Abstract
RNF6 is a little-studied ring finger protein. In the present study, we found that RNF6 was overexpressed in various leukemia cells and that it accelerated leukemia cell proliferation, whereas knockdown of RNF6 delayed tumor growth in xenografts. To find out the mechanism of RNF6 overexpression in leukemia, we designed a series of truncated constructs of RNF6 regulatory regions in the luciferase reporter system. The results revealed that the region between −144 and −99 upstream of the RNF6 transcription start site was critical and that this region contained a PBX1 recognition element (PRE). PBX1 modulated RNF6 expression by binding to the specific PRE. When PRE was mutated, RNF6 transcription was completely abolished. Further studies showed that PBX1 collaborated with PREP1 but not MEIS1 to modulate RNF6 expression. Moreover, RNF6 expression could be suppressed by doxorubicin, a major anti-leukemia agent, via down-regulating PBX1. This study thus suggests that RNF6 overexpression in leukemia is under the direction of PBX1 and that the PBX1/RNF6 axis can be developed as a novel therapeutic target of leukemia.
Keywords: leukemia, oncogene, small interfering RNA (siRNA), transcription factor, tumor cell biology, ring finger protein, RNF6, pre-B-cell leukemia homeobox 1, leukemia, promoter modulation
Introduction
The ring finger protein 6 (RNF6) belongs to the largest RING ubiquitin ligase family, and it is mapped to chromosome band 13q12.2, a harbor of several critical tumor suppressor genes (1). RNF6 is believed to be a tumor suppressor because of its chromosomal location and somatic mutations in esophageal squamous cell carcinomas (2), but confirmative evidence is not available. In contrast, recent studies suggest that RNF6 is probably an oncogene. RNF6 is found at a high level in prostate cancers. As a ubiquitin ligase, RNF6 interacts with androgen receptor (AR)3 and mediates atypical polyubiquitination chains at Lys-6 and Lys-27, thus promoting the transcriptional activity of AR by facilitating its binding to the coactivators (3). By modulating AR function, RNF6 promotes prostate cancer cell growth. In contrast, mutations and specific knockdown of RNF6 alter AR transcriptional activity and delay prostate cancer growth in xenograft models (3). RNF6 is also elevated in cisplatin-resistant human lung adenocarcinoma cells (4). Therefore, RNF6 probably plays a critical role in tumorigenesis and chemoresistance. However, the studies on RNF6 are very limited, and the biological functions and modulation of RNF6 are largely unknown.
In the present study, we evaluated the RNF6 function in leukemia cells and found that RNF6 is overexpressed in leukemia cells and contributes to leukemia cell proliferation. Furthermore, RNF6 overexpression in leukemia is found to be modulated by the transcription factor PBX1, the pre-B-cell leukemia homeobox 1.
Experimental Procedures
Cells, Culture, and Chemicals
Leukemia (OCI-AML2, HL60, NB4, THP1, K562, Jurkat, and preB-697) and multiple myeloma (H929, KMS11, LP1, OCI-My5, OPM2, and RPMI-8226) cell lines were purchased from American Type Culture Collection (Manassas, VA). Multiple myeloma, leukemia, and HEK293 cells were maintained in Iscove's modified Dulbecco's medium, RPMI 1640 medium, and Dulbecco's high glucose modified Eagle's medium, respectively. All media were supplemented with 10% fetal bovine serum, 100 μg/ml penicillin, and 100 units/ml streptomycin.
The primary leukemia cells and normal adult bone marrow samples were collected from the Department of Hematology, the First Affiliated Hospital of Soochow University. The primary blood cells were applied to isolate mono-nuclear cells by Lympholyte® Cell Separation Media according to the manufacturer's instructions (Cedarlane, Burlington, Ontario, Canada) as described previously (5). The collection and use of human tissues for this study were approved by the Institutional Review Board of Soochow University. Informed consent was obtained in accordance with the Declaration of Helsinki. Doxorubicin (DOX) and 5-amino 8-hydroxyquinoline (5AHQ) were purchased from Sigma-Aldrich.
