Spatial regulation of Cdc42 activity is essential to maintain polarized growth. Fission yeast Rga6 is a new Cdc42 GAP that collaborates with Rga4, the only known Cdc42 GAP, in the spatial restriction of active Cdc42 at the cell tip. Both GAPs localize preferentially at the nongrowing areas of the membrane in different clusters.
Abstract
Active Cdc42 is essential for the establishment of polarized growth. This GTPase is negatively regulated by the GTPase-activating proteins (GAPs), which are important for the spatial specificity of Cdc42 function. Rga4 is the only GAP described as negative regulator of fission yeast Cdc42. We report here that Rga6, another fission yeast Cdc42 GAP, shares some functions with Rga4. Cells lacking Rga6 are viable but slightly shorter and broader than wild type, and cells lacking Rga6 and Rga4 simultaneously are rounded. In these cells, active Cdc42 is observed all around the membrane. These additive effects indicate that both GAPs collaborate in the spatial regulation of active Cdc42. Rga6 localizes to the plasma membrane, forming clusters different from those formed by Rga4. A polybasic region at the Rga6 C-terminus is responsible for its membrane localization. Rga6-GFP fluorescence decreases considerably at the growing tips, and this decrease is dependent on the actin cables. Of note, in the absence of Rga6, the amplitude of active Cdc42 oscillations at the tips decreases, and less GTP-Cdc42 accumulates at the new end of the cells. We propose that Rga6 collaborates with Rga4 to spatially restrict active Cdc42 at the cell tips and maintain cell dimensions.
INTRODUCTION
Cdc42 GTPase regulates cell polarity in all eukaryotic organisms from yeast to mammalian cells (Etienne-Manneville, 2004). This GTPase regulates actin cytoskeleton organization and participates in secretion and endocytosis, processes necessary for polarized growth (Heasman and Ridley, 2008; Harris, 2011). As do other GTPases, Cdc42 cycles between two states, active, GTP-bound and inactive, GDP-bound Cdc42. These states are spatial and temporarily regulated by activators named guanine nucleotide exchange factors (GEFs) and inactivators named GTPase-activating proteins (GAPs) and GDP dissociation inhibitors (GDIs).
Cells of the fission yeast Schizosaccharomyces pombe have a cylindrical shape; they grow by tip extension in a monopolar or bipolar manner and maintain a constant diameter. The cell poles are defined by the complex Tea1-Tea4 delivered by the microtubules (Mata and Nurse, 1997; Martin et al., 2005). Tea4 binds to the formin For3 and recruits the type I phosphatase Dis2 to the poles. In turn, this phosphatase allows the membrane binding of Pom1 kinase that regulates morphogenesis, bipolar growth, and cytokinesis (Bahler and Pringle, 1998; Hachet et al., 2011).
S. pombe Cdc42 is an essential protein required for polarized growth, which determines cell dimensions (Kelly and Nurse, 2011). Cdc42 localizes to fission yeast plasma membrane and endomembranes, and when is constitutively active, it promotes a nonpolarized phenotype giving rise to round cells (Miller and Johnson, 1994; Bendezu et al., 2015). S. pombe has two known Cdc42 GEFs—Scd1 and Gef1—with different but overlapping functions, since double deletion of these GEFs is lethal (Coll et al., 2003). Both localize to the tips and division area (Rincon et al., 2007), where active, GTP-bound Cdc42 is observed (Miller and Johnson, 1994; Tatebe et al., 2008). Scd1 is a major Ras1 target (Chang et al., 1994). Cells lacking Scd1 are rounded and have endocytosis defects (Murray and Johnson, 2001). In contrast, Gef1 deletion results in slightly thinner cells with defects in the establishment of bipolar growth and cytokinesis (Coll et al., 2003). Rga4 is the only GAP described for S. pombe Cdc42 (Das et al., 2007), and it is also a GAP for Rho2 (Soto et al., 2010). Rga4 localizes to the lateral membranes and is excluded from the growth areas. This exclusion depends at least in part on the DYRK kinase Pom1, which interacts with Rga4 through its C-terminal region (Tatebe et al., 2008), and on the Dis2 phosphatase (Kokkoris et al., 2014). Cells lacking Rga4 are 10% larger in volume and grow with a cell diameter greater than that of wild-type cells, yet retain the cylindrical shape and polarized growth (Das et al., 2007). Scd1 and Rga4 seem to act additively to define the dimensions of the growth zone, since the lack of both proteins gives rise to round cells (Kelly and Nurse, 2011).
Active Cdc42 requires adaptor proteins to signal to target proteins. Thus Scd2 adapter recruits Scd1, allowing further activation of Cdc42 and the binding of the kinase Pak1 (Endo et al., 2003; Wheatley and Rittinger, 2005). Pob1, another adapter protein from the Boi family, is required for formin For3 activation by Cdc42 (Rincón et al., 2009) and for the regulation of secretion (Bendezu and Martín, 2011; Estravis et al., 2011). For3 originates the interphase actin cables that further stabilize the axis of polarity by concentrating the secretion of components involved in Cdc42 activation and in surface growth (Kelly and Nurse, 2011; Bonazzi et al., 2015). Active Cdc42 localization is also regulated by the NDR kinase Orb6 and by Rad24, which restrict Gef1 localization to the poles (Das et al., 2009; 2015). All of these molecules promote the formation of a polarity axis by restricted activation of Cdc42. On the other hand, most polarity models predict negative mechanisms that ensure global inhibition of Cdc42 activity at the cortex (Sohrmann and Peter, 2003; Wedlich-Soldner et al., 2003; Das et al., 2012). In this study, we characterize the function of fission yeast Rga6. Our results indicate that Rga6 is a newly described Cdc42 GAP that collaborates with Rga4 in the spatial regulation of Cdc42 and the control of cell dimensions.
RESULTS
Rga6 is a Cdc42 and Rho2 GAP
The gene rga6+ is one of the 10 putative genes coding for Rho GAPs in the S. pombe genome. It encodes a protein with 733 amino acid residues and a molecular mass of 80.78 kDa. In silico structural analysis of Rga6 shows a central Rho GAP domain (amino acids 329–547), a serine-rich (SR) region before the GAP (amino acids 187–253), and a polybasic region (PBR) at the C-terminus of the protein (amino acids 700–733; Figure 1A). To determine the specificity of Rga6 activity toward S. pombe Rho GTPases, we first performed yeast two-hybrid assays to detect binding of Rga6 to constitutively active Rho GTPases. The results of the β-galactosidase assay used as reporter suggested that Rga6 interacts with active Rho2 and Cdc42 (Figure 1B). We then analyzed the amount of GTP-bound Cdc42 and Rho2 proteins in cells with or without Rga6. Cells expressing hemagglutinin (HA)-tagged versions of Cdc42 and Rho2 GTPases were crossed with rga6Δ cells. The total amount of HA-tagged versions of Cdc42 and Rho2 GTPases from wild-type and rga6Δ cell extracts was determined by Western blot, and the GTP-bound proteins were pulled down from the extracts with glutathione S-transferase (GST)–Cdc42/Rac interactive binding (CRIB) motif from Pak2 (for Cdc42 pull down) and GST-C21 Rho-binding domain (RBD) from Rhotekin (for Rho2 pull down). Cells lacking rga6+ showed an increase in GTP-bound Cdc42 (Figure 1C) and also in GTP-bound Rho2 (Figure 1D). In contrast, no increase in GTP-Rho1 or GTP-Rho3, used as controls, was observed in rga6Δ cells (Supplemental Figure S1A). These results suggest that Rga6 negatively regulates both Cdc42 and Rho2 activities in vivo.
FIGURE 1:
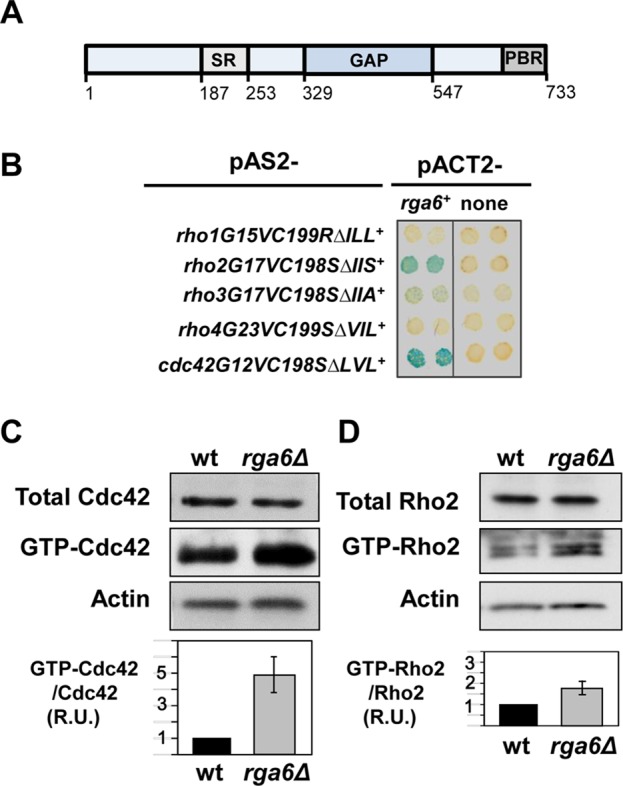
Rga6 is a Rho2 and Cdc42 GAP. (A) Schematic representation of Rga6 protein, indicating the serine-rich region (SR), GAP domain, and polybasic region (PBR) at the C-terminus. (B) β-Galactosidase analysis of the two-hybrid interaction between rga6+ and the constitutively active versions of rho1, rho2, rho3, rho4, and cdc42 lacking the coding sequence of the three C-terminal amino acids. (C, D) Levels of total and GTP-bound Cdc42 (C) and Rho2 (D) in rga6Δ strain. Extracts from wild-type and rga6Δ cells expressing HA-cdc42+, and HA-rho2+ were pulled down with GST-CRIB or GST-C21RBD, respectively, and blotted with anti-HA antibody (middle). Total HA-Rho proteins in the extracts were visualized by Western blot using anti-HA antibody (top). Actin was visualized in the same extracts as loading control (bottom). The ratio of GTP-bound to total protein is represented as a bar graph and shows the average and SD of four different experiments for each GTPase.
