Abstract
Major facilitator superfamily domain containing 2A (MFSD2A) was recently characterized as a sodium-dependent lysophosphatidylcholine transporter expressed at the blood-brain barrier endothelium. It is the primary route for importation of docosohexaenoic acid and other long-chain fatty acids into fetal and adult brain and is essential for mouse and human brain growth and function. Remarkably, MFSD2A is the first identified major facilitator superfamily member that uniquely transports lipids, implying that MFSD2A harbors unique structural features and transport mechanism. Here, we present three three-dimensional structural models of human MFSD2A derived by homology modeling using MelB- and LacY-based crystal structures and refined by biochemical analysis. All models revealed 12 transmembrane helices and connecting loops and represented the partially outward-open, outward-partially occluded, and inward-open states of the transport cycle. In addition to a conserved sodium-binding site, three unique structural features were identified as follows: a phosphate headgroup binding site, a hydrophobic cleft to accommodate a hydrophobic hydrocarbon tail, and three sets of ionic locks that stabilize the outward-open conformation. Ligand docking studies and biochemical assays identified Lys-436 as a key residue for transport. It is seen forming a salt bridge with the negative charge on the phosphate headgroup. Importantly, MFSD2A transported structurally related acylcarnitines but not a lysolipid without a negative charge, demonstrating the necessity of a negatively charged headgroup interaction with Lys-436 for transport. These findings support a novel transport mechanism by which lysophosphatidylcholines are “flipped” within the transporter cavity by pivoting about Lys-436 leading to net transport from the outer to the inner leaflet of the plasma membrane.
Keywords: brain metabolism, drug transport, lysophospholipid, membrane protein, polyunsaturated fatty acid (PUFA), X-ray crystallography, blood-brain barrier, docosahexaenoic acid, membrane transporter, microcephaly
Introduction
Brain and eye contain membrane phospholipids that are enriched in the ω-3 fatty acid docosohexaenoic acid (DHA).3 It is widely accepted that DHA is important for brain and eye function and brain development (1, 2), although mechanisms for DHA function in these tissues are not well defined. Nonetheless, despite the high level of DHA enrichment in phospholipids of brain and eye, these organs do not synthesize DHA but must import DHA from the blood. The mechanism by which DHA and other conditionally essential and essential fatty acids cross the blood-brain barrier has been a long-standing mystery. Recently, we identified major facilitator Superfamily domain containing 2a (MFSD2A, also known as NLS1) as the primary transporter by which the brain obtains DHA. Importantly, MFSD2A does not transport unesterified DHA but transports DHA in the chemical form of lysophosphatidylcholine (LPC) that is synthesized by the liver and circulate largely on albumin (3). This is consistent with biochemical evidence that the brain does not transport unesterified fatty acids (4) and that LPC is the preferred carrier of DHA to the brain (5, 6).
MFSD2A is a sodium-dependent transporter that is part of the major facilitator superfamily (MFS) of proteins. Members of this family with elucidated structures have 12 transmembrane domains composed of two evolutionarily duplicated six transmembrane units (7). Transporting an LPC is a unique feature of MFSD2A, because most members of this family transport water-soluble and minimally polar substrates such as sugars (GLUT, MelB, and LacY) and amino acids (TAT1). MFSD2A transport is not limited to LPCs containing DHA, as it can transport LPCs containing a variety of fatty acyl chains, with a higher specificity for LPCs with unsaturated fatty acyl chains with a minimum chain length of 14 carbons (6, 8). Crystal structures have been solved for more than a dozen members of the MFS family, with more than 19 structures, including that of melibiose permease (MelB) of Salmonella typhimurium (9), lactose permease (LacY) of Escherichia coli (10), glycerol 3-phosphate transporter of E. coli (11), and the mammalian glucose transporters 1, 3, and 5 (GLUT1, GLUT3, and GLUT5) (12–14). A common transport mechanism has emerged from both biochemical and structural analyses of MFSs, in which they transport via a rocker-switch, alternating access mechanism (7, 15). In the rocker-switch model, rigid-body relative motion of the N- and C-terminal domains renders the substrate-binding site alternatively accessible from either side of the membrane.
MFSD2A is highly expressed at the blood-brain barrier in both mouse and human (6, 16). Mfsd2a-deficient mice (KO) have significantly reduced brain DHA as a result of a 90% reduction in brain uptake of LPC containing DHA as well as other LPCs. The most prominent phenotype of Mfsd2a KO mice is microcephaly, and KO mice additionally exhibit motor dysfunction and behavioral disorders, including anxiety and memory and learning deficits (6). In line with the mouse KO phenotypes, human patients with partially or completely inactivating mutations in MFSD2A presented with severe microcephaly, intellectual disability, and motor dysfunction (8, 16). Plasma LPCs are significantly elevated in both KO mice and human patients with MFSD2A mutations, consistent with reduced uptake at the blood-brain barrier. Taken together, these findings demonstrate that LPCs are essential for normal brain development and function in mouse and humans.
The fact that MFSD2A transports a lysolipid, a non-canonical substrate for an MFS protein, might indicate unique structure features and a novel transport mechanism. However, no structural information or mechanism of transport of MFSD2A is known. Human MFSD2A is composed of 530 amino acids, with two glycosylation sites at Asn-217 and Asn-227. MFSD2A is evolutionarily conserved from teleost fish to humans. Although not a functional ortholog of bacterial MFS transporters, MFSD2A shares 25 and 26% amino acid sequence identity with S. typhimurium MelB (9, 17), and LacY from E. coli (10), respectively. Given the high conservation of the MFS fold, the use of homology modeling to gain insight into the structure of S. typhimurium MelB, for example, has proven to be highly accurate and largely consistent with subsequent x-ray crystal data (9, 18). Here, we take advantage of two recently derived high resolution x-ray crystal structures of S. typhimurium MelB (9) and a high resolution x-ray crystal structure of LacY (10) to generate three predictive structural models of human MFSD2A. These models reveal the following three unique regions critical for function: an LPC headgroup binding site, a hydrophobic cleft occupied by the LPC fatty acyl tail, and three sets of ionic locks. These structural features indicate a novel mechanism of transport for LPCs.
Experimental Procedures
Reagents and Buffers
Non-radiolabeled lysophosphatidylcholines were purchased from Avanti Polar Lipids. Non-radiolabeled acylcarnitines and cetyltrimethylammonium bromide were purchased from Sigma. Radiolabeled 1-palmitoyl 2-lysophosphocholine (LPC [3H]palmitate), 1-oleoyl 2-lysophosphocholine (LPC [14C]oleate), [14C]CTAB, [14C]octanoylcarnitine, [3H]palmitoylcarnitine, and [3H]acetylcarnitine were purchased from American Radiolabeled Chemicals.
Lysophospholipids received in chloroform (non-labeled) or ethanol/toluene (radiolabeled) were completely dried under a nitrogen gas stream and were solubilized in 12% fatty acid-free BSA (Sigma), which was dissolved in 150 mm NaCl. To prepare the LPC [14C]oleate and LPC oleate mixture, 25 μCi of LPC [14C]oleate (specific activity 55 mCi mmol−1) were dried and dissolved in 3 ml of 20 mm non-labeled LPC oleate/BSA. The LPC [3H]palmitate and LPC palmitate mixture was prepared by dissolving 25 μCi of LPC [3H]palmitate (specific activity 60 Ci mmol−1) in 4 ml of 20 mm LPC palmitate. The [14C]CTAB and CTAB mixture were prepared by dissolving 10 μCi of [14C]CTAB (specific activity 56 mCi mmol−1) in 1 ml of 10 mm CTAB. The [3H]acetylcarnitine and acetylcarnitine mixture was prepared by dissolving 50 μCi of [3H]acetylcarnitine (specific activity 80 Ci mmol−1) in 4 ml of 104 mm acetylcarnitine. The [14C]octanoylcarnitine and octanoylcarnitine mixture were prepared by dissolving 25 μCi of [14C]octanoylcarnitine (specific activity 50 mCi mmol−1) in 4 ml of 74 mm octanoylcarnitine. The [3H]palmitoylcarnitine and palmitoylcarnitine mixture were prepared by dissolving 25 μCi of [3H]palmitoylcarnitine (specific activity 60 Ci mmol−1) in 4 ml of 10 mm palmitoylcarnitine.
