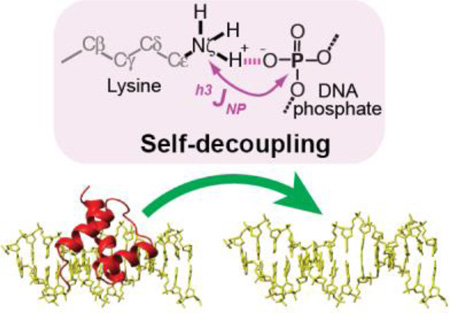
Abstract
The residence times of molecular complexes in solution are important for understanding biomolecular functions and drug actions. Here we show that NMR data of intermolecular hydrogen-bond scalar couplings can yield information on the residence times of molecular complexes in solution. The molecular exchange of binding partners via the breakage and reformation of a complex causes self-decoupling of intermolecular hydrogen-bond scalar couplings, and this self-decoupling effect depends on the residence time of the complex. For protein–DNA complexes, we investigated the salt-concentration dependence of intermolecular hydrogen-bond scalar couplings between the protein side-chain 15N and DNA phosphate 31P nuclei, from which the residence times were analyzed. The results were consistent with those obtained by 15Nz-exchange spectroscopy. This self-decoupling-based kinetic analysis is unique in that it does not require any different signatures for the states involved in the exchange, whereas such conditions are crucial for kinetic analyses by typical NMR and other methods.
Graphical Abstract
In solution, molecular complexes formed via noncovalent interactions typically undergo dynamic equilibria involving dissociation and association. The residence times of molecular complexes are important for our understanding of biomolecular functions and for the development of effective drugs.1–3 Various methods, such as fluorescence and NMR spectroscopy and surface plasmon resonance, can be used to determine the residence time of a complex. Regardless of the method used, the determination of residence times usually requires the observation of a transition between distinct states (e.g., free and bound states or two different complexes) that exhibit distinct physiochemical signatures (e.g., different NMR chemical shifts). In this paper, we demonstrate a unique NMR approach that does not require distinct states with different NMR chemical shifts, yet does provide information on the residence times of molecular complexes.
This approach utilizes NMR scalar couplings across intermolecular hydrogen bonds. Hydrogen-bond scalar couplings were discovered in the late 1990s, initially for the hydrogen bonds of nucleic-acid base pairs and protein secondary structures (e.g., as reviewed in ref. 4). This type of scalar coupling represents direct evidence of the presence of hydrogen bonds and provides structural and dynamic information on hydrogen bonding. Hydrogen-bond scalar couplings have also been observed for the intermolecular hydrogen bonds formed at the molecular interfaces of nucleic acids (as reviewed in refs 5–6), protein–nucleic acid complexes,7–11 and other protein–ligand complexes12–13.
First, we consider how the residence time of a complex is related to the apparent values of the intermolecular hydrogen-bond scalar coupling constants measured by quantitative J-modulation spin-echo difference experiments.7,14 This type of experiment involves two sub-experiments, with (B) and without (A) intensity modulation by evolution of the hydrogen-bond scalar coupling hJ during the constant-time period 2T (an example is shown in the Supporting Information [SI]). From the signal intensities IA and IB in the spectra of sub-experiments A and B, the magnitude of the coupling constant is determined by:
| [1] |
.
The following consideration shows that this yields the intrinsic magnitude |hJ| only when the “self-decoupling” effect15–17 is negligible. The evolution of the density operator ρ is given by:
| [2] |
for the sub-experiment A and
| [3] |
for the sub-experiment B, where t1 represents the time for chemical shift evolution for the F1 dimension; L, a Liouvillian matrix; SN and SP, the rotation matrices for 180° pulses for coupled nuclei. In the case of intermolecular h3JNP coupling between the 15N and 31P nuclei, a basis for the density operators can be defined as a column vector of [Nx, 2NyPz]. For this basis, the Liouvillian matrix L and the rotation matrices SN and SP are as follows:
| [4] |
| [5] |
, where R2,N represents the 15N transverse relaxation rate; J, the intrinsic hydrogen-bond scalar coupling; R1,P, the 31P longitudinal relaxation rate; and k, a first-order rate constant for molecular exchange. Eq. 5 assumes 180° pulses along x. The mean residence time τ of the molecular complex is given by:
| [6] |
.
