Abstract
Polysialic acid is an oncofetal glycopolymer, added to the glycans of a small group of substrates, that controls cell adhesion and signaling. One of these substrates, neuropilin-2, is a VEGF and semaphorin co-receptor that is polysialylated on its O-glycans in mature dendritic cells and macrophages by the polysialyltransferase ST8SiaIV. To understand the biochemical basis of neuropilin-2 polysialylation, we created a series of domain swap chimeras with sequences from neuropilin-1, a protein for which polysialylation had not been previously reported. To our surprise, we found that membrane-associated neuropilin-1 is polysialylated at ∼50% of the level of neuropilin-2 but not polysialylated when it lacks its cytoplasmic tail and transmembrane region and is secreted from the cell. This was not the case for neuropilin-2, which is polysialylated when either membrane-associated or soluble. Evaluation of the soluble chimeric proteins demonstrated that the meprin A5 antigen-μ tyrosine phosphatase (MAM) domain and the O-glycan-containing linker region of neuropilin-2 are necessary and sufficient for its polysialylation and serve as better recognition and acceptor sites in the polysialylation process than those regions of neuropilin-1. In addition, specific acidic residues on the surface of the MAM domain are critical for neuropilin-2 polysialylation. Based on these data and pull-down experiments, we propose a model where ST8SiaIV recognizes and docks on an acidic surface of the neuropilin-2 MAM domain to polysialylate O-glycans on the adjacent linker region. These results together with those related to neural cell adhesion molecule polysialylation establish a paradigm for the process of protein-specific polysialylation.
Keywords: glycosylation, protein trafficking (Golgi), protein-protein interaction, sialic acid, sialyltransferase, neuropilin, polysialic acid, polysialylation, polysialyltransferase
Introduction
Polysialic acid (polySia)2 is a glycopolymer consisting of 8 to >100 α2,8-linked sialic acid residues (1). It is synthesized on the N-linked or O-linked glycans of a very specific set of cell surface proteins by two Golgi-localized polysialyltransferases (polySTs), ST8SiaII and ST8SiaIV (2, 3). Polysialylated proteins include the neural cell adhesion molecule (NCAM) (4), neuropilin-2 (NRP-2) (5), synaptic cell adhesion molecule 1 (6), the CD36 scavenger receptor in human milk (7), the α subunit of the voltage-dependent sodium channel (8), and the polySTs themselves (9).
PolySia has been shown to be crucially important for the development of the nervous system, synaptic plasticity and cell migration in the adult nervous system, and the regeneration of damaged nerves and tissues, and it is also up-regulated in many types of late stage cancers, where it is suggested to promote cancer metastasis (reviewed in Refs. 2, 3, and 10). Notably, work by Tanaka et al. (11) demonstrates that a substantial proportion of late stage non-small cell lung cancers express polySia, but not NCAM, suggesting that other substrates are polysialylated in these cancers. These functions of polySia are due to its abilities to serve as an anti-adhesive, a reservoir for biologically significant ligands, and a signaling modulator (reviewed in Ref. 2). Due to its large size and negative charge, polySia binds water and increases the hydrodynamic radius of the proteins it modifies (12). As a result, the presence of polySia abrogates adhesion mediated by it carriers as well as by other nearby adhesion molecules (13). PolySia has also been shown to bind to neurotransmitters, neurotrophins, and growth factors, serving as a reservoir for these molecules and altering their availability for receptor binding and downstream signaling (reviewed in Ref. 2).
NRPs are well established co-receptors for VEGF and class 3 semaphorins and also bind other growth factors (14). These interactions serve to modulate the signaling mediated by these ligands' primary receptors (15). There are two neuropilins, NRP-1 and NRP-2, that share 44% homology at the amino acid level. Mice lacking NRP-1 die between embryonic days 10 and 13.5 due to defects in vascular development and angiogenesis that cause severe hemorrhage. In contrast, NRP-2 knock-out mice are viable and have a milder phenotype characterized by defects in lymphatic system development and axon guidance (reviewed in Ref. 16). The expression of both NRP-1 and NRP-2 is observed in a variety of cancers, including non-small cell lung cancers, neuroblastoma, gliomas, astrocytomas, and colorectal, pancreatic, and breast cancers, where they are important for the proliferation, survival, and migration of cancer cells (reviewed in Refs. 14, 16, and 17). Notably, many of these cancers also express polySia (10).
Curreli et al. (5) showed that O-glycans of NRP-2 are polysialylated by ST8SiaIV in mature dendritic cells. Dendritic cells are antigen-presenting cells that migrate to the lymph node to activate T-lymphocytes (18). This group presented evidence suggesting that the presence of polySia on dendritic cell NRP-2 prevents T-lymphocyte activation and proliferation. However, later work by Bax et al. (19) and Vega and colleagues (20) suggested that NRP-2 polySia binds chemokine (C-C motif) ligand 21 (CCL21), presents this chemokine to the chemokine (C-C motif) receptor 7 (CCR7) on mature dendritic cells, and stimulates their migration to the lymph node to promote T-lymphocyte activation. Recent work by Kiermaier et al. (21) indicates that the dendritic cell CCR7 chemokine receptor itself is polysialylated and suggests that CCR7-linked polySia binds the CCL21 chemokine, releasing its autoinhibition to allow receptor binding. Recently, NRP-2 was also reported to be polysialylated in macrophages and microglia (22, 23). In microglia, polysialylated NRP-2 is predominantly localized in the Golgi and only moves to the cell surface when these cells are activated by lipopolysaccharide (23).
Our laboratory and others have demonstrated that polysialylation is a protein-specific event that requires an initial protein-protein interaction between polyST and substrate prior to glycan modification (2, 24–26). Using NCAM as a model substrate, we showed that the first fibronectin type III repeat (FN1) is required for the polysialylation of N-glycans in the adjacent immunoglobulin domain (Ig5) (24). A three-residue acidic patch on the surface of the FN1 domain plays a primary role in NCAM recognition, binding, and polysialylation by ST8SiaIV (25, 27). A secondary interaction site in the Ig5 domain is also required for optimum NCAM polysialylation (28).
The precise biochemical determinants of NRP-2 polysialylation are not known. In this work, we investigate the sequence requirements for NRP-2 polysialylation using NRP-2 chimeric and mutant proteins. We find that the NRP-2 meprin A5 protein-μ tyrosine phosphatase (MAM) domain is required for the ST8SiaIV recognition and the polysialylation of O-glycans in the adjacent linker region. Additionally, we unexpectedly find that, unlike other generic glycoproteins, membrane-associated NRP-1 can also be polysialylated when ectopically expressed with ST8SiaIV in COS cells, but less robustly than NRP-2.
Experimental Procedures
Tissue culture media and reagents, including DMEM, FBS, and penicillin and streptomycin, were purchased from Fisher. BioWhittaker serum-free medium was purchased from Lonza (Walkersville, MD). Lipofectin transfection reagent and Opti-MEM I medium, mouse monoclonal IgG2a anti-V5 epitope tag antibody (catalogue no. R960-25), and DAPI were purchased from Invitrogen. Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and Invitrogen. The anti-polySia 12F8 rat monoclonal IgM antibody (catalogue no. 556325) was purchased from BD Biosciences. HRP-conjugated anti-human IgG (H+L) (catalogue no. W403B, lot 0000042550) was purchased from Promega (Madison, WI). Myc tag (9B11) mouse mAb (IgG2a detecting N- or C- terminal Myc tag) magnetic bead conjugates (catalogue no. 5698) and rabbit polyclonal anti-Myc antibody (catalogue no. 2272S, lot 6) were purchased from Cell Signaling Technology (Danvers, MA). Peptide N-glycosidase F (PNGase F) was purchased from New England Biolabs (Ipswich, MA). The cDNAs for full-length human neuropilin-1 and neuropilin-2 were generous gifts from Dr. Nicholas Stamatos (University of Maryland School of Medicine, Baltimore, MD). pcDNA4-NRP2-hFc vector was a kind gift from Dr. Ken Kitajima (Nagoya University, Nagoya, Japan). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). The QuikChangeTM site-directed mutagenesis kit and Pfu DNA polymerase were obtained from Agilent Technologies (Santa Clara, CA). The In-Fusion HD cloning kit was obtained from Clontech. DNA purification kits were purchased from Qiagen (Valencia, CA). Precision Plus ProteinTM standards and 4–15% Mini-Protean TGX precast gels were purchased from Bio-Rad. Protein A-Sepharose beads were obtained from GE Healthcare. Protease inhibitors were purchased from Roche Applied Science. HRP- and FITC-conjugated secondary antibodies were purchased from Jackson ImmunoResearch (catalogue nos. 115-035-146, 111-035-144, 115-095-062, and 112-035-020) (West Grove, PA). Nitrocellulose membranes were obtained from Schleicher & Schuell. SuperSignal West Pico chemiluminescence reagent was acquired from Thermo Fisher (Waltham, MA). HyBlot CL® autoradiography film was obtained from Denville Scientific (Denville, NJ). All other chemicals and reagents were purchased from Sigma and Fisher.
Construction of NRP-2ΔCF and NRP-2ΔLCF Proteins
In the pcDNA3.1/V5-His B vector, the NRP-2 cDNA sequence is flanked by HindIII and XbaI restriction sites. For the construction of the NRP-2ΔCF construct, a unique KpnI restriction site was introduced after Cys592 at the beginning of the NRP-2 linker region using primer set 1 (Table 1). The NRP-2ΔCF portion was excised using restriction enzymes KpnI and XbaI and ligated into pcDNA3.1/V5-His B vector at these sites. To insert the signal sequence, the first 22 amino acids of NRP-2 were amplified to include a HindIII site at the N terminus and a KpnI site at the C terminus using primer set 2 (Table 1), and the amplified fragment was ligated into the pcDNA3.1 NRP-2ΔCF vector between the HindIII and KpnI sites. For the construction of the NRP-2ΔLCF construct, NRP-2 sequences from the MAM domain to the cytoplasmic tail were amplified with a KpnI site on the N terminus and a XbaI site on the C terminus using primer set 3 (Table 1). The pcDNA3.1 NRP-2ΔCF vector with the inserted signal sequence was then digested with KpnI and XbaI, and the amplified NRP-2ΔCF sequence was inserted between the signal sequence and the V5 tag.
TABLE 1.
