Abstract
Cardiac tissue is an excitable system that can support complex spatiotemporal dynamics, including instabilities (arrhythmias) with lethal consequences. While over the last two decades optical mapping of excitation (voltage and calcium dynamics) has facilitated the detailed characterization of such arrhythmia events, until recently, no precise tools existed to actively interrogate cardiac dynamics in space and time. In this work, we discuss the combined use of new methods for space‐ and time‐resolved optogenetic actuation and simultaneous fast, high resolution optical imaging of cardiac excitation waves. First, the mechanisms, limitations and unique features of optically induced responses in cardiomyocytes are outlined. These include the ability to bidirectionally control the membrane potential using depolarizing and hyperpolarizing opsins; the ability to induce prolonged sustained voltage changes; and the ability to control refractoriness and the shape of the cardiac action potential. At the syncytial tissue level, we discuss optogenetically enabled experimentation on cell–cell coupling, alteration of conduction properties and termination of propagating waves by light. Specific attention is given to space‐ and time‐resolved application of optical stimulation using dynamic light patterns to perturb ongoing activation and to probe electrophysiological properties at desired tissue locations. The combined use of optical methods to perturb and to observe the system can offer new tools for precise feedback control of cardiac electrical activity, not available previously with pharmacological and electrical stimulation. These new experimental tools for all‐optical electrophysiology allow for a level of precise manipulation and quantification of cardiac dynamics comparable in robustness to the computational setting, and can provide new insights into pacemaking, arrhythmogenesis and suppression or cardioversion.
Abbreviations
- ATP
anti‐tachycardia pacing
- BZ
Belousov‐Zhabotinski reaction
- ChR2
Channelrhodopsin‐2
- GECI
genetically‐encoded calcium indicators
- GEVI
genetically‐encoded voltage indicators
Towards all‐optical cardiac electrophysiology
‘If you want to understand a complex (biological) system, you have to interfere with it very delicately and precisely … likely using new molecular biology tools.’
Francis Crick
Francis Crick's insight is that of a (brilliant) biologist capturing something that physicists, mathematicians and engineers have known for a long time – to understand a complex system and be able to control its behaviour, passive observations are not sufficient: active interrogation is needed. This is especially true for the brain and the heart, both complex systems with spatiotemporal dynamics that cannot be easily inferred from the activity of their individual components (cells). While over the last two decades optical mapping of excitation (voltage and calcium dynamics) (Efimov et al. 2004) has facilitated the detailed characterization of cardiac arrhythmias, until recently no precise tools existed to actively interrogate and manipulate cardiac dynamics in space and time. Optogenetic actuation (stimulation or inhibition by light) uses genetically expressed light‐sensitive ion channels (opsins) (Nagel et al. 2003) to provide the means to perturb electrical activity ‘very delicately and precisely’. When combined with high‐resolution optical imaging, it allows for contactless all‐optical electrophysiology (Entcheva, 2013; Hochbaum et al. 2014), and the possibility for feedback control (Miesenböck, 2009). These methodologies give experimentalists a level of control over macroscopic emergent phenomena that is normally associated with computer models, effectively enabling ‘live simulations’ that can help unravel mechanisms of arrhythmias. Translationally, they can be used to achieve cell‐type selectivity, very high levels of parallelism (throughput), and long‐term in vivo observation and manipulation. All of these features can significantly improve drug discovery and cardiotoxicity testing, phenotyping and optimization of stem‐cell (patient‐derived) cardiomyocytes, as well as permit potential cell type‐specific in vivo uses for control of cardiac electrical function.
Experimental neuroscience has been transformed (Adamantidis et al. 2015) since the first demonstratration of optical control of neurons (Zemelman et al. 2002) and the introduction of the versatile excitatory opsin channelrhodopsin‐2 (ChR2) (Nagel et al. 2003; Boyden et al. 2005). The arsenal of available technical tools has grown exponentially, and the technology has been adopted for use in intact animals, including recent sophisticated all‐optical approaches for manipulation and imaging of single cells within the brain (Packer et al. 2014). In contrast, cardiac applications of optogenetics have remained relatively limited after a few early reports (Arrenberg et al. 2010; Bruegmann et al. 2010; Abilez et al. 2011; Jia et al. 2011). Beyond transgenic animals, viral transduction has been used in neonatal and adult cardiomyocytes in vitro (Williams et al. 2013; Ambrosi & Entcheva, 2014; Bingen et al. 2014; Park et al. 2014), as well as in rodent hearts (Nussinovitch & Gepstein, 2015; Vogt et al. 2015) to facilitate optical rhythm control (pacing or suppression of activity) – the most obvious cardiac application of optogenetics.