Rga6 collaborates with Rga4 in the control of S. pombe cell dimensions
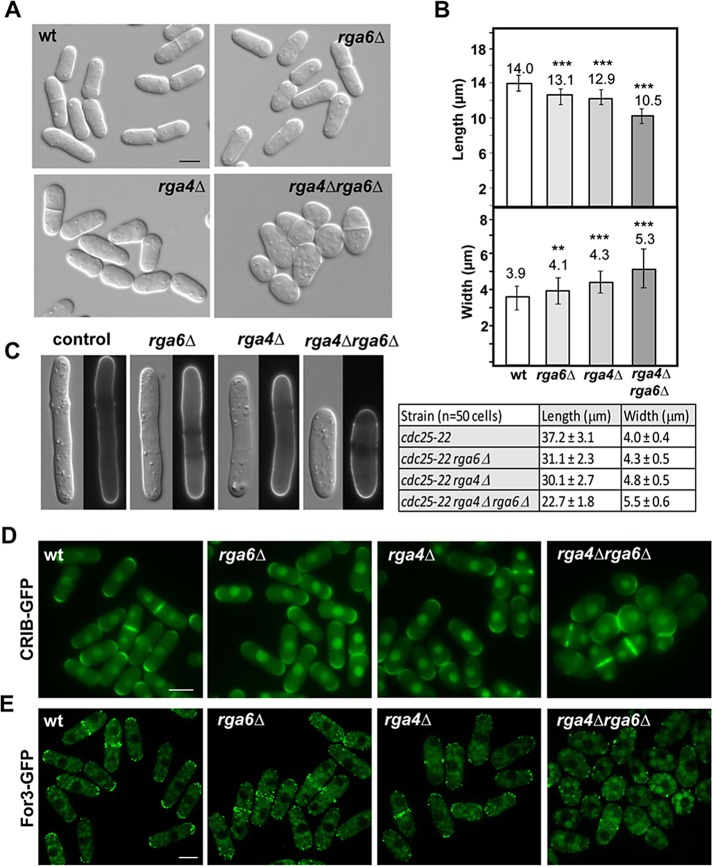
As previously reported, rga6+ is not essential (Nakano et al., 2001). However, cells lacking Rga6 displayed mild morphological defects (Figure 2A); they were ∼10% shorter than wild type (13.1 ± 0.7 μm [n = 80] vs. 14.02 ± 0.8 μm [n = 80] at the time of division; p < 0.0001, Student’s t test) and slightly wider (4.12 ± 0.4 μm [n = 80] vs. 3.95 ± 0.3 μm [n = 80]; p < 0.001, Student’s t test; Figure 2B). Although less pronounced, this phenotype was similar to that caused by the absence of Rga4 (Figure 2, A and B), the only Cdc42-GAP described until now, which plays a role in the regulation of the growth area (Das et al., 2007; Tatebe et al., 2008). Moreover, the rga4Δ rga6Δ double-mutant strain showed an additive effect of the two GAPs. Thus rga4Δ rga6Δ cells were rounded, shorter, and wider than either rga4Δ or rga6Δ single mutants (Figure 2, A and B). The lack of epistasis suggests that Rga4 and Rga6 collaborate but act independently in the control of cell dimensions. To see whether the rounded rga4Δ rga6Δ cells were still polarized, we constructed the single and double GAP mutants in a cdc25-22 background and synchronized the cells in G2 by cdc25-22 arrest at 36°C during 4 h (Moreno et al., 1991). In this condition, cells grow but do not divide, and we observed more pronounced polarized growth differences, particularly in cell length (Figure 2C). Simultaneous elimination of both GAPs did not cause complete loss of polarity, since growth still occurred over wider but delimited areas (Figure 2C).
FIGURE 2:
Morphology and cell dimensions of S. pombe strains lacking rga6+. (A) Differential interference contrast (DIC) images of wild-type, rga6Δ, rga4Δ, and rga4Δ rga6Δ double-mutant cells. Bar, 5 μm. (B) Cell dimensions of the indicated strains. Bars represent the mean (80 cells of each strain) of the cell length (top) and the cell diameter (bottom). Error bars represent SD. **p < 0.001, ***p < 0.0001; Student’s t test. (C) DIC and fluorescence images from Calcofluor-stained cdc25-22, cdc25-22 rga6Δ, cdc25-22 rga4Δ, and cdc25-22 rga4Δ rga6Δ cells blocked in G2 phase of the cell cycle by growing at 36°C for 4 h. Cell dimensions are indicated in the table. (D) Fluorescence images of wild-type, rga6Δ, rga4Δ, and rga4Δ rga6Δ cells expressing CRIB-GFP. Bar, 5 μm. (E) Fluorescence images of wild-type, rga6Δ, rga4Δ, and rga4Δ rga6Δ cells expressing For3-GFP. Bar, 5 μm.
As shown in Figure 1, Rga6 negatively regulates Cdc42 and Rho2. However, deletion of Rho2 does not suppress the morphological phenotype of rga6Δ cells, and rga4Δ rga6Δ rho2Δ triple mutant cells are as rounded as rga4Δ rga6Δ cells (Supplemental Figure S1B). Therefore these morphological changes are not due to increased Rho2 activity in the absence of Rga4 and Rga6 but instead to changes in Cdc42 activity.
Active GTP-bound Cdc42 appears concentrated at the plasma membrane of growing cell ends when observed using the Cdc42/Rac interactive binding (CRIB) domain of Gic2 bound to GFP (Tatebe et al., 2008). CRIB-GFP observation revealed that active Cdc42 was restricted to the wider-growing poles in either rga4Δ or rga6Δ single mutant cells and extended around different areas of the membrane in rga4Δ rga6Δ rounded cells (Figure 2D). Consequently, the formin For3, which assembles polarized actin cables and is localized by active Cdc42 (Martin et al., 2007; Rincón et al., 2009), was less concentrated to the tips of rga4Δ or rga6Δ cells and was completely dispersed throughout the cell cortex of the rga4Δ rga6Δ double mutant strain (Figure 2E). Other polarity markers independent of Cdc42, such as Tea1-GFP, do not extend around the membranes of rga4Δ rga6Δ rounded cells (Supplemental Figure S1C), suggesting that Rga6 plays an additive role with Rga4 in restricting Cdc42 activation to cell tips.