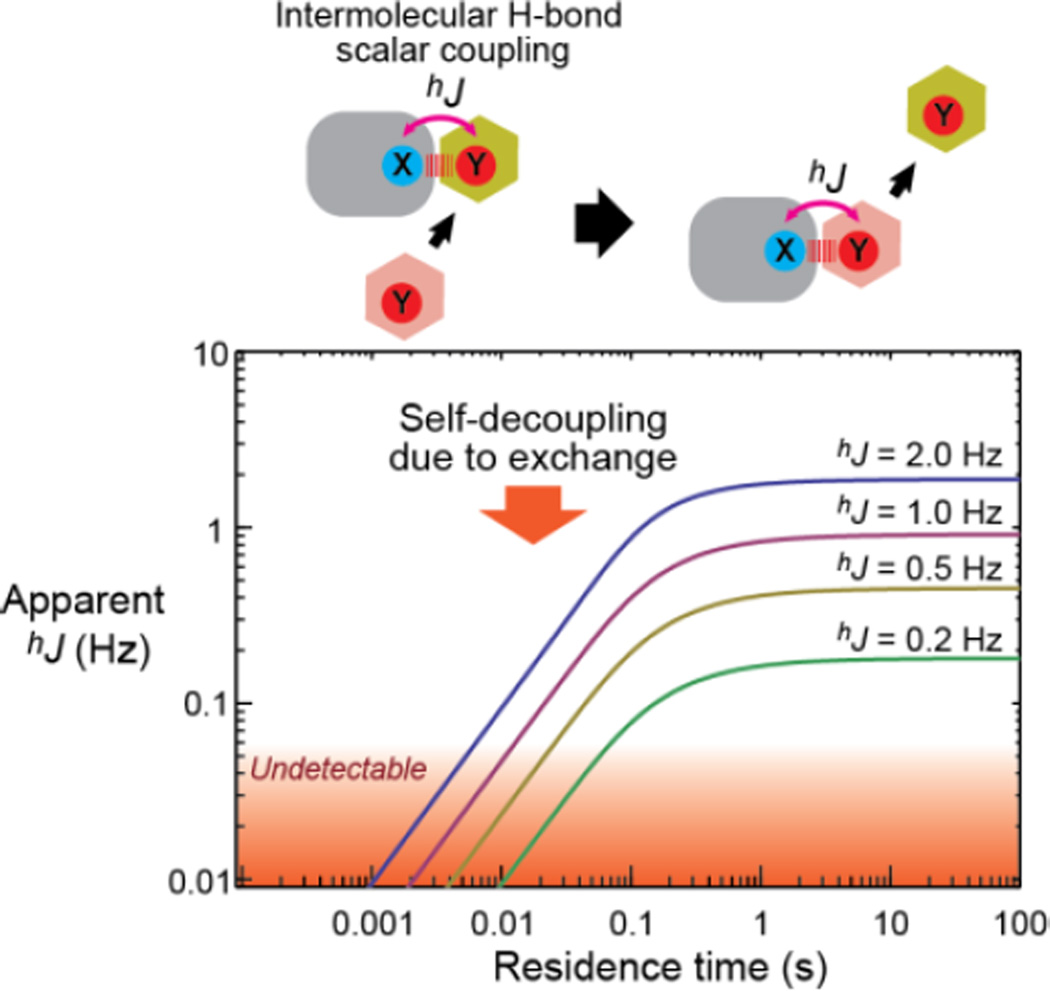
Figure 1 shows the apparent constant hJapp calculated with Eqs. 1–6 as a function of the residence time of a complex. When the residence time is short, making k >> 2π|hJ | (i.e., fast exchange on a scalar coupling timescale rather than a chemical shift timescale), the apparent hJapp constant approximates zero (Figure 1). This represents self-decoupling due to molecular exchange.