Optical pacing in the intact heart in freely moving animals still has not been realized to date; in addition to the typical challenges associated with gene therapy, unsolved problems also include light access to desired locations. Two possible solutions (Ambrosi et al. 2014) are (1) microendoscopy with fibre optic conduits (Klimas & Entcheva, 2014), and (2) use of implantable miniaturized devices, which include light sources and photodiodes (Xu et al. 2014). Both of these solution have the potential to enable all‐optical electrophysiology in vivo. Suppression of electrical activity, including termination of cardiac arrhythmias (cardioversion/defibrillation) by light, can theoretically be realized by either globally applying a depolarizing optical perturbation with sufficient magnitude (Bingen et al. 2014) or hyperpolarizing tissues using inhibitory opsins. In the latter case, the possibility for ‘anode‐break’ excitation (resetting sodium channels by hyperpolarization) requires the capture of the whole tissue without escape areas, more so than when depolarizing pulses are used. Furthermore, in practice, the available inhibitory opsins based on ion pumps (halorhodopsin, ArchT, etc.), despite their ability to reduce action potential duration, cannot effectively control activity in multicellular cardiac preparations at physiologically tolerated light levels, in our own experience and as reported by others (Park et al. 2014). This is due to the mechanism of the pump, which moves a single ion for each photon, resulting in a relatively small hyperpolarizing current. A new and highly anticipated family of hyperpolarizing ion channel‐based opsins, ACRs, was reported very recently (Govorunova et al. 2015), with substantially higher photocurrent. The new ACRs appear to be a viable partner to ChR2 for true bidirectional control of electrical potential.
Optical mapping, usually performed using synthetic dyes for membrane voltage or cytoplasmic calcium, offers a high‐resolution view of cardiac activity. In cardiomyocyte monolayer cultures, an alternative, non‐invasive dye‐free imaging modality also has been applied (Hwang et al. 2004; Burton et al. 2015), albeit in only a few studies. Among other advantages, dye‐free imaging works at all wavelengths, which simplifies its integration with spectrally restricted opsins used for actuation. For in vivo applications and long‐term monitoring and manipulation, the all‐optical electrophysiological approach is best realized by combining optogenetic actuators and optogenetic sensors (Hochbaum et al. 2014). Genetically encoded calcium indicators (GECIs), mostly based on calmodulin sensing of calcium ions, led the way for in vivo use; the latest generation of GCaMP6 provides excellent sensitivity (Chen et al. 2013). Spectrally, the combination of GCaMP sensors with ChR2‐based actuators is problematic, but red‐shifted variants in either the GECI or the actuator enable such combined use. Genetically encoded voltage indicators (GEVIs) have progressed in terms of sensitivity and speed of kinetics to faithfully capture action potential morphology (Dugué et al. 2012). Now, their use has been demonstrated in whole‐heart imaging in transgenic mice (Chang Liao et al. 2015), and several groups work on new and improved GEVIs that are combinable with optogenetic actuators (Hochbaum et al. 2014; Gong et al. 2015).
Clinical (therapeutic) use of all‐optical technologies, and optogenetic actuation in particular, is uncertain but cannot be ruled out (Adamantidis et al. 2015); several start‐up companies are pursuing such options for neuropsychiatric and metabolic disorders. Current electrode‐based technologies for cardiac rhythm management are excellent and any optogenetic approach would need to demonstrate significant energy and safety benefits, possibly by targeting specific cell types (Boyle et al. 2013), to be competitive. The transformative potential of optogenetics‐inspired all‐optical electrophysiological technology lies in the new capabilities it brings as a basic science tool. A recent example includes the experimental optical probing of the critical mass of cardiac tissue needed for ectopic activity in the intact mammalian heart (Zaglia et al. 2015). At the cellular level, light can be used to optogenetically shape the action potential (Park et al. 2014), including corrections at the tissue level in pathological conditions (Karathanos et al. 2014). Furthermore, communication between distinct cell types in the heart, e.g. fibroblasts and cardiomyocytes or neurons and cardiomyocytes, can be probed by optogenetic means (Ambrosi et al. 2014; Wengrowski et al. 2015).
