Abstract
RNase R is a 3′ to 5′ hydrolytic exoribonuclease that has the unusual ability to digest highly structured RNA. The enzyme possesses an intrinsic, ATP-dependent RNA helicase activity that is essential in vitro for efficient nuclease activity against double-stranded RNA substrates, particularly at lower temperatures, with more stable RNA duplexes, and for duplexes with short 3′ overhangs. Here, we inquired whether the helicase activity was also important for RNase R function in vivo and for RNA metabolism. We find that strains containing a helicase-deficient RNase R due to mutations in its ATP-binding Walker motifs exhibit growth defects at low temperatures. Most importantly, cells also lacking polynucleotide phosphorylase (PNPase), and dependent for growth on RNase R, grow extremely poorly at 34, 37, and 42 °C and do not grow at all at 31 °C. Northern analysis revealed that in these cells, fragments of 16S and 23S rRNA accumulate to high levels, leading to interference with ribosome maturation and ultimately to cell death. These findings indicate that the intrinsic helicase activity of RNase R is required for its proper functioning in vivo and for effective RNA metabolism.
Keywords: bacteria, Escherichia coli (E. coli), ribonuclease, ribosomal ribonucleic acid (rRNA) (ribosomal RNA), RNA degradation, RNA helicase
Introduction
RNase R and its homologues are processive, 3′ to 5′ exoribonucleases present in organisms from bacteria to higher eukaryotes (1). These enzymes play important roles in many aspects of RNA metabolism including mRNA and stable RNA turnover (2, 3) and rRNA maturation (4). In contrast to most exoribonucleases, RNase R is particularly adept at degrading highly structured RNA molecules (2, 5–7), and defining the mechanism by which it carries out this process is highly important (8, 9). Escherichia coli RNase R contains an intrinsic RNA helicase activity (10), and recent work from our laboratory characterized this helicase and defined its relation to overall nuclease activity (8, 9). In the E. coli protein, we also identified Walker A and Walker B motifs that are responsible for ATP binding and consequent RNA helicase activity, and found that they are conserved in most mesophilic bacterial genera, but are absent from thermophilic bacteria (8), suggesting that the helicase is important for RNase R function. This conclusion was reinforced by biochemical analyses showing that the helicase activity is essential for efficient nuclease activity in vitro, particularly at lower temperatures, with duplexes containing short 3′ overhangs, and with more stable RNA duplexes (8, 9). Whether the helicase activity is also important for nuclease activity in vivo has been unclear.
It is known that RNase R can complement the essential function of CsdA, a DEAD-box RNA helicase, during cold shock (11), implying that the helicase activity of RNase R can function in vivo. Moreover, this complementation activity persists even when the nuclease activity of RNase R has been eliminated by mutation. However, all of these studies were carried out under conditions in which RNase R was overexpressed, raising the question of whether the helicase ever functions physiologically in vivo independent of the nuclease activity of RNase R. On the other hand, because the helicase activity is important for efficient RNase R nuclease activity in vitro, it was of considerable interest for our understanding of RNase R function to determine whether the helicase also affects RNase R catalysis in vivo.
In this study, we examine how the helicase activity affects RNase R function and RNA metabolism in vivo. We find that mutation of either the Walker A or Walker B motifs, which eliminates the helicase activity (8), leads to growth defects even though all other RNA helicases are present. Most importantly, we show that mutant strains lacking the RNase R helicase activity, and also lacking polynucleotide phosphorylase (PNPase),2 cannot grow at 31 °C, and grow poorly even at 37 and 42 °C. In addition, these cells accumulate high levels of rRNA fragments. Because earlier studies from our laboratory had shown that only PNPase or RNase R is able to degrade such rRNA fragments (3), these data indicate that RNase R does not function properly in vivo when its intrinsic RNA helicase activity is eliminated. Based on these findings, we conclude that the helicase activity plays an essential role in RNase R function and in RNA metabolism.