Construction and mutagenesis primers
| Primer Set | Mutant | Template | Primer sequence |
|---|---|---|---|
| 1 | NRP-2 KpnI insertion after Cys592 | NRP-2 | 5′-GAGGTGCTGGGCTGTGGTACCATGGACTGGACAG-ACTCC-3′ |
| 5′-GGAGTCTGTCCAGTCCATGGTACCACAGCCCAGC-ACCTC-3′ | |||
| 2 | NRP-2 signal sequence amplification including HindIII and KpnI sites | NRP-2 | 5′-GTTAAGCTTATGGATATGTTTCCTCTCACCTGGG-3′ |
| 5′-AATGGTACCGCCTCTCACTTGGTGTCTTGA-3′ | |||
| 3 | MAM domain amplification with KpnI and XbaI sites | NRP-2 | 5′-CGGGGTACCTCGGGATTCAATTGCAACTTC-3′ |
| 5′-CCTTCTAGAGCTGCCTCGGAGCAGCACTTT-3′ | |||
| 4 | NRP-2 EcoRV insertion between Pro641 and Ser642 | NRP-2 | 5′-GCAGCTCCCTGATATCTCGGGATTCAATTG-3′ |
| 5′-CGTCGAGGGACTATAGAGCCCTAAGTTAAC-3′ | |||
| 5 | Nrp-2 NheI insertion between Glu802 and Pro803 | NRP-2 | 5′-GAACTGCATGGAAGCTAGCCCCATCTCGGC-3′ |
| 5′-CTTGACGTACCTTCGATCGGGGTAGAGCCG-3′ | |||
| 6 | Nrp-1 EcoRV insertion between Pro644 and Thr645 | NRP-1 | 5′-CAATCAGAGTTTCCAGATATCACATATGGTTTT-AAC-3′ |
| 5′-GTTAGTCTCAAAGGTCTATAGTGTATACCAAAA-TTG-3′ | |||
| 7 | Nrp-1 NheI insertion between Lys811 and Pro812 | NRP-1 | 5′-GAAGATTGTGCAAAAGCTAGCCCAGCAGACCTG-3′ |
| 5′-CTTCTAACACGTTTTCGATCGGGTCGTCTGGAC-3′ | |||
| 8 | NRP-1Δ2 remove EcoRV | NRP-1Δ2 + restriction sites | 5′-CAATCAGAGTTTCCATCGGGATTCAATTG-3′ |
| 5′-GTTAGTCTCAAAGGTAGCCCTAAGTTAAC-3′ | |||
| 9 | NRP-1Δ2 remove NheI | NRP-1Δ2 + restriction sites | 5′-GAACTGCATGGAACCAGCAGACCTG-3′ |
| 5′-CTTGACGTACCTTGGTCGTCTGGAC-3′ | |||
| 10 | NRP-2Δ1 remove EcoRV | NRP-2Δ1 + restriction sites | 5′-GCAGCTCCCTACATATGGTTTTAAC-3′ |
| 5′-CGTCGAGGGATGTATACCAAAATTG-3′ | |||
| 11 | NRP-2Δ1 remove NheI | NRP-2Δ1 + restriction sites | 5′--GAAGATTGTGCAAAACCCATCTGCGC--3′ |
| 5′-CTTCTAACACGTTTTGGGTAGACGCG-3′ | |||
| 12 | NRP-2Δ1 (to remove stop codon inserted during MAM swap at Glu622) | NRP-2Δ1 | 5′-CCACCGAAGAGGAGGCCACAGAGTG-3′ |
| 5′-GGTGGCTTCTCCTCCGGTGTCTCAC-3′ | |||
| 13 | Amplification of ectodomains to make NRP-Fc constructs | NRP-1 and NRP-1Δ2 | 5′-AGACCCAAGCTGGCCATGGAGAGGGGGCTGCCGCTCCTC-3′ |
| 5′-AGACACCCTCCCTCCGGGGTCTAAGGTCTTCAACACATT-3′ | |||
| 14 | Amplification of ectodomains to make NRP-Fc constructs | NRP-2Δ1 | 5′-GAGACCCAAGCTGGCCATGGATATGTTTCCTCTCACCTGG-3′ |
| 5′-AGACACCCTCCCTCCGGGATCCAGGGTGTACAGCCAGCTC-3′ | |||
| 15 | Amplification of pcDNA4-hFc vector for In-Fusion | pcDNA4-NRP-2-hFc | 5′-GCTCTAGAGGGCCCGCGGTTCGAAGGTA-3′ |
| 5′-AGTCTAGAGGGCCCGCGGTTC-3′ | |||
| 16 | Insertion of splice consensus sequence | NRP-2-Fc and NRP-2Δ1-Fc | 5′-CACCCTGGATCCCACAGGTAAGTGGAGGGAG-GGTGT-3′ |
| 5′-ACACCCTCCCTCCACTTACCTGTGGGATCCAGGGTG-3′ | |||
| 17 | Insertion of splice consensus sequence | NRP-1-Fc and NRP-1Δ2-Fc | 5′-AGACCTTAGACCCCACAGGTAAGTGGAGGGAGGGTGTC-3′ |
| 5′-GACACCCTCCCTCCACTTACCTGTGGGGTCTAAGGTCT-3′ | |||
| 18 | Amplification of NRP-1 linker region through the cytoplasmic tail for NRP-2ΔLM1 construct | NRP-1 | 5′-GAGGTGCTGGGCTGTGAAGTGGAAGCCCCTACAGCTGGA-3′ |
| 5′-CGGGCCCTCTAGAGCTGCCTCCGAATAAGTACTCTGTG-3′ | |||
| 19 | Amplification of NRP-2 linker region through the cytoplasmic tail for NRP-1ΔLM2 construct | NRP-2 | 5′-GAGCTGCTGGGCTGTGACTGGACAGACTCCAAGCCCACG-3′ |
| 5′-CGGGCCCTCTAGAGCTGCCTCGGAGCAGCACTTTTGGT-3′ | |||
| 20 | Amplification of NRP-2 sequences except the linker region through the cytoplasmic tail sequences in pcDNA3.1/V5-His B vector | NRP-2 | 5′-ACAGCCCAGCACCTCCAGCCGCATCCCA-3′ |
| 5′-GCTCTAGAGGGCCCGCGGTTCGAAGGTA-3′ | |||
| 21 | Amplification of NRP-1 sequences except the linker region through the cytoplasmic tail in pcDNA3.1/V5-His B vector | NRP-1 | 5′-ACAGCCCAGCAGCTCCATTCTGAGCCCCA-3′ |
| 5′-GCTCTAGAGGGCCCGCGGTTCGAAGGTA-3′ | |||
| 22 | Deletion of extra cytosine base | NRP-1ΔLM2 | 5′-GACAACCACCCCCTACCCCACCGAAG-3′ |
| 5′-CTTCGGTGGGGTAGGGGGTGGTTGTC-3′ | |||
| 23 | Amplification of ectodomains to make NRP-Fc constructs | NRP-2ΔLM1 | 5′-GAGACCCAAGCTGGCCATGGATATGTTTCCTCT-CACCTGG-3′ |
| 5′-GACACCCTCCCTCCACTTACCTGTGGGGTCTAAGGTCT-3′ | |||
| 24 | Amplification of ectodomains to make NRP-Fc constructs | NRP-1ΔLM2 | 5′-AGACCCAAGCTGGCCATGGAGAGGGGGCTGCCGCTCCTC-3′ |
| 5′-ACACCCTCCCTCCACTTACCTGTGGGATCCAGGGTG-3′ | |||
| 25 | Inverse PCR of NRP-2-Fc to eliminate MAM domain | NRP-2-Fc | 5′-CCTCCACTTACCTGTAGGGAGCTGCAAATCTTTGTCATCCTCA-3′ |
| 5′-GGAGGGAGGGTGTCTGCTGGAA-3′ | |||
| 26 | NRP-2 E652A | NRP-2-Fc | 5′-CTTCGATTTCCTCGCGGAGCCCTGTGG-3′ |
| 5′-GAAGCTAAAGGAGCGCCTCGGGACACC-3′ | |||
| 27 | NRP-2 E653A | NRP-2-Fc | 5′-CGATTTCCTCGAGGCGCCCTGTGGTTGG-3′ |
| 5′-GCTAAAGGAGCTCCGCGGGACACCAACC-3′ | |||
| 28 | NRP-2 E652A/E653A | NRP-2-Fc | 5′-CTTCGATTTCGCGGCGCCCTGTGGTTGG-3′ |
| 5′-GAAGCTAAAGCGCCGCGGGACACCAACC-3′ | |||
| 29 | NRP-2 D683A | NRP-2-Fc | 5′-GACGTTTCCAGATGCCAGGAATTTCTTGCG-3′ |
| 5′-CTGCAAAGGTCTACGGTCCTTAAAGAACGC-3′ |
Construction of V5-tagged NRP-1 and NRP-2 MAM Domain Swap Chimeras
To create the NRP-1 and NRP-2 chimeric proteins with their MAM domains swapped (NRP-2Δ1 and NRP-1Δ2), the NRP-2 and NRP-1 MAM domains were flanked with a unique EcoRV restriction site on the N terminus and unique NheI restriction site on the C terminus, which were inserted by site-directed mutagenesis using primer sets 4–7 (Table 1), respectively. The individual domains were extracted by restriction enzyme digestion. The NRP-1 MAM domain was inserted between newly engineered EcoRV and NheI sites in the NRP-2 cDNA to obtain the NRP-2Δ1 construct, whereas the NRP-2 MAM domain was inserted between engineered EcoRV and NheI sites in the NRP-1 cDNA to obtain the NRP-1Δ2 construct. The EcoRV and NheI restriction sites flanking the MAM domains were removed from both the chimeric mutants using primer sets 8–11 (Table 1) by site-directed mutagenesis. During the removal of the restriction sites from NRP-2Δ1, a single nucleotide change at amino acid Glu622 introduced a stop codon, which was removed by site-directed mutagenesis using primer set 12.
Construction of Fc-tagged NRP-1, NRP-2, and Their Chimeras
The pcDNA4-NRP2-hFc vector was a kind gift from Dr. Ken Kitajima (Nagoya University, Nagoya, Japan), and we constructed NRP-1-Fc, NRP-1Δ2-Fc, and NRP-2Δ1-Fc in this vector. Extracellular portions of NRP-1 and the NRP-2Δ1 and NRP-1Δ2 chimeras were amplified from the full-length constructs to include a 15-bp overhang complementary to both the pcDNA4 vector on the N terminus and the human antibody Fc sequence on the C terminus using primer set 13 for NRP-1 and NRP-1Δ2 and primer set 14 for NRP-2Δ1 (Table 1). The In-Fusion homology-directed cloning kit was used according to the manufacturer's protocol to construct the Fc-tagged proteins. Briefly, the pcDNA4-hFc vector was amplified using primer set 15 (Table 1). The amplified vector and insert fragments were mixed. A splice consensus sequence was inserted between the NRP sequences and Fc sequence, which contains introns, to ensure splicing within the Fc portion. This sequence, ACAGGTAAGT, was inserted by site-directed mutagenesis using primer set 16 for NRP-2-Fc and NRP-2Δ1-Fc constructs and primer set 17 for the NRP-1 and NRP-1Δ2-Fc constructs (Table 1).
Construction of Fc-tagged NRP-1 and NRP-2 Linker MAM (LM) Domain Chimeric Proteins
NRP-2ΔLM1 and NRP-1ΔLM2 constructs were initially made as full-length, V5-tagged constructs in pcDNA3.1/V5-His B vector. For the NRP-2ΔLM1 construct, NRP1 sequences from the linker through the cytoplasmic tail were amplified with primer set 18 (Table 1), which includes 15-bp overhangs complementary to the NRP2 coagulation factor 5/8 homology (F5/8)-2 region on the N terminus and to the pcDNA3.1/V5-His B, including the V5 tag, on the C terminus. For the NRP-1ΔLM2 construct, NRP2 sequences from the linker through the cytoplasmic tail were amplified with primer set 19 (Table 1), which includes 15-bp overhangs complementary to the NRP1 F5/8-2 region on the N terminus and to the pcDNA3.1/V5-His B, including the V5 tag, on the C terminus. The In-Fusion homology-directed cloning kit was used according to the manufacturer's protocol to construct the chimeric proteins. The pcDNA3.1/V5-His B vector containing NRP2 sequences was amplified using primer set 20 (Table 1), and the same vector containing NRP1 sequences was amplified using primer set 21 (Table 1). An extra cytosine base was erroneously introduced during construction of the NRP-1ΔLM2 construct and was deleted by site-directed mutagenesis using primer set 22 (Table 1). For Fc-tagged NRP-2ΔLM1 and NRP-1ΔLM2 constructs, ectodomains of these chimeras were amplified using primer set 23 (Table 1) for NRP-2ΔLM1 and primer set 24 (Table 1) for NRP-1ΔLM2. These primers inserted splice consensus sequence in one step and included 15-bp complementary sequences on both sides for the pcDNA4-hFc vector. The amplified pcDNA4-hFc vector, generated as described above, was then mixed with the above fragments, and the In-Fusion protocol was followed using the manufacturer's guidelines.
