ABSTRACT
The opportunistic pathogen Acinetobacter baumannii is able to persist in the environment and is often multidrug resistant (MDR), causing difficulties in the treatment of infections. Here, we show that the two-component system AdeRS, which regulates the production of the AdeABC multidrug resistance efflux pump, is required for the formation of a protective biofilm in an ex vivo porcine mucosal model, which mimics a natural infection of the human epithelium. Interestingly, deletion of adeB impacted only on the ability of strain AYE to form a biofilm on plastic and only on the virulence of strain Singapore 1 for Galleria mellonella. RNA-Seq revealed that loss of AdeRS or AdeB significantly altered the transcriptional landscape, resulting in the changed expression of many genes, notably those associated with antimicrobial resistance and virulence interactions. For example, A. baumannii lacking AdeRS displayed decreased expression of adeABC, pil genes, com genes, and a pgaC-like gene, whereas loss of AdeB resulted in increased expression of pil and com genes and decreased expression of ferric acinetobactin transport system genes. These data define the scope of AdeRS-mediated regulation, show that changes in the production of AdeABC mediate important phenotypes controlled by AdeRS, and suggest that AdeABC is a viable target for antimicrobial drug and antibiofilm discovery.
IMPORTANCE
Acinetobacter baumannii is a nosocomial pathogen and is an increasing problem in hospitals worldwide. This organism is often multidrug resistant, can persist in the environment, and forms a biofilm on environmental surfaces and wounds. Overproduction of efflux pumps can allow specific toxic compounds to be pumped out of the cell and can lead to multidrug resistance. This study demonstrates the role of the A. baumannii efflux pump AdeB, and its regulator AdeRS, in multidrug resistance, epithelial cell killing, and biofilm formation. Deletion of the genes encoding these systems led to increased susceptibility to antibiotics, decreased biofilm formation on biotic and abiotic surfaces, and decreased virulence. Our data suggest that inhibition of AdeB could prevent biofilm formation or colonization in patients by A. baumannii and provides a good target for drug discovery.
INTRODUCTION
Acinetobacter baumannii is an opportunistic pathogen that commonly causes nosocomial infections, such as ventilator-associated pneumonia and skin, soft tissue, wound, and bloodstream infections (1). There are several factors that contribute to the environmental persistence of, and infection by, this organism. These include its propensity to resist desiccation and survive in the hospital environment (2, 3) and its ability to form biofilms on both medical devices and biological surfaces (4–7). A. baumannii isolates are often resistant to high concentrations of antimicrobial drugs because of both intrinsic and acquired mechanisms, such as increased production of multidrug resistance (MDR) efflux pump proteins (8–10).
Bacterial cells often exist as a biofilm, where cells are attached to a surface and enclosed in an extracellular matrix that can be composed of polysaccharides, extracellular DNA, and protein. Biofilms are significantly more resistant to antimicrobial treatment, host immune responses, desiccation, and UV light, which enables them to persist in harsh environments, including the hospital setting. Growth on medical devices and tissue surfaces can lead to biofilm formation and increase the risk of bloodstream and respiratory infections, as the biofilm acts as a reservoir of bacteria (11). In vitro studies have shown that biofilms can survive antibiotic concentrations of up to 1,000 times the MIC for a planktonic culture, and in vivo, bacteria that survive antibiotic exposure in a biofilm state can cause recurrence of infection once antibiotic treatment is stopped (12, 13). It is hypothesized that biofilm resistance to antimicrobial therapy is due to multiple factors: the slow penetration of the biofilm by the antibiotic, the heterogeneity of the biofilm creating an altered microenvironment in which antibiotics are less effective, the differentiation of biofilm cells into a protected slow-growing state including a large proportion of “persister cells,” and changed gene expression due to this altered growth state (12–15). Bacteria in a sessile state exhibit proteomic profiles distinct from those of their planktonic counterparts, suggesting that these cells differ functionally and physically (16). Analysis of the transcriptome of A. baumannii in planktonic and biofilm states has revealed distinct and specific expression patterns and demonstrated the complexity of the networks regulating biofilm formation (17–20). Increased expression of resistance-nodulation-division (RND) efflux pump genes in biofilm versus planktonic states was also seen (17). Although several genes involved in biofilm pathways have been identified in A. baumannii (4–7), relatively little is known about the factors controlling biofilm formation or their contribution to virulence in this pathogen. Expression of factors involved in biofilm formation can vary widely, depending on the model system used (18–21). Therefore, it is essential that clinically relevant model systems be used to evaluate the microbial factors that contribute to infectious biofilm formation.
Bacterial MDR efflux pumps can extrude a broad range of substrates from the cell, conferring resistance to numerous antibiotics, biocides, dyes, and detergents. Increased expression of MDR efflux pump genes such as Acinetobacter RND efflux pump genes adeABC leads to MDR and is commonly seen in clinical isolates of A. baumannii (8, 22). Changes in the expression levels of these MDR efflux pump genes are often due to mutations in the genes encoding the two-component system (TCS) AdeRS, which regulates adeABC (22). Mutations in adeRS can cause overproduction of AdeABC and lead to MDR (8, 22). Deletion of either adeR or adeS from isolates overexpressing adeABC results in increased susceptibility to antimicrobials and other substrates of this pump (22). Bacterial TCSs have previously been shown to be involved in the regulation of various functions, such as growth, competence, metabolism, adaptation to starvation, osmoregulation, and expression of toxins (23).
In this study, we investigated the effect of deletion of adeRS and adeB on antimicrobial susceptibility and the abilities of two A. baumannii strains to form a biofilm on abiotic and biotic surfaces and cause infection in Galleria mellonella. The transcriptomes of A. baumannii strains AYE (24) and Singapore 1 (S1) (25) and adeRS and adeB mutants thereof were determined by RNA sequencing (RNA-Seq). This allowed a distinction to be made between the effects of adeB deletion and those of adeRS deletion. Multiple genes encoding proteins with potential functions including drug resistance, biofilm formation, and virulence, such as those encoding RND efflux pumps, type IV pili, DNA uptake channels, and acinetobactin transport systems, were differentially expressed in these mutants.
RESULTS
Choice of strains for this study.
In this study, we used A. baumannii strain AYE as a well-characterized isolate that is MDR because of an Ala94Val mutation in adeS that has been previously associated with increased expression of adeABC (10, 26). A clinical isolate from Singapore, S1, that was more susceptible to antimicrobials than strain AYE was and had no mutation in adeS and therefore did not express a high level of adeABC was also studied. Together, these strains represent clones that are internationally successful (strain AYE is sequence type 1 [ST1] of international clone I, and S1 is ST40 and representative of strains causing infections in Southeast Asia) (see Fig. S1 in the supplemental material). Type strain ATCC 19606 (ST52) was included as a control in the ex vivo biofilm experiments as a strain that has a well-characterized biofilm phenotype (21, 27). Deletion of adeRS was carried out by a markerless deletion method (28). It was hypothesized that the phenotype displayed by AYEΔadeRS might be due to decreased production of the tripartite MDR RND efflux pump AdeABC (as observed by RNA-Seq). To distinguish between the role of AdeRS and that of AdeABC in drug resistance, biofilm formation, and virulence in A. baumannii, an AdeB efflux pump deletion mutant of strain AYE and an AdeAB efflux pump deletion mutant of strain S1 were created. Gene deletion was confirmed by PCR and Sanger sequencing.
Deletion of adeRS reduces susceptibility to antimicrobials and dyes and reduces levels of efflux in AYE.
