Abstract
Here we describe the expression of two novel transcripts, Ende (AK014119) and Npe (AK084355), during early mouse embryogenesis. Ende mRNA was first detected at embryonic day (E) 7.0 in a small population of epiblast cells in the distal half of the embryo. At E7.5, Ende was expressed by newly formed definitive endoderm cells in the proximal half of the embryo, and was not expressed in extra-embryonic endoderm. This expression pattern then changed to the ventral aspect of the developing foregut pocket and the entire hindgut pocket at E8.0–8.5, before becoming restricted to the foregut overlying the heart and the posterior most hindgut. By E9.0 Ende expression was also observed in the posterior half of the ventral neural tube. Thus, Ende was expressed dynamically and in specific populations of the definitive endoderm from E7.0 to E8.5. We found Npe expression to be restricted to the node/posterior notochord region at the distal tip of the embryo between E7.0 and E8.0. By E9.5, Npe expression was observed in the posterior most population of dorsal hindgut cells and notochord cells. Given their expression in mouse definitive endoderm populations, Ende and Npe will be valuable tools to study formation and development of this tissue.
Keywords: mouse, embryo, gastrulation, definitive endoderm, node, posterior notochord, serial analysis of gene expression, SAGE, AK014119, AK084355, Ende, Npe, Pyy, Hex
1. Results and Discussion
The definitive endoderm is one of the three germ layers of the embryo that are generated during gastrulation and gives rise to the organs of the respiratory and gastrointestinal tracts including the lungs, liver, pancreas and intestine (Wells and Melton, 1999). Progress in the understanding of definitive endoderm formation and patterning has lagged behind that of the mesoderm and ectoderm, particularly due to the lack of genetic markers specific to this tissue. Understanding the development of the definitive endoderm is crucial for future in vitro based approaches to regenerative medicine for diabetes or liver diseases.
We previously described a systematic screen for genes expressed in the definitive endoderm using Serial Analysis of Gene Expression (SAGE) libraries (Hou et al., 2007). Three mouse definitive endoderm LongSAGE libraries were compared against more than 200 mouse LongSAGE libraries from various embryonic stages and tissues, successfully identifying several genes as new markers for the definitive endoderm, including Nephrocan (Nepn) and Peptide YY (Pyy) (Hou et al., 2007). This study, however, limited itself to screening for sequences that mapped to transcript databases, and omitted a list of non-annotated, uncharacterized sequences, potentially representing novel genes enriched for expression in the definitive endoderm. Thus, we extended this SAGE study and analyzed tag sequences representing non-annotated transcripts to identify candidates expressed in the definitive endoderm. From this analysis we identified two ESTs, AK014119 and AK084355, showing enrichment for definitive endoderm. AK014119 was renamed endoderm enriched or Ende and AK084355 was renamed notochord posterior end or Npe for the sake of simplicity.
Ende lies on chromosome 8 and is 9.8kb upstream of Trim60 and 48kb downstream of a non-coding RNA, EST BC030870. Ende has a predicted hypothetical protein from a short 240bp open reading frame (http://www.ncbi.nlm.nih.gov/nuccore/74182885). Rapid amplification of 5′ cDNA end (5′ RACE) was done to define the start of the transcript in E8.5 mouse embryo. The RACE product for Ende indicated that the transcript begins in exon 2 of the annotated gene model at nucleotide +246, although transcriptome sequencing of purified hepatoblasts suggested that the 5′ end of the transcript may extend into intron 1 (Supplementary Fig. 1). The RACE result was confirmed by reverse transcription-polymerase chain reaction (RT-PCR) (data not shown). Interestingly, we did not find any evidence that the predicted first exon is part of the transcript that is expressed in E8.5 mouse embryo or purified hepatoblasts.
Npe lies on chromosome 12 and is 68kb downstream of an EST DV039614, and 217kb upstream of Sp4, and lacks a definitive open reading frame (NCBI ORF finder, http://pga.mgh.harvard.edu/web_apps/web_map/start). We were unable to generate a RACE product for Npe, likely due to the highly restricted and lower level of expression compared to Ende. Since Npe has no definitive coding region and displays no homology to known classes of non-coding RNAs such as tRNA and rRNA, the transcript was bioinformatically folded to the most thermodynamically stable secondary structure using mFOLD (http://mfold.bioinfo.rpi.edu/; (Zuker, 2003)). Short hair-pin loops from the secondary structure were then analysed with a miRNA precursor prediction program MiPred (http://www.bioinf.seu.edu.cn/miRNA/; (Jiang et al., 2007)) to explore the potential of Npe as a miRNA gene. The algorithm predicted several potential miRNA precursors along the transcript suggesting Npe maybe a non-coding miRNA gene (data not shown). Ende was also put through the same analysis, but did not produce any predicted miRNAs. More studies will be needed to explore the coding or non-coding potential of these two ESTs. However, in this study, we report the expression patterns of Ende and Npe during post-implantation stages of mouse development.