Perhaps the most exciting aspect of all‐optical electrophysiology is the ability to deliver dynamic space–time patterns of light for stimulation or suppression, and the possibility for real‐time feedback control of macroscopic emergent properties, i.e. cardiac waves of excitation (Fig. 1). Unlike current strategies for rhythm control of the heart, which are limited by the spatial restrictions of electrical perturbation, the all‐optical approach offers the possibility for a conceptually new ‘wave control’ or ‘wave steering’, demonstrated for the first time recently (Burton et al. 2015) (Fig. 2). Wave control goes beyond the simple initiation and termination of excitation waves (pacing and defibrillation/cardioversion) and the associated frequency modulation. Instead, using all‐optical means, one can focus on manipulation of the macroscopic (emergent) properties, i.e. to quite literally dynamically reshape excitation waves with light. The flexibility of such wave control yields, for the first time, an experimental platform to rapidly test anti‐arrhythmic strategies, likely to be faster than can be performed in computer models.
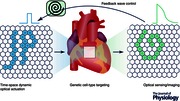
Figure 1.
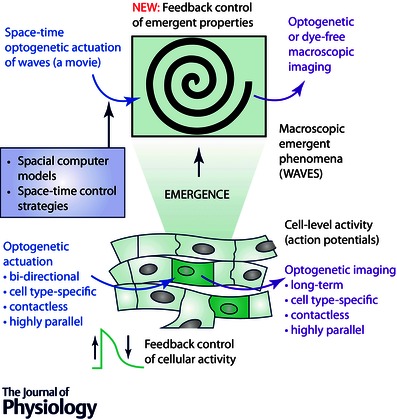
Control of emergent cardiac tissue‐level function by all‐optical technology
Figure 2. Optical control of spiral wave chirality in cardiac monolayer .
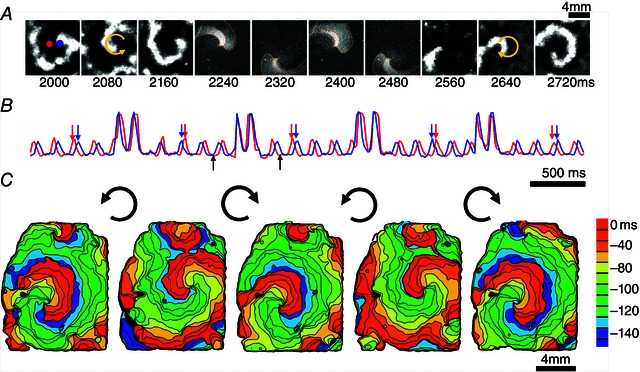
A, snapshots from an ongoing counter‐clockwise spiral wave (frames 2000–2160), an optically applied computer‐generated clockwise spiral wave (frames 2240–2480) and the persisting spiral wave post‐chirality reversal (frames 2560–2720). B, temporal traces of activity from the indicated red and blue pixels showing four light‐controlled chirality reversals (computer‐generated spirals were faster and imposed at random phase for less than two rotations, as seen in the four higher‐intensity transients; black arrows indicate the time period presented in panel A; red and blue arrows indicate the switch of order of excitation at the chosen locations due to chirality reversal. C, activation maps for the initial spiral wave and the four resultant spirals after each of the chirality reversals by light. Note the slight difference in spiral wave tip location and in rotation frequency for clockwise vs. counter‐clockwise spirals. Reproduced with permission from Burton et al. (2015).
