Whole-body and liver-specific ATF4–knockout mice are used to evaluate the role of ATF4 transcription factor in the unfolded protein response (UPR). ATF4 directs a small subset of UPR gene expression, and deletion of ATF4 in the liver triggers enhanced oxidative stress, disrupts cholesterol metabolism, and enhances cell death.
Abstract
Disturbances in protein folding and membrane compositions in the endoplasmic reticulum (ER) elicit the unfolded protein response (UPR). Each of three UPR sensory proteins—PERK (PEK/EIF2AK3), IRE1, and ATF6—is activated by ER stress. PERK phosphorylation of eIF2 represses global protein synthesis, lowering influx of nascent polypeptides into the stressed ER, coincident with preferential translation of ATF4 (CREB2). In cultured cells, ATF4 induces transcriptional expression of genes directed by the PERK arm of the UPR, including genes involved in amino acid metabolism, resistance to oxidative stress, and the proapoptotic transcription factor CHOP (GADD153/DDIT3). In this study, we characterize whole-body and tissue-specific ATF4-knockout mice and show in liver exposed to ER stress that ATF4 is not required for CHOP expression, but instead ATF6 is a primary inducer. RNA-Seq analysis indicates that ATF4 is responsible for a small portion of the PERK-dependent UPR genes and reveals a requirement for expression of ATF4 for expression of genes involved in oxidative stress response basally and cholesterol metabolism both basally and under stress. Consistent with this pattern of gene expression, loss of ATF4 resulted in enhanced oxidative damage, and increased free cholesterol in liver under stress accompanied by lowered cholesterol in sera.
INTRODUCTION
The endoplasmic reticulum (ER) is a central hub for protein and lipid metabolism, and disruptions in ER homeostasis can trigger the unfolded protein response (UPR). The UPR features translational and transcriptional control mechanisms that collectively serve to enhance protein folding and assembly, thereby expanding the capacity of the ER to process proteins slated for the secretory pathway (Walter and Ron, 2011; Baird and Wek, 2012; Baird et al., 2014). In addition, the ER is instrumental for intermediary and complex lipid metabolism (Fu et al., 2012). Among lipids, prudent regulation of cholesterol synthesis is critical, as cholesterol helps to order phospholipids in membranes. UPR sensory proteins, such as the type 1 ER transmembrane protein kinase PERK (EIF2AK3/PEK), are activated by disruptions in protein folding or membrane compositions. The ensuing PERK phosphorylation of the α subunit of eIF2 (eIF2α-P) represses global protein synthesis, thereby reducing influx of nascent polypeptides into the overloaded ER. Coincident with global translational control, eIF2α-P enhances preferential translation of a subset of stress-related mRNAs, including ATF4 (CREB2), a transcriptional activator of UPR target genes (Harding et al., 2003; Vattem and Wek, 2004; Dey et al., 2010). Because eIF2α-P induces ATF4 translational expression in response to a range of environmental and physiological stresses in addition to those afflicting the ER, the ATF4-directed regulatory scheme has been referred to as the integrated stress response (Harding et al., 2003; Baird and Wek, 2012).
ATF4 serves to enhance transcriptional expression of genes involved in amino acid metabolism and resistance to oxidative stress (Barbosa-Tessmann et al., 2000; Harding et al., 2003; Roybal et al., 2005; Kilberg et al., 2012). Furthermore, ATF4 functions to induce transcription of another transcription factor, CHOP (GADD153/DDIT3), and ATF4 and CHOP independently or in combination are believed to coordinate key facets of the UPR transcriptional and translational control directed by PERK (Wang et al., 1996; Ma et al., 2002; Harding et al., 2003; Marciniak et al., 2004; Su and Kilberg, 2008; Han et al., 2013; Teske et al., 2013). For example, together ATF4 and CHOP induce the expression of GADD34, which serves to direct feedback dephosphorylation of eIF2α-P (Connor et al., 2001; Novoa et al., 2001; Brush et al., 2003; Marciniak et al., 2004; Harding et al., 2009). ATF4 and CHOP also function in a feedforward loop to induce expression of a related transcription factor ATF5, and this transcription network is central for determining cell fate in response to ER stress (Teske et al., 2013). It is important to emphasize that the ATF4-directed transcriptome has been defined largely by analyses of cultured cells, especially mouse embryo fibroblast (MEF) cells, and an important question is whether ATF4 is the primary effector of gene expression evoked by eIF2α-P during ER stress in vivo.
In addition to PERK, two other UPR sensors, ATF6 and IRE1, function to regulate transcriptional expression. Upon ER stress, ATF6 is trafficked from the ER to the Golgi, where it is proteolytically cleaved, releasing the N-terminal ATF6 segment, a transcription factor of genes involved in the folding and trafficking of proteins slated for the secretory pathway (Haze et al., 1999; Ye et al., 2000; Chen et al., 2002; Shen et al., 2002; Wu and Kaufman, 2006; Wu et al., 2007). IRE1 facilitates cytosolic splicing of XBP1 mRNA, allowing translation of active XBP1s, which enhances transcriptional expression of genes that participate in protein folding, degradation of unfolded or misfolded proteins, and membrane expansion and renewal (Sidrauski and Walter, 1997; Tirasophon et al., 1998; Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2002, 2003; Yamamoto et al., 2007; Hetz et al., 2011).
The UPR sensors PERK, ATF6, and IRE1 pathways are often viewed as functioning in parallel, and each branch of the UPR is suggested to have the potential for different effects on cell viability (Schroder and Kaufman, 2006; Wu et al., 2007; Walter and Ron, 2011). For example, PERK is posited to promote cell death by enhancing CHOP expression, whereas IRE1 is suggested to promote cell survival during ER stress (Lin et al., 2007, 2009; Wu et al., 2007). The premise of distinct sensory pathways in the UPR is challenged by emerging findings that there is cross-regulation between the UPR branches. This is illustrated by the report that loss of PERK in cultured cells or livers of mice or deletion of ATF4 in cultured MEF cells substantially ablated activation of ATF6 and reduced expression of XBP1s during ER stress (Teske et al., 2011). This cross-talk in the UPR can also regulate PERK, with, for example, XBP1s inducing transcriptional expression of p58IPK, which is reported to lower PERK phosphorylation of eIF2α and translational control (Lee et al., 2003; Todd et al., 2008). Therefore it is likely that the UPR represents an interconnected network of regulatory proteins that function together to modulate gene expression and help to restore protein homeostasis during the progression of the UPR (Brewer, 2014).
In this study, we evaluated the central role of ATF4 in controlling transcriptional expression directed by PERK. To determine the functional roles of ATF4 in the UPR, we measured the UPR transcriptome in mice deleted for ATF4 in the liver and compared those changes with gene expression patterns altered by depletion of ATF4 in cultured cells. Using molecular, cellular, and biochemical assays, we found that basal expression of ATF4 lowered oxidative stress, and ATF4 contributed to cholesterol homeostasis in the liver independent of stress. Of importance, we showed that ATF4 was required for only a subset of PERK-dependent genes in vivo. Distinct from loss of PERK in the liver, we found that deletion of ATF4 in the liver was not required for induction of either UPR transcription factor CHOP or ATF6 during ER stress. Furthermore, deletion of ATF4 showed a 10-fold increase in hepatocyte cell death in response to ER stress. Although significant, the level of cell death resulting from deletion of ATF4 in the liver was only a fraction of the cell death determined for PERK deficiency.
RESULTS
UPR signaling varies upon ATF4 loss in different cell types
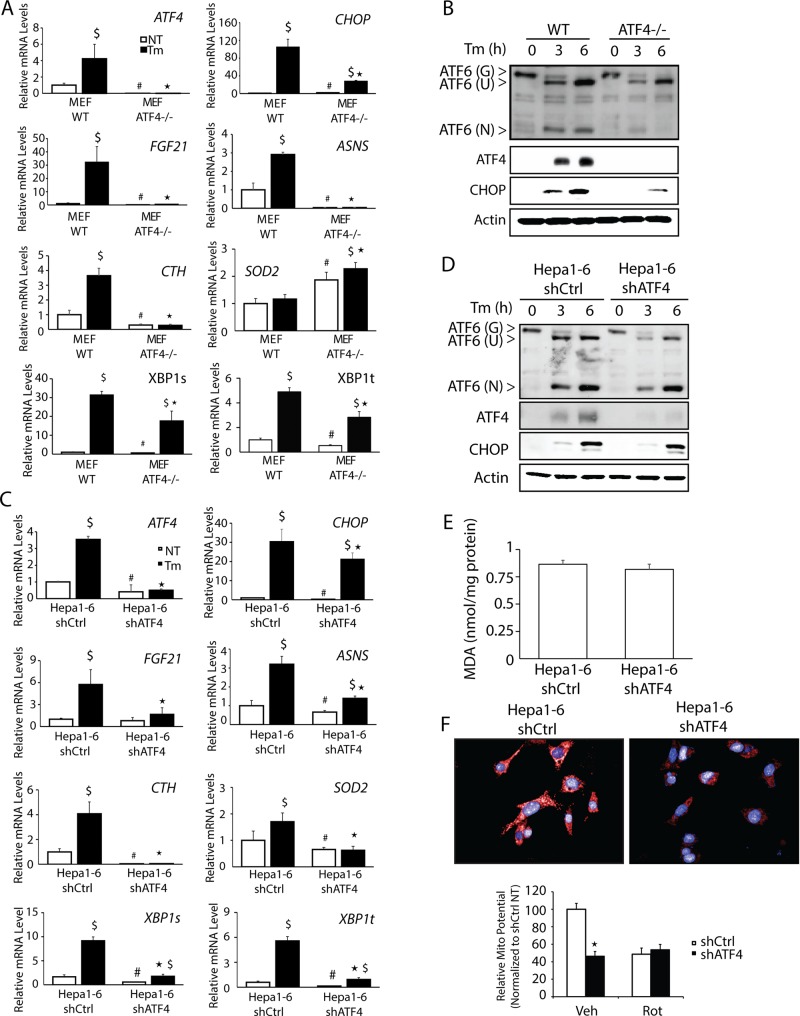
UPR studies featuring MEF cells subjected to pharmacological induction of ER stress indicated that ATF4 directs transcriptional expression of genes involved in amino acid metabolism, oxidative stress reduction, and control of apoptosis (Harding et al., 2003). To address the role of ATF4 in the UPR in hepatocytes, we depleted ATF4 expression in the mouse hepatoma cell line Hepa1-6 using short hairpin RNA (shRNA) and compared the induction of key UPR genes with that of MEF cells deleted for ATF4 (Figure 1, A–D). There was a significant reduction in ATF4 mRNA and protein in the shATF4 cells compared with control after 3 or 6 h of treatment with 2 μM tunicamycin, an inhibitor of N-glycosylation of proteins in the ER and potent inducer of ER stress (Figure 1, C and D). Known ATF4-target genes involved in amino acid metabolism, including ASNS, encoding asparagine synthetase, which catalyzes the conversion of aspartate to asparagine, and FGF21, which serves in the metabolic adaptation to fasting, were also reduced (Barbosa-Tessmann et al., 2000; Chen et al., 2004; De Sousa-Coelho et al., 2012; Woo et al., 2013). ATF4, through heterodimerization with C/EBPγ, can also provide for resistance to oxidative stress by enhancing the expression of several genes in the glutathione pathway, including CTH, encoding cystathionine γ-lyase (Dickhout et al., 2012; Huggins et al., 2015). Loss of C/EBPγ or ATF4 has been shown to reduce levels of glutathione (GSH) in MEF cells (Dickhout et al., 2012; Huggins et al., 2015). There were increased levels of ASNS, FGF21, and CTH mRNAs in both the Hepa1-6 and MEF cells treated with tunicamycin, and this induction was significantly ablated upon loss of ATF4 (Figure 1, A and C). Emphasizing the importance of cross-regulation in the UPR, ATF4 was also required for full induction of XBP1t mRNA and its spliced variant XBP1s during ER stress (Figure 1, A and C).