1.1. Expression of Ende and Npe was enriched in definitive endoderm LongSAGE libraries
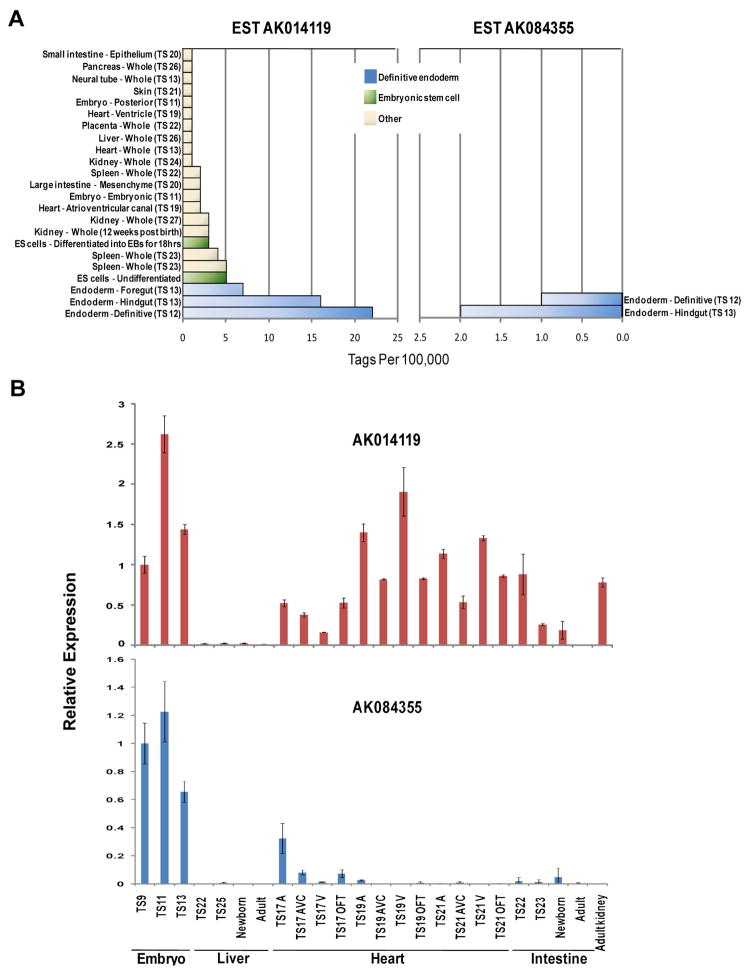
To examine the expression patterns of Ende and Npe, we screened 202 LongSAGE gene expression libraries generated for the Mouse Atlas of Gene Expression project (www.mouseatlas.org; (Siddiqui et al., 2005)). These libraries cover a wide variety of tissues and stages from the fertilized egg to the adult mouse. Ende expression was detected at greater than or equal to 1 tag per 100,000 total tags in 23 out of 202 LongSAGE libraries (Fig. 1A). Highest levels of Ende mRNA were detected in the definitive endoderm of 0- to 6- somite stage embryos (Theiler stage [TS] 12; embryonic day (E) 8.0), and in the foregut and hindgut definitive endoderm at 6- to 12- somites (TS13, E8.5) libraries. From E10.5 (TS17) onwards, Ende transcripts were also detected in the developing spleen, heart, kidney, and endodermal derived organs such as liver, intestine and pancreas. However, levels of Ende mRNA in these libraries were less than 3 tags per 100,000 as opposed to the higher levels observed during definitive endoderm formation. Post embryogenesis, Ende transcripts were observed in the kidney, pituitary gland, brain, urogenital sinus and testis libraries at less than 1 tag per 100,000 tags (data not shown). Interestingly, Ende transcripts were observed in mouse embryonic stem (ES) cells. In contrast, Npe was only expressed in 2 of the libraries at greater than or equal to 1 tag per 100,000 tags (Fig. 1A); the whole definitive endoderm at TS12 and the hindgut endoderm at TS13. Npe transcripts were also detected in a posterior neuropore, brain and ovary library (data not shown), but at less than 1 tag per 100,000 tags. No tags for Ende or Npe were observed in libraries corresponding to the pre-implantation stages of the embryo. Of note, we do not see similar expression patterns for the genes neighboring Ende or Npe in the SAGE data, suggesting that they are independently regulated.
Figure 1. Expression of Ende and Npe during mouse embryogenesis.