Three-dimensional Structure Prediction
The amino acid sequence of human MFSD2A was retrieved from the UNIPROT database (Q8NA29). Homology models of MFSD2A were generated in four steps. First, the primary sequence of MFSD2A was submitted to the PSI-BLAST server at NCBI to search for suitable template structures (19). MelB from S. typhimurium (Protein Data Bank code 4M64) is the most closely related sequence of known structure, sharing a 25% sequence identity to MFSD2A. In that crystal structure MelB was determined in two conformations representing an outward-partially occluded (chain A) and a partially outward-open (chain B) state of MelB. The crystal structures of both chains were used as templates. Besides MelB, another MFS transporter, LacY from E. coli, shares 18% sequence identity to MFSD2A. The crystal structure of LacY determined in the inward-open state (Protein Data Bank code 2V8N) was also used as a template. Second, a sequence alignment between MFSD2A and each of the three templates was computed by MUSCLE (Multiple Sequence Comparison by Log Expectation) (20). Third, a total of 500 homology models were generated with the standard “automodel” class in MODELLER on the basis of each template (21). Finally, each set of 500 models was evaluated by the discrete optimized protein energy score, and the best ranking model was selected (22), resulting in three models in total.
Structural Validation
In two initial models of MFSD2A based on MelB crystal structure, three polar side chains (Ser-339, Thr-341, and Thr-343) on helix VIII face the membrane exterior. To make these side chains point toward the internal cavity, we manually rotated helix VIII in these two outward-open models, relaxed it and neighboring helices using MODELLER, and optimized side chains in these helices using PLOP (23), thereby creating refined models of the partial outward-open and outward-partially occluded states.
Next, we generated a chemical library of 25 molecules for docking validation of these five MFSD2A models. This library contains seven known substrates of MFSD2A, including LPC, LPE, lysoplasmalogen, lyso-platelet-activating factor, platelet-activating factor, lysosphingomyelin, and miltefosine (6) and substrate analogs that are known or predicted not to be transported by MFSD2A. Docking validation was then performed with DOCK version 3.6 (24) to evaluate the initial and refined models for their ability to recognize the known substrates. Each docked compound was scored and ranked by the docking energy function that is the sum of van der Waals, Poisson-Boltzmann electrostatic, and ligand desolvation penalty terms.
Transport Assays
Transport assays were performed using HEK293 cells as described previously (6). Briefly, plasmids encoding wild-type or mutant human MFSD2A were transfected into HEK293 cells. Uptake assays of radiolabeled ligands, including LPC-[14C]DHA, LPC-[14C]oleate, LPC-[3H]palmitate, [3H]palmitoylcarnitine, [14C]octanoylcarnitine, and [3H]acetylcarnitine, were performed after 18 h of transfection. Radiolabeled ligands were diluted in the transport buffer (5 mm KCl, 10 mm Hepes, pH 7.4, 150 mm NaCl). Uptake activity is expressed in disintegrations/min per well. We found that acylcarnitines were toxic to cells; hence, we reduced the incubation time for experiments involving acylcarnitines to 20 min instead of the usual 30 min.
Immunoblotting and Immunofluorescence Microscopy
Immunoblotting and immunofluorescence microscopy for MFSD2A were carried out as described previously using a rabbit polyclonal antibody against MFSD2A in HEK293 cells (6).
Mutagenesis of MFSD2A
Human MFSD2A in pSport6 (Open Biosystems) was amplified using primers hMFSD2ABamHI and hMFSD2AXbaI and cloned into pcDNA3.1 in the BamHI and XbaI sites. Mutagenesis of various sites in hMFSD2A was carried out by PCR. Next, the mutated cDNA was cloned into pcDNA3.1 and sequenced to verify the mutation.
Results
Overall Architecture of Human MFSD2A
A number of atomic resolution x-ray structures of MFS protein family members have been published. As noted previously by Guan and co-workers (9), S. typhimurium MelB exhibits ∼54% similarity with human MFSD2A. Specifically, S. typhimurium MelB exhibits 26% sequence identity with MFSD2A, and E. coli LacY exhibits 25% sequence identity with MFSD2A. Using the three published atomic resolution x-ray structures of S. typhimurium MelB and E. coli LacY as structural templates, we constructed three initial models of MFSD2A (Fig. 1, A and B) by homology modeling using the program MODELLER (25). The available high resolution structures of S. typhimurium MelB and E. coli LacY that were used to generate human MFSD2A models represent the protein in different states of the transport cycle, with the ligand-binding site partially opened toward the extracellular surface (MelB Mol-B) (9), with the ligand-binding site open toward the extracellular surface but partially occluded (MelB Mol-A) (9), and with the ligand-binding site open toward the inner surface (LacY) (10). The models were refined using the Protein Local Optimization Program (PLOP). PLOP is a refinement program that optimizes side chains, loop prediction, and prediction of helix positions and orientation by a combination of algorithms based on energy minimization (26). The models were also refined using the program, Side Chain with Rotamer Library, which predicts side chain conformations by sampling a backbone-based rotamer library and taking into account hydrogen bonding and van der Waals interactions (27).
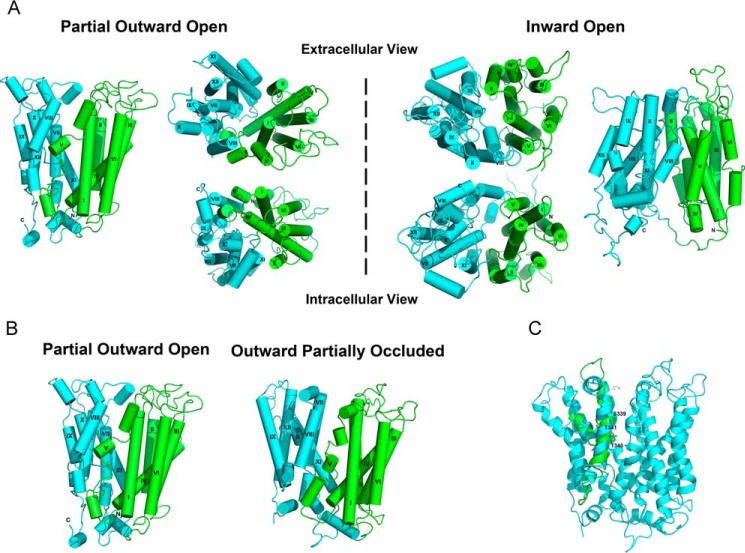
FIGURE 1.
Homology models of MFSD2A. The N- and C-terminal domains are shown in green and cyan, respectively. The helices are labeled with roman numerals. A, overall structure of MFSD2A in the partial outward-open (left of dashed line) and inward-open states (right of dashed line). B, overall structure of MFSD2A in the partial outward-open and outward-partially occluded states. C, rotation of helix VIII in the outward-partially occluded model of MFSD2A to achieve a more likely alignment of the polar side chains of Ser-339, Thr-341, and Thr-343 (labeled in the refined model) toward the protein interior and away from the hydrophobic core of the lipid bilayer. We manually rotated helix VIII in the outward-partially occluded model of MFSD2A. The refined model is shown in cyan and the initial model is shown in green.
Close examination of these homology models showed that the register of residues in helix VIII needed to be modified by half a turn such that three juxtaposed polar side chains (Ser-339, Thr-341, and Thr-343) would project toward the protein interior and not the hydrophobic core of the lipid bilayer. Therefore, manual adjustments were performed (Fig. 1C) as described under “Experimental Procedures.” The structural model was further refined and tested using site-directed mutagenesis and biochemical analyses described below to produce the final refined models presented here.
MFSD2A has an overall structure that is similar to S. typhimurium MelB and E. coli LacY. Viewed parallel to the membrane, MFSD2A has a heart shape. Viewed from the cytoplasmic or extracellular surfaces, it has an oval shape (Fig. 1A). Two of the models were built from two available crystal structures of S. typhimurium MelB, representing the transporter in the partial outward and outward-partially occluded states. Comparing these two models, we found that they are similar in their global fold, but in particular helix V is broken into four segments in the outward-partially occluded state, relative to three segments in the partial outward state (Fig. 1B). Like many other MFS members, MFSD2A is made up of two six-helix bundles connected by a central cytoplasmic loop. The interface of the two halves form an internal cavity lined by helices. Studying the charge distribution of MFSD2A, there is a concentration of charges at the top and bottom surfaces of the protein, where interaction with the hydrophobic membrane core is minimized and interaction with the hydrophilic intracellular or extracellular environments is maximized. There is a particular concentration of positively charged residues on the cytoplasmic side (Fig. 2), which supports the positive inside rule that controls the transporter's membrane topology (28). Moreover, there are only four charged residues within the central cavity, namely Asp-90, Asp-93, andAsp-97 on helix II and Lys-436 on helix XI, that lie midway along the transmembrane helices.
FIGURE 2.
