Abstract
Objective
Cholesterol homeostasis is fundamental to human health, and is thus, tightly regulated. MicroRNAs exert potent effects on biological pathways, including cholesterol metabolism, by repressing genes with related functions. We reasoned that this mode of pathway regulation could be exploited to identify novel genes involved in cholesterol homeostasis.
Approach and Results
Here, we identify oxysterol binding protein-like 6 (OSBPL6) as a novel target of two miRNA hubs regulating cholesterol homeostasis: miR-33 and miR-27b. Characterization of OSBPL6 revealed that it is transcriptionally regulated in macrophages and hepatocytes by LXR and in response to cholesterol loading, and in mice and non-human primates by Western diet feeding. OSBPL6 encodes the OSBPL-related protein 6 (ORP6), which contains dual membrane- and ER-targeting motifs. Subcellular localization studies showed that ORP6 is associated with the endolysosomal network and ER, suggesting a role for ORP6 in cholesterol trafficking between these compartments. Accordingly, knockdown of OSBPL6 results in aberrant clustering of endosomes and promotes the accumulation of free cholesterol in these structures, resulting in reduced cholesterol esterification at the ER. Conversely, ORP6 overexpression enhances cholesterol trafficking and efflux in macrophages and hepatocytes. Moreover, we show that hepatic expression of OSBPL6 is positively correlated with plasma levels of HDL-cholesterol in a cohort of 200 healthy individuals, whereas its expression is reduced in human atherosclerotic plaques.
Conclusions
These studies identify ORP6 as a novel regulator of cholesterol trafficking that is part of the miR-33 and miR-27b target gene networks that contribute to the maintenance of cholesterol homeostasis.
Keywords: MicroRNA, Cholesterol Homeostasis, Lipids and Lipoproteins
INTRODUCTION
Cholesterol is a vital component of mammalian cells, where it regulates membrane fluidity and lipid raft-dependent signal transduction, and it is also the essential precursor of steroid hormones, bile acids and vitamin D1. As such, cholesterol homeostasis is tightly regulated at both systemic and cellular levels. Importantly, cellular cholesterol homeostasis is dependent on the maintenance of a cholesterol gradient, with the highest concentrations in the plasma membrane and the lowest in the endoplasmic reticulum (ER), the site of regulation of cholesterol synthesis1. As a result, cholesterol internalized through the uptake of plasma lipoproteins such as low density lipoproteins (LDL) and de novo synthesized cholesterol are rapidly redistributed via both vesicular and non-vesicular mechanisms involving lipid transport proteins to maintain this cholesterol gradient. For example, cholesterol and cholesterol esters in endocytosed LDL particles are carried from early endosomes to sorting endosomes, where LDL dissociates from the LDL receptor (LDLR) and the latter is recycled back to the plasma membrane via the endocytic recycling compartment2. In turn, early endosomes mature to late endosomes before fusing with lysosomes. Within the acidic pH of late endosomes/lysosomes, LDL-associated cholesterol esters are hydrolyzed by lysosomal acid lipase, and the arising free cholesterol can efflux from these compartments to the plasma membrane and ER. Upon reaching the ER, lipoprotein-derived cholesterol results in feedback inhibition of cholesterol synthesis and uptake by inhibiting the sterol regulatory element-binding protein (SREBP) pathway, leading to decreased transcription of genes responsible for cholesterol synthesis (eg., 3-hydroxy-3-methylglutaryl-CoA; HMGCR) and uptake (eg., LDLR). Excess cholesterol is esterified by the ER-resident acyl-CoA:cholesterol acyltransferase (ACAT) and stored in cytosolic lipid droplets (LDs)2, 3.
Elimination of excess cellular cholesterol is important to prevent cell toxicity and development of atherosclerosis and associated metabolic disorders. Lipid-droplet stored cholesterol can be removed through a process that requires hydrolysis of cholesterol esters to free cholesterol for subsequent efflux via the ATP-binding cassette transporters A1 (ABCA1) and G1 (ABCG1) to apolipoprotein A-I (apoA1) and high density lipoproteins (HDL), respectively4. This represents the first step in the reverse cholesterol transport (RCT) pathway, through which cholesterol is removed from peripheral cells and returned to the liver for excretion. The liver X receptors (LXR) are key transcriptional regulators of the response to cholesterol excess by coordinating the transcription of numerous genes within the RCT pathway, including ABCA1 and ABCG1. Despite extensive study of these pathways, many questions remain about how cholesterol is trafficked between different cellular compartments and how these processes are regulated.
The recent discovery of microRNAs (miRNAs) has increased our understanding of the molecular mechanisms regulating cholesterol homeostasis5, 6. miRNAs exert post-transcriptional control of gene expression by promoting the degradation of target mRNAs and/or inhibiting mRNA translation. Notably, miRNAs can fine tune biological pathways by repressing numerous genes with related functions, with potent cumulative effects. Examples of this orchestrated mode of gene regulation include miR-33a/b and miR-27b, which act as regulatory hubs that repress genes involved in lipid metabolism7-13. miR-33a/b are co-expressed with the SREBF1/2 genes and coordinately repress genes involved in cholesterol efflux, RCT and HDL biogenesis (ABCA1, ABCG1, ABCB11, NPC1 and ATP8B1)9, 12-14, as well as fatty acid oxidation (HADHB, CPT1A, CROT, AMPK)10, 11, 15. miR-27b represses genes involved in cholesterol efflux and HDL metabolism (ABCA1, ANGPTL3), as well as triglyceride metabolism (LPL, GPAM)7, 8. By repressing multiple steps in cellular lipid metabolism, these miRNAs exert rapid and potent effects on sterol homeostasis. We thus hypothesized that study of miR-33a/b- and miR-27b-regulated gene networks may uncover novel genes involved in cellular cholesterol homeostasis. Using this approach, we identified oxysterol binding protein-like 6 (OSBPL6) as a novel target gene in the miR-33a/b and miR-27b network that contributes to cholesterol homeostasis.
OSBPL6, also known as oxysterol-binding protein-related protein (ORP) 6 is a poorly characterized member of the ORP family of cytoplasmic sterol binding proteins, known to function in sterol transport, endosomal trafficking and signaling16. This family includes 12 members that are characterized by an OSBP-related ligand binding domain at the C-terminus that binds cholesterol and oxysterols16. Many ORPs also contain a pleckstrin homology domain at the N-terminus, which interacts with phosphatidylinositol phosphates (PIPs) for membrane targeting, while certain members of this class also possess an ER-targeting phenylalanine in an acidic tract (FFAT)-motif, which interacts with integral membrane proteins of the ER called vesicle-associated membrane protein-associated proteins (VAPs)16. By targeting both the ER and other organelle membranes, ORPs can localize to membrane contact sites (MCSs), defined as close appositions between the ER and other organelles, which may aid in non-vesicular lipid transfer16. Although little is known about ORP6 function, this gene resides in a locus on chromosome 2 that has been linked to premature coronary artery disease17 and to variations in HDL-cholesterol (HDL-C) levels18, suggesting a potential role in cholesterol metabolism. We show herein that OSBPL6 is an LXR-responsive gene that is upregulated in response to increased cellular and dietary cholesterol levels in mice and non-human primates, and contributes to the maintenance of cholesterol homeostasis by regulating cellular cholesterol trafficking and efflux.