FIGURE 1:
Hepa1-6 cells demonstrate ATF4-independent CHOP expression. (A) WT and ATF4−/− MEF cells were treated with 2 μM tunicamycin (Tm) for 6 h or given no stress treatment (NT), and levels of ATF4, CHOP, FGF21, ASNS, CTH, SOD2, XBP1t, and XPB1s mRNAs were determined by qPCR. (B) WT and ATF4−/− MEF cells were treated with 2 μM tunicamycin for 3 or 6 h, and ATF4 and CHOP levels were evaluated by immunoblot analyses using specific antibodies. Levels of mRNAs (C) and proteins (D) were measured in in Hepa1-6 shCtrl and Hepa1-6 shATF4 cells by qPCR and immunoblot analyses, respectively. (E) MDA analysis on Hepa1-6 shCtrl and shATF4 cells was performed using BioVision Lipid Peroxidation Kit. (F) Hepa1-6 and Hepa1-6 shATF4 cells were plated on glass-bottom plates and then evaluated using MitoTracker Red. Top, images of the red fluorescent dye that stains mitochondria in live cells; bottom, quantitation indicated by histograms. *p < 0.05 indicates statistical significance with respect to the treated samples between the two cell types, #p < 0.05 indicates statistical significance with respect to the untreated samples between the two cell types, and $p < 0.05 indicates statistical significance between untreated sample vs. ER stress for each cell type.
Our comparison between ATF4-directed gene expression in Hepa1-6 and MEF cells also showed key differences between the two cell types. ATF4 is required for full cleavage to the active N-terminal version ATF6(N) in MEF cells treated with tunicamycin (Teske et al., 2011). As previously shown, the loss of ATF4 in the MEF cells led to a reduction in full-length ATF6 protein by 67% at 6 h, with a more pronounced 97% reduction of ATF6(N). However, the role of ATF4 in the activation of ATF6(N) was largely diminished in Hepa1-6 cells (Figure 1, B and D). In the case of SOD2, encoding superoxide dismutase 2, which serves to clear reactive oxygen species (ROS) from mitochondria, deletion of ATF4 in MEF cells led to higher levels of SOD2 mRNA in both basal and ER stress conditions, whereas loss of ATF4 expression in Hepa1-6 cells led to a substantial reduction of SOD2 transcripts in both stressed and nonstressed conditions.
Given the reported antioxidation role of ATF4 in MEF cells (Harding et al., 2003), we also addressed whether there was generation of oxidative stress in the Hepa1-6 cells upon loss of ATF4. We assessed the oxidation status in shATF4 and control cells by measuring malondialdehyde (MDA), which is generated by lipid peroxidation. Depletion of ATF4 in Hepa1-6 cells grown DMEM, which features high levels of glucose, did not affect the accumulation of MDA (Figure 1E). Furthermore, we measured mitochondrial function by using the MitoTracker Red assay to measure mitochondrial membrane potential. There was a greater-than-twofold reduction in the mitochondrial potential upon depletion of ATF4, which was similar to the lowered levels by treatment with rotenone (Rot), a potent inhibitor of mitochondrial respiration (Figure 1F).
CHOP is considered to be a canonical downstream effector of the PERK/ATF4 pathway, and, as expected, we observed a sharp increase in CHOP mRNA and protein levels in MEF cells treated with tunicamycin, which was significantly reduced upon deletion of ATF4 (Figure 1, A and B). However, in Hepa1-6 cells, the substantial increase in CHOP transcripts observed during ER stress was only modestly reduced upon depletion of ATF4, and there was full induction of CHOP protein even with depleted ATF4 expression (Figure 1, C and D). These results suggest that many features of UPR gene expression networks described in MEF cells cannot be applied to every cell type and that UPR regulators other than ATF4 can contribute to CHOP expression.
Loss of ATF6 in Hepa1-6 cells reduces CHOP expression
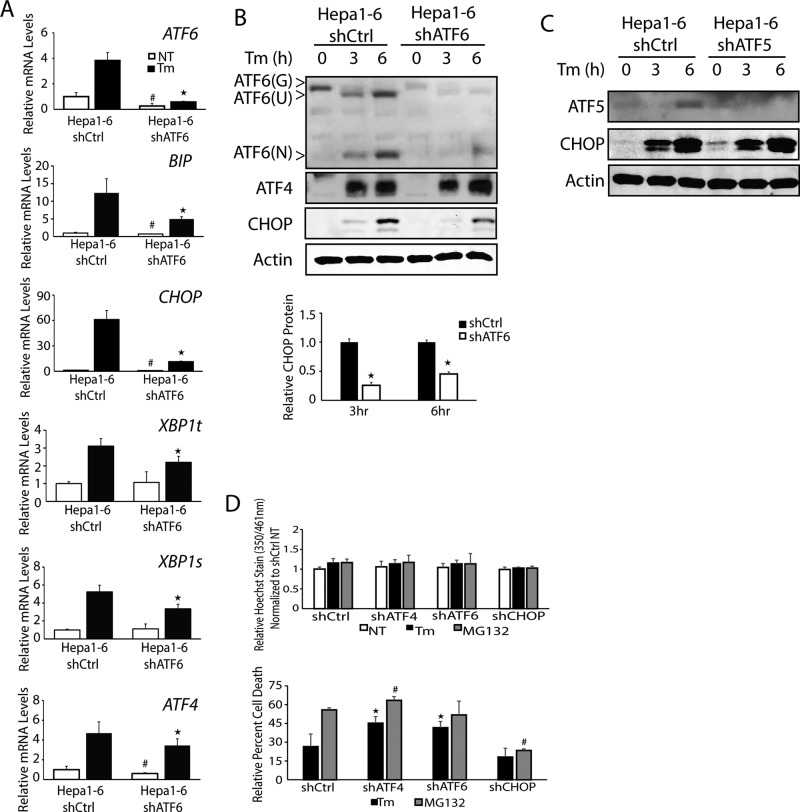
A previous study of CHO and COS-1 cell lines suggested that ATF6 can contribute to CHOP expression by an ER-stress element situated in the CHOP promoter (Ma et al., 2002). We addressed the role played by ATF6 in CHOP expression in Hepa1-6 cells by depleting ATF6 using shRNA, followed by treatment with 2 μM tunicamycin. Loss of ATF6 led to reductions in the expression of well-defined ATF6-target genes, including ER chaperone BiP (GRP78/HSPA5) and XBP1t and its spliced variant XBP1s (Figure 2A; Yoshida et al., 2001; Baumeister et al., 2005). ATF4 mRNA levels were also modestly reduced in the ATF6-depleted cells treated with tunicamycin, but there was no measurable reduction in ATF4 protein levels (Figure 2, A and B). Of importance, depletion of ATF6 sharply lowered the induction of CHOP mRNA and protein upon ER stress in the Hepa1-6 cells. The CHOP protein levels were reduced by 76 and 51% at 3 and 6 h, respectively. These results suggest that ATF6 can be a major contributor to CHOP expression, especially during ER stress. To address whether other ATF transcription factors regulated by PERK can effect CHOP expression, we depleted ATF5 expression in Hepa1-6 cells using shRNA and found no changes in induced expression of CHOP protein by ER stress (Figure 2C). These findings indicate that the requirement of ATF6 for induced CHOP expression in the Hepa1-6 hepatocytes is not broadly shared among other transcription factors controlled by PERK.
FIGURE 2:
ATF6 facilitates induced CHOP expression in liver cells subjected to ER stress. (A) Hepa1-6 shCtrl and Hepa1-6 shATF6 cells were treated with 2 μM Tm for 6 h or given no stress treatment (NT), and the levels of ATF4, CHOP, ATF6, XBP1t, XBP1s, and BiP mRNAs were determined by qPCR. (B) Hepa1-6 shCtrl and Hepa1-6 shATF6 cells were treated with 2 μM Tm for 3 or 6 h, and ATF4, ATF6, and CHOP levels were evaluated by immunoblot analyses. CHOP levels were quantified using ImageJ based on three independent experiments for the 3- and 6-h time points. (C) Hepa1-6 shCtrl and Hepa1-6 shATF5 cells were treated with 2μM Tm for 3 or 6 h, and levels of ATF5 and CHOP proteins were measured by immunoblot analyses. (D) Hepa1-6 shCtrl, shATF4, shATF6, and shCHOP cells were treated with 2 μM Tm or 1 μM MG132 for 24 h or given no stress treatment. Cell proliferation was analyzed via Hoechst fluorescence, and cell death was measured by calcein fluorescence. *p < 0.05 indicates statistical significance with respect to the treated samples between the indicated cell types, and #p < 0.05 indicates statistical significance with respect to the untreated samples between the cell types.