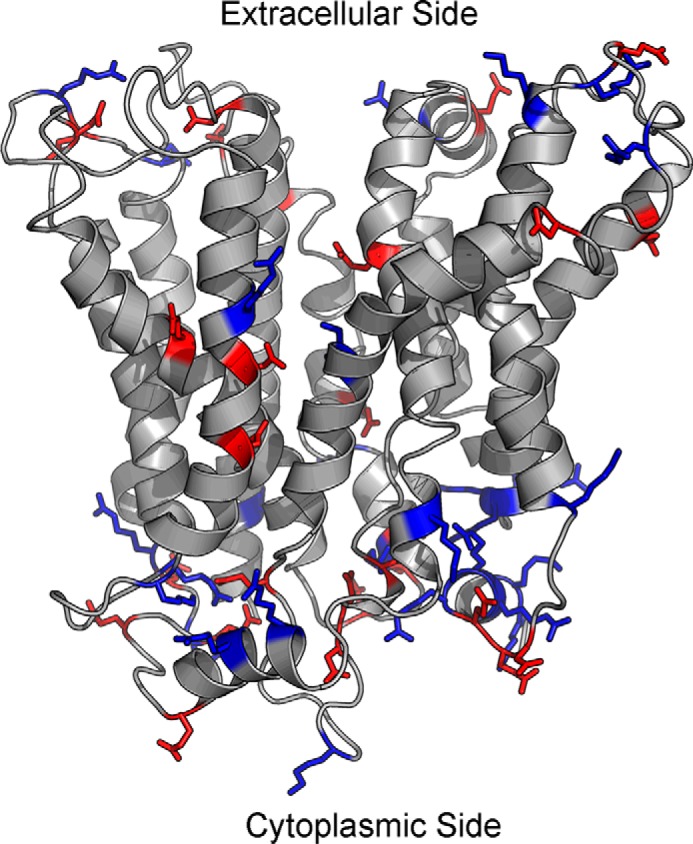
Distribution of positive and negative charges in the outward-partially occluded model of MFSD2A. Majority of the charged residues are located at the top and bottom surfaces of the protein, where there is minimal contact with the hydrophobic lipid bilayer, and maximal contact with the hydrophilic extracellular or intracellular environment. There are more positively charged residues on the cytoplasmic surface of MFSD2A. Blue, positively charged residues; red, negatively charged residues.
Taken together, the three models are consistent with a rocker-switch mechanism. The N-terminal domain remains rigid and rotates between the outward-open and inward-open states around a centrally located translocation pathway. The C-terminal domain undergoes a greater degree of local rearrangements, especially in transmembrane helix XI, as access to the substrate-binding site switches from the outer surface to the inner surface (Fig. 1A). Switching from the outward-open to the inward-open conformation, helix XI undergoes a helix break and the outer segment shifts inward to occlude access to the translocation pathway. To a smaller degree, helices I–IV similarly shift inward on the outer membrane surface to occlude access to the translocation pathway. Because the largest structural differences are seen between the partial outward-open and inward-open models, we will mainly focus the remainder of these studies on these two structurally distinct states.
Sodium-binding Site
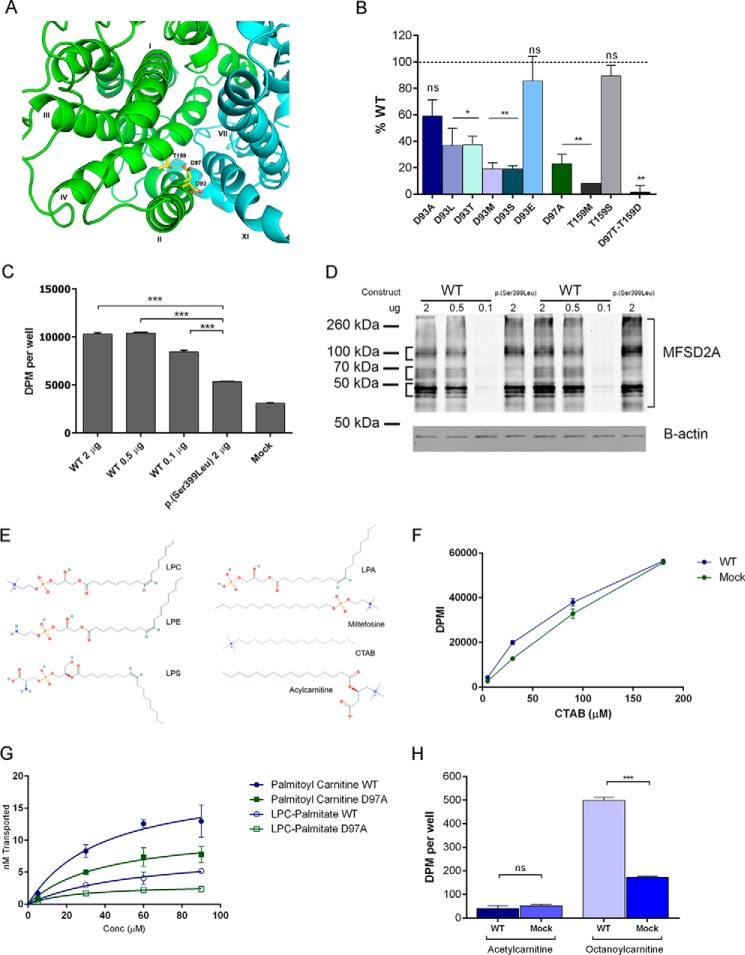
Both MelB and MFSD2A utilize sodium electrochemical gradients to drive the uphill translocation of substrates. The sodium-binding site has been well characterized in MelB (29–36). Of the cluster of residues identified, three are conserved in human MFSD2A, Asp-93, Asp-97, and Thr-159 (Fig. 3A). We recently found Thr-159 to be mutated in a consanguineous family in Libya, and this p.(T159L) inactivating mutation resulted in microcephaly, spastic quadriparesis, and death within the first few years of life (16). The MelB structures indicate that the orthologous Thr-121 forms a hydrogen bond with the orthologous Asp-59 and is essential for MelB transport (9). We interrogated the sodium-binding site in MFSD2A by mutating these three conserved residues. We found that all three residues are critical for normal MFSD2A function, and individually mutating them results in loss of transport activity as determined by a radiolabeled ligand transport assay we previously established to measure LPC transport (6) (Fig. 3B). In this transport assay, the rate of cellular uptake of radiolabeled ligand is measured in cells transfected with plasmids expressing MFSD2A mutants relative to wild-type MFSD2A. A titration of the expression levels of MFSD2A showed that expression levels of MFSD2A in this assay are in great excess such that only titration down to a very low level of expression (0.1 μg of transfected DNA) resulted in a significant decrease in transport activity, likely the result of fewer cells being transfected (Fig. 3, C and D). Therefore, the variations in MFSD2A expression observed by Western blot analysis for all tested mutant MFSD2A constructs had no impact on the rate of substrate transport (supplemental Fig. S1). Additionally, we demonstrated that key mutants with decreased transport function had normal cell surface localization (supplemental Fig. S2).
FIGURE 3.
Sodium-binding site of MFSD2A and the structural requirements for transport. A, sodium-binding site of MFSD2A viewed from the extracellular side of the partial outward-open model with the N- and C-terminal halves in green and cyan, respectively. B, transport of LPC-[14C]oleate after 30 min by HEK293 cells transiently expressing MFSD2A constructs with indicated mutations in residues in the sodium-binding site. Data are expressed as percent activity relative to wild-type MFSD2A. C, transport of LPC-[14C]oleate after 30 min following transfection of HEK293 cells with indicated quantities of WT MFSD2A or S339L plasmids. Mock and S339L transfected cells served as background control and negative control, respectively. D, MFSD2A expression levels in response to concentration of DNA plasmid transfected into HEK293 cells. HEK293 cells were transfected with indicated quantities of plasmid. 18 h after transfection, cells were harvested in RIPA buffer containing protease inhibitor. Western blot was conducted, probing for expression of MFSD2A, and of B-actin as a loading control. S339L was used as a positive control. E, structures of lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), lysophosphatidylserine (LPS), lysophosphatidic acid (LPA), miltefosine, cetyltrimethylammonium bromide (CTAB), and acylcarnitine. F, concentration-dependent transport of [14C]CTAB after 30 min by wild-type MFSD2A. Mock transfected cells served as a negative control. G, comparison of transport capacity of MFSD2A for LPC-[3H]palmitate and [3H]palmitoylcarnitine. H, transport of 100 μm [3H]acetylcarnitine and [14C]octanoylcarnitine after 20 min by wild-type MFSD2A and mock expressing HEK293 cells. Experiments in B, C, D, and F–H were performed twice in triplicate. Data in B, C, and F–H are expressed as mean ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
Of the sodium-binding site mutants tested, the only mutations that are well tolerated are T159S and D93E. These results indicated that the functional groups important for sodium binding are the hydroxyl group and carboxylic acid group on Thr-159 and Asp-93, respectively. These findings are in agreement with their respective proposed roles of hydrogen bonding and sodium binding (9).