Interfacing models and experiments in cardiac electrophysiology
Electrophysiology, due to the complexity of the observed phenomena, has been a field that is not only receptive to, but, in fact, necessitates input from computational modelling. Real‐time interfacing between models and experiments is common. In the dynamic clamp approach (Prinz et al. 2004), through real‐time feedback, a cell can be subjected to computer model‐produced ionic currents, and its action potential can be transformed into the electrophysiological response of a cell with a different phenotype and/or disease state. However, using intracellular electrodes to control membrane potential in a macroscopic, spatially extended tissue is not feasible, as it will require millions of electrodes. In contrast, the all‐optical electrophysiological approach can easily be applied to populations of cells in a contactless manner and the availability of bidirectionally actuating opsins can potentially control action potential dynamics at millions of locations simultaneously. If technical issues with light access are resolved, this optogenetic technique can be realized in vivo as well. Furthermore, the control by light does not have to be limited to cell‐level properties per se; rather the target of control can be emergent tissue‐level phenomena, i.e. wave control, as discussed above. For example, a spiral wave of excitation can be manipulated by a dynamic light pattern without knowledge of the membrane potential of each cell within the tissue, by manipulating key properties of the macroscopic wave (Burton et al. 2015). Here, fast phenomenological models that simulate cardiac wave dynamics can be used to generate light patterns and to enable real‐time feedback experiments (Fig. 2). As the light application (the irradiance) and the actual change in the membrane potential are linked by non‐linear opsin currents, it is imperative to consider the biophysical response of the opsins. Computational work in this area has advanced to provide insights for future experiments (Abilez et al. 2011; Boyle et al. 2013; Williams et al. 2013; Karathanos et al. 2014), including at the whole heart level.
Simple experimental models of cardiac excitation
Simplified experimental models have been used throughout the history of cardiac research in an effort to make the structural and functional complexity of the intact heart manageable. Biological models, such as tissue slices and cardiac monolayer cultures, have helped uncover fundamental principles relevant to the management, control and prevention of cardiac arrhythmias. Even non‐living systems have been helpful because they have offered the opportunity to test experimental strategies in a well‐controlled setting. For example, the oscillating Belousov–Zhabotinski (BZ) reaction can act as a test‐bed for linking basic excitable media theory to the more complex case of the living heart. The 2D BZ reaction can support target and spiral waves (Winfree, 1972), which share many characteristics with excitation/contraction waves in cardiac tissue. Indeed, insights from studies on the interaction of spirals with higher frequency sources in the BZ reaction (Krinsky & Agladze, 1983) were applied to experiments where spirals were rapidly paced in cardiac tissue (Davidenko et al. 1995), a strategy at the heart of anti‐tachycardia pacing (ATP) therapy.
Chemists have a tool in their arsenal that has not been available to cardiac researchers until now: the BZ reaction can be controlled with light. Light‐sensitive variants of the BZ reaction have been exploited to investigate how pattern formation in excitable media reacts to spatially complex external perturbations. These experiments involve projecting static or dynamic patterned light on the BZ reaction, and, in feedback experiments, light is varied as a function of the captured dynamics. These tools have been leveraged to show that the trajectory and periodicity of spiral waves in two‐dimensional media can be manipulated by periodic forcing (Steinbock et al. 1993) and that wave direction and speed can be precisely controlled using feedback (Sakurai et al. 2002). Indeed, the level of control afforded by light sensitivity has allowed the BZ reaction to be used as a reaction‐diffusion computer (de Lacy Costello et al. 2009). A light‐sensitized cardiac syncytium can similarly be turned into a living simulator/computer that can run multiple scenarios of wave perturbation by light in search of effective anti‐arrhythmia therapies. In many aspects, this can present a faster and more realistic alternative to computer models exploring control of cardiac wave dynamics.