Construction of NRP-2 ΔMAM-Fc Construct
To remove the MAM domain from the pcDNA4-NRP-2-Fc construct, we amplified vector and NRP sequences from the Fc portion to the NRP-2 linker region and in this way eliminated the MAM sequences, using primer set 25 (Table 1). A primer with sequences complementary to those of the NRP-2 linker region and with a 15-bp overhang complementary to the Fc portion was used to amplify a linear fragment that was then used to regenerate the circular plasmid by homologous recombination.
NRP-2 Mutagenesis
Mutagenesis reactions were carried out using the Stratagene QuikChangeTM site-directed mutagenesis kit, according to the manufacturer's protocol. The primers designed and used for this purpose include primer set 26 (Table 1) for NRP-2-Fc E652A, primer set 27 (Table 1) for NRP-2-Fc E653A, primer set 28 (Table 1) for NRP-2-Fc E652A/E653A, and primer set 29 (Table 1) for NRP-2-Fc D683A. Isolated clones were sequenced by the DNA Sequencing Facility of the Research Resources Center at the University of Illinois (Chicago, IL) and confirmed on SnapGene Viewer version 2.7.2 software (GSL Biotech, Chicago, IL) for accuracy.
Transfection of COS-1 Cells for Immunofluorescence Localization
COS-1 cells were maintained in DMEM with 10% FBS and 1% pencillin/streptomycin and grown in a 37 °C, 5% CO2 incubator. They were plated on 12-mm glass coverslips in 24-well plates and incubated overnight at 37 °C. At ∼50–70% confluence, cells in each well were then transfected with 500 ng of NRP-1, NRP-2, chimeric protein, or deletion mutant cDNAs and 3 μl of Lipofectin in 300 μl of Opti-MEM I and incubated at 37 °C for 6 h, according to the manufacturer's protocol. After 6 h, 1 ml of DMEM, 10% FBS, 1% pencillin/streptomycin was added to each well and was kept for further incubation at 37 °C in 5% CO2 for 20 h.
Analysis of NRP-1, NRP-2, and Chimeric Protein Localization by Indirect Immunofluorescence Microscopy
After 20 h post-transfection, COS-1 cells expressing NRP proteins were washed twice with 1 ml of phosphate-buffered saline (PBS). Cells were fixed and permeabilized with 1 ml of ice-cold methanol. Cells were again washed twice with PBS and blocked for 1 h at room temperature in 1 ml of blocking buffer (5% normal goat serum in PBS). Cells were then incubated with a 1:250 dilution of anti-V5 epitope tag antibody in blocking buffer for 2 h and then washed twice for 5 min with PBS. The cells were then incubated with a 1:100 dilution of FITC-conjugated goat anti-mouse IgG secondary antibody in blocking buffer for 45 min and washed twice with PBS for 5 min. Next, the cells were incubated with a 1:2000 dilution of DAPI in blocking buffer for 5 min in the dark and then washed twice with PBS for 5 min. After washing, coverslips were then rinsed in deionized H2O and mounted on glass microscope slides using 20 μl of mounting medium (15% (w/v) Vinol 205 polyvinyl alcohol, 33% (w/v) glycerol, 0.1% sodium azide in PBS, pH 8.5). Cells were visualized and imaged with a Zeiss LSM 700 inverted confocal microscope, equipped with an AxioCam digital microscope camera using a ×100 oil immersion objective at room temperature. Images were acquired using Zen software by Zeiss and processed with ImageJ software (National Institutes of Health).
Transfection of COS Cells for Immunoprecipitation and Immunoblotting
COS-1 cells or COS-7 cells were maintained in DMEM with 10% FBS and 1% penicillin/streptomycin and grown on 100-mm tissue culture plates in a 37 °C, 5% CO2 incubator. At 80–90% confluence, cells were transfected using 3 μg of V5- or Fc-tagged NRP cDNA and ST8SiaIV-Myc cDNA (cloning of ST8SiaIV cDNA into the pcDNA3.1 Myc/HisB expression was described previously (29)) and 30 μl of Lipofectin in 3 ml of Opti-MEM I and incubated at 37 °C in a 5% CO2 incubator, according to the manufacturer's protocol. After a 6-h incubation, 7 ml of DMEM, 10% FBS, 1% pencillin/streptomycin was added to each plate and incubated for an additional 18–24 h.
Immunoprecipitation of V5-tagged NRP Proteins and Chimeras
Eighteen hours post-transfection, the cells were washed with 1× PBS and lysed in 500 μl of immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 0.1% SDS). A 50-μl aliquot of the lysate was reserved and boiled (100 °C) with Laemmli sample buffer (62.5 mm Tris-HCl, pH 6.8, 25% glycerol, 2% SDS, 0.01% bromphenol blue) containing 10% β-mercaptoethanol to remove polySia and assess the relative expression of these proteins. The remaining lysate was precleared with 50 μl of protein A-Sepharose beads (50% suspension in PBS) for 1 h at 4 °C, and NRP proteins were immunoprecipitated with 2 μl of anti-V5 epitope tag antibody overnight at 4 °C with rotation. Samples were then rotated with 50 μl of protein A-Sepharose beads for 1 h and washed four times with immunoprecipitation buffer. Samples were then resuspended in 50 μl of Laemmli sample buffer containing 10% β-mercaptoethanol, heated at 65 °C (to retain polySia) for 8 min, and separated on a 4–15% precast polyacrylamide gel (Bio-Rad) at 110 V for 1 h.
PNGase F Treatment of Immunoprecipitated Proteins and Proteins in Cell Lysates
To remove N-glycans from immunoprecipitated proteins, protein A-Sepharose beads bound to the anti-V5 antibody and NRP-2 proteins were incubated with 1500 units of PNGase F in G7 buffer (New England Biolabs), 0.5% Nonidet P-40 for 3 h at 37 °C (24, 30). For PNGase F treatment of cell lysates, 100 μl of cell lysate was incubated with 1500 units of PNGase F in G7 buffer, 0.5% Nonidet P-40 at 37 °C overnight.
Immunoprecipitation of Fc-tagged NRP Proteins
Medium containing secreted Fc-tagged proteins was harvested 22–24 h post-transfection and incubated with an 80-μl slurry of 50% protein A-Sepharose beads overnight at 4 °C. The medium was discarded, and the beads were washed four times with the immunoprecipitation buffer. Forty percent of the beads were heated to 100 °C with 32 μl of Laemmli sample buffer containing 10% β-mercaptoethanol to remove polySia and assess relative protein expression. The remaining beads were heated with 48 μl of Laemmli sample buffer containing 10% β-mercaptoethanol at 65 °C for 8 min, and proteins were separated as described above.
Immunoblot Analysis of the Expression and Polysialylation of NRP Proteins
Following SDS-PAGE, proteins were transferred to a nitrocellulose membrane at 100 V for 1 h at 4 °C. Membranes were blocked for 1 h at room temperature in blocking buffer (5% nonfat dry milk in Tris-buffered saline, 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.1% Tween 20 (TBST)). To evaluate protein polysialylation, membranes were incubated overnight at 4 °C with a 1:1000 dilution of 12F8 anti-polySia antibody in 2% nonfat dry milk in Tris-buffered saline, pH 8.0, to detect polysialylation. After two 15-min washes, these membranes were incubated for 1 h at 4 °C with HRP-conjugated goat anti-rat IgM, diluted 1:5000 in blocking buffer. The specificity of the commercially available 12F8 anti-polySia antibody was confirmed by the disappearance of the immunoblot signal using endoneuraminidase N, a sialidase specific for polySia (data not shown).
To evaluate V5-tagged NRP protein expression levels, membranes were incubated for 1 h at 4 °C with a 1:5000 dilution of anti-V5 in blocking, washed twice with TBST, for 15 min each, and then incubated again for 1 h at 4 °C in 1:5000-diluted HRP-conjugated goat anti-mouse IgG in blocking buffer. To evaluate the expression of Fc-tagged proteins, the membranes were blocked in blocking buffer overnight and were incubated with the HRP-conjugated anti-human IgG (1:5000) in high salt TBST (500 mm NaCl, 150 mm Tris-HCl, pH 8.0, and 0.1% Tween 20) for 45 min. All membranes were washed with high salt TBST four times, each for 15 min. Immunoblots were then developed using the SuperSignal West Pico chemiluminescence kit and HyBlot CL® autoradiography film. To quantify changes in polysialylation between NRP-2, NRP-1, and their respective mutants, we used ImageJ software and compared the ratio of polysialylated to loading control for each protein with the value for wild type NRP-2 set to 100%. Mean and S.D. were calculated.
Pull-down Experiments to Assess NRP-2-ST8SiaIV Binding
ST8SiaIV-Myc and NRP-2-Fc or NRP-2 ΔMAM-Fc proteins were expressed individually in COS-7 cells using Lipofectin transfection reagent, as described above. For NRP-2-Fc- and NRP-2 ΔMAM-Fc-expressing cells, 7 ml of serum-free BioWhittakerTM medium was added after 6 h of transfection, and incubation was continued overnight in a 37 °C, 5% CO2 incubator. ST8SiaIV-expressing cells were lysed in 500 μl of immunoprecipitation buffer, and lysates from two 100-mm plates were combined. Lysates were then rotated overnight with anti-Myc magnetic beads at 4 °C. Medium containing NRP-2-Fc or NRP-2 ΔMAM-Fc was harvested 24 h post-transfection. A 1-ml aliquot of medium was rotated with protein A-Sepharose beads overnight at 4 °C to recover secreted proteins and assess their relative secretion/expression. These beads were washed four times with immunoprecipitation buffer and then boiled in Laemmli sample buffer containing 10% β-mercaptoethanol for 7 min prior to SDS-PAGE. To assess binding of the polyST to the NRP2 proteins, ST8SiaIV-loaded magnetic beads were then washed with co-immunoprcipitation buffer (50 mm HEPES, 100 mm NaCl, 1% Triton X-100, pH 7.2) and were added to the serum-free medium containing NRP-2-Fc or NRP-2 ΔMAM-Fc. As a control, anti-Myc magnetic beads were added to the medium containing NRP-2-Fc or NRP-2 ΔMAM-Fc to assess nonspecific binding. After 2 h of rotation at 4 °C, the beads were washed four times with the co-immunoprecipitation buffer. All of the samples were resuspended in Laemmli sample buffer containing 10% β-mercaptoethanol and boiled for 7 min. Proteins were separated by SDS-PAGE, and immunoblotting was performed as described above. To quantify changes in binding between NRP-2-Fc and NRP-2 ΔMAM-Fc, we used ImageJ software and compared the ratio of bound protein to secreted protein for the NRP-2 ΔMAM-Fc mutant versus wild type NRP-2-Fc (normalized to 100%). Mean and S.D. were calculated.
Results
To determine the sequence requirements for NRP-2 polysialylation, we created a series of domain deletion mutants, chimeric proteins, and point mutants and evaluated their polysialylation when co-expressed with ST8SiaIV (Fig. 1). NRPs are type I cell surface glycoproteins with two complement homology (CUB) domains, two F5/8 domains, and one MAM domain (Fig. 1). The CUB domains and possibly the first F5/8 (F5/8-1) domain, are important for the interaction of NRPs with semaphorins, whereas F5/8 domains are important for the interaction with VEGFs (reviewed in Ref. 16). The MAM domain has been suggested to have a role in dimerization of NRPs (31); however, other reports suggest the importance of the CUB and F5/8 domains as well as the transmembrane region in NRP dimerization (32–34). Recently, Rollenhagen et al. (35) showed that NRP-2 is polysialylated on O-glycans located in the linker region between the second F5/8 (F5/8-2) and MAM domains. The locations of the threonine residues carrying the O-glycans that are polysialylated in the NRP-2 linker are shown in Fig. 1B. Based on our laboratory's findings with NCAM polysialylation, we hypothesized that the MAM domain may be critical for the polysialylation of O-glycans in the adjacent linker region.