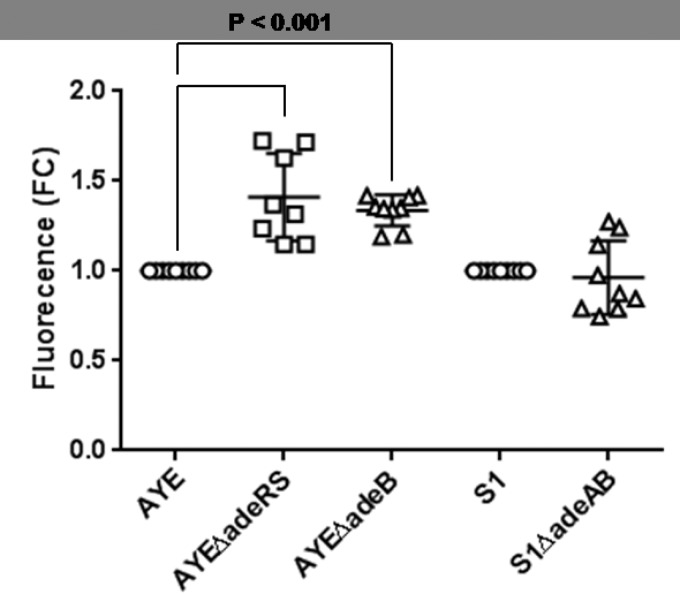
Growth curves, determined by measuring the optical density (OD) of the culture over time, showed no difference in growth rate between strain AYE and the adeRS mutant (see Fig. S2 in the supplemental material). However, deletion of adeRS resulted in a change in the drug resistance profile of strain AYE. There was a decrease in the MICs of imipenem, kanamycin, gentamicin, ciprofloxacin, tetracycline, tigecycline, chloramphenicol, and ethidium bromide upon the deletion of adeRS from strain AYE such that the phenotype of the mutant was similar to that of an adeB mutant of the same strain (Table 1). Although some of these changes were only 2-fold, which is considered to be the margin of error of this method, these changes were consistent in multiple experiments and were confirmed by growth kinetics in the presence of antibiotics (see Fig. S3 in the supplemental material). We have previously showed that accumulation of the dye Hoechst 33342 (H33342) is a good indication of efflux activity in A. baumannii (25). Accumulation of H33342 in strain AYEΔadeRS was 40% greater than that in strain AYE (P < 0.05), indicating reduced levels of efflux in the mutant (Fig. 1). Similar results were seen with efflux of ethidium bromide (data not shown).
TABLE 1 .
MICs of dyes and antibiotics used in this study
| Dye or antibiotica |
MICb (µg/ml) for: |
||||
|---|---|---|---|---|---|
| AYE | AYEΔadeRS | AYEΔadeB | S1 | S1ΔadeAB | |
| AMP | >1,024 | >1,024 | >1,024 | 16 | 16 |
| TAZ | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 |
| IMP | 1.5 | 0.75 | 0.75 | 0.125 | 0.125 |
| MER | 0.25 | 0.25 | 0.125 | 0.94 | 0.94 |
| KAN | 1,024 | 512 | 256 | 1 | 1 |
| GEN | 128 | 8 | 4 | 0.25 | 0.25 |
| NOR | 128 | 128 | 128 | 2 | 2 |
| CIP | 128 | 32 | 32 | 0.25 | 0.25 |
| COL | 1 | 1 | 1 | 0.5 | 0.5 |
| TET | 256 | 128 | 128 | 4 | 4 |
| TIG | 1 | 0.25 | 0.25 | 0.12 | 0.12 |
| CHL | 512 | 256 | 256 | 128 | 128 |
| CCCP | 32 | 32 | 16 | 16 | 16 |
| PAβN | 1,024 | 1,024 | 512 | 512 | 128 |
| EtBrb | 512 | 256 | 128 | 256 | 256 |
AMP, ampicillin, TAZ, ceftazidime, IMP, imipenem; MER, meropenem; KAN, kanamycin, GEN, gentamicin, NOR, norfloxacin, CIP, ciprofloxacin, COL, colistin, TET, tetracycline, TIG, tigecycline, CHL, chloramphenicol; CCCP, carbonyl cyanide m-chlorophenylhydrazone; PAβN, phenylalanine-arginine beta-napthalymide; EtBr, ethidium bromide.
Values in bold indicate significant differences between the mutant and parental strains.
FIG 1 .
Accumulation of H33342 as determined by fluorescence. Markers show H33342 fluorescence at steady state compared with that of the parental strain in nine individual biological replicates, with horizontal lines representing the mean and vertical lines and whiskers showing the standard error of the mean. FC, fold change.
Deletion of adeRS in AYE results in decreased biofilm formation on mucosal tissue.
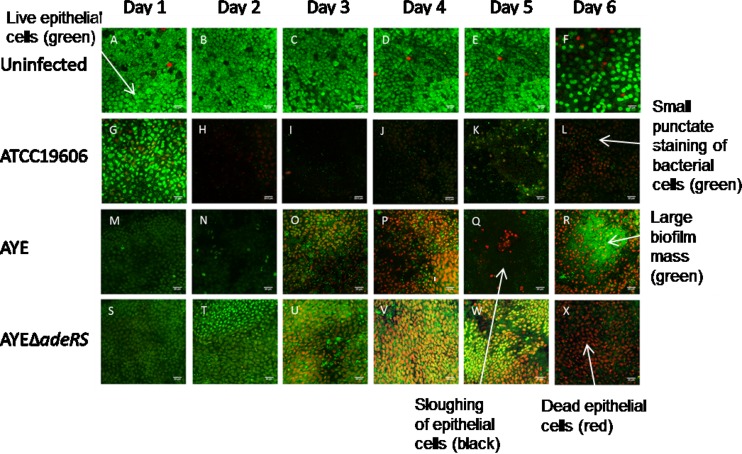
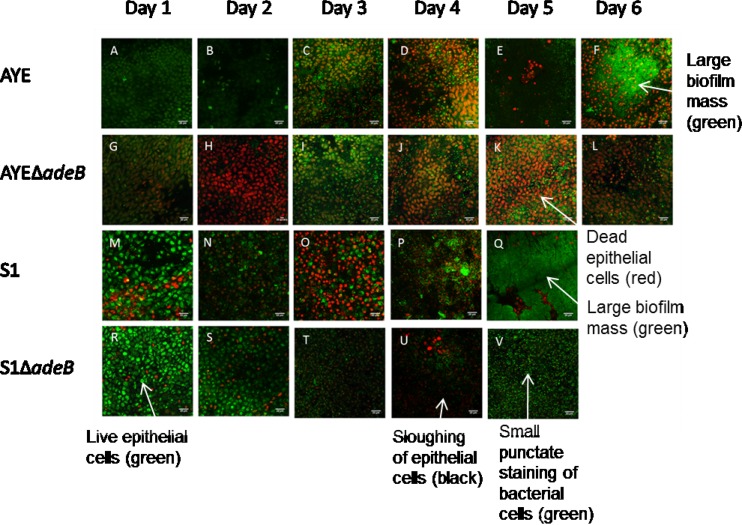
We used an ex vivo porcine vaginal mucosa (PVM) model to mimic a biofilm infection of human mucosa (20, 29, 30). To determine whether strain AYE showed a biofilm phenotype similar to that of a previously characterized biofilm-forming type strain of A. baumannii ATCC 19606, which has been shown to form a robust biofilm on both plastic and human skin equivalents (4, 21, 27), attachment of strain AYE and ATCC 19606 cells to PVM was imaged over 6 days by LIVE/DEAD staining and confocal laser scanning microscopy (Fig. 2). Viability of the tissue at 6 days was confirmed by imaging of uninfected tissue over the same time course. Uninfected tissue stained green, indicating live, intact cells with tight cell junctions (Fig. 2A to F). Epithelial cells and bacteria were distinguishable from each other on the basis of the size of the punctate staining, which could be clearly viewed by confocal microscopy. Over the 6-day time course, strains AYE and ATCC 19606 showed similar biofilm phenotypes. At 1 day postinoculation, loss of mucosal integrity was evidenced by rounding of epithelial cells and adherent bacteria (small, bright green, punctate staining) were visible on the tissue (Fig. 2G and M). By 3 days postinoculation, epithelial cell death (red) was observed and the number of bacteria visualized was greater (Fig. 2I and O). Black areas indicated sloughing of cells, as the extracellular matrix does not stain, and some biofilm formation was visible with ATCC 19606. By 6 days postinoculation, a large biofilm mass of both strains was visible, few dead epithelial cells remained, and most of the tissue was covered by biofilm (Fig. 2L and R).
FIG 2 .