Refined Rules for LPC Transport
Although the overall structure of MFSD2A is similar to other members of the MFS family, MFSD2A is likely unique in its substrate binding pocket because it transports a polar lipid. Most MFS proteins transport hydrophilic substrates such as amino acids and sugars. However, MFSD2A transports LPCs, which have a zwitterionic phosphocholine headgroup, and a hydrophobic hydrocarbon tail having a minimum of 14 carbons. We previously determined that a phosphocholine headgroup, but not the glycerol backbone and carbonyl group, is an essential chemical feature for substrate transport by MFSD2A (6). For example, lysolipid-like molecules such as miltefosine, which has an 18 carbon alkane chain attached to a phosphocholine headgroup, can compete for LPC transport (6). In addition to LPC, MFSD2A could transport LPE and lysophosphatidylserine, but not lysophosphatidic acid (6), indicating specific headgroup structure requirements for transport. Common to LPC, LPE, and lysophosphatidylserine is a zwitterionic headgroup (Fig. 3E), with a negatively charged phosphate group and a positively charged amine group. An isolated negatively charged phosphate group is insufficient for transport, as illustrated by the inability of lysophosphatidic acid to compete for LPC transport (6).
Based on these findings it is not known whether the phosphate group is obligate for transport or more simply a negative charge is critical. To first address these questions, we tested transport of CTAB. CTAB is a lysolipid-like compound similar to miltefosine, but lacking a negatively charged headgroup (Fig. 3E). Importantly, MFSD2A was unable to transport CTAB (Fig. 3F), demonstrating that an isolated positively charged trimethylamine is insufficient for transport, definitively indicating the obligate nature of a zwitterionic headgroup for transport. To determine whether the LPC phosphate group is a required ligand feature or can be replaced by a negative charge carried by a carboxyl group, we tested the ability of MFSD2A to transport acylcarnitines. Acylcarnitines have a similar structure to LPC, with a four-carbon backbone, a negatively charged carboxyl group, a positively charged trimethylamine group (at neutral pH), and a fatty acyl chain esterified to the hydroxyl group at carbon 3 (Fig. 3E). The negatively charged carboxyl group, but absent phosphate group, will allow us to test whether a negatively charged headgroup is required for transport. Human MFSD2A was able to transport palmitoylcarnitine having a 16-carbon acyl chain (Fig. 3G) and octanoylcarnitine (eight carbon acyl chain) but not acetylcarnitine (two carbon acyl chain) (Fig. 3H). These findings are consistent with the requirement of a minimal degree of hydrophobicity for ligand transport (6, 37). In addition, these data indicate that a negative charge in the headgroup is required, but there are no structural or spatial requirements for a phosphate group.
Because MFSD2A could transport acylcarnitines, we tested the relative transport capacity of palmitoylcarnitine and LPC-palmitate. Interestingly, MFSD2A was found to have a higher transport capacity for palmitoylcarnitine relative to LPC-palmitate (Fig. 3G). Acylcarnitines are found at low micromolar concentrations in human plasma (5 μm), 2 orders of magnitude lower than LPCs. Furthermore, the pool of acylcarnitines in human plasma is primarily composed of acetylcarnitine, which is not transported by MFSD2A (5, 8, 16, 38). These findings make it unlikely that long-chain acylcarnitines represent a physiological ligand for MFSD2A.
LPC Headgroup Binding Site
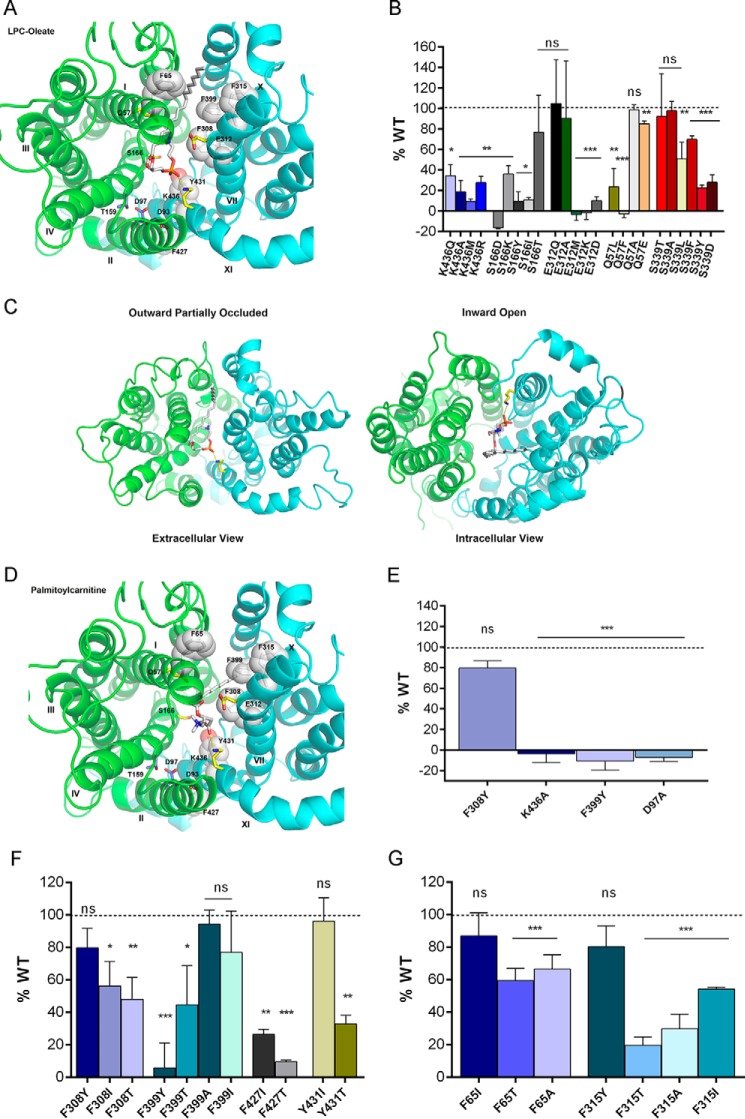
Based on our homology structural models, 13 polar or charged side chains project into the central cavity, bound by the two six-transmembrane subunits, that likely delineates the translocation pathway (Fig. 4A). We hypothesized that these residues are candidates for interactions with the zwitterionic headgroup of LPC. Of these residues, Lys-436, Ser-166, and Glu-312 were found to be crucial for normal transport (Fig. 4B). Altering the charge or size of side chains at these positions reduced or abolished transport activity of MFSD2A (Fig. 4B), without affecting protein cell surface localization and expression levels (supplemental Fig. S1 and S2). As mentioned earlier, we manually rotated helix VIII to achieve an expected alignment of three juxtaposed polar side chains away from the hydrophobic lipid bilayer. To validate this resultant refined outward-partially occluded model, we interrogated the abilities of this model and the model without this rotated helix VIII to dock and rank a set of 25 molecules using the program DOCK 3.6. This set of molecules included known physiological ligands of MFSD2A, known non-physiological ligands, and molecules predicted not to be transported by MFSD2A because of insufficiently long hydrophobic fatty acyl chains (supplemental Fig. S3). Ranking was based on the DOCK score of various docking poses of the ligands, which is a summation of van der Waals, Poisson-Boltzmann electrostatic, and ligand desolvation penalty terms (24, 39). Although both the initial and refined models were able to discriminate between ligands and non-ligands, only the refined model was able to rank physiological ligands above non-physiological ligands, thus discerning physiological from non-physiological ligands (supplemental Fig. S3).
FIGURE 4.
Headgroup binding site and hydrophobic cleft of MFSD2A. A, headgroup binding site (yellow sticks) and hydrophobic cleft (white sticks and spheres) viewed from the extracellular side of the outward-partially occluded model. LPC-oleate is shown docked. B, transport of LPC-[14C]oleate after 30 min by HEK293 cells transiently expressing MFSD2A constructs with indicated mutations in residues proposed to be involved in headgroup binding. C, docking of LPC-oleate in the outward-partially occluded and inward-open models. In both models, the negatively charged phosphate group remains in close proximity to the positively charged side chain of Lys-436. In the outward-partially occluded model, the side chain of Lys-436 is pointing outward, and LPC-oleate lies within the translocation pathway with its fatty acyl chain projecting toward the extracellular surface of MFSD2A. In the inward-open model, the side chain of Lys-436 is pointing downwards, whereas LPC-oleate lies within the translocation pathway with its fatty acyl chain projecting toward the cytoplasmic surface of MFSD2A. D, palmitoylcarnitine shown docked as in A, indicating similar residue interactions as seen with LPC-oleate docking. E, transport of [3H]palmitoylcarnitine after 20 min by HEK293 cells transiently expressing MFSD2A constructs with indicated mutations. F and G, transport of LPC-[14C]oleate after 30 min by HEK293 cells transiently expressing MFSD2A constructs with indicated mutations in residues in the proposed hydrophobic cleft. Experiments in B and E–G were performed twice in triplicate. Data in B and E–G are expressed as the mean ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
Docking of the physiological substrate LPC-oleate in the outward-partially occluded model revealed that the negatively charged phosphate group was in close proximity to the positively charged side chain of Lys-436 (Fig. 4A). Mutation of Lys-436 to non-polar residues (Ala and Met) or polar side chain (Glu) or even Arg that has a positive charge but a different size and conformation resulted in complete loss of function (Fig. 4B). These data indicated that the specific positioning and positive charge of the side chain of Lys-436 is critical for transport function and points to this residue as likely forming a salt bridge with the negatively charged phosphate group of LPC. Comparing the docking positions of LPC-oleate and the orientation of Lys-436 in the outward-partially occluded and inward-open models, interaction between Lys-436 and the headgroup was maintained in both models. However, Lys-436 is oppositely oriented in the inward-open model pointing downward in the transport cleft and the fatty acyl tail of the LPC ligand appears flipped in the hydrophobic cleft pointing toward the intracellular surface (Fig. 4C). These findings suggest that Lys-436 acts as a pivot about which the LPC molecule was flipped from one membrane to the other. Similar to LPC-oleate, docking of palmitoylcarnitine indicated that its carboxyl group is in close proximity to Lys-436 (Fig. 4D). Consistent with this structural model, mutation of Lys-436 also abolished transport of palmitoylcarnitine (Fig. 4E), supporting a likely role of Lys-436 in binding to the negatively charged LPC headgroup.