RT-PCR
The polymerase chain reaction was performed in a 25-μl reaction system containing 12.5 μl of 2×Easy Taq SuperMix (TransGen Biotechnology, Beijing, China), 1 μl of cDNA, 0.5 μl of each primer, and 10.5 μl of sterile pure H2O. The primers used were as follows: RNF6, forward 5′-CATCAGTGGCTCTTCGGTCA-3′and reverse 5′-ATGCTCATAGTGCCTGGTGG-3′; GAPDH, forward 5′-AGTCCACTGGCGTCTTCA-3′ and reverse 5′-CTCCGACGCCTGCTTCACCA-3′. Reaction cycling conditions were 3 min at 95 °C, followed by 30 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 40 s, and 1 cycle at 72 °C for 10 min. Products were analyzed on 2% agarose gels.
Quantitative Real-time PCR (qRT-PCR)
Total RNA was extracted using the TRIzol® Reagent (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized from equal quantities of total RNA using the EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotechnology). To determine the mRNA level of RNF6, qRT-PCR was performed using Luminaris Color HiGreen High ROX qPCR Master Mix (Thermo Fisher Scientific) with StepOnePlusTM real-time PCR system (Applied Biosystems, Foster City, CA). The primers used were as follows: RNF6, forward 5′-AGAAGATGGCAGCAAGAGCG-3′ and reverse 5′-TCAAGTCAGGCTGAGATGCTAGT-3′; GAPDH, forward 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse 5′-TGGTGAAGACGCCAGTGGA-3′. The primers for PBX1 were: 5′-AGGACATCGGGGACATTTTAC-3′ and 5′-CATTAAACAAGGCAGGCTTCA-3′.
Preparation of RNF6 Lentivirus
The full-length RNF6 gene was amplified by PCR with primers, forward 5′-CCCGGAATTCATGAATCAGTCTAGATCGAGATCAG-3′ and reverse 5′-AAATATGCGGCCGCTTACCCATTGTTTGCTATGTTAGACCC-3′. The underlined sequences were recognized with EcoRI and NotI, respectively. RNF6 gene was then inserted between EcoRI and NotI of the pCDH lentiviral vector (System Biosciences, Mountain View, CA).
The lentivirus-delivered shRNAs against RNF6 (shRNF6) and the negative control (shNC) were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China). The target sequences of shRNF6#1 and shRNF6#2 were 5′-TCAGGCAATTACCTTGCAT-3′ and 5′-ATAACAGTTCCTCTTCGTA-3′, respectively.
The viral particles were prepared with a standard method according to the manufacturer's instructions and included control and package plasmids (Shanghai GeneChem Co., Ltd.). The individual plasmids were co-transfected into HEK293T cells with the calcium precipitate method. Viruses were obtained 72 h later from transfected cells and were applied to infect the appropriate cells.
Cell Growth and Viability
K562 cells infected with RNF6 or shRNF6 or scramble lentivirus were cultured for 0–6 days at a start density of 2 × 104 cells/well in a 24-well plate. The viable cells were evaluated by trypan blue exclusion staining as described previously (6).
Immunoblotting
Whole cell lysates were prepared as described previously (5). Equal amounts (30 μg) of total proteins were subjected to SDS-PAGE separation, followed by immunoblotting analyses with specific antibodies including monoclonal antibodies against RNF6 (Thermo Fisher), PBX1 (Santa Cruz Biotechnology), FLAG, Myc, HA (Medical & Biological Laboratories, Tokyo, Japan), PARP (Cell Signaling Technology Co.), PBX-regulating protein 1 (PREP1) (Signalway Biotechnology Co. Ltd., Nanjing, China), and GAPDH (Abgent, Suzhou, China). Anti-mouse IgG and anti-rabbit IgG horseradish peroxidase-conjugated antibody were purchased from R&D Systems.
