)00317-6. [DOI] [PubMed] [Google Scholar]
- Sandblad L, Busch KE, Tittmann P, Gross H, Brunner D, Hoenger A. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006;127:1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Schek HT, 3rd, Gardner MK, Cheng J, Odde DJ, Hunt AJ. Microtubule assembly dynamics at the nanoscale. Curr Biol. 2007;17:1445–1455. doi: 10.1016/j.cub.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelden E, Wadsworth P. Observation and quantification of individual microtubule behavior in vivo: microtubule dynamics are cell-type specific. J Cell Biol. 1993;120:935–945. doi: 10.1083/jcb.120.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui SS. Metazoan motor models: kinesin superfamily in C. elegans. Traffic. 2002;3:20–28. doi: 10.1034/j.1600-0854.2002.30104.x. [DOI] [PubMed] [Google Scholar]
- Srayko M, Kaya A, Stamford J, Hyman AA. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. 2006(Feb 11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell. 2011;43:764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Grego S, Salmon ED, Mitchison TJ. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell. 2002;13:3614–3626. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R, Popov A, Kinoshita K, Ashford AJ, Rybina S, Pozniakovsky A, Mayer TU, Walczak CE, Karsenti E, Hyman AA. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- VanBuren V, Cassimeris L, Odde DJ. Mechanochemical model of microtubule structure and self-assembly kinetics. Biophys J. 2005;89:2911–2926. doi: 10.1529/biophysj.105.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B, Akhmanova A, Straube A. Regulation of microtubule dynamic instability. Biochem Soc Trans. 2009;37:1007–1013. doi: 10.1042/BST0371007. [DOI] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J Cell Biol. 1992;118:1097–1108. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P. Regional regulation of microtubule dynamics in polarized, motile cells. Cell Motil Cytoskeleton. 1999;42:48–59. doi: 10.1002/(SICI)1097-0169(1999)42:1<48::AID-CM5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, John KA, Janas JA, Newey SE, Van Aelst L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron. 2006;51:727–739. doi: 10.1016/j.neuron.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J Cell Biol. 1997;139:417–434. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberg RC, Deery WJ, Dickinson PJ. Tubulin-nucleotide interactions during the polymerization and depolymerization of microtubules. Biochemistry. 1976;15:4248–4254. doi: 10.1021/bi00664a018. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J Cell Biol. 2003;161:845–851. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Tsutsui H, Takahashi M, Ishibashi Y, Tagawa H, Imanaka-Yoshida K, Saeki Y, Takeshita A. Role of microtubules in the viscoelastic properties of isolated cardiac muscle. J Mol Cell Cardiol. 1998;30:1841–1853. doi: 10.1006/jmcc.1998.0747. [DOI] [PubMed] [Google Scholar]
- Zanic M, Widlund P, Hyman A, Howard J. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat Cell Biol. 2013;15:688–693. doi: 10.1038/ncb2744. [DOI] [PubMed] [Google Scholar]
- Zhang D, Jin N, Rhoades RA, Yancey KW, Swartz DR. Influence of microtubules on vascular smooth muscle contraction. J Muscle Res Cell Motil. 2000;21:293–300. doi: 10.1023/a:1005600118157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.