CHOP can direct the expression of genes involved in protein homeostasis, in particular those promoting protein folding, such as HSPA1B and DNAJ4, and a number of aminoacyl tRNA-synthetases, including WARS (Marciniak et al., 2004; Marciniak and Ron, 2006; Han et al., 2013; Teske et al., 2013). By contrast, during prolonged ER stress triggered by pharmacological agents, CHOP can instead trigger cell death through expression of proapoptotic genes such as BIM or GADD34 (Marciniak et al., 2004; Marciniak and Ron, 2006; Puthalakath et al., 2007; Oslowski and Urano, 2011; Teske et al., 2013)
We wished to determine whether depletion of the upstream regulator ATF6 or ATF4 altered Hepa1-6 survival upon treatment with either tunicamycin or another stress agent, the proteasome inhibitor MG132. As expected, Hepa1-6 cells depletion of CHOP showed protective effects upon either stress treatment as judged by lowered calcein-AM labeling (Figure 2D). Lowered expression of either ATF4 or ATF6 led to significant increases in cell death upon ER stress of 20 and 15%, respectively, compared with control cells. These findings are consistent with the protective effects attributed to the ATF4 and ATF6 transcription factors in the UPR (Harding et al., 2003; Wu et al., 2007).
ATF4 and ATF6 contribute to induced CHOP expression during nutrient stress
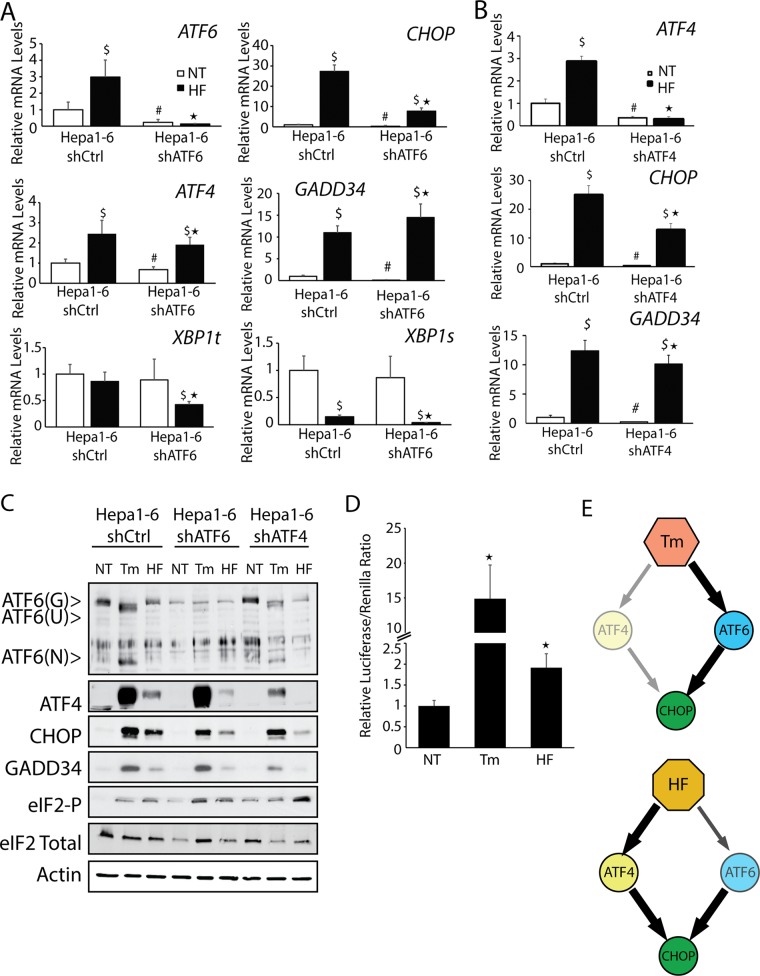
We next addressed whether ATF4 and ATF6 contribute to CHOP expression in response to a stress that does not directly perturb the ER. Halofuginone has been shown to deplete the charging of tRNAPro, leading to activation of GCN2, another eIF2 kinase, which can in turn enhance the transcriptional and translational expression of ATF4 and CHOP (Sundrud et al., 2009; Peng et al., 2012). Treatment of Hepa1-6 cells with 50 nM halofuginone for 6 h led to significant increases in ATF4 and CHOP mRNAs and proteins (Figure 3, A–C). Loss of either ATF6 or ATF4 substantially lowered both CHOP mRNA and protein (Figure 3, A–C). Expression of GADD34 mRNA and protein was largely dependent on ATF4, with further increases in GADD34 induction upon depletion of ATF6. Despite there being robust eIF2α-P and induction of ATF4, CHOP, and GADD34 mRNAs in response to halofuginone, the increase in ATF4, CHOP, and GADD34 proteins was reproducibly less than with tunicamycin treatment (Figure 1C). This difference in the levels of induced CHOP and ATF4 proteins may be a consequence of the diminished Pro-tRNAPro available for protein synthesis during halofuginone treatment.
FIGURE 3:
ATF6 contributes to CHOP expression in multiple stresses. Hepa1-6 cells expressing shCtrl, shATF6, or shATF4 were treated with 2 μM Tm or 50 nM halofuginone (HF) for 6 h or given no stress treatment (NT), as indicated. (A, B) Levels of the indicated gene transcripts were measured by qPCR. (C) The amounts of the indicated proteins were measured by immunoblot analyses. (D) Hepa1-6 cells were cotransfected with 5x ATF6 Binding Element Luciferase construct and Renilla. After 24 h, cells were treated with 2 μM Tm or 50 nM HF, and prepared, and firefly luciferase normalized for Renilla was measured for prepared lysates. (E) Model of the roles of ATF4 and ATF6 on expression of CHOP during ER stress and cytosolic stress triggered by HF treatment. *p < 0.05 indicates statistical significance between ER stress–treated cells, #p < 0.05 indicates significance for untreated cells, and $p < 0.05 indicates significance for untreated cells.
Consistent with the idea that the treatment regimen of halofuginone did not elicit ER stress, there was no increase in the levels of XBP1t mRNA, and the amount of XBP1s transcript was in fact substantially lower in Hepa1-6 cells exposed to halofuginone. ATF6 was suggested to contribute to CHOP expression, and there was a threefold increase in ATF6 mRNA levels during halofuginone treatment. Although we did not measure appreciable activated ATF6(N) protein upon halofuginone treatment by immunoblot analyses, there was a twofold increase in ATF6-directed transcription as measured by a luciferase reporter assay, which was significant although reduced compared with the ∼15-fold enhancement during tunicamycin treatment (Figure 3D). These results suggest a model in which nutrient stress afflicting the cytosol induces both ATF4 and ATF6 to contribute to increased CHOP expression. By comparison, ER stress triggers a pathway that is largely dependent upon ATF6 for CHOP expression (Figure 3E).
ATF4 is dispensable for induced expression of CHOP in the liver during ER stress
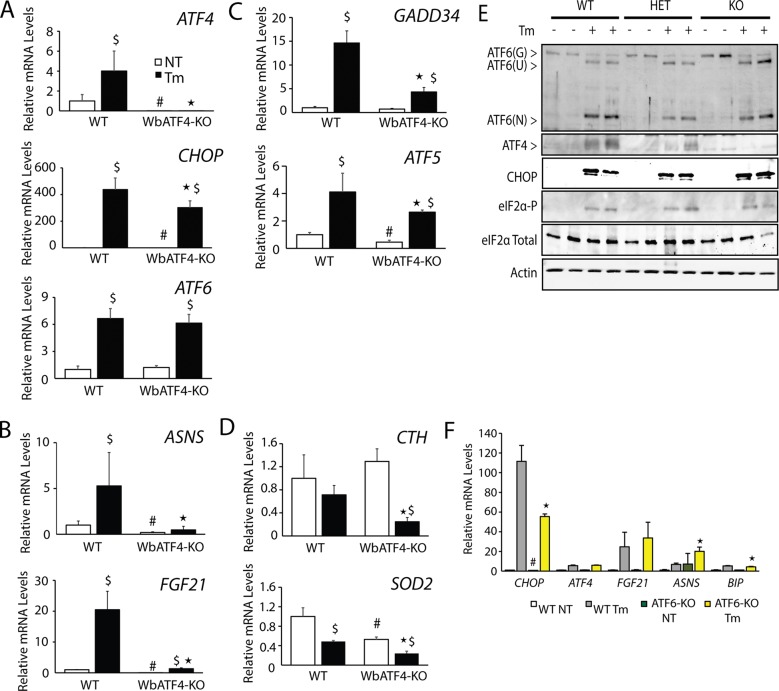
The identification of ATF4-independent induction of CHOP expression in Hepa1-6 cells subjected to ER stress raises the question of whether this pattern of gene expression also occurs in vivo. We initially characterized whole-body ATF4-depleted mice (WbATF4-KO), but breeding of these animals can be problematic (Masuoka and Townes, 2002), and we subsequently extended our analysis to mice with a liver-specific ATF4 deletion (LsATF4-KO). Mice expressing Cre from the albumin promoter were bred to ATF4 gene floxed mice in a manner similar to the liver-specific PERK knockout that our laboratories previously described (Teske et al. 2011). WbATF4-KO (Figure 4) and LsATF4-KO (Figure 5) mice and their wild-type (WT) controls were treated with 1 mg/kg tunicamycin or 0.3% dimethyl sulfoxide (DMSO) vehicle administered via an intraperitoneal (IP) injection for 6 h. As expected, ATF4 mRNA and protein were induced upon tunicamycin treatment in WT livers, and ATF4 was minimally expressed in either knockout model (Figures 4, A and E, and 5, A and E). Consistent with the loss of ATF4 transcriptional activity, there was significant reduction in the expression of ATF4-target genes ASNS, FGF21, and CTH in the WbATF4-KO and LsATF4-KO livers (Figures 4, B and D, and 5, B and D).
FIGURE 4:
CHOP expressed independently of ATF4 in whole-body ATF4-knockout livers. WT and WbATF4-KO were injected IP with 1 mg/kg body weight Tm or vehicle indicating no treatment (NT) for 6 h. (A–D) Livers were collected, and mRNA levels were measured by qPCR for the indicated genes. (B) ASNS and FGF21 represent ATF4-target genes. (C) GADD34 and ATF5 highlight genes regulated by both ATF4 and CHOP. (D) CTH and SOD2 were analyzed to assess genes involved in mitochondrial function and oxidative stress. (E) The indicated proteins were measured by immunoblot analyses using lysates prepared from livers derived from mice treated with DMSO (–) or Tm (+) for 6 h. (F) Measurement of mRNAs from microarray data derived from livers of whole-body ATF6-knockout mice and their WT littermates (Wu et al., 2007). *p < 0.05 indicates statistical significance among ER stress–treated livers, #p < 0.05 indicates statistical significance among untreated liver tissues, and $p < 0.05 indicates significance with respect to the untreated and treated tissues.