In docking studies, both Gln-57 and Glu-312 were noted to flank the positively charged trimethylamine group of the substrate headgroup of LPC-oleate (Fig. 4A). However, mutational analysis of these two residues was not conclusive (Fig. 4B), suggesting that the stabilizing interactions of these residues with the substrate are either redundant or that the only requirement for these side chains is that they are sterically unobtrusive.
Interestingly, mutation of Ser-166 is disease-causing in humans. p.(S166L) is an inactivating mutation found in a consanguineous family in Egypt that causes a lethal microcephaly syndrome (16). Indeed, mutation of Ser-166 to residues with uncharged, positively charged, negatively charged, or hydrophobic side chains resulted in abolished transport activity (Fig. 4B). The only mutation that is well tolerated is S166T, indicating the importance of a hydroxyl group on the amino acid side chain. However, based on the docking analysis with the current available structures, Ser-166 is not visualized to be in close proximity with the ligand headgroup in the outward-open conformation. A third, partially inactivating, mutation was found in residue Ser-339. p.(S339L) was found in an extended consanguineous family in Pakistan to cause a non-lethal microcephaly syndrome (8). In these homology models, the side chain of Ser-339 also projects inward toward the translocation pathway. Mutation of this residue presented a mixed picture. Conversion to Thr or Ala is well tolerated. However, a mutation to Leu, Asp, Tyr, or Phe causes a reduction in transport function (Fig. 4B). Similarly, docking studies did not reveal an interaction of Ser-339 with the ligand headgroup in the outward-partially occluded conformation. Our models are built on three crystal structures, which represent only three states in the transport cycle. With this limitation, we cannot rule out the possibility that Ser-166 or Ser-339 interacts with the ligand headgroup in a different transport state. Another possible role of these residues is in helix packing in specific transport states.
Hydrophobic Cleft for Acyl Chain Interactions
A unique feature of the physiological MFSD2A ligand, LPC, is a hydrophobic hydrocarbon tail. Consistent with transporting a lysolipid, we have identified a “greasy patch” of bulky hydrophobic amino acids that extends through the central cavity of MFSD2A, forming what we termed a hydrophobic cleft (Fig. 4A). We hypothesize that this cleft stabilizes the fatty acyl tail, allowing it to “slide” unhindered through the transporter along with headgroup movement. Mutational analysis showed that the hydrophobicity of residues Phe-308, Phe-399, Phe-427, and Tyr-431 is critical for transport activity of MFSD2A (Fig. 4F). Subtle mutation of Phe-399 into a tyrosine, which only has an additional polar hydroxyl group, abolished transport activity of MFSD2A. Mutation of Phe-308, Phe-399, Phe-427, or Tyr-431 to the polar residue Thr also reduced transport activity of MFSD2A (Fig. 4F). For Phe-308 and Phe-427, even mutation to isoleucine, which has a hydrophobic but smaller side chain, resulted in decreased transport activity (Fig. 4F). Based on docking analysis of LPC in the outward-partially occluded model, the fatty acyl tail lies in close proximity to Phe-65 and Phe-315. Interrogating these residues, we found that mutation of Phe-65 into a polar Thr or the small non-polar residue Ala reduced transport activity, but a mutation to a hydrophobic isoleucine does not affect transport activity (Fig. 4G). Mutation of Phe-315 to Thr, Ala, or Ile reduced transport activity (Fig. 4G), but a mutation to tyrosine is well tolerated. Identical hydrophobic amino acid side chain interactions with the acyl chain of palmitoylcarnitine and the requirement for transport of palmitoylcarnitine were noted (Fig. 4, D and E), consistent with the importance of the hydrophobic cleft in ligand transport.
By volumetric analysis, we determined that the proposed translocation pocket has a volume of 3680 and 2016 Å3 in the outward-open and inward-open conformations, respectively. LPCs occupy a volume of ∼500 Å3, depending on the specific side chain (i.e. 16:0, 495 Å3, 18:1, 445 Å3, and 20:5, 520 Å3). These calculations indicate that the hydrophobic pocket is much larger in volume than required to accommodate LPCs. In total, these data indicate that a hydrophobic cleft is critical for transporting LPCs, but they do not point to a specific interaction with the LPC-acyl chain and residues within the hydrophobic cleft that would explain higher affinity for transporting LPCs with polyunsaturated acyl chains, such as DHA, over LPCs with saturated acyl chains, such as palmitate (8). Sequence alignment and superimposition of the outward-partially occluded structure of MFSD2A and MelB-MolA indicate the uniqueness of the hydrophobic ligand binding cleft of Msfd2a on an otherwise conserved MFS fold (supplemental Fig. S4, A and B).
Ionic Locks and a Charged C-terminal Helix
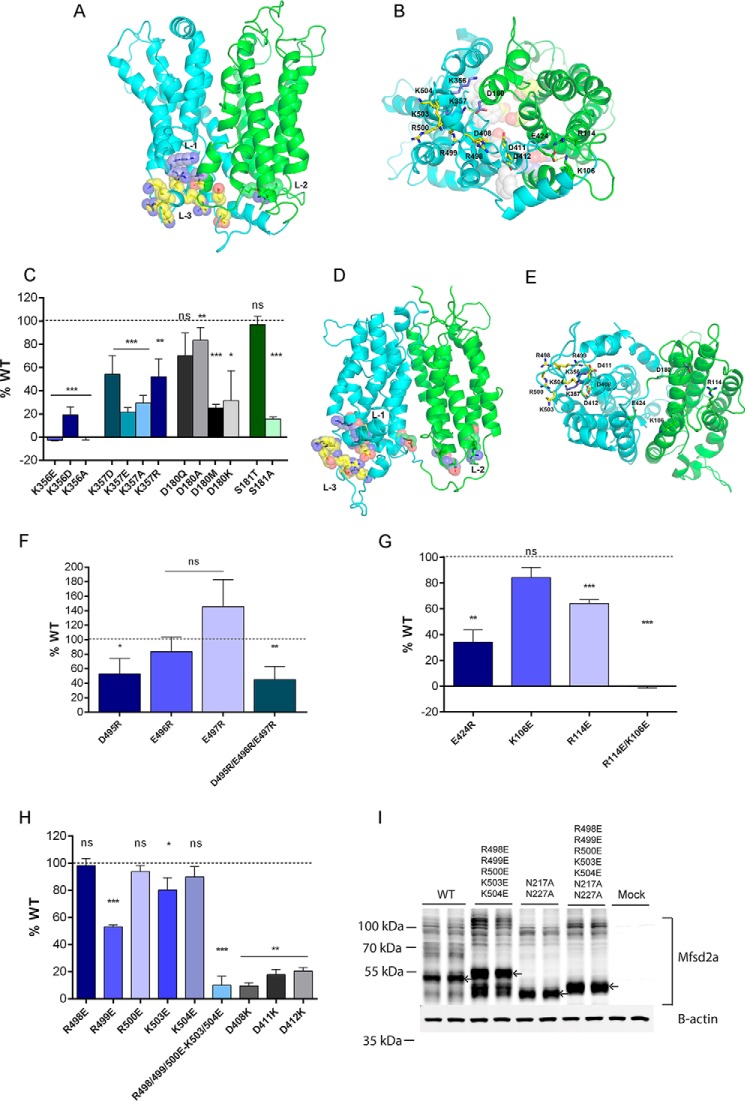
The structural models of MelB have revealed the existence of inter-domain electrostatic interactions, termed ionic locks, proposed to stabilize specific conformational states of the transport cycle (9). This feature is also found in several other MFS transporters (9, 14). Examining the human MFSD2A models for the existence of similar inter-domain ionic side chain interactions, we identified two clusters of oppositely charged residues residing separately on the N- and C-terminal domains that are in close proximity in the outward-partially occluded model but far apart in the inward-open model. At the first ionic lock (L-1), Lys-356 and Lys-357 on the C-terminal domain are clustered with Asp-180 and Ser-181 located on the N-terminal domain (Fig. 5, A and B). Mutational analysis of these residues revealed that these specific residues are critical for normal transport function (Fig. 5C). Mutating either Lys-356 or Lys-357 to Asp, Glu, or Ala resulted in loss of transport function (Fig. 5C). Mutating Asp-180 to Met, Lys, or Ala reduced transport function, but mutation to Gln was well tolerated (Fig. 5C). Mutating Ser-181 to Ala abolished transport activity but replacement of the residue with Thr, which similarly has a hydroxyl group, is well tolerated (Fig. 5C). In the inward-open model, Lys-356 and Lys-357 are situated at a distance from Asp-180 (Fig. 5, D and E). Interestingly, in this conformation Lys-356 and Lys-357 are instead clustered with the C-terminal residues Asp-495, Glu-496, and Glu-497. Mutational analysis of these three residues individually indicated that D495R mutation resulted in a significant reduction in transport, whereas no significant reduction in transport was found for E496R or E497R (Fig. 5F). Combined mutation of all three residues resulted in a similar inhibition of transport as observed for D495R, indicating that this residue is critical for transport (Fig. 5F) and that multiple redundant ionic interactions among these three negatively charged side chains with Lys-356 and Lys-357 are likely necessary for stabilizing the inward-open conformation.