Another promising research area that can leverage optogenetic stimulation involves understanding the diverse effects of noise on cardiac wave propagation. Macroscopic propagating wavefronts are thought to be relatively immune to heterogeneities at the cell level (Spach & Heidlage, 1995) and stochastic activity of individual channels is attenuated at the tissue level (Romero et al. 2009). On the other hand, small stochastic variations can be amplified in cardiac systems due to nonlinear processes: a few channels can push the tissue response over the action potential activation threshold (Lerma et al. 2006), stochastic ryanodine channel activity generates calcium sparks that in turn may trigger ectopic beats, and bi‐stable solutions for action potential shape can result in heterogeneities that may lead to macroscopic re‐entrant waves (Maoz et al. 2009). The light‐sensitive BZ reaction has proved to be a valuable experimental system for investigating such stochastic processes, as noise can be added in a controlled fashion by imposing randomized patterns of light on the substrate. Experiments have demonstrated the presence of noise‐aided propagation (wave speed increases in two‐dimensions in the presence of stochastic noise), and noise‐dependent drift of pre‐existing spiral waves (Sagués et al. 2007). All‐optical approaches have now enabled similar experiments in cardiac tissues (Burton et al. 2015). They may provide insights into wave dynamics in the damaged myocardium with increased heterogeneities, as well as a deeper understanding of cardiac arrhythmias and strategies for their control.
High‐throughput and remote experimentation
Classic electrophysiology is inherently very low throughput (manual handling of one cell at a time) because of the need for contact. Despite new technological developments to increase throughput (Dunlop et al. 2008), automated probing of multiple cells and/or spatial locations remains limited. All‐optical electrophysiology offers instant parallelism, as it allows for simultaneous actuation and sensing at millions of locations. Further benefits of the method include the ability to handle not only single cells but also three‐dimensional structures (engineered tissue or tissue slices), as well as to probe cardiac cells within the intact heart, as currently pursued in brain. Immediate applications for parallelism can be found in preclinical cardiotoxicity testing, which is demanded for all new drugs, as well as in high‐throughput characterization of stem cell products for regenerative medicine, e.g. human induced pluripotent stem cell‐derived cardiomyocytes (iPSC‐CMs) (Bellin et al. 2012). These are areas in which there are at present no suitable solutions. All‐optical technology lends itself to automation (Klimas et al. 2015), and can indeed be quite impactful in streamlining and speeding up the drug development process.
Long‐term monitoring and/or stimulation are highly desirable in stem cell research during the derivation and differentiation of stem cell‐derived cardiomyocytes. The contactless nature of all‐optical tools offers a simple high‐throughput solution. Furthermore, miniaturization and automation make it possible to bring such tools within the cell culture incubator. Overall, this new methodology represents a step towards remote experimentation.
The term ‘remote experimentation’ refers to experimental set‐ups that can be accessed in real‐time by users far from the host site (e.g. via the internet). Remote experimentation has been successfully implemented in the physical sciences: researchers can, for example, control equipment in large telescopes without leaving their offices. In addition to increasing access to valuable scientific resources, remote experimentation helps address issues of reproducibility and transparency that are increasingly common across small life‐science laboratories (Anon, 2014). A similarly useful remote access system for wet‐lab experimentation would have to contend with the need for direct, hands‐on access to equipment, reagents and biological models. This is especially true in the cardiac sciences, which often involve complex experimental protocols on short‐lived biological preparations that require time and expertise to master. All‐optical methods offer potential solutions to several of these issues, and may prove to be a disruptive technology, enabling true remote access experimentation.
After an initial setting up of an experiment (be it cell culture, cardiac tissue or intact animal with implanted actuator and sensor) at a laboratory location with the proper expertise and rigorous quality control, all‐optical electrophysiological approaches can offer both contactless actuation and detection of cardiac activity, in a highly parallel (high‐throughput) manner (Entcheva, 2013). In principle, an all‐optical computer‐controlled stimulation and detection system can be built inside an environmentally controlled chamber, and a valuable preparation (cell or tissue slice culture) can then be used in a shared manner productively and continuously for days or weeks. This will permit multiple independent researchers around the world to perform experiments on the same preparation, reaping benefits in improved accessibility, data reproducibility and transparency, as well as reduction of animal use. The approach would be in the same spirit as popular initiatives towards ‘open science’, where researchers put raw data in public repositories and even share their lab books electronically, but here the experimental platform itself can be made open and accessible in real time. This is particularly relevant to limited‐access biological samples such as human patient‐derived cardiomyocytes (Bellin et al. 2012), which can be cultured and experimented on continuously using the all‐optical methods discussed here.