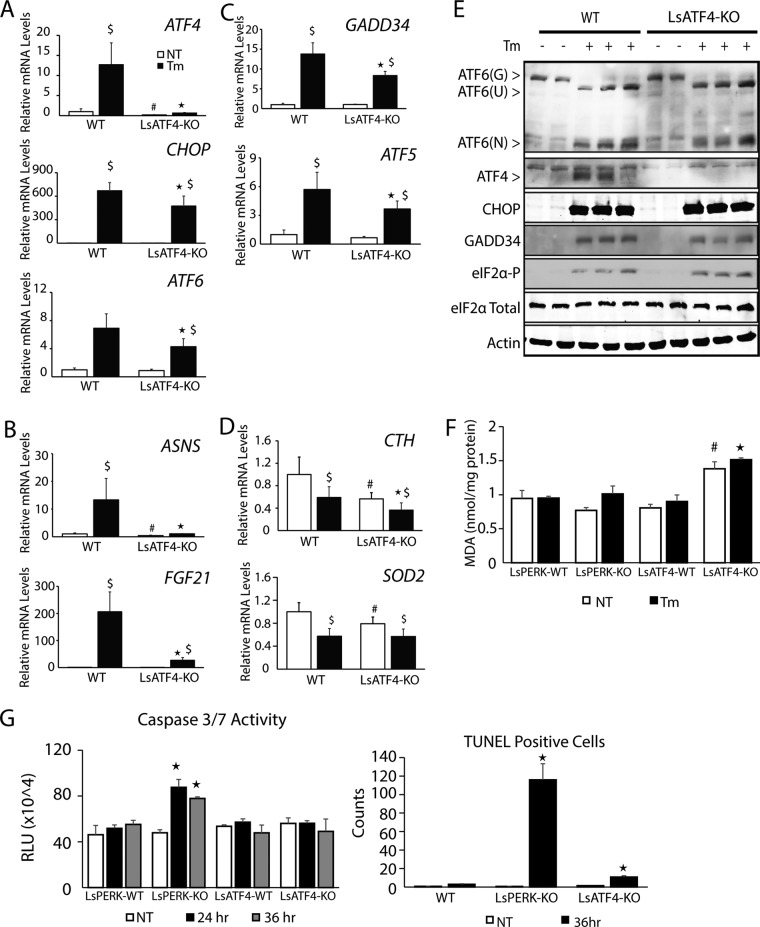
FIGURE 5:
CHOP is expressed independently of ATF4 in liver-specific ATF4-knockout mice. WT and LsATF4-KO were injected IP with 1 mg/kg body weight Tm or vehicle indicating no treatment (NT) for 6 h. (A–D) Livers were collected, and mRNA levels for the indicated genes were measured by qPCR. (B) ASNS and FGF21 are indicated ATF4-target genes. (C) GADD34 and ATF5 highlight genes regulated by both ATF4 and CHOP. (D) CTH and SOD2 were analyzed to assess genes involved in mitochondrial function and oxidative stress. (E) Immunoblot analyses of the indicated proteins were carried out in livers obtained from mice treated with DMSO (−) or Tm (+) for 6 h. (F) Liver samples from WT and LsATF4-KO mice treated with TM for 24 h were compared for levels of MDA via fluorescence using the Lipid Peroxidation Kit from BioVision. (G) LsPERK-WT, LsPERK-KO, LsATF4-WT, and LsATF4-KO mice were treated with IP injection of 1 mg/kg tunicamycin or vehicle for 24 or 36 h, and caspase 3/7 activity was measured. Histological samples were also assessed for TUNEL-positive cells after 36 h of Tm treatment, and the presented results are the average counts of eight different histological sections. *p < 0.05 indicates statistical significance among cells subjected to ER stress, #p < 0.05 indicates significance among untreated cells, and $p < 0.05 indicates significance with respect to the untreated vs. treated tissues.
We next addressed the requirement for ATF4 for in vivo induction of key UPR gene markers. Activation of ATF6(N) protein in the liver during ER stress was largely independent of ATF4 function, and there was only modest reduction of induced ATF6 mRNA in WbATF4-KO and LsATF4-KO compared with WT (Figure 4, A and E, and 5, A and E). Of importance, there was only minimal reduction of induced CHOP mRNA upon ER stress in the ATF4-deleted livers and full induction of CHOP protein independent of ATF4 function. Expression at WT levels of ATF6 and CHOP can also be seen in the heterozygous mice for the ATF4-KO (Figure 4E). Transcriptional expression of GADD34 and ATF5 depends on both CHOP and ATF4, and consistent with retention of CHOP function in the ATF4-knockout models, there was a partial induction of GADD34 and ATF5 mRNAs compared with WT (Figures 4C and 5C). Taken together, these results are similar to our analyses with Hepa1-6 hepatoma cells and suggest that in the liver, CHOP and ATF6 expression is uncoupled from ATF4 in the UPR. Of importance, reported microarray analyses of livers prepared from WT and ATF6α−/− mice showed that loss of ATF6 significantly reduced CHOP mRNA expression basally and upon IP injection with tunicamycin (Wu et al., 2007; Figure 4F), a finding consistent with our UPR analyses in Hepa1-6 cells (Figure 2A).
As previously seen in the Hepa1-6 shATF4 cells, expression of CTH and SOD2 mRNA was reduced in the absence of ATF4 (Figures 4D and 5D). LsATF4-KO mice also displayed elevated levels of MDA independent of stress (Figure 5F). Of importance, this resistance to oxidative stress in vivo can be conferred by basal expression of ATF4, and this protection can occur independent of PERK, as the LsPERK-KO did not show significant differences in MDA levels compared with WT.
LsPERK-KO livers were previously shown to trigger cell death during extended periods of ER stress (Teske et al., 2011). There was increased caspase 3/7 cleavage activity in LsPERK-KO mice upon treatment with 1 mg/kg tunicamycin for 24 or 36 h (Figure 5G). By comparison, LsATF4-KO livers showed no change in the caspase activity compared with WT. Livers prepared from LsPERK-KO treated with tunicamycin also exhibited a robust increase in terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)–positive cells, whereas LsATF4-KO showed a more moderate but still significant 10× increase compared with WT (Figure 5G). These results indicate that PERK provides robust protection of livers exposed to ER stress, whereas ATF4 plays a more moderate role.
RNA sequencing analysis of LsATF4-KO mice
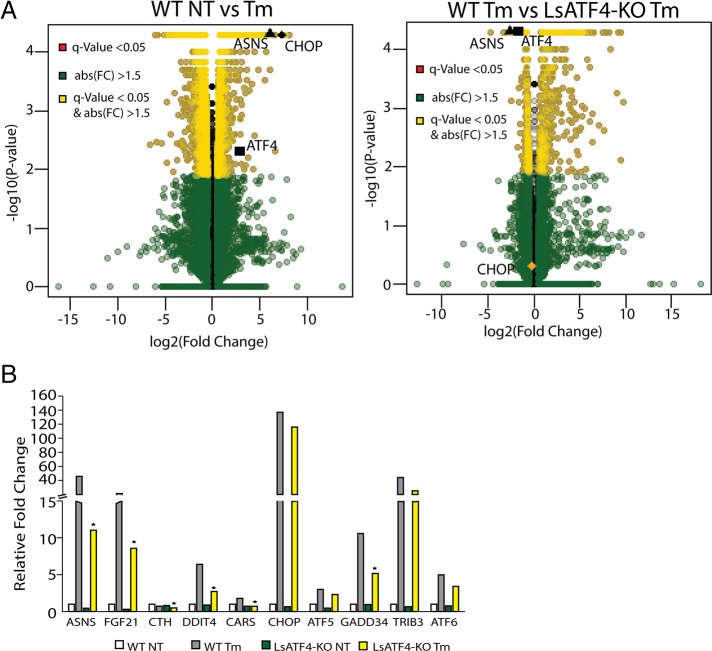
To address the role of ATF4 in the genome-wide regulation of the UPR transcriptome, we carried out RNA sequencing (RNA-Seq) analysis using livers prepared from WT and LsATF4-KO mice treated with 1 mg/kg tunicamycin or vehicle for 6 h. The RNA-Seq featured single-end RNA sequencing with 100–base pair reads; ∼32 million reads were collected for each mouse, with three animals per group. Reads were mapped to the mouse mm9 genome using TopHat (Trapnell et al., 2009, 2012), and expression levels were characterized using Cufflinks, Cuffmerge, and Cuffdiff (Trapnell et al., 2010, 2012). Results were first analyzed by a volcano plot comparing the log2 of fold change to the −log10 of the q value (Figure 6A). There were 4717 gene transcripts showing a significant change in WT liver upon ER stress (>1.5-fold, p < 0.05, false discovery rate [FDR] < 0.05), with 2271 genes showing an increase and 2446 a decrease in expression. A total of 352 genes, or 7.5% of the total UPR changes, were significantly affected by loss of ATF4.
FIGURE 6:
ATF4 is required for expression of a subset of UPR genes. (A) RNA-Seq analyses measuring levels of gene transcripts from WT and LsATF4-KO mice treated with 1 mg/kg Tm or vehicle (NT) for 6 h. Volcano plot of WT NT vs. Tm, and WT Tm vs. LsATF4-KO Tm. (B) Levels of UPR gene transcripts by RNA-Seq analysis, which are organized into ATF4-dependent groups (ASNS, FGF21, CTH, DDIT4, and CARS) and genes bound by both ATF4 and CHOP (CHOP, ATF5, GADD34, TRIB3, ATF6).
Key UPR target genes ATF4, CHOP, and ASNS were each significantly induced in the WT liver (Figure 6A). Consistent with our quantitative PCR (qPCR) findings, the levels of ASNS, FGF21, and CTH were significantly lowered in the LsATF4-KO liver. By comparison, induced CHOP expression was not significantly affected by loss of ATF4 (Figure 6B). The limited role of ATF4 in the UPR is noteworthy, given our earlier genome-wide analysis that nearly 50% of the gene transcripts showing significant changes in liver exposed to tunicamycin were dependent on PERK (Teske et al., 2011). We conclude that ATF4 is required for induction of only a portion of PERK-dependent genes. This finding suggests that there are other PERK-driven events that are critical for regulation of the UPR-directed gene expression.
It is noteworthy that many genes required ATF4 for full expression during basal conditions, suggesting that ATF4 plays a significant role in directing the transcriptome in seemingly nonstressed conditions in the liver. There were 385 genes that were altered by the loss of ATF4 during vehicle treatment. Pathway analysis of ATF4-targeted genes using PANTHER was performed on gene expression with a significant q value between WT and LsATF4-KO livers during either basal or ER stress conditions (Tables 1 and 2; Mi and Thomas, 2009; Mi et al., 2013). The analysis was performed by using the PANTHER statistical overrepresentation test, which involves comparing a user input list of genes against a reference list containing all genes. These lists were then subdivided on the basis of Gene Ontology (GO) classification, and then a binomial test was performed to determine overrepresentation of the user list of genes compared with the expected number if they occurred at random. Under basal conditions, several categories demonstrated greater-than-fivefold enrichment, including those representing fatty acid biosynthetic processes and fatty acid metabolic processes (Table 1). During ER stress, we identified three pathways with fold enrichment greater than five, including steroid biosynthetic process, cholesterol metabolic process, and sterol metabolic process (Table 2). Given that the top three pathways affected during ER stress–included genes involved in sterol and cholesterol metabolism, we next experimentally addressed the effect of ATF4 loss on cholesterol.