METHODS AND MATERIAL
Methods and material are available in the online-only Data Supplement.
RESULTS
OSBPL6 is a target of the lipid metabolism regulating miRNAs, miR-33 and miR-27b
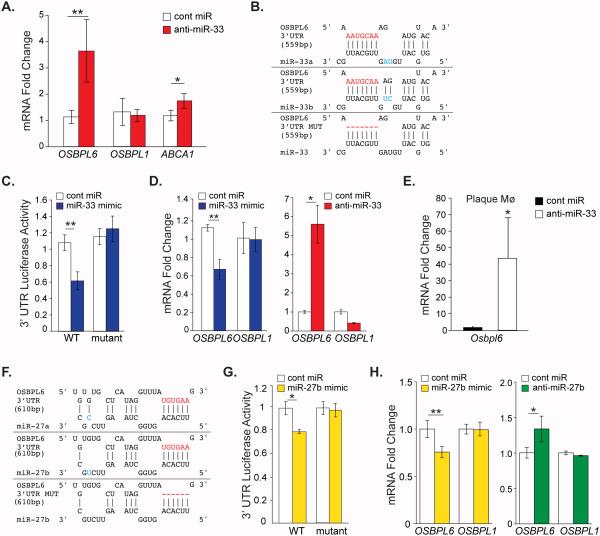
To identify novel target genes regulated by miR-33 in a model relevant to humans, we performed gene expression profiling of the livers of African green monkeys treated with control or miR-33-targeting antisense oligonucleotides for 4 weeks as previously reported10. This study identified OSBPL6 as among the most strongly derepressed miR-33 seed-hexamer matched target genes in the livers of anti-miR-33-treated monkeys10. As shown in Figure 1A, using quantitative PCR (qPCR) we show that anti-miR-33 treatment increased hepatic OSBPL6 mRNA by 3.5 fold in monkeys treated with anti-miR-33 compared to control anti-miR. By contrast, expression of OSBPL1, another OSBPL family member lacking miR-33 binding sites in its 3’UTR was similar between the two groups (Figure 1A). Selective derepression of OSBPL6 and the miR-33 target gene ABCA1 was also observed in the livers of C57BL/6 mice treated with a lentiviral miR-33 inhibitor (Supplemental Figure 1A) and in atherosclerotic Ldlr−/− mice treated for 4 weeks with anti-miR-33 oligonucleotides (Supplemental Figure 1B) compared to control treatments. Conversely, overexpression of miR-33 in C57BL/6 mice repressed hepatic Osbpl6 and Abca1, but not Osbpl1, mRNA levels (Supplemental Figure 1A), indicating that miR-33 targeting of OSBPL6 is conserved in mice and non-human primates. miRNA target prediction algorithms identified a single miR-33a/b binding site in the 3’UTR of human OSBPL6 (Figure 1B) that is conserved in mice. To confirm miR-33 targeting of the OSBPL6 3’UTR at this site, we performed 3’UTR-luciferase reporter assays in HEK293 cells transfected with control or miR-33 mimics. miR-33a/b overexpression reduced OSBPL6-3’UTR-luciferase activity by approximately 40%, and this repression was reversed by mutating the miR-33a/b binding site (Figure 1B, C).
Figure 1. OSBPL6 is a novel miR-33 and miR-27b target gene.
A) Relative hepatic expression of OSBPL6, OSBPL1 and ABCA1 mRNA in African green monkeys on chow diet treated with control miR or anti-miR-33 oligonucleotides for 4 wks (n=6/group). B, F) Predicted binding sites for miR-33a/b (B) or miR-27a/b (F) in the 3’UTR of human wild type (WT) OSBPL6 and with the binding site mutated (MUT) using RNA-Hybrid. C, G) Activity of WT or mutant OSBPL6 3’UTR-luciferase reporter constructs in HEK293 cells transfected with miR-33 mimics (C) or miR-27b mimics (G). D, H) Relative expression of OSBPL6 and OSBPL1 mRNA determined by qPCR in THP-1 macrophages transfected with miR-33 mimics or inhibitors (D) or miR-27b mimics or inhibitors (G). C-D,G,H Data are expressed as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 compared to control treatment. E) Expression of Osbpl6 mRNA in plaque macrophages isolated from Ldlr−/− mice fed a Western diet (14 wk) and treated with control miR or anti-miR-33 oligonucleotides for 4 wks (n=4/group). Data are expressed as mean ± SD. *P < 0.05, **P < 0.01 compared to control.
As miR-33 has been shown to play important roles in cholesterol homeostasis of macrophages as well as hepatocytes, we next investigated whether miR-33 regulates expression of OSBPL6 in human THP-1 macrophages treated with miR-33 mimics or inhibitors. Overexpression of miR-33 repressed, while inhibition of miR-33 increased OSBPL6 but not OSBPL1 mRNA in THP-1 macrophages (Figure 1D). Consistent with this, isolation of macrophages from aortic sinus plaques of Ldlr−/− mice treated for 4 weeks with anti-miR-33 showed that Osbpl6 mRNA levels were markedly increased compared to those in plaque macrophages from control anti-miR treated mice (Figure 1E). Immunofluorescence staining of aortic sinus plaques from Ldlr−/− mice confirmed abundant intracellular ORP6 staining localized to MOMA-2-positive macrophages (Supplemental Figure 1C), supporting an important role in this cell type.
During our analysis of the 3’UTR of OSBPL6 we noted that this gene also contains a predicted binding site for miR-27b (Figure 1F), a miRNA recently identified as a hub that regulates genes involved in lipid metabolism7, 8. To confirm miR-27b targeting of OSBPL6, we performed 3’UTR-luciferase reporter assays. Transfection of HEK293 cells with miR-27b mimics reduced OSBPL6-3’UTR-luciferase activity by approximately 20% compared to control mimic, and this repression was abrogated by mutating the predicted miR-27b binding site (Figure 1G). Furthermore, transfection of human THP-1 macrophages with miR-27b mimics reduced OSBPL6 mRNA levels, but not those of OSBPL1 (Figure 1H), which is not a predicted miR-27b target. Conversely, transfection of human THP-1 macrophages with an inhibitor of miR-27b increased OSBPL6 mRNA levels (Figure 1H). Together, these data establish OSBPL6 as a bona fide target of both miR-33 and miR-27b. Notably, miR-33 and miR-27b have also been reported to both target the cholesterol transporter ABCA18, 12-14, suggesting that these lipid-miRNAs may repress overlapping gene networks involved in cholesterol homeostasis.
