Abstract
Laryngeal squamous cell carcinoma (LSCC) remains a highly morbid and fatal disease. Historically, it has been a model example for organ preservation and treatment stratification paradigms. Unfortunately, survival for LSCC has stagnated over the past few decades. As the era of next-generation sequencing and personalized treatment for cancer approaches, LSCC may be an ideal disease for consideration of further treatment stratification and personalization. Here, we will discuss the important history of LSCC as a model system for organ preservation, unique and potentially targetable genetic signatures of LSCC, and methods for bringing stratified, personalized treatment strategies to the 21st century.
Keywords: Head and neck cancer, laryngeal squamous cell carcinoma, genetics, targeted therapy, personalized medicine
Introduction
Laryngeal squamous cell carcinoma (LSCC) remains a prevalent disease, accounting for over 150, 000 new cases annually across the world1. Previous clinical trials in LSCC demonstrated the potential for non-surgical, organ-preservation treatment options for LSCC, with similar survival rates to surgery2,3. While these initial organ-preserving paradigms have gradually become the predominant treatment choice for LSCC4, no new treatment options have surfaced in the ensuing decades. For recurrent LSCC after chemotherapy, radiation, or surgery, treatments are limited. This is particularly concerning given the continued poor survival in advanced or recurrent LSCC, where 5-year survival is less than 50%5 and has not improved in decades6.
Whole exome and genome sequencing studies have recently provided valuable insight into dysregulated pathways and potential drivers of disease in multiple cancers, including head and neck cancers7-10. These early studies have identified novel genetic mutations and pathway dysregulations across a variety of head and neck cancers. Importantly, LSCCs have constituted a significant portion of the tumors in these studies.
Cancer treatment is entering an exciting new era, combining the information gained from next-generation sequencing studies with targeted therapeutics to allow for models of personalized cancer care. Indeed, cancer sequencing and targeted therapy trials are being launched globally, and with some encouraging initial results11-13. LSCC may prove to be an ideal model for further investigation into personalized targeted therapies given its successful history in response to nonsurgical techniques, previous paradigms for treatment stratification14, and the need to improve survival in this important cohort.
Here, we will discuss the important history of LSCC as a model system for organ preservation, current knowledge of the genomic landscapes, targeted therapies for LSCC, and potential strategies for developing stratified, personalized treatment strategies for LSCC.
Historical treatment of LSCC
For early stage LSCC, single modality therapy (surgery or radiation) achieves cure for a majority of patients. However, patients with locally advanced disease had historically required total laryngectomy followed by adjuvant radiation as the gold standard treatment. Unfortunately, many of these surgeries are accompanied by significant morbidity and many patients are left with significant swallowing difficulties, communication difficulties, and poor cosmetic outcomes15. Thus, in the 1990s, investigations began into equally effective but less morbid therapies.
As a result, the Veterans Affairs (VA) Laryngeal Cancer Study2 was performed. In this prospective randomized controlled study, 332 patients with advanced LSCC were stratified between induction chemotherapy (3 cycles of cisplatin and 5-FU) followed by definitive radiation vs. laryngectomy followed by postoperative radiation. Patients in the chemotherapy group were assessed after 2 cycles of chemotherapy; those that showed clinical response to therapy went on to receive one final cycle of chemotherapy followed by radiation. Those that had no response to therapy or disease progression after 2 cycles went on to immediate laryngectomy and then post-operative radiation. There was no difference in two-year survival between the chemotherapy and surgery groups, and laryngeal function was preserved in 64% of the patients in the chemotherapy group. This study established that organ preservation in LSCC was a feasible goal of treatment, while still providing equivalent overall survival.
These findings were confirmed with data from a randomized study in Europe (EORTC trial 24891)16, where patients with cancers of the hypopharynx underwent either induction chemotherapy consisting of cisplatin and 5-FU followed by irradiation, or surgical resection followed by post-operative radiotherapy. In this study, again overall survival was equivalent, and laryngeal preservation was achieved in greater than 50% of patients after 5 years. A third study (RTOG 91-11)17 compared concurrent chemotherapy and radiation, induction chemotherapy followed by radiation, and standard radiation therapy. This study found that laryngeal preservation was significantly higher in patients receiving concurrent chemoradiation. It is important to note this study did exclude large volume T4 tumors with cartilage invasion or extension into the base of the tongue. Finally, investigators at the University of Michigan studied the utility of a single cycle of induction chemotherapy in LSCC as stratification for further treatment in a phase 2 clinical trial14. Over 75% of patients had response to induction chemotherapy, and overall larynx preservation was achieved in 70% of patients. This study verified that paradigms of treatment stratification could be utilized in LSCC.
These trials together demonstrated the efficacy of combined chemotherapy and radiation therapy in treating locally advanced LSCC while maintaining the functional status of the larynx. Additionally, they showed that treatment with induction chemotherapy did not increase complications for surgical treatment or radiotherapy administered afterwards. Finally, although there was no benefit in overall survival, a significant reduction in the rate of distant metastasis was shown in the chemotherapy group as compared with primary surgery or radiation therapy alone2,16-19.
Although there was significant improvement in organ preservation gained by treatment with induction chemotherapy, unfortunately overall outcomes in LSCC remain poor. In the European study, disease free survival at 5 years remained at 25% and 27% for the chemotherapy arm and immediate surgery arm respectively16. Additionally, patients who responded poorly to chemotherapy were likely to respond poorly to radiation20. As such, research began into molecular markers to predict radiosensitivity and chemo sensitivity in order to better personalize therapy and more accurately predict which patients would be eligible for laryngeal preservation.
Several studies have evaluated various molecular biomarkers in an attempt to better predict a response to therapy. Malecki21 looked at EGFR, p53, and Ki-67, which are biomolecular markers found to be altered in patients with head and neck squamous cell carcknoma (HNSCC). In his retrospective trial, only patients without the presence of EGFR expression were noted to have a significantly improved response to induction chemotherapy. In LSCC specifically, it has been recently found that levels of BAK, a gene involved in apoptosis, is associated with response to induction chemotherapy. The same study identified cyclin D1 as a predictor of LSCC overall and disease-specific survival, and overexpression of EGFR as associated with risk of death22.
These biomarker studies have led to clinical trials to evaluate novel therapies with the potential to improve outcomes in LSCC. For examples, we previously showed that AT-101, which inhibits the anti-apoptotic genes Bcl-2 and Bcl-XL, effectively blocks proliferation in LSCC models23 and have now initiated an ongoing trial specifically targeted LSCC evaluating the use of AT-101, in combination with induction chemotherapy with platinum and docetaxel (NCT01633541). Further combinations of traditional chemotherapy, radiation and targeted therapies may be applicable for LSCC. While traditional biomarker studies have been limited in identifying additional targetable options, recent whole-genome sequencing studies have shed more light into potential key pathways in LSCC.
Genetic landscape of LSCCs
Along with the possibility of identifying additional biomarkers of LSCCs, genomic sequencing offers the potential to identify drivers of tumor genesisand targets for new therapy. Initial exome sequencing studies have already produced valuable insights into the underlying genetic processes, nominating multiple pathways as potential targets for LSCC treatment.