Construction of the Truncated RNF6 Regulatory Regions and Mutants
Genomic DNA was extracted from HEK293 cells according to the manufacturer's protocol (Beyotime Biotechnology Institute, Nantong, China). The regulatory sequences of RNF6 were predicted by the UCSC Genome Browser website, and a series of truncated RNF6 regulatory regions or site-directed mutants was amplified by PCR referenced by the TFSearch website. These RNF6 constructs were then inserted into the pGL4 vector (Promega Corp., Madison, WI). The primers used for PCR amplification of truncated regulatory region of RNF6 are shown in Table 1. The mutation primers for the core regulatory region (−354 to −99) of RNF6 were: 1) for mutated PBX1/GATA, forward 5′-CTCCGCTGGCTGGTCCATGGTTGGGAT-3′, and reverse 5′-ATCCCAACCATGGACCAGCCAGCGGAG-3′; and 2) for mutated PBX1, forward 5′-GCTGGCTGGTGATCGGTTGGGATG-3′ and reverse 5′-CATCCCAACCGATCACCAGCCAGC-3′. The bold sequence indicates the mutated nucleotide.
TABLE 1.
The primers for PCR amplification of the regulatory regions of RNF6
| Regulatory sequence region | Primer direction | Sequences (5′-3′) |
|---|---|---|
| −2000/+2108 | Forward | GGGGTACCTTTTCCATGGGAAAAGAGGGCAAGG |
| −2000/+2108 | Reverse | TGCAGGATATCCCTGAGATTCCTGGCTTTCTGTTCAAAC |
| −1500/+2108 | Forward | GGGGTACCACACTGGTAGCTCATTGATGCTAAGG |
| −1500/+2108 | Reverse | TGCAGGATATCCCTGAGATTCCTGGCTTTCTGTTCAAAC |
| −1000/+2108 | Forward | GGGGTACCTACTGGGATGAGGAGGGAGGATCGCT |
| −1000/+2108 | Reverse | TGCAGGATATCCCTGAGATTCCTGGCTTTCTGTTCAAAC |
| −500/+2108 | Forward | GGGGTACCGGCGTGGCTGTCGGGAAAGAAGGGCT |
| −500/+2108 | Reverse | TGCAGGATATCCCTGAGATTCCTGGCTTTCTGTTCAAAC |
| −365/+2108 | Forward | GGGGTACCTCCGGAGGCGCGGCGGCAAGCCTATC |
| −365/+2108 | Reverse | TGCAGGATATCCCTGAGATTCCTGGCTTTCTGTTCAAAC |
| −200/+2108 | Forward | GGGGTACCACGCCGGGGAAAGCAGGCCATC |
| −200/+2108 | Reverse | TGCAGGATATCCCTGAGATTCCTGGCTTTCTGTTCAAAC |
| +1/+2108 | Forward | GGGGTACCTTTATTTATCGTAGTGGGGATCGTC |
| +1/+2108 | Reverse | TGCAGGATATCCCTGAGATTCCTGGCTTTCTGTTCAAAC |
| −2000/−1 | Forward | GGGGTACCTTTTCCATGGGAAAAGAGGGC |
| −2000/−1 | Reverse | TGCAGGATATCGTGGGCTCAAGAGGCCCAACCAAC |
| −500/−99 | Forward | GGGGTACCGGCGTGGCTGTCGGGAAAGAAG |
| −500/−99 | Reverse | TGCAGGATATCTGCATCCCAACCAATCACCAGCCAGCGG |
| −365/−99 | Forward | GGGGTACCTCCGGAGGCGCGGCGGCAAGCCTATC |
| −365/−99 | Reverse | TGCAGGATATCTGCATCCCAACCAATCACCAGCCAGCGG |
| −323/−99 | Forward | GGGGTACCGCCTCCAGCACCCGAGAGAAC |
| −323/−99 | Reverse | TGCAGGATATCTGCATCCCAACCAATCACCAGCCAGCGG |
| −200/−99 | Forward | GGGGTACCACGCCGGGGAAAGCAGGCCATC |
| −200/−99 | Reverse | TGCAGGATATCTGCATCCCAACCAATCACCAGCCAGCGG |
| −365/−108 | Forward | GGGGTACCTCCGGAGGCGCGGCGGCAAGCCTATC |
| −365/−108 | Reverse | TGCAGGATATCACCAATCACCAGCCAGCGGAGCG |
| −365/−144 | Forward | GGGGTACCTCCGGAGGCGCGGCGGCAAGCCTATC |
| −365/−144 | Reverse | TGCAGGATATCGTTAACCAATCGGAGAAGAGATGGGTGG |