Figure 1.
Exchange-induced self-decoupling of intermolecular hydrogen-bond scalar coupling hJ. The graph shows the theoretical relationship between the residence time of a complex and the apparent hJ constants (on a logarithmic scale for each axis). The curves were obtained with Eqs. 1–6 together with R1,P = 1 s−1 and the intrinsic hJ constants indicated.
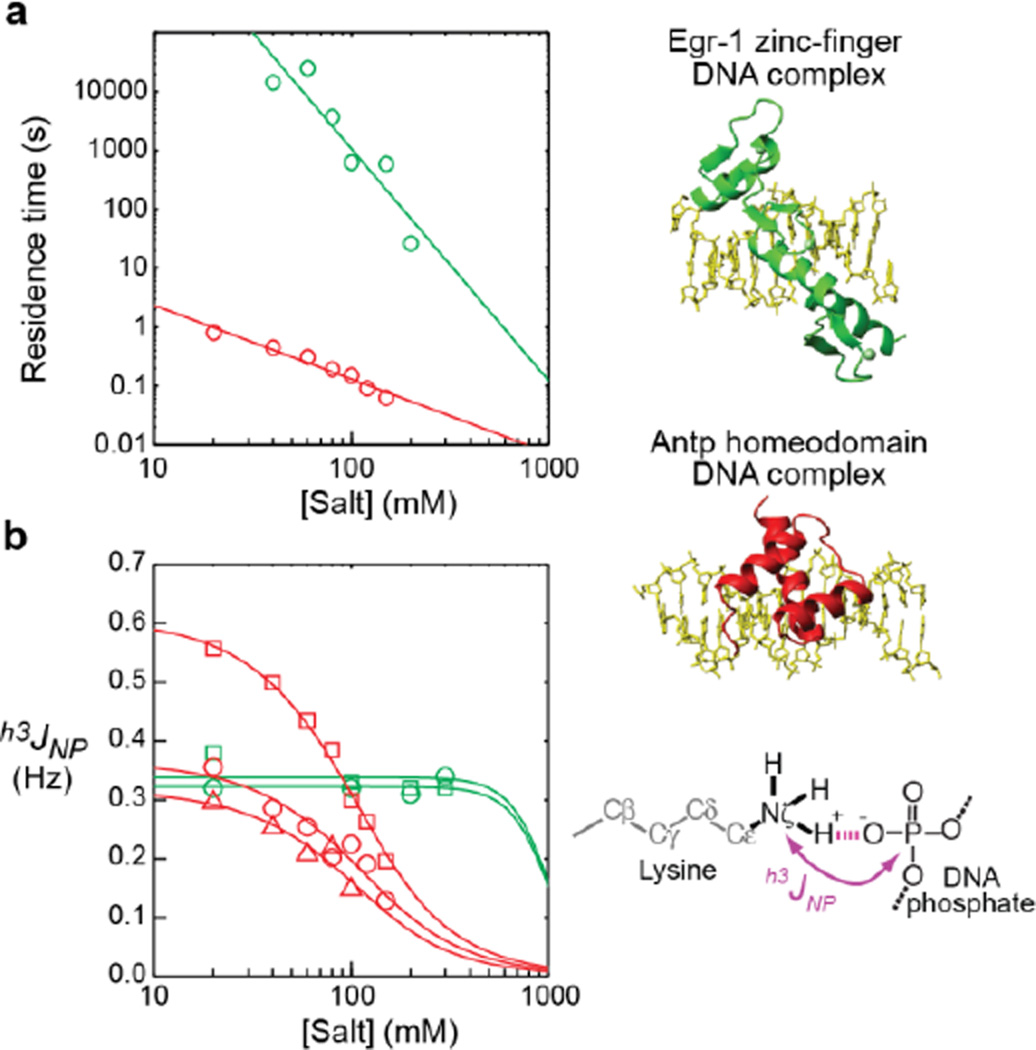
We have examined this theoretical relationship between the intermolecular hydrogen-bond scalar couplings hJapp and the residence times for protein–DNA complexes. We can alter the residence times of the complexes by changing the ionic strength. Figure 2a shows the residence times for the specific protein–DNA complexes of the Antp homeodomain (with C39S mutation)11 and the Egr-1 zinc-finger (with T23K/Q60E mutations)18 proteins with their target DNA at various concentrations of salt. The residence times were measured by 15Nz-exchange spectroscopy19–20 (for Antp) or a fluorescence-based kinetic assay (for Egr-1), as described in the SI. For these systems, the residence time is defined as the inverse of the apparent first-order rate constant for translocation of the protein from one target DNA duplex to another. This rate constant does not necessarily correspond to a dissociation rate constant because proteins can transfer from one DNA duplex to another via the “intersegment transfer” (also known as the “direct transfer”) mechanism without going through the intermediary of free protein.21 It is known that homeodomains can undergo intersegment transfer between specific DNA duplexes.22 For Egr-1, the intersegment transfer between two target DNA duplexes is virtually negligible,23 though this protein can undergo extremely efficient intersegment transfer between nonspecific DNA sites via the “search mode” during the target search process.24–27 Due to this difference, the residence time of the Antp–DNA complex is far shorter than that of the Egr-1–DNA complex, although these complexes exhibit a comparable dissociation constant (Kd) at the same ionic strength.11,27 Figure 2a shows that for each complex, the logarithm of the residence time is linearly dependent on the logarithm of the salt concentration, as expected from previous theoretical and experimental studies.20,25,28