OSBPL6 is induced in response to increased cellular and dietary cholesterol
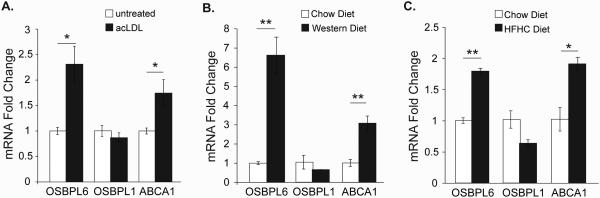
To investigate a role for OSBPL6 in cholesterol homeostasis, we tested its regulation by cholesterol in vitro and in vivo. Cholesterol loading of THP-1 macrophages with acetylated LDL (acLDL) to load cells with cholesterol led to a two-fold induction of OSBPL6 mRNA, but not OSBPL1 mRNA, as measured by qPCR (Figure 2A). The acLDL-induced increase in OSBPL6 mRNA was comparable to that observed for the cholesterol-responsive gene ABCA1 (Figure 2A), suggesting that OSBPL6 is regulated by cellular cholesterol content. Notably, miR-33 and miR-27b levels decrease in THP-1 macrophages loaded with acLDL, coincident with an increase in OSBPL6 mRNA (13 and Supplemental Figure 1D). To assess the regulation of OSBPL6 in vivo, we fed mice and non-human primates diets enriched in cholesterol. Ldlr−/− mice showed a 6-fold increase in hepatic Osbpl6 mRNA after 14 weeks of Western diet compared to chow diet feeding (Figure 2B). Similarly, there was a marked increase in OSBPL6 mRNA levels in the livers of African green monkeys fed a high-fat/high-cholesterol diet for 10 weeks compared to chow fed monkeys (Figure 2C). Together, these data indicate that expression of OSBPL6 is upregulated by cellular and dietary cholesterol content.
Figure 2. OSBPL6 mRNA is induced in response to cellular and dietary cholesterol.
Relative expression of OSBPL6, OSBPL1 and ABCA1 mRNA by qPCR analysis in A) THP1 macrophages treated with acLDL for 24 hours; B) Livers of Ldlr−/− mice (n=5) fed a chow or a Western diet for 14 weeks; C) Livers of African green monkeys (n=5/group) fed a chow or high-fat/high-cholesterol (HFHC) diet for 10 weeks. Data are expressed as mean ± SD. Data in (A) are representative of three independent experiments. *P < 0.05, **P < 0.01 compared to untreated group or chow diet.
OSBPL6 is an LXR transcriptional target
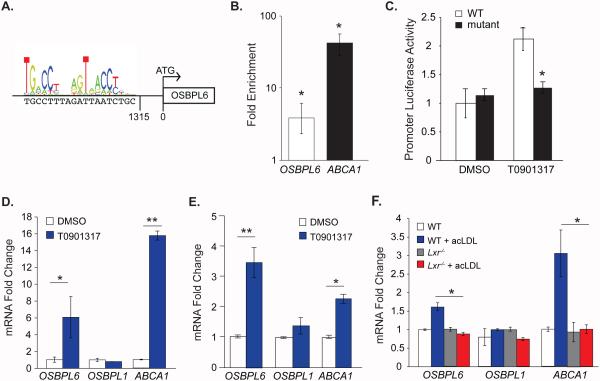
LXRα/β are transcription factors that are activated in response to elevated cholesterol levels in multiple cells types and induce the expression of an array of genes involved in cholesterol trafficking, efflux and excretion. To investigate whether LXRα/β play a role in regulating OSBPL6 transcription, we examined the promoter sequence of OSBPL6 and identified an LXR response element (LXRE) approximately 1300 base pairs upstream of the start codon (Figure 3A). To test whether this LXRE is functional, we performed a chromatin immunoprecipitation assay in macrophages. As shown in figure 3B, LXR is bound to the promoter region of OSBPL6 and the known LXR-target gene ABCA1, suggesting that OSBPL6 may be a direct transcriptional target of LXR. Consistent with this, the LXR agonist T0901317 induced expression of a human OSBPL6 promoter-luciferase reporter construct, but not when the putative LXRE was mutated (Figure 3C). Furthermore, T0901317 treatment of THP-1 macrophages (Figure 3D) and hepatocytic HepG2 cells (Figure 3E) increased OSBPL6 mRNA in these cell types, compared to vehicle treated cells. Finally, to confirm the specificity of this regulation, we treated wild type and Lxr−/− mouse bone marrow derived macrophages with acLDL and measured Osbpl6 mRNA levels by qPCR. Loading macrophages with acLDL increased Osbpl6 and Abca1 mRNA levels in wild type but not Lxr−/− macrophages (Figure 3F), confirming a key role for LXR in the regulation of Osbpl6 in response to cellular cholesterol loading.
Figure 3. OSBPL6 expression is regulated by the Liver X Receptor (LXR).
A) LXR response element (LXRE) in the promoter of human OSBPL6 as predicted by the JASPAR database. B) Relative enrichment of OSBPL6 and ABCA1 in LXRα chromatin IP from THP-1 cells treated with 10 μM LXRα agonist (T0901317). Data are normalized to control IgG. C) Activity of the wild type (WT) or LXRE-mutant OSBPL6 promoter-luciferase reporter in HEK293T cells treated with 10 μM T0901317 or DMSO. (D-F) Relative expression of OSBPL6, OSBPL1 and ABCA1 mRNA determined by qPCR in (D) THP-1 cells or (E) HepG2 cells treated with 10 μM T0901317 or vehicle control (DMSO) or (F) bone marrow derived macrophages from wild-type (WT) and Lxr−/− mice treated with 50 μg/mL acLDL or untreated. Data are expressed as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 compared to control treatment.
ORP6 localizes to endocytic vesicles and traffics cholesterol to the ER
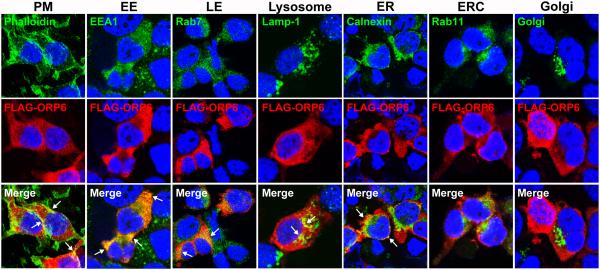
ORP6 contains a pleckstrin homology membrane-targeting domain, as well as a FFAT-motif predicted to interact with VAPs in the ER, suggesting a potential function in cholesterol movement between the ER and endosomal or plasma membranes19. To investigate whether ORP6 plays a role in intracellular cholesterol trafficking, we first investigated its subcellular localization. Due to a lack of specific and high affinity ORP6 antibodies, we generated an N-terminal FLAG-tagged human OSBPL6 plasmid and used an anti-FLAG antibody to detect FLAG-ORP6 in transfected HEK293 cells. First, we confirmed the expression of OSBPL6/ORP6 at the mRNA and protein levels, detecting a band for FLAG-ORP6 at the appropriate molecular weight (Supplemental Figure 2A). Consistent with a previous report demonstrating the presence of ORP6 on the ER19, immunofluorescent staining showed that FLAG-ORP6 co-localized with the ER marker calnexin (Figure 4). In addition, we observed abundant FLAG-ORP6 staining on the plasma membrane, which is enriched in F-actin filaments, as well as in EEA1-positive early endosomes, in Rab7-positive late endosomes and Lamp1-positive lysosomes (Figure 4). There was minimal overlap between FLAG-ORP6 and the endocytic recycling compartment (Rab11-positive) and no overlap with the Golgi apparatus (Figure 4). By fixing cells after a short permeabilization with saponin to release cytosol and facilitate the observation of membrane-bound proteins, we confirmed that FLAG-ORP6 primarily localized to membranes of early endosomes and ER (Supplemental Figure 3). Using this technique, we again observed some staining of plasma membrane and membranes of late endosomes and lysosomes, but not the endocytic recycling compartment and Golgi apparatus (Supplemental Figure 3). This dual targeting of ORP6 to the endolysosomal and ER compartments is consistent with a potential role in cholesterol trafficking to or from the ER.