A: Transcript distribution for Ende and Npe in Mouse Atlas of Gene Expression SAGE libraries. Transcripts are represented as tag counts for Ende and Npe, normalized to library size and expressed as tags per 100,000 total tags. B: qRT-PCR analysis of Ende and Npe during early post-implantation stages in the mouse embryo, the liver, heart, intestine and kidney. Theiler stage (TS), atria (A), ventricle (V), atrioventricular canal (AVC), outflow tract (OFT)
Expression of Ende and Npe in the whole embryo and in various tissues of the embryo was confirmed using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) (Fig. 1B). Ende and Npe expression was observed in the whole embryo samples between TS9 (E6.5) and TS13 (E8.5) Interestingly, Ende was also observed to be differentially expressed in the heart at different spatial and temporal points, in the intestine at different stages of development, and in the adult kidney (Fig. 1B). In contrast, Npe expression was only observed in the whole embryo samples. Overall, the expressional analysis of Ende and Npe was consistent between qRT-PCR and SAGE.
1.2. Dynamic expression of Ende in the definitive endoderm
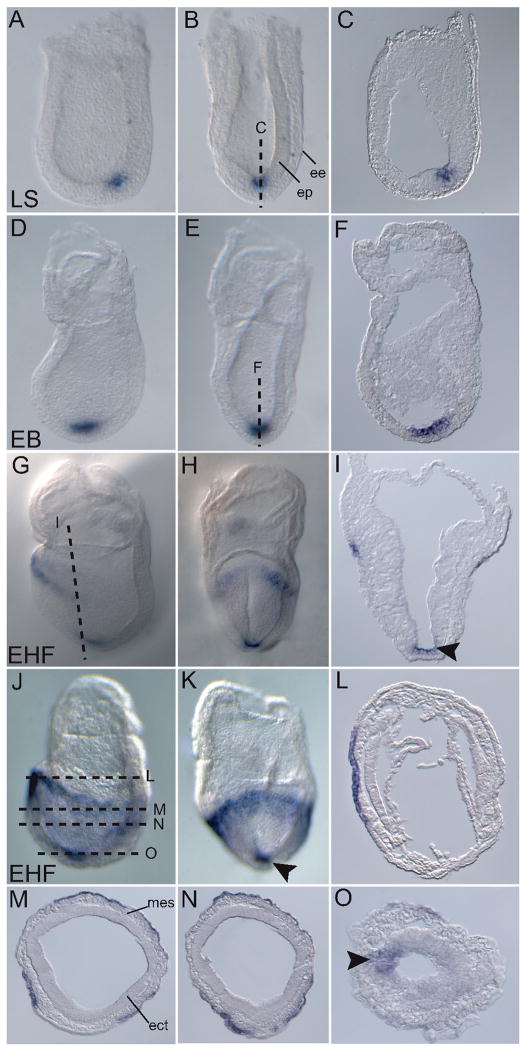
Since Ende showed highest levels of gene expression between TS9–TS13 (E6.5 –E8.5) in the SAGE analysis, we focused our expression analysis on these embryonic time periods using whole mount in situ hybridization (WISH). Ende was first observed in a small population of epiblast cells in the distal region of the embryo at the late streak stage (E7.25) (Fig. 2A, B, C). The expression was specific to the epiblast and was excluded from the endoderm layer that surrounds the epiblast cells (Fig. 2A, B, C). The expression in the distal epiblast remained (Fig. 2D, E, F) until the early headfold stages (E7.75) when cells in the anterior-most region of the embryo also began to express Ende (Fig. 2G, H). As the embryo developed, this expression domain expanded as a band around the proximal half of the embryo, from the anterior towards the posterior (Fig. 2J, K). The distal-most Ende expression remained, representing a population of cells in the midline of the ectodermal layer (arrowheads in Fig. 2I, K, O). Sections of the early headfold embryos showed that Ende expression in the proximal part of the embryo was specific to the outer layer of definitive endoderm and was excluded from mesoderm and ectoderm (Fig. 2L, M, N, O).
Figure 2. Expression of Ende during gastrulation.