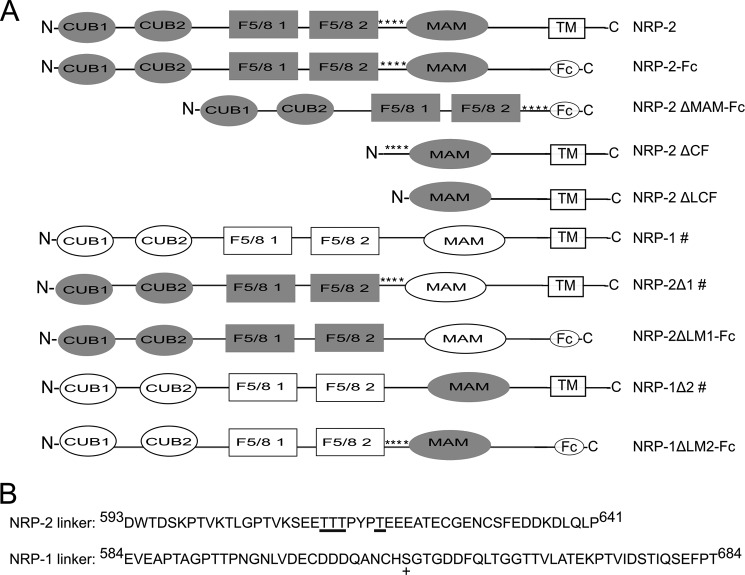
FIGURE 1.
Schematic representation of NRP-1, NRP-2, their MAM and LM chimeras, and domain deletion mutants. A, neuropilin ectodomains are composed of two CUB domains, two F5/8 domains, and a MAM domain. Fc-tagged proteins used in this study lack the transmembrane regions (TM) and cytoplasmic tails of the NRPs, and these sequences are replaced with the human IgG Fc fragment. Both the membrane-associated form with the transmembrane regions and cytoplasmic tail and the soluble Fc form of NRP-2 are shown. Other proteins that are expressed as both membrane-associated and soluble Fc forms are shown as the membrane-associated form and indicated (#). The locations of the threonine residues that carry the polysialylated O-glycans in the NRP-2 linker are indicated (****) in constructs containing these sequences. B, the sequences of the NRP-1 and NRP-2 linker regions between the F5/8-2 and MAM domains are shown. These differ with respect to length and the presence of potential O-glycosylation sites. PolySia is found on the O-glycans occupying four Thr residues in the linker of NRP-2 (these residues are underlined). The serine and threonine residues in the NRP-1 linker that are O-glycosylated are unknown; however, the Ser612 that carries the glycosaminoglycan chain in the NRP-1 linker is indicated (+).
The MAM Domain and the Adjacent Linker Region Are Minimally Required for NRP-2 Polysialylation
To begin to test our hypothesis, we first evaluated which NRP-2 sequences are minimally required for polysialylation. To do this, we made NRP-2 domain deletion mutants lacking the CUB and F5/8 domains (NRP-2 ΔCF) and lacking these domains plus the linker between the MAM domain and the F5/8-2 domain (NRP-2 ΔLCF) (Fig. 1). These V5-tagged mutants and wild type NRP-2-V5 were co-expressed with ST8SiaIV-Myc in COS-1 cells, an aliquot of the cell lysate was removed to evaluate protein expression, and the remaining sample was split in two to verify that the polySia observed on the mutant proteins was on O-glycans, as in the wild type NRP-2 protein. Specifically, we treated one-half of the sample with PNGase F, which removes all N-linked glycans but not O-linked glycans (30). The polysialylation of untreated and PNGase F-treated samples was then evaluated by immunoprecipitation with the anti-V5 tag antibody and immunoblotting with the 12F8 anti-polySia antibody. Immunoprecipitated samples were incubated at 65 °C in Laemmli sample buffer, 10% β-mercaptoethanol to retain the heat-sensitive polySia (Fig. 2, top), whereas aliquots reserved to assess protein expression levels were heated at 100 °C in Laemmli sample buffer, 10% β-mercaptoethanol to remove polySia for more accurate quantification (Fig. 2, bottom left), as described under “Experimental Procedures.” Polysialylated NRP-2 migrated as a broad band extending from 140 kDa to well over 250 kDa (Fig. 2, top, NRP-2). NRP-2ΔCF was also polysialylated, but at a much lower level, migrating as a broad band from 130 to 170 kDa (Fig. 2, top, NRP-2ΔCF). In contrast, the NRP-2 ΔLCF protein that lacked the sites of polysialylation in the linker was unpolysialylated (Fig. 2, top, NRP-2ΔLCF). PNGase F treatment did not alter the anti-polySia antibody recognition of either NRP-2 or NRP-2ΔCF, demonstrating that the polySia on both of these proteins is on PNGase F-insensitive O-glycans (Fig. 2, top, +PNGase F).
FIGURE 2.
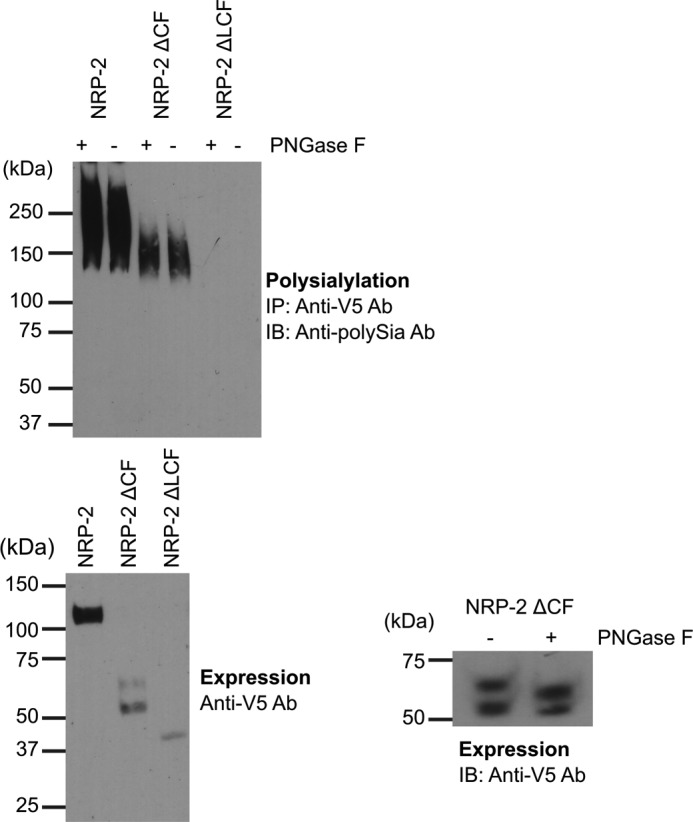
The MAM domain and adjacent linker region are minimally sufficient for NRP-2 polysialylation. Top, NRP proteins were immunoprecipitated from lysates of COS-1 cells transiently expressing ST8SiaIV-Myc and V5-tagged NRP-2 and its domain deletion mutants using an anti-V5 epitope tag antibody. The N-glycans were removed in one-half of each sample by treatment of protein A-Sepharose·antibody·NRP protein complex with PNGase F (+PNGase F), as described under “Experimental Procedures.” Polysialylation of both PNGase F-treated and untreated proteins was assessed by immunoblotting (IB) these immunoprecipitated (IP) proteins with the 12F8 anti-polySia antibody (top). Bottom left, prior to immunoprecipitation, aliquots of cell lysates were heated to 100 °C to remove polySia and immunoblotted with an anti-V5 antibody to evaluate relative expression levels of NRP-2 proteins. Bottom right, the N-glycosylation status of the NRP-2ΔCF doublet was assessed by PNGase F treatment of lysates from expressing cells and immunoblotting with the anti-V5 epitope tag antibody.
Based on our experience with the polysialylation of the NCAM Ig5-FN1 tandem (24), we were surprised that the NRP-2ΔCF protein exhibited such low polysialylation. A number factors could have contributed to this, including a low expression level, underglycosylation, mislocalization (retention in the endoplasmic reticulum), or the requirement for additional sequences. First, its clear that NRP-2ΔCF is expressed at much lower levels than NRP-2 (Fig. 2, bottom left). In addition, the unpolysialylated NRP-2ΔCF migrates as a 54/62 kDa doublet with the lower band predominating (Fig. 2, bottom left). This may reflect an underglycosylation of a large proportion of the NRP-2ΔCF protein. Possibly, in the absence of the more N-terminal domains, the structure or positioning of the linker region may have changed, making O-glycosylation of this region inefficient.
O-Glycosylation is difficult to analyze by enzymatic digestion. However, we identified two predicted N-glycosylation sites in the NRP-2ΔCF protein: one in the linker region with the O-glycans that are polysialylated and one in the membrane-proximal region. Treatment of the unpolysialylated NRP-2ΔCF protein with PNGase F and evaluation of its migration on SDS gels demonstrated that there were no N-glycans on the 54 kDa band (no change in molecular mass following treatment) and at least one N-glycan and another modification on the 62 kDa band (a change in molecular mass but not a reduction to 54 kDa) (Fig. 2, bottom right). This other modification on the 62 kDa band is probably O-glycosylation. These results suggest that a large proportion of the NRP-2ΔCF protein may lack the O-glycans that serve as acceptors for polySia and that this could in part explain the lower levels of NRP-2ΔCF polysialylation relative to the full-length protein.
An underglycosylation of the NRP-2ΔCF mutant could be the result of its complete or partial retention in the endoplasmic reticulum. However, immunofluorescence localization of NRP-2 and the two domain deletion mutants showed that the staining patterns of the wild type protein and NRP-2ΔCF are essentially identical with both localized on the cell surface with some internal staining as expected for proteins transiting the secretory pathway (Fig. 3). In contrast, the NRP-2ΔLCF protein exhibited a strong internal, reticular staining pattern that suggested that much of this protein was localized in the endoplasmic reticulum (Fig. 3, ΔLCF). Although we expected that the ΔLCF would not be polysialylated because it lacks the O-glycosylated linker region, we could not draw any definitive conclusion about this largely mislocalized protein. In sum, the NRP-2ΔCF mutant exhibited weaker polysialylation relative to wild type NRP-2 than we expected, and this can be explained, at least in part, by its lower overall expression and underglycosylation.
FIGURE 3.
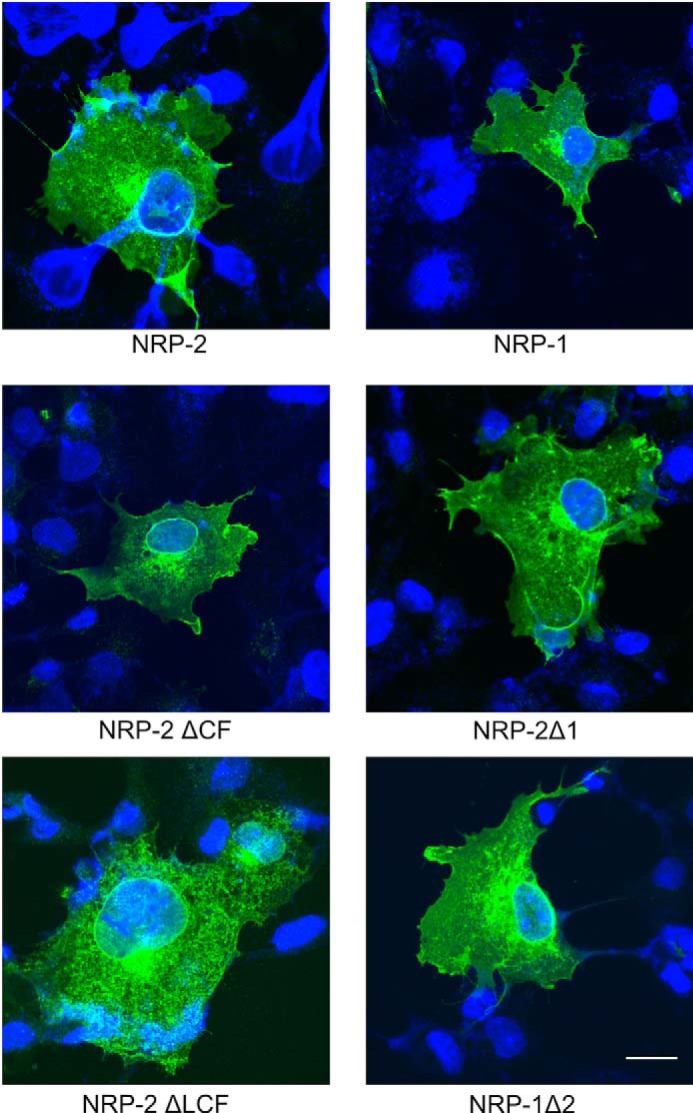
Localization of NRP domain deletion mutants and MAM domain chimera. V5-tagged wild type and mutant NRP proteins were transiently expressed in COS-1 cells. Following methanol fixation and permeabilization, protein cellular localization was determined by indirect immunofluorescence microscopy using an anti-V5 antibody and an FITC-conjugated goat anti-mouse antibody (green) with DAPI staining (blue) indicating the location of the nucleus, as described under “Experimental Procedures.” Bar, 20 μm.
