Abstract
Objective
Although early proof-of-concept studies of somatic in vivo genome editing of the mouse ortholog of proprotein convertase subtilisin/kexin type 9 (Pcsk9) in mice have established its therapeutic potential for the prevention of cardiovascular disease, the unique nature of genome-editing technology—permanent alteration of genomic DNA sequences—mandates that it be tested in vivo against human genes in normal human cells with human genomes in order to give reliable preclinical insights into the efficacy (on-target mutagenesis) and safety (lack of off-target mutagenesis) of genome-editing therapy before it can be used in patients.
Approach and Results
We used a clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated 9 (Cas9) genome-editing system to target the human PCSK9 gene in chimeric liver-humanized mice bearing human hepatocytes. We demonstrated high on-target mutagenesis (approaching 50%), greatly reduced blood levels of human PCSK9 protein, and minimal off-target mutagenesis.
Conclusions
This work yields important information on the efficacy and safety of CRISPR-Cas9 therapy targeting the human PCSK9 gene in human hepatocytes in vivo, and it establishes humanized mice as a useful platform for the preclinical assessment of applications of somatic in vivo genome editing.
Keywords: Gene therapy, PCSK9, lipids and lipoprotein metabolism, molecular biology
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas) 9 systems have elicited enormous interest from the biomedical community due to their versatile use in research applications and, perhaps more so, due to their therapeutic potential in addressing human diseases.1 As with any novel therapeutic approach, before any given CRISPR-Cas9 application can be used in the human body, it will need to undergo extensive preclinical testing. CRISPR-Cas9 and other genome-editing tools present an unusual challenge in that the target is DNA sequence in the human genome. While animal models such as rodents and non-human primates offer opportunities to assess the physiological consequences of in vivo therapies, they do not allow for accurate assessment of on-target and off-target mutagenesis by a CRISPR-Cas9 application targeted against a human gene, due to lack of conservation across genomes. What are needed are preclinical models in which the somatic in vivo targeting of human genes in normal human cells (i.e., not tumor cells) with human genomes can be performed.
Due to ease of delivery to the organ, as well as the diversity of grievous genetic disorders involving the organ, the liver has emerged as an early target of preclinical genome-editing applications.2–8 Accordingly, we sought to establish the feasibility of using chimeric liver-humanized mice to assess for on-target and off-target effects of CRISPR-Cas9 in vivo. There are several mouse models in which endogenous hepatocytes can be replaced with primary human hepatocytes. Perhaps the best established is the Fah−/−Rag2−/−Il2rg−/− (FRG KO) mouse,9–11 in which withholding of a specific drug in the diet [2-(2-nitro-4-fluoromethylbenzoyl)-1,3-cyclohexanedione; NTBC] results in the death of endogenous mouse hepatocytes from accumulation of a toxic metabolite [due to the Fah (fumarylacetoacetate hydrolase) deficiency], and which are immunocompromised (Rag2−/−Il2rg−/−) so as to accept transplanted human hepatocytes that can complement the deficient mouse liver function and rescue the animals.
Using the FRG KO mouse model, we targeted the human PCSK9 gene because it is a prime therapeutic target in the prevention of cardiovascular disease and because the mouse Pcsk9 gene has previously been the focus of successful in vivo genome-editing studies.5,6 A demonstration of the efficacy and safety of human PCSK9-targeting therapy in vivo would provide a strong rationale for further preclinical studies with the aim of ultimately bringing to the clinic a one-shot CRISPR-Cas9 “vaccination” for the reduction of low-density lipoprotein cholesterol and cardiovascular risk.
MATERIALS AND METHODS
Materials and Methods are available in the online-only Data Supplement.
RESULTS AND DISCUSSION
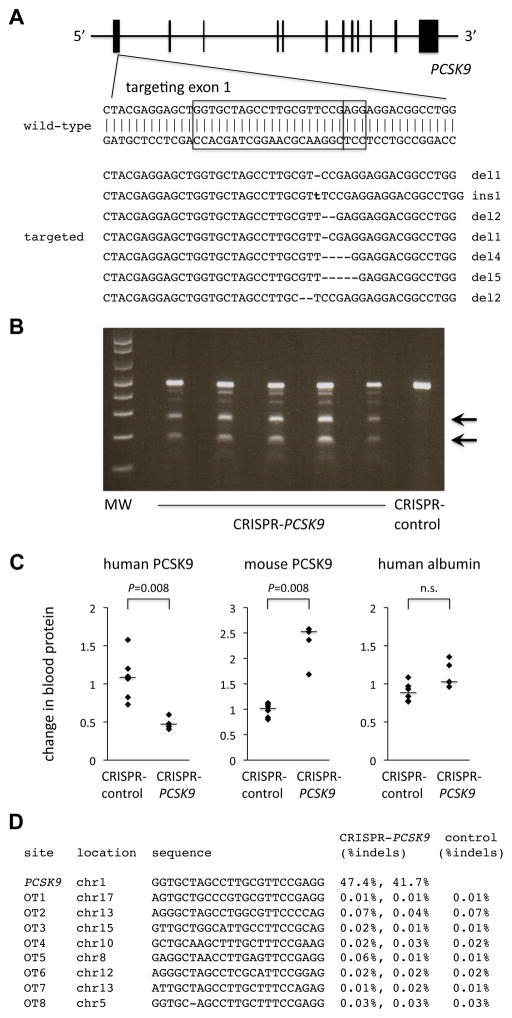
Using primary human hepatocytes, we generated FRG KO mice with varying degrees of engraftment at least five months after transplantation (10% to 60% reconstitution of liver with human cells as judged by human serum albumin levels in the mice11). We generated adenoviruses bearing Streptococcus pyogenes Cas9 and either a guide RNA targeting a sequence in the first coding exon of the human PCSK9 gene (CRISPR-PCSK9) (Figure 1A) or a guide RNA targeting a control sequence (CRISPR-control). Of note, the 20-nt PCSK9 protospacer has six mismatches with the orthologous sequence in the mouse genome, and no NGG- or NAG-adjacent sequence in the human genome has less than four mismatches with the PCSK9 protospacer (Supplemental Figures I and II).