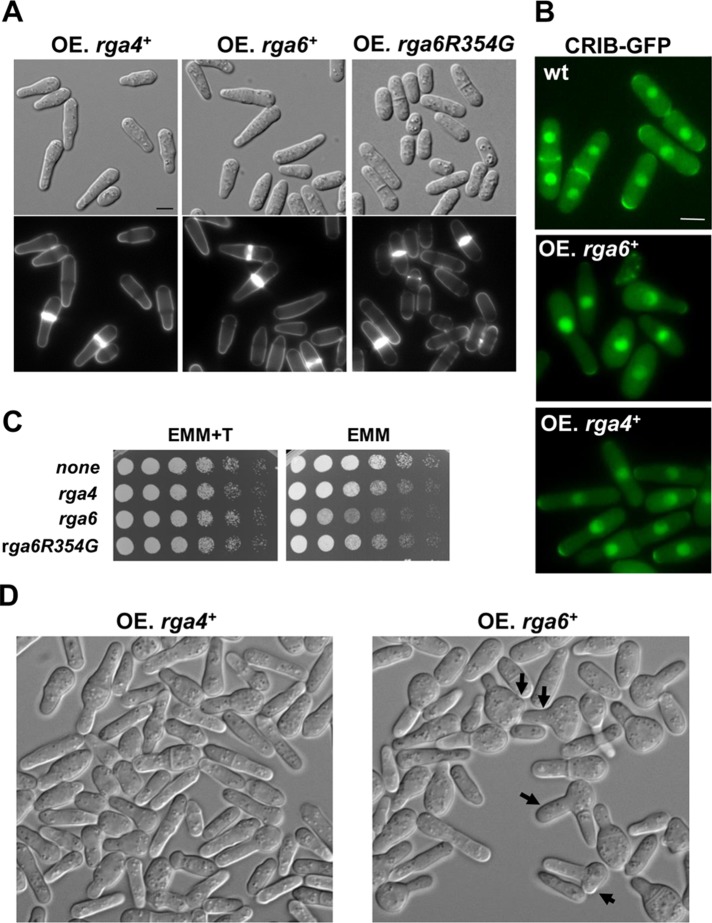
Overexpression of Rga6 alters the morphology of cells
S. pombe cells transformed with pREP1-rga6+, which carries the thiamine-repressible nmt1 promoter, and grown in the absence of thiamine to overexpress Rga6 were monopolar, and their growing tips were longer and thinner than those of wild-type cells (Figure 3A). This morphology is similar to the one caused by overexpression of Rga4 (Das et al., 2007; Figure 3A). To verify that the effect of Rga6 overexpression in growth dimensions was due to its Cdc42-GAP function, we mutated amino acid residue 354 corresponding to the arginine that is conserved in all GAP proteins and is essential for their activity (Bourne, 1997). Overexpression of the rga6R354G allele did not cause any alteration of the cell morphology (Figure 3A). The width of the growing tip in cells overexpressing Rga6 was 2.31 ± 0.28 versus 3.76 ± 0.29 μm in cells overexpressing Rga6R354G and 3.94 ± 0.31 μm in wild-type cells (50 cells of each type; p < 0.0001, Student’s t test). Therefore the GAP activity of Rga6 is required for its morphogenetic function. Fluorescence analysis of cells carrying CRIB-GFP showed that Cdc42 activation was restricted to the thin growing tip of cells overexpressing either rga6+ or rga4+ (Figure 3B), suggesting that both GAPs might have a similar function as negative regulators of Cdc42. Overexpression of either GAP integrated under the control of the nmt1 promoter was detrimental for the cells grown in solid medium for longer times (Figure 3C). Of interest, Rga6 overexpression at longer times caused the cells to swell and in some cases (10%, 158 cells) form new growth zones, whereas Rga4 overexpression caused some cells to swell, but they never formed new growth zones (Figure 3D). To understand the origin of the swelling, we recorded cells with integrated nmt1-rga6+ by time lapse during 8 after 24 h of growth in medium without thiamine. Rounded cells originated upon division of the monopolar cells and derived from the daughter cell carrying the nongrowing pole. Eventually, these rounded cells formed a new thin growing area, generating skittle-like cells. Some cells did not separate after septation and formed a new growing zone, generating a branch in the skittle-like cell (Supplemental Figure S2A). These observations suggest that even if there is not a cell end mark in the rounded cells, the formation of a new growing area can still eventually occur. We considered the possibility that Rga6 overexpression could displace Rga4 from the lateral membranes and might originate the swelling of the cells by local exclusion of this GAP (Kokkoris et al., 2014). However, this does not seem to be the cause of the ectopic growth, since Rga4-GFP localized correctly to the sides of the cells overexpressing Rga6 (Supplemental Figure S2B).
FIGURE 3:
Rga6 overexpression alters cell morphology and is detrimental for cell growth. (A) DIC (top) and fluorescence images from Calcofluor-stained cells (bottom) expressing pREP1-rga4+, pREP1-rga6+, and pREP1-rga6R354G. Cells grown for 18 h in the absence of thiamine. Bar, 5 μm. (B) Localization of active Cdc42, using CRIB-GFP as a probe, in the same cells grown for 22 h without thiamine. Bar, 5 μm. (C) Growth in solid medium of wild-type cells transformed with the pREP1 empty or carrying rga4+, rga6+, or rga6R354G as inserts. Exponentially growing cells were spotted at OD600 = 4 and serial 1:4 dilutions in EMM with (+T) or without thiamine and incubated at 28ºC for 3 d. (D) DIC images from cells expressing rga4+ (left) and rga6+ (right) integrated at the leu1+ locus under the control of nmt1 promoter and grown at 28°C for 30 h in EMM without thiamine. Arrows indicate the presence of swollen and branched cells.
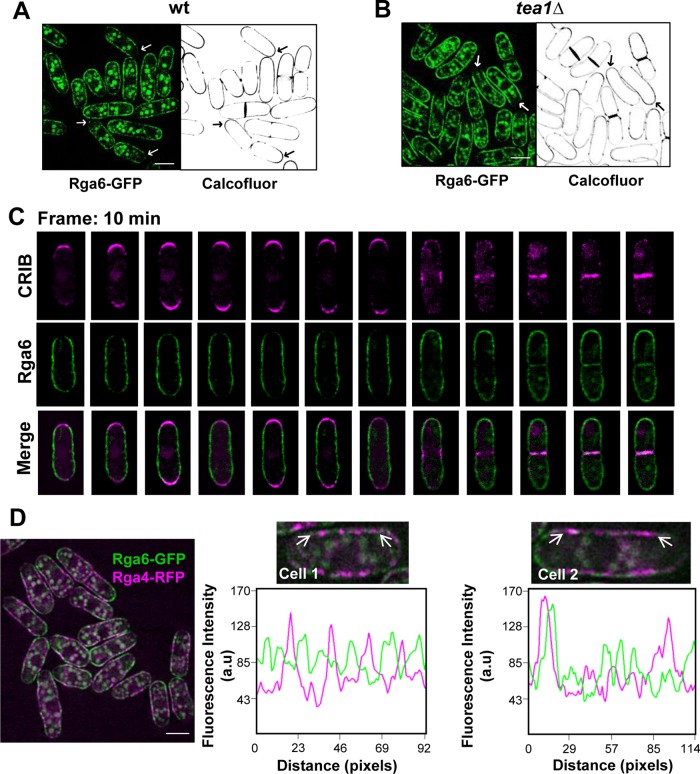
Rga6 preferentially localizes to nongrowing tips and lateral areas of the plasma membrane
Rga6 cellular distribution was analyzed by using cells carrying GFP-tagged Rga6 expressed under the control of its own promoter. Cells carrying either rga6+-GFP or GFP-rga6+ were morphologically normal, suggesting that both fusion proteins were functional. The GFP-fluorescence pattern observed in these cells indicated that Rga6 formed cortical clusters along the plasma membrane and these clusters were reduced at the growing tips, as shown by observation of Rga6-GFP in Calcofluor-stained cells (Figure 4A). We also visualized Rga6-GFP in monopolar growing mutants such as tea1Δ (Mata and Nurse, 1997), corroborating that Rga6 clusters diminished at the growing tip (Figure 4B). Moreover, overexpression of GFP-labeled Rga6 showed that this GAP localized to the membrane in a uniform pattern, since protein clusters at the membrane were no longer distinguished, but a reduction of Rga6 fluorescence at the thin growing tips was still observed (Supplemental Figure S2C). These results suggest that the phenotype caused by Rga6 overexpression was not due to ectopic localization of the protein.
FIGURE 4:
Rga6 preferentially localizes to nongrowing tips and lateral areas of the plasma membrane. Fluorescence images from cells expressing Rga6-GFP and stained with Calcofluor. Arrows show examples of growing tips with stronger Calcofluor signal and less abundant Rga6-GFP. (A) Wild type, (B) tea1Δ. (C) Time-lapse images (single focal planes) taken with 10-min interval from cells expressing GFP-Rga6 and CRIB-td-Tomato show Rga6 decrease at the growing areas. (D) Fluorescence microscopy from cells carrying integrated versions of Rga6-GFP and Rga4-RFP. Right, two enlarged cells, with their corresponding fluorescence line scans made along a region (between the two arrows) in the plasma membrane represented below to show that both Cdc42-GAPs localize to distinct nodes. Bar, 5 μm.
Time-lapse analysis performed in cells carrying GFP-Rga6 and CRIB-td-Tomato detailed the dynamics of Rga6 and showed a reduction of GFP fluorescence in one or both tips coincident with monopolar or bipolar cell growth. During early cytokinesis, Rga6 appeared in both poles and reduced its concentration in the center of the cell, unlike active Cdc42, which disappeared from the poles and concentrated in the division area. At the end of cytokinesis, Rga6 moved to the new membrane that ultimately formed the nongrowing tip upon cell separation. During this period, Rga6 and CRIB-td-Tomato were detected in the same area (Figure 4C).
Rga4 also localizes to the nongrowing lateral plasma membrane, where it forms clusters and is excluded from the growing areas (Das et al., 2007; Tatebe et al., 2008). To see whether both GAPs, Rga4 and Rga6, were in the same membrane complexes, we used cells carrying Rga6-GFP and Rga4-RFP. Colocalization of the clusters formed by the two Cdc42 GAPs was rarely observed (Figure 4D). Therefore Rga4 and Rga6 form part of different complexes in the membrane.