FIGURE 5.
Ionic locks. A, N- and C-terminal domains are shown in green and cyan, respectively. Three sets of ionic locks are represented as spheres, with lock 1 (L-1) in purple, lock 2 (L-2) in green, and lock 3 (L-3) in yellow in the outward-partially occluded model. B, three sets of ionic locks are shown as sticks, as viewed from the cytoplasmic surface of the outward-partially occluded model, with L-1 in purple, L-2 in green, and L-3 in yellow. C, transport of LPC-[14C]oleate after 30 min by HEK293 cells transiently expressing MFSD2A constructs with indicated mutations in L-1 residues. D, N- and C-terminal domains are shown in green and cyan, respectively. Three sets of ionic locks are represented as spheres, with lock 1 (L-1) in purple, lock 2 (L-2) in green, and lock 3 (L-3) in yellow in the inward-open model. E, three sets of ionic locks are shown as sticks, as viewed from the cytoplasmic surface of the inward-open model, with L-1 in purple, L-2 in green, and L-3 in yellow. In the inward-open model, the residues involved in locks L-1, L-2, and L-3 are not in close proximity as they were in the outward-partially occluded model. F, mutational analysis of residues of L-1. Transport of LPC-[14C]oleate after 30 min by HEK293 cells transiently transfected with MFSD2A constructs with indicated mutations in residues involved in L-1. G, transport of LPC-[14C]oleate after 30 min by HEK293 cells transiently expressing MFSD2A constructs with indicated mutations in L-2 residues. H, transport of LPC-[14C]oleate after 30 min by HEK293 cells transiently expressing MFSD2A constructs with indicated mutations in L-3 residues. I, Western blot analysis of MFSD2A mutants and wild-type MFSD2A indicating a mobility shift in the R498E/R499E/R500E/K503E/K504E mutant relative to wild-type MFSD2A. R498E/R499E/R500E/K503E/K504E mutant with additionally mutated glycosylation sites, N217A/N227A, exhibited a mobility shift relative to non-glycosylated MFSD2A. Arrows indicate MFSD2A bands that have shifted. Experiments in C and F–I were performed twice in triplicate. Data in C and F–H are expressed as the mean ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
At the second ionic lock (L-2), the C-terminal domain residue Glu-424 and N-terminal domain residues Lys-106 and Arg-114 are arranged in a tight cluster in the outward-partially occluded model (Fig. 5, A and B). Replacing Glu-424 with Lys significantly reduced transport activity (Fig. 5G). Mutating Lys-106 or Arg-114 individually reduced transport activity to a lesser extent then E424R, but mutating both Lys-106 and Arg-114 abolished transport activity (Fig. 5G), suggesting that Glu-424 interacts with Lys-106 and Arg-114 in a redundant manner to stabilize the outward-open conformation.
A related feature of MFSD2A is a mobile polybasic C-terminal α-helix composed of five consecutive positively charged residues Arg-498, Arg-499, Arg-500, Lys-503, and Lys-504 (L-3). In the outward-partially occluded model, this helix is held tightly against the transmembrane domains of helices IX and XII (Fig. 5, A and B). In the inward-open model, the helix unravels and moves away from the transmembrane domains (Fig. 5, D and E). Studying the outward-partially occluded model, this helix comes into close contact with a negatively charged surface comprising Asp-408, Asp-411, and Asp-412. We hypothesized that similar to the first two inter-domain ionic locks, this intra-domain electrostatic interaction stabilizes the C-terminal domain in the outward-open conformation. With the exception of Arg-499 and Lys-503, mutating these positive charged residues individually to Glu does not affect MFSD2A transport activity (Fig. 5H). However, mutating them in concert abolished transport activity (Fig. 5H) and caused a conformational change as indicated by an upward shift in mobility of MFSD2A on SDS-PAGE (Fig. 5I). This upward mobility shift of the mutant was not due to alterations in glycosylation of MFSD2A because combined mutation of glycosylation sites (Asn-217 and Asn-227) with mutation of these four basic residues still resulted in an upward mobility shift relative to MFSD2A mutated only in the glycosylation sites (Fig. 5I). We have not identified any likely inter-domain locks stabilizing the inward-open conformation. This suggests that the outward-open state is energetically less favorable, requiring ionic locks to stabilize and favor conversion to this state in the transport cycle.
Discussion
MFSD2A is a sodium-dependent lysophosphatidylcholine transporter essential for human brain growth and function (40). MFSD2A is the only known MFS member or secondary transporter that transports a lipid. In line with its unique function, this study has identified three unique structural features based on a combination of homology structural modeling and biochemical analysis as follows: 1) a unique headgroup binding site; 2) a hydrophobic cleft for acyl chain binding, and 3) three sets of ionic locks that stabilize the outward-open conformation. Drawing together these findings with studies of the mechanism of transport of other MFS family members, we propose the following alternating-access mechanism for LPC transport (Fig. 6). In the first steps, LPC inserts itself into the outer leaflet of the membrane and diffuses laterally into the transporter's hydrophobic cleft. As MFSD2A undergoes conformational changes from the outward-open to the inward-open conformation, the zwitterionic headgroup is inverted from the outer membrane leaflet to the inner membrane leaflet along a translocation pathway within the transporter, interacting with specific polar and charged residues lining the path. Because LPCs are hydrophobic phospholipids, it is unlikely that they will partition out of the transporter into the aqueous environment of the cytoplasm. We propose that the flipped LPC exits the transporter laterally into the membrane environment of the inner leaflet. This model of LPC flipping requires further biochemical proof. Of particular interest is the visualization of the interaction of the negatively charged phosphate headgroup of LPC with Lys-436 that is maintained in both outward-open and inward-open conformations. The side chain of Lys-436 is seen to be pointing in the upward direction in the outward-open conformation but is pointing downward into the translocation cleft in the inward-open conformation. These findings suggest that the Lys-436 acts as a tether to push or pivot the headgroup down into the translocation cavity, although the N and C termini of MFSD2A rock and switch from outward-open to inward-open. Interestingly, Lys-436 is orthologous to the residue Lys-377 in the melibiose transporter of S. typhimurium. Based on the S. typhimurium MelB crystal structure, Lys-377 has been predicted to be involved in binding melibiose and in forming a hydrogen bond with Tyr-120, likely separating the sodium-binding site from the central hydrophilic cavity (9). In a recent molecular dynamic simulation of E. coli MelB, Lys-377 was noted to interact differently with residues involved in the sodium-binding site (Asp-55, Asp-59, and Asp-124) in the presence or absence of a sodium ion and was thought to be critical for the spatial organization of the sodium-binding site (41). Similarly, in our refined models of MFSD2A, Lys-436 is localized in close proximity to the sodium-binding site residue, Asp-93, and the central translocation pathway where it has been identified by docking studies to interact with the charged headgroup of LPC. We hypothesize that Lys-436 may shuttle between the two binding sites, communicating and coordinating the occupancy status of the two sites.
FIGURE 6.