Time course of biofilm development on mucosa, observed by LIVE/DEAD staining and confocal laser microscopy. Uninfected epithelia are live (green) and intact throughout. Red, rounded epithelial cells indicate epithelial cell death. Small, punctate green staining indicates bacterial cells, and large, green-staining masses indicate bacterial biofilm. Black areas depict the exposed extracellular matrix. Arrows indicate examples of live and dead epithelial cells, bacterial cells, epithelial cell sloughing, and biofilm masses. (A to F) Uninfected epithelial cells. (G to L) Growth of strain ATCC 19606 on mucosal tissue. (M to R) Growth of parental strain AYE on mucosal tissue. (S to X) Growth of strain AYEΔadeRS on mucosal tissue.
To investigate whether AdeRS is important for biofilm formation on biotic surfaces, the growth of strains AYE and AYEΔadeRS on PVM was measured. There was a rapid increase in the number of adherent cells on PVM up to the 1-day time point, followed by a slow but steady increase between 1 and 6 days (see Fig. S4 in the supplemental material). There was no significant difference between the numbers of adherent cells of the parental and mutant strains on days 1 to 6. However, LIVE/DEAD staining and confocal laser scanning microscopy showed a difference between the impact on the epithelial tissue of strains AYE and AYEΔadeRS (Fig. 2). At 3 days postinoculation, strain AYE-infected tissue displayed epithelial cell death (red) (Fig. 2O), and by 6 days postinoculation, large biofilm masses and epithelial cell sloughing were visible, as described above (Fig. 2R). However, for AYEΔadeRS, although epithelial cell death was evident and the tissue exhibited some epithelial cell sloughing, in contrast to coincubation with AYE, many dead epithelial cells were still visible and a biofilm had not covered the tissue. AYEΔadeRS cells appeared as single attached cells, with no biofilm observed (Fig. 2V to X). These data suggest a critical role for AdeRS in mucosal biofilm infections and host cell cytotoxicity.
Deletion of adeRS in AYE results in no change in biofilm formation on plastic.
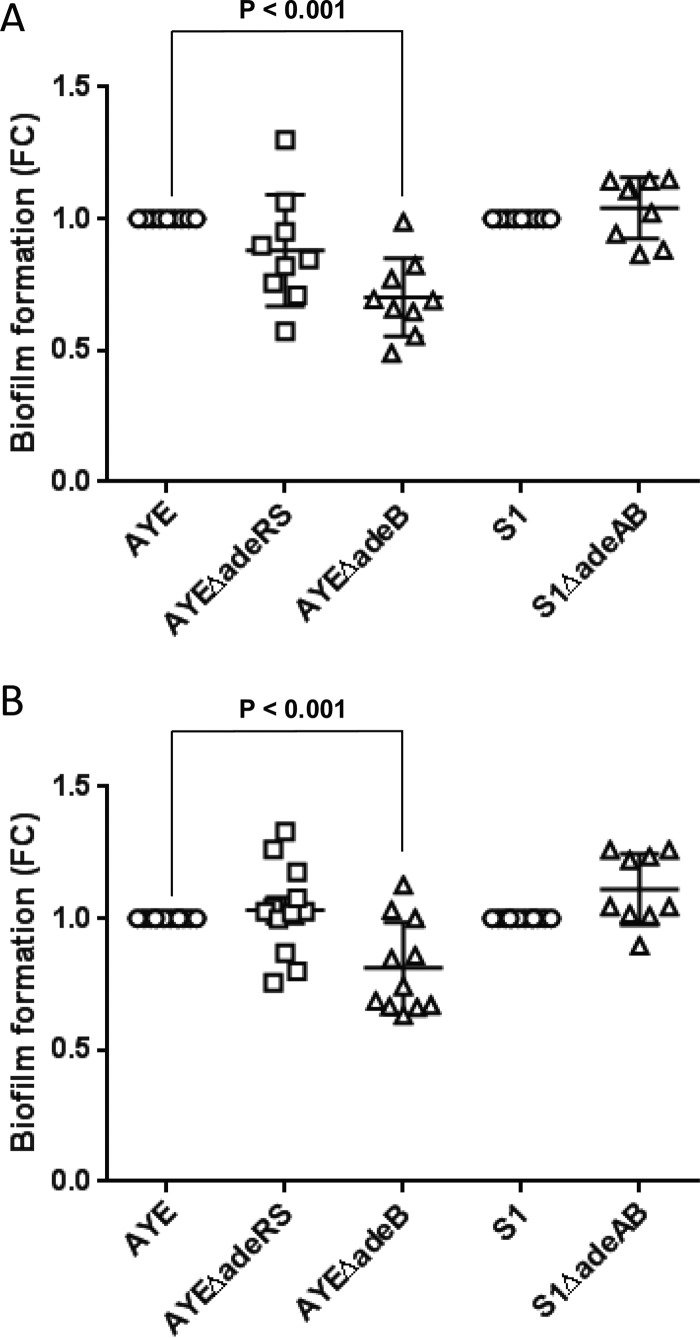
To investigate whether lack of AdeRS conferred a change in biofilm formation on an abiotic surface, we grew the parental strain AYE and the adeRS deletion mutant on plastic pegs for 8 h. At that time point, the biofilm formed by strain AYE was similar to that of previously characterized biofilm-forming strain ATCC 19606 (a range of time frames had been tested to determine the optimum length of time for biofilm growth to be measured accurately; data not shown). Pegs were then washed, and crystal violet staining was used to quantify biofilm mass. In this in vitro model, compared with the parental strain, there was no significant change in the biofilm mass produced by the adeRS deletion mutant at either 30°C or 37°C (Fig. 3). There was also no detectable change in the amount of pellicle formed in static cultures incubated for 48 h (data not shown).
FIG 3 .
Biofilm formation on plastic pegs at 30°C and 37°C as determined by crystal violet staining. Panels: A, 30°C; B, 37°C. Markers show fold change (FC) in OD600 compared with the parental strain in individual biological replicates, with horizontal lines representing the mean and vertical lines and whiskers showing the standard error of the mean.
AYEΔadeRS is as virulent as AYE in G. mellonella.
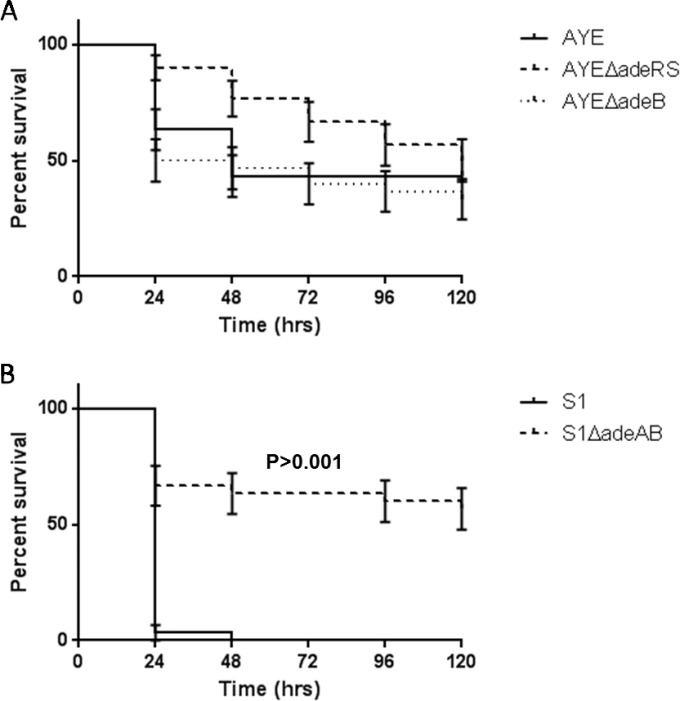
Previous studies have shown a positive correlation between virulence in the G. mellonella infection model and mammalian models (31, 32). Furthermore, G. mellonella has been established as a good model system to study A. baumannii pathogenesis, as larvae can be maintained at 37°C and have both a cellular and a humoral immune response (33). In the G. mellonella model, compared with the parental strain, AYEΔadeRS showed a small but nonsignificant change in virulence at an infectious dose of 106 CFU (Fig. 4).
FIG 4 .
Kaplan-Meier survival curve showing the virulence of individual isolates in G. mellonella. (A) adeRS and adeB mutants of strain AYE. (B) adeAB mutant of strain S1. Data show percent survival (n = 30) of G. mellonella after inoculation with 106 CFU of bacteria. Error bars represent the standard error of the mean.