The Rga6 polybasic C-terminal region is necessary and sufficient for membrane localization
Rga6 primary sequence includes 733 residues and does not have any defined domain except for the RhoGAP (amino acid residues 329–547). There is a serine-rich region (amino acid residues 187–253) before the GAP domain, and a polybasic region (amino acid residues 700–733) at the protein C-terminus. To analyze the function of the different Rga6 regions in the membrane localization of Rga6, we made several truncations of the protein that were tagged with GFP at the C-terminus (Figure 5A). Full-length Rga6 and the different truncations were introduced by integrative transformation into the leu1+ locus of rga6Δ strain, and Western blot analysis of the transformants cells showed that the protein levels of all the truncations were considerably higher than the level of the full protein (Figure 5B). The constructs expressing Rga6-∆N1-GFP and Rga6-∆N2-GFP, lacking 186 and 259 N-terminal amino acid residues, respectively, reached the membrane and were able to suppress the morphology defects caused by rga6Δ (Figure 5, C and D). Of interest, we observed a reduction of Rga6-∆N1-GFP fluorescence at the growing tip but not as pronounced as in Rga6-GFP, and we observed no reduction in cells with Rga6-∆N2-GFP (Figure 5C). These results suggest that the N-terminal part of the protein is necessary for the modulation of Rga6 concentration at the growing area. Because Rga6-∆N2-GFP lacks the serine-rich region (SRs), we made a strain carrying Rga6∆SR-GFP in which only the SR was deleted. We still observed a reduction of Rga6∆SR-GFP fluorescence at the growing tip (unpublished data). In summary, different parts of the Rga6 N-terminus (amino acid residues 1–259), including the SR region, are required for decreased concentration of Rga6 at the growing tips.
FIGURE 5:
The Rga6 polybasic C-terminal region is necessary for its membrane localization. (A) Schematic representation of several Rga6 protein truncations. (B) Expression levels of wild-type Rga6-GFP and the truncations shown in A tagged with GFP at the C-terminal region, determined by Western blot. Actin levels were used as loading control (bottom). (C) Localization of the Rga6-GFP and the GFP-tagged protein truncated versions. The exposition time of the Rga6-GFP picture is two times longer than the pictures corresponding to Rga6-GFP truncations to allow a better comparison of the localization. Bar, 5 μm. (D) Box plots representing the cellular dimensions (length, left; and width, right) of strains carrying the indicated truncated Rga6 versions compared with the wild-type and rga6Δ cells. The black dot in each box represents the median, the upper and lower limits of the box represent the 25th and 75th percentiles, respectively, and the whiskers represent the minimum and maximum values. ***p < 0.0001; n.s., not significant difference relative to wild type; Student’s t test.
Rga6ΔC1-GFP, which lacks the 33 amino acid residues at the C-terminus of the protein, was cytoplasmic, and cells carrying this truncation have phenotypes similar to rga6Δ cells (Figure 5, C and D). Thus the polybasic C-terminal region was necessary for protein localization to the membrane. We also fused GFP to the polybasic C-terminal region and observed that it was sufficient to localize it to the membrane (Supplemental Figure S3A).
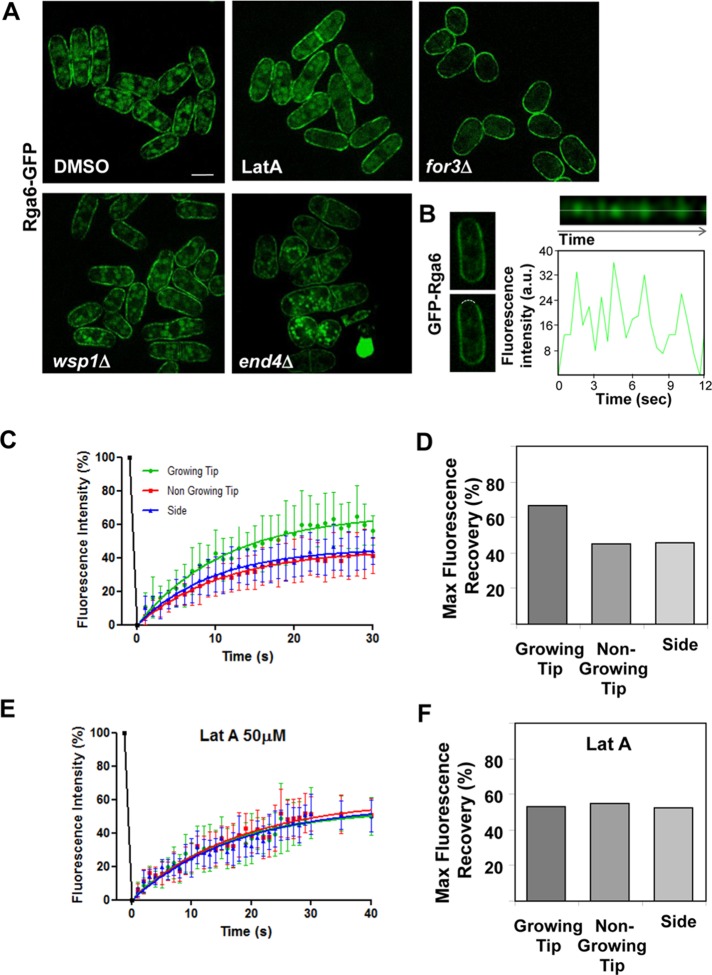
Rga6 reduced concentration at growing ends requires actin cables
Rga6 localization was independent of microtubules, since depolymerization of tubulin with carbendazim (MBC) did not cause a change in Rga6 localization (Supplemental Figure S3B). We also examined whether Rga6 localization depended on actin polymerization, using 50 μM latrunculin A (Lat A), which causes actin disassembly. In these conditions, Rga6 was still localized to the membrane of the cells, but the clusters appeared uniformly extended, and there was no Rga6 reduction at the growing tips after 15 min of Lat A treatment (Figure 6A). These results indicate that actin polymerization is required for the decrease of Rga6 at the growing tip. S. pombe actin forms cables and patches during interphase, which participate in polarized secretion and endocytosis, respectively (Mishra et al., 2014). For3 is the only S. pombe formin that makes actin cables during interphase (Feierbach and Chang, 2001). Analysis of Rga6-GFP in for3Δ cells showed clusters of Rga6 extended uniformly along the plasma membrane (Figure 6A). Actin patches are formed by many branched actin filaments nucleated by the Arp2/3 complex (Gachet and Hyams, 2005). Wsp1 is a nucleation-promoting factor that stimulates Arp2/3 complex (Lee et al., 2000), and End4/Sla2 is an adaptor protein that couples actin patches to the membrane in endocytic sites (Iwaki et al., 2004; Skruzny et al., 2012). In both wsp1Δ and end4Δ mutant strains, which have endocytosis defects, Rga6-GFP clusters localized along the plasma membrane but were reduced at the growing tips, as occurs in wild-type cells (Figure 6A). Therefore the reduced Rga6 concentration at the growing tips is likely due to polarized secretion mediated by For3 nucleated actin cables and not by endocytosis. Corroborating these results, the effect of Rga6 overexpression in for3Δ cells was different from that in wild-type cells and made for3Δ cells rounded. On the other hand, Rga4 localization to the lateral membranes is not dependent on actin cables (Das et al., 2007; Tatebe et al., 2008), and rga4+ overexpression was still able to promote polarized growth in for3Δ cells (Supplemental Figure S3C).
FIGURE 6:
Rga6 protein is highly dynamic, and its reduction at the growing tips is mediated by actin cables. (A) Rga6-GFP localization in wild-type cells treated with DMSO or 50 μM Lat A during 15 min, in for3Δ, wsp1Δ, and end4Δ mutant cells. Bar, 5 μm. (B) Kymograph (top) of a region (8 pixels) of the growing tip from a single cell expressing GFP-Rga6 (left) showing the fluorescence fluctuations over time. This cell was selected from the time-lapse sequence of images taken with a 0.5-s interval (Supplemental Video S1). The linescan below the image represents the total fluorescence levels along the time in the central region of the kymograph shown above (dotted line). (C) Fluorescence recovery curves of GFP-Rga6 after photobleaching of different regions (growing or nongrowing tip and side) at the plasma membrane. Best-fit curves derived from the mean intensity values of 22 cells. (D) Maximal fluorescence recovery represented as percentage of the original fluorescence from confocal FRAP measurements of GFP-Rga6 at different plasma membrane regions. (E) Fluorescence recovery curves of GFP-Rga6 treated with Lat A 50 μM for 30 min before photobleaching experiments. Best-fit curves derived from the mean intensity values of nine cells (Prism software). (F) Maximal fluorescence recovery represented as percentage of the original fluorescence from confocal FRAP measurements of GFP-Rga6 cells treated with Lat A.