Figure 4. ORP6 localizes to early endosomes, lysosomes, and the ER.
Immunofluorescence staining for FLAG (ORP6) and the indicated organelle markers (PM: plasma membrane, EE: early endosome, LE: late endosome, ER: endoplasmic reticulum, ERC: endocytic recycling compartment) in HEK293 cells transfected with FLAG-tagged ORP6. Co-localization is shown in the merged images as yellow and is indicated by arrows.
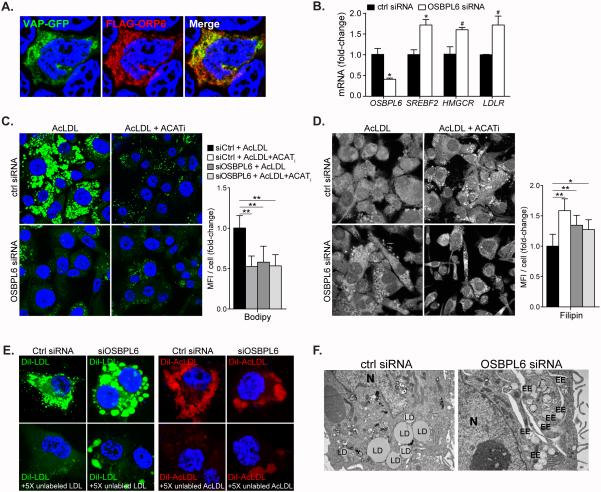
ORP proteins have been observed at MCSs between the ER and various organelles; for example ORP2 localizes at LD-ER MCSs and ORP1L at late endosome-ER MCSs, where they facilitate the transport of cholesterol through these compartments16. Therefore, we investigated whether ORP6 co-localized with the ER integral protein VAPA at MCSs in HEK293 cells co-expressing VAPA-GFP and FLAG-ORP6. Indeed, after saponin permeabilization to release cytosol, FLAG-ORP6 co-localized with VAPA (Figure 5A), suggesting a role for ORP6 in mediating sterol transfer between the ER and other compartments. To investigate whether ORP6 regulates lipid transport to the ER, we used an OSBPL6-targeted siRNA to silence ORP6 expression. OSBPL6-siRNA reduced the expression of OSBPL6 mRNA and ORP6 protein by more than 70% as compared to control non-targeting siRNA (Supplemental Figure 2C). Consistent with less trafficking of cholesterol to the ER, ORP6 knockdown in THP-1 macrophages was associated with increased activation of SREBP2, as evidenced by elevated expression of SREBF2 and its downstream regulated genes HMGCR and LDLR - genes that are normally induced in the cholesterol-depleted state (Figure 5B).
Figure 5. ORP6 regulates cholesterol trafficking.
A) Co-localization of ORP6 and VAPA in HEK cells co-expressing FLAG-ORP6 and VAPA-GFP. B) Gene expression in THP-1 macrophages treated with control or OSBPL6 siRNA. Data are mean ± SEM of one experiment representative of three independent experiments. #P ≤ 0.1, *P < 0.05, **P < 0.005. C) BODIPY (neutral lipid) and D) Filipin (free cholesterol) staining in THP-1 cells transfected with control or OSBPL6 targeted siRNAs and loaded with acLDL (50 μg/mL) in the presence or absence of an inhibitor of acyl-CoA cholesterol acyltransferase (ACATi) to prevent the esterification of cholesterol in the ER. Quantification is shown at right; fluorescence images from an average of nine different fields were quantified by averaging the mean fluorescence intensity from an average of 70 cells (C) or 45 cells (D) selected randomly from each field by using image J. Data are the mean ± SEM of two independent experiments. *P < 0.05, **P < 0.005 (ANOVA). E) THP-1 macrophages were incubated with DiI-labeled LDL or AcLDL (10μg/mL) in the presence or absence of unlabeled lipoprotein in 5X excess (50μg/mL) for 4h to visualize internalized lipoproteins in cells treated with control or OSBPL6 siRNA. F) Electron microscopy of THP-1 macrophages treated with control or OSBPL6 siRNA and loaded with AcLDL (50μg/mL) for 24h.
Next, we transfected THP-1 macrophages with OSBPL6-siRNA or control siRNA and examined if ORP6 knock-down affects cellular cholesterol distribution upon cholesterol loading of the cells. For this, THP-1 cells were treated with acLDL in the presence or absence of an ACAT inhibitor (ACATi) to prevent the esterification of cholesterol in the ER. As expected, control siRNA-transfected macrophages treated with acLDL and stained with BODIPY to identify neutral lipid showed a punctate pattern indicative of the formation of cholesterol ester-rich LDs and the addition of ACATi reduced the formation of BODIPY-positive LDs upon acLDL loading (Figure 5C). Notably, ORP6 knock-down in THP-1 cells treated with acLDL led to a phenotype similar to that observed with the ACATi treatment, whereby THP-1 cells transfected with OSBPL6-siRNA showed reduced LD genesis. Furthermore, like ACATi, ORP6 silencing led to the formation of filipin-positive punctate structures indicative of free cholesterol accumulation in endocytic vesicles (Figure 5D), which is reminiscent of the cholesterol trafficking defect in NPC deficient fibroblasts. Together these data implicate ORP6 in the transport of cholesterol from endosomes to the ER for esterification and storage.
To test whether SREBP2 activation and endosomal cholesterol accumulation in cells where ORP6 was knocked down might be due to defective lipoprotein uptake, we quantified lipoprotein endocytosis in THP-1 macrophages treated with control or OSBPL6-siRNA and found comparable uptake of radiolabeled LDL and acLDL (Supplemental Figure 4). However, there was a striking phenotypic difference between control-treated and OSBPL6-siRNA treated THP-1 cells incubated with DiI-labeled lipoproteins. In control siRNA-treated cells, there were multiple small endocytic vesicles resulting from the uptake of the fluorescently labeled lipoproteins (Figure 5E). In contrast, in macrophages where ORP6 was knocked down, there appeared to be a fused endocytic network (Figure 5E). In both cases, DiI-labeled lipoprotein internalization could be competed using an excess of unlabeled lipoproteins. Morphological analysis of acLDL-loaded macrophages by electron microscopy showed the presence of clustered early endosomes in cells where ORP6 was knocked down (Figure 5F). Moreover, this was associated with a reduction in cytosolic LDs in those lipid-loaded cells as compared to control siRNA-treated cells (Figure 5F). Collectively, these experiments support a role for ORP6 in cholesterol trafficking from early endosomes to the ER.