Figure 1.
CRISPR-Cas9 targeting of PCSK9 in human hepatocytes in vivo. A, Targeting of a sequence in exon 1 of the human PCSK9 gene. The boxes indicate the 20-bp sequence matching the protospacer and the 3-bp protospacer-adjacent motif (PAM). The seven targeted sequences shown below the wild-type sequence reflect the seven most common mutations (in descending order of frequency) detected by deep sequencing of the locus in human hepatocytes targeted in vivo in chimeric liver-humanized mice (see Supplemental Dataset for more information). B, Surveyor assays performed with genomic DNA from liver samples taken from mice 4 days after receiving an adenovirus expressing Cas9 and the PCSK9 guide RNA (CRISPR-PCSK9) or a control adenovirus (CRISPR-control). Arrows show the cleavage products resulting from the Surveyor assays; the intensity of the cleavage product bands relative to the uncleaved product band corresponds to the mutagenesis rate. C, Relative changes in blood human PCSK9 protein levels, blood mouse PCSK9 protein levels, and blood human albumin levels (post-treatment divided by pre-treatment levels) in chimeric liver-humanized mice receiving CRISPR-control virus (n = 6 mice) or CRISPR-PCSK9 virus (n = 5 mice). The bars indicate the median values for the relative changes within the groups. The Mann–Whitney U test was performed to compare the relative changes in the two groups. D, Indel rates at on-target and off-target sites from next-generation DNA sequencing of liver samples from post-treatment mice.
We administered CRISPR-PCSK9 virus (n = 5) or CRISPR-control virus (n = 6) to FRG KO mice. After four days, we found by Surveyor assay that all of the CRISPR-PCSK9 mice had substantial mutagenesis at the on-target site (Figure 1B). We performed deep sequencing of PCR amplicons from liver samples of two of the CRISPR-PCSK9 mice and identified indels in 47% and 42% of the sequence reads. This degree of mutagenesis is quite concordant with previous studies of CRISPR-Cas9 delivered by adenovirus or adeno-associated virus (AAV).5–7 More than three-quarters of the identified indels were either 1-bp or 2-bp insertions or deletions at the expected CRISPR-Cas9 cleavage site (3 nt upstream of the 3′ end of the protospacer) (Figure 1A, Supplemental Dataset).
Consistent with these DNA-level changes, we found that post-treatment blood levels of human PCSK9 protein—produced and secreted specifically by engrafted human hepatocytes—were reduced on average by 52% compared to pre-treatment levels (P = 0.007; Figure 1C and Supplemental Figure III). This is a mildly greater effect than might be expected from the degree of on-target mutagenesis; of note, the mutagenesis detected by sequencing is almost certainly underestimated since next-generation DNA sequencing of small PCR amplicons fails to capture larger indels. Notably, we found that post-treatment blood levels of mouse PCSK9 protein were increased more than two-fold compared to pre-treatment levels (P = 0.002; Figure 1C and Supplemental Figure III), suggesting a compensatory mechanism at work within the mouse hepatocytes still present in the transplanted FRG KO mice. Presumably as a consequence, total cholesterol levels were not significantly changed (data not shown). Human albumin levels were unchanged (Figure 1C and Supplemental Figure III), confirming the stability of the engrafted human hepatocytes in the mice with CRISPR-PCSK9 treatment.
We used deep sequencing to assess for off-target mutagenesis in the human genome at eight top candidate off-target sites by sequence similarity identified by the CRISPR Design server12 and COSMID server13 (Supplemental Figures I and II). Given the concordance in the degree of on-target mutagenesis observed among the mice in the CRISPR-PCSK9 cohort by Surveyor assay, we felt that off-target data from two CRISPR-PCSK9 mice and one CRISPR-control mouse would be representative of the entire cohort. There was no detectable off-target mutagenesis out of the range of background indel rates resulting from errors inherent in PCR amplification and next-generation DNA sequencing (Figure 1D).