TABLE 1:
Pathway analysis using PANTHER of altered genes in WT and LsATF4-KO livers in nonstressed conditions.
| GO category | Mus musculus (reference) | Uploaded | Expected | Fold enrichment | ± | p value |
|---|---|---|---|---|---|---|
| Regulation of liquid surface tension (GO: 0050828) | 55 | 9 | 0.95 | >5 | + | 1.50E-04 |
| Cell–matrix adhesion (GO: 0007160) | 88 | 12 | 1.52 | >5 | + | 1.50E-05 |
| Fatty acid biosynthetic process (GO: 0006633) | 61 | 8 | 1.05 | >5 | + | 3.08E-03 |
| Locomotion (GO: 0040011) | 63 | 8 | 1.09 | >5 | + | 3.87E-03 |
| Fatty acid metabolic process (GO: 0006631) | 252 | 23 | 4.35 | >5 | + | 4.12E-08 |
| Steroid metabolic process (GO: 0008202) | 198 | 15 | 3.42 | 4.39 | + | 5.98E-04 |
| Macrophage activation (GO: 0042116) | 173 | 13 | 2.98 | 4.36 | + | 2.99E-03 |
| Homeostatic process (GO: 0042592) | 208 | 15 | 3.59 | 4.18 | + | 1.07E-03 |
| Receptor-mediated endocytosis (GO: 0006898) | 217 | 15 | 3.74 | 4.01 | + | 1.76E-03 |
| Cell–cell adhesion (GO: 0016337) | 362 | 22 | 6.24 | 3.52 | + | 1.20E-04 |
| Cell adhesion (GO: 0007155) | 550 | 31 | 9.49 | 3.27 | + | 3.06E-06 |
| Biological adhesion (GO: 0022610) | 577 | 31 | 9.95 | 3.11 | + | 8.88E-06 |
| Generation of precursor metabolites and energy (GO: 0006091) | 290 | 15 | 5 | 3 | + | 4.44E-02 |
| Lipid metabolic process (GO: 0006629) | 966 | 47 | 16.66 | 2.82 | + | 5.65E-08 |
| Endocytosis (GO: 0006897) | 380 | 18 | 6.55 | 2.75 | + | 3.13E-02 |
| Immune system process (GO: 0002376) | 1480 | 50 | 25.53 | 1.96 | + | 1.13E-03 |
| Developmental process (GO: 0032502) | 2468 | 75 | 42.57 | 1.76 | + | 2.01E-04 |
| Localization (GO: 0051179) | 2788 | 76 | 48.09 | 1.58 | + | 8.12E-03 |
| Cell communication (GO: 0007154) | 3175 | 85 | 54.77 | 1.55 | + | 4.81E-03 |
| Cellular process (GO: 0009987) | 7033 | 158 | 121.31 | 1.3 | + | 1.12E-02 |
TABLE 2:
Pathway analysis using PANTHER of regulated genes in WT and LsATF4-KO livers during treatment with tunicamycin.
| GO category | Mus musculus (reference) | Uploaded | Expected | Fold enrichment | ± | p value |
|---|---|---|---|---|---|---|
| Steroid biosynthetic process | 90 | 18 | 3.2 | >5 | + | 6.08E-05 |
| Cholesterol metabolic process | 92 | 17 | 3.27 | >5 | + | 4.79E-04 |
| Sterol metabolic process | 99 | 18 | 3.52 | >5 | + | 2.52E-04 |
| Cellular hormone metabolic process | 74 | 13 | 2.63 | 4.94 | + | 2.84E-02 |
| Renal system process | 77 | 13 | 2.74 | 4.75 | + | 4.32E-02 |
| Steroid metabolic process | 194 | 29 | 6.89 | 4.21 | + | 1.67E-06 |
| Striated muscle cell development | 115 | 17 | 4.09 | 4.16 | + | 1.01E-02 |
| Lipid homeostasis | 111 | 16 | 3.94 | 4.06 | + | 2.79E-02 |
| Renal system development | 217 | 31 | 7.71 | 4.02 | + | 1.19E-06 |
| Organic hydroxy compound biosynthetic process | 142 | 20 | 5.05 | 3.96 | + | 2.56E-03 |
| Hormone metabolic process | 121 | 17 | 4.3 | 3.95 | + | 1.97E-02 |
| Gland morphogenesis | 114 | 16 | 4.05 | 3.95 | + | 3.88E-02 |
| Positive regulation of lipid metabolic process | 122 | 17 | 4.33 | 3.92 | + | 2.19E-02 |
| Negative regulation of cellular response to growth factor stimulus | 115 | 16 | 4.09 | 3.92 | + | 4.32E-02 |
| Kidney development | 197 | 27 | 7 | 3.86 | + | 4.22E-05 |
| Lipid transport | 206 | 28 | 7.32 | 3.83 | + | 2.64E-05 |
| Regulation of transmembrane transporter activity | 172 | 23 | 6.11 | 3.76 | + | 8.59E-04 |
| Organic acid biosynthetic process | 217 | 29 | 7.71 | 3.76 | + | 2.02E-05 |
| Carboxylic acid biosynthetic process | 217 | 29 | 7.71 | 3.76 | + | 2.02E-05 |
| Regulation of ion transmembrane transporter activity | 167 | 22 | 5.93 | 3.71 | + | 2.06E-03 |
| Monocarboxylic acid biosynthetic process | 146 | 19 | 5.19 | 3.66 | + | 1.56E-02 |
| Small-molecule biosynthetic process | 316 | 40 | 11.23 | 3.56 | + | 1.01E-07 |
| Regulation of transporter activity | 182 | 23 | 6.47 | 3.56 | + | 2.27E-03 |
| Lipid localization | 233 | 29 | 8.28 | 3.5 | + | 9.35E-05 |
Loss of ATF4 leads to alterations in the cholesterol pathways
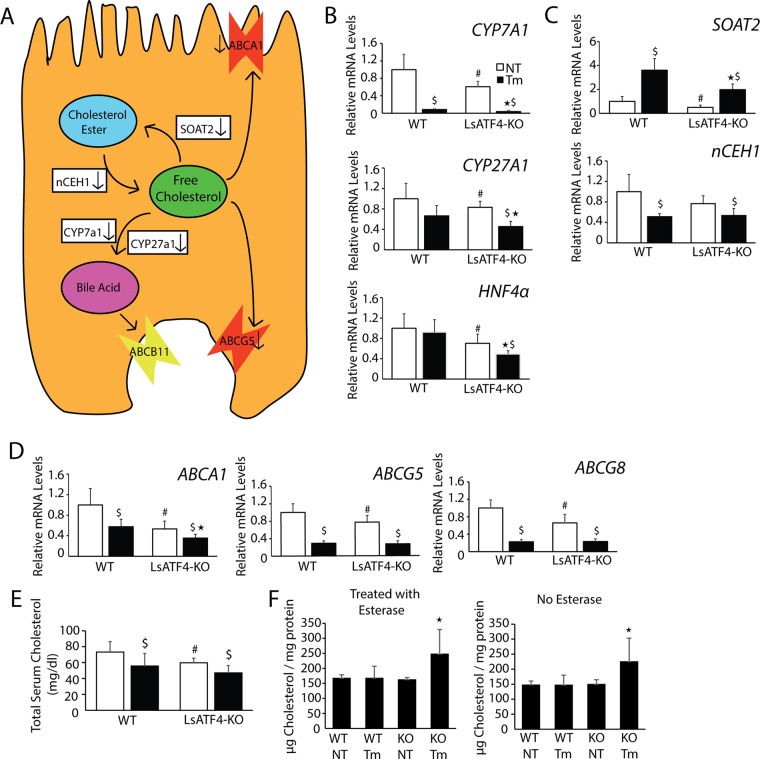
Our RNA-Seq analysis suggested a role for ATF4 in metabolism of cholesterol and related bile acids (Figure 7, A–D). ATF4 was required for expression of several key genes involved in bile acid synthesis, which represents one pathway for disposing of free cholesterol in hepatocytes. Among these synthetic genes, CYP7A1, encoding cholesterol 7α-hydoxylase, and CYP27A1, encoding sterol 27-hydoxylase, were significantly lowered in LsATF4-KO cells in both basal and ER stress conditions (Figure 7, A and B). Expression of HNF4α, a gene that enhances transcription of CYP7A1 (De Fabiani et al., 2001; Kir et al., 2012), was also significantly reduced in LsATF4-KO livers. ATF4 is also required for full induction of SOAT2, which facilitates conversion of free cholesterol to esterified cholesterol (Figure 7, A and C; Cases et al., 1998; Zhao et al., 2008). Free cholesterol can be damaging to cells, and injury can be prevented by cholesterol esterification (Kellner-Weibel et al., 1998; Feng et al., 2003). nCEH1 was significantly reduced in response to ER stress independent of ATF4. Expression of genes responsible for exporting free cholesterol was also altered in the LsATF4-KO mice. ABCG5 and ABCG8 encode transporters that function on the enterocyte side of the hepatocyte, which export free cholesterol (Figure 7A; Lu et al., 2001). Expression of both genes was significantly lowered during ER stress, and loss of ATF4 further lowered the amounts of ABCG5 and ABCG8 mRNAs during both basal conditions (Figure 7D). ABCA1 encodes a transporter of free cholesterol from hepatocytes to the plasma, and consistent with a previous report (Rohrl et al., 2014), ABCA1 mRNA was reduced during ER stress. Deletion of ATF4 further reduced ABCA1 expression independent of stress (Figure 7D). Together these findings suggest that ATF4 contributes to expression of key genes involved in cholesterol metabolism and transport.