Deletion of adeRS causes differential expression of functional gene groups in A. baumannii AYE.
To understand the role of the TCS AdeRS in the regulation of the expression of genes involved in antimicrobial drug resistance, biofilm formation, and virulence in A. baumannii strain AYE, changes in gene expression in the TCS deletion mutant AYEΔadeRS were identified by RNA-Seq (see Tables S1 to S3 in the supplemental material).
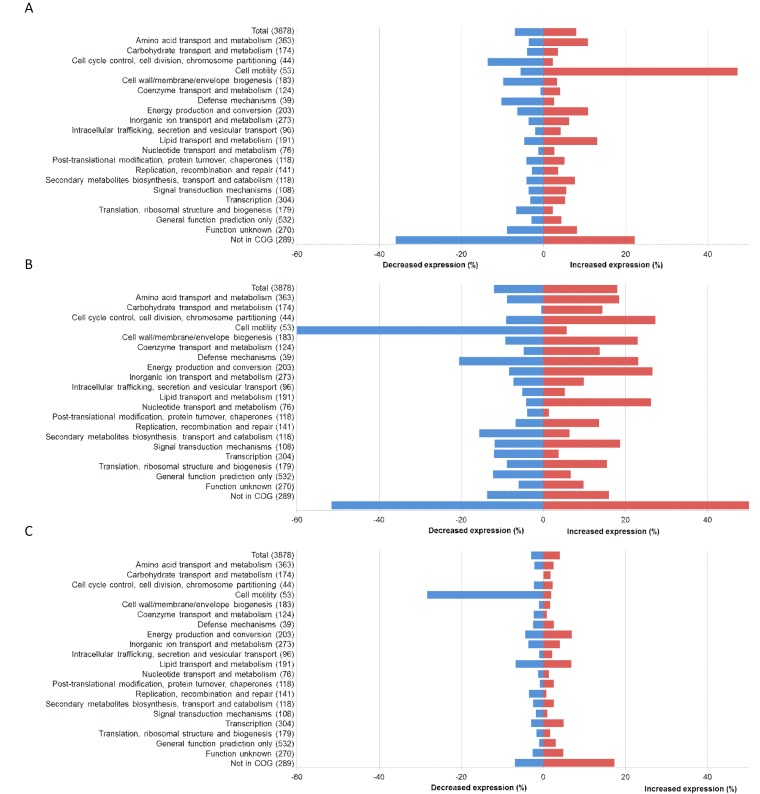
Loss of AdeRS had a large impact upon the transcriptome of strain AYE, with the differential expression of 579 genes (308 genes with increased expression and 271 with decreased expression) (P < 0.05). Differentially expressed genes were categorized into clusters of orthologous groups (COGs) (34), and correlations with the phenotypic changes seen in AYEΔadeRS were sought (Fig. 5). First, differential expression of genes known to confer antimicrobial resistance was identified. These included the RND efflux pump genes adeABC, which showed 128-, 91-, and 28-fold reductions in expression, respectively. One putative MDR RND efflux pump (ABAYE3036) had 1.9-fold increased expression, while the expression of another (ABAYE1796) was decreased by 1.5-fold. Genes encoding products with known and potential virulence functions, such as pili (35) and acinetobactin transport systems (36), had significant (P < 0.05) changes in expression levels in AYEΔadeRS. For instance, there was increased expression of genes that encode motility, such as competence genes comB, comC, comF, comL, comM, comN comO, and comQ (2.5- to 9.8-fold). These genes putatively encode DNA uptake channels, and deletion of comEC has been shown to reduce DNA uptake, motility, and virulence in G. mellonella (37). There was also increased expression of pilB, pilC, pilD, pilG, pilH, pilI, pilJ, pilT, pilU, and pilZ (1.7- to 8-fold). These genes encode type IV pili, which participate in processes such as natural transformation and twitching motility (37, 38). Type IV pili have also been associated with the ability of A. baumannii to form a biofilm on plastic (39). Alterations in the expression of other biofilm genes included the decreased expression of a pgaC-like gene (2-fold) that putatively encodes a protein involved in the synthesis of cell-associated poly-β-(1-6)-N-acetylglucosamine, which is required for biofilm formation on abiotic surfaces (7), and the decreased expression of five putative biofilm genes, ABAYE0792, -1395, -1397, -1470, and -1473 (1.7- to 2.5-fold). In addition, multiple putative transport protein, outer membrane protein, and transcriptional regulator genes that lacked a comprehensive annotation showed differential expression.
FIG 5 .
RNA-Seq results displayed by COG functions. Panels: A, AYEΔadeRS; B, AYEΔadeB; C, S1ΔadeAB.
Type IV pili are required for motility in A. baumannii, and inactivation of pilT has previously been shown to reduce twitching motility (37, 38). To determine whether the increased expression of motility genes correlated with altered motility, twitching motility and swarming experiments were carried out with 1% Mueller-Hinton agar and 0.3% Luria-Bertani (LB) agar, respectively (40). Strain AYE displayed twitching motility, typical of international clone I, but did not display swarming motility, like the majority of A. baumannii strains. However, no altered twitching motility or swarming phenotype was detected in strain AYEΔadeRS.
Deletion of adeB reduced efflux in AYE and S1 but only increased drug susceptibility in AYE.
Growth curves, determined by measuring the OD of the culture over time, showed no difference in growth rate between either strains AYE and AYEΔadeB or S1 and S1ΔadeAB (see Fig. S1 in the supplemental material). To determine whether deletion of adeB conferred a change in the drug resistance profile of strain AYE, the MICs of commonly used antibiotics, dyes, and biocides for strains AYE, AYEΔadeB, S1, and S1ΔadeAB were determined. There was a decrease in the MICs of meropenem, imipenem, kanamycin, gentamicin, ciprofloxacin, tetracycline, tigecycline, chloramphenicol, and ethidium bromide upon the deletion of adeB from strain AYE (Table 1). All 2-fold changes were confirmed by growth kinetics in the presence of antibiotics (see Fig. S3 in the supplemental material). AYEΔadeB had altered susceptibility to many of the same drugs as AYEΔadeRS, but the MICs of kanamycin, gentamicin, and ethidium bromide were lower after the deletion of adeB. Deletion of adeAB from S1 only resulted in a decrease in the MIC of phenylalanine-arginine β-naphthylamide (PAβN; an inhibitor of AcrB and MexB, homologues of AdeB), although the MICs of most drugs for S1 were much lower than those for strain AYE.
Compared with the steady-state accumulation level of the dye H33342 in the parental strain, that in AYEΔadeB was increased by 33% (P ≤ 0.05) (Fig. 1), indicating reduced levels of efflux in this efflux pump gene deletion mutant. There was no difference between the accumulation of H33342 in S1ΔadeAB and that in its parental strain, S1; this was consistent with the minimal changes in MICs (Fig. 1).
Deletion of adeB resulted in decreased biofilm formation of AYE on plastic.
To investigate whether AdeABC contributes to A. baumannii biofilm formation, we grew parental strains AYE and S1 and their isogenic efflux pump mutants on plastic pegs for 8 h and quantified the biofilm masses as described previously. Compared with the biofilm mass produced by the respective parental strains in this in vitro model, there was a significant decrease (P < 0.05) in that produced by AYEΔadeB at both 30°C and 37°C (Fig. 3). However, there was no change in biofilm formation by S1ΔadeAB at either temperature (Fig. 3). There was also no change in the amount of pellicle formed in static cultures of AYEΔadeB and S1ΔadeAB incubated for 48 h (data not shown).
Deletion of adeB from AYE and adeAB from S1 results in decreased biofilm formation on mucosal tissue.