Lateral views (A, D, G, J), frontal views (B, E, H, K), and transverse sections (C, F, I, L-O) of representative embryos expressing Ende at the indicated stages. At the LS stage Ende was expressed in a small population of epiblast cells in the distal half of the embryo, and was absent from the surrounding extra-embryonic endoderm cells (A–C). At early head fold stages Ende was expressed in the anterior most definitive endoderm, and then spread along the proximal half of the embryo to the lateral and posterior definitive endoderm (G–N). Expression in the midline ectoderm in distal tip of embryo was maintained (arrowheads in I, K, O). Late streak (LS), early bud (EB), early head fold (EHF), epiblast (ep), extra-embryonic endoderm (ve), ectoderm (ect), mesoderm (mes)
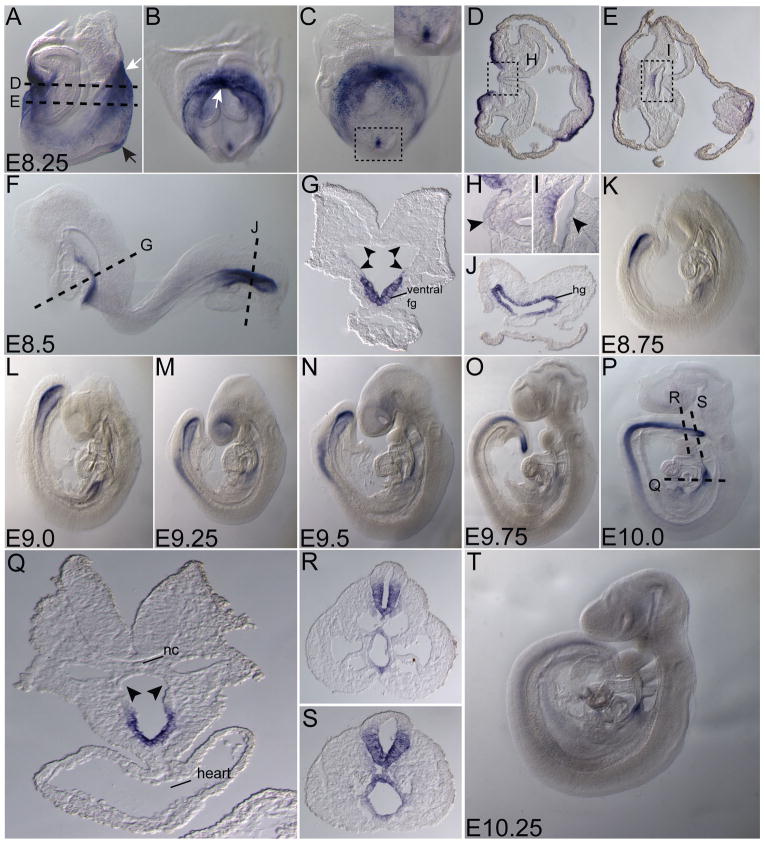
At early somite stages (E8.25), Ende was highly expressed in both the developing foregut and hindgut pockets (white arrows in Fig. 3A, B). Sections of the early somite embryo showed Ende expression enriched in the ventral foregut lip and the ventral part of the foregut invagination. Expression was absent from the dorsal midline and flanking cells of the foregut pocket (Fig. 3D, E; arrowheads in H, I). Ende expression in midline ectoderm was maintained, and symmetrically expressed in the node region of the embryo (black arrow Fig. 3A; insert in Fig. 3C). As the embryo developed to 10–12 somites (E8.5), Ende continued to be expressed in both the foregut and hindgut pockets. Sections of the embryos illustrated the expression of Ende in the entire hindgut pocket, while in the foregut, Ende expression was restricted to the ventral region and was absent from the dorsal and lateral aspects (arrowheads in Fig. 3G; Fig. 3J). By E9.25, cells of the neural tube in the posterior region began to express Ende (Fig. 3M). Sections of an E10.0 embryo showed that expression was restricted to the ventral half of the neural tube (Fig. 3R, S). At this stage Ende was maintained in a region of the foregut overlying the heart (Fig. 3P, Q). The expression in foregut, hindgut and neural tube were observed as late as E10.25, at reduced levels (Fig. 3T).
Figure 3. Expression of Ende in the definitive endoderm and neural tissue.
Lateral views (A, F, K–P, T), frontal view (B), posterior view (C) and transverse sections (D, E, G–J, Q–S) of representative embryos expressing Ende at the indicated stages. At E8.25, Ende expression was enriched in the foregut pocket and the presumptive hindgut of the embryo (white arrows in A, B). Expression in the ectoderm at the distal part of the embryo was maintained (black arrow in A), and was absent from the dorsal midline and flanking cells of the developing foregut (D, E; arrowheads in H, I). At E8.5, Ende expression continued to be specific to the ventral aspect of the foregut and absent in the remainder of the foregut (F; arrowheads in G), while expression was present throughout the hindgut (F, J). By E9.25 cells of the neural tube began to express Ende. As the embryo developed towards E10.0 Ende expression remained in the foregut overlying the heart, absent in the remainder of the foregut (P–S; arrowheads in Q), and was expressed in ventral part of the neural tube in the posterior half of the embryo (N–P, R, S). As late as E10.25 expression in the neural tube, hindgut and foregut were observed at reduced levels was significantly reduced (T). Foregut (fg), hindgut (hg), notochord (nc), embryonic day (E). E8.0 (1–4 somites), E8.5 (8–12 somites)
Recent fate-mapping studies have helped identify developmental fates of definitive endoderm cells during gastrulation (Tam et al., 2003; Tam et al., 2007; Franklin et al., 2008). Consistent with the fate-mapping analyses, the definitive endoderm cells expressing Ende under the head folds of the early head fold embryo are destined for the ventral aspects of the foregut, which was observed at later stages of embryos expressing Ende (Fig. 2G; Fig. 3). In contrast, expression of Ende in the small population of epiblast cells overlying the anterior end of the primitive streak at E8.25 does not appear to be related to the expression in the neural tube at E9.0 since there is no continuous expression pattern. The fate of the epiblast expressing cells is unclear since detailed fate mapping studies have not systematically examined this population in the mouse. Interestingly, Ende expression has some similarities to that of Thyrotropin releasing hormone, identified as being enriched in the foregut definitive endoderm in our previous SAGE analysis (Hou et al., 2007; McKnight et al., 2007). Future functional studies will help identify any shared regulatory mechanisms between the two.