Figure 2.
Salt concentration-dependence data for the sequence-specific DNA complexes of the Antp homeodomain (C39S mutant) (red) and the Egr-1 zinc-finger (Q32E/T23K mutant) (green). (a) Residence times measured for the protein–DNA complexes. The residence times in these graphs were measured by 15Nz-exchange spectroscopy for the Antp complex and by a fluorescence anisotropy-based kinetic method for the Egr-1 complex, as described in the SI. The data are shown on logarithmic scales in both axes. (b) Intermolecular hydrogen-bond scalar coupling h3JNP between protein side-chain NH3+ and DNA phosphate groups. The red data points are Lys46 (square), Lys55 (circle), and Lys57 (triangle) of the Antp homeodomain–DNA complex. The green points are Lys23 (square) and Lys79 (circle) of the Egr-1 zinc-finger(Q32E/T23K mutant)–DNA complex. The solid curves represent the best-fit curves with Eqs. 1–6 and log τ = alog[Salt]+b. Further details of the curve-fitting calculations are included in the SI.
For these complexes, we measured the intermolecular hydrogen-bond scalar couplings between 15N and 31P nuclei across the contact ion pairs of lysine (Lys) side-chain NH3+ and DNA phosphate groups at various ionic strengths (Figure 2b). Owing to the extremely slow 15N transverse relaxation of the Lys side-chain NH3+ groups (typically, R2,N < 3 s−1), the magnitude of the h3JNP coupling constants can be measured precisely for the Lys side chains that are in contact with DNA phosphate groups.7,9,11,29 The h3JNP coupling constants for the Antp–DNA complex exhibited a strong dependence on the salt concentration between 20 and 150 mM, whereas those for the Egr-1–DNA complex exhibited no significant dependence on the salt concentration up to 300 mM (Figure 2b). Importantly, these observations are at least qualitatively consistent with the theoretical expectation from Figures 1 and 2a.