Replacing the NRP-2 MAM Domain with That of NRP-1 Reduces Its Polysialylation
To specifically evaluate the role of the MAM domain in NRP-2 polysialylation, we created chimeric proteins using sequences from NRP-2 and NRP-1. The NRP-1 MAM domain shares 35% sequence identity with that of NRP-2, but the polysialylation of NRP-1 has not been reported. We swapped the MAM domains of these two proteins with the expectation that replacing the NRP-2 MAM domain with that of NRP-1 (NRP-2Δ1) would substantially reduce or eliminate polysialylation, whereas replacing the NRP-1 MAM domain with that of NRP-2 (NRP-1Δ2) may actually allow NRP-1 polysialylation if appropriate glycan acceptors are present (Fig. 1).
NRP-1, NRP-2, and the two chimeric proteins were co-expressed in COS-1 cells with ST8SiaIV-Myc, and their polysialylation was evaluated by immunoprecipitation, followed by immunoblotting with the 12F8 anti-polySia antibody (Fig. 4A). We found that the NRP-2Δ1 protein did exhibit lower polysialylation than wild type NRP-2 (18 ± 11% (S.D.), p < 0.0001). However, to our surprise, we observed that NRP-1 was polysialylated by co-expressed ST8SiaIV, although, taking into account differences in expression level, its polysialylation was about 50% of that of NRP-2 (47 ± 9% (S.D.), p = 0.0002). Peptide N-glycosidase F digestion of polysialylated NRP-1 confirmed the location of polySia on O-glycans (data not shown). Replacing the NRP-1 MAM domain with that of NRP-2 led to an increase in polysialylation of this protein (to 77 ± 22% (S.D.) (p = 0.08) (not significant) of NRP-2, or a 30% increase from NRP-1 (p = 0.03)).
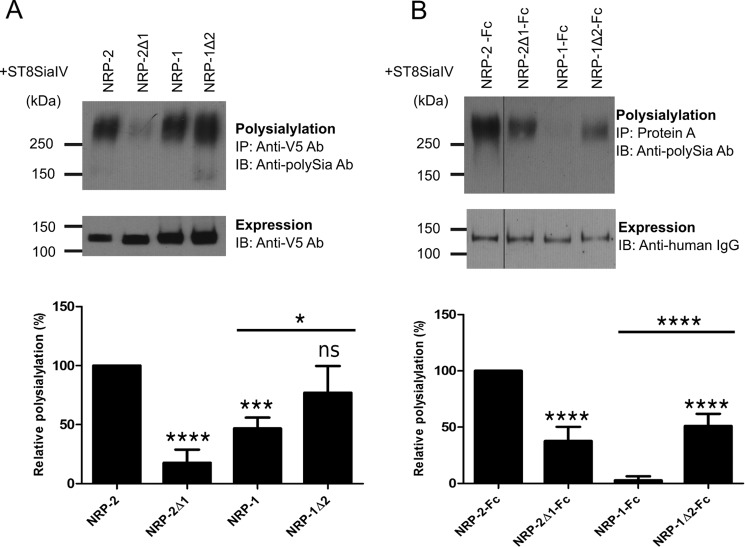
FIGURE 4.
The MAM domain is critical for NRP-2 polysialylation. A, V5-tagged full-length NRP proteins were co-expressed with ST8SiaIV-Myc in COS-1 cells. NRP proteins were immunoprecipitated (IP) from cell lysates using an anti-V5 antibody, and their polysialylation was assessed using the anti-polySia 12F8 antibody (top). Relative expression levels were determined by immunoblotting (IB) a boiled aliquot of each cell lysate using an anti-V5 antibody (bottom). B, soluble, Fc-tagged NRP chimeric proteins were transiently expressed with ST8SiaIV-Myc in COS-1 cells. The proteins were precipitated from the cell medium using protein A-Sepharose beads. Polysialylation was assessed by immunoblotting with the anti-polySia 12F8 antibody (top). Relative protein expression levels were determined by removing the bound protein from half of the protein A-Sepharose beads by boiling and immunoblotting with an HRP-linked anti-human IgG (bottom). The line separating NRP-2-Fc and NRP-2Δ1-Fc reflects the movement of the NRP-2-Fc lane from the same gel for the purposes of direct comparison with NRP-2Δ1-Fc. Quantification of the experiments shown in A and B was performed as described under “Experimental Procedures” with data from 5 and 7 different experiments, respectively, with error bars representing S.D. Statistical analysis was performed using unpaired Student's t tests. *, 0.01 < p < 0.05; ***, 0.0001 < p < 0.001; ****, p ≤ 0.0001; ns, p ≥ 0.05 with respect to wild type NRP-2, which is normalized to 100%. Other comparisons are indicated by a line above the compared bars in the graph.
To determine whether the differences in trafficking and localization of the chimeric proteins impacted their polysialylation, we localized these proteins following expression in COS-1 cells. Wild type NRP-1 and NRP-2 are localized to the cell surface and in internal compartments, including the perinuclear Golgi region (Fig. 3). However, whereas NRP-2Δ1 and NRP-1Δ2 showed considerable surface staining, we also observed higher internal staining, suggesting that these proteins may take longer to fold than their wild type counterparts. These differences in localization suggest that the polysialylation observed for the chimeric proteins may be an underestimate.
Differences in Intracellular Trafficking May Impact Levels of NRP Polysialylation
It was very surprising that NRP-1 was polysialylated in our COS-1 cell system, because unlike NRP-2, this protein has not been reported to be polysialylated in vivo. We did not expect that ST8SiaIV overexpression per se would lead to aberrant NRP-1 polysialylation, because previous work demonstrated that high expression of polySTs in COS-1 cells does not lead to aberrant polysialylation of non-substrate endogenous or co-expressed glycoproteins (36).3 However, the high level expression of ST8SiaIV might lead to its localization in compartments beyond the Golgi, such as in the endosomal system. A high level of polyST expression in endosomes combined with repeated recycling of a substrate through these compartments could lead to the polysialylation of a weaker substrate to levels similar to that observed for a stronger substrate. Could this be the case for NRP-1 (weaker substrate) versus NRP-2 (stronger substrate)?
Both NRP-1 and NRP-2 are co-receptors for growth factors and semaphorins and as such are expected to be endocytosed and recycled to the cell surface during the course of receptor activation and signal transduction. Sequences in their cytoplasmic tails are required for endocytic trafficking and recycling (37). The endocytic compartments are linked to the late Golgi/trans-Golgi network, where sialyltransferases, including polySTs, reside. Previous work by Duncan and Kornfeld (38) demonstrated that mannose 6-phosphate receptors are endocytosed from the cell surface to an internal compartment, where they are resialylated. Notably, it was previously shown that NRP-1 undergoes continuous endocytosis and recycling even in the absence of the VEGF receptor or VEGF (39). Less is known about NRP-2 endocytic trafficking. However, a study by Okon et al. (40) indicates that NRP-1 and NRP-2 are not found in the same endocytic vesicles. We wondered whether some or all of the observed NRP-1 polysialylation might reflect multiple exposures to highly expressed ST8SiaIV during the course of its continuous endocytosis and recycling.
To test this idea, we created soluble Fc-tagged NRP-1 and NRP-2 and chimeric proteins with the idea that this would ensure that each would pass through ST8SiaIV-containing compartments one time. Fc-tagged proteins were co-expressed with ST8SiaIV in COS-1 cells and recovered from cell medium using protein A-Sepharose beads, and polysialylation was assessed by immunoblotting using the 12F8 anti-polySia antibody. NRP-2Δ1-Fc exhibited 38 ± 13% (S.D.) (p < 0.0001) of the polysialylation of NRP-2-Fc, slightly higher than that seen for the membrane-associated protein (Fig. 4, compare A and B) but again demonstrating the importance of the MAM domain in NRP-2 polysialylation. In contrast, the polysialylation profile of the soluble, Fc-tagged NRP-1 proteins was significantly different from that of their membrane-associated forms. Little to no polysialylation of NRP-1-Fc was observed, and replacing the NRP-1 MAM domain with that of NRP-2 in the NRP-1Δ2-Fc protein led to a 51 ± 11% (S.D.) (p < 0.0001) increase in its polysialylation (Fig. 4B). These results highlight the importance of the NPR-2 MAM domain for polysialylation but also suggest that the continuous recycling of NRP-1 between the cell surface and internal compartments containing ST8SiaIV could enhance the polysialylation of this membrane-associated protein.
The MAM Domain and the Adjacent Linker Region Are Necessary and Sufficient for NRP-2 Polysialylation
In Fig. 4, we noticed that replacing the MAM domain of NRP-2 with that of NRP-1 did not eliminate polysialylation completely. Likewise, replacing the MAM domain of NRP-1 with that of NRP-2 did not bring the polysialylation of the NRP-1Δ2-Fc protein to the level of NRP-2-Fc. These observations suggest that sequences in NRP-2 outside the MAM domain may also be important. The linker region between NRP-2 MAM and the second F5/8 domain contains sites for O-glycan addition that are modified with polySia in the wild type protein (Fig. 1) (35). The inability of the NRP-1Δ2-Fc protein to attain the level of polysialylation observed for NRP-2-Fc may reflect differences in the O-glycosylation status of this region in NRP-2 and NRP-1 or possibly the absence of sequences that allow secondary ST8SiaIV interactions, as seen for NCAM Ig5 (28).
To address the role of this linker region, we constructed LM chimeras and analyzed their polysialylation by ST8SiaIV (Figs. 1 and 5). Replacing both the NRP-2 MAM domain and linker region with those sequences from NRP-1 almost completely eliminated polysialylation of NRP-2-Fc (Fig. 5, compare NRP-2-Fc, NRP-2Δ1-Fc, and NRP-2ΔLM1-Fc). Similarly, replacing the MAM domain and linker region of NRP-1 with those sequences from NRP-2 resulted in robust polysialylation to levels seen for NRP-2-Fc (Fig. 5, compare NRP-1-Fc, NRP-1Δ2-Fc, and NRP-1ΔLM2-Fc). These results indicate that together, the MAM domain and the linker between this domain and the F5/8-2 domain are necessary and sufficient for NRP-2 polysialylation.
FIGURE 5.