Experimental Procedures
Materials
Mutagenic primers were synthesized and purified by Sigma Genosys. Bacteriophage T4 polynucleotide kinase was purchased from New England Biolabs, Inc., [γ-32P]ATP was from PerkinElmer Life Sciences, nylon membrane was from GE Healthcare Life Sciences, and ExpressHyb hybridization solution was from Clontech. All chemicals were reagent grade.
Bacterial Strains
All strains used in this study were derivatives of E. coli MG1655(Seq)* I−, which was considered to be wild type for this study. MG1655(Seq)*, an rph+ derivative of MG1655, was constructed by Donald Court (NCI, National Institutes of Health, Bethesda, MD) and provided by Kenneth Rudd (University of Miami, FL). The RNase I− derivatives were constructed by recombineering (12, 13). Mutant strains lacking the processive exoribonucleases, RNase R and PNPase, were generated by phage P1-mediated transduction using rnr::kan and pnp200, respectively, as described previously (14). Site-directed mutagenesis of conserved amino acid residues in the RNase R Walker A motif (K736A) and Walker B motif (D168A,D169A) in the chromosome were performed using oligonucleotides T1 and T2, respectively (supplemental Table 1), and the protocol described previously (15). Recombinants were selected by PCR with primers S1, S2 and S3, S4, respectively (supplemental Table 1). Aspartic acid 272 of RNase R was changed to asparagine using oligonucleotide T3 (supplemental Table 1), and recombinants were selected by PCR with primers S5 and S6 (supplemental Table 1). All the chromosomal mutations were confirmed by DNA sequencing. Mutant strains lacking PNPase and carrying Walker A (K736A) or Walker B (D168A,D169A) motif mutations were generated by P1-mediated transduction. Mutant strains were confirmed by kanamycin resistance. The E. coli MG1655*Δ6 helicase strain was kindly provided by Dr. Chaitanya Jain, University of Miami (16). The Δrnr derivative of the E. coli MG1655*Δ6 helicase strain was generated by phage P1-mediated transduction with rnr::kan and was confirmed by kanamycin resistance.
Measurement of Bacterial Growth
For measurement of cell doubling times, overnight cultures were subcultured into fresh rich medium (LB medium) or minimal medium (1× M9 salts, 0.2% glucose). Antibiotics were added as required at the following concentrations: 50 mg/ml kanamycin, 20 mg/ml chloramphenicol, and 15 mg/ml tetracycline. Cultures were incubated with shaking at various temperatures as indicated to an A600 of ∼0.2. Samples were taken at regular time intervals to measure cell density during exponential phase growth, and doubling times were calculated from the data points obtained.
RNA Preparation
Overnight cultures were subcultured into fresh LB medium and incubated with shaking at various temperatures as indicated to an A600 = 0.6. Total RNA was prepared by hot phenol/chloroform treatment of cell pellets and ethanol precipitation (17). RNA was resolved in a 1.2% agarose gel, and then stained with ethidium bromide.
Northern Blotting Analysis
RNA was resolved on a 1.2% agarose gel in 1× TAE buffer (40 mm Tris acetate, 1 mm EDTA) and transferred to a nylon membrane using 1× TAE transfer solution by downward capillary transfer for 3 h. DNA oligonucleotide probes complementary to either 16S rRNA (16S(18–34), 5′-GCGTTCAATCTGAGCCATG-3′; 16S(790–812), 5′-CGGCGTGGACTACCAGGGTATC-3′; 16S(1520–1540), 5′-AGGAGGTGATCCAACCGCAGG-3′) or 23S rRNA (23S(35–55), 5′-CCTTCATCGCCTCTGACTGCC-3′; 23S(1501–1521), 5′-CACGCCTCAGCCTTGATTTTC-3′; 23S(2881–2901), 5′-GTTAAGCCTCACGGTTCATTA-3′) were 32P-labeled at their 5′ ends using T4 polynucleotide kinase and [γ-32P]ATP. Probes were annealed to the transferred RNA using ExpressHyb hybridization solution by overnight incubation. Bands were visualized by autoradiography.