High-speed time-lapse experiments on cells expressing GFP-Rga6 showed that this protein is very dynamic along the plasma membrane and seems to move by lateral diffusion (Supplemental Video S1). The fluorescence intensity of GFP-Rga6 at the growing tip fluctuates rapidly (Figure 6B), suggesting high mobility and unsteady localization of the protein in this area. To analyze in detail GFP-Rga6 mobility at the plasma membrane, we performed fluorescence recovery after photobleaching (FRAP) experiments (Figure 6C). An identical 3.5 μm2 circular region was bleached on one of the cell tips or the cell side. Of interest, the fluorescence recovery rate was similar at both growing and nongrowing tips, with recovery halftimes of 7.8 and 7.6 s (R-square = 0.98 and 0.99, respectively) and slightly faster at the cell sides, with recovery halftimes of 6.6 s (R-square = 0.98). On the other hand, there was a higher recovery of the initial fluorescence at the growing tip than at the nongrowing tip or the cell sides (Figure 6D). These results indicate the existence of a larger mobile fraction of Rga6 in the growing area. We also performed FRAP after treatment of the cells with 50 μM Lat A for 30 min (Figure 6, E and F). As mentioned before, in these conditions, there is no decrease of Rga6 clusters at the growing tips. The rate and level of fluorescence recovery decreased, and the recovery halftimes were similar in both the tips and the cell sides (11.6, 12.1, and 12.4 s, respectively; R-square = 0.94, 0.95, and 0.94), suggesting that polymerized actin not only mediated the decrease of Rga6 concentration at the growing tip, but it also increased Rga6 exchangeable fraction at the tip.
Photobleaching of GFP-Rga6 over the entire cell ends caused a significant fluorescence recovery decrease compared with the recovery observed when a smaller region of the end was photobleached (Supplemental Figure S4, A and B). In addition, photobleaching at two distant lateral areas resulted in partial fluorescence recovery at those zones, but the whole GFP-Rga6 fluorescence at the membrane decreased (Supplemental Figure S4C). Finally, photobleaching of GFP-Rga6 over contiguous zones at the cell sides and the cell tip caused almost complete absence of fluorescence recovery at the tip (Supplemental Figure S4D). Together these observations suggest a major contribution of membrane lateral diffusion to Rga6 dynamics.
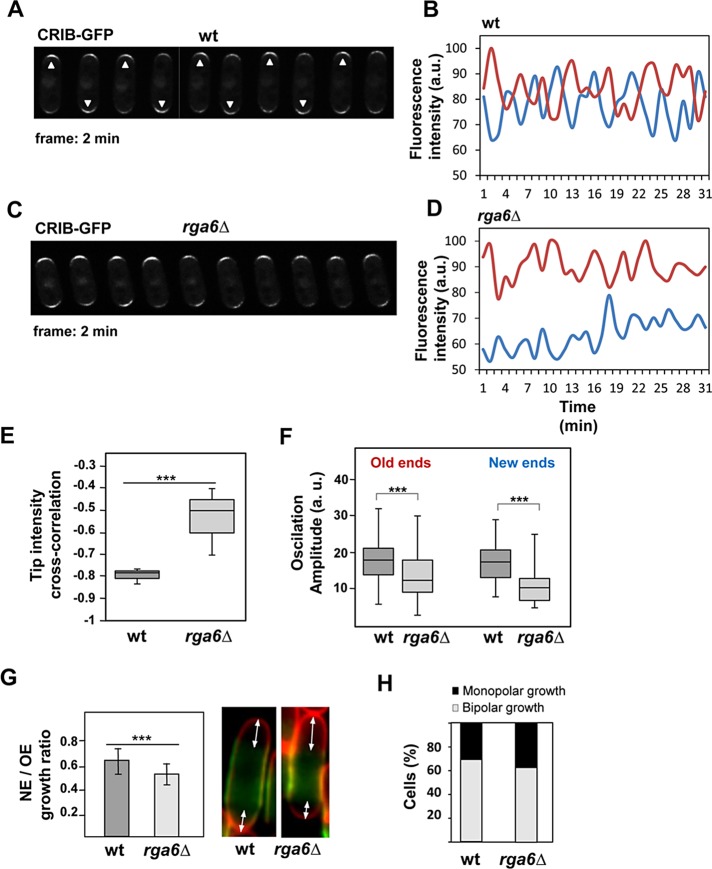
Rga6 elimination decreases the amplitude of active Cdc42 oscillations at cell tips and Cdc42-GTP concentration at the new end
Fission yeast active Cdc42, as detected with CRIB-GFP, displays oscillatory dynamics anticorrelated at the two cell tips. Oscillations indicate the presence of positive and negative feedback regulations, and the anticorrelation indicates competition for active Cdc42 or its regulators (Das et al., 2012). Rga4 is localized at the cell sides and does not reach the growing tip (Tatebe et al., 2008; Das et al., 2009). Thus it has a major role in the control of cell diameter, but it is unlikely that Rga4 regulates Cdc42 dynamics at the cell tips. Indeed, rga4Δ mutant cells display normal, mostly symmetrical oscillations (Das et al., 2012). In contrast, Rga6 reaches the growing cells tips and might participate in the negative regulation needed for the active Cdc42 oscillatory dynamics. To test this hypothesis, we analyzed CRIB-GFP oscillations in wild-type and rga6Δ cells. CRIB-GFP oscillated with a period of 4 min at the tips of wild-type cells (Figure 7, A and B) in an anticorrelated manner between the two cell tips (cross-correlation coefficient r = −0.78 ± 0.03), consistent with previous reports (Das et al., 2012; Scheffler et al., 2014). In cells without Rga6 (Figure 7, C and D), CRIB-GFP still oscillated with a periodicity similar to that of wild-type cells, but the anticorrelation between the tips was considerably reduced (cross-correlation coefficient r = −0.54 ± 0.11, p < 0.0001; Figure 7E), and the amplitude of the oscillations decreased significantly, mainly at the new end (Figure 7F). In addition, the intensity of CRIB-GFP fluorescence was always substantially lower at the new ends of rga6Δ cells (Figure 7, C and D). The cell growth at both ends was measured in bipolar wild- type and rga6Δ cells (100 each; see Materials and Methods), and a marked reduction of the growth at the new end was found in cells lacking Rga6 compared with wild-type cells (Figure 7G). We also detected a mild increase in monopolar cells in rga6Δ cultures with respect to wild- type cultures (38 and 31% monopolar cells, respectively; 240 cells of each type; Figure 7H).
FIGURE 7:
The amplitude of Cdc42-GTP oscillations at the cell tips decreases in cells lacking rga6+. (A, C) Time-lapse fluorescence images at 2-min intervals of wild-type (A) and rga6Δ (C) bipolar cells carrying CRIB-GFP. Arrowheads in A point to the brightest cell end in each time-point image. (B, D) Quantification of CRIB-GFP fluorescence intensity at the old (red) and new (blue) ends from cell images of wild-type (B) and rga6Δ (D) bipolar cells taken at 1-min intervals. a.u., arbitrary units. (E) Box plot showing the cross-correlation of CRIB-GFP tip intensities in wild-type (n = 10) and rga6Δ cells (n = 10; p < 0.0001; Student’s t test). (F) Box plot representing the quantification of the oscillation amplitude of CRIB-GFP fluorescence at the old and new ends in wild-type cells (n = 50) and rga6Δ mutant cells (n = 65; p < 0.0005 for the old ends and p < 0.0001 for the new ends; Student’s t test). In the box plots in E and F, the medial horizontal line represents the median, the upper and lower limits of the box represent the 25th and 75th percentiles, respectively, and the whiskers represent the minimum and maximum values. (G) Ratio of new end (NE) growth vs. old end (OE) growth in wild-type and rga6Δ bipolar cells as determined in Calcofluor-stained cells previously treated with FITC-lectin and cultured for 150 min after having been washed. Right, two example cells with Calcofluor in red and lectin in green. Arrows mark the distance measurements performed in 100 cells from each type. (H) Percentage of monopolar and bipolar cells in an asynchronous exponentially growing culture at 28°C of wild-type and rga6Δ cells (240 for each type). Cells with septa were not counted.
The proposed model of Cdc42-GTP dynamics predicts a lack of a symmetric attractor for cells with reduced negative feedback (Das et al., 2012). Cells lacking Rga6 behave like cells having reduced negative regulation of Cdc42 at the tip, with reduced Cdc42 fluctuations, increased old, tip-bound GTP-Cdc42, and decreased active Cdc42 symmetry. Taken together, these results suggest that Rga6 is part of the negative regulation of Cdc42 at the growing cell tip.
Rga6 function depends on the Cdc42 GEF Scd1
Rga4 and the Scd1 GEF control Cdc42 activity independently. Thus scd1Δ and rga4Δ have additive effects, and double scd1Δ rga4Δ cells are round and nonpolarized (Kelly and Nurse, 2011). To see whether Rga6 was also independent of Scd1, we performed a similar genetic analysis with the mutant cells grown in 12 mM hydroxyurea (HU) for 5 h to better appreciate the morphology changes. Whereas scd1Δ rga4Δ cells remained round when incubated in HU, scd1Δ rga6Δ cells slightly elongated like the scd1Δ parental strain (Figure 8A). In addition, overexpression of rga6+did not change the morphology of scd1Δ cells, whereas overexpression of rga4+ caused the appearance of some pointy growing ends in the scd1Δ cells (Figure 8B). These results suggest that Rga6 negative regulation of Cdc42 depends on the Scd1 signaling pathway, and therefore Rga6 levels do not affect cell dimensions when Scd1 is absent. Alternatively, these results could be explained if Rga6 has a minor role in morphology and the effects of deletion or overexpression of this GAP cannot overcome the effect of scd1 deletion, which already displays a loss of polarity. We also observed that Rga6-GFP fluorescence was extended around the membrane in scd1Δ cells, with no decrease of the fluorescence at the growing pole even in cells maintained for 5 h in HU to allow polarization of scd1Δ cells (Figure 8C). Therefore Scd1 is necessary for the reduction of Rga6 concentration at the growing tip. In contrast, Rga4 remains localized to the cell sides of scd1Δ cells (Kelly and Nurse, 2011).