Common mutations and copy number variations
Initial exome-sequencing studies by Agrawal et al.7 and Stransky et al.8 contained some LSCCs in their chosen HNSCC cohort (n=2 and n=15, respectively), but the smaller sample size did not give a broad view of genetic alterations in LSCC. The Cancer Genome Atlas (TCGA) has now whole-genome sequenced 29 HNSCC tumor-normal pairs (low coverage, 30x) and whole exome sequenced 279 HNSCC tumor-normal pairs (high coverage), of which 72 are primary LSCCs9. These LSCC samples are predominantly Caucasian (n=57, 79.2%), male (n=58, 80.6%), and older (mean age=61). Additionally, most patients had a smoking history (n=50, 69.4%) and were diagnosed at Stage III or IV (n=55, 76.4%)9, with very few epidemiologically low risk patients in the cohort24. The initial studies by Agrawal et al.7 and Stransky et al.8 had similar cohort characteristics. Publicly available databases compiling clinical, mutation, and copy number data were used for the analysis of this manuscript25,26.
Previously, evaluating the existence of distinct mutation profiles in LSCCs from other subsites has been limited from lack of power. The significant contribution of the HNSCC cohort from the TCGA has allowed the question to begin to be addressed. Many genes that are frequently mutated are common to all HNSCC subsites such as TP53, CDKN2A, FAT1, and NOTCH1 (Table 1). CASP8, a gene whose product plays a central role in the cell carrying out apoptosis, is frequently mutated in other HNSCCs. However, CASP8 has significantly less mutations in LSCCs compared to the other subsites (P<0.005). Studies have suggested that CASP8 mutations indicate a distinct molecular profile of SCCs27,28, but this subset does not seem to exist in LSCCs. Additionally, mutations in PIK3CA trend towards occurring more frequently in LSCCs than other subsites (P=0.058), and copy number amplification of 3q26 which contains PIK3CA is found at significantly higher frequency in LSCCs compared to other HNSCCs (P<0.001, Table 2). Additionally, amplifications of 3q28 and 9q34 occur at significantly higher frequency in LSCCs (P<0.001, P<0.007).
Table1.
Common genetic mutations
| Gene | TCGA n=71 (%) | Stransky n=15 (%) | Total LSCC n=88 (%) | Total (non-LSCC) n=237 (%) |
| Frequently mutated genes in LSCC samples. The total (non-SCC) column represents mutations from oral cavity, oral pharynx, and hypopharynx samples. Only HPV-negative samples are included. *P=0.058, ** P<0.005 between total LSCC and non-LSCC samples. | ||||
| TP53 | 64 (88.9) | 9 (60.0) | 75 (85.2) | 188 (79.3) |
| CDKN2A | 17 (23.6) | 1 (6.7) | 19 (21.6) | 53 (22.4) |
| PIK3CA | 18 (25.0) | 1 (6.7) | 19 (21.6)* | 31 (13.1) |
| FAT1 | 14 (19.4) | 4 (26.7) | 18 (20.4) | 53 (22.4) |
| NOTCH1 | 13 (18.1) | 2 (13.3) | 15 (17.0) | 44 (18.6) |
| CASP8 | 1 (1.4) | 0 (0) | 1 (1.1)** | 29 (12.2) |
Table2.
Frequent copy number variations
| Cytoband (Gene) | CNV | Larynx n=72, % | Non-larynx n=172, % |
| Common copy number variations (CNV), either amplifications or deletions, in HPV negative samples of the TCGA cohort. * P<0.05. | |||
| 3q26 (PIK3CA, SOX2) | Amp | 37.5* | 12.8 |
| 11q13 (CCND1, FGF3/4/19) | Amp | 36.1 | 29.1 |
| 9p21 (CDKN2A/B) | Del | 31.9 | 32.0 |
| 3q28 (TP63, ETV5) | Amp | 34.7* | 12.8 |
| 8q24 (MYC, PTK2) | Amp | 16.7 | 12.2 |
| 7p12 (EGFR) | Amp | 12.5 | 12.2 |
| 9q34 (NOTCH1, TRAF2) | Amp/Del | 4.2*/1.4 | 0/0 |
While it is clear that many of the aberrations in HNSCC are common across subsites, differences are beginning to emerge as more LSCC samples are being sequenced. By sequencing more LSCCs, we will begin to understand if these differences are due to anatomical subsite or epidemiologic variation between tumors. Likewise, as genome-wide information continues to become available, no doubt distinct subsets of molecular mechanisms will be identified.
Human papilloma virus (HPV) in LSCC
The link between HPV status and HNSCC has been well established at some anatomical subsites29,30; for example, HPV appears to be a common initiating event in oropharyngeal SCCs . The oncogenic potential of high-risk HPVs in LSCCs is not as clear. Studies with larger sample sizes (where n>80) over the past 30 years have shown HPV prevalence in LSCC tumors from as low as 1% to as high as 50%31. However, studies have also found HPV DNA in up to 19% of normal laryngeal mucosa samples32-35, indicating that a significant portion of HPV positive LSCCs that have been reported may be latent HPV infections, and more specific techniques should be used to truly determine HPV positivity. In contrast to many of the PCR-based studies, the HNSCC TCGA project relied on multiple methods of detection including RNA-sequencing and identified only one HPV positive LSCC case (1.4%)9. The low prevalence of HPV in this study indicates that HPV rates may have been historically overestimated in some LSCCs cohorts with similar epidemiology to the TCGA cohort, and unfortunately the low number of samples detected in LSCC will make it difficult to study the extent of the oncogenic role of HPV in LSCC until more studies are performed. This significant variance between studies is most likely due to both the method of detection and the differential rates of HPV infection in each region. For example, HPV DNA PCR assays are capable of detecting very few copies of the viral DNA and some have argued that these assays are too sensitive, able to pick up viral DNA from a transient infection rather than an integrated event35,36.
Consequently, many additional assays have been developed that rely on detecting common downstream events of HPV biology or directly sequencing across genomic insertion sites. For example, when HPV integrates into the genome, early genes E6 and E7 are highly expressed36. E7 then inactivates pRb, causing increased levels of p16INK4A which can be detected by immunohistochemistry37. Thus, using the downstream protein expression of p16 as a surrogate marker for HPV has become a widely acceptable method and clinically relevant method38. Likewise, a second method of detecting HPV integration is to directly sequence the genomic breakpoints between the viral and human genomes. This method is usually cost prohibitive at this point, due to the high cost of whole genome sequencing, but may become more routine in the future. Regardless as the methods for rapid detection and location of HPV insertion sites in the genome improve, so will our understanding of the prevalence and pathogenic role for this virus in LSCC.
Translating genetics into targeted therapies
The potential to improve patient survival by using genetic information to match optimal treatments can be seen in a growing number of successes in other cancers: imatinib for chronic myelogenous leukemia39, trastuzumab for breast cancer with ERBB2 amplification40, and erlotinib and gefitinib for lung cancers that express mutant EGFR41. Here, we will review a few specific molecular lesions that are altered in a large percentage of LSCC cases and have a similar potential for molecularly driven clinical trials.
PI3K pathway
PIK3CA mutations and amplifications frequently occur in LSCCs. PIK3CA encodes p110α, the alpha catalytic subunit to the phosphoinositide 3-kinase (PI3K) which plays a central role in pathways involved in cell growth, survival, and metabolism42. PI3K receives signals from activated receptor tyrosine kinases such as EGFR and VEGFRs, and phosphorylates the lipid PIP2 on the cell membrane to create PIP3. AKT is then activated by PIP3, resulting in a downstream cascade through multiple effectors including GSK-3 and mTOR (Figure 1). This pathway has been noted to be frequently overactive in other cancers including gastric, breast, and lung43, and developing therapies targeting this pathway are underway44.
Figure1.