Complementation Analysis
E. coli MG1655* WT, E. coli MG1655*Δrnr, E. coli MG1655* Δ6helicase, and E. coli MG1655*Δrnr Δ6helicase were used. Plasmid pET44 containing RNase R WT and its mutant derivatives were used for the complementation analysis. Leaky expression of the inserted genes in this vector system in the absence of isopropyl-1-thio-β-d-galactopyranoside was used to keep overexpression low. After transformation, each strain was streaked on LB plates and incubated at different temperatures for various time periods, as indicated in the figures.
Results
In earlier studies, we found that in vitro the intrinsic helicase activity of RNase R is important for effective nucleolytic activity against dsRNA substrates, particularly at lower temperatures, with duplexes having short 3′ overhangs, and with more stable duplexes (8, 9). The studies presented below examine the role of the helicase activity in RNase R function in vivo.
Growth of RNase R Walker Motif Mutant Strains
To examine the importance of the RNase R helicase activity, we mutated conserved amino acids within the Walker A (Lys-736) or Walker B (Asp-168 and Asp-169) ATP-binding motifs to alanine residues. These mutations were previously shown to inactivate the helicase activity (8). For initial analysis of the mutant strains carrying each of these mutations in the chromosome, we measured their growth rates in rich medium and compared them with those of an RNase R deletion strain and with WT. Strains containing either the Walker A or Walker B mutant motifs grew essentially the same as the RNase R deletion strain at all temperatures between 42 and 15 °C (Table 1). Moreover, although all the mutant strains grew similarly to the WT at 42 and 37 °C, their growth rates slowed relative to WT at 31 °C, and the growth defect became even more pronounced as the temperature decreased further (Table 1). These data indicate that the presence of RNase R is most important at lower temperatures and that the absence of helicase activity is as deleterious to cell growth as complete removal of RNase R.
TABLE 1.
Growth rates of RNase R Walker motif mutant strains in rich medium
Overnight cultures of the wild-type strain, MG*1655 I−, and its derivatives carrying RNase R gene deletion or a mutation in the Walker A or Walker B motifs were subcultured into rich medium and grown at temperatures between 42 and 15 °C. Periodic measurements of A600 were made within the exponential growth phase. Cell doubling times and standard deviations were calculated based on data from four replicate experiments. * denotes p < 0.01 and ** denotes p < 0.0001 by a two-tailed Student's t test as compared with the WT strain.
| Growth temperature | Doubling time (min) |
|||
|---|---|---|---|---|
| WT | R− | R736A | R D168A,D169A | |
| 42 °C | 22 ± 2 | 23 ± 2 | 22 ± 3 | 22 ± 2 |
| 37 °C | 26 ± 2 | 27 ± 2 | 28 ± 2 | 28 ± 2 |
| 31 °C | 41 ± 2 | 45 ± 2 | 48 ± 3* | 47 ± 3* |
| 25 °C | 74 ± 3 | 114 ± 4 | 123 ± 4** | 119 ± 4** |
| 20 °C | 121 ± 3 | 197 ± 4 | 213 ± 5** | 207 ± 4** |
| 15 °C | 235 ± 5 | 396 ± 5 | 417 ± 6** | 409 ± 6** |
We also determined the doubling times of WT and mutant strains in M9/glucose minimal medium at different temperatures. As shown in Table 2, all the growth rates were dramatically slowed in minimal medium as compared with rich medium, but the relative rates remained the same. The Walker motif mutants grew the same as the RNase R deletion strain at all temperatures examined. At higher temperatures (37 and 42 °C), the mutant strains had doubling times almost the same as that of the WT strain, but grew more slowly as the temperature decreased, particularly at 25 and 15 °C. Based on these data, it appears that the intrinsic helicase activity is an important component for the proper functioning of RNase R in vivo, and that its role becomes more pronounced at lower temperatures, consistent with our previous findings that the helicase activity is essential for efficient degradation of double-stranded RNA in vitro (8, 9).
TABLE 2.