FIGURE 8:
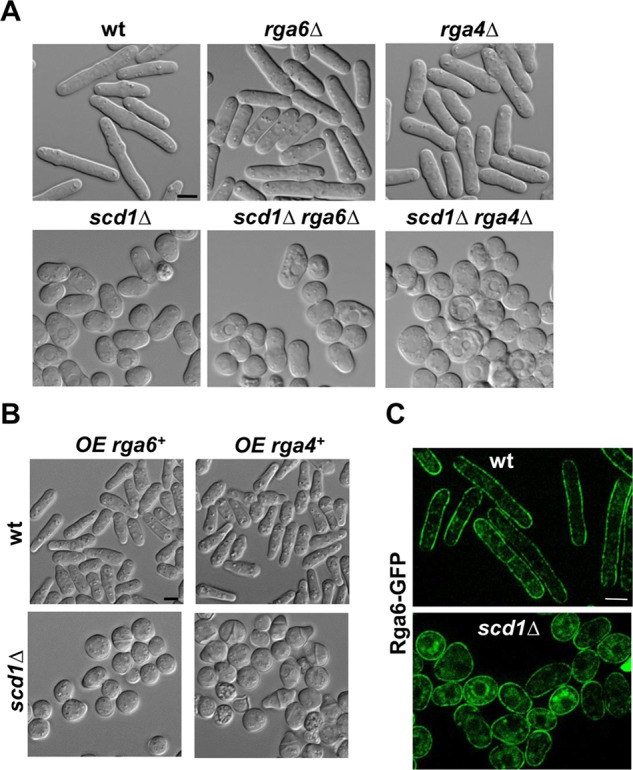
Rga6 effect on cell morphology depends on the Cdc42 GEF Scd1. (A) DIC images from wild-type, rga6Δ, rga4Δ, scd1Δ, scd1Δ rga6Δ, and scd1Δ rga4Δ cells treated with 12 mM HU for 5 h. (B) DIC images from wild-type (top) and scd1Δ mutant (bottom) strains expressing rga6+ (left) or rga4+ (right) from the nmt1 promoter. Cells grown in EMM without thiamine for 22 h at 28°C. (C) Rga6-GFP localization in wild-type and scd1Δ cells treated with 12 mM HU for 5 h. Bar, 5 μm.
DISCUSSION
Cdc42 GTPase localizes to all cell membranes (Estravis et al., 2011; Bendezu et al., 2015). However, active Cdc42, as detected using CRIB-GFP, is restricted to the growing areas (Tatebe et al., 2008). Rga4 was the only Cdc42-GAP described so far, and it seems to be restricted to the cell sides, preventing Cdc42 activation there. In this report, we provided genetic and molecular evidence for the functional interaction between the fission yeast Rga6, a member of the Rho-GAP protein family, and the GTPase Cdc42. Attempts to find the S. pombe orthologues of Saccharomyces cerevisiae GAPs have been made based on the primary structures of the proteins and their GAP domains (Nakano et al., 2001). Rga6 has been paired to Bem3 and Sac7. However, although the primary structure might reflect some conservation among the protein motifs, conclusions about functional conservation of the GAPs in these two evolutionarily distant yeasts cannot be drawn because most GAPs display activity toward multiple Rho proteins, and different GAPs might regulate the same Rho protein acting in different signaling pathways (Tcherkezian and Lamarche-Vane, 2007). The biochemical data demonstrate that Rga6 modulates the level of GTP-bound Cdc42 and Rho2 in vivo. However, the morphological alterations caused by rga6+ deletion occur even in rho2Δ cells, suggesting that this GTPase does not participate in the Rga6 regulation of S. pombe dimensions and that changes in active Cdc42 cause the morphological alterations in these cells. Of interest, Rga4, which causes similar but stronger phenotypes than Rga6 when eliminated or overproduced, is also a GAP for both Cdc42 and Rho2 GTPases (Tatebe et al., 2008; Soto et al., 2010).
We show here that Rga6 collaborates with Rga4 in the regulation of cell dimensions mediated by Cdc42. Both GAPs preferentially localize as small clusters in the nongrowing areas of the plasma membrane, but they are not in the same complexes. Rga6 reaches the growing tip, and the regulation of its localization and function is different from that of Rga4. The polybasic C-terminal region of Rga6 is necessary and sufficient for its localization to the membrane in a uniform manner, and membrane localization is required for the function of Rga6. A polybasic region is also required for the membrane binding of Rga4 (Tatebe et al., 2008). However, in Rga4, as in most GAPs having a polybasic region required for localization, this region precedes the GAP domain (Karimzadeh et al., 2012), whereas in Rga6, it is located at the C-terminus. The N-terminal half of the protein does not seem to play a major role in Rga6 function but is required for the decreased concentration of Rga6 at the growing tips. Furthermore, we observed an increase in the protein levels of the different Rga6 truncations compared with the entire Rga6. It is tempting to propose that Rga6, once it reaches the membrane, is regulated by degradation, and the N-terminal region of the protein is necessary for this degradation.
It was recently suggested that an actin-independent system may assemble polarity domains with a minimal size, and then the fusion of secretory vesicles transported along actin cables may extend these domains to scale them to the tip curvature (Abenza et al., 2015; Bonazzi et al., 2015). In agreement with this hypothesis, our data show here that actin cables are required for the decrease in Rga6 concentration at the growing tip, and this in turn might help to extend active Cdc42 domains. Indeed, rga6+ overexpression decreases the width of the polarized growth area in wild-type cells, whereas in for3Δ cells, it causes complete loss of polarity, and the cells become round. In contrast, rga4+ overexpression was still able to promote polarized growth in for3Δ cells. Probably this difference is due to the fact that For3 is required for Rga6 reduction at the growing tip, and Rga6, but not Rga4, is spread through all of the plasma membrane in for3Δ cells. A possible explanation for these results is that actin cables made by For3 are necessary for the efficient transport to the tip of some molecules, such as Scd1, that regulate the decrease of Rga6 at the tip.
The analysis of GTP-bound Cdc42 in rga6Δ cells revealed the physiological relevance of the Rga6-mediated regulation of Cdc42 activity. According to the proposed model of Cdc42-GTP dynamics (Das et al., 2015), growth at the new end is due to GTP-Cdc42 redistribution, and increased amounts of active Cdc42 could overcome the competition at the cell tips and give rise to more symmetrical GTP-Cdc42 concentration and more bipolar cells (Das et al., 2015). However, the model also predicts that decreased negative feedback leads to the accumulation of most Cdc42 activity at one single tip and will cause monopolar growth (Das et al., 2012). It is possible that Rga6 participates in the negative feedback regulation of Cdc42 at the tip, and thus the lack of Rga6 dampens Cdc42 fluctuations. As a consequence, lack of Rga6 increases old tip-bound GTP-Cdc42 and decreases active Cdc42 symmetry. The effect of Rga6 deletion on Cdc42 dynamics is not strong enough to cause a severe monopolar growth, as does the lack of Pak1 activity (Das et al., 2012), but its deletion can cause the observed slightly wider diameter and decreased growth at the new end.
Rga4 regulation of Cdc42 is independent of Scd1 (Kelly and Nurse, 2011). In contrast, Rga6 decrease at the growing tips is Scd1 dependent. In addition, Rga6 deletion or overexpression had no effect in the absence of Scd1. These results support that Rga6 negative regulation of Cdc42 might participate in the negative feedback regulation that is mainly mediated by the Scd1-Scd2-Pak1 complex (Das et al., 2012).
In summary, our results demonstrate that Rga6 is a Cdc42-GAP that spatially regulates the Cdc42 activity in collaboration with Rga4 and it contributes to the maintenance of the polarized growth and cell shape.
MATERIALS AND METHODS
Strains, growth conditions, and genetic methods
Standard S. pombe media and genetic manipulations were used as described (Moreno et al., 1991). All of the strains used were isogenic to wild-type strains 972 h− and 975 h+, and they are described in Supplemental Table S1. The strains were constructed by either tetrad dissection or random spore germination. Cells were grown in rich medium (YES) or minimal medium (EMM) supplemented with the necessary requirements. Escherichia coli DH5α was used as host for propagation of plasmids. Cells were grown in LB medium supplemented with 50 μg/ml ampicillin when appropriate. Solid media contained 2% agar.
Recombinant DNA methods
All DNA manipulations were carried out by established methods. Enzymes were used according to the recommendations of the suppliers. S. pombe was transformed by the lithium acetate method (Ito et al., 1983). The nmt1+ promoter–containing vector pREP1 (Forsburg and Sherman, 1997) was used for the overexpression of rga6+, which was induced by growing the cells transformed with this plasmid in the absence of thiamine for at least 16 h. For overexpression of the GAPs during longer times, the open reading frame (ORF) of rga4+, rga6+, or GFP-rga6+ under control of the nmt1 promoter was integrated at the leu1+ locus using the pJK148 vector.