To determine the role of AdeABC in biofilm formation on biotic surfaces, the growth of strains AYE and S1 and their isogenic efflux pump mutants on PVM was measured as described previously. S1 showed growth similar to that of strain AYE in terms of both adherent cell counts (see Fig. S4 in the supplemental material) and biofilm imaging (Fig. 6). However, S1 was able to form a robust biofilm more rapidly than strain AYE and a thick biofilm mat could be seen by day 5 (Fig. 6Q), at which point the experiment was stopped. There was no significant difference in the number of adherent cells on the tissue between either strains AYE and AYEΔadeB or S1 and S1ΔadeAB (see Fig. S4). However, compared with the respective parental strains, both efflux pump mutants showed a defect in biofilm formation when imaged by confocal microscopy. On days 5 and 6, S1 and AYE, respectively, formed large biofilm masses with extensive epithelial cell sloughing (Fig. 6F and Q), whereas although the mutants were able to cause epithelial cell death, only individual bacterial cells were observed to be attached to the mucosal tissue and less sloughing was evident (Fig. 6L and V). These data suggest that the AdeABC efflux pump plays a key role in biofilm formation on mucosal tissue and host cell cytotoxicity and that the decreased production of this MDR efflux pump may be responsible for the similar phenotype seen in strain AYEΔadeRS.
FIG 6 .
Time course of biofilm development on mucosa observed by LIVE/DEAD staining and confocal laser microscopy. Uninfected epithelia are live (green) and intact throughout. Red, rounded epithelial cells indicate epithelial cell death. Small, punctate green staining indicates bacterial cells, and large, green-staining masses indicate bacterial biofilm. Black areas depict the exposed extracellular matrix. Arrows indicate examples of live and dead epithelial cells, bacterial cells, epithelial cell sloughing, and biofilm masses. (A to F) Growth of parental strain AYE on mucosal tissue. (G to L) Growth of strain AYEΔadeB on mucosal tissue. (M to Q) Growth of parental strain S1 on mucosal tissue. (R to V) Growth of strain S1ΔadeAB on mucosal tissue. Images are representative of at least three repeated experiments.
Deletion of adeAB from S1 results in attenuated virulence in G. mellonella.
S1ΔadeAB displayed attenuated virulence in the G. mellonella model, compared with that of S1. After infection with S1, all larvae were dead by day 2, whereas 60% were still alive on day 5 after infection with the mutant (Fig. 4). Strain AYE was less virulent than S1 in G. mellonella, and there was no significant difference in the killing of larvae by strains AYE and AYEΔadeB (Fig. 4). This suggests a strain-specific role for AdeABC in the virulence of A. baumannii.
Deletion of adeB causes differential gene expression in A. baumannii AYE and S1.
Compared with strain AYE, AYEΔadeB had increased expression of 693 genes and decreased expression of 477 genes (P < 0.05). Compared with S1, S1ΔadeAB had increased expression of 164 genes and decreased expression of 119 genes (P < 0.05). There were 108 genes with changed expression in both adeB mutants. Genes with increased expression in AYEΔadeB included those encoding products with potential antibiotic resistance functions such as a tetracycline resistance protein, TetA (1.4-fold); membrane fusion protein (MFP) AdeT (1.4-fold), which is associated with MDR by active efflux in A. baumannii (41); a putative porin protein associated with imipenem resistance (ABAYE0924) (4.3-fold); and two putative MDR efflux systems (ABAYE1777, ABAYE3036) (1.4- and 3-fold). There was decreased expression of genes encoding a putative tetracycline resistance protein (ABAYE2235) (1.7-fold) and the lipid phosphoethanolamine transferase EptA (2.5-fold), which is associated with colistin resistance (42–44). Six β-lactamase genes (ABAYE0825, -1940, -2122, -2456, -3619, and -3623) also had altered expression (2.2- to 50-fold) in AYEΔadeB. The only gene related to drug resistance that displayed differential expression, albeit only a small increase, in S1ΔadeAB was that for a putative MDR efflux system (ABAYE3036) (1.1-fold) that had increased expression in all three deletion mutants. Similar to strain AYEΔadeRS, genes encoding products with known and potential virulence functions such as pili (35) and acinetobactin transport systems (36) also had significant changes in expression levels in adeB deletion mutants. The ferric acinetobactin transport system operon bauDCEBA, which encodes proteins required for persistence and virulence in the host (36, 45), had decreased expression in AYEΔadeB (1.9- to 3.7-fold). The most striking change observed in both efflux pump mutants was in cell motility genes. The competence (com) genes which, as previously mentioned, are associated with DNA uptake, motility, and virulence, had significantly decreased expression in AYEΔadeB and S1ΔadeAB (1.7- to 10-fold), in contrast to the significant increase in the expression of these genes in strain AYEΔadeRS (Fig. 5). Likewise, there was a significant decrease in the expression of the type IV pilus genes, which are involved in natural transformation, twitching motility, and biofilm formation. One other putative biofilm-associated gene (ABAYE0792) showed increased expression (3-fold) in AYEΔadeB, and no further genes with an annotated biofilm function were changed in S1ΔadeAB. However, multiple putative transcriptional-regulator-encoding genes (araC, lysR, and tetR family) and outer membrane protein-encoding genes that lacked a comprehensive annotation had differential expression in both mutants.
DISCUSSION
We have shown that deletion of the TCS AdeRS is associated with decreased antibiotic resistance, epithelial cell killing, and biofilm formation on mucosal tissue. We hypothesized that AdeRS regulates the expression of genes, including the RND MDR efflux system genes adeABC, that are required for these functions. In strain AYE, deletion of adeRS led to significant changes in the global transcriptional landscape, with the changed expression of 579 genes, including 128-, 91-, and 28-fold changes in adeA, adeB, and adeC expression, respectively. We show here that, in this strain, deletion of adeB produces a phenotype similar to that caused by the deletion of adeRS, consistent with the hypothesis that the phenotype of AdeRS mutants is due to the decreased expression of this RND MDR efflux pump. However, deletion of adeB from an unrelated strain, S1, resulted in reduced epithelial cell killing and decreased biofilm formation on PVM but, in contrast to deletion in strain AYE, had little effect on antibiotic resistance and resulted in a reduction in virulence in G. mellonella. This suggests that AdeABC has a strain-specific role in A. baumannii and may perform different functions in different strains.
AdeABC has a well-defined role in resistance to antimicrobials in A. baumannii BM4587 (8, 46, 47). In our study, deletion of adeRS or adeB from strain AYE resulted in a decrease in the MICs of several antibiotics that have been previously described as substrates of this pump (46, 47). This indicates that the downregulation of adeABC in strain AYEΔadeRS is responsible for the change in antibiotic susceptibility in this strain. Changes in MICs were more pronounced upon the deletion of adeB than upon the deletion of adeRS, which suggests that although deletion of adeRS results in an up to 128-fold decrease in the expression of adeABC, the efflux pump is still transcribed, albeit at a low level. We hypothesize that this is also responsible for the difference in the amounts of biofilm formed by these strains on plastic. While deletion of adeB from strain AYE resulted in a significant decrease in biofilm formation at both 30°C and 37°C, deletion of adeRS had no detectable effect. We hypothesize that low levels of expression of adeABC in strain AYEΔadeRS are sufficient to maintain biofilm function, whereas inactivation of the pump in AYEΔadeB significantly reduces the ability to form a biofilm.
Deletion of adeAB from a strain with a different background, clinical isolate S1, also resulted in a phenotype different from that seen in the strain AYE mutant: increased susceptibility to PAβN only. We hypothesize that the limited impact upon susceptibility to antimicrobial drugs after inactivation of AdeAB in S1 is a result of little change in the expression of adeABC between S1 and its mutant. Although adeB was expressed in S1, the level of expression was 16-fold lower in S1 than in strain AYE, as compared by RNA-Seq. This may explain the modest impact of the deletion of adeAB from S1 upon this strain’s susceptibility to antibiotics and why it is more susceptible to antibiotics than strain AYE is. Deletion of adeAB from S1 also had no significant effect on biofilm formation on plastic; this may also be due to the small impact of deleting a gene expressed at a low level.