Growth rates of RNase R Walker motif mutant strains in minimal medium
Overnight cultures of the strains used in Table 1 were subcultured into M9/glucose minimal medium and grown at the indicated temperatures. Periodic measurements of A600 were made within the exponential growth phase. Cell doubling times and standard deviations were calculated based on data from five replicate experiments. * denotes p < 0.05 and ** denotes p < 0.0001 by a two-tailed Student's t test as compared with the WT strain.
| Growth temperature | Doubling time (min) |
|||
|---|---|---|---|---|
| WT | R− | R736A | R D168A,D169A | |
| 42 °C | 79 ± 5 | 81 ± 6 | 80 ± 5 | 80 ± 5 |
| 37 °C | 63 ± 4 | 65 ± 5 | 67 ± 5 | 66 ± 5 |
| 31 °C | 119 ± 6 | 126 ± 6 | 129 ± 7* | 127 ± 6* |
| 25 °C | 135 ± 6 | 183 ± 7 | 189 ± 7** | 184 ± 6** |
| 15 °C | 963 ± 8 | 1402 ± 10 | 1421 ± 11** | 1410 ± 11** |
Growth of RNase R Walker Motif Mutant Strains Also Lacking PNPase
In E. coli, in addition to RNase R, PNPase also participates in degradation of structured RNA (2, 18). Cells become inviable only when both nucleases are absent. Thus, to more accurately assess the specific role of the helicase activity for RNase R function, we introduced a PNP− mutation into the strains already mutated in their Walker A or Walker B motifs, and then measured growth of the resulting strains (Table 3). Cells lacking PNPase grew more slowly than those lacking RNase R at all temperatures examined, and the growth differential became more pronounced at lower temperatures, indicating that PNPase is required for normal cell growth, especially at temperatures below ∼25 °C. However, removal of the RNase R helicase activity by mutation of either Walker motif together with the absence of PNPase had a profound effect on cell growth. Cells were able to grow only at temperatures at or above 34 °C, and they grew extremely slowly as compared with cells lacking just PNPase; at 31 °C or below, cells were unable to grow. It should be noted that cells lacking both RNase R and PNPase are inviable even at the elevated temperatures (3). These data clearly demonstrate that the helicase activity is required for the proper functioning of RNase R in vivo. Even at higher temperatures, conditions in which RNase R lacking helicase activity functions relatively normally in vitro (8), the role of RNase R is severely compromised in vivo.
TABLE 3.
Growth rates in rich medium of strains mutated in an RNase R Walker motif and lacking PNPase
Overnight cultures of the indicated strains were subcultured into rich medium and grown measured at temperatures between 42 and 15 °C. Periodic measurements of A600 were made within the exponential growth phase. Cell doubling times and standard deviations were calculated based on data from five replicate experiments. RA and RB indicate the R736A and the D168A,D169A Walker A and Walker B mutants, respectively. Data for the R− column were taken from Table 1 for comparison purposes. * denotes p < 0.0001 by a two-tailed Student's t test as compared with the PNP− strain.
| Growth temperature | Doubling time (min) |
|||
|---|---|---|---|---|
| R − | PNP− | PNP− RA | PNP− RB | |
| 42 °C | 23 ± 2 | 26 ± 2 | 71 ± 4* | 68 ± 4* |
| 37 °C | 27 ± 2 | 31 ± 2 | 103 ± 5* | 98 ± 5* |
| 34 °C | 34 ± 2 | 42 ± 3 | 315 ± 6* | 286 ± 7* |
| 31 °C | 45 ± 2 | 52 ± 3 | No growth | No growth |
| 25 °C | 114 ± 4 | 136 ± 5 | No growth | No growth |
| 20 °C | 197 ± 4 | 260 ± 5 | No growth | No growth |
| 15 °C | 396 ± 5 | 613 ± 7 | No growth | No growth |
We also examined the doubling times of the strains lacking PNPase together with a Walker motif mutation in M9/glucose minimal medium and compared their growth with an RNase R deletion mutant strain and with a PNPase deletion mutant strain (Table 4). As before, cells grew more slowly in minimal medium, and as was observed in rich medium, cells containing a non-functional Walker motif, and also lacking PNPase, were unable to grow at 31 °C or below. Moreover, they grew more slowly than cells lacking RNase R or PNPase, even at 37 and 42 °C, temperatures more optimal for growth. From these observations, it is apparent that the RNase R helicase activity is essential for normal growth and viability of cells lacking PNPase, and therefore, these data also support the conclusion that the intrinsic helicase activity is an essential component for RNase R function in vivo.