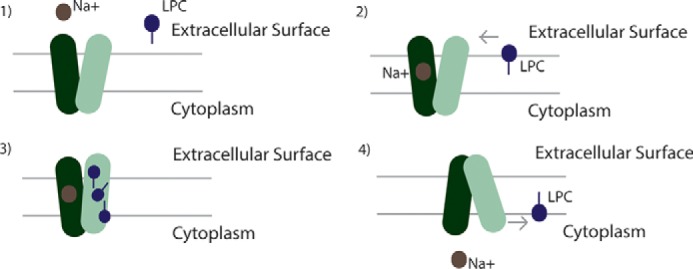
Scheme for Na+/LPC symport by MFSD2A. The following four steps in the transport reaction cycle are shown: 1) MFSD2A is in the outward-open conformation; 2) sodium ion(s) bind to the sodium-binding site. LPC inserts into the outer leaflet of the membrane bilayer and diffuses laterally into the central cavity of MFSD2A via a hydrophobic cleft; 3) MFSD2A undergoes conformational changes that pivots the LPC headgroup bound by Lys-436 through the translocation pathway from the outer leaflet to the inner leaflet of the membrane; 4) LPC, being a hydrophobic lipid, exits the transporter laterally into the inner leaflet of the membrane.
Interestingly, there is a distinct mobility shift in MFSD2A bands on SDS-PAGE between wild-type MFSD2A and the L-3 mutant (R498E, R499E, R500E, K503E, and K504E) (Fig. 5I) that is not seen when each of the residues are mutated individually (supplemental Fig. S1). These findings are consistent with a conformational change in the L-3 mutant. Given that the L-3 ionic lock is visualized in the outward-partially occluded model, we hypothesize that the loss of the L-3 ionic lock results in MFSD2A being trapped in an energetically more favorable inward-open conformation, resulting in the loss of transport function (Fig. 5H).
Patients with the partially inactivating mutation p.(S399L) exhibited significant increases specifically in plasma LPCs having monounsaturated (18:1–92%, p = 0.004) and polyunsaturated LPCs (18:2, 20:4, 20:3–254%, p = 0.002; 117%, p = 0.007, and 238%, p = 0.002), but not in the most abundant LPC-saturated LPCs (C16:0, C18:0) (8). This is consistent with a greater specificity of MFSD2A for LPCs with unsaturated fatty acyl chains (6). One possibility for increased affinity is a greater specificity for the hydrophobic cleft. However, this seems unlikely given that MFSD2A can transport LPCs with a variety of fatty acyl chains having a multitude of potential conformations. It is unlikely that the hydrophobic cleft as defined in our studies binds the fatty acyl tail sufficiently tightly to provide this specificity. Another possible explanation for this acyl chain specificity is related to the mobility of the acyl tail in the membrane. It is known that phospholipids with unsaturated fatty acyl chains disrupt the packing of the bilayer, resulting in greater lateral membrane fluidity (42). Therefore, one possible mechanism for LPC specificity is that LPCs with unsaturated fatty acyl chains have greater lateral mobility in the membrane, increasing the Ka value for interacting with the transport cleft of MFSD2A.
Another important structural feature of the physiological ligand, LPC, is a minimum acyl chain length of 14 carbons is required for transport by MFSD2A. A possible explanation for this requirement is that the hydrocarbon chain must extend beyond the cleft, protruding into the hydrophobic milieu of the phospholipid bilayer core. This interaction of the fatty acyl tail with the acyl chains of the membrane bilayer may provide a hydrophobic force strong enough to pull the molecule through and out of the transporter as the LPC headgroup partitions into the inner leaflet of the membrane. A similar scenario is seen in the Sec translocon where a hydrophobic transmembrane domain of a protein partitions laterally from the Sec61p complex channel into the lipid bilayer (43, 44). This proposal that the ω-carbon of the fatty acyl chain sticks out of the MFSD2A pocket is consistent with the observation that MFSD2A can transport nitrobenzoxadiazole or Topfluor when these moieties are attached to the ω-carbon of the LPC fatty acyl tail (1).
Other known transmembrane phospholipid transporters include flippases, floppases, and scramblases. Flippases and floppases utilize ATP to drive the uphill transport of aminophospholipids from the outer to the inner leaflet and specific substrates from the inner to the outer leaflet, respectively (45–47). Scramblases are less well understood, facilitating transport of substrates in either direction down concentration gradients upon activation. Although the substrates are similar, several differences make comparisons between MFSD2A and phospholipid transporters of limited relevance. First, the shapes of the substrates differ in shape and size; lysophospholipids are smaller and conical, whereas phospholipids are cylindrical. Second, unlike flippases and floppases, MFSD2A is a secondary transporter, utilizing a sodium electrochemical gradient to drive the transport of lysophospholipids from one leaflet to the other. Third, the overall structure of MFS members is different from P4-ATPases and ATP-binding cassette transporters. Consequently, the mechanism of action between MFSD2A and flippases such as P4-ATPases and ATP-binding cassette transporters or floppases is expected to differ.
Being expressed at the blood-brain barrier, MFSD2A is a potential conduit for drug delivery to the brain. The blood-brain barrier is highly impermeable, protecting the brain from blood-derived molecules, pathogens, and toxins. However, its impermeability poses a challenge for pharmacological treatment of brain diseases. It has been predicted that 98% of small molecule drugs are excluded from the brain by the blood-brain barrier (48). Currently, most drugs used to treat brain diseases are lipid-soluble small molecules with a molecular mass of less than 400 Da (49). A small number of drugs traverse the blood-brain barrier by carrier-mediated transport. An example of this is levodopa, a treatment for Parkinson disease, which is a precursor of the neurotransmitter dopamine. Levodopa is transported across the blood-brain barrier by the large neutral amino acid transporter, LAT1 (50). Our findings here provide a further refinement of understanding of the structure-activity relationship of LPCs to their transport and educate the search and design of drugs that can be transported by MFSD2A. Candidates for transport, whether as a drug itself or as an LPC scaffold, must have a zwitterionic headgroup, but not necessarily a phosphate, and a minimal threshold of hydrophobic character. As the binding pocket is several times larger than LPC, it is sterically feasible to attach a small molecule drug onto LPC or LPC-like scaffolds for delivery across the blood-brain barrier.
In summary, these studies represent a first structural model of human MFSD2A based on homology modeling and biochemical interrogation. We expect that this model will serve as a foundation for the future development of x-ray crystal structures of the protein, which would provide further insight into the structure and function of this physiologically important transporter required for human brain growth and function.
Author Contributions
D. L. S., D. Q. Y. Q., and H. F. designed research; D. Q. Y. Q. and H. F. performed research; L. N. N. performed experiments for Fig. 3H; D. Q. Y. Q. and H. F. performed structural modeling and analysis. D. Q. Y. Q., D.L.S., and H. F. wrote the paper.
Supplementary Material
This work was supported by Singapore Ministry of Health National Medical Research Council Grant CBRG/069/2014, Singapore Ministry of Education Tier2 Grant MOE2014-T2-2-018 (to D. L. S.), Biomedical Research Council of A*STAR (to H. F.), and the President's Graduate Fellowship Award from National University of Singapore (to D. Q. Y. Q.). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. 1–4.
- DHA
- docosohexaenoic acid
- LPC
- lysophosphatidylcholine
- MFS
- major facilitator superfamily
- LPE
- lysophosphatidylethanolamine
- CTAB
- cetyltrimethylammonium bromide.