OSBPL6 regulates cellular cholesterol efflux
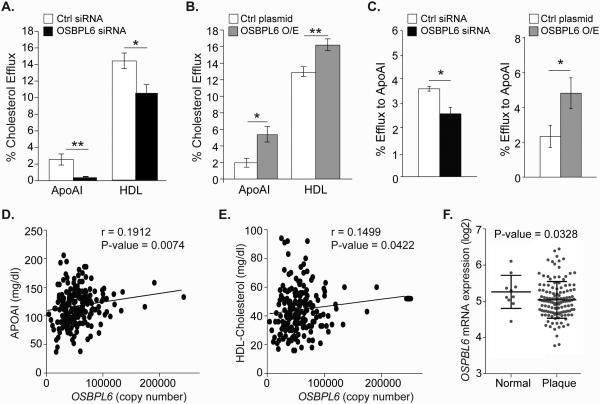
As endocytic flux has also been shown to be required for cellular cholesterol efflux, we next measured the effect of OSBPL6 gain- or loss-of-function on cholesterol efflux to the cholesterol acceptors apoA1 and HDL in THP-1 macrophages. THP-1 macrophages transfected with OSBPL6-siRNA showed reduced cholesterol efflux to both apoA1 and HDL (Figure 6A). Conversely, transfection of THP-1 cells with an OSBPL6 expression plasmid increased OSBPL6 mRNA and ORP6 protein (Supplemental Figure 2) and enhanced cholesterol efflux to both apoA1 and HDL (Figure 6B). Similarly, overexpression of FLAG-ORP6 in HEK293 cells stably expressing ABCA1 increased cholesterol efflux to apoA1 (Supplemental Figure 2B). As hepatic cholesterol efflux via ABCA1 regulates the formation of nascent HDL particles, we next investigated whether ORP6 also modulates cholesterol efflux in HepG2 cells. Indeed, ORP6 silencing reduced and ORP6 overexpression increased cholesterol efflux from hepatic cells to apoA1 (Figure 6C). Furthermore, quantification of hepatic OSBPL6 mRNA expression in a cohort of 200 healthy individuals revealed a positive correlation with plasma levels of apoA1 (Figure 6D) and HDL-C (Figure 6E). As cholesterol efflux is known to be atheroprotective, we next measured the expression of OSBPL6 mRNA in healthy versus diseased human carotid arteries. Notably, we show that OSBPL6 mRNA is reduced in arterial plaque samples as compared to healthy controls (Figure 6F), coincident with an increase in miR-27b (Supplemental Figure 5) and miR-33a/b20 levels. Collectively, these data identify OSBPL6/ORP6 as a novel regulator of cellular cholesterol trafficking and efflux.
Figure 6. OSBPL6 regulates cholesterol efflux, is correlated with plasma levels of HDL and apoAI and its expression is reduced in atherosclerosis.
A-C) Cholesterol efflux to apoAI or HDL by THP-1 cells (A-B) or to apoAI by HepG2 cells (C) that are transfected with a siRNA against OSBPL6 or control siRNA or an expression plasmid for OSBPL6. Dot plots showing direct relationship between human hepatic expression of OSBPL6 and plasma (D) HDL and (E) APOAI levels. Data are expressed as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 compared to control treatment. F) OSBPL6 mRNA expression (log2) in healthy arteries (Normal) or carotids from patients with atherosclerosis (Plaque) from the Biobank of Karolinska Endarterectomies (BiKE).
DISCUSSION
Cholesterol homeostasis is fundamental to human health and therefore has been extensively studied. However, despite the advances in our knowledge since the pioneering work of Brown and Goldstein on the molecular basis of cholesterol homeostasis2, many aspects of intracellular cholesterol transport remain poorly understood. Cholesterol rapidly traffics in cells both in vesicles such as endosomes, as well as via non-vesicular mechanisms involving lipid transport proteins. These highly dynamic processes are essential to the maintenance of the cholesterol gradient in the cell, and studies of inherited human mutations in the LDLR, PCSK9, NPC1/2 and ABCA1 genes have provided important insights into the pathways regulating cholesterol uptake, trafficking and efflux. Cholesterol homeostasis is a complex process that requires the coordination of sensors (SCAP-Insig, SREBPs), transcriptional regulators (LXRα/β, SREBPs) and effectors (ABCA1, ABCG1). An added level of complexity is provided by the more recent discovery of miRNAs, such as miR-33 and miR-27b, that can fine-tune cholesterol homeostasis by targeting multiple genes in this pathway, such as targeting of the cholesterol efflux genes NPC1, ABCA1, ABCG1 by miR-3313 and targeting of genes in the reverse cholesterol transport pathway ABCA1 and ANGPTL3 by miR-27b7. By analyzing the gene networks targeted by miR-33 and miR-27b, we identified ORP6 as a novel regulator of cholesterol homeostasis and demonstrate a role for this cytosolic protein in the endolysomal transport of cholesterol between the plasma membrane and the ER, with important implications on cholesterol efflux8, 12-14.
ORPs comprise a large family of cytosolic proteins that bind sterols (cholesterol or oxysterols) and PIPs. Several members of this family have been shown to associate peripherally with the cellular membrane of various organelles and to function in lipid trafficking and signaling. For example, ORP1L regulates late endosomal trafficking by acting as an effector for the GTPase Rab7 and appears to modulate LXR-ligand interactions affecting the expression of genes controlled by this transcription factor21, 22. ORP1L on late endosomes undergoes a conformational change upon binding to cholesterol; at low cholesterol concentrations, its FFAT-motif is exposed allowing it to interact with the ER protein-VAPA, whereas at high cholesterol concentrations ORP1L detaches from the ER and tethers late endosomes to lysosomes, thereby enabling their fusion and facilitating efficient cargo delivery between these compartments23. As another example, ORP2 localizes at the LD-ER interface, and in complex with VAPs at these MCSs, ORP2 regulates LD neutral lipid turnover24. Interestingly, the ORP2-LD association is dependent on sterol binding, whereby binding of ORP2 to oxysterols inhibits its association with LDs and impairs triglyceride and cholesterol ester metabolism25. ORP4 was shown to bind to vimentin through its lipid-binding domain and suggested to play a role in lipid transport between endosomes/lysosomes and the ER, and cholesterol esterification26. Unlike other family members, ORP5 and ORP8 are anchored to the ER by a C-terminal transmembrane domain and act to tether the plasma membrane with the ER to mediate mediate phosphatidylinositol 4-phosphate (PI4P) and phosphatidylserine (PS) countertransport between these compartments27. In contrast to these other ORP family members, ORP6 has not been extensively studied and its function is to date unknown.