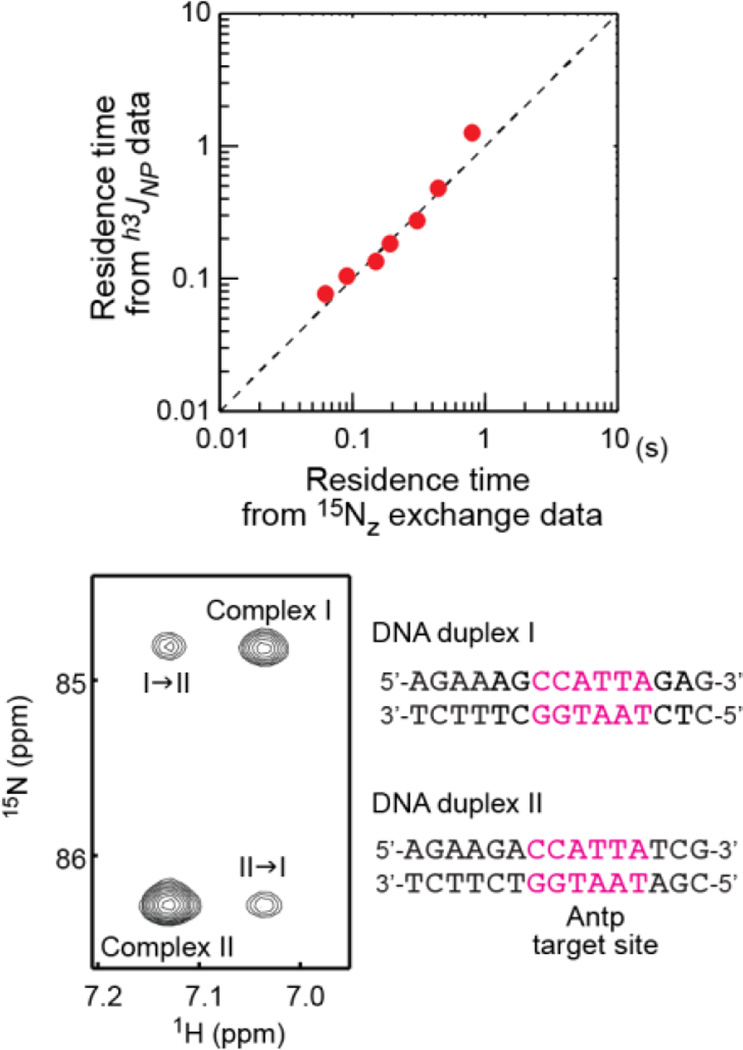
For the Egr-1–DNA complex, the h3JNP data and Figure 1 qualitatively suggest that the residence time of this complex is longer than (2π|h3JNP|)−1 in the range of 20–300 mM KCl. For the Antp–DNA complex, the salt-dependent h3JNP data provide more quantitative information on the residence time of the Antp–DNA complex. Assuming that the residence time τ satisfies log τ = alog[Salt]+b (as observed in Figure 2a), we conducted a nonlinear least-squares fit of the salt-dependent h3JNP data using Eqs 1–6. For the Antp–DNA complex, the calculations were performed via a global fit of the experimental data for three residues (with Lys46, Lys55, and Lys57), as described in the SI. The parameters R1,P and hJ, which were individually defined for each residue, and the global parameters a and b were optimized. The best-fit curves (red solid lines in Figure 2b) showed excellent agreement with the experimental salt-dependent h3JNP data. Furthermore, as shown in Figure 3, the residence times from this analysis were in good agreement with those from the 15Nz-exchange experiment in which the exchange process of two complexes with different DNA duplexes was analyzed.
Figure 3.
Comparison of the residence times determined from the salt-dependent h3JNP data with those from the 15Nz-exchange data for the Antp homeodomain-DNA complex. Assuming that the residence time τ satisfies log τ = alog[Salt]+b, the parameters a, b, R1,P, and hJ were optimized via nonlinear least-squares fitting with Eqs 1–6 to the salt-dependent h3JNP data (Figure 2b). In the 15Nz-exchange experiment, the exchanges between two DNA complexes (one with the duplex I and the other with duplex II; the sequences are shown) were investigated through the analysis of the time-dependence of auto and exchange cross peaks, as previously described.22 The signals from the Arg-5 Nε-Hε groups are shown.
