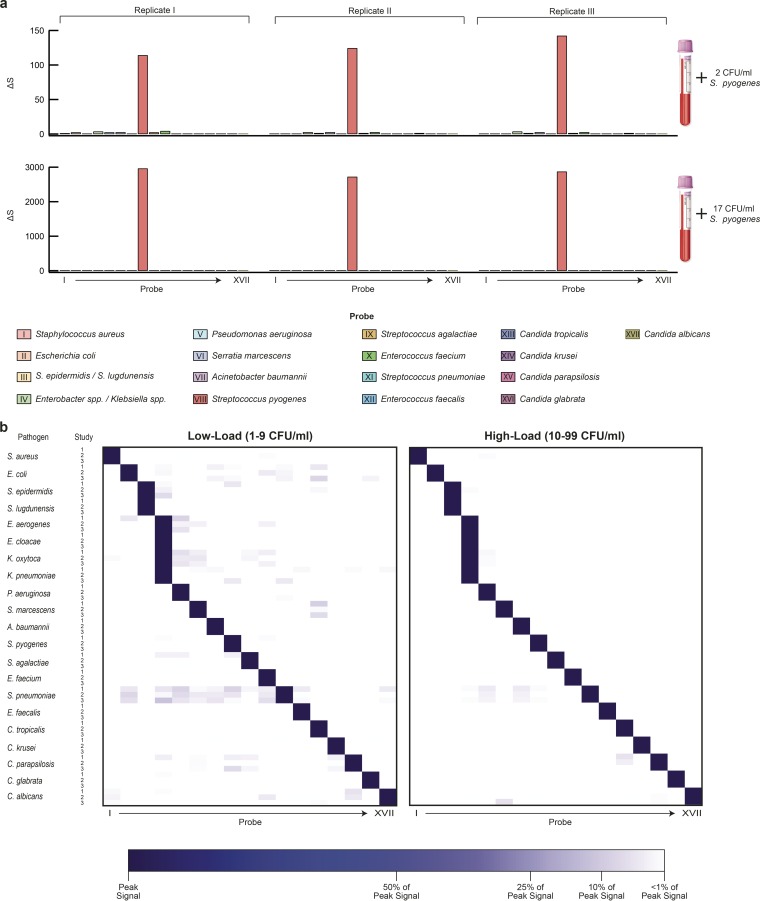
FIG 3 .
Direct blood PID validation. The performance of the integrated PID assay was tested using 1.5 ml blood spiked at both low (1 to 9 CFU/ml) and high (10 to 99 CFU/ml) pathogen loads. Processing steps included (i) removal of blood hDNA, (ii) extraction of gDNA, (iii) PCR amplification, and (iv) γPNA-based detection of amplicons. (a) Representative results for a triplicate study with S. pyogenes blood spiked at 2 CFU/ml (top) and 17 CFU/ml (bottom). Of the 17 γPNA probes, only probe VIII, specific for S. pyogenes, generated a significant signal, while signals generated from all other γPNAs were negligible. (b) Heat maps comparing results from blood spiked with either low loads (left panel) or high loads (right panel) of panel organisms. Signals were normalized to the target signal to facilitate visualization. Note that in all cases, off-target signal was negligible, even at the <10-CFU/ml level. Each pathogen/dilution combination was tested in triplicate.