1.3. Npe expression is observed in the node/posterior notochord region and the posterior-most hindgut of the embryo
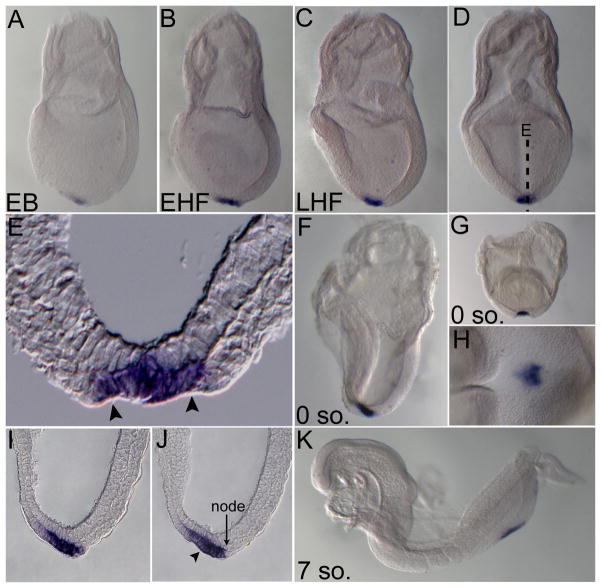
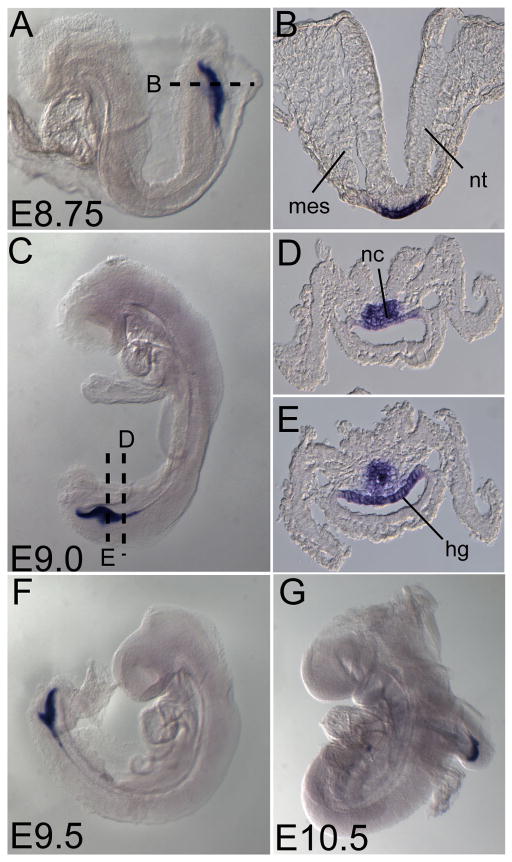
In contrast to Ende, Npe expression was much more restricted during early development and was first detected at the early bud stage in the distal tip of the embryo (Fig. 4A). This expression was maintained as development proceeded (Fig. 4B–D, F, G, H, K). Sections of the late head fold and zero somite embryos revealed that Npe was expressed in the region morphologically distinguished by the node and the posterior notochord (arrowheads in Fig. 4E, J, delineate the posterior notochord). The node is composed of a disordered array of organizer cells that give rise to the axial mesoderm tissues such as the notochord, which are important in patterning of the embryonic axes, and are able to induce axis structures when transplanted ectopically (Beddington, 1994; Sander and Faessler, 2001; Robb and Tam, 2004). Until recently the node was thought to encompass the small indent at the distal tip of the embryo where vectorial fluid flow termed “nodal flow” generated by motile monocilia gives rise to a proposed morphogen gradient resulting in left-right axis generation in the mouse (Nonaka et al., 1998; Nonaka et al., 2002). However, it was recently shown that the cilia harboring indent is the posterior end of the emerging notochord and that the true node/organizer lies immediately posterior to it (Hirokawa et al., 2006; Blum et al., 2007). Npe was observed to be strongly and symmetrically expressed in cells within the domain bordered by the node and extending into the posterior notochord (Fig. 4H, I, J). At 7–10 somite stages Npe expression migrated to the posterior end of the embryo, likely following the passive migration of the posterior notochord and the node upon the elongation of the embryonic axis (Fig. 4K). From E8.75 onward Npe expression was observed in the developing posterior-most hindgut and notochord (Fig. 5A, C, F, G). Sections of an E9.0 embryo revealed expression of Npe was restricted to the dorsal aspect of the hindgut and a population of poorly defined notochord cells (Fig. 5D, E). By E10.5 Npe expression was seen in the posterior-most tip of the hindgut.