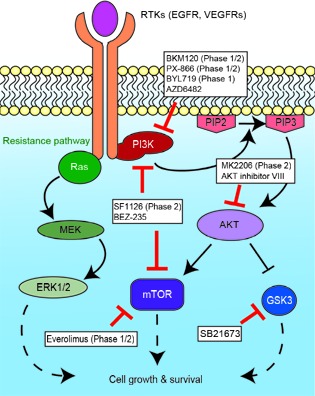
Key components of the PI3K pathway with possible therapeutic targets. Drugs targeting individual components are either in trials as noted, or were effective in vitro with cell lines containing PI3KCA mutations. The RAS/MEK/ERK pathway, which has been noted to play a role in resistance to PI3K-targeted therapies, is shown.
The majority of mutations found in PIK3CA have been defined as 'hotspot' mutations, where the specific amino acid residue is recurrently altered in multiple tumortypes45. These hotspot mutations, such as E542K, E545K, and H1047L/R, have functional consequences of increasing the lipid activity resulting in overactive AKT signaling and downstream effector pathway activation46. The over activation of the PI3K pathway in these cancer cells could make the cells reliant on these signals47. For example, Garnett et al.48 found that PIK3CA mutations were a significant biomarker of sensitivity for several drugs targeting the PI3K pathway after screening over 600 cancer cell lines, including 23 HNSCC lines, against 130 drugs at clinical and preclinical stages. HNSCC cell lines with hotspot PIK3CA mutations demonstrated sensitivity to PI3K/mTOR inhibitors compared to PIK3CA wildtype cells, in both in vitro49 and in vivo models50. These preclinical results are now being tested in early clinical trials for patients with a variety of advanced cancers, including HNSCCs. In a phase 1 trial, patients containing PIK3CA mutations had significantly greater partial response rates to PI3K/AKT/mTOR therapy (6/17, 35%) than those without PIK3CA mutations (6/241, 6%)51. A following early-phase trial indicated that only the H1047R mutation predicted partial response (6/16, 38%) compared to other PIK3CA mutations (5/50, 10%) or PIK3CA wildtype (23/174, 13%)52. However, this study also noted that other hotspot PIK3CA mutations, such as E542K and E545K, had a strong association with KRAS mutations whereas the H1047R mutation did not. As members of the Ras signaling pathway (KRAS, HRAS) have been known to mediate resistance to PI3K inhibition53,54, it is unsurprising that patients with both gene mutations may not respond to PI3K-targeting monotherapies. Notably, KRAS mutations are rare in HNSCCs55-57, and there are no KRAS mutations present in the recent exome sequenced LSCCs7-9. HRAS mutations occur with more prevalence50, and of the 2 HRAS mutations in sequenced LSCCs both occur in tumors with additional PIK3CA hotspot mutations11,12. However, 68.4% (13/19) of the PIK3CA mutations in LSCCs are hotspot mutations without Ras mutations, and PI3K-targeted therapies could be a well-matched choice for this patient population.
In contrast, amplification of 3q26 with the PIK3CA gene has not been found to indicate sensitivity to PI3K-targeted therapy48,49. It is still unclear how the amplification of the PIK3CA gene affects the signaling of the PI3K pathway. While it has been shown that amplification of PIK3CA correlates with increased mRNA and protein expression of p110α58, it does not necessarily lead to increased levels of phosphorylated Akt and mTOR as would be expected for increased pathway activation49. Given the significant amplification of 3q26 in LSCCs specifically, it is crucial to understand the effects this amplification has on tumorigenesis whether PIK3CA or another nearby gene is the cause.
EGFR & HER2
The important role that the epidermal growth factor receptor (EGFR) plays in HNSCCs has been known for several decades59,60 as it has been shown to be overexpressed in >90% of HNSCCs. A tyrosine kinase receptor, EGFR belongs to the ERBB family of cell-surface receptors. Upon ligand binding to the receptor, EGFR homodimerizes or heterodimerizes with other ERBB family receptors such as HER2 and initiates a signaling cascade61. Potential activated pathways include Ras-MEK and PI3K/AKT/mTOR as discussed above, as well as signal transducers and activators of transcription (STATs). EGFR signaling can contribute to tumorigenesis by driving cell proliferation, evasion of apoptosis, angiogenesis, and metastasis62.
Consistent with the molecular role of EGFR, cetuximab, a monoclonal antibody targeting EGFR, is currently the only approved targeted molecular therapeutic for HNSCCs. The combination of cetuximab and radiation therapy has been shown to extend patient survival by 19.3 months compared to radiation alone in patients with recurrent or metastatic disease63. However, contrasting the clear story of EGFR mutations in lung adenocarcinomas predicting sensitivity to EGFR-targeted therapies41, there are still no biomarkers that predict response to cetuximab.
Part of the reason for the lack of biomarkers in HNSCC as compared to lung adenocarcinomas may be due to the differential genetic lesions. On contrast to lung cancers where EGFR mutation is a common event, EGFR mutations are rare in HNSCCs (13/279, 4.7%)9, while amplifications have been reported to vary between 10%-30%64. In HNSCC TCGA data, LSCCs had a similar frequency of amplification as other subsites at around 12% (Table 2). However, amplification of EGFR correlated with worse overall survival in LSCCs specifically65. Additionally, in a phase 2 trial advanced LSCC patients received a single cycle of induction chemotherapy before stratification into surgery and radiation or chemoradiation treatments. Here, EGFR expression predicted increased risk of death22. While there is no evidence for any biological difference in EGFR signaling between HNSCC subsites, the prognostic role of EGFR in LSCCs specifically suggests an especially critical role of this receptor and pathway.
Activating similar pathways is the HER2 receptor which heterodimerizes with EGFR as well as other members of the ERBB family. While HER2 amplifications seem rare (3/72, 4.2%)9, experiments in LSCC cell lines have shown response to anti-HER2 therapy in models with HER2 over-expression66. Targeting HER2 in this distinct subset of patients looks promising as research continues, and significant improvements to patient survival might be made through focusing on targeting this pathway for LSCCs.
Notch signaling
The frequency of NOTCH1 mutations in HNSCCs was surprising when first discovered, additionally so as many of the mutations were predicted to be loss-of-function7. Traditionally, the Notch signaling pathway has been studied in an oncogenic role as activating mutations in NOTCH1 have been shown to significantly contribute to tumorigenesis for several malignances including chronic lympocytic leukemia (CLL)67 and prostate cancer68. However, solid tumors such as lung squamous cell carcinoma, cutaneous squamous cell carcinoma69, and HNSCC display loss-of-function mutations indicative of Notch signaling has playing a role as a tumor suppressor.
The Notch signaling pathway is a direct cell-cell communication network, where a signaling cell displays a ligand that binds and activates the receptor on the receiving cell membrane. There are four receptors, NOTCH1-4, which upon activation are cleaved by gamma secretase, following which the intracellular domain (ICD) of the receptor is translocated to the nucleus resulting in transcriptional activation of target genes.
In keratinocytes, it has been shown that Notch activity controls cell cycle exit as well as commitment to differentiation70, where loss of NOTCH1 promotes tumorigenesis71. Importantly, the loss of Notch signaling leads to an accumulation of β-catenin expression and an increase in Wnt pathway activity72, and Wnt signaling has been shown to have an oncogenic role in multiple cancers73. The possibility of addiction to Wnt signaling resulting from a loss of Notch signaling creates the opportunity for therapeutic intervention. Indeed, the PORCN inhibitor (this gene palmitoylates WNT ligands enabling their secretion into the tumor niche) called WNT974 has shown inhibition of growth of HNSCC models with loss-of-function mutations in NOTCH174. Accordingly, we have now opened a phase 2 clinical trial for metastatic HNSCC patients that will be enriched for NOTCH-deficient cancer to receive WNT974 (NCT02649530). As 17% of LSCCs contained mutations in NOTCH1, WNT974 is a potential targeted therapeutic that will be evaluated for further clinical advancement.