TABLE 4.
Growth rate in minimal medium of strains mutated in a Walker motif and lacking RNase R
Overnight cultures of the same strains as in Table 3 were subcultured into M9/glucose minimal medium and grown measured at the indicated temperatures. Periodic measurements of A600 were made within the exponential growth phase. Cell doubling times and standard deviations were calculated based on data from five replicate experiments. RA and RB indicate the R736A and the D168A,D169A Walker A and Walker B mutants, respectively. Data for the R− column were repeated from Table 2 for comparison purpose. * denotes p < 0.0001 by a two-tailed Student's t test as compared with the PNP− strain.
| Growth temperature | Doubling time (min) |
|||
|---|---|---|---|---|
| R − | PNP− | PNP− RA | PNP− RB | |
| 42 °C | 81 ± 6 | 84 ± 5 | 142 ± 6* | 136 ± 6* |
| 37 °C | 65 ± 5 | 67 ± 5 | 153 ± 7* | 149 ± 6* |
| 31 °C | 126 ± 6 | 135 ± 7 | No growth | No growth |
| 25 °C | 183 ± 7 | 202 ± 8 | No growth | No growth |
RNA Metabolism in Cells Lacking PNPase and Mutated in an RNase R Walker Motif
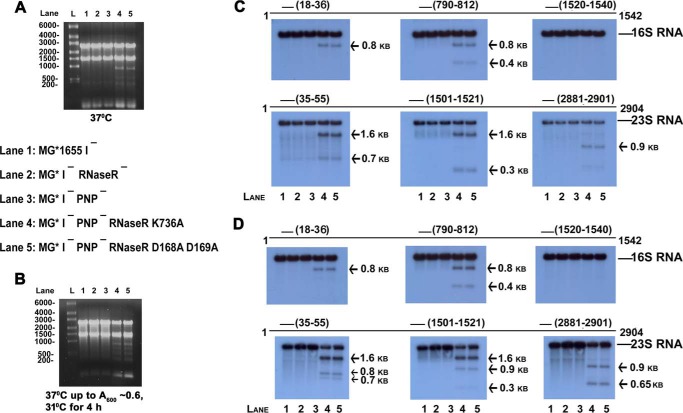
In previous studies, we found that strains lacking PNPase and RNase R are inviable because they accumulate large amounts of 23S and 16S rRNA fragments (3). Here, we examined whether the poor growth observed for strains missing PNPase and carrying an RNase R Walker motif mutation was also due to an inability to degrade rRNA fragments. To examine this point, cells were grown in LB medium to an A600 = 0.6 at either 37 °C or 37 °C and then switched to 31 °C, a temperature at which these cells cease growth. Total RNA was isolated and analyzed on an agarose gel. As shown in Fig. 1, A and B, several new RNA fragments, detectable by ethidium bromide staining, appeared in cells lacking PNPase and also containing an RNase R Walker motif mutation that was absent in the WT, RNase R deletion mutant, or PNPase deletion mutant strains.
FIGURE 1.
Northern analysis of RNA from MG1655* I− and mutant derivatives lacking RNase R, PNPase, and the RNase R helicase. Cultures of the indicated strains were grown in LB medium to an A600 = 0.6. Total RNA was isolated and resolved in a 1.2% agarose gel. A, cultures were grown at 37 °C; gels were stained with ethidium bromide. L denotes RNA ladder. B, cultures were grown at 37 °C to an A600 = 0.6, and then shifted to 31 °C for 4 h; gels were stained with ethidium bromide. C, Northern analysis using probes specific for 16S and 23S rRNA of the gel shown in panel A. D, Northern analysis using probes specific for 16S and 23S rRNA of the gel shown in panel B. Arrows indicate the positions of the major rRNA fragments. Lines and numbers above the gels indicate the positions of the probes on the 16S and 23S RNA sequences.