Figure 4. Expression of Npe in the node and posterior notochord region during and post gastrulation.
Lateral views (A–C, F, K), posterior views (D, G), ventro-distal view (H), transverse section (E), and sagittal sections of panel F (I, J) of representative embryos expressing Npe at indicated stages. From early bud to the somite stages, Npe was expressed in the posterior notochord (boundary marked by arrowheads in E, J) and node region in the distal tip of the embryo (A–K). At 7 somites, Npe was expressed in the posterior of the embryo (K). Early bud (EB), early head fold (EHF), late head fold (LHF), somite (so.). 0 somites (E8.0), 7 somites (E8.5).
Figure 5. Expression of Npe in posterior-most hindgut and notochord cells.
Lateral views (A, C, F, G) and transverse sections (B, D, E) of representative embryos expressing Npe at indicated stages. Npe marked the posterior-most part of the developing hindgut (A–G). At E9.0–10.5, Npe was also expressed by a poorly defined group of posterior-most notochord cells and the dorsal aspect of the posterior-most hindgut (C–G). Somite (so.), mesoderm (mes), neural tube (nt), notochord (nc), hindgut (hg), embryonic day (E)
1.4. Expression of Ende and Npe overlaps with known markers during definitive endoderm development
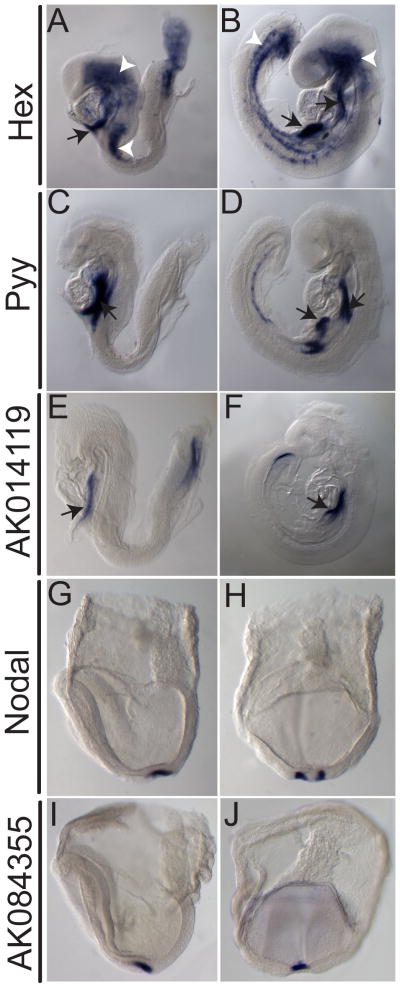
To determine more precisely, the domains of expression for Ende and Npe, we compared the expression patterns to previously described markers. During early definitive endoderm development, the expression domain of Ende overlaps with Hex and Pyy, two established markers of specific populations of definitive endoderm (Fig. 6A–F) (Thomas et al., 1998; Hou et al., 2007). At E8.5, Hex is expressed in the involuting ventral foregut endoderm, clusters of hemangioblast cells, and the allantois (Fig. 6A) (Thomas et al., 1998). By E9.5 Hex expression in the foregut is resolved to two distinct populations representative of thyroid and liver primordia, while increased numbers of Hex expressing hemangioblasts are observed throughout the lateral mesoderm and cranial mesenchyme (Fig. 6B). At E8.5, Pyy is strongly expressed in ventral and lateral foregut, and as the embryo enters organogenesis this expression is maintained in the pancreas/liver bud region (Fig. 6C, D). At E8.5, the foregut expression of Ende is more restricted than that of Pyy, and extends further anteriorly than that of Hex (Fig. 6E, F). Consistent with this pattern, at E9.5, Ende is expressed in the liver bud and foregut-midgut junction but is absent from the pancreatic buds. Npe expression was compared with Nodal, a marker of the node region of the embryo (Zhou et al., 1993). Nodal, an important signaling factor during gastrulation, is expressed in the crown cells in the node region of the embryo at E7.5 (Fig 6G, H) (Zhou et al., 1993; Nonaka et al., 2002). Npe expression is restricted to the central cells of posterior notochord and thus appears to complement that of Nodal in the node region (Fig. 6G–J).