In our study, only for AYEΔadeB was a significant biofilm defect observed in the in vitro abiotic model. A similar observation was previously shown in an adeB deletion mutant of drug-susceptible A. baumannii BM4587 (47). However, all three of the mutants tested in our study showed reduced biofilm formation on mucosal tissue. The same phenotype was observed in AYEΔadeRS and AYEΔadeB, with a significant decrease in epithelial cell killing and biofilm formation seen in both. We hypothesize that the downregulation of AdeABC in the former is responsible for this biofilm defect. This suggests that regulation of adeABC is an important function of AdeRS in the ability of A. baumannii to form a biofilm on biotic surfaces. Although there was no change in the number of adherent cells on the mucosal tissue upon the deletion of adeRS or adeB, there was a clear difference in the structure of the biofilm when imaged by LIVE/DEAD staining and confocal laser scanning microscopy. This is a phenomenon that has been previously observed in Staphylococcus aureus (20). This suggests that while AdeB does not affect the initial attachment to tissue, cells lacking this efflux pump are unable to form a mature biofilm on mucosal tissue. Similar observations have been made in Salmonella efflux pump mutants, which are able to adhere to surfaces but are not able to produce a mature biofilm (48). PVM is made up of stratified squamous epithelium, similar in structure to human mucosal surfaces (29). Furthermore, the growth characteristics of A. baumannii on the PVM were similar to those observed in a three-dimensional human skin equivalent model (26). Therefore, differences in the ability of A. baumannii mutants to form a biofilm in the PVM model may have implications for respiratory and wound infections.
Despite a change in the expression of known (and putative) genes associated with virulence in A. baumannii, such as the acinetobactin iron acquisition system and pilin genes, deletion of adeRS or adeB from strain AYE had no effect on virulence in G. mellonella. It is possible that AYE does not express the adeABC efflux genes in vivo. In contrast, deletion of adeAB from S1 greatly reduced virulence. The MFP-encoding gene adeA is also partially deleted from strain S1, which may account for this difference. Previous studies with other species have shown that some MFPs, such as AcrA, can form a complex with multiple different RND components, such as AcrB and AcrF (49, 50). The presence of AdeA in strain AYE may allow it to interact with other proteins and so compensate for the lack of AdeB and ameliorate any impact upon virulence in this model. However, to our knowledge, so far, no other A. baumannii efflux pumps have been shown to play a role in virulence. Therefore, our data indicate a strain-specific role for the AdeABC efflux pump in virulence. A correlation between pathogenicity in G. mellonella and in humans has previously been observed with A. baumannii (33). Therefore, our data suggest that for some A. baumannii strains, such as S1, AdeABC may be required for infection of humans. The finding that the major RND MDR efflux pump in A. baumannii can also play a fundamental role in its ability to infect its host further underscores a role for MDR efflux pumps in the basic biology of pathogenic bacteria.
Using transcriptome profiling, we have demonstrated that deletion of AdeRS results in altered transcription patterns compared to those of the parental strain, thus highlighting its wider role in gene regulation in A. baumannii. Deletion of AdeRS modulates the expression of many genes, notably those associated with MDR, epithelial cell killing, and biofilm formation. Our data suggest that downregulation of the AdeABC efflux pump genes in the AYE adeRS deletion mutant is responsible for the increase in susceptibility to antimicrobials and the decrease in biofilm formation observed in this strain. This is the first time, to our knowledge, that this TCS has been associated with the regulation of biofilm formation in A. baumannii. Furthermore, we have shown a role for the AdeABC efflux pump in biofilm formation on plastic and mucosal tissue in an ex vivo model. These data suggest that inhibition of MDR efflux pumps may be a useful strategy to help prevent or treat colonization in patients by A. baumannii. Finally, we demonstrate that two AdeB efflux pump deletion mutations in different strains produced different virulence phenotypes in a G. mellonella model, highlighting the strain variability of A. baumannii. This suggests that broad conclusions about the roles of specific genes and proteins in the species should not be drawn from the study of single strains.
MATERIALS AND METHODS
Strains, media, and chemicals.
A. baumannii strain AYE is an MDR isolate that carries a mutation in adeS and is a member of epidemic clone II. A. baumannii S1 (DR00539/06) is a more drug-sensitive clinical isolate provided by the Network for Antimicrobial Resistance Surveillance (Singapore) and is representative of isolates causing infections in Singaporean hospitals (51). Strains AYEΔadeRS, AYEΔadeB, and S1ΔadeAB are unmarked deletion mutants created by a previously described method for generating markerless deletions in A. baumannii (28). Briefly, DNA fragments upstream and downstream of the genes of interest were amplified and ligated into a tellurite-resistant (sacB+ xylE+) suicide vector, pMo130-Telr. A two-step selection process was then used to select for deletion mutants. Isolates S2 (DR01138/06), R2 (TTSH6013 654325/06), and R3 (DM01800/06) are representative Singapore clinical isolates used to compare the levels of adeB expression (28). Strain ATCC 19606 (obtained from the American Type Culture Collection, Manassas, VA) was used throughout as a reference strain with a previously characterized biofilm phenotype (4). All strains were routinely cultured on LB agar or tryptic soy agar II containing 5% sheep blood and in LB or Todd-Hewitt broth. All chemicals were from Sigma-Aldrich (Poole, Dorset, United Kingdom).
Measurement of bacterial growth.
Bacterial strains were grown with aeration in LB broth at 37°C overnight. A total of 200 µl of culture was added to each well of a clear 96-well microtiter tray (MTT) at an inoculum density of 104 CFU/ml. OD at 600 nm (OD600) was measured over 16 h in a BMG FLUOstar OPTIMA (BMG, United Kingdom) at 37°C. The difference in the final OD600 at 16 h was calculated, and values returning a P value of <0.05 from a Student t test were taken as significant.
RNA-Seq.
RNA was extracted from a minimum of three biological replicates of each strain with a Qiagen RNAprotect Bacteria Reagent and RNeasy minikit and DNase treated with an Ambion TURBO DNA-free kit. RNA libraries were prepared and sequenced with an Illumina MiSeq or HiSeq at ARK Genomics, Roslin Institute, University of Edinburgh, University of Birmingham, or BGI Genomics, Hong Kong. Raw sequences were assessed for quality with FASTQC and processed. Alignments with a strain AYE reference genome (52) were performed with bowtie2. A bed file of the gene loci was generated from the gff annotation, and bedtools was used to count tags overlapping the regions of interest. Raw tag counts per sample were scale normalized to the sample by using the lowest number of tags within each data set. Counts were converted to log2 and quantile normalized within each series for comparisons within each data set. Pairwise comparisons of the normalized tag counts were performed by linear modeling (Bioconductor limma package). A raw P cutoff value of 0.05 was used to produce a list of changed genes that could be verified by phenotypic testing. No fold change cutoff was used.
Measurement of efflux activity.
Efflux activity was determined by measuring the accumulation of H33342. The H33342 (bis-benzimide) assay was carried out as described previously (53). Steady-state fluorescence values were taken and converted to fold changes compared to the parental strain. The differences between parent and mutant strains were calculated, and values returning a P value of <0.05 from a Student t test were taken as significant.
Determination of antimicrobial susceptibility.
The MICs of ampicillin, ceftazidime, kanamycin, gentamicin, norfloxacin, ciprofloxacin, colistin, tetracycline, tigecycline, chloramphenicol, carbonyl cyanide m-chlorophenylhydrazone, PAβN, and ethidium bromide were determined by the agar doubling dilution method according to British Society for Antimicrobial Chemotherapy standard methodology (54). MICs of imipenem and meropenem were determined by E test (bioMérieux, Hampshire, United Kingdom). Clinically relevant resistance was defined as a MIC above the recommended breakpoint according to the EUCAST guidelines (55). All experiments were carried out in triplicate.
Measurement of biofilm formation on plastic.
Biofilm formation on plastic was measured with an adapted version of a previously described method (56). Wells of an MTT were inoculated with 100 µl of diluted culture, and a sterile, 96-well polystyrene PCR plate was placed into the MTT. Plates were incubated for 8 h at either 30°C or 37°C. Pegs were washed and stained with 1% crystal violet for 15 min. Dye was solubilized in 70% ethanol, and OD600 was measured with a BMG FLUOstar Optima. OD600 was converted to fold change compared to the parental strain in order to compare results between experiments. The differences between parent and mutant strains were calculated, and values returning a P value of <0.05 from a Student t test were taken as significant.
Measurement of biofilm formation on PVM tissue.