In conclusion, emerging all‐optical technologies offer significant advantages when compared with conventional electrical and pharmacological methods for controlling cardiac activity. In addition to enabling new strategies for rhythm control and pacing, all‐optical methods can be leveraged for high throughput studies at the cell level, and give researchers access to fundamental properties of cardiac excitation at the tissue level. Given the rapid pace of developments in this area, all‐optical techniques may eventually impact the cardiac sciences to the same extent that they have the neurosciences. Indeed, in some cases, optogenetic approaches already give cardiac scientists a level of control that is normally associated with computational models. These methods have the potential to change the way we think about control of cardiac function by expanding what is possible to achieve experimentally.
Additional information
Competing interests
The authors do not have any competing interests.
Author contributions
Both authors have approved the final version of the manuscript and agree to be accountable for all aspects of the work. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This work was supported by the BHF Centre of Research Excellence, Oxford (RE/08/004) and MR/K015877/1 (G.B.), as well as NIH R01 HL111649 and NSF‐Biophotonics grant 1511353 (E.E.).
Biographies
Emilia Entcheva is a Professor of Biomedical Engineering at George Washington University (GWU). She holds a PhD in Biomedical Engineering from the University of Memphis and BS and MS degrees in Electrical Engineering from the Technical University in Sofia, Bulgaria. After a postdoctoral fellowship at Johns Hopkins University, she started her own laboratory at Stony Brook in 2001 and recently moved to GWU. Her group has contributed to the development of new tools for optical mapping of excitation in cardiac cells and computational modelling of cardiac electrophysiology, and has recently spearheaded efforts in cardiac optogenetics.

Gil Bub investigates the interplay of activity between neuron and cardiac cells in cell culture, as well as new technologies for imaging excitable cells. He did his graduate work at McGill University, Canada on the dynamics of cardiac monolayers, and is currently University Research Lecturer in Oxford's Department of Physiology Anatomy and Genetics.
This review was presented at the symposium “Cardiac Arrhythmias: Challenges for Diagnosis and Treatment. A symposium in honour of George Ralph Mines (1886–1914)”, which took place at McGill University, Montreal, QC, Canada, between 6–7 November 2014.
References
- Abilez OJ, Wong J, Prakash R, Deisseroth K, Zarins CK & Kuhl E (2011). Multiscale computational models for optogenetic control of cardiac function. Biophys J 101, 1326–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamantidis A, Arber S, Bains JS, Bamberg E, Bonci A, Buzsáki G, Cardin JA, et al (2015). Optogenetics: 10 years after ChR2 in neurons–views from the community. Nat Neurosci 18, 1202–1212. [DOI] [PubMed] [Google Scholar]
- Ambrosi CM & Entcheva E (2014). Optogenetic control of cardiomyocytes via viral delivery. Methods Mol Biol 1181, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosi CM, Klimas A, Yu J & Entcheva E (2014). Cardiac applications of optogenetics. Prog Biophys Mol Biol 115, 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon (2014). Journals unite for reproducibility. Nature 515, 7. [DOI] [PubMed] [Google Scholar]
- Arrenberg AB, Stainier DYR, Baier H & Huisken J (2010). Optogenetic control of cardiac function. Science 330, 971–974. [DOI] [PubMed] [Google Scholar]
- Bellin M, Marchetto MC, Gage FH & Mummery CL (2012). Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol 13, 713–726. [DOI] [PubMed] [Google Scholar]