A previous screen of ORP family members found that ORP6 is the only ORP that is upregulated in cholesterol-loaded macrophages28. We confirmed here that OSBPL6 expression is induced in macrophages enriched in cholesterol in vitro, and showed that ORP6 is expressed in macrophage foam cells present in atherosclerotic plaques in mice. Furthermore, OSBPL6 expression is upregulated in the livers of both mice and non-human primates in response to dietary cholesterol. Consistent with this, we show that the OSBPL6 gene is transcriptionally regulated by the LXR transcription factors that are activated in response to increased cellular cholesterol and coordinate the expression of genes involved in cholesterol trafficking, efflux and excretion. Through the coordinated actions of both LXR-mediated transcriptional regulation and miRNA-mediated post-transcriptional repression, ORP6 levels appear to be tightly controlled in response to sterol levels. Elevated cellular cholesterol levels lead to activation of LXR, which transcriptionally activates ORP6, while simultaneous inhibition of SREBP2 activity results in reduced expression of SREBF2 and consequently miR-33a, which is encoded in an intron of the SREBF2 gene. Therefore, reduced levels of miR-33 under these conditions would result in OSBPL6 de-repression and increased ORP6 levels. These findings add to our understanding of the pathways mediating cellular cholesterol mobilization and of the mechanisms by which miR-33 and miR-27b regulate cholesterol efflux, which was previously thought to be primarily through targeting of ABCA18, 12-14. This work highlights how miRNAs, through seemingly small effects (10-30%) on individual, related genes, can have a substantial cumulative impact on a biological pathway. Importantly, this work also suggests that miRNA-controlled networks can be interrogated to enable the discovery of novel regulators of conserved pathways, as we have shown here for OSBPL6. Although it is well established that miR-33 works in concert with SREBP2 to maintain cellular cholesterol homeostasis, the role of miR-27b in this process is less well understood. Our findings indicate that miR-27b and miR-33 are upregulated in parallel by cholesterol loading in macrophages and in human carotid artery plaques, suggesting that these two miRNAs may work together to regulate macrophage efflux and RCT.
Like other OSBP and ORP family members, ORP6 contains a conserved sterol-binding domain and a pleckstrin homology domain that facilitates interactions with PIPs enriched in membranes. In addition, the subclass III ORPs, which include ORP6, ORP3 and ORP7, have FFAT-motif that are predicted to interact with VAPs that anchor numerous proteins to the ER. Indeed we show using a FLAG-ORP6 construct that ORP6 localizes to both the endolysosomal compartments and the ER, specifically co-localizing with VAPA at the ER. This dual targeting supports a potential role for ORP6 in sterol transport between membranes, and we show that silencing of ORP6 impairs cholesterol movement from the endolysosomal network to the ER for esterification, resulting in a clustered endosomal reticular network and the accumulation of free cholesterol. This phenotype is similar to that seen when cells are treated with an inhibitor of the ER-resident ACAT enzyme that blocks cholesterol esterification and formation and also in cells lacking functional NPC1. In NPC1−/− macrophages, free cholesterol accumulates in late endosomes/lysosomes and cannot be exported out of these compartments to be effluxed from the cells29, 30, despite the fact that lipoprotein-derived cholesterol esters are properly hydrolyzed to generate free cholesterol. Interestingly, a previous study of fibroblasts taken from an individual with an NPC1 mutation reported an increase in ORP6 expression31 suggesting that this may represent a compensatory attempt to transport cholesterol out of the endosomal compartment. In future studies, it will be of interest to further investigate whether ORP6 can bind other sterols and/or PIPs in addition to cholesterol and 25-hydroxycholesterol32, and to determine how ligand binding affects ORP6 localization to distinct cellular compartments.
Cholesterol transport from endosomes/lysosomes to the plasma membrane also contributes to the pool of free cholesterol available for efflux via ABC transporters. This transport pathway is poorly understood, but likely involves multiple components, such as the ORPs and steroidogenic acute regulatory protein-related lipid transfer proteins. Using gain- and loss-of-function approaches we show that ORP6 contributes to cellular cholesterol efflux to apoA1 and HDL. In hepatocytes, ABCA1-mediated efflux plays an essential role in the biogenesis of HDL, and in macrophages, ABCA1- and ABCG1-mediated cholesterol efflux is important for protection from atherosclerosis4. Together, these cholesterol efflux pathways contribute to RCT of cholesterol to the liver, which is thought to underlie the inverse association of HDL-C levels with coronary artery disease risk4. Notably, the OSBPL6 gene lies in a locus on human chromosome 2 that has been associated with premature coronary artery disease in the absence of hyperlipidemia17 and to variations in plasma HDL-C levels18. Our findings that ORP6 is involved in the pathway leading to cholesterol efflux and that hepatic levels of OSBPL6 mRNA are positively correlated with plasma levels of HDL-C and apoA1 in humans suggest that OSBPL6 is a strong candidate gene in this risk locus. While the observed correlations between OSBPL6 and apoAI/HDL are low, these significant relationships have important biological relevance when considering the multitude of factors that influence human circulating apoA1 and HDL-C.
Supplementary Material
Supplemental Figure I. OSBPL6 is a novel miR-33 target gene.
Relative hepatic expression of OSBPL6, OSBPL1 and ABCA1 mRNA levels determined by qPCR in A) C57BL/6 mice fed chow diet and infected with control miR, anti-miR-33 or miR-33 lentiviruses for 6 days (n=5/group), and B) Ldlr−/−mice fed a Western diet (14 wk) and treated with control miR or anti-miR-33 oligonucleotides for 4 wks (n=10/group). C) Immunofluorescence staining of aortic sinus plaques from Ldlr−/− mice fed a Western diet (8 wk) for the macrophage marker MOMA-2 (red) or OSBPL6 (green). (100x magnification). Data are expressed as mean ± SD. *P < 0.05, **P < 0.01 compared to control. D) OSBPL6 mRNA in human THP-1 macrophages treated with AcLDL (50μg/mL) for 2h. Data are the mean ± SEM of three independent experiments. **P ≤ 0.005.
Supplemental Figure II. A) Anti-FLAG western blot detecting the FLAG-OSBPL6 fusion protein. B) 3H-cholesterol efflux from HEK293 cells expressing ABCA1 and FLAG-ORP6. Data are mean and SEM and are representative of two independant experiments. *P < 0.05. C-D) Relative expression of OSBPL6 mRNA and OSBPL6 protein in (C) THP-1 cells transfected with control or OSBPL6 targeted siRNAs, or (D) control or OSBPL6 expression plasmids.
*P < 0.05, **P < 0.01 compared to control treatment. Data are expressed as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 compared to control treatment.
Supplemental Figure III. A, B) Immunofluorescence staining for the FLAG-ORP6 fusion protein, in HEK293T cells expressing FLAG-ORP6 fixed either directly (A) or after a short permeabilization with saponin to release cytosol (B) Data are representative of two independant experiments.