Theoretically, the self-decoupling effect should become significant when the interconversions between the α and β spin states of the coupling partners (in the current case, 31P nuclei) occur at a rate faster than 2π|J|.15–17 Such interconversions can be caused by longitudinal relaxation of the coupling partner or by molecular exchange (i.e., large R1,P or k in Eq. 4). Intramolecular dynamics that transiently break the hydrogen bond within the residence time should certainly attenuate hJ coupling,4,30 but do not necessarily cause self-decoupling because the α and β spin states of the coupling partner remain the same. In fact, our previous NMR and molecular dynamics studies showed that the Lys 15NH3+ groups that exhibit sizable h3JNP coupling with the DNA 31P nuclei undergo dynamic transitions on a sub-nanosecond timescale between the contact ion-pair and solvent-separated ion-pair states.7,9,31
In conclusion, our current work demonstrates that intermolecular hydrogen-bond scalar coupling data can provide information on the residence times of molecular complexes in solution. The observation of intermolecular hydrogen-bond scalar couplings is possible only if the residence time is sufficiently long (> ~10−2 s); otherwise, complete self-decoupling occurs as shown in Figure 1. Qualitatively, when an intermolecular hydrogen-bond scalar coupling is observed with a magnitude comparable to that of the intrinsic coupling constant hJ, the residence time of the complex should be longer than (2π|hJ|)−1. The intrinsic values of hydrogen-bond scalar couplings can be calculated from structural information by quantum chemical (e.g., DFT) calculations or from the empirical relationship between the coupling constants and the hydrogen-bond geometry.4,32–33 As demonstrated above, detailed analysis of the self-decoupling of intermolecular hydrogen-bond scalar couplings can provide more quantitative information about the residence time of the complex. For quantitative applications of h3JNP, the analyzable range of residence time is ~ 0.001 – 1 s, provided that | h3JNP | ≤ ~4 Hz (as estimated by DFT calculations7,33). For systems for which the ionic-strength dependence cannot be used, temperature dependence could be used under the assumption that the molecular exchange rates obey the Arrhenius equation (or the Kramers theorem on mean first passage times34). Thus, although self-decoupling is typically regarded as a nuisance in NMR scalar coupling measurements,15–17 the quantitative analysis of self-decoupling presents a useful tool for kinetic analysis. This self-decoupling-based method is unique because it does not require different signatures for the states involved in the exchange, although such conditions are typically crucial for other methods. For example, kinetic analyses by other NMR methods (e.g., relaxation dispersion, CEST, exchange spectroscopy)35–36 require different chemical shifts for the interconverting states. The self-decoupling-based method can provide kinetic information even if the exchange is not accompanied by any change in the chemical shifts.
Experimental Methods
Chemically synthesized DNA oligonucleotides were purchased from the Integrated DNA Technologies, Inc. (Coralville, IA) and purified by anion-exchange chromatography. Isotopically labeled proteins were expressed in E. coli and purified by chromatographic methods. The protein–DNA complexes were prepared as previously described.11,18 Intermolecular hydrogen-bond scalar couplings between protein side-chain 15N and DNA phosphate 31P nuclei were measured using a Bruker Avance III 600 MHz spectrometers equipped with a 1H/13C/15N/31P QCI cryogenic probe, as described.7 The 15Nz-exchange spectroscopy data were obtained using a Bruker Avance III 800 MHz spectrometer equipped with a 1H/13C/15N TCI cryogenic probe. Further experimental details are included in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by Grants R01-GM107590 and R01-GM105931 from the National Institutes of Health (to J.I.). We thank Dr. Tianzhi Wang for the maintenance of the NMR instruments at the Sealy Center for Structural Biology and Molecular Biophysics.
Footnotes
Supporting Information: The following file is available free of charge: Experimental details (PDF)
The authors declare no competing financial interests.