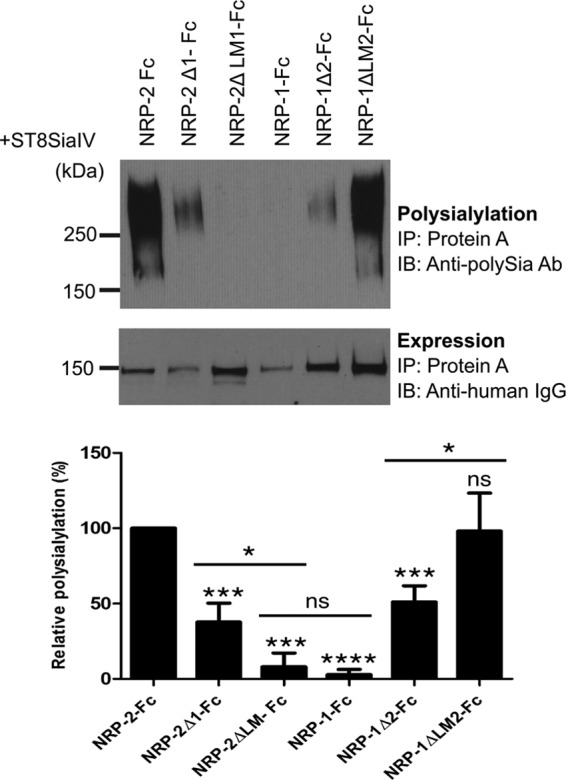
Both the MAM domain and the adjacent linker region between MAM and F5/8 domain are necessary and sufficient for polysialylation. The Fc-linked NRP MAM and LM chimeras were coexpressed with ST8SiaIV in COS-1 cells. The proteins were precipitated (IP) from the cell medium using protein A-Sepharose beads. Polysialylation of these proteins was assessed by immunoblotting (IB) using the anti-polySia 12F8 antibody (top). Relative protein expression levels were determined by removing the bound protein from half of the protein A-Sepharose beads by boiling and immunoblotting with an HRP-linked anti-human IgG (bottom). Quantification of the experimental results was performed as described under “Experimental Procedures” with data from four different experiments with error bars representing S.D. Statistical analysis was performed using unpaired Student's t tests. *, 0.01 < p < 0.05; ***, 0.0001 < p < 0.001; ****, p ≤ 0.0001; ns, p ≥ 0.05 with respect to wild type NRP-2, which is normalized to 100%. Other comparisons are indicated by a line above the compared bars in the graph.
The NRP-2 MAM Domain Is Required for ST8SiaIV Recognition
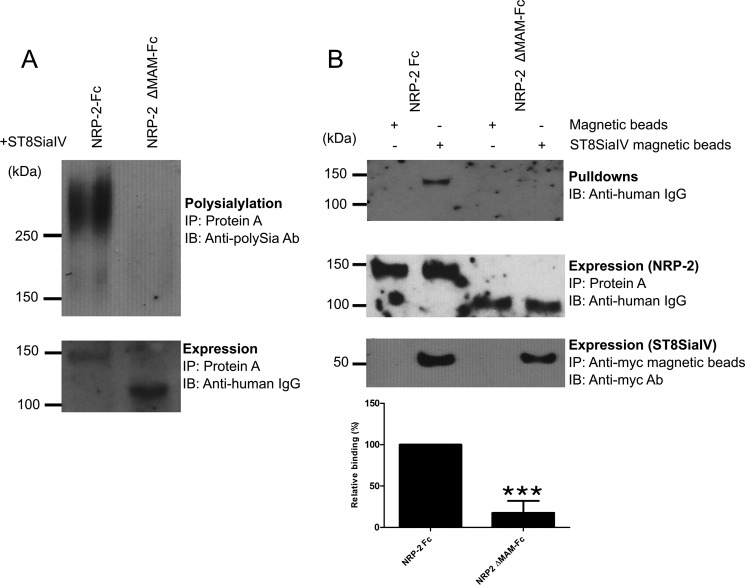
Our model of protein-specific polysialylation suggests that a polyST first recognizes and binds a protein determinant on the substrate prior to polysialylation of its glycans. We predict that this is the basis for the protein specificity of polysialylation and that it probably promotes the initial polymerization process until the length of the growing polySia chain makes the polyST-substrate protein-protein interaction impossible. Our work with NCAM demonstrated that its FN1 domain is required for polyST recognition and subsequent polysialylation. To evaluate the role of the NRP-2 MAM domain in ST8SiaIV recognition, we created a new mutant in which the MAM domain was deleted from soluble NRP-2-Fc (NRP-2 ΔMAM-Fc) (Figs. 1 and 6A). Evaluation of the polysialylation of the NRP-2 ΔMAM-Fc demonstrated that it was not polysialylated by ST8SiaIV (Fig. 6A). To determine whether this protein was able to be recognized and bind ST8SiaIV, we devised a pull-down experiment using ST8SiaIV-Myc bound to anti-Myc magnetic beads and serum-free medium containing secreted NRP-2-Fc and NRP-2 ΔMAM-Fc. Binding of these soluble proteins to anti-Myc magnetic beads in the absence of ST8SiaIV was used as a control. We observed that neither NRP-2-Fc nor NRP-2 ΔMAM-Fc bound to the anti-Myc magnetic beads, and NRP-2 ΔMAM-Fc demonstrated substantially less binding than NRP-2-Fc to the ST8SiaIV-Myc-loaded anti-Myc magnetic beads (17 ± 14% (S.D.), p = 0.0005) (Fig. 6B). Taken together, these results suggest that the NRP-2 MAM domain contains sequences essential for ST8SiaIV recognition and polysialylation.
FIGURE 6.
The NRP-2 MAM domain is essential for NRP-2 recognition as well as polysialylation by ST8SiaIV. A, the MAM domain was deleted from the NRP-2-Fc construct (NRP-2ΔMAM-Fc, Fig. 1). NRP-2-Fc and NRP-2ΔMAM-Fc were expressed with ST8SiaIV in COS-7 cells and precipitated (IP) from the medium using protein A-Sepharose beads, and their expression (bottom) and polysialylation (top) were assessed as described under “Experimental Procedures.” IB, immunoblotting. B, ST8SiaIV, NRP-2-Fc, and NRP-2ΔMAM-Fc were individually expressed in COS-7 cells cultured in serum-free medium. Anti-Myc magnetic beads were loaded with ST8SiaIV-Myc. Media containing either NRP-2-Fc or NRP-2ΔMAM-Fc were incubated with either anti-Myc magnetic beads (control) or ST8SiaIV-bound anti-Myc magnetic beads. Proteins bound to the magnetic beads were immunoblotted with HRP-linked anti-human IgG to determine the proportion of NRP-2-Fc or NRP-2ΔMAM-Fc bound to ST8SiaIV-Myc (top). An aliquot from the medium was incubated with protein A-Sepharose and was immunoblotted with HRP-linked anti-human IgG to assess the relative expression levels of NRP-2-Fc and NRP-2ΔMAM-Fc (middle). The amount of ST8SiaIV in complex with the NRP-2-Fc proteins was determined by immunoblotting these complexes with the anti-Myc antibody (bottom). Quantification of the experimental results was performed as described under “Experimental Procedures” with data from three different experiments with error bars representing S.D. Statistical analysis was performed using unpaired Student's t tests. ***, 0.0001 < p < 0.001 with respect to wild type NRP-2, which is normalized to 100%.
Glu652 and Glu653 on the Surface of the NRP-2 MAM Domain Are Critical for Polysialylation
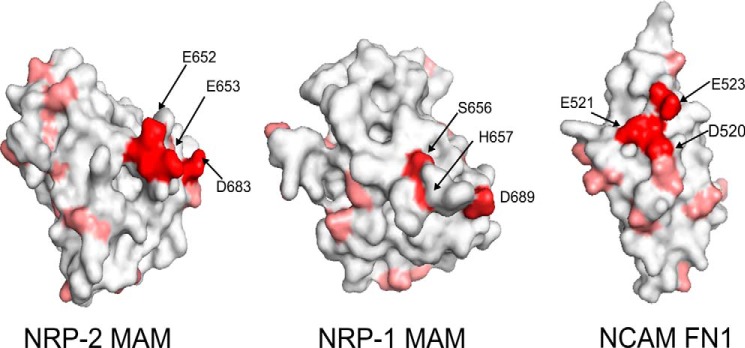
Previous work in our laboratory demonstrated the importance of an acidic patch on the surface of the FN1 domain of NCAM for polyST recognition and NCAM polysialylation (27, 28). To identify acidic residues on the surface of NRP MAM domains, we constructed homology models of both NRP-1 and NRP-2 MAM domains using the SWISS-MODEL modeling server (41–44) and the meprin A MAM domain as a template (Protein Data Bank entry 4GWN) (45). This MAM domain exhibits 26% sequence identity with the NRP-2 MAM domain and 25% identity with the NRP-1 MAM domain. The two NRP MAM domains share 35% sequence identity. Our model of the NRP-2 MAM domain revealed several acidic residues but not the three-residue acidic patch observed in NCAM FN1 (Fig. 7). However, the NRP-2 MAM domain does possess two adjacent acidic residues, Glu652 and Glu653, which are missing from the analogous surface of the NRP-1 MAM domain (replaced by Ser656 and His657). In addition, Asp683 is another acidic residue near Glu652 and Glu653 in the NRP-2 folded structure. Interestingly, when NRP-1 and NRP-2 sequences are aligned, Asp683 in the NRP-2 sequence matches with Asp689 in the NRP-1 MAM domain. We mutated these residues to alanines in NRP-2-Fc and compared the polysialylation of the mutants with that of the wild type protein (Fig. 8). Individually replacing Glu652 or Glu653 with alanine (E652A and E653A) resulted in decreases in polysialylation to 59 ± 20% (S.D.) (p = 0.001) and 39 ± 16% (S.D.) (p = 0.0003) of that of the wild type enzyme, respectively (Fig. 8, compare NRP-2-Fc, E652A, and E653A). Replacing both acidic residues with alanine led to a larger decrease in polysialylation to 15 ± 11% (S.D.) (p = 0.0003) of that seen for the wild type protein (Fig. 8, compare NRP-2-Fc and E652A/E653A). In contrast, replacing Asp683 with alanine had no effect on NRP-2-Fc polysialylation (Fig. 8, D683A). These results suggested that Glu652 and Glu653 are important for NRP-2 polysialylation and may be part of a recognition site for the ST8SiaIV. We tested this possibility by comparing the ST8SiaIV binding of NRP-2-Fc with that of NRP-2-Fc E652A/E653A using the pull-down assay described above. Unfortunately, we saw no substantial decrease in ST8SiaIV binding for the E652A/E653A mutant (data not shown). This might not be so surprising because binding of NCAM, as assayed by co-immunoprecipitation, was only marginally decreased when the FN1 acidic patch was replaced by arginine residues, a mutation that eliminated NCAM polysialylation (25).
FIGURE 7.
Structural modeling shows the distribution of acidic residues on the NCAM FN1 and NRP-2 and NRP-1 MAM domains. The structure of the NRP MAM domains was modeled on meprin A MAM domain (Protein Data Bank entry 4GWN (45)) using the SWISS-MODEL homology modeling server. Analysis of these models reveals two adjacent acidic residues, Glu652 (E652) and Glu653 (E653), on the surface of the NRP-2 MAM domain (left), which are not present on the surface of the NRP-1 MAM domain (right). Instead, Ser656 (S656) and His657 (H657) in the linear sequence of NRP-1 replace Glu652 and Glu653 in NRP-2. A similar acidic patch in the NCAM FN1 domain (Protein Data Bank entry 2HAZ (48)) was shown to be critical for the polysialylation of NCAM (Asp520, Glu521, Glu523).
FIGURE 8.