Supplemental Figure IV. ORP6 expression does not regulate lipoprotein endocytosis.
A) Lipoprotein endocytosis in human THP-1 macrophages treated with control siRNA or OSBPL6 siRNA. Cells were incubated with 3H-cholesterol-containing LDL or acLDL (50 μg/mL; 0.5 μCi/mL) for 6h prior to cellular radioactivity quantification. Data are the mean ± SEM of two independent experiments. n.s. = non significant.
Supplemental Figure V. miR-27b expression in human atherosclerotic plaques.
miR-27b expression in healthy arteries (Normal) or carotids from patients with atherosclerosis (Plaque) from the Biobank of Karolinska Endarterectomies (BiKE).
SIGNIFICANCE.
Cholesterol is a vital component of mammalian cell membranes and is essential to human health. Dysregulated cellular cholesterol levels can lead to multiple pathologies, including atherosclerosis and coronary artery disease and, as such, molecular mechanisms have evolved to tightly regulate cholesterol homeostasis. However, many aspects of intracellular cholesterol transport remain poorly understood. Recently, miRNAs such as miR-33 and miR-27b have been characterized important fine-tuners of cholesterol homeostasis by repressing multiple genes in related pathways such as cholesterol efflux or reverse cholesterol transport. Here we studied the network of miR-33 and miR-27b target genes and identified ORP6 as a novel regulator of intracellular cholesterol trafficking and efflux to HDL and its protein component apoA1. Finally, we showed that levels of OSBPL6 mRNA in the liver are positively correlated with plasma levels of HDL in humans, suggesting an important role for OSBPL6/OPR6 in cholesterol homeostasis.
ACKNOWLEDGMENTS
We would like to acknowledge Regulus Therapeutics, Inc. for providing the control and anti-miR-33 oligonucleotides and Alice F. Liang from the NYU OCS Microscopy Core for assistance with the electron microscopy. We thank Jonathan Smith (Cleveland Clinic) for kindly providing HEK293 cells stably expressing ABCA133. We thank Ekaterina Chernogubova for technical assistance in quantification of miR-27b in human arteries.
Sources of funding - Support for this work came from the National Institutes of Health (R01HL108182 to K.J.M; R01HL119047 to K.J.M. and K.J.R.; R01HL117226 to MG, R00HL088528 to RET; T32HL098129 to E.J.H, and T32AI07180 to MH), the German Research Foundation (DFG) as part of the CRC 1123 (Project B1) (L.M.H, D.T), the American Heart Association (13POST14490016 to B.R.; 14POST20180018 to C.v.S.), and Canadian Institutes of Health Research (M.O.). The BiKE study was conducted with support from the Swedish Heart and Lung Foundation, the Swedish Research Council, Uppdrag Besegra Stroke, the Strategic Cardiovascular Programs of Karolinska Institutet and Stockholm County Council, the Stockholm County Council, the Foundation for Strategic Research and the European Commission (CarTarDis, AtheroRemo, VIA and AtheroFlux projects).
NONSTANDARD ABBREVIATIONS AND ACRONYMS
- ACAT
Acyl-CoA:cholesterol acyltransferase
- ACATi
ACAT inhibitor
- APOA1
Apolipoprotein A-I
- ABCA1
ATP-binding cassette transporter A1
- ABCG1
ATP-binding cassette transporter G1
- ER
Endoplasmic reticulum
- FFAT-motif
Phenylalanine in an acidic tract motif
- HDL
High density lipoprotein
- HDL-C
HDL-cholesterol
- LD
Lipid droplet
- LDL
Low density lipoprotein
- LDLR
LDL receptor
- LXR
Liver X receptor
- LXRE
LXR response element
- MCS
Membrane contact site
- miRNA
microRNA
- ORP6
OSBPL-related protein 6 (protein)
- OSBPL6
Oxysterol binding protein-like 6 (RNA transcript)
- PIP
Phosphatidylinositol phosphate
- RCT
Reverse cholesterol transport
- SREBP
Sterol regulatory element-binding protein
- VAP
Vesicle-associated membrane protein-associated protein
Footnotes
Disclosures - None
REFERENCES
- 1.Maxfield FR, van Meer G. Cholesterol, the central lipid of mammalian cells. Current opinion in cell biology. 2010;22:422–429. doi: 10.1016/j.ceb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein JL, Brown MS. The ldl receptor. Arteriosclerosis, thrombosis, and vascular biology. 2009;29:431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeon TI, Osborne TF. Srebps: Metabolic integrators in physiology and metabolism. Trends in endocrinology and metabolism: TEM. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yvan-Charvet L, Wang N, Tall AR. Role of hdl, abca1, and abcg1 transporters in cholesterol efflux and immune responses. Arteriosclerosis, thrombosis, and vascular biology. 2010;30:139–143. doi: 10.1161/ATVBAHA.108.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouimet M, Moore KJ. A big role for small rnas in hdl homeostasis. Journal of lipid research. 2013;54:1161–1167. doi: 10.1194/jlr.R036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun X, Feinberg MW. Microrna-management of lipoprotein homeostasis. Circulation research. 2014;115:2–6. doi: 10.1161/CIRCRESAHA.114.304228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. Microrna-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang M, Wu JF, Chen WJ, et al. Microrna-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in thp-1 macrophages. Atherosclerosis. 2014;234:54–64. doi: 10.1016/j.atherosclerosis.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. Mir-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO molecular medicine. 2012;4:882–895. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davalos A, Goedeke L, Smibert P, et al. Mir-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of mir-33 from an srebp2 intron inhibits cholesterol export and fatty acid oxidation. The Journal of biological chemistry. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. Microrna-33 and the srebp host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. Mir-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marquart TJ, Allen RM, Ory DS, Baldan A. Mir-33 links srebp-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rottiers V, Najafi-Shoushtari SH, Kristo F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N, Mostoslavsky R, Naar AM. Micrornas in metabolism and metabolic diseases. Cold Spring Harbor symposia on quantitative biology. 2011;76:225–233. doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olkkonen VM, Li S. Oxysterol-binding proteins: Sterol and phosphoinositide sensors coordinating transport, signaling and metabolism. Progress in lipid research. 2013;52:529–538. doi: 10.1016/j.plipres.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Nsengimana J, Samani NJ, Hall AS, Balmforth AJ, Mangino M, Yuldasheva N, Maqbool A, Braund P, Burton P, Bishop DT, Ball SG, Barrett JH. Enhanced linkage of a locus on chromosome 2 to premature coronary artery disease in the absence of hypercholesterolemia. Eur J Hum Genet. 2007;15:313–319. doi: 10.1038/sj.ejhg.5201752. [DOI] [PubMed] [Google Scholar]