FIGURE 7:
Loss of ATF4 leads to increase in free cholesterol within the liver. (A) ATF4-targeted gene functions in hepatocyte cholesterol metabolism. (B–F) WT and LsATF4-KO mice were injected IP with 1 mg/kg Tm or vehicle (NT) for 6 h. Levels of the indicated mRNAs were measured by qPCR (B–D) or cholesterol content analysis (E, F). (B–D) Genes involved in bile acid synthesis (CYP7A1, CYP27A1, HNF4α), cholesterol esterification (SOAT2), or cholesterol export (ABCA1, ABCG5, ABCG8) were significantly reduced due to ATF4 loss. (E, F) WT and LsATF4-KO livers treated with Tm or vehicle (NT) were analyzed for total cholesterol content in the serum (E) and within tissue (F). *p < 0.05 indicates statistical significance with respect to the ER-treated tissues, #p < 0.05 indicates significance among untreated tissues, and $p < 0.05 indicates significance between respective untreated ER stress tissues.
We next determined whether there are changes in free cholesterol levels upon loss of ATF4 and ER stress. There were lowered levels of serum cholesterol in the LsATF4-KO mice compared with WT (Figure 7E), a finding consistent with that reported for WbATF4-KO (Xiao et al., 2013). Furthermore, cholesterol was lowered in both WT and LsATF4-KO mice upon treatment with tunicamycin (Figure 7E). Although there was a trend for the LsATF4-KO mice to exhibit lower total cholesterol during ER stress than with WT, it did not reach statistical significance. We also measured the levels of free and total cholesterol in liver tissue (Figure 7F). Liver tissue from LsATF4-KO mice during stress showed significantly increased levels of free cholesterol, based on assays in the absence of esterase that represents free cholesterol. Integration of ATF4-directed gene expression in cholesterol metabolism will be considered further in the Discussion.
DISCUSSION
In this study, we used biochemical and RNA-Seq analysis to determine that ATF4 is required for only 7.5% of genes regulated by ER stress, compared with nearly 50% in an LsPERK-KO model (Teske et al., 2011). This subset includes genes involved in amino acid metabolism, resistance to oxidative stress, and cholesterol metabolism. Deletion of ATF4 alters genes that are required for the conversion of cholesterol to bile acid (CYP7A1), esterification of cholesterol (SOAT2), and transport from the hepatocyte (ABCA1), and when ATF4 loss is coupled with ER stress, it results in an increase in free cholesterol within hepatocytes (Figure 7; Fu et al., 2012; Rohrl et al., 2014). Of importance, ATF4 has a role in gene expression during basal conditions, with 385 genes altered by the loss of ATF4 in the absence of apparent stress (Table 1). The consequences of ATF4 in basal gene expression are illustrated by the finding that deletion of ATF4 in the liver is required for prevention of oxidative stress (Figure 5F), a finding consistent with a report on cultured MEF cells (Harding et al., 2003). In cultured Hepa1-6 cells, we also found a linkage between basal ATF4 expression and mitochondrial membrane potential (Figure 1F). Finally, loss of ATF4 in cultured Hepa1-6 cells subjected to ER stress led to an increase in cell death, and there was also significant cell death in the livers of LsATF4-KO mice as measured by TUNEL, although the amount of cell death was much less than in PERK deficiency (Figures 2D and 5G).
ATF6 is a primary inducer of CHOP expression in the liver during ER stress
Although certain characteristics were shared between cultured MEF and Hepa1-6 cells and liver in vivo, there were also striking differences. In contrast to MEF cells, ATF4 was not required for induced CHOP mRNA and protein in liver exposed to ER stress (Figures 4 and 5). This ATF4-independent expression of CHOP was also seen in the Hepa1-6 cell line (Figure 1), for which ATF6 was a necessary transcription factor for CHOP expression during ER stress (Figure 2). Induction of CHOP expression depended on both ATF4 and ATF6 during halofuginone treatment, which inhibits tRNA charging in the cytosol. It is noteworthy that there is a more modest activation of ATF6 during halofuginone exposure than with tunicamycin, although ATF6 still plays a role in the expression of CHOP.
ATF6 was previously shown to be a component of CHOP expression through an endoplasmic reticulum stress response element situated in its promoter (Ma et al., 2002). The role of ATF6 has been overshadowed by the requirement in MEF culture cells on ATF4 binding for the amino acid response element within the CHOP promoter (Bruhat et al., 2002; Ma et al., 2002) and the idea that the three arms of the UPR operate independently. This switch from ATF4- to ATF6-dependent CHOP expression may result in part from the timing of activation of ATF4 protein versus ATF6 or the availability of their binding partners. ATF6 is rapidly cleaved and activated upon ER stress, whereas ATF4 requires preferential translation, and its induction occurs later in the UPR. These findings support the idea that PERK/ATF6 cross-talk is a primary driver of CHOP expression within the liver (Figure 3E).
ATF4 is responsible for a small subset of PERK-dependent genes during ER stress
RNA-Seq analysis suggests an expanded role for ATF4-directed gene expression in the liver not subjected to any apparent stress. A total of 385 genes were significantly altered by the loss of ATF4 basally, which exceeded those regulated during ER stress. The ATF4-target genes expressed during basal conditions are represented by metabolic processes of fatty acids and steroids and cell adhesion (Table 1). ATF4 was previously shown to promote metastasis in a tumor environment, but it is also required basally (Dey et al., 2015). Deletion of ATF4 significantly changed the expression of 352 genes among the 4717 genes altered during ER stress. The unexpected small subset of genes requiring ATF4 in the UPR compared with those involving PERK emphasizes that PERK can facilitate multiple transcriptional control networks, including those for ATF6 and XBP1 (Teske et al., 2011). In addition, ATF4 and CHOP ChIP-Seq analyses indicated that induced CHOP can play a direct role in the expression of genes that are also targeted by ATF4 (Han et al., 2013). Thus the PERK/ATF6/CHOP arm can alleviate loss of ATF4 for transcriptional regulation during ER stress by robustly activating CHOP.
Deletion of ATF4 in the liver demonstrates its role in prevention of oxidative stress independent of stress (Figure 5F). There was a lack of a difference in MDA levels in cultured Hepa1-6 cells depleted of ATF4 (Figure 1E). Loss of ATF4 in the Hepa1-6 cells did lead to a striking reduction in mitochondrial membrane potential (Figure 1F), and reduced ROS production via oxidative phosphorylation might help to explain the absence of observed oxidative stress in Hepa1-6 cells depleted of ATF4. Rapidly proliferating cells cultured in media with high glucose concentration can proceed by aerobic glycolysis (Vander Heiden et al., 2009); however, we did not observe changes in oxidative stress in the ATF4-depleted Hepa1-6 cells even when cultured in medium with lower amounts of glucose (unpublished data). It was reported that ATF4 contributes to increased H2S sulfhydration of certain metabolic proteins during ER stress, altering their activity (Gao et al., 2015). The H2S sulfhydration occurs on several proteins within glycolysis and the tricarboxylic acid cycle, leading to an increase in glycolysis and a decrease in oxidative phosphorylation, which can be restored through CTH inhibition (Gao et al., 2015). This connection between ATF4 and energy metabolism, together with the reduced mitochondrial membrane potential seen in the Hepa1-6 cells, warrants further investigation.
ATF4 is involved in cholesterol metabolism
Our biochemical and RNA-Seq analysis established the importance of ATF4 in the expression of genes important in cholesterol metabolism and transport (Figure 7, B–D). Loss of ATF4 led to decreased expression of SOAT2, a gene that was also identified as an ATF4 and CHOP target by ChIP-Seq analyses (Han et al., 2013), suggesting a lowered conversion of free cholesterol to cholesterol esters in the livers of LsATF4-KO mice (Figure 7, A and C). Free cholesterol would also be converted to bile acids at a reduced rate in the ATF4-deleted livers due to reduced expression of genes encoding CYP7A1 and CYP27A1 (Figure 7B). Export of free cholesterol would also be expected to be adversely affected in LsATF4-KO mice (Figure 7A). Previous studies suggested that ER stress results in ∼50% reduction in ABCA1 mRNA levels and decreased plasma cholesterol concentration (Rohrl et al., 2014). We determined that deletion of ATF4 results in a similar 50% reduction in ABCA1 mRNA basally, which is further reduced under stress (Figure 7D). We also showed that ER stress triggered decreased serum cholesterol, which is further lowered both basally and during stress upon loss of ATF4 (Figure 7E). Changes in gene expression and cholesterol levels may be a direct consequence of ATF4 transcriptional regulation or indirect through downstream UPR effectors. Collectively, these ATF4-directed changes in expression of genes involved in cholesterol metabolism and transport help to explain the increased free cholesterol load within liver tissues of the LsATF4-KO mice (Figure 7F).
MATERIALS AND METHODS
Animals
Animal protocols were approved by the Institutional Animal Care and Use Committees at Rutgers University and the Indiana University School of Medicine. WbATF4-KO mice were as described (Masuoka and Townes, 2002) and obtained from Jackson Laboratory (Bar Harbor, ME). C57BL/6J mice homozygous for the LoxP allele of ATF4 (ATF4fl/fl; Ebert et al., 2012) were bred with C57BL/6J mice heterozygous for the Cre recombinase gene under an albumin gene promoter (AlbCre) to create a liver-specific knockout of ATF4, LsATF4-KO (AlbCre−* ATF4fl/fl). Cre-negative mice from these litters expressed WT levels of ATF4 and were used as controls. Genotyping showed efficient ATF4 gene deletion, and proteins level reductions were seen in response to tunicamycin treatment (Figure 5, A and E). Female adult mice (five to eight per treatment group) aged 3–6 mo were individually housed in plastic cages with soft bedding, maintained on 12-h light/dark cycles, and freely provided tap water and commercial pelleted diet (5001 Laboratory Rodent Diet; LabDiet, St. Louis, MO). Mice received IP injections of tunicamycin at a dose of 1 mg/kg body weight, whereas control mice were given a vehicle consisting of a solution of 0.3% DMSO in phosphate-buffered saline (PBS). Mice were killed by decapitation using a rodent guillotine at 6, 24, and 36 h after treatment, as indicated. Livers were rinsed with ice-cold PBS and then snap frozen in liquid nitrogen or fixed in 4% paraformaldehyde. Mice homozygous for the LoxP allele of PERK (PERK/fl) were previously described (Teske et al., 2011) and bred with transgenic mice heterozygous for AlbCre to create a liver-specific knockout of PERK, LsPERK-KO (AlbCre−* PERK/fl). Cre-negative mice from these litters expressed WT levels of PERK and were used as controls.