The deletion of rga6+ was generated by PCR-base gene targeting as described (Bähler et al., 1998). Genomic versions of rga6+tagged with GFP fused to the 5′ or 3′ end of the ORF were generated by cloning the corresponding ORF and the GFP sequence, plus 500 base pairs of the 5′ and 3′ flanking sequences into a KS BlueScript. The 5′ and 3′ flanking sequences, as well as the ORF, were amplified from the fission yeast genomic DNA using the appropriate primers. Truncated rga6ΔN or rga6ΔC was generated by amplifying and cloning the corresponding ORF fragment into a KS Bluescript plasmid carrying the enhanced GFP sequence in-frame and the 5′ and 3′ flanking rga6+ sequences. All of the previous resulting constructs were cloned into the integrative vector pJK148, which was then cut with Tth11I and integrated at the leu1+ locus of the leu1-32 ura4-D18 rga6Δ strain.
The rga6 GAP-dead mutant was generated by PCR site-directed mutagenesis using the appropriate primers and a KS Bluescript plasmid containing the rga6+ ORF as template. Substitution of the arginine residue 354 for glycine was confirmed by DNA sequencing.
For the two-hybrid analysis, the rga6+ ORF was cloned into the NdeI-XmaI sites of pAS2. The ORFs of the Rho GTPases coding genes, excluding the 3′ region that codes for the prenylatable domain, were cloned into the NcoI-XmaI sites of pACT2. S. cerevisiae Y190 (MATa gal4 gal80 his3 trp1-901 ura3-52 leu2-3,-112 URA3::GAL–<lacZ, lys2::GAL (UAS)–<HIS3 cyhr) cells were transformed and grown on plates without leucine, tryptophan, and histidine and supplemented with 40 mM 3-aminotriazole. β-Galactosidase activity was analyzed in the transformant colonies as described (Coll et al., 2003).
Pull down of GTP-bound Cdc42 and Rho2
GTP-bound Rho proteins were analyzed as described (Calonge et al., 2003) using a Rho-GTP pull-down assay. Briefly, cell extracts from wild-type or rga6Δ cells expressing HA-cdc42+ or HA-rho2+ from their own promoter were obtained by mechanical breakage of the cells using glass beads and FastPrep. Cells were resuspended in 500 μl of lysis buffer (50 mM Tris-HCl, pH 7.5, 20 mM NaCl, 0.5% NP-40, 10% glycerol; 0,1 mM dithiothreitol, 1 mM NaF, and 2 mM MgCl2 containing 100 μM p-aminophenyl methanesulfonyl fluoride, leupeptin, and aprotinin). A 10-μg amount of either GST-CRIB or GST-RBD, previously obtained from E. coli DNA expression, purified, and coupled to glutathione–Sepharose beads, was used to precipitate the GTP-bound Rho GTPases from 2 mg of the total cell lysates. The extracts were incubated with the beads for 2 h at 4°C, washed four times, and blotted against anti-HA high-affinity monoclonal antibody (Clone 3F10; Roche Molecular Biochemicals, Mannheim, Germany) to detect the corresponding GTP-bound HA-Rho protein. The total amounts of Rho proteins from the extracts were determined by Western blot using the anti-HA monoclonal antibody.
Detection of total expression levels of Rga6 truncations
Cell extracts from exponentially growing cells were obtained using 20% trichloroacetic acid, acid-washed glass beads (G8772; Sigma-Aldrich, St. Louis, MO), and a FastPrep FP120 (Bio 101; Savant Instruments Ltd., Hyderabad, India). After centrifugation, the pellet was neutralized with 1 M Tris, pH 9, and resuspended in 2× Laemmli buffer. Equal amounts of total protein were resolved in SDS–PAGE gels and transferred to Hybond-ECL membranes. The different Rga6 GFP-tagged truncations were detected using an anti-GFP monoclonal antibody (Living Colors JL-8; Clontech laboratories, Mountain View, CA) at 1:3000 dilution. Actin was used as loading control and detected with anti-actin monoclonal antibody (C-4 clone; Millipore, Darmstadt, Germany) at 1:10,000 dilution.
Microscopy techniques and image analysis
Fluorescence images were captured on an inverted microscope (model IX71; Olympus America, Center Valley, PA) equipped with a PlanApo 100×/1.40 IX70 objective and a Personal DeltaVision system (Applied Precision). Images were captured using a CoolSnap HQ2 monochrome camera (Photometrics, Tucson, AZ) and softWoRx 5.5.0 imaging software (Applied Precision). Images were then corrected by three-dimensional deconvolution (conservative ratio, 10 iterations, and medium noise filtering) using softWoRx imaging software. ImageJ (National Institutes of Health, Bethesda, MD) was using for image processing. For Calcofluor staining of S. pombe cell wall and septum, exponentially growing cells were harvested, resuspended in medium with Calcofluor (0.1 mg/ml), and observed in a microscope with the corresponding ultraviolet filter.
For time-lapse imaging, 0.3 ml of log-phase cell cultures was placed in a well from a μ-Slide eight-well (Ibidi, Planegg/Martinsried, Germany) previously coated with 10 μl of 1 mg/ml soybean lectin (Sigma-Aldrich). Cells were left 1 min to attach to the well bottom, and culture medium was removed carefully. Then, cells were washed three times with the same medium, and finally 0.3 ml of fresh medium was added (Cortés et al., 2012). Experiments were made at 25 or 28ºC, and single middle planes were taken at the time points stated in each figure.
High-speed imaging was done on a spinning disk confocal microscope (Olympus IX81; with a spinning module) equipped with a 100×/1.40 PlanApo oil immersion objective, on the stream time acquisition mode, controlled by MetaMorph 7.7 software. Kymographs of the growing tip of cells expressing GFP-Rga6 and subsequent linescan analysis to quantify fluorescence intensities were made with MetaMorph 7.7 software.
FRAP experiments were performed on a spinning-disk confocal microscope. A 3.5-μm2 circular region of the cell tips or the cell side was bleached with a sequence of three high-intensity laser iterations after two prebleach acquisitions. Postbleaching images were taken every 1 s over a period of 30 s. In the Lat A–treated cells, the postbleaching acquisition was 10 s longer in order to reach the fluorescence recovery “plateau,” and images were taken every 5 s during this extra period.
For FRAP analysis, intensities of bleached regions during recovery were corrected for out-of-cell background intensity. In addition, bleaching due to subsequent imaging was corrected by normalizing the intensities at each time point to the average intensity at the unbleached cell. These intensities were then normalized with the first postbleaching time point corresponding to 0% and the prebleaching time point corresponding to 100%. Best-fit curves derived from the mean intensity-normalized values of n cells were created using GraphPad Prism software, and the half-times, mobile fractions, and R-square values were obtained from these curves. Student’s t test was applied to determine the statistical significance of the differences between the data.
To analyze active Cdc42 oscillations at the tips, time-lapse experiments were performed with wild-type and rga6Δ cells expressing CRIB-GFP. Cells were placed in μ-Slide eight-wells coated with soybean lectin as described and imaged every 1 min over a 45-min period using a spinning-disk confocal microscope. Bipolar cells were selected (according to cell length and tip shape), and the average CRIB-GFP intensity at the tips was measured using ImageJ software. Intensities from both tips were normalized to the maximum intensity and plotted against the time. Period and amplitude between oscillations were calculated from the plots, the period being the time between two subsequent maxima of intensity, and the amplitude being the difference between maximum and minimum intensities within the oscillation.
Other methods
Fluorescein isothiocyanate (FITC)–conjugated Bandeiraea simplicifolia lectin was used to visualize and measure growth zones. Briefly, cells were grown in EMM at 28ºC, resuspended in EMM plus 5 μg/ ml FITC-lectin (L-9381; Sigma-Aldrich), and stained in the dark for 15 min at the same temperature; then they were washed twice in EMM and incubated further in EMM in the dark for 150 min. Cells were then counterstained with Calcofluor and observed.
Plate assays for growth were performed by spotting the appropriate dilutions of log phase–growing cells on solid EMM with or without thiamine containing 2% (wt/vol) bacto-agar. Incubation was done at 28ºC. All experiments were repeated at least three times.
Supplementary Material
Acknowledgments
We thank S. Rincón, J. Encinar, B. Santos, and R. R. Daga for useful comments. We also thank D. M. Posner for language revision of the manuscript. We are very grateful to K. Nakano, K. Shiozaki, T. Toda, R. Daga, and T. Pollard for generous gifts of strains and plasmids. This work was supported by Grants BFU2013-43439-P and BIO2015-69958-P from the Ministry of Economy and Competitiveness, Spain, and Grant CSI037U14 from Junta de Castilla y León, Spain.
Abbreviations used:
- CRIB
Cdc42/Rac interactive binding
- FRAP
fluorescence recovery after photobleaching
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- Lat A
latrunculin A
- NDR
nuclear dbf2-related
- PBR
polybasic region
- RBD
Rho-binding domain
- RFP
red fluorescent protein
- SR
serine-rich
- tdTomato
tandem dimer Tomato
- wt
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-12-0818) on March 9, 2016.