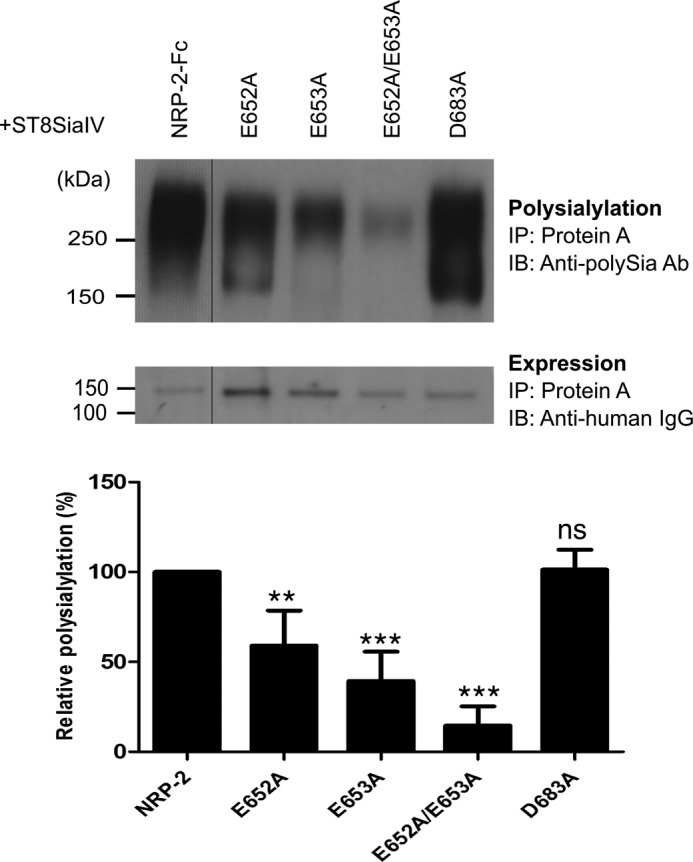
Glu652 and Glu653 on the surface of NRP-2 MAM domain are important for the polysialylation of NRP-2. Acidic residues Glu652 and Glu653 were mutated in NRP-2-Fc. As a control, Asp683 was also mutated to alanine in NRP-2-Fc. NRP-2-Fc and its mutants were expressed with ST8SiaIV in COS-1 cells, recovered from the cell medium using protein A-Sepharose beads (IP), and immunoblotted (IB) with the anti-polySia 12F8 antibody to evaluate the effect of the MAM domain mutations on NRP-2 polysialylation (top). Relative protein expression levels were determined by removing the bound protein from an aliquot of the protein A-Sepharose beads by boiling and immunoblotting with an HRP-linked anti-human IgG (bottom). The line separating NRP-2-Fc and the E652A mutant reflects the removal of an extraneous lane so that the mutants and NRP-2-Fc can be more directly compared. Quantification of the experimental results was performed as described under “Experimental Procedures” with data from four different experiments with error bars representing S.D. Statistical analysis was performed using unpaired Student's t tests. **, 0.001 < p < 0.01; ***, 0.0001 < p < 0.001; ns, p ≥ 0.05 with respect to wild type NRP-2, which is normalized to 100%.
Taken together, these data demonstrate that the MAM domain of NRP-2 plays a primary role in polyST recognition and NRP-2 polysialylation, that acidic residues in the MAM domain are critical for polysialylation, and that secondary interactions with the adjacent linker region containing the O-glycans that are polysialylated may contribute to enzyme recognition and NRP-2 polysialylation.
Discussion
In this study, we have evaluated the role of the MAM domain and adjacent sequences in NRP-2 polysialylation. Using chimeric and mutant proteins, we have provided evidence that the MAM domain plays a primary role in NRP-2 polysialylation and that sequences in this domain are critical for ST8SiaIV recognition (Figs. 4 and 6). In addition, our data suggest that the adjacent linker region, which contains the O-glycans that are polysialylated, may also possess a secondary interaction site for ST8SiaIV, because elimination of both the linker and the MAM regions is necessary to completely eliminate polysiaylation (Fig. 5). This would be analogous to what is observed for NCAM polysialylation, where an acidic surface patch on the FN1 domain forms the primary interaction site, and the adjacent Ig5 domain, which contains the N-glycans that are polysialylated, serves as a secondary or stabilizing interaction site (28).
Based on our work on the sequences in NCAM and the polySTs required for substrate recognition and polysialylation, we looked for an acidic surface patch on the NRP-2 MAM domain that could, as in NCAM FN1, mediate polyST recognition. Two adjacent acidic residues, Glu652 and Glu653 on the surface of NRP-2 MAM, are reminiscent of Asp520 and Glu521 that constitute two of the three residues forming the acidic patch on NCAM FN1 (27). Replacing Glu652 and Glu653 with alanines substantially decreased NRP-2 polysialylation, suggesting that they may play a role in polyST recognition. However, evaluation of the NRP-2 E652A/E653A mutant in pull-down studies did not show a substantial decrease in ST8SiaIV binding. This was similar to what we saw in our evaluation of NCAM acidic patch mutants and ST8SiaIV binding. Both the elimination of the NCAM FN1 domain and the NRP-2 MAM domain substantially decreased or eliminated their interactions with ST8SiaIV (Fig. 6) (25). However, replacing the three FN1 acidic patch residues with arginine, a change that eliminated NCAM polysialylation, had only a modest impact on NCAM-ST8SiaIV interaction (25). At the time we suspected that other residues in FN1 might be participating in the binding, but later evidence suggested that residues in the adjacent Ig5 domain probably formed a secondary binding site (25, 28). This could be the case for the NRP-2 linker, or alternatively, in the absence of Glu652 and Glu653, a non-productive binding interaction may occur that does not optimally position ST8SiaIV to polysialylate NRP-2 O-glycans.
A suprising result in this study was our finding that membrane-associated NRP-1 was polysialylated by co-expressed ST8SiaIV (Fig. 4). The polysialylation of membrane-associated NRP-1 was weaker than that of NRP-2, suggesting deficiencies in the ability of the ST8SiaIV to recognize the NRP-1 MAM domain, fewer O-glycans to polysialylate in its linker region, and/or a weaker secondary interaction site in its linker region.
The importance of the integrity of the modular recognition-modification unit in polysialylation is illustrated by previous work with NCAM chimeras generated with analogous domains from the olfactory cell adhesion molecule (OCAM), an unpolysialylated protein with a domain structure identical to that of NCAM. Although the FN1 domain of OCAM could partially replace that of NCAM to support polyST recognition and polysialylation, and N-glycans were present in appropriate locations on the OCAM Ig5 domain, other sequences in the OCAM Ig5 domain served to block a stable interaction and OCAM polysialylation (28, 29). In addition, the FN1 domain of OCAM had a weak polyST recognition site with only one of the three acidic patch residues found in the NCAM FN1 present; inserting the other two acidic residues substantially increased OCAM polysialylation when the blocking sequences in the OCAM Ig5 domain were removed (28). We also attempted to recreate an acidic patch in NRP-1-Fc to see if we could promote its polysialylation without success (data not shown).
The linker regions of NRP-1 and NRP-2 are different with respect to their length and the density and arrangement of threonine residues that could serve as O-glycosylation and polysialylation sites (see Fig. 1B). Rollenhagen et al. (35) have identified four clustered threonine residues (Thr613, Thr613, Thr614, and Thr619) as the attachment sites for the O-glycans that are polysialylated (Fig. 1B, underlined residues). In NRP-1, the linker is shorter, and the threonine residues are more spread out and would not be expected to be positioned relative to the MAM domain like those in NRP-2. In addition, NRP-1 is modified by a glycosaminoglycan chain (heparan sulfate or chondroitin sulfate) attached to Ser612 (46) (Fig. 1B). It would be tempting to speculate that the presence of this glycosaminoglycan chain might block polyST access to O-glycans in this region. However, replacing this serine with alanine had no impact on full-length NRP-1 polysialylation (data not shown).
In addition to the finding that membrane-associated NRP-1 was polysialylated by co-expressed ST8SiaIV, another remarkable finding in this study was that a soluble Fc-tagged version of NRP-1 was not polysialylated. How can changes in membrane association influence the process of polysialylation? NRPs are highly active in terms of cellular trafficking. Cytoplasmic tails of NRPs associate with the adaptor protein synectin/GAIP-interacting protein C terminus, which couples NRPs to intracellular trafficking and cytoskeletal assembly machinery (37). These interactions have been shown to govern the fate of VEGF receptors upon stimulation. NRP-1, irrespective of expression of VEGF receptors, undergoes internalization, and its own recycling and that of associated receptors is controlled by Rab11 interactions mediated by synectin/GAIP-interacting protein C terminus (39). Intracellular trafficking of NRP-2 has been less well studied, but a report by Okon et al. (40) shows that the two NRPs are found in different Rab vesicles. Work by Bae et al. (47) has also suggested that the two NRPs have different intracellular fates due to differences in their trafficking.
Surprised to see NRP-1 polysialylated by co-expressed ST8SiaIV, we hypothesized that membrane-associated NRP-1 uniquely recycles from the surface to a ST8SiaIV-containing compartment (Golgi or an associated endosomal compartment), allowing multiple interactions with ST8SiaIV and increasing its potential for polysialylation, despite weaker recognition elements in its MAM domain and fewer or less optimally positioned O-glycans in its linker region. We reasoned that creating soluble NRP-1 and NRP-2 proteins would equalize their exposure to ST8SiaIV by allowing only one pass through the secretory pathway. We observed that NRP-2-Fc continued to be polysialylated by co-expressed ST8SiaIV, whereas NRP-1-Fc was not (Fig. 4B). Adding the NRP-2 MAM domain to the soluble NRP-1-Fc led to polysialylation equivalent to ∼50% of NRP-2-Fc (NRP-1Δ2-Fc), and adding both the NRP-2 MAM domain and O-glycosylated linker region allowed polysialylation essentially equivalent to that of NRP-2-Fc (NRP1ΔLM2-Fc). Taken together, these results suggest that neither the MAM domain nor the linker region of NRP-1 is optimal for polysialylation and raise the question of whether NRP-1 is polysialylated in vivo and, if so, how its recycling controls its polysialylation.
In summary, these results demonstrate the importance of the NRP-2 MAM domain for polyST recognition and then polysialylation of the O-glycans in the adjacent linker region, thereby substantiating the protein-specific polysialylation paradigm. This work raises additional questions about the NRP-2 MAM interaction surface, the possibility of NRP-1 polysialylation in vivo, and the role of polySia in NRP-2, and possibly NRP-1, signaling.
Author Contributions
G. P. B., N. R. J. F., and K. J. C. conceived and planned the study. G. P. B. and N. R. J. F. created the chimeric and mutant proteins. G. P. B. performed the experiments and, together with K. J. C., wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Gerd Prehna for assistance with the modeling of MAM domains and Dr. Helena Palka-Hamblin, Navika Shukla, and Masaya Hane for helpful discussions.
This work was supported by National Institutes of Health Grant RO1 GM101949 (to K. J. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
G. Bhide, unpublished data.
- polySia
- polysialic acid
- polyST
- polysialyltransferase
- NCAM
- neural cell adhesion molecule
- NRP
- neuropilin
- MAM
- meprin A5 antigen-μ tyrosine phosphatase domain
- CCL21
- chemokine (C-C motif) ligand 21
- CCR7
- chemokine (C-C motif) receptor 7
- FN1
- fibronectin type III repeat 1 of NCAM
- PNGase F
- peptide N-glycosidase F
- CUB
- complement C1r/C1s, Uegf, Bmp1
- F5/8
- factor 5/8 homology
- OCAM
- olfactory cell adhesion molecule
- LM
- linker MAM.