Figure 6. Comparison of Ende and Npe with known markers during definitive endoderm development.
Lateral views (A–F, G, I) and posterior views (H, J) of representative embryos expressing indicated markers at stages E7.5 (G–J) E8.5 (A, C, E) and E9.5 (B, D, F). Hex expression is seen in the ventral foregut (white arrows in A), and clusters of hemangioblasts (white arrowheads in A) at E8.5. At E9.5 expression of Hex in the foregut resolves to two regions of the foregut, namely the thyroid and liver bud regions (white arrows in B), and a few more hemangioblast cells (white arrowheads in B). Strong Pyy expression at E8.5 is seen in populations of the ventral foregut (white arrow in C). At E9.5 this foregut expression remains in the ventral foregut, and is also seen in the liver/pancreas region (white arrows in D). Expression of Ende overlapped at the foregut with Pyy and Hex (E, F). Nodal expression seen in the crown cells of the node region (G, H). Npe was observed to complement the expression pattern of Nodal(I, J).
The acquisition of novel genetic markers of the definitive endoderm is crucial in understanding the formation and morphogenesis of this tissue during development. Markers such as Ende and Npe that mark specific populations of the definitive endoderm will be useful in building more accurate fate maps for this tissue. In addition, these markers will be useful during mouse mutant analyses of gastrulation and definitive endoderm formation. Finally, in combination with other markers, Ende and Npe can be used to define and isolate cell populations produced during in vitro differentiation of ES cells to definitive endoderm. Thus, Ende and Npe are valuable markers of the definitive endoderm to address important questions regarding mouse definitive endoderm formation and patterning.
2. Experimental Procedures
2.1. Mouse strains and dissection
Embryos were obtained from ICR mice and considered E0.5 at noon of the day of detection of the vaginal plug. Embryos were dissected in Dulbecco’s phosphate buffered saline (PBS; Stem Cell Technologies Inc), fixed overnight at 4°C in 4% paraformaldehyde, dehydrated through a graded methanol series and stored at −20°C.
2.2. Serial analysis of gene expression library analysis
SAGE libraries from the Mouse Atlas of Gene Expression project (www.mouseatlas.org) have been previously described (Siddiqui et al., 2005). Data were analyzed using DiscoverySpace 4 software (www.bcgsc.ca/platform/bioinfo/software/ds) (Robertson et al., 2007).
2.3. RNA extraction, cDNA synthesis and qRT-PCR
Embryos were placed into Trizol (Invitrogen) and stored at −80°C. RNA was extracted using MaxTract phase-lock gels (Qiagen) following the manufacturer’s instructions. cDNA was synthesized as previously described (McKnight et al., 2007). PCR was performed with Hot Star Taq polymerase (Invitrogen) in PCR buffer and dNTPs. The sequences of primers used are as follows (forward and reverse):
Ende; 5′-GGACAGATACTGCGATTTACAGACG-3′ and 5′CCATCCTGAGGTTGTTTTTCCG-3′,
Npe;5′CCTATCAATCAGCATACACCACAGG - 3′ and 5′-GGAATCGCTGTTGTGTTCAGGTAAC-3′
qRT-PCR analysis was performed on an ABI PRISM 7300 Sequence Detection System using the SYBR Green PCR Master Mix (ABI). The PCR consisted of 12.5 μl of SYBR Green PCR Master Mix, 1 μl of 10 mM forward and reverse primers, 10.5 μl water, and 1 μl template cDNA in a total volume of 25 μl. Cycling was performed using the default conditions of ABI 7300 SDS Software 1.3.1: 2 min at 95°C, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. The relative expression of each gene was normalized against Gapdh. The primers used are as follows: (forward and reverse):
Ende; 5′ – CCTCTAATTTCCTTGGCTCTACC -3′ and 5′-AAAAGCCATCCACGGTACTC-3′
Npe; 5′ – GCTAAAACGAAAGGGAATGGG- 3′
5′-CAGGCAGAATTGAAACATGGG -3′
2.4. Probe construction and WISH
The murine cDNA clones for Ende, Npe, Pyy, and Nodal were obtained by RT-PCR. The Hex in situ clone was obtained from R. Beddington. The cDNA clones were used as a template for generating Digoxigenin – UTP RNA probes (Roche). Briefly, RNA probes were in vitro transcribed using 1 μg of linearized template and 20 Units of SP6, T3 or T7 RNA polymerase (Roche). RNA probes were purified using G-50 sephadex columns (Amersham) and stored at −80°C. Whole mount in situ hybridization (WISH) was performed on embryos ranging from E6.0 to E9.0 as previously described (Wilkinson and Nieto, 1993). Embryos were incubated in BM purplealkaline phosphate substrate (Roche) at 4°C or room temperature for colour development.