In contrast to inactivating NOTCH mutations, a small subset of studies have also reported that over-activation of Notch signaling can contribute to HNSCC75. As LSCCs have a significant amplification of NOTCH1 compared to other HNSCC subsites (P<0.007, Table 2), Notch signaling may also act as an oncogene for a defined subset. A role as both an oncogene and tumor suppressor suggests Notch signaling can have a bimodal effect in HNSCCs, dependent on timing and order of mutations. These roles will need to be further elucidated to directly target Notch signaling or any of its modulators.
Cyclin D1 (CCND1)
The CCND1 gene encodes cyclin D1, a member of a highly conserved cyclin family. Cyclin D1 regulates cyclin-depending kinases (CDKs) 4 and 6 which control the G1/S phase transition of the cell cycle. The amplification or gain of 11q13, which contains CCND1, is a frequent event in LSCCs specifically (36.1%, Table 2). Importantly, high expression of cyclin D1 correlated with increased risk of death in advanced LSCC patients22. The efficacy of CDK inhibitors to prevent cell cycle progression by overexpressed cyclin D1 is an area of active investigation in LSCC. Currently, multiple clinical trials investigating CDK4/6 inhibitors in HNSCC are underway76, and some of these inhibitors such as palbociclib have already shown efficacy in other cancers77. For example, in one phase 1 trial of recurrent and/or metastatic HNSCC, the CDK4/6 inhibitor LEE011 is being evaluated in combination with cetuximab and the results will soon be published (NCT02429089). The extent of correlation between high expression of cyclin D1 and patient response to CDK inhibitors will be critical to clarifying the potential biomarker role of CCND1 amplification status. Consequently, much research remains for CCND1 amplified HNSCCs.
Immunotherapy
An additional novel treatment option for LSCC that has rapidly advanced in recent years involved immune modulating agents. Immune dysregulation and escape have been increasingly recognized as a hallmark of cancer and potential therapeutic target over the past several years78. It is believed that the adaptive immune system recognizes and eliminates pre-malignant cells. Progressive derangements in the immune system driven by transformed cells gradually leads to immune escape and widespread tumor proliferation79. Observed derangements include inflammatory cytokine expression and activation of inflammatory transcription factors in tumor cells80,81. LSCC and other subsites of HNSCC have been demonstrated to be markedly immunosuppressive via numerous mechanisms, including downregulation of antigen presenting via human leucocyte antigen (HLA) class I molecules9,82-84, development of T-cell tolerance to overexpressed antigens85,86, inhibitory cytokine production87,88, and increased programmed death-ligand 1 (PD-L1)/programmed death-1 (PD-1) expression89-91.
Based on these preclinical findings, numerous potential targets and interventions have been proposed. Monoclonal antibodies targeting cytotoxic T-lymphocyte-associated protein-4 (CTLA-4) (ipilimumab), PD-1 (pembrolizumab), or PD-L1 (nivolumab) have been developed with the goal of manipulating mechanisms of tumor escape and eliciting an adaptive immune system response targeted towards the tumor92. Therapy in patients refractory to standard therapy has been well-tolerated and yielded favorable response rates93-100. However, of more excitement, there are a few patients that appear to achieve lasting complete disease response. These durable responses are being observed in patients who previously would have had a rapidly terminal disease. PD-1/PD-L1 inhibitors have now been approved for metastatic non-small cell lung cancer, renal cell cancer, and melanoma.
Given the clinical efficacy in other malignancies and the immunologic underpinnings, immunotherapy is an avenue of significant interest in LSCC. Various modalities are under development, including vaccine therapy and targeted monoclonal antibodies. Although many of these trials are ongoing, preliminary data has been presented of the Phase 1/2 KEYNOTE-012, where a cohort of 132 HNSCC patients with unresectable recurrent or metastatic tumors were treated with the PD-1 inhibitor pembrolizumab. This cohort was heavily pretreated with 59% of patients having received 2 or more previous lines of therapy for recurrent or metastatic disease. The overall response rate was observed to be 24.8% with an additional 24.8% achieving stable disease. At the time of the interim report, the median duration of response was not reached and 86% of responding patients appeared to have an ongoing response101. Correlative analysis suggested that an inflamed genotype gene expression was able to predict 6 month progression free survival and response to anti-PD-1 therapy102.
A few trials are examining the incorporation of immunotherapy in the management of locoregional HNSCC, including LSCC. These include neoadjuvant vaccine administration (NCT02002182, NCT02609386), concomitant cetuximab and ipilimumab with intensity modulated radiation therapy (IMRT) (NCT01860430), and addition of nivolumab to concomitant cisplatin and IMRT (RTOG3504). Although the majority of trials are targeting recurrent or metastatic HNSCC (all subsites), the preliminary promise of immunotherapy in advanced and recurrent HNSCC cases suggest that LSCC patients could benefit from this novel therapeutic approach.
Overcoming challenges for targeted therapy
While targeted therapies have had several clinical successes in other cancers, these are often in tumors with relatively few "actionable" aberrations. In contrast to tumors with low genetic complexity, the relatively high number genomic alterations in LSCCs coupled with the complex level of intra-tumor genetic heterogeneity will make distilling the critical pathways disrupting tumor growth difficult to identify. Moving forward, additional LSCC genetic information and models for tumors associated with both under-represented epidemiologic-risk groups and genetic landscapes are needed to improve our ability to predict the response of tumors to genetic lesion-matched therapeutics.
As mentioned above, the available genetic information for LSCC tumors is currently largely limited to previously untreated, stage III/IV tumors from Caucasian patients with a history of tobacco and/or alcohol use. Genetic information from untreated early stage (I/II) LSCC tumors would be beneficial to isolating initial aberrational events that drive tumorigenesis. Likewise, as patients are most likely to enroll in personalized medicine trials with advanced or recurrent tumors, genomic landscapes of advanced tumors following relapse from frontline therapy would be the most beneficial for designing novel interventional strategies. Unfortunately, large sequencing studies of tumors from previously treated, advanced LSCCs have not yet been published, with the exception of a few small cohorts demonstrating that advanced and recurrent LSCCs typically have higher numbers of genetic aberrations than untreated counterparts10. These studies are critical because they demonstrate proof-of-principle that the genomic landscape of LSCCs evolve with therapeutic course. Thus, an important goal for the immediate future is to build a comprehensive understanding of the genetic and molecular interaction between the highly recurrent disruptive genomic events found at each stage of tumor progression.
Importantly, several LSCC models already exist that can be used to dissect the genetic and molecular mechanisms of LSCC pathogenesis, but these are also limited to a few epidemiologic subsets and few genetic landscapes. For example, there are LSCC cell line models from primary, metastatic, and recurrent tumors103, including two pairs of primary and metastatic cell lines from the same patient104 and an HPV-positive line105 (Table 3). While the value of these existing cell lines is clear, more models of various stages and pre/post-treatment status consistent with normal interventional progression are still needed to fully explore therapeutic responses at various points in the normal pathogenic course.
Table3.