References
- 1. Sato C., and Kitajima K. (2013) Disialic, oligosialic and polysialic acids: distribution, functions and related disease. J. Biochem. 154, 115–136 [DOI] [PubMed] [Google Scholar]
- 2. Colley K. J., Kitajima K., and Sato C. (2014) Polysialic acid: biosynthesis, novel functions and applications. Crit. Rev. Biochem. Mol. Biol. 49, 498–532 [DOI] [PubMed] [Google Scholar]
- 3. Schnaar R. L., Gerardy-Schahn R., and Hildebrandt H. (2014) Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 94, 461–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finne J. (1982) Occurrence of unique polysialosyl carbohydrate units in glycoproteins of developing brain. J. Biol. Chem. 257, 11966–11970 [PubMed] [Google Scholar]
- 5. Curreli S., Arany Z., Gerardy-Schahn R., Mann D., and Stamatos N. M. (2007) Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J. Biol. Chem. 282, 30346–30356 [DOI] [PubMed] [Google Scholar]
- 6. Galuska S. P., Rollenhagen M., Kaup M., Eggers K., Oltmann-Norden I., Schiff M., Hartmann M., Weinhold B., Hildebrandt H., Geyer R., Mühlenhoff M., and Geyer H. (2010) Synaptic cell-2,8. Proc. Natl. Acad. Sci. U.S.A. 107, 10250–10255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yabe U., Sato C., Matsuda T., and Kitajima K. (2003) Polysialic acid in human milk. CD36 is a new member of mammalian polysialic acid-containing glycoprotein. J. Biol. Chem. 278, 13875–13880 [DOI] [PubMed] [Google Scholar]
- 8. James W. M., and Agnew W. S. (1987) Multiple oligosaccharide chains in the voltage-sensitive Na channel from Electrophorus electrius: evidence for α-2,8-linked polysialic acid. Biochem. Biophys. Res. Commun. 148, 817–826 [DOI] [PubMed] [Google Scholar]
- 9. Close B. E., and Colley K. J. (1998) In vivo autopolysialylation and localization of the polysialyltransferases PST and STX. J. Biol. Chem. 273, 34586–34593 [DOI] [PubMed] [Google Scholar]
- 10. Falconer R. A., Errington R. J., Shnyder S. D., Smith P. J., and Patterson L. H. (2012) Polysialyltransferase: a new target in metastatic cancer. Curr. Cancer Drug Targets 12, 925–939 [DOI] [PubMed] [Google Scholar]
- 11. Tanaka F., Otake Y., Nakagawa T., Kawano Y., Miyahara R., Li M., Yanagihara K., Nakayama J., Fujimoto I., Ikenaka K., and Wada H. (2000) Expression of polysialic acid and STX, a human polysialyltransferase, is correlated with tumor progression in non-small cell lung cancer. Cancer Res. 60, 3072–3080 [PubMed] [Google Scholar]
- 12. Rutishauser U. (2008) Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 9, 26–35 [DOI] [PubMed] [Google Scholar]
- 13. Johnson C. P., Fujimoto I., Rutishauser U., and Leckband D. E. (2005) Direct evidence that neural cell adhesion molecule (NCAM) polysialylation increases intermembrane repulsion and abrogates adhesion. J. Biol. Chem. 280, 137–145 [DOI] [PubMed] [Google Scholar]
- 14. Prud'homme G. J., and Glinka Y. (2012) Neuropilins are multifunctional coreceptors involved in tumor initiation, growth, metastasis and immunity. Oncotarget 3, 921–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neufeld G. (2002) The neuropilins multifunctional semaphorin and VEGF receptors that modulate axon guidance and angiogenesis. Trends Cardiovasc. Med. 12, 13–19 [DOI] [PubMed] [Google Scholar]
- 16. Pellet-Many C., Frankel P., Jia H., and Zachary I. (2008) Neuropilins: structure, function and role in disease. Biochem. J. 411, 211–226 [DOI] [PubMed] [Google Scholar]
- 17. Grandclement C., and Borg C. (2011) Neuropilins: a new target for cancer therapy. Cancers 3, 1899–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Friedl P., den Boer A. T., and Gunzer M. (2005) Tuning immune responses: diversity and adaptation of the immunological synapse. Nat. Rev. Immunol. 5, 532–545 [DOI] [PubMed] [Google Scholar]
- 19. Bax M., van Vliet S. J., Litjens M., García-Vallejo J. J., and van Kooyk Y. (2009) Interaction of polysialic acid with CCL21 regulates the migratory capacity of human dendritic cells. PLoS One 4, e6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rey-Gallardo A., Delgado-Martín C., Gerardy-Schahn R., Rodríguez-Fernández J. L., and Vega M. A. (2011) Polysialic acid is required for neuropilin-2a/b-mediated control of CCL21-driven chemotaxis of mature dendritic cells and for their migration in vivo. Glycobiology 21, 655–662 [DOI] [PubMed] [Google Scholar]
- 21. Kiermaier E., Moussion C., Veldkamp C. T., Gerardy-Schahn R., de Vries I., Williams L. G., Chaffee G. R., Phillips A. J., Freiberger F., Imre R., Taleski D., Payne R. J., Braun A., Förster R., Mechtler K., Mühlenhoff M., Volkman B. F., and Sixt M. (2016) Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 351, 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stamatos N. M., Zhang L., Jokilammi A., Finne J., Chen W. H., El-Maarouf A., Cross A. S., and Hankey K. G. (2014) Changes in polysialic acid expression on myeloid cells during differentiation and recruitment to sites of inflammation: role in phagocytosis. Glycobiology 24, 864–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Werneburg S., Mühlenhoff M., Stangel M., and Hildebrandt H. (2015) Polysialic acid on SynCAM 1 in NG2 cells and on neuropilin-2 in microglia is confined to intracellular pools that are rapidly depleted upon stimulation. Glia 63, 1240–1255 [DOI] [PubMed] [Google Scholar]
- 24. Close B. E., Mendiratta S. S., Geiger K. M., Broom L. J., Ho L. L., and Colley K. J. (2003) The minimal structural domains required for neural cell adhesion molecule polysialylation by PST/ST8Sia IV and STX/ST8Sia II. J. Biol. Chem. 278, 30796–30805 [DOI] [PubMed] [Google Scholar]
- 25. Thompson M. G., Foley D. A., Swartzentruber K. G., and Colley K. J. (2011) Sequences at the interface of the fifth immunoglobulin domain and first fibronectin type III repeat of the neural cell adhesion molecule are critical for its polysialylation. J. Biol. Chem. 286, 4525–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Angata K., Suzuki M., McAuliffe J., Ding Y., Hindsgaul O., and Fukuda M. (2000) Differential biosynthesis of polysialic acid on neural cell adhesion molecule (NCAM) and oligosaccharide acceptors by three distinct alpha 2,8-sialyltransferases, ST8Sia IV (PST), ST8Sia II (STX), and ST8Sia III. J. Biol. Chem. 275, 18594–18601 [DOI] [PubMed] [Google Scholar]
- 27. Mendiratta S. S., Sekulic N., Lavie A., and Colley K. J. (2005) Specific amino acids in the first fibronectin type III repeat of the neural cell adhesion molecule play a role in its recognition and polysialylation by the polysialyltransferase ST8Sia IV/PST. J. Biol. Chem. 280, 32340–32348 [DOI] [PubMed] [Google Scholar]
- 28. Thompson M. G., Foley D. A., and Colley K. J. (2013) The polysialyltransferases interact with sequences in two domains of the neural cell adhesion molecule to allow its polysialylation. J. Biol. Chem. 288, 7282–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Foley D. A., Swartzentruber K. G., Thompson M. G., Mendiratta S. S., and Colley K. J. (2010) Sequences from the first fibronectin type III repeat of the neural cell adhesion molecule allow O-glycan polysialylation of an adhesion molecule chimera. J. Biol. Chem. 285, 35056–35067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maley F., Trimble R. B., Tarentino A. L., and Plummer T. H. (1989) Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180, 195–204 [DOI] [PubMed] [Google Scholar]
- 31. Nakamura F., Tanaka M., Takahashi T., Kalb R. G., and Strittmatter S. M. (1998) Neuropilin-1 extracellular domains mediate semaphorin D/III-induced growth cone collapse. Neuron 21, 1093–1100 [DOI] [PubMed] [Google Scholar]
- 32. Appleton B. A., Wu P., Maloney J., Yin J., Liang W. C., Stawicki S., Mortara K., Bowman K. K., Elliott J. M., Desmarais W., Bazan J. F., Bagri A., Tessier-Lavigne M., Koch A. W., Wu Y., Watts R. J., and Wiesmann C. (2007) Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding. EMBO J. 26, 4902–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vander Kooi C. W., Jusino M. A., Perman B., Neau D. B., Bellamy H. D., and Leahy D. J. (2007) Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Natl. Acad. Sci. U.S.A. 104, 6152–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sawma P., Roth L., Blanchard C., Bagnard D., Crémel G., Bouveret E., Duneau J. P., Sturgis J. N., and Hubert P. (2014) Evidence for new homotypic and heterotypic interactions between transmembrane helices of proteins involved in receptor tyrosine kinase and neuropilin signaling. J. Mol. Biol. 426, 4099–4111 [DOI] [PubMed] [Google Scholar]
- 35. Rollenhagen M., Buettner F. F., Reismann M., Jirmo A. C., Grove M., Behrens G. M., Gerardy-Schahn R., Hanisch F. G., and Mühlenhoff M. (2013) Polysialic acid on neuropilin-2 is exclusively synthesized by the polysialyltransferase ST8SiaIV and attached to mucin-type O-glycans located between the b2 and c domain. J. Biol. Chem. 288, 22880–22892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Close B. E., Tao K., and Colley K. J. (2000) Polysialyltransferase-1 autopolysialylation is not requisite for polysialylation of neural cell adhesion molecule. J. Biol. Chem. 275, 4484–4491 [DOI] [PubMed] [Google Scholar]
- 37. Berger P., and Ballmer-Hofer K. (2011) The reception and the party after: how vascular endothelial growth factor receptor 2 explores cytoplasmic space. Swiss Med. Wkly. 141, w13318. [DOI] [PubMed] [Google Scholar]
- 38. Duncan J. R., and Kornfeld S. (1988) Intracellular movement of two mannose 6-phosphate receptors: return to the Golgi apparatus. J. Cell Biol. 106, 617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ballmer-Hofer K., Andersson A. E., Ratcliffe L. E., and Berger P. (2011) Neuropilin-1 promotes VEGFR-2 trafficking through Rab11 vesicles thereby specifying signal output. Blood 118, 816–826 [DOI] [PubMed] [Google Scholar]
- 40. Okon I. S., Coughlan K. A., Zhang C., Moriasi C., Ding Y., Song P., Zhang W., Li G., and Zou M. H. (2014) Protein kinase LKB1 promotes RAB7-mediated neuropilin-1 degradation to inhibit angiogenesis. J. Clin. Invest. 124, 4590–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guex N., Peitsch M. C., and Schwede T. (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30, S162–S173 [DOI] [PubMed] [Google Scholar]
- 42. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 43. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kiefer F., Arnold K., Künzli M., Bordoli L., and Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arolas J. L., Broder C., Jefferson T., Guevara T., Sterchi E. E., Bode W., Stöcker W., Becker-Pauly C., and Gomis-Rüth F. X. (2012) Structural basis for the sheddase function of human meprin metalloproteinase at the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 109, 16131–16136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shintani Y., Takashima S., Asano Y., Kato H., Liao Y., Yamazaki S., Tsukamoto O., Seguchi O., Yamamoto H., Fukushima T., Sugahara K., Kitakaze M., and Hori M. (2006) Glycosaminoglycan modification of neuropilin-1 modulates VEGFR2 signaling. EMBO J. 25, 3045–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bae D., Lu S., Taglienti C. A., and Mercurio A. M. (2008) Metabolic stress induces the lysosomal degradation of neuropilin-1 but not neuropilin-2. J. Biol. Chem. 283, 28074–28080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mendiratta S. S., Sekulic N., Hernandez-Guzman F. G., Close B. E., Lavie A., and Colley K. J. (2006) A novel α-helix in the first fibronectin type III repeat of the neural cell adhesion molecule is critical for N-glycan polysialylation. J. Biol. Chem. 281, 36052–36059 [DOI] [PubMed] [Google Scholar]