2.5. Histology and imaging
Following WISH, embryos were processed through a graded PBT:Glycerol series to 80% glycerol and imaged. After imaging, the embryos were passed through a graded series of sucrose treatments to 80% sucrose, and incubated in 80% sucrose: Tissue-Tek OCT (1:1) embedding medium (Sakura Finetek) overnight at 4°C. Embryos were embedded in 100% OCT and frozen at −80°C. Cryosections at 10 μm were placed on Superfrost Plus glass slides (VWR) and allowed to dry overnight. Slides were washed twice in PBS for 5 minutes and mounted using 60% glycerol and glass coverslips (VWR).
Supplementary Material
Acknowledgments
We would like to thank Anita M. Charters for the initial bioinformatics analysis of the serial analysis of gene expression data, Sam Lee for the liver cDNA, Eric Xu and Pavle Vrljicak for the heart cDNA. This work was supported by Genome British Columbia, Genome Canada and Canadian Institutes of Health Research (CIHR MOP-89806). PAH is a Michael Smith Foundation for Health Research Senior Scholar.
References
- Beddington RS. Induction of a second neural axis by the mouse node. Development. 1994;120:613–620. doi: 10.1242/dev.120.3.613. [DOI] [PubMed] [Google Scholar]
- Blum M, Andre P, Muders K, Schweickert A, Fischer A, Bitzer E, Bogusch S, Beyer T, van Straaten HW, Viebahn C. Ciliation and gene expression distinguish between node and posterior notochord in the mammalian embryo. Differentiation; research in biological diversity. 2007;75:133–146. doi: 10.1111/j.1432-0436.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- Franklin V, Khoo PL, Bildsoe H, Wong N, Lewis S, Tam PP. Regionalisation of the endoderm progenitors and morphogenesis of the gut portals of the mouse embryo. Mechanisms of development. 2008;125:587–600. doi: 10.1016/j.mod.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Tanaka Y, Okada Y, Takeda S. Nodal flow and the generation of left-right asymmetry. Cell. 2006;125:33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Hou J, Charters AM, Lee SC, Zhao Y, Wu MK, Jones SJ, Marra MA, Hoodless PA. A systematic screen for genes expressed in definitive endoderm by Serial Analysis of Gene Expression (SAGE) BMC developmental biology. 2007;7:92. doi: 10.1186/1471-213X-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Wu H, Wang W, Ma W, Sun X, Lu Z. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–44. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight KD, Hou J, Hoodless PA. Dynamic expression of thyrotropin-releasing hormone in the mouse definitive endoderm. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236:2909–2917. doi: 10.1002/dvdy.21313. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Shiratori H, Saijoh Y, Hamada H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature. 2002;418:96–99. doi: 10.1038/nature00849. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–37. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Robb L, Tam PP. Gastrula organiser and embryonic patterning in the mouse. Seminars in cell & developmental biology. 2004;15:543–554. doi: 10.1016/j.semcdb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Robertson N, Oveisi-Fordorei M, Zuyderduyn S, Varhol R, Fjell C, Marra M, Jones S, Siddiqui A. DiscoverySpace: an interactive data analysis application. Genome biology. 2007;8:R6. doi: 10.1186/gb-2007-8-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander K, Faessler PE. Introducing the Spemann-Mangold organizer: experiments and insights that generated a key concept in developmental biology. The International journal of developmental biology. 2001;45:1–11. [PubMed] [Google Scholar]
- Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, Bohacec S. A mouse atlas of gene expression: large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proc Natl Acad Sci USA. 2005;102:18485–18490. doi: 10.1073/pnas.0509455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, Kanai-Azuma M, Kanai Y. Early endoderm development in vertebrates: lineage differentiation and morphogenetic function. Current opinion in genetics & development. 2003;13:393–400. doi: 10.1016/s0959-437x(03)00085-6. [DOI] [PubMed] [Google Scholar]
- Tam PP, Khoo PL, Lewis SL, Bildsoe H, Wong N, Tsang TE, Gad JM, Robb L. Sequential allocation and global pattern of movement of the definitive endoderm in the mouse embryo during gastrulation. Development (Cambridge, England) 2007;134:251–260. doi: 10.1242/dev.02724. [DOI] [PubMed] [Google Scholar]
- Thomas PQ, Brown A, Beddington RS. Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development. 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- Wells JM, Melton DA. Vertebrate endoderm development. Annual Review of Cell and Developmental Biology. 1999;15:393–410. doi: 10.1146/annurev.cellbio.15.1.393. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods in Enzymology. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sasaki H, Lowe L, Hogan BL, Kuehn MR. Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–7. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.