REFERENCES
- 1.Copeland RA, Pompliano DL, Meek TD. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 2.Lu H, Tonge PJ. Drug-target residence time: critical information for lead optimization. Curr. Opin. Chem. Biol. 2010;14:467–474. doi: 10.1016/j.cbpa.2010.06.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tummino PJ, Copeland RA. Residence time of receptor-ligand complexes and its effect on biological function. Biochemistry. 2008;47:5481–5492. doi: 10.1021/bi8002023. [DOI] [PubMed] [Google Scholar]
- 4.Grzesiek S, Cordier F, Jaravine V, Barfield M. Insights into biomolecular hydrogen bonds from hydrogen bond scalar couplings. Prog. Nucl. Magn. Reson. Spectrosc. 2004;45:275–300. [Google Scholar]
- 5.Dallmann A, Sattler M. Detection of hydrogen bonds in dynamic regions of RNA by NMR spectroscopy. Curr. Protoc. Nucleic Acid Chem. 2014;59:72221. doi: 10.1002/0471142700.nc0722s59. [DOI] [PubMed] [Google Scholar]
- 6.Majumdar A, Patel DJ. Identifying hydrogen bond alignments in multistranded DNA architectures by NMR. Acc. Chem. Res. 2002;35:1–11. doi: 10.1021/ar010097+. [DOI] [PubMed] [Google Scholar]
- 7.Anderson KM, Esadze A, Manoharan M, Bruschweiler R, Gorenstein DG, Iwahara J. Direct observation of the ion-pair dynamics at a protein-DNA interface by NMR spectroscopy. J. Am. Chem. Soc. 2013;135:3613–3619. doi: 10.1021/ja312314b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson KM, Nguyen D, Esadze A, Zandrashvili L, Gorenstein DG, Iwahara J. A chemical approach for site-specific identification of NMR signals from protein side-chain NH3+groups forming intermolecular ion pairs in protein-nucleic acid complexes. J. Biomol. NMR. 2015;62:1–5. doi: 10.1007/s10858-015-9909-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, Esadze A, Zandarashvili L, Nguyen D, Montgomery Pettitt B, Iwahara J. Dynamic equilibria of short-range electrostatic interactions at molecular interfaces of protein-DNA complexes. J. Phys. Chem. Lett. 2015;6:2733–2737. doi: 10.1021/acs.jpclett.5b01134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu AZ, Majumdar A, Jiang F, Chernichenko N, Skripkin E, Patel DJ. NMR detection of intermolecular N-H•••N hydrogen bonds in the human T cell leukemia virus-1 Rex peptide-RNA aptamer complex . J. Am. Chem. Soc. 2000;122:11226–11227. [Google Scholar]
- 11.Zandarashvili L, Nguyen D, Anderson KM, White MA, Gorenstein DG, Iwahara J. Entropic enhancement of protein-DNA affinity by oxygen-to-sulfur substitution in DNA phosphate. Biophys. J. 2015;109:1026–1037. doi: 10.1016/j.bpj.2015.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Löhr F, Mayhew SG, Rüterjans H. Detection of scalar couplings across NH•••OP and OH•••OP hydrogen bonds in a flavoprotein. J. Am. Chem. Soc. 2000;122:9289–9295. [Google Scholar]
- 13.Mishima M, Hatanaka M, Yokoyama S, Ikegami T, Walchli M, Ito Y, Shirakawa M. Intermolecular31P-15N and31P-1H scalar couplings across hydrogen bonds formed between a protein and a nucleotide. J. Am. Chem. Soc. 2000;122:5883–5884. [Google Scholar]
- 14.Bax A, Vuister GW, Grzesiek S, Delaglio F, Wang AC, Tschudin R, Zhu G. Measurement of homonuclear and heteronuclear J-couplings from quantitative J-correlation. Methods Enzymol. 1994;239:79–105. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- 15.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR sprectroscopy: principles and practice. 2nd. Burlignton: Elsevier Academic Press; 2007. [Google Scholar]
- 16.Harbison GS. Interference between J-couplings and cross-relaxation in solution NMR Spectroscopy - consequences for macromolecular structure determination. J. Am. Chem. Soc. 1993;115:3026–3027. [Google Scholar]
- 17.Kuboniwa H, Grzesiek S, Delaglio F, Bax A. Measurement of HN-Hα J-couplings in calcium-free calmodulin using New 2D and 3D water-flip-back methods. J. Biomol. NMR. 1994;4:871–878. doi: 10.1007/BF00398416. [DOI] [PubMed] [Google Scholar]