Biofilm formation on mucosal tissue was measured as described previously (29). Briefly, specimens of normal PVM were excised from animals at slaughter and washed in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (Invitrogen), penicillin (50 IU/ml; MP Biomedicals, Solon, OH), streptomycin (50 µg/ml; MP Biomedicals), and amphotericin B (2.5 μg/ml; HyClone, Logan, UT); 5-mm tissue explants were cut; and excess muscle was trimmed away. Tissue explants were washed in serum- and antibiotic-free medium three times. Explants were then placed mucosal side up on a 0.4-µm cell culture insert (BD Bioscience, San Jose, CA, USA) in six-well plates containing fresh serum- and antibiotic-free RPMI 1640 medium. Explants were inoculated with 105 CFU of bacteria and incubated at 37°C. Adherent and planktonic cells were counted at 24, 48, 72, 96, 120, and 144 h. The difference in the number of adherent cells between parent and mutant strains was calculated, and values returning a P value of <0.05 from a Student t test were taken as significant. For imaging, explants were stained with a FilmTracer LIVEDEAD Biofilm Viability kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The LIVE/DEAD stain consists of SYTO9, which stains live cells green, and propidium iodide (PI), which stains dead cells red. PI penetrates only cells with damaged membranes, and once it is in close proximity to SYTO9, it quenches the green signal, so only the red is visible. This allows live versus dead cells to be detected. These dyes are nonspecific nucleic acid dyes that do not discern between prokaryotic and eukaryotic cells (29, 57–59). After staining, specimens were gently washed three times in Hanks’ balanced salt solution and transferred to glass slides. A coverslip with a 1-mm spacer (Electron Microscopy Sciences, Hatfield, PA) was then applied, and specimens were imaged on an Olympus FluoView 1000 BX2 (Olympus America Corporation, Center Valley, PA) with a 60× oil immersion objective. Images were captured and processed with FluoView software (Olympus America Corporation, Center Valley, PA).
Measurement of motility.
Swarming and twitching motility was measured as described previously (40). Briefly, a single colony was used to inoculate petri dishes containing 0.3% LB agar or 1% Mueller-Hinton agar. Plates were incubated overnight at 37°C and examined for growth.
Measurement of virulence in G. mellonella.
Survival in G. mellonella was assayed as previously described (60). Bacteria were injected into G. mellonella larvae in an inoculum of 106 CFU. Larvae were incubated statically at 37°C in petri dishes, and the number of dead larvae was scored periodically. All experiments were carried out in triplicate, and statistical analysis was carried out with GraphPad Prism 5 to produce Kaplan-Meier survival curves. The statistical significance of differences between parent and mutant strain survival curves was calculated with a log rank test. P values of <0.05 were taken as significant.
Ethics statement.
No animals were used in this study. Porcine tissue is not subject to regulation, as it is a by-product of the slaughter of animals for food.
Microarray data accession numbers.
The RNA-Seq data obtained in this study were submitted to ArrayExpress (accession no. E-MTAB-4047, E-MTAB-4049, and E-MTAB-4071).
SUPPLEMENTAL MATERIAL
Relationships among 615 A. baumannii isolates based on multilocus sequence typing data (Pasteur scheme), as calculated by the BURST algorithm. Circles indicate major international groups responsible for the majority of diseases. Arrows indicate the strains used in this study. ATCC 19606 represents a singleton ST (ST52) and is not in this network. Download
Growth at 37°C, as determined by OD600. Download
A representative example of confirmation of MIC changes for AYEΔadeRS and AYEΔadeB by measurement of growth kinetics in the presence of tetracycline, as determined by OD600. Download
Adherent bacterial growth on mucosa, as determined by bacterial cell counts. (A) adeRS and adeB mutants of strain AYE. (B) adeAB mutant of S1. Download
Gene expression data from the complete transcriptome analysis of A. baumannii AYE by RNA-Seq, showing differentially expressed genes (P < 0.05) in AYEΔadeRS compared with AYE.
Gene expression data from the complete transcriptome analysis of A. baumannii AYE by RNA-Seq, showing differentially expressed genes (P < 0.05) in AYEΔadeB compared with AYE.
Gene expression data from the complete transcriptome analysis of A. baumannii S1 by RNA-Seq, showing differentially expressed genes (P < 0.05) in AYEΔadeAB compared with S1.
ACKNOWLEDGMENTS
We thank the Network for Antimicrobial Resistance Surveillance, Singapore, for the donation of strain S1.
Funding Statement
This article presents independent research funded by the National Institute for Health Research Surgical Reconstruction and Microbiology Research Centre (NIHR SRMRC) (partnership between University Hospitals Birmingham NHS Foundation Trust, the University of Birmingham and the Royal Centre for Defence Medicine, UK). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The views expressed in this article are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnotes
Citation Richmond GE, Evans LP, Anderson MJ, Wand ME, Bonney LC, Ivens A, Chua KL, Webber MA, Sutton JM, Peterson ML, Piddock LJV. 2016. The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. mBio 7(2):e00430-16. doi:10.1128/mBio.00430-16.
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol 36:1938–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catalano M, Quelle LS, Jeric PE, Di Martino A, Maimone SM. 1999. Survival of Acinetobacter baumannii on bed rails during an outbreak and during sporadic cases. J Hosp Infect 42:27–35. doi: 10.1053/jhin.1998.0535. [DOI] [PubMed] [Google Scholar]
- 4.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 6.Goh HM, Beatson SA, Totsika M, Moriel DG, Phan M-D, Szubert J, Runnegar N, Sidjabat HE, Paterson DL, Nimmo GR, Lipman J, Schembri MA. 2013. Molecular analysis of the Acinetobacter baumannii biofilm-associated protein. Appl Environ Microbiol 79:6535–6543 doi: 10.1128/aem.01402-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litrán T. 2009. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon EJ, Courvalin P, Grillot-Courvalin C. 2013. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother 57:2989–2995. doi: 10.1128/AAC.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roca I, Espinal P, Vila-Farrés X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol 3:148. doi: 10.3389/fmicb.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 12.Stewart PS, William Costerton J. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. doi: 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 13.Mah TF, O’Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 14.Conlon BP, Rowe SE, Lewis K. 2015. Persister cells in biofilm associated infections. Adv Exp Med Biol 831:1–9. doi: 10.1007/978-3-319-09782-4_1. [DOI] [PubMed] [Google Scholar]
- 15.de la Fuente-Núñez C, Reffuveille F, Fernández L, Hancock RE. 2013. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol 16:580–589. doi: 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Cabral MP, Soares NC, Aranda J, Parreira JR, Rumbo C, Poza M, Valle J, Calamia V, Lasa I, Bou GN. 2011. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J Proteome Res 10:3399–3417. doi: 10.1021/pr101299j. [DOI] [PubMed] [Google Scholar]
- 17.Rumbo-Feal S, Gómez MJ, Gayoso C, Álvarez-Fraga L, Cabral MP, Aransay AM, Rodríguez-Ezpeleta N, Fullaondo A, Valle J, Tomás M, Bou G, Poza M. 2013. Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS One 8:e72968. doi: 10.1371/journal.pone.0072968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson GG, Moreau-Marquis S, Stanton BA, O’Toole GA. 2008. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun 76:1423–1433. doi: 10.1128/IAI.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson MJ, Lin YC, Gillman AN, Parks PJ, Schlievert PM, Peterson ML. 2012. Alpha-toxin promotes Staphylococcus aureus mucosal biofilm formation. Front Cell Infect Microbiol 2:64. doi: 10.3389/fcimb.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Breij A, Gaddy J, van der Meer J, Koning R, Koster A, van den Broek P, Actis L, Nibbering P, Dijkshoorn L. 2009. CsuA/BABCDE-dependent pili are not involved in the adherence of Acinetobacter baumannii ATCC 19606T to human airway epithelial cells and their inflammatory response. Res Microbiol 160:213–218. doi: 10.1016/j.resmic.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 48:3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West AH, Stock AM. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26:369–376. doi: 10.1016/S0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 24.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JM, Raoult D, Médigue C, Weissenbach J, Cruveiller S. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richmond GE, Chua KL, Piddock LJ. 2013. Efflux in Acinetobacter baumannii can be determined by measuring accumulation of H33342 (bis-benzimide). J Antimicrob Chemother 68:1594–1600 doi: 10.1093/jac/dkt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornsey M, Loman N, Wareham DW, Ellington MJ, Pallen MJ, Turton JF, Underwood A, Gaulton T, Thomas CP, Doumith M, Livermore DM, Woodford N. 2011. Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J Antimicrob Chemother 66:1499–1503. doi: 10.1093/jac/dkr168. [DOI] [PubMed] [Google Scholar]