LSCC cell line models
| Cell Line | Age | Gender | TNM | Stage | Subsite | Type of Lesion | HPV status |
| Patient-derived cell lines are from LSCC patients at University of Michigan Comprehensive Cancer Center. Paired cell lines (-10, -17) are derived from subsequent cancers from the same patient. | |||||||
| Paired | |||||||
| UMSCC-10A | 57 | M | T3N0M0 | III | True cord | Primary | - |
| UMSCC-10B | 58 | M | T3N1M0 | III | Lymph node | Metastasis | - |
| UMSCC-17A | 47 | F | T1N0M0 | I | Supraglottis | Primary | - |
| UMSCC-17B | 47 | F | T1N0M0 | I | Soft tissue | Metastasis | - |
| Primary | |||||||
| UMSCC-11A | 65 | M | T2N2aM0 | IV | Epiglottis | Primary | - |
| UMSCC-23 | 36 | F | T2 N0M0 | II | Supraglottis | Primary | - |
| UMSCC-28 | 61 | F | T1 N0M0 | I | True cord | Primary | - |
| UMSCC-41 | 78 | M | T2N1M0 | III | Arytenoid | Primary | - |
| UMSCC-81B | 53 | M | T2 N0M0 | II | True cord | Primary | - |
| UMSCC-105 | 51 | M | T4 N0M0 | IV | True cord | Primary | Positive |
| Recurrent and metastatic | |||||||
| UMSCC-12 | 71 | M | T2N1M0 | III | Larynx | Recurrence | - |
| UMSCC-13 | 60 | M | T3N0M0 | III | Esophagus | Recurrence | - |
| UMSCC-25 | 50 | M | T3N0M0 | III | Neck | Metastasis | - |
In addition to cell lines, models in which surgically excised tissue from patients is implanted into immunocompromised mice called patient-derived xenograft (PDX) models have recently gained traction as powerful tools for assessing therapeutic responses in pre-clinical studies. In fact, HNSCC studies using PDX models in this manner have already been used to support the translation of targeted strategies into clinical trials106, supporting the utility of these models. Early studies have indicated that HNSCC PDX models represent parental tumors by histology107,108, gene expression profiles109, single-nucleotide polymorphisms110, copy number variants110,111 and proteome profiles112 and there also has not been an indication that engraftment of the tumor is biased by either genetic or clinical factors, including HNSCC subsite113. Unfortunately the expense as well as the variable grafting rate (30%-80% reported for HNSCC) is currently limiting the wide-spread use114, but once the PDX model is established, the tumor can be propagated and expanded into additional mice for parallel, sequential, and long-term therapeutics experiments115. Moving forward, the establishment and characterization of LSCC PDX models from both untreated and pretreated tumors will be essential for the advancement of therapeutics for different epidemiologic and genetic subsets of LSCC.
Algorithms for integrating LSCC organ preservation/treatment
As noted above, LSCC historically has been a unique and successful model for treatment selection and for organ preservation. However, with the current evolving state of genetics and targeted agents in cancer, we will need to revisit treatment algorithms for this disease.
While existing selection paradigms have been focused on objective clinical response to induction chemotherapy14, future goals would be to identify these potential responders to organ-preservation therapy without the need for an induction chemotherapy cycle. Through next generation sequencing studies, we have already identified specific genetic pathways that may be of interest for targeted therapy in LSCC (Figure 2)76. Further incorporating patient genetic information into treatment algorithms for LSCC could serve to further stratify and improve patient outcomes (Figure 3). As the cost and turnaround for targeted next-generation sequencing improves, valuable time could be saved to initiation of definitive treatment, and patients could be treated more specifically and effectively.
Figure2.
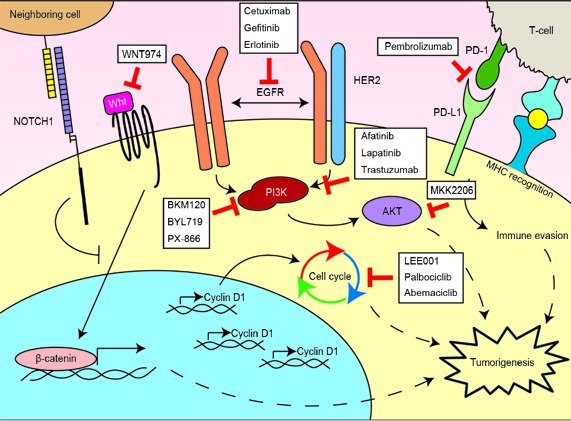
Major oncogenic mechanisms in LSCC and therapeutic opportunities. Dysregulated pathways common to LSCCs with targeted therapies in clinical trials for HNSCCs are shown76. WNT974 targets PORCN thereby blocking Wnt ligand secretion from neighboring cells.
Figure3.
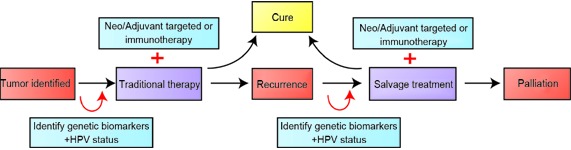
Decision algorithm for treating LSCC patients. Current practice is shown with black arrows, with traditional treatments such as surgery/radiation/chemotherapy. The red arrows incorporate HPV status and patient genetics to add targeted treatments or immunotherapy to improve cure rate.
Specific issues should be considered as treatment algorithms are adapted to include new agents and genomic sequencing. An important decision in use of targeted therapies is whether they should be used irrespective of mutational status (as cetuximab is currently used in HNSCC), or whether targeted therapies should be employed only in those patients with genetic aberrations. Additionally, protocols will need to be designed and implemented investigating these agents in different clinical scenarios (i.e. neoadjuvant vs. adjuvant, monotherapy vs. combination therapy, early vs. late stage tumors, primary vs. recurrent tumors, Figure 3). Likewise, we must consider when and where to add immunotherapy into LSCC treatment algorithms. In a similar fashion to targeted therapy, we must identify predictive biomarkers to allow for treatment stratification102.
Another key group of LSCC patients in need of additional treatment options are those with recurrent disease after both chemoradiation and surgery. These patients have poor outcomes (5-year overall survival 49% and disease-free survival 58%; our unpublished data). Moreover, these recurrences are often untreatable, as patients will have exhausted all other avenues of treatments. Currently, there are no other available options for these patients, and their care is often palliative. Interestingly, patients with recurrent HNSCCs may have different mutational signatures10. Thus, identifying these patients (through predictive genetic biomarkers) and intervening with additional therapies (targeted agents, immunotherapy) earlier in their disease-course may lead to modified treatment algorithms and improve outcomes.
Currently, targeted therapy clinical trials are aimed towards recurrent and advanced stage cancers. In the future, the possibility of expanding these agents to early-stage tumors will be important. Potentially, early-stage LSCC may be more responsive to targeted therapies, given they may have a lower overall mutational burden, and fewer potential targetable dysregulated driver mutations.
As with all next-generation sequencing trials, ethical considerations must be addressed116. Thus, future programs for personalized medicine in LSCC should have well-established guidelines for pretest counseling on disclosure of genetic information, and have genetic counselors actively involved throughout the process.
Conclusions
With the increasing implementation of next-generation sequencing and personalized medicine protocols for cancer, LSCCs may be a particularly useful and successful model disease for novel treatment paradigms. Given the long history and relative success of LSCC and organ-preservation protocols, there will be an inevitable evolution towards adopting targeted and immune modulating agents for this disease. While identification of prognostic genetic biomarkers, therapeutic targets and models to perform molecular studies specific to LSCC remains incomplete, this field is rapidly advancing. Ultimately, these novel strategies will increasingly be investigated and applied to LSCC, which will hopefully improve both organ preservation and overall survival for patients with this disease.