References
- 1. Salem N. Jr., Litman B., Kim H. Y., and Gawrisch K. (2001) Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids 36, 945–959 [DOI] [PubMed] [Google Scholar]
- 2. Bazan N. G. (2009) Neuroprotectin D1-mediated anti-inflammatory and survival signaling in stroke, retinal degenerations, and Alzheimer's disease. J. Lipid Res. 50, S400–S405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baisted D. J., Robinson B. S., and Vance D. E. (1988) Albumin stimulates the release of lysophosphatidylcholine from cultured rat hepatocytes. Biochem. J. 253, 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Edmond J., Higa T. A., Korsak R. A., Bergner E. A., and Lee W. N. (1998) Fatty acid transport and utilization for the developing brain. J. Neurochem. 70, 1227–1234 [DOI] [PubMed] [Google Scholar]
- 5. Lagarde M., Bernoud N., Brossard N., Lemaitre-Delaunay D., Thiès F., Croset M., and Lecerf J. (2001) Lysophosphatidylcholine as a preferred carrier form of docosahexaenoic acid to the brain. J. Mol. Neurosci. 16, 201–204 [DOI] [PubMed] [Google Scholar]
- 6. Nguyen L. N., Ma D., Shui G., Wong P., Cazenave-Gassiot A., Zhang X., Wenk M. R., Goh E. L., and Silver D. L. (2014) MFSD2A is a transporter for the essential ω-3 fatty acid docosahexaenoic acid. Nature 509, 503–506 [DOI] [PubMed] [Google Scholar]
- 7. Law C. J., Maloney P. C., and Wang D. N. (2008) Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62, 289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alakbarzade V., Hameed A., Quek D. Q., Chioza B. A., Baple E. L., Cazenave-Gassiot A., Nguyen L. N., Wenk M. R., Ahmad A. Q., Sreekantan-Nair A., Weedon M. N., Rich P., Patton M. A., Warner T. T., Silver D. L., and Crosby A. H. (2015) A partially inactivating mutation in the sodium-dependent lysophosphatidylcholine transporter MFSD2A causes a non-lethal microcephaly syndrome. Nat. Genet. 47, 814–817 [DOI] [PubMed] [Google Scholar]
- 9. Ethayathulla A. S., Yousef M. S., Amin A., Leblanc G., Kaback H. R., and Guan L. (2014) Structure-based mechanism for Na+/melibiose symport by MelB. Nat. Commun. 5, 3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guan L., Mirza O., Verner G., Iwata S., and Kaback H. R. (2007) Structural determination of wild-type lactose permease. Proc. Natl. Acad. Sci. U.S.A. 104, 15294–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lemieux M. J., Song J., Kim M. J., Huang Y., Villa A., Auer M., Li X. D., and Wang D. N. (2003) Three-dimensional crystallization of the Escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Protein Sci. 12, 2748–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng D., Xu C., Sun P., Wu J., Yan C., Hu M., and Yan N. (2014) Crystal structure of the human glucose transporter GLUT1. Nature 510, 121–125 [DOI] [PubMed] [Google Scholar]
- 13. Deng D., Sun P., Yan C., Ke M., Jiang X., Xiong L., Ren W., Hirata K., Yamamoto M., Fan S., and Yan N. (2015) Molecular basis of ligand recognition and transport by glucose transporters. Nature 526, 391–396 [DOI] [PubMed] [Google Scholar]
- 14. Nomura N., Verdon G., Kang H. J., Shimamura T., Nomura Y., Sonoda Y., Hussien S. A., Qureshi A. A., Coincon M., Sato Y., Abe H., Nakada-Nakura Y., Hino T., Arakawa T., Kusano-Arai O., et al. (2015) Structure and mechanism of the mammalian fructose transporter GLUT5. Nature 526, 397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaback H. R., Smirnova I., Kasho V., Nie Y., and Zhou Y. (2011) The alternating access transport mechanism in LacY. J. Membr. Biol. 239, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guemez-Gamboa A., Nguyen L. N., Yang H., Zaki M. S., Kara M., Ben-Omran T., Akizu N., Rosti R. O., Rosti B., Scott E., Schroth J., Copeland B., Vaux K. K., Cazenave-Gassiot A., Quek D. Q., et al. (2015) Inactivating mutations in MFSD2A, required for ω-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nat. Genet. 47, 809–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Angers M., Uldry M., Kong D., Gimble J. M., and Jetten A. M. (2008) MFSD2A encodes a novel major facilitator superfamily domain-containing protein highly induced in brown adipose tissue during fasting and adaptive thermogenesis. Biochem. J. 416, 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yousef M. S., and Guan L. (2009) A 3D structure model of the melibiose permease of Escherichia coli represents a distinctive fold for Na+ symporters. Proc. Natl. Acad. Sci. U.S.A. 106, 15291–15296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 22. Shen M. Y., and Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sherman W., Day T., Jacobson M. P., Friesner R. A., and Farid R. (2006) Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 49, 534–553 [DOI] [PubMed] [Google Scholar]
- 24. Mysinger M. M., and Shoichet B. K. (2010) Rapid context-dependent ligand desolvation in molecular docking. J. Chem. Inf. Model. 50, 1561–1573 [DOI] [PubMed] [Google Scholar]
- 25. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., and Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 26. Jacobson M. P., Pincus D. L., Rapp C. S., Day T. J., Honig B., Shaw D. E., and Friesner R. A. (2004) A hierarchical approach to all-atom protein loop prediction. Proteins 55, 351–367 [DOI] [PubMed] [Google Scholar]
- 27. Krivov G. G., Shapovalov M. V., and Dunbrack R. L. Jr. (2009) Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77, 778–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heijne G. (1986) The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 5, 3021–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pourcher T., Deckert M., Bassilana M., and Leblanc G. (1991) Melibiose permease of Escherichia coli: mutation of aspartic acid 55 in putative helix II abolishes activation of sugar binding by Na+ ions. Biochem. Biophys. Res. Commun. 178, 1176–1181 [DOI] [PubMed] [Google Scholar]
- 30. Pourcher T., Zani M. L., and Leblanc G. (1993) Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. I. Effect on Na+-dependent transport and binding properties. J. Biol. Chem. 268, 3209–3215 [PubMed] [Google Scholar]
- 31. Zani M. L., Pourcher T., and Leblanc G. (1993) Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. II. Effect on cationic selectivity and coupling properties. J. Biol. Chem. 268, 3216–3221 [PubMed] [Google Scholar]
- 32. Hama H., and Wilson T. H. (1994) Replacement of alanine 58 by asparagine enables the melibiose carrier of Klebsiella pneumoniae to couple sugar transport to Na+. J. Biol. Chem. 269, 1063–1067 [PubMed] [Google Scholar]
- 33. Zani M. L., Pourcher T., and Leblanc G. (1994) Mutation of polar and charged residues in the hydrophobic NH2-terminal domains of the melibiose permease of Escherichia coli. J. Biol. Chem. 269, 24883–24889 [PubMed] [Google Scholar]
- 34. Franco P. J., and Wilson T. H. (1999) Arg-52 in the melibiose carrier of Escherichia coli is important for cation-coupled sugar transport and participates in an intrahelical salt bridge. J. Bacteriol. 181, 6377–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuzaki S., Weissborn A. C., Tamai E., Tsuchiya T., and Wilson T. H. (1999) Melibiose carrier of Escherichia coli: use of cysteine mutagenesis to identify the amino acids on the hydrophilic face of transmembrane helix 2. Biochim. Biophys. Acta 1420, 63–72 [DOI] [PubMed] [Google Scholar]
- 36. Franco P. J., Jena A. B., and Wilson T. H. (2001) Physiological evidence for an interaction between helices II and XI in the melibiose carrier of Escherichia coli. Biochim. Biophys. Acta 1510, 231–242 [DOI] [PubMed] [Google Scholar]
- 37. Breckenridge W. C., Gombos G., and Morgan I. G. (1972) The lipid composition of adult rat brain synaptosomal plasma membranes. Biochim. Biophys. Acta 266, 695–707 [DOI] [PubMed] [Google Scholar]
- 38. Mihalik S. J., Goodpaster B. H., Kelley D. E., Chace D. H., Vockley J., Toledo F. G., and DeLany J. P. (2010) Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity 18, 1695–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shoichet B. K. (2004) Virtual screening of chemical libraries. Nature 432, 862–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Betsholtz C. (2014) Physiology: Double function at the blood-brain barrier. Nature 509, 432–433 [DOI] [PubMed] [Google Scholar]
- 41. Fuerst O., Lin Y., Granell M., Leblanc G., Padrós E., Lórenz-Fonfría V. A., and Cladera J. (2015) The melibiose transporter of Escherichia coli: critical contribution of Lys-377 to the structural organization of the interacting substrate-binding sites. J. Biol. Chem. 290, 16261–16271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barton P. G., and Gunstone F. D. (1975) Hydrocarbon chain packing and molecular motion in phospholipid bilayers formed from unsaturated lecithins. Synthesis and properties of 16 positional isomers of 1,2-dioctadecenoyl-sn-glycero-3-phosphorylcholine. J. Biol. Chem. 250, 4470–4476 [PubMed] [Google Scholar]
- 43. Heinrich S. U., Mothes W., Brunner J., and Rapoport T. A. (2000) The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell 102, 233–244 [DOI] [PubMed] [Google Scholar]
- 44. Van den Berg B., Clemons W. M. Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., and Rapoport T. A. (2004) X-ray structure of a protein-conducting channel. Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 45. Sanyal S., and Menon A. K. (2009) Flipping lipids: why an' what's the reason for? ACS Chem. Biol. 4, 895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clark M. R. (2011) Flippin' lipids. Nat. Immunol. 12, 373–375 [DOI] [PubMed] [Google Scholar]
- 47. Holthuis J. C., and Menon A. K. (2014) Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48–57 [DOI] [PubMed] [Google Scholar]
- 48. Pardridge W. M. (2005) The blood-brain barrier: bottleneck in brain drug development. NeuroRx 2, 3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pardridge W. M. (2012) Drug transport across the blood-brain barrier. J. Cereb. Blood Flow Metab. 32, 1959–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gomes P., and Soares-da-Silva P. (1999) l- DOPA transport properties in an immortalized cell line of rat capillary cerebral endothelial cells, RBE 4. Brain Res. 829, 143–150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.