- Bingen BO, Engels MC, Schalij MJ, Jangsangthong W, Neshati Z, Feola I, Ypey DL, Askar SFA, Panfilov AV, Pijnappels DA, et al (2014). Light‐induced termination of spiral wave arrhythmias by optogenetic engineering of atrial cardiomyocytes. Cardiovasc Resc 104, 194–205. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G & Deisseroth K (2005). Millisecond‐timescale, genetically targeted optical control of neural activity. Nat Neurosci 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Boyle PM, Williams JC, Ambrosi CM, Entcheva E & Trayanova NA (2013). A comprehensive multiscale framework for simulating optogenetics in the heart. Nat Commun 4, 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruegmann T, Malan D, Hesse M, Beiert T, Fuegemann CJ, Fleischmann BK & Sasse P (2010). Optogenetic control of heart muscle in vitro and in vivo. Nat Methods 7, 897–900. [DOI] [PubMed] [Google Scholar]
- Burton RAB, Klimas A, Ambrosi CM, Tomek J, Corbett A, Entcheva E & Bub G (2015). Optical control of excitation waves in cardiac tissue. Nat Photonics 9, 813–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Liao M‐L, de Boer TP, Mutoh H, Raad N, Richter C, Wagner E et al (2015). Sensing cardiac electrical activity with a cardiac myocyte‐targeted optogenetic voltage indicator. Circ Res 117, 401–412. [DOI] [PubMed] [Google Scholar]
- Chen T‐W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K & Kim DS (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidenko JM, Salomonsz R, Pertsov AM, Baxter WT & Jalife J (1995). Effects of pacing on stationary reentrant activity: theoretical and experimental study. Circ Res 77, 1166–1179. [DOI] [PubMed] [Google Scholar]
- De Lacy Costello B, Toth R, Stone C, Adamatzky A & Bull L (2009). Implementation of glider guns in the light‐sensitive Belousov‐Zhabotinsky medium. Phys Rev E 79, 026114. [DOI] [PubMed] [Google Scholar]
- Dugué GP, Akemann W & Knöpfel T (2012). A comprehensive concept of optogenetics. Prog Brain Res 196, 1–28. [DOI] [PubMed] [Google Scholar]
- Dunlop J, Bowlby M, Peri R, Vasilyev D & Arias R (2008). High‐throughput electrophysiology: an emerging paradigm for ion‐channel screening and physiology. Nat Rev Drug Discov 7, 358–368. [DOI] [PubMed] [Google Scholar]
- Efimov IR, Nikolski VP & Salama G (2004). Optical imaging of the heart. Circ Res 95, 21–33. [DOI] [PubMed] [Google Scholar]
- Entcheva E (2013). Cardiac optogenetics. Am J Physiol Heart Circ Physiol 304, H1179–H1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S & Schnitzer MJ (2015). High‐speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350, 1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Sineshchekov OA, Janz R, Liu X & Spudich JL (2015). Natural light‐gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, Kapoor V, Zou P, Kralj JM, Maclaurin D, Smedemark‐Margulies N, Saulnier JL, Boulting GL, Straub C, Cho YK, Melkonian M, Wong GK, Harrison DJ, Murthy VN, Sabatini BL, Boyden ES, Campbell RE & Cohen AE (2014). All‐optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods 11, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Yea K & Lee KJ (2004). Regular and alternant spiral waves of contractile motion on rat ventricle cell cultures. Phys Rev Lett 92, 198103. [DOI] [PubMed] [Google Scholar]
- Jia Z, Valiunas V, Lu Z, Bien H, Liu H, Wang H‐Z, Rosati B, Brink PR, Cohen IS & Entcheva E (2011). Stimulating cardiac muscle by light cardiac optogenetics by cell delivery. Circ Arrhythmia Electrophysiol 4, 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanos TV, Boyle PM & Trayanova NA (2014). Optogenetics‐enabled dynamic modulation of action potential duration in atrial tissue: feasibility of a novel therapeutic approach. Europace 16 Suppl 4, iv69–iv76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimas A, Yu J, Ambrosi CM, Williams JC, Bien H & Entcheva E (2015). OptoDyCE: Automated system for high‐throughput all‐optical dynamic cardiac electrophysiology. bioRxiv, doi: http://dx.doi.org/10.1101/023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimas A & Entcheva E (2014). Toward microendoscopy‐inspired cardiac optogenetics in vivo: technical overview and perspective. J Biomed Opt 19, 080701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinsky VI & Agladze KI (1983). Interaction of rotating waves in an active chemical medium. Phys D Nonlinear Phenom 8, 50–56. [Google Scholar]