Acknowledgements
J. Chad Brenner received funding from NIH (Grants No. U01DE025184 and P30: CA046592 S1). Andrew C. Birkeland and Rebecca Hoesli received support from University of Michigan Otolaryngology Resident Research (Grant No. T32DC005356). Megan L. Ludwig was supported by NIH (Grant No. T-32-GM007315).
Conflict of interest statement
J. Chad Brenner has previously collaborated with Novartis on the development of WNT974 for NOTCH-deficient HNSCC. The other authors have no conflicts of interest to disclose.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61: 69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Induction chemotherapy plus radiation compared with surgery plus radiation in patients with advanced laryngeal cancer. The department of veterans affairs laryngeal cancer study group. N Engl J Med. 1991;324: 1685–90. doi: 10.1056/NEJM199106133242402. [DOI] [PubMed] [Google Scholar]
- Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, et al. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N Engl J Med. 2003;349: 2091–8. doi: 10.1056/NEJMoa031317. [DOI] [PubMed] [Google Scholar]
- Megwalu UC, Sikora AG. Survival outcomes in advanced laryngeal cancer. JAMA Otolaryngol Head Neck Surg. 2014;140: 855–60. doi: 10.1001/jamaoto.2014.1671. [DOI] [PubMed] [Google Scholar]
- Edge SB, American Joint Committee on C, American Cancer S. Ajcc cancer staging handbook from the ajcc cancer staging manual. New York: Springer, 2010.
- Olsen KD. Reexamining the treatment of advanced laryngeal cancer. Head Neck. 2010;32: 1–7. doi: 10.1002/hed.21294. [DOI] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in notch1. Science. 2011;333: 1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333: 1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517: 576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg ML, Goh G, Chiosea SI, Bauman JE, Freilino ML, Zeng Y, et al. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J Clin Invest. 2016;126: 169–80. doi: 10.1172/JCI82066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingrid Breuskin, Caroline Even, Ecaterina Ileana, Christophe Massard, Naima Lezghed, Ludovic Lacroix, et al. Molecular screening for cancer treatment optimization in head and neck cancer (moscato 01): A prospective molecular triage trial; interim analysis of 78 patients with recurrent or metastatic head and neck cancers. American Head and Neck Society Translational Meeting 2015.
- Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015;1: 466–74. doi: 10.1001/jamaoncol.2015.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, et al. Personalized oncology through integrative high-throughput sequencing: A pilot study. Sci Transl Med. 2011;3: 111ra21. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urba S, Wolf G, Eisbruch A, Worden F, Lee J, Bradford C, et al. Single-cycle induction chemotherapy selects patients with advanced laryngeal cancer for combined chemoradiation: A new treatment paradigm. J Clin Oncol. 2006;24: 593–8. doi: 10.1200/JCO.2005.01.2047. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL. Larynx preservation. Curr Opin Oncol. 2012;24: 218–22. doi: 10.1097/CCO.0b013e3283523c95. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL, Andry G, Chevalier D, Luboinski B, Collette L, Traissac L, et al. Laryngeal preservation with induction chemotherapy for hypopharyngeal squamous cell carcinoma: 10-year results of eortc trial 24891. Ann Oncol. 2012;23: 2708–14. doi: 10.1093/annonc/mds065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forastiere AA, Zhang Q, Weber RS, Maor MH, Goepfert H, Pajak TF, et al. Long-term results of rtog 91-11: A comparison of three nonsurgical treatment strategies to preserve the larynx in patients with locally advanced larynx cancer. J Clin Oncol. 2013;31: 845–52. doi: 10.1200/JCO.2012.43.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denaro N, Russi EG, Lefebvre JL, Merlano MC. A systematic review of current and emerging approaches in the field of larynx preservation. Radiother Oncol. 2014;110: 16–24. doi: 10.1016/j.radonc.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Vainshtein JM, Wu VF, Spector ME, Bradford CR, Wolf GT, Worden FP. Chemoselection: A paradigm for optimization of organ preservation in locally advanced larynx cancer. Expert Rev Anticancer Ther. 2013;13: 1053–64. doi: 10.1586/14737140.2013.829646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings CW, Flint P. Cummings otolaryngology-head & neck surgery. Philadelphia, Penn: Elsevier Mosby. 2010.
- Malecki K, Glinski B, Mucha-Malecka A, Rys J, Kruczak A, Roszkowski K, et al. Prognostic and predictive significance of p53, egfr, ki-67 in larynx preservation treatment. Rep Pract Oncol Radiother. 2010;15: 87–92. doi: 10.1016/j.rpor.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford CR, Kumar B, Bellile E, Lee J, Taylor J, D'Silva N, et al. Biomarkers in advanced larynx cancer. Laryngoscope. 2014;124: 179–87. doi: 10.1002/lary.24245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora MJ, Bauer JA, Verhaegen M, Belbin TJ, Prystowsky MB, Taylor JC, et al. Anti-oxidant treatment enhances anti-tumor cytotoxicity of (-)-gossypol. Cancer Biol Ther. 2008;7: 767–76. doi: 10.4161/cbt.7.5.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillman BN, Yanik M, Birkeland AC, Liu C, Hovelson DH, Cani AK, et al. Fibroblast growth factor family aberrations as a putative driver of head and neck squamous cell carcinoma in an epidemiologically low-risk patient as defined by targeted sequencing. Head Neck. 2016 Feb 5. doi: 10.1002/hed.24292. [Epub ahead of print]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2: 401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci Signal. 2013;6: pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3: 770–81. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat Commun. 2013; 4: 2873.
- Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, et al. Improved survival of patients with human papillomavirus–positive head and neck squamous cell carcinoma in a prospective clinical trial. Journal of the National Cancer Institute. 2008;100: 261–9. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- Vokes EE, Agrawal N, Seiwert TY. HPV-associated head and neck cancer. J Natl Cancer Inst. 2015;107: djv344. doi: 10.1093/jnci/djv344. [DOI] [PubMed] [Google Scholar]
- Torrente MC, Rodrigo JP, Haigentz M, Dikkers FG, Rinaldo A, Takes RP, et al. , Dikkers FG, Rinaldo A, Takes RP, et al. Human papillomavirus infections in laryngeal cancer. Head Neck. 2011;33: 581–6. doi: 10.1002/hed.21421. [DOI] [PubMed] [Google Scholar]
- Garcia-Milian R, Hernandez H, Panade L, Rodriguez C, Gonzalez N, Valenzuela C, et al. Detection and typing of human papillomavirus DNA in benign and malignant tumours of laryngeal epithelium. Acta Otolaryngol. 1998;118: 754–8. doi: 10.1080/00016489850183313. [DOI] [PubMed] [Google Scholar]
- Fukushima K, Ogura H, Watanabe S, Yabe Y, Masuda Y. Human papillomavirus type 16 DNA detected by the polymerase chain reaction in non-cancer tissues of the head and neck. Eur Arch Otorhinolaryngol. 1994;251: 109–12. doi: 10.1007/BF00179903. [DOI] [PubMed] [Google Scholar]
- Rihkanen H, Peltomaa J, Syrjanen S. Prevalence of human papillomavirus (hpv) DNA in vocal cords without laryngeal papillomas. Acta Otolaryngol. 1994;114: 348–51. doi: 10.3109/00016489409126068. [DOI] [PubMed] [Google Scholar]
- Smith EM, Summersgill KF, Allen J, Hoffman HT, McCulloch T, Turek LP, et al. Human papillomavirus and risk of laryngeal cancer. Ann Otol Rhinol Laryngol. 2000;109: 1069–76. doi: 10.1177/000348940010901114. [DOI] [PubMed] [Google Scholar]
- Wiest T, Schwarz E, Enders C, Flechtenmacher C, Bosch FX. Involvement of intact hpv16 e6/e7 gene expression in head and neck cancers with unaltered p53 status and perturbed prb cell cycle control. Oncogene. 2002;21: 1510–7. doi: 10.1038/sj.onc.1205214. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat Rev Cancer. 2002;2: 342–50. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- Robinson M, Sloan P, Shaw R. Refining the diagnosis of oropharyngeal squamous cell carcinoma using human papillomavirus testing. Oral Oncol. 2010;46: 492–6. doi: 10.1016/j.oraloncology.2010.02.013. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the bcr-abl tyrosine kinase in chronic myeloid leukemia. New England Journal of Medicine. 2001;344: 1031–7. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of her2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20: 719–26. doi: 10.1200/JCO.20.3.719. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J of Med. 2004;350: 2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7: 606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the pik3ca gene in human cancers. Science. 2004;304: 554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Engelman JA. Targeting pi3k signalling in cancer: Opportunities, challenges and limitations. Nat Rev Cancer. 2009;9: 550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34: 155–63. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102: 802–7. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti D, Trusolino L. Oncogene addiction as a foundational rationale for targeted anti-cancer therapy: Promises and perils. EMBO Mol Med. 2011;3: 623–36. doi: 10.1002/emmm.201100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483: 570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar T, Byers LA, Ng PK, Mills GB, Peng S, Diao L, et al. A comprehensive evaluation of biomarkers predictive of response to pi3k inhibitors and of resistance mechanisms in head and neck squamous cell carcinoma. Mol Cancer Ther. 2014;13: 2738–50. doi: 10.1158/1535-7163.MCT-13-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the pi3k pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3: 761–9. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, Hong DS, et al. Pik3ca mutations in patients with advanced cancers treated with pi3k/akt/mtor axis inhibitors. Mol Cancer Ther. 2011;10: 558–65. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. Pik3ca mutation h1047r is associated with response to pi3k/akt/mtor signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73: 276–84. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of pi3k and mek inhibitors to treat mutant kras g12d and pik3ca h1047r murine lung cancers. Nat Med. 2008;14: 1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle NT, Lemos R, Wipf P, Yacoub A, Mitchell C, Siwak D, et al. , Wipf P, Yacoub A, Mitchell C, Siwak D, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor px-866 whereas oncogenic ras is a dominant predictor for resistance. Cancer Res. 2009;69: 143–50. doi: 10.1158/0008-5472.CAN-07-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough WG, Shores C, Witsell DL, Weissler MC, Fidler ME, Gilmer TM. Ras mutations and expression in head and neck squamous cell carcinomas. Laryngoscope. 1994;104: 1337–47. doi: 10.1288/00005537-199411000-00005. [DOI] [PubMed] [Google Scholar]
- Matsuda H, Konishi N, Hiasa Y, Hayashi I, Tsuzuki T, Tao M, et al. Alterations of p16/cdkn2, p53 and ras genes in oral squamous cell carcinomas and premalignant lesions. J Oral Pathol Med. 1996;25: 232–8. doi: 10.1111/j.1600-0714.1996.tb01377.x. [DOI] [PubMed] [Google Scholar]
- Bissada E, Abboud O, Abou Chacra Z, Guertin L, Weng X, Nguyen-Tan PF, et al. Prevalence of k-ras codons 12 and 13 mutations in locally advanced head and neck squamous cell carcinoma and impact on clinical outcomes. Int J Otolaryngol. 2013;2013: 848021. doi: 10.1155/2013/848021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woenckhaus J, Steger K, Werner E, Fenic I, Gamerdinger U, Dreyer T, et al. Genomic gain of pik3ca and increased expression of p110alpha are associated with progression of dysplasia into invasive squamous cell carcinoma. J Pathol. 2002;198: 335–42. doi: 10.1002/path.1207. [DOI] [PubMed] [Google Scholar]
- Ozanne B, Richards CS, Hendler F, Burns D, Gusterson B. Over-expression of the egf receptor is a hallmark of squamous cell carcinomas. J Pathol. 1986;149: 9–14. doi: 10.1002/path.1711490104. [DOI] [PubMed] [Google Scholar]
- Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger rna are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53: 3579–84. [PubMed] [Google Scholar]
- Hynes NE, Lane HA. Erbb receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer. 2005;5: 341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- Holbro T, Civenni G, Hynes NE. The erbb receptors and their role in cancer progression. Exp Cell Res. 2003;284: 99–110. doi: 10.1016/S0014-4827(02)00099-X. [DOI] [PubMed] [Google Scholar]
- Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354: 567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11: 9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- Birkeland AC, Ludwig ML, Meraj TS, Brenner JC, Prince ME. The tip of the iceberg: Clinical implications of genomic sequencing projects in head and neck cancer. Cancers (Basel) 2015;7: 2094–109. doi: 10.3390/cancers7040879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland AC, Yanik M, Tillman BN, Scott MV, Foltin SK, Mann JE, et al. Identification of targetable her2 aberrations in head and neck squamous cell carcinoma. JAMA Oncol. 2015; In press.
- Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, et al. Analysis of the chronic lymphocytic leukemia coding genome: Role of notch1 mutational activation. J Exp Med. 2011;208: 1389–401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, et al. Jagged1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64: 6854–7. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. Loss-of-function mutations in notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A. 2011;108: 17761–6. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff BJ, Qin JZ, Chaturvedi V, Denning MF, Bonish B, Miele L. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through nf-kappab and ppargamma. Cell Death Differ. 2002;9: 842–55. doi: 10.1038/sj.cdd.4401036. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33: 416–21. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, et al. Notch post-translationally regulates [beta]-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13: 1244–51. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012; 4:
- Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, et al. Targeting wnt-driven cancer through the inhibition of porcupine by lgk974. Proc Natl Acad Sci U S A. 2013;110: 20224–9. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Gaykalova DA, Ochs MF, Mambo E, Arnaoutakis D, Liu Y, et al. Activation of the notch pathway in head and neck cancer. Cancer Res. 2014;74: 1091–104. doi: 10.1158/0008-5472.CAN-13-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland AC, Brenner JC. Personalizing medicine in head and neck squamous cell carcinoma: The rationale for combination therapies. Med Res Arch. 2015; 3. doi: 10. 18103/mra. v0i3. 77.
- Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, her2-negative, advanced breast cancer (paloma-1/trio-18): A randomised phase 2 study. Lancet Oncol. 2015;16: 25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144: 646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nat immunol. 2002;3: 991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL. Deficiency of activated stat1 in head and neck cancer cells mediates tap1-dependent escape from cytotoxic t lymphocytes. Cancer Immunol Immunother. 2011;60: 525–35. doi: 10.1007/s00262-010-0961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz MS, Srivastava RM, Andrade Filho PA, Egloff AM, Wang L, Seethala RR, et al. Shp2 is overexpressed and inhibits pstat1-mediated apm component expression, T-cell attracting chemokine secretion, and ctl recognition in head and neck cancer cells. Clin Cancer Res. 2013;19: 798–808. doi: 10.1158/1078-0432.CCR-12-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Kono K, Maruyama T, Watanabe M, Kawaguchi Y, Kamimura K, et al. Downregulation of hla class i molecules in the tumour is associated with a poor prognosis in patients with oesophageal squamous cell carcinoma. Br J Cancer. 2008;99: 1462–7. doi: 10.1038/sj.bjc.6604715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Shigyo H, Ishii H, Katayama A, Miyokawa N, Harabuchi Y, et al. Hla class i antigen down-regulation in primary laryngeal squamous cell carcinoma lesions as a poor prognostic marker. Cancer Res. 2006;66: 9281–9. doi: 10.1158/0008-5472.CAN-06-0488. [DOI] [PubMed] [Google Scholar]
- Ferris RL, Whiteside TL, Ferrone S. Immune escape associated with functional defects in antigen-processing machinery in head and neck cancer. Clin Cancer Res. 2006;12: 3890–5. doi: 10.1158/1078-0432.CCR-05-2750. [DOI] [PubMed] [Google Scholar]
- Pak AS, Wright MA, Matthews JP, Collins SL, Petruzzelli GJ, Young MR. Mechanisms of immune suppression in patients with head and neck cancer: Presence of CD34(+) cells which suppress immune functions within cancers that secrete granulocyte-macrophage colony-stimulating factor. Clin Cancer Res. 1995;1: 95–103. [PubMed] [Google Scholar]
- Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: Linking inflammation and cancer. J Immunol. 2009;182: 4499–506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebreel A, Mistry D, Loke D, Dunn G, Hough V, Oliver K, et al. Investigation of interleukin 10, 12 and 18 levels in patients with head and neck cancer. J Laryngol Otol. 2007;121: 246–52. doi: 10.1017/S0022215106002428. [DOI] [PubMed] [Google Scholar]
- Cheng F, Wang HW, Cuenca A, Huang M, Ghansah T, Brayer J, et al. A critical role for stat3 signaling in immune tolerance. Immunity. 2003;19: 425–36. doi: 10.1016/S1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- Lyford-Pike S, Peng S, Young GD, Taube JM, Westra WH, Akpeng B, et al. Evidence for a role of the pd-1:Pd-l1 pathway in immune resistance of hpv-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73: 1733–41. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badoual C, Hans S, Merillon N, Van Ryswick C, Ravel P, Benhamouda N, et al. Pd-1-expressing tumor-infiltrating t cells are a favorable prognostic biomarker in hpv-associated head and neck cancer. Cancer Res. 2013;73: 128–38. doi: 10.1158/0008-5472.CAN-12-2606. [DOI] [PubMed] [Google Scholar]
- Jie HB, Schuler PJ, Lee SC, Srivastava RM, Argiris A, Ferrone S, et al. Ctla-4(+) regulatory t cells increased in cetuximab-treated head and neck cancer patients suppress nk cell cytotoxicity and correlate with poor prognosis. Cancer Res. 2015;75: 2200–10. doi: 10.1158/0008-5472.CAN-14-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay AG. Immune checkpoint blockade immunotherapy to activate anti-tumour t-cell immunity. Br J Haematol. 2013;162: 313–25. doi: 10.1111/bjh.12380. [DOI] [PubMed] [Google Scholar]
- Gettinger SN, Horn L, Gandhi L, Spigel DR, Antonia SJ, Rizvi NA, et al. Overall survival and long-term safety of nivolumab (anti-programmed death 1 antibody, bms-936558, ono-4538) in patients with previously treated advanced non-small-cell lung cancer. J Clin Oncol. 2015;33: 2004–12. doi: 10.1200/JCO.2014.58.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373: 23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JS, D'Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-ctla-4 treatment (checkmate 037): A randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16: 375–84. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- Ribas A, Puzanov I, Dummer R, Schadendorf D, Hamid O, Robert C, et al. Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (keynote-002): A randomised, controlled, phase 2 trial. Lancet Oncol. 2015;16: 908–18. doi: 10.1016/S1470-2045(15)00083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373: 123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372: 2018–28. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373: 1803–13. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372: 2521–32. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- Seiwert SY, Haddad RI, Gupta S, Mehra R, Tahara M, Berger R, et al. Antitumor activity and safety of pembrolizumab in patients (PTS) with advanced squamous cell carcinoma of the head and neck (SCCHN): Preliminary results from keynote-012 expansion cohort. J Clin Oncol. 2015; 33 (suppl): abstr LBA6008.
- SSeiwert TY, Burtness B, Weiss J, Eder JP, Yearley J, Murphy E, et al. Inflamed-phenotype gene expression signatures to predict benefit from the anti-pd-1 antibody pembrolizumab in pd-l1+ head and neck cancer patients. J Clin Oncol. 2015; 33 (suppl): abstr 6017.
- Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, et al. Genotyping of 73 um-scc head and neck squamous cell carcinoma cell lines. Head Neck. 2010;32: 417–26. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey TE, Van Dyke DL, Worsham MJ, Bradford CR, Babu VR, Schwartz DR, et al. Characterization of human laryngeal primary and metastatic squamous cell carcinoma cell lines um-scc-17a and um-scc-17b. Cancer Res. 1989;49: 6098–107. [PubMed] [Google Scholar]
- Chinn SB, Walline HM, McHugh JB, Prince ME, Carey TE. Prevalence of a hpv laryngeal squamous cell carcinoma and a novel cell line. Otolaryngol Head Neck Surg. 2012;147:173–4. doi: 10.1177/0194599811431663. [DOI] [PubMed] [Google Scholar]
- Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9: 338–50. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17: 1514–20. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukopoulos P, Kanetaka K, Takamura M, Shibata T, Sakamoto M, Hirohashi S. Orthotopic transplantation models of pancreatic adenocarcinoma derived from cell lines and primary tumors and displaying varying metastatic activity. Pancreas. 2004;29: 193–203. doi: 10.1097/00006676-200410000-00004. [DOI] [PubMed] [Google Scholar]
- Zhao X, Liu Z, Yu L, Zhang Y, Baxter P, Voicu H, et al. Global gene expression profiling confirms the molecular fidelity of primary tumor-based orthotopic xenograft mouse models of medulloblastoma. Neuro Oncol. 2012;14: 574–83. doi: 10.1093/neuonc/nos061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy J, Ulyanov A, Brennan R, Wu G, Pounds S, Zhang J, et al. Analysis of mdm2 and mdm4 single nucleotide polymorphisms, mrna splicing and protein expression in retinoblastoma. PLoS One. 2012;7: e42739. doi: 10.1371/journal.pone.0042739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc. 2007;2: 247–50. doi: 10.1038/nprot.2007.25. [DOI] [PubMed] [Google Scholar]
- Li H, Wheeler S, Park YS, Ju Z, Thomas SM, Fichera M, et al. Proteomic characterization of head and neck cancer patient-derived xenografts. Mol Cancer Res. 2016;14: 278–86. doi: 10.1158/1541-7786.MCR-15-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Milo GE, Shuler CF, Schuller DE. Xenograft growth and histodifferentiation of squamous cell carcinomas of the pharynx and larynx. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1996;81: 197–202. doi: 10.1016/S1079-2104(96)80415-X. [DOI] [PubMed] [Google Scholar]
- Siolas D, Hannon GJ. Patient-derived tumor xenografts: Transforming clinical samples into mouse models. Cancer Res. 2013;73: 5315–9. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhia KA, Hadley AM, Haluska P, Scott CL. Prioritizing therapeutic targets using patient-derived xenograft models. Biochim Biophys Acta. 2015;1855: 223–34. doi: 10.1016/j.bbcan.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland AC, Uhlmann WR, Brenner JC, Shuman AG. Getting personal: Head and neck cancer management in the era of genomic medicine. Head Neck. 2015 May 20. doi: 10. 1002/hed. 24132. [Epub ahead of print].