- 18.North KE, Martin LJ, Dyer T, Comuzzie AG, Williams JT, Framingham Heart S. Hdl cholesterol in females in the framingham heart study is linked to a region of chromosome 2q. BMC genetics. 2003;4(Suppl 1):S98. doi: 10.1186/1471-2156-4-S1-S98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehto M, Tienari J, Lehtonen S, Lehtonen E, Olkkonen VM. Subfamily iii of mammalian oxysterol-binding protein (osbp) homologues: The expression and intracellular localization of orp3, orp6, and orp7. Cell Tissue Res. 2004;315:39–57. doi: 10.1007/s00441-003-0817-y. [DOI] [PubMed] [Google Scholar]
- 20.Karunakaran D, Thrush AB, Nguyen MA, et al. Macrophage mitochondrial energy status regulates cholesterol efflux and is enhanced by anti-mir33 in atherosclerosis. Circulation research. 2015;117:266–278. doi: 10.1161/CIRCRESAHA.117.305624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson M, Lehto M, Tanhuanpaa K, Cover TL, Olkkonen VM. The oxysterol-binding protein homologue orp1l interacts with rab7 and alters functional properties of late endocytic compartments. Molecular biology of the cell. 2005;16:5480–5492. doi: 10.1091/mbc.E05-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan D, Jauhiainen M, Hildebrand RB, Willems van Dijk K, Van Berkel TJ, Ehnholm C, Van Eck M, Olkkonen VM. Expression of human osbp-related protein 1l in macrophages enhances atherosclerotic lesion development in ldl receptor-deficient mice. Arteriosclerosis, thrombosis, and vascular biology. 2007;27:1618–1624. doi: 10.1161/ATVBAHA.107.144121. [DOI] [PubMed] [Google Scholar]
- 23.van der Kant R, Fish A, Janssen L, Janssen H, Krom S, Ho N, Brummelkamp T, Carette J, Rocha N, Neefjes J. Late endosomal transport and tethering are coupled processes controlled by rilp and the cholesterol sensor orp1l. Journal of cell science. 2013;126:3462–3474. doi: 10.1242/jcs.129270. [DOI] [PubMed] [Google Scholar]
- 24.Weber-Boyvat M, Kentala H, Peranen J, Olkkonen VM. Ligand-dependent localization and function of orp-vap complexes at membrane contact sites. Cellular and molecular life sciences : CMLS. 2015;72:1967–1987. doi: 10.1007/s00018-014-1786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hynynen R, Suchanek M, Spandl J, Back N, Thiele C, Olkkonen VM. Osbp-related protein 2 is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids. Journal of lipid research. 2009;50:1305–1315. doi: 10.1194/jlr.M800661-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, JeBailey L, Ridgway ND. Oxysterol-binding-protein (osbp)-related protein 4 binds 25-hydroxycholesterol and interacts with vimentin intermediate filaments. The Biochemical journal. 2002;361:461–472. doi: 10.1042/0264-6021:3610461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P. Intracellular transport. Pi4p/phosphatidylserine countertransport at orp5- and orp8-mediated er-plasma membrane contacts. Science. 2015;349:428–432. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehto M, Laitinen S, Chinetti G, Johansson M, Ehnholm C, Staels B, Ikonen E, Olkkonen VM. The osbp-related protein family in humans. Journal of lipid research. 2001;42:1203–1213. [PubMed] [Google Scholar]
- 29.Boadu E, Nelson RC, Francis GA. Abca1-dependent mobilization of lysosomal cholesterol requires functional niemann-pick c2 but not niemann-pick c1 protein. Biochimica et biophysica acta. 2012;1821:396–404. doi: 10.1016/j.bbalip.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Wang MD, Franklin V, Sundaram M, Kiss RS, Ho K, Gallant M, Marcel YL. Differential regulation of atp binding cassette protein a1 expression and apoa-i lipidation by niemann-pick type c1 in murine hepatocytes and macrophages. The Journal of biological chemistry. 2007;282:22525–22533. doi: 10.1074/jbc.M700326200. [DOI] [PubMed] [Google Scholar]
- 31.Reddy JV, Ganley IG, Pfeffer SR. Clues to neuro-degeneration in niemann-pick type c disease from global gene expression profiling. PloS one. 2006;1:e19. doi: 10.1371/journal.pone.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suchanek M, Hynynen R, Wohlfahrt G, Lehto M, Johansson M, Saarinen H, Radzikowska A, Thiele C, Olkkonen VM. The mammalian oxysterol-binding protein-related proteins (orps) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. The Biochemical journal. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith JD, Waelde C, Horwitz A, Zheng P. Evaluation of the role of phosphatidylserine translocase activity in abca1-mediated lipid efflux. The Journal of biological chemistry. 2002;277:17797–17803. doi: 10.1074/jbc.M201594200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure I. OSBPL6 is a novel miR-33 target gene.
Relative hepatic expression of OSBPL6, OSBPL1 and ABCA1 mRNA levels determined by qPCR in A) C57BL/6 mice fed chow diet and infected with control miR, anti-miR-33 or miR-33 lentiviruses for 6 days (n=5/group), and B) Ldlr−/−mice fed a Western diet (14 wk) and treated with control miR or anti-miR-33 oligonucleotides for 4 wks (n=10/group). C) Immunofluorescence staining of aortic sinus plaques from Ldlr−/− mice fed a Western diet (8 wk) for the macrophage marker MOMA-2 (red) or OSBPL6 (green). (100x magnification). Data are expressed as mean ± SD. *P < 0.05, **P < 0.01 compared to control. D) OSBPL6 mRNA in human THP-1 macrophages treated with AcLDL (50μg/mL) for 2h. Data are the mean ± SEM of three independent experiments. **P ≤ 0.005.
Supplemental Figure II. A) Anti-FLAG western blot detecting the FLAG-OSBPL6 fusion protein. B) 3H-cholesterol efflux from HEK293 cells expressing ABCA1 and FLAG-ORP6. Data are mean and SEM and are representative of two independant experiments. *P < 0.05. C-D) Relative expression of OSBPL6 mRNA and OSBPL6 protein in (C) THP-1 cells transfected with control or OSBPL6 targeted siRNAs, or (D) control or OSBPL6 expression plasmids.
*P < 0.05, **P < 0.01 compared to control treatment. Data are expressed as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 compared to control treatment.
Supplemental Figure III. A, B) Immunofluorescence staining for the FLAG-ORP6 fusion protein, in HEK293T cells expressing FLAG-ORP6 fixed either directly (A) or after a short permeabilization with saponin to release cytosol (B) Data are representative of two independant experiments.
Supplemental Figure IV. ORP6 expression does not regulate lipoprotein endocytosis.
A) Lipoprotein endocytosis in human THP-1 macrophages treated with control siRNA or OSBPL6 siRNA. Cells were incubated with 3H-cholesterol-containing LDL or acLDL (50 μg/mL; 0.5 μCi/mL) for 6h prior to cellular radioactivity quantification. Data are the mean ± SEM of two independent experiments. n.s. = non significant.
Supplemental Figure V. miR-27b expression in human atherosclerotic plaques.
miR-27b expression in healthy arteries (Normal) or carotids from patients with atherosclerosis (Plaque) from the Biobank of Karolinska Endarterectomies (BiKE).