Cell culture and lentivirus shRNA knockdown
WT and ATF4−/− MEF cells (Harding et al., 2003) and mouse hepatoma Hepa1-6 cells were cultured in DMEM (4.5 g/l glucose) supplemented with 1× nonessential amino acids (SH3023.01; HyClone), 1× MEM essential amino acids (SH30598.01; HyClone), and 50 μM β-mercaptoethanol, as described previously (Harding et al., 2003). shRNA-directed knockdowns of ATF4, ATF5, ATF6, and CHOP, were performed using a lentivirus delivery system featuring Addgene third-generation plasmids and mission shRNA clones (Sigma-Aldrich, St. Louis, MO) for generation of the viral particles. The shRNAs used in this study were as follows: ATF4, TRCN0000071726, TRCN0000071727; ATF5, TRCN0000075556, TRCN0000075553; ATF6, TRCN0000321326, TRCN0000321328; CHOP, TRCN0000103709, TRCN0000305677; and control Ctrl, TRC2-puro SHC201V. Cultured cells were treated with 2 μM tunicamycin, 1 μM MG132, or 50 nM halofuginone for the indicated times. Cultured cells that were treated with halofuginone were first washed with PBS to remove DMEM before addition of medium containing dialyzed fetal bovine serum (FBS) and subsequent addition of halofuginone.
Immunoblot analyses
Cellular lysates were prepared from cultured MEF and Hepa1-6 cells using RIPA-buffered solution containing 50 mM Tris-HCl (pH 7.5), 150 mM sodium chloride, 1% Nonidet P-40, 0.1% SDS, 100 mM sodium fluoride, 17.5 mM β-glycerophosphate, 0.5% sodium deoxycholate, and 10% glycerol that was supplemented with EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland). Liver samples were prepared as previously described (Bunpo et al., 2009). Proteins in the cell lysates were separated by SDS–PAGE, followed by immunoblot analysis using chemiluminescence as previously described (Teske et al., 2011). Primary and secondary antibodies used in immunoblot analyses include those from Abcam (Cambridge, United Kingdom) recognizing eIF2α-P (ab32157) and from Cell Signaling Technology (Danvers, MA) for actin (#8457). Antibodies for GADD34 (sc-8327) and CHOP (sc-7351) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The eIF2α total antibody was a kind gift from the laboratory of Scot Kimball (Pennsylvania State College of Medicine, Hershey, PA). ATF6 antibody was prepared in rabbits using the mouse ATF6 N-terminal amino acids 6–207 as the antigen and used at a concentration of 1:250 in 5% milk powder in PBS with 0.2% Tween 20, as previously described (Teske et al., 2011). ATF4 and ATF5 antibodies were prepared against the corresponding recombinant human proteins, which were affinity purified. Secondary antibodies were purchased from Bio-Rad (Hercules, CA). The Odyssey infrared imaging system (LI-COR, Lincoln, NE) was used for actin, eIF2α total, and CHOP antibodies. Protein levels were measured by densitometry using the ImageJ (Schneider et al., 2012).
Reverse transcription and real-time PCR
Total RNA was extracted from cultured Hepa1-6 and MEF cells and from liver tissues by using TRIzol reagent (Life Technologies, Grand Island, NY) following the manufacturer’s instructions. Reverse transcription was performed with 1 μg of each RNA sample using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Levels of the indicted mRNAs were determined by qPCR. Primers used for SYBR Green are listed in Supplemental Table S1. Amplification and detection were measured on the Realplex2 Master Cycler (Eppendorf, Hauppauge, NY). All experiments were performed with biological and technical triplicates. Results were generated using the comparative Ct method and are expressed as fold change relative to the untreated control.
Histology
Tissues were fixed in 4% paraformaldehyde, frozen, and then sectioned at 10 μM using a cryostat. TUNEL assays were performed according to the manufacturer’s instructions (Trevigen TACS 2 TdT-Blue Label In Situ Apoptosis Detection Kit; R&D Systems, Minneapolis, MN). TUNEL-positive cells were measured from equal-size sections of livers derived from similar location within the tissues. Digital images of the selected areas were prepared at 200× and imported into Scion Image for Windows (Scion, Frederick, MD), and TUNEL-positive cells were manually marked and counted.
RNA-Seq analysis
WT and LsATF4-KO mice were injected IP with 1 mg/kg tunicamycin for 6 h, or vehicle and total RNA was prepared with TRIzol reagent (Life Technologies) following the manufacturer’s instructions. RNA sequencing was performed using an Illumina HiSeq2000 at the Sulzberger Genome Center, Columbia University (New York, NY), with single-end reads and a read length of 100 base pairs. Real-time analysis was used for base calling. Fastq files were mapped to the mouse genome (NCBI37/mm9) using TopHat (version 2.0.4). Mapped reads were then assembled via Cufflinks (version 2.0.2) with the default settings. Assembled transcripts were then merged using the Cuffmerge program with the reference genome. Analysis of mRNA levels was carried out using the Cuffdiff program, with samples being grouped by treatment condition, giving four conditions and three replicates per group. Samples were considered if they demonstrated a fold change between WT no stress treatment (NT) and WT tunicamycin (TM) of ≥1.5 and if their q value, an FDR-adjusted p value with FDR set at 0.05, was ≤0.05. Venn diagrams were created with the use of the Venn diagram program (Venny 2.0.2) available at bioinfogp.cnb.csic.es/tools/venny/. Volcano plots comparing log10(statistical relevance) to log2(fold change) were generated using R (version 3.1.1), using the base plotting system and calibrate library. RNA-Seq data are deposited at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under the series number GSE76771.
Luciferase assays
Luciferase assays were carried out in six-well plates with the Dual Luciferase Reporter Assay System according to the manufacturer’s instructions (Promega, Madison, WI). The plasmid p5XATF6GLC3 used for measuring ATF6 activity plasmid was provided by Ron Prywes (Columbia University) and was previously described (Wang et al., 2000). This reporter contains five ATF6-binding elements fused to the firefly luciferase reporter gene and was cotransfected into Hepa1-6 cells along with a Renilla-expressing plasmid for normalization. Measurements were determined as the relative light units (RLUs) of the firefly luciferase to the Renilla luciferase.
Cholesterol analysis
Cholesterol content was measured in lysates prepared from liver tissues by using the Amplex Red Cholesterol Assay Kit (Molecular Probes, Eugene, OR). Cholesterol standards, liver lysate samples, and resorufin positive control were placed onto a 96-well plate in a volume of 50 μl (100 μl for the resorufin) per well before the addition of the working solution of 300 μM Amplex Red reagent. Plates were then incubated for 30 min at 37°C and protected from light sources. Resorufin, the product of the Amplex Red reagent, was then measured in a Synergy H1 (BioTek, Winooski, VT) fluorescence microplate reader using excitation of 530 nm and emission detection of 590 nm. Fluorescence intensity was then used to calculate micrograms of cholesterol-based values from the standard curve, which were normalized for protein concentrations determined for lysates.
Cell survival assays
The Calcein AM Viability Dye (eBioscience, San Diego, CA) was used to measure cell survival. This assay was performed in 96-well plates and evaluated according to manufacturer’s instructions using a Synergy H1 reader with excitation wavelength of 495 nm and emission detection at 515 nm. We cultured 5 × 103 cells in DMEM supplement with 10% FBS, penicillin (100 IU/ml), and streptomycin (100 μg/ml) in a final concentration of 2 μM tunicamycin, 1 μM MG132, or no stress agent for 24 h. Calcein-AM dye was then added for 30 min at room temperature before measurement of fluorescence. The caspase 3/7 assay was performed using the Caspase-Glo 3/7 Kit (Promega). This assay was performed in 96-well plates and done as previously described (Liu et al., 2004). Livers were homogenized in a hypotonic extraction buffer (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 5 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, 1 mM phenylmethylsulfonyl fluoride, and 1 μg/ml aprotinin, leupeptin, and pepstatin). The livers were then centrifuged at 13,000 rpm for 30 min at 4°C. Equal volumes of diluted extract (10 μg/ml) and Caspase-Glo reagent were plated in the 96-well plates before 1-h incubation at room temperature. The results were read using a Synergy H1 plate reader.
MDA analysis
The Lipid Peroxidation (MDA) Colorimetric/Fluorometric Assay kit from BioVision (Milpitas, CA) was used to measure the natural byproduct of lipid peroxidation, MDA. Liver tissue was obtained from LsPERK-WT, LsPERK-KO, LsATF4-WT, and LsATF4-KO mice that were treated with 0.3% DMSO or 1 mg/kg tunicamycin for 24 h before livers were collected. A 10-mg amount of liver tissue was prepared in MDA lysis buffer and then sonicated before centrifugation. Cells were collected with the MDA lysis buffer and sonicated. Cell lysate protein was calculated via Lowery assay to allow normalization of samples per milligram of protein used. The MDA-TBA adduct was colorimetrically detected at 532 nm, and MDA concentration was adjusted according to the mass of liver tissue used for each sample and the standard curve. Samples were measured on a Synergy H1 fluorescence microplate reader.
MitoTracker analysis
We plated 15,000 Hepa1-6 shCtrl and Hepa1-6 shATF4 cells per well in a 96-well plate. Cells were treated with vehicle or 2 μM rotenone for 6 h. The medium was removed, and 100 μl of MitoTracker Red (ThermoFisher, Waltham, MA) was added in serum-free DMEM plus 10 μg/ml Hoechst stain (ThermoFisher). The cells were incubated at 37°C for 1 h before removing the probes and washing twice with DMEM. The cells were then imaged using spinning disk confocal microscopy using Opera (PerkinElmer, Waltham, MA) and quantified using the program Columbus as previously described (Willy et al., 2015).
Statistics
All data are shown as mean ± SD and from at least three independent experiments. Statistical significance was determined using the two-tailed Student’s t test. Multiple testing between groups was assessed using a one-way analysis of variance followed by a post hoc Tukey honest significant difference test to compare multiple groups. *p < 0.05 indicates statistical significance with respect to the treated samples between the two cell types; #p < 0.05 indicates statistical significance with respect to the untreated samples between the two cell types; and $p < 0.05 indicates statistical significance between untreated sample versus ER stress for each cell type.
Supplementary Material
Acknowledgments
We thank Brian Teske, Thomas Baird, Jeffrey Elmendorf, and Howard Masuoka for helpful discussions. This study was supported by National Institutes of Health Grants GM049164 (R.C.W.), HD070487 (T.G.A.), and AR059115 (C.M.A.), the Ralph W. and Grace M. Showalter Research Trust Fund (R.C.W.), Diabetes and Obesity Training Grant Diabetes Training Grant T32DK064466 (M.E.F.), and Veterans Affairs Grants BX00976 and RX001477 (C.M.A.).