Our results indicate that chimeric liver-humanized mice can be used as a platform to assess for on-target and off-target mutagenesis from CRISPR-Cas9 delivered to human hepatocytes in vivo by a somatic approach. Of note, AAV is regarded as a safer vehicle than adenovirus for therapeutic applications in humans, as it is better tolerated by the immune system, but it has a much more limited cargo size that is less conducive to the use of S. pyogenes Cas9 and large, strong, tissue-specific promoters. Although CRISPR-Cas9 has been adapted for use in AAV by means of a smaller Cas9 protein from Staphylococcus aureus,6,7 the AAV serotypes that efficiently transduce mouse hepatocytes in vivo do not target human hepatocytes in vivo well, which means that AAV will need to be optimized before use in genome-editing applications in human liver, perhaps through the development of novel capsid proteins.14 At present, the primary advantages of adenovirus for the testing of CRISPR-Cas9 applications in chimeric liver-humanized mice are that it efficiently targets human hepatocytes in vivo and that it allows for direct comparisons of safety and efficacy of large proteins intended to improve on-target specificity, such as FokI-Cas9 fusion proteins.15,16
Although the chimeric liver-humanized FRG KO mouse, being immunocompromised, does not model the immune consequences of using viral vectors to heterologously express bacterial Cas9 protein in the liver, this could potentially be addressed in the future by double humanization of FRG KO mice with respect to both the liver and the hematopoietic system.17 Finally, we note the inherent challenge of assessing for off-target mutagenesis throughout the genome in a targeted organ with billions of cells. While we can rule out >0.1% events at a number of candidate off-target sites with deep sequencing, a more sophisticated approach will be needed to assess for rare events across the genome. Unbiased screening methods have been developed for use in cell lines in vitro,6,18–20 and such methods will need to be adapted for use in living animals.
As these various issues are successfully addressed, we anticipate that studies in humanized animals will become an important component of the preclinical assessment of applications of somatic in vivo genome editing. Our specific results establish the efficacy of and suggest a favorable safety profile of CRISPR-Cas9 therapy targeting the human PCSK9 gene in authentic human hepatocytes in vivo and support further development of the therapy with the goal of eventual clinical use for long-term protection against cardiovascular disease.
Supplementary Material
SIGNIFICANCE.
Although early proof-of-concept studies of somatic in vivo genome editing in mice highlighted the therapeutic potential of CRISPR-Cas9 to target PCSK9 and other disease-related genes, the unique nature of genome-editing technology—permanent alteration of DNA sequences—argues for it to be tested in vivo against human genes in normal human cells (i.e., not tumor cells) with human genomes in order to give reliable preclinical insights into the efficacy (on-target mutagenesis) and safety (lack of off-target mutagenesis) of a CRISPR-Cas9 therapy before it can be used in patients. We describe the use of chimeric liver-humanized mice as a means to do this for the human PCSK9 gene in human hepatocytes in vivo, and we thereby establish the efficacy of CRISPR-Cas9 in human hepatocytes.
Acknowledgments
SOURCES OF FUNDING
This work was supported by the Howard Hughes Medical Institute Medical Research Fellows Program (A.R.); the Shanghai Institutes for Biological Sciences Fellowship (Y5Y1X41491) (Y.Z.); the American Heart Association (Q.D.); the Hundred Talents Program of the Chinese Academy of Sciences (Q.D.); the Shanghai Pujiang Program (15PJ1409200) (Q.D.); the Harvard Stem Cell Institute (K.M.); and grants R01-HL118744, R01-GM104464, and R01-DK099571 from the United States National Institutes of Health (NIH) (K.M.).
ABBREVIATIONS
- CRISPR-Cas9
clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas) 9
- PCSK9
proprotein convertase subtilisin/kexin type 9
- AAV
adeno-associated virus
Footnotes
DISCLOSURES
None.
References
- 1.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Haurigot V, Doyon Y, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barzel A, Paulk NK, Shi Y, Huang Y, Chu K, Zhang F, Valdmanis PN, Spector LP, Porteus MH, Gaensler KM, Kay MA. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML, Wilson JM. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016 Feb 1; doi: 10.1038/nbt.3469. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin H, Song CQ, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016 Feb 1; doi: 10.1038/nbt.3471. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, Kay MA, Finegold M, Grompe M. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bissig KD, Wieland SF, Tran P, Isogawa M, Le TT, Chisari FV, Verma IM. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120:924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis EC, Naugler WE, Parini P, Mörk LM, Jorns C, Zemack H, Sandblom AL, Björkhem I, Ericzon BG, Wilson EM, Strom SC, Grompe M. Mice with chimeric livers are an improved model for human lipoprotein metabolism. PLoS One. 2013;8:e78550. doi: 10.1371/journal.pone.0078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: a Web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol Ther Nucleic Acids. 2014;2:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lisowski L, Dane AP, Chu K, Zhang Y, Cunningham SC, Wilson EM, Nygaard S, Grompe M, Alexander IE, Kay MA. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson EM, Bial J, Tarlow B, Bial G, Jensen B, Greiner DL, Brehm MA, Grompe M. Extensive double humanization of both liver and hematopoiesis in FRGN mice. Stem Cell Res. 2014;13(3 Pt A):404–412. doi: 10.1016/j.scr.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.