REFERENCES
- Abenza JF, Couturier E, Dodgson J, Dickmann J, Chessel A, Dumais J, Salas RE. Wall mechanics and exocytosis define the shape of growth domains in fission yeast. Nat Commun. 2015;6:8400. doi: 10.1038/ncomms9400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–1370. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bendezu FO, Martin SG. Actin cables and the exocyst form two independent morphogenesis pathways in the fission yeast. Mol Biol Cell. 2011;22:44–53. doi: 10.1091/mbc.E10-08-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, Vincenzetti V, Vavylonis D, Wyss R, Vogel H, Martin SG. Spontaneous Cdc42 polarization independent of GDI-mediated extraction and actin-based trafficking. PLoS Biol. 2015;13:e1002097. doi: 10.1371/journal.pbio.1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi D, Haupt A, Tanimoto H, Delacour D, Salort D, Minc N. Actin-based transport adapts polarity domain size to local cellular curvature. Curr Biol. 2015;25:2677–2683. doi: 10.1016/j.cub.2015.08.046. [DOI] [PubMed] [Google Scholar]
- Bourne HR. G proteins. The arginine finger strikes again. Nature. 1997;389:673–674. doi: 10.1038/39470. [DOI] [PubMed] [Google Scholar]
- Calonge TM, Arellano M, Coll PM, Pérez P. Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol Microbiol. 2003;47:507–518. doi: 10.1046/j.1365-2958.2003.03312.x. [DOI] [PubMed] [Google Scholar]
- Chang EC, Barr M, Wang Y, Jung V, Xu HP, Wigler MH. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- Coll PM, Trillo Y, Ametzazurra A, Pérez P. Gef1p, a new guanine nucleotide exchange factor for Cdc42p, regulates polarity in Schizosaccharomyces pombe. Mol Biol Cell. 2003;14:313–323. doi: 10.1091/mbc.E02-07-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés JC, Sato M, Munoz J, Moreno MB, Clemente-Ramos JA, Ramos M, Okada H, Osumi M, Durán A, Ribas JC. Fission yeast Ags1 confers the essential septum strength needed for safe gradual cell abscission. J Cell Biol. 2012;198:637–656. doi: 10.1083/jcb.201202015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Drake T, Wiley DJ, Buchwald P, Vavylonis D, Verde F. Oscillatory dynamics of Cdc42 GTPase in the control of polarized growth. Science. 2012;337:239–243. doi: 10.1126/science.1218377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Nunez I, Rodriguez M, Wiley DJ, Rodriguez J, Sarkeshik A, Yates JR, 3rd, Buchwald P, Verde F. Phosphorylation-dependent inhibition of Cdc42 GEF Gef1 by 14-3-3 protein Rad24 spatially regulates Cdc42 GTPase activity and oscillatory dynamics during cell morphogenesis. Mol Biol Cell. 2015;26:3520–3534. doi: 10.1091/mbc.E15-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Wiley DJ, Chen X, Shah K, Verde F. The conserved NDR kinase Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42. Curr Biol. 2009;19:1314–1319. doi: 10.1016/j.cub.2009.06.057. [DOI] [PubMed] [Google Scholar]
- Das M, Wiley DJ, Medina S, Vincent HA, Larrea M, Oriolo A, Verde F. Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol Biol Cell. 2007;18:2090–2101. doi: 10.1091/mbc.E06-09-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shirouzu M, Yokoyama S. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J Biol Chem. 2003;278:843–852. doi: 10.1074/jbc.M209714200. [DOI] [PubMed] [Google Scholar]
- Estravis M, Rincón SA, Santos B, Pérez P. Cdc42 regulates multiple membrane traffic events in fission yeast. Traffic. 2011;12:1744–1758. doi: 10.1111/j.1600-0854.2011.01275.x. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S ( Cdc42—the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Sherman DA. General purpose tagging vectors for fission yeast. Gene. 1997;191:191–195. doi: 10.1016/s0378-1119(97)00058-9. [DOI] [PubMed] [Google Scholar]
- Gachet Y, Hyams JS. Endocytosis in fission yeast is spatially associated with the actin cytoskeleton during polarised cell growth and cytokinesis. J Cell Sci. 2005;118:4231–4242. doi: 10.1242/jcs.02530. [DOI] [PubMed] [Google Scholar]
- Hachet O, Berthelot-Grosjean M, Kokkoris K, Vincenzetti V, Moosbrugger J, Martin SG. A phosphorylation cycle shapes gradients of the DYRK family kinase Pom1 at the plasma membrane. Cell. 2011;145:1116–1128. doi: 10.1016/j.cell.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Harris SD. Cdc42/Rho GTPases in fungi: variations on a common theme. Mol Microbiol. 2011;79:1123–1127. doi: 10.1111/j.1365-2958.2010.07525.x. [DOI] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T, Tanaka N, Takagi H, Giga-Hama Y, Takegawa K. Characterization of end4+, a gene required for endocytosis in Schizosaccharomyces pombe. Yeast. 2004;21:867–881. doi: 10.1002/yea.1134. [DOI] [PubMed] [Google Scholar]
- Karimzadeh F, Primeau M, Mountassif D, Rouiller I, Lamarche-Vane N. A stretch of polybasic residues mediates Cdc42 GTPase-activating protein (CdGAP) binding to phosphatidylinositol 3,4,5-trisphosphate and regulates its GAP activity. J Biol Chem. 2012;287:19610–19621. doi: 10.1074/jbc.M112.344606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly FD, Nurse P. Spatial control of Cdc42 activation determines cell width in fission yeast. Mol Biol Cell. 2011;22:3801–3811. doi: 10.1091/mbc.E11-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkoris K, Gallo Castro D, Martin SG. The Tea4-PP1 landmark promotes local growth by dual Cdc42 GEF recruitment and GAP exclusion. J Cell Sci. 2014;127:2005–2016. doi: 10.1242/jcs.142174. [DOI] [PubMed] [Google Scholar]
- Lee WL, Bezanilla M, Pollard TD. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J Cell Biol. 2000;151:789–800. doi: 10.1083/jcb.151.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, McDonald WH, Yates JR, 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell. 2005;8:479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Martin SG, Rincón SA, Basu R, Pérez P, Chang F. Regulation of the formin for3p by cdc42p and bud6p. Mol Biol Cell. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast. Cell. 1997;89:939–950. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Miller PJ, Johnson DI. Cdc42p GTPase is involved in controlling polarized growth in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Huang J, Balasubramanian MK. The yeast actin cytoskeleton. FEMS Microbiol Rev. 2014;38:213–227. doi: 10.1111/1574-6976.12064. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murray JM, Johnson DI. The Cdc42p GTPase and its regulators Nrf1p and Scd1p are involved in endocytic trafficking in the fission yeast Schizosaccharomyces pombe. J Biol Chem. 2001;276:3004–3009. doi: 10.1074/jbc.M007389200. [DOI] [PubMed] [Google Scholar]
- Nakano K, Mutoh T, Mabuchi I. Characterization of GTPase-activating proteins for the function of the Rho-family small GTPases in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2001;6:1031–1042. doi: 10.1046/j.1365-2443.2001.00485.x. [DOI] [PubMed] [Google Scholar]
- Rincon S, Coll PM, Pérez P. Spatial regulation of Cdc42 during cytokinesis. Cell Cycle. 2007;6:1687–1691. doi: 10.4161/cc.6.14.4481. [DOI] [PubMed] [Google Scholar]
- Rincón SA, Ye Y, Villar-Tajadura MA, Santos B, Martin SG, Pérez P. Pob1 participates in the Cdc42 regulation of fission yeast actin cytoskeleton. Mol Biol Cell. 2009;20:4390–4399. doi: 10.1091/mbc.E09-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler K, Recouvreux P, Paoletti A, Tran PT. Oscillatory AAA+ ATPase Knk1 constitutes a novel morphogenetic pathway in fission yeast. Proc Natl Acad Sci USA. 2014;111:17899–17904. doi: 10.1073/pnas.1407226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzny M, Brach T, Ciuffa R, Rybina S, Wachsmuth M, Kaksonen M. Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc Natl Acad Sci USA. 2012;109:E2533–2542. doi: 10.1073/pnas.1207011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M, Peter M. Polarizing without a clue. Trends Cell Biol. 2003;13:526–533. doi: 10.1016/j.tcb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Soto T, Villar-Tajadura MA, Madrid M, Vicente J, Gacto M, Pérez P, Cansado J. Rga4 modulates the activity of the fission yeast cell integrity MAPK pathway by acting as a Rho2 GTPase-activating protein. J Biol Chem. 2010;285:11516–11525. doi: 10.1074/jbc.M109.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Nakano K, Maximo R, Shiozaki K. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr Biol. 2008;18:322–330. doi: 10.1016/j.cub.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Altschuler S, Wu L, Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- Wheatley E, Rittinger K. Interactions between Cdc42 and the scaffold protein Scd2: requirement of SH3 domains for GTPase binding. Biochem J. 2005;388:177–184. doi: 10.1042/BJ20041838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.