Abbreviations used:
- DMSO
dimethyl sulfoxide
- ER
endoplasmic reticulum
- GO
Gene Ontology
- GSH
glutathione
- HF
halofuginone
- IP
intraperitoneal
- LsATF4-KO
liver-specific ATF4 knockout
- LsPERK-KO
liver-specific PERK knockout
- MDA
malondialdehyde
- MEF
mouse embryonic fibroblast
- qPCR
quantitative PCR
- RNA-Seq
RNA sequencing
- ROS
reactive oxygen species
- Rot
rotenone
- shRNA
short hairpin RNA
- Tm
tunicamycin
- TUNEL
terminal deoxynucleotidyl transferase dUTP
- UPR
unfolded protein response
- WbATF4-KO
whole body ATF4 knockout
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0039) on April 13, 2016.
REFERENCES
- Baird TD, Palam LR, Fusakio ME, Willy JA, Davis CM, McClintick JN, Anthony TG, Wek RC. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKalpha. Mol Biol Cell. 2014;25:1686–1697. doi: 10.1091/mbc.E14-02-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Tessmann IP, Chen C, Zhong C, Siu F, Schuster SM, Nick HS, Kilberg MS. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J Biol Chem. 2000;275:26976–26985. doi: 10.1074/jbc.M000004200. [DOI] [PubMed] [Google Scholar]
- Baumeister P, Luo S, Skarnes WC, Sui G, Seto E, Shi Y, Lee AS. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol Cell Biol. 2005;25:4529–4540. doi: 10.1128/MCB.25.11.4529-4540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JW. Regulatory crosstalk within the mammalian unfolded protein response. Cell Mol Life Sci. 2014;71:1067–1079. doi: 10.1007/s00018-013-1490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhat A, Averous J, Carraro V, Zhong C, Reimold AM, Kilberg MS, Fafournoux P. Differences in the molecular mechanisms involved in the transcriptional activation of CHOP and asparagine synthetase in response to amino acid deprivation or activation of the unfolded protein response. J Biol Chem. 2002;277:48107–48114. doi: 10.1074/jbc.M206149200. [DOI] [PubMed] [Google Scholar]
- Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation factor 2. Mol Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG. GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent L-asparaginase. J Biol Chem. 2009;284:32742–32749. doi: 10.1074/jbc.M109.047910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing of XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Cases S, Novak S, Zheng YW, Myers HM, Lear SR, Sande E, Welch CB, Lusis AJ, Spencer TA, Krause BR, et al. ACAT-2, a second mammalian acyl-CoA:cholesterol acyltransferase. Its cloning, expression, and characterization. J Biol Chem. 1998;273:26755–26764. doi: 10.1074/jbc.273.41.26755. [DOI] [PubMed] [Google Scholar]
- Chen H, Pan YX, Dudenhausen EE, Kilberg MS. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. J Biol Chem. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- Chen X, Shen J, Prywes R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem. 2002;277:13045–13052. doi: 10.1074/jbc.M110636200. [DOI] [PubMed] [Google Scholar]
- Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol. 2001;21:6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fabiani E, Mitro N, Anzulovich AC, Pinelli A, Galli G, Crestani M. The negative effects of bile acids and tumor necrosis factor-alpha on the transcription of cholesterol 7alpha-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem. 2001;276:30708–30716. doi: 10.1074/jbc.M103270200. [DOI] [PubMed] [Google Scholar]
- De Sousa-Coelho AL, Marrero PF, Haro D. Activating transcription factor 4-dependent induction of FGF21 during amino acid deprivation. Biochem J. 2012;443:165–171. doi: 10.1042/BJ20111748. [DOI] [PubMed] [Google Scholar]
- Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem. 2010;285:33165–33174. doi: 10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Sayers CM, Verginadis II, Lehman SL, Cheng Y, Cerniglia GJ, Tuttle SW, Feldman MD, Zhang PJ, Fuchs SY, et al. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. J Clin Invest. 2015;125:2592–2608. doi: 10.1172/JCI78031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickhout JG, Carlisle RE, Jerome DE, Mohammed-Ali Z, Jiang H, Yang G, Mani S, Garg SK, Banerjee R, Kaufman RJ, et al. Integrated stress response modulates cellular redox state via induction of cystathionine gamma-lyase: cross-talk between integrated stress response and thiol metabolism. J Biol Chem. 2012;287:7603–7614. doi: 10.1074/jbc.M111.304576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert SM, Dyle MC, Kunkel SD, Bullard SA, Bongers KS, Fox DK, Dierdorff JM, Foster ED, Adams CM. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J Biol Chem. 2012;287:27290–27301. doi: 10.1074/jbc.M112.374777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, Sweeney M, Rong JX, Kuriakose G, Fisher EA, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012;15:623–634. doi: 10.1016/j.cmet.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M, Diatchenko L, Kabil O, Willard B, Banerjee R, et al. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife. 2015;4:e10067. doi: 10.7554/eLife.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Scheuner D, Chen JJ, Kaufman RJ, Ron D. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2alpha) dephosphorylation in mammalian development. Proc Natl Acad Sci USA. 2009;106:1832–1837. doi: 10.1073/pnas.0809632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Cell Biol. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- Huggins CJ, Mayekar MK, Martin N, Saylor KL, Gonit M, Jailwala P, Kasoji M, Haines DC, Quinones OA, Johnson PF. C/EBPgamma is a critical regulator of cellular stress response networks through heterodimerization with ATF4. Mol Cell Biol. 2015;36:693–713. doi: 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner-Weibel G, Jerome WG, Small DM, Warner GJ, Stoltenborg JK, Kearney MA, Corjay MH, Phillips MC, Rothblat GH. Effects of intracellular free cholesterol accumulation on macrophage viability: a model for foam cell death. Arterioscler Thromb Vasc Biol. 1998;18:423–431. doi: 10.1161/01.atv.18.3.423. [DOI] [PubMed] [Google Scholar]
- Kilberg MS, Balasubramanian M, Fu L, Shan J. The transcription factor network associated with the amino acid response in mammalian cells. Adv Nutr. 2012;3:295–306. doi: 10.3945/an.112.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kir S, Zhang Y, Gerard RD, Kliewer SA, Mangelsdorf DJ. Nuclear receptors HNF4alpha and LRH-1 cooperate in regulating Cyp7a1 in vivo. J Biol Chem. 2012;287:41334–41341. doi: 10.1074/jbc.M112.421834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Li C, Chen Y, Burnett C, Liu XY, Downs S, Collins RD, Hawiger J. Nuclear import of proinflammatory transcription factors is required for massive liver apoptosis induced by bacterial lipopolysaccharide. J Biol Chem. 2004;279:48434–48442. doi: 10.1074/jbc.M407190200. [DOI] [PubMed] [Google Scholar]
- Lu K, Lee MH, Hazard S, Brooks-Wilson A, Hidaka H, Kojima H, Ose L, Stalenhoef AF, Mietinnen T, Bjorkhem I, et al. Two genes that map to the STSL locus cause sitosterolemia: genomic structure and spectrum of mutations involving sterolin-1 and sterolin-2, encoded by ABCG5 and ABCG8, respectively. Am J Hum Genet. 2001;69:278–290. doi: 10.1086/321294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318:1351–1365. doi: 10.1016/s0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuoka HC, Townes TM. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood. 2002;99:736–745. doi: 10.1182/blood.v99.3.736. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslowski CM, Urano F. The binary switch that controls the life and death decisions of ER stressed beta cells. Curr Opin Cell Biol. 2011;23:207–215. doi: 10.1016/j.ceb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Robertson L, Gallinetti J, Mejia P, Vose S, Charlip A, Chu T, Mitchell JR. Surgical stress resistance induced by single amino acid deprivation requires Gcn2 in mice. Sci Transl Med. 2012;4:118ra111. doi: 10.1126/scitranslmed.3002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Rohrl C, Eigner K, Winter K, Korbelius M, Obrowsky S, Kratky D, Kovacs WJ, Stangl H. Endoplasmic reticulum stress impairs cholesterol efflux and synthesis in hepatic cells. J Lipid Res. 2014;55:94–103. doi: 10.1194/jlr.M043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal CN, Hunsaker LA, Barbash O, Vander Jagt DL, Abcouwer SF. The oxidative stressor arsenite activates vascular endothelial growth factor mRNA transcription by an ATF4-dependent mechanism. J Biol Chem. 2005;280:20331–20339. doi: 10.1074/jbc.M411275200. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. Divergent roles of IRE1alpha and PERK in the unfolded protein response. Curr Mol Med. 2006;6:5–36. doi: 10.2174/156652406775574569. [DOI] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Su N, Kilberg MS. C/EBP homology protein (CHOP) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. J Biol Chem. 2008;283:35106–35117. doi: 10.1074/jbc.M806874200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundrud MS, Koralov SB, Feuerer M, Calado DP, Kozhaya AE, Rhule-Smith A, Lefebvre RE, Unutmaz D, Mazitschek R, Waldner H, et al. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 2009;324:1334–1338. doi: 10.1126/science.1172638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske BF, Fusakio ME, Zhou D, Shan J, McClintick JN, Kilberg MS, Wek RC. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol Biol Cell. 2013;24:2477–2490. doi: 10.1091/mbc.E13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske BF, Wek SA, Bunpo P, Cundiff JK, McClintick JN, Anthony TG, Wek RC. The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol Biol Cell. 2011;22:4390–4405. doi: 10.1091/mbc.E11-06-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires the bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream open reading frames regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang XZ, Lawson B, Brewer J, Zinszner H, Sanjay A, Mi LJ, Boorsterin R, Kreibich G, Hendershot L, Ron D. Signals from the stress endoplasmic reticulum induce C/EBP homologous protein (CHOP/GADD153) Mol Cell Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Willy JA, Young HA, Stevens JL, Masuoka HC, Wek RC. CHOP links endoplasmic reticulum stress to NF-kB activation in the pathogenesis of nonalcoholic steatophepatitis. Mol Biol Cell. 2015;26:2190–2204. doi: 10.1091/mbc.E15-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YC, Xu A, Wang Y, Lam KS. Fibroblast growth factor 21 as an emerging metabolic regulator: clinical perspectives. Clin Endocrinol (Oxf) 2013;78:489–496. doi: 10.1111/cen.12095. [DOI] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Xiao G, Zhang T, Yu S, Lee S, Calabuig-Navarro V, Yamauchi J, Ringquist S, Dong HH. ATF4 protein deficiency protects against high fructose-induced hypertriglyceridemia in mice. J Biol Chem. 2013;288:25350–25361. doi: 10.1074/jbc.M113.470526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamotot A, Okada T, Mori K. XPB1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zhao B, Song J, Ghosh S. Hepatic overexpression of cholesteryl ester hydrolase enhances cholesterol elimination and in vivo reverse cholesterol transport. J Lipid Res. 2008;49:2212–2217. doi: 10.1194/jlr.M800277-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.