More detailed examination was carried out by Northern analysis using multiple probes complementary to 16S or 23S rRNA. As shown in Fig. 1, C and D, the RNA molecules that accumulated were, in fact, fragments of both 16S and 23S rRNA, and were present at a much lower level in the WT, the RNase R deletion mutant, and the PNPase deletion mutant strains as compared with cells lacking both PNPase and the RNase R helicase activity. Based on these data, we conclude that cells lacking PNPase and mutated in either Walker motif behave essentially the same with regard to accumulation of rRNA fragments as cells that lack both PNPase and RNase R activity (3). These findings support the conclusion that in the absence of its RNA helicase activity, RNase R is relatively inactive in vivo.
Helicase-deficient RNase R Does Not Complement Cells Lacking Other RNA Helicases
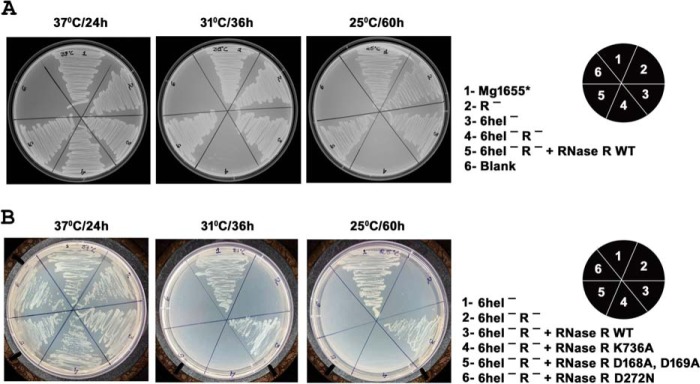
As reported earlier (3), cells require either active RNase R or active PNPase to maintain cell viability, and as shown above, growth cannot be maintained by RNase R alone at temperatures of 31 °C or below if the enzyme lacks its intrinsic helicase activity. A similar situation occurs with PNPase. Thus, in cells lacking RNase R, in which growth is dependent on the presence of active PNPase, growth ceased at temperatures of 31 °C or lower when the six known RNA helicases in E. coli were also absent (Fig. 2A, 6Hel−, R− strain). Cells lacking only RNase R or only the six RNA helicases could grow at all the temperatures tested (Fig. 2A). These data show that PNPase also requires RNA helicase activity to function in vivo.
FIGURE 2.
Growth and complementation analysis of stains lacking the RNase R helicase activity or DEAD-box RNA helicases. Cells were grown on LB plates at the temperatures and for the times indicated. A, growth of E. coli Mg1655*, Mg1655*Δrnr, Mg1655*Δ6helicase, Mg1655*Δrnr,Δ6helicase, and Mg1655*Δrnr,Δ6helicase containing pET44rnr was measured. B, complementation of Mg1655*Δrnr,Δ6helicase by pET44 containing WT rnr or mutated rnr genes. Leaky expression of the inserted genes in the plasmid system was used; isopropyl-1-thio-β-d-galactopyranoside was not added to the plates.
As an additional test of the importance of the RNase R helicase activity for RNase R function, we examined the ability of RNase R to complement the growth of the 6Hel−, R− strain at lower temperatures. As shown in Fig. 2, A and B, growth was restored to the 6Hel−, R− strain when the WT RNase R gene was introduced. However, growth did not occur if the complementing RNase R gene was mutated in either its Walker A or its Walker B motif (Fig. 2B), which eliminates the intrinsic helicase activity. These data confirm that RNase R does not function in vivo at 31 °C or below when its RNA helicase activity is absent. Moreover, cells do not grow at 31 or 25 °C when the complementing RNase R gene lacks nuclease activity (D272N) despite the fact that this RNase R still retains helicase activity (8). Thus, the helicase activity of RNase R, by itself, is unable to complement the six missing helicases.