- 18.Zandarashvili L, Esadze A, Vuzman D, Kemme CA, Levy Y, Iwahara J. Balancing between affinity and speed in target DNA search by zinc-finger proteins via modulation of dynamic conformational ensemble. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E5142–E5149. doi: 10.1073/pnas.1507726112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 20.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 21.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 22.Iwahara J, Clore GM. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J. Am. Chem. Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 23.Takayama Y, Sahu D, Iwahara J. NMR studies of translocation of the Zif268 protein between its target DNA Sites. Biochemistry. 2010;49:7998–8005. doi: 10.1021/bi100962h. [DOI] [PubMed] [Google Scholar]
- 24.Esadze A, Iwahara J. Stopped-flow fluorescence kinetic study of protein sliding and intersegment transfer in the target DNA search process. J. Mol. Biol. 2014;426:230–244. doi: 10.1016/j.jmb.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esadze A, Kemme CA, Kolomeisky AB, Iwahara J. Positive and negative impacts of nonspecific sites during target location by a sequence-specific DNA-binding protein: origin of the optimal search at physiological ionic strength. Nucleic Acids Res. 2014;42:7039–7046. doi: 10.1093/nar/gku418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zandarashvili L, Vuzman D, Esadze A, Takayama Y, Sahu D, Levy Y, Iwahara J. Asymmetrical roles of zinc fingers in dynamic DNA-scanning process by the inducible transcription factor Egr-1. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1724–E1732. doi: 10.1073/pnas.1121500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zandarashvili L, White MA, Esadze A, Iwahara J. Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1. FEBS Lett. 2015;589:1748–1753. doi: 10.1016/j.febslet.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohman TM, DeHaseth PL, Record MT., Jr Analysis of ion concentration effects of the kinetics of protein-nucleic acid interactions. Application to lac repressor-operator interactions. Biophys. Chem. 1978;8:281–294. doi: 10.1016/0301-4622(78)80011-8. [DOI] [PubMed] [Google Scholar]
- 29.Zandarashvili L, Esadze A, Iwahara J. NMR studies on the dynamics of hydrogen bonds and ion pairs involving lysine side chains of proteins. Adv. Protein Chem. Struct. Biol. 2013;93:37–80. doi: 10.1016/B978-0-12-416596-0.00002-6. [DOI] [PubMed] [Google Scholar]
- 30.Jaravine VA, Alexandrescu AT, Grzesiek S. Observation of the closing of individual hydrogen bonds during TFE-induced helix formation in a peptide. Protein Sci. 2001;10:943–950. doi: 10.1110/ps.48501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwahara J, Esadze A, Zandarashvili L. Physicochemical properties of ion pairs of biological macromolecules. Biomolecules. 2015;5:2435–2463. doi: 10.3390/biom5042435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barfield M. Structural dependencies of interresidue scalar coupling h3JNC and donor 1H chemical shifts in the hydrogen bonding regions of proteins. J. Am. Chem. Soc. 2002;124:4158–4168. doi: 10.1021/ja012674v. [DOI] [PubMed] [Google Scholar]
- 33.Czernek J, Brüschweiler R. Geometric dependence of 3hJ(31P-15N) and 2hJ(31P-1H) scalar couplings in protein-nucleotide complexes. J. Am. Chem. Soc. 2001;123:11079–11080. doi: 10.1021/ja011618r. [DOI] [PubMed] [Google Scholar]
- 34.Zwanzig R. Nonequilibrium statistical mechanics. New York: Oxford University Press; 2001. [Google Scholar]
- 35.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Palmer AG., 3rd Chemical exchange in biomacromolecules: past, present, and future. J. Magn. Reson. 2014;241:3–17. doi: 10.1016/j.jmr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.