- Lerma C, Krogh‐Madsen T, Guevara M & Glass L (2006). Stochastic aspects of cardiac arrhythmias. J Stat Phys 128, 347–374. [Google Scholar]
- Maoz A, Krogh‐Madsen T & Christini DJ (2009). Instability in action potential morphology underlies phase 2 reentry: a mathematical modeling study. Heart Rhythm 6, 813–822. [DOI] [PubMed] [Google Scholar]
- Miesenböck G (2009). The optogenetic catechism. Science 326, 395–399. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P & Bamberg E (2003). Channelrhodopsin‐2, a directly light‐gated cation‐selective membrane channel. Proc Natl Acad Sci USA 100, 13940–13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinovitch U & Gepstein L (2015). Optogenetics for in vivo cardiac pacing and resynchronization therapies. Nat Biotechnol 33, 750–754. [DOI] [PubMed] [Google Scholar]
- Packer AM, Russell LE, Dalgleish HWP & Häusser M (2014). Simultaneous all‐optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat Methods 12, 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SA, Lee S‐R, Tung L & Yue DT (2014). Optical mapping of optogenetically shaped cardiac action potentials. Sci Rep 4, 6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz AA, Abbott LF & Marder E (2004). The dynamic clamp comes of age. Trends Neurosci 27, 218–224. [DOI] [PubMed] [Google Scholar]
- Romero L, Pueyo E, Fink M & Rodríguez B (2009). Impact of ionic current variability on human ventricular cellular electrophysiology. Am J Physiol Heart Circ Physiol 297, H1436–H1445. [DOI] [PubMed] [Google Scholar]
- Sagués F, Sancho JM & García‐Ojalvo J (2007). Spatiotemporal order out of noise. Rev Mod Phys 79, 829–882. [Google Scholar]
- Sakurai T, Mihaliuk E, Chirila F & Showalter K (2002). Design and control of wave propagation patterns in excitable media. Science 296, 2009–2012. [DOI] [PubMed] [Google Scholar]
- Spach MS & Heidlage JF (1995). The stochastic nature of cardiac propagation at a microscopic level: electrical description of myocardial architecture and its application to conduction. Circ Res 76, 366–380. [DOI] [PubMed] [Google Scholar]
- Steinbock O, Zykov V & Müller S (1993). Control of spiral‐wave dynamics in active media by periodic modulation of excitability. Nature 366, 322–324. [Google Scholar]
- Vogt CC, Bruegmann T, Malan D, Ottersbach A, Roell W, Fleischmann BK & Sasse P (2015). Systemic gene transfer enables optogenetic pacing of mouse hearts. Cardiovasc Res 106, 338–343. [DOI] [PubMed] [Google Scholar]
- Wengrowski AM, Wang X, Tapa S, Posnack NG, Mendelowitz D & Kay MW (2015). Optogenetic release of norepinephrine from cardiac sympathetic neurons alters mechanical and electrical function. Cardiovasc Res 105, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JC, Xu J, Lu Z, Klimas A, Chen X, Ambrosi CM, Cohen IS & Entcheva E (2013). Computational optogenetics: empirically‐derived voltage‐ and light‐sensitive channelrhodopsin‐2 model. PLoS Comput Biol 9, e1003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfree AT (1972). Spiral waves of chemical activity. Science 175, 634–636. [DOI] [PubMed] [Google Scholar]
- Xu L, Gutbrod SR, Bonifas AP, Su Y, Sulkin MS, Lu N, Chung HJ, Jang KI, Liu Z, Ying M, Lu C, Webb RC, Kim JS, Laughner JI, Cheng H, Liu Y, Ameen A, Jeong JW, Kim GT, Huang Y, Efimov IR & Rogers JA (2014). 3D multifunctional integumentary membranes for spatiotemporal cardiac measurements and stimulation across the entire epicardium. Nat Commun 5, 3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaglia T, Pianca N, Borile G, Da Broi F, Richter C, Campione M, Lehnart SE, Luther S, Corrado D, Miquerol L & Mongillo M (2015). Optogenetic determination of the myocardial requirements for extrasystoles by cell type‐specific targeting of ChannelRhodopsin‐2. Proc Natl Acad Sci USA 112, E4495–E4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemelman BV, Lee GA, Ng M & Miesenböck G (2002). Selective photostimulation of genetically chARGed neurons. Neuron 33, 15–22. [DOI] [PubMed] [Google Scholar]