- 27.de Breij A, Haisma EM, Rietveld M, El Ghalbzouri A, van den Broek PJ, Dijkshoorn L, Nibbering PH. 2012. Three-dimensional human skin equivalent as a tool to study Acinetobacter baumannii colonization. Antimicrob Agents Chemother 56:2459–2464. doi: 10.1128/AAC.05975-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amin IM, Richmond GE, Sen P, Koh TH, Piddock LJ, Chua KL. 2013. A method for generating marker-less gene deletions in multidrug-resistant Acinetobacter baumannii. BMC Microbiol 13:158. doi: 10.1186/1471-2180-13-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson MJ, Parks PJ, Peterson ML. 2013. A mucosal model to study microbial biofilm development and anti-biofilm therapeutics. J Microbiol Methods 92:201–208. doi: 10.1016/j.mimet.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson MJ, Scholz MT, Parks PJ, Peterson ML. 2013. Ex vivo porcine vaginal mucosal model of infection for determining effectiveness and toxicity of antiseptics. J Appl Microbiol 115:679–688. doi: 10.1111/jam.12277. [DOI] [PubMed] [Google Scholar]
- 31.Miyata S, Casey M, Frank DW, Ausubel FM, Drenkard E. 2003. Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect Immun 71:2404–2413. doi: 10.1128/IAI.71.5.2404-2413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jander G, Rahme LG, Ausubel FM. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol 182:3843–3845. doi: 10.1128/JB.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahar O, Goffer T, Burdman S. 2009. Type IV pili are required for virulence, twitching motility, and biofilm formation of Acidovorax avenae subsp. citrulli. Mol Plant Microbe Interact 22:909–920. doi: 10.1094/MPMI-22-8-0909. [DOI] [PubMed] [Google Scholar]
- 36.Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilharm G, Piesker J, Laue M, Skiebe E. 2013. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J Bacteriol 195:4146–4153. doi: 10.1128/JB.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360–13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, Davies BW. 2014. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5:e01313–14. doi: 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH. 2011. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol Lett 323:44–51. doi: 10.1111/j.1574-6968.2011.02362.x. [DOI] [PubMed] [Google Scholar]
- 41.Srinivasan VB, Rajamohan G, Pancholi P, Marcon M, Gebreyes WA. 2011. Molecular cloning and functional characterization of two novel membrane fusion proteins in conferring antimicrobial resistance in Acinetobacter baumannii. J Antimicrob Chemother 66:499–504. doi: 10.1093/jac/dkq469. [DOI] [PubMed] [Google Scholar]
- 42.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock RE. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mihara K, Tanabe T, Yamakawa Y, Funahashi T, Nakao H, Narimatsu S, Yamamoto S. 2004. Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology 150:2587–2597. doi: 10.1099/mic.0.27141-0. [DOI] [PubMed] [Google Scholar]
- 46.Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454 Antimicrob Agents Chemother 45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon E-J, Chabane YN, Goussard S, Snesrud E, Courvalin P, Dé E, Grillot-Courvalin C. 2015. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 6:e00309-15. doi: 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baugh S, Ekanayaka AS, Piddock LJ, Webber MA. 2012. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J Antimicrob Chemother 67:2409–2417. doi: 10.1093/jac/dks228. [DOI] [PubMed] [Google Scholar]
- 49.Elkins CA, Nikaido H. 2003. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J Bacteriol 185:5349–5356. doi: 10.1128/JB.185.18.5349-5356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith HE, Blair JM. 2014. Redundancy in the periplasmic adaptor proteins AcrA and AcrE provides resilience and an ability to export substrates of multidrug efflux. J Antimicrob Chemother 69:982–987. doi: 10.1093/jac/dkt481. [DOI] [PubMed] [Google Scholar]
- 51.Koh TH, Tan TT, Khoo CT, Ng SY, Tan TY, Hsu LY, Ooi EE, Van Der Reijden TJ, Dijkshoorn L. 2012. Acinetobacter calcoaceticus-Acinetobacter baumannii complex species in clinical specimens in Singapore. Epidemiol Infect 140:535–538. doi: 10.1017/S0950268811001129. [DOI] [PubMed] [Google Scholar]
- 52.Kersey PJ, Allen JE, Christensen M, Davis P, Falin LJ, Grabmueller C, Hughes DS, Humphrey J, Kerhornou A, Khobova J, Langridge N, McDowall MD, Maheswari U, Maslen G, Nuhn M, Ong CK, Paulini M, Pedro H, Toneva I, Tuli MA, Walts B, Williams G, Wilson D, Youens-Clark K, Monaco MK, Stein J, Wei X, Ware D, Bolser DM, Howe KL, Kulesha E, Lawson D, Staines DM. 2014. Ensembl genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res 42:D546–D552. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richmond GE, Chua KL, Piddock LJ. 2013. Efflux in Acinetobacter baumannii can be determined by measuring accumulation of H33342 (bis-benzimide). J Antimicrob Chemother 68:1594–1600. doi: 10.1093/jac/dkt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 55.EUCAST 2015. The European Committee on Antimicrobial Susceptibility Testing breakpoint tables for interpretation of MICs and zone diameters, version 5.0. European Committee on Antimicrobial Susceptibility Testing, Basel, Switzerland: http://www.eucast.org/clinical_breakpoints/. [Google Scholar]
- 56.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Gorle AK, Ainsworth TD, Heimann K, Woodward CE, Grant Collins J, Richard Keene F. 2015. RNA and DNA binding of inert oligonuclear ruthenium(II) complexes in live eukaryotic cells. Dalton Trans 44:3594–3603. doi: 10.1039/C4DT02575J. [DOI] [PubMed] [Google Scholar]
- 58.Haimovich B, Tanaka JC. 1995. Magainin-induced cytotoxicity in eukaryotic cells: kinetics, dose-response and channel characteristics. Biochim Biophys Acta 1240:149–158. doi: 10.1016/0005-2736(95)00204-9. [DOI] [PubMed] [Google Scholar]
- 59.Tsai YJ, Lin YC, Wu WB, Chiu PH, Lin BJ, Hao SP. 2013. Biofilm formations in nasopharyngeal tissues of patients with nasopharyngeal osteoradionecrosis. Otolaryngol Head Neck Surg 148:633–636. doi: 10.1177/0194599812474971. [DOI] [PubMed] [Google Scholar]
- 60.Wand ME, Müller CM, Titball RW, Michell SL. 2011. Macrophage and Galleria mellonella infection models reflect the virulence of naturally occurring isolates of B. pseudomallei, B. thailandensis and B. oklahomensis. BMC Microbiol 11:11. doi: 10.1186/1471-2180-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relationships among 615 A. baumannii isolates based on multilocus sequence typing data (Pasteur scheme), as calculated by the BURST algorithm. Circles indicate major international groups responsible for the majority of diseases. Arrows indicate the strains used in this study. ATCC 19606 represents a singleton ST (ST52) and is not in this network. Download
Growth at 37°C, as determined by OD600. Download
A representative example of confirmation of MIC changes for AYEΔadeRS and AYEΔadeB by measurement of growth kinetics in the presence of tetracycline, as determined by OD600. Download
Adherent bacterial growth on mucosa, as determined by bacterial cell counts. (A) adeRS and adeB mutants of strain AYE. (B) adeAB mutant of S1. Download
Gene expression data from the complete transcriptome analysis of A. baumannii AYE by RNA-Seq, showing differentially expressed genes (P < 0.05) in AYEΔadeRS compared with AYE.
Gene expression data from the complete transcriptome analysis of A. baumannii AYE by RNA-Seq, showing differentially expressed genes (P < 0.05) in AYEΔadeB compared with AYE.
Gene expression data from the complete transcriptome analysis of A. baumannii S1 by RNA-Seq, showing differentially expressed genes (P < 0.05) in AYEΔadeAB compared with S1.