Discussion
The studies presented here clearly show that the intrinsic RNA helicase activity of RNase R is essential for the proper functioning of RNase R in vivo, even at a temperature (37 °C) at which its catalytic activity is relatively unaffected in vitro (8, 9). Thus, in the absence of the helicase activity, cells grow extremely poorly, especially when PNPase is also missing such that growth is dependent on a fully active RNase R. Cells lacking both PNPase and RNase R are unable to grow, and accumulate massive amounts of rRNA fragments (3). The fact that cells lacking PNPase, and containing a helicase-deficient RNase R, exhibit the identical phenotype shows that RNase R without its helicase activity is essentially non-functional in vivo. The slow rate of growth exhibited by these cells at 34–42 °C is likely due to a low level of residual RNase R nuclease activity combined with the greater “breathing” of RNA duplexes at the higher temperatures. However, growth is so slow under these conditions that any residual nuclease activity must be quite low.
It is interesting that in vitro the nuclease activity of helicase-deficient RNase R is relatively normal at 37 °C (8), whereas it is markedly deficient in vivo at this temperature. We believe that this difference is due to the much wider range of potential duplex substrates present in vivo as compared with the few synthetic substrates used to assess catalytic activity in the in vitro experiments. Moreover, duplex RNAs are probably stabilized in vivo as a consequence of their association with proteins, polyamines, and divalent cations as well as the very different ionic conditions that exist in cells as compared with those in the in vitro assays.
Based on our findings, it is reasonable to conclude that exoribonucleases would require the assistance of an RNA helicase in vivo to digest through structured RNA. This helicase activity may be intrinsic to the nuclease, as in the case of RNase R, or it may be a distinct protein as occurs with PNPase in its association with components of the RNA degradosome (19, 20). Interestingly, although bioinformatic analysis indicated that most RNase Rs contain Walker A and Walker B motifs indicative of an intrinsic helicase activity, the RNase Rs of some organisms lack these motifs (8). For example, RNase Rs in thermophilic bacteria do not contain Walker motifs, suggesting that they lack an intrinsic RNA helicase activity, and raising the possibility that the activity is not necessary for cells living in a high temperature environment in which duplex RNA structures might be less stable. On the other hand, the RNase R of the psychrophilic bacterium, Pseudomonas syringae, also lacks Walker motifs, and this nuclease is part of a degradosome containing an RNA helicase (21). Perhaps an intrinsic RNA helicase is insufficient to open the stable RNA duplexes expected to be present in an organism surviving at low temperatures, and only an energy-driven helicase suffices. Further work will be needed to answer these questions.
One aspect of the RNase R helicase activity that is not yet fully understood is its ability to complement the cold shock function of CsdA, a DEAD-box helicase (11). CsdA is essential at low temperatures, and we believe this is because it associates with PNPase and is required for PNPase to function under these conditions (22, 23). In the absence of CsdA, PNPase would be unable to function properly, thereby leading to the same impaired growth as is observed for a PNP− strain. The single chromosomal copy of rnr is unable to provide sufficient activity to overcome the absence of PNPase activity, but RNase R can complement when overexpressed (10). This scenario would explain the earlier complementation data. What has not been easy to explain is how RNase R lacking its nuclease activity is still able to complement a csdA deletion (11). We have now found in vitro that at high levels, nuclease-deficient RNase R can complement because it converts a duplex substrate to single strands, which then are easily digested by PNPase and RNase R (data not shown). If a similar situation occurs in vivo, such a mechanism would allow PNPase and the WT RNase R present in the strain to function in the absence of CsdA and lead to the apparent complementation by nuclease-deficient RNase R that was observed (11).
The work presented in this study greatly increases our appreciation of the importance of the RNA helicase activity to RNase R function. Taken together with our recent study (9) explaining how the helicase facilitates RNase R action on structured RNA, we now have a much better understanding of the mechanism and role of RNase R in RNA metabolism.
Author Contributions
S. T. H. and M. P. D. designed the experiments. S. T. H. performed the experiments. S. T. H. and M. P. D. analyzed the data and wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Chaitanya Jain for the Δ6helicase-deficient strain and Dr. Arun Malhotra for critical comments on the manuscript.
This work was supported by Grant GM 16317 (to M. P. D.) from the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table 1.
- PNPase
- polynucleotide phosphorylase.
References
- 1. Zuo Y., and Deutscher M. P. (2001) Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheng Z. F., and Deutscher M. P. (2005) An important role for RNase R in mRNA decay. Mol. Cell 17, 313–318 [DOI] [PubMed] [Google Scholar]
- 3. Cheng Z. F., and Deutscher M. P. (2003) Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl. Acad. Sci. U.S.A. 100, 6388–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sulthana S., and Deutscher M. P. (2013) Multiple exoribonucleases catalyze maturation of the 3′ terminus of 16S ribosomal RNA (rRNA). J. Biol. Chem. 288, 12574–12579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vincent H. A., and Deutscher M. P. (2006) Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 281, 29769–29775 [DOI] [PubMed] [Google Scholar]
- 6. Vincent H. A., and Deutscher M. P. (2009) The roles of individual domains of RNase R in substrate binding and exoribonuclease activity: the nuclease domain is sufficient for digestion of structured RNA. J. Biol. Chem. 284, 486–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vincent H. A., and Deutscher M. P. (2009) Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J. Mol. Biol. 387, 570–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hossain S. T., Malhotra A., and Deutscher M. P. (2015) The helicase activity of ribonuclease R is essential for efficient nuclease activity. J. Biol. Chem. 290, 15697–15706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hossain S. T., Malhotra A., and Deutscher M. P. (2016) How RNase R degrades structured RNA: role of the helicase activity and the S1 domain. J. Biol. Chem. 291, 7877–7887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Awano N., Rajagopal V., Arbing M., Patel S., Hunt J., Inouye M., and Phadtare S. (2010) Escherichia coli RNase R has dual activities, helicase and RNase. J. Bacteriol. 192, 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Awano N., Xu C., Ke H., Inoue K., Inouye M., and Phadtare S. (2007) Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J. Bacteriol. 189, 5808–5815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Datta S., Costantino N., and Court D. L. (2006) A set of recombineering plasmids for gram-negative bacteria. Gene 379, 109–115 [DOI] [PubMed] [Google Scholar]
- 14. Basturea G. N., Zundel M. A., and Deutscher M. P. (2011) Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA 17, 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang W., and Deutscher M. P. (2010) A novel mechanism for ribonuclease regulation: transfer-messenger RNA (tmRNA) and its associated protein SmpB regulate the stability of RNase R. J. Biol. Chem. 285, 29054–29058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jagessar K. L., and Jain C. (2010) Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA 16, 1886–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang S., and Deutscher M. P. (1992) Sequence and transcriptional analysis of the Escherichia coli rnt gene encoding RNase T. J. Biol. Chem. 267, 25609–25613 [PubMed] [Google Scholar]
- 18. Khemici V., and Carpousis A. J. (2004) The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 51, 777–790 [DOI] [PubMed] [Google Scholar]
- 19. Liou G. G., Chang H. Y., Lin C. S., and Lin-Chao S. (2002) DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J. Biol. Chem. 277, 41157–41162 [DOI] [PubMed] [Google Scholar]
- 20. Lin P. H., and Lin-Chao S. (2005) RhlB helicase rather than enolase is the β-subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)-exoribonucleolytic complex. Proc. Natl. Acad. Sci. U.S.A. 102, 16590–16595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Purusharth R. I., Madhuri B., and Ray M. K. (2007) Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3′ end processing of 16 and 5 S ribosomal RNA. J. Biol. Chem. 282, 16267–16277 [DOI] [PubMed] [Google Scholar]
- 22. Yamanaka K., and Inouye M. (2001) Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 183, 2808–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prud'homme-Généreux A., Beran R. K., Iost I., Ramey C. S., Mackie G. A., and Simons R. W. (2004) Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a 'cold shock degradosome'. Mol. Microbiol. 54, 1409–1421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.