| −365/−158 | Forward | GGGGTACCTCCGGAGGCGCGGCGGCAAGCCTATC |
| −365/−158 | Reverse | TGCAGGATATCGAAGAGATGGGTGGCTCGAAGGATGGC |
Plasmids and siRNA Transfection
Plasmids were transiently transfected into HEK293 or K562 cells by Lipofectamine® 2000 (Invitrogen) according to the manufacturer's instructions. siRNAs of PBX1 or control were synthesized by Shanghai GenePharma (Shanghai, China). The specific sequences targeting PBX1 were as follows: siPBX1 sense 5′-CCAUCCAGAUGCAGCUCAATT-3′ and antisense 5′-UUGAGCUGCAUCUGGAUGGTT-3′; siNC sense 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense 5′-ACGUGACACGUUCGGAGAATT-3′. PREP1 siRNAs were purchased from RiboBio (Guangzhou, China). siRNAs were transfected into K562 cells using riboFECTTM CP (RiboBio) according to the manufacturer's instructions.
Dual-Luciferase Reporter Assays
The constructs of RNF6 regulatory sequences or mutants, along with the internal control vector Renilla, were transfected into HEK293 cells. Luciferase assays were performed after 36 h using the Dual-Luciferase® Reporter Assay System (Promega) as described previously (7). Firefly luciferase activity was normalized to the Renilla expression for each sample.
ChIP
The ChIP assay was performed according to the manufacturer's instructions (Millipore). Briefly, HEK293, OCI-AML2, and K562 cells were fixed with 1% formaldehyde, followed by cell lysis and sonication to shear genomic DNA. After centrifugation, the clarified supernatants were incubated with a specific anti-PBX1 antibody or anti-rabbit IgG for 24 h at 4 °C, followed by precipitation with protein A beads. The fragment of the −234/−1 RNF6 regulatory region from HEK293 cells was identified by regular RT-PCR. The −234/−1 RNF6 regulatory sequence from OCI-AML2 and K562 cells were identified by qRT-PCR. The primers used were as follows: forward 5′-GGAAAGATCTGGGTCCCAC-3′ and reverse 5′-GTGGGCTCAAGAGGCCCAAC-3′.
Cycloheximide Chase Assay
To evaluate whether PBX1 regulates the stability of RNF6 protein, K562 cells were transfected with 25 nm siPBX1. Sixty hours later, cells were treated with cycloheximide (50 μg/ml, Sigma-Aldrich) for a specific time period before being lysed by immunoblotting.
Co-immunoprecipitation
HEK293 cells were co-transfected with HA-PBX1 and Myc-PREP1 plasmids for 24 h, followed by incubation with DOX (0, 2, and 4 μm) or 5AHQ (0, 20, and 40 μm) for 12 h. Whole cell lysates were then prepared for co-immunoprecipitation. Firstly, the cell lysates were incubated with an anti-HA or anti-Myc antibody overnight at 4 °C, followed by incubation with protein A+G-Sepharose beads (Beyotime Biotechnology) for 4 h. The co-precipitated proteins were identified by immunoblotting analysis against specific antibodies.
Xenograft Studies
K562 cells (2 × 107) were subcutaneously inoculated into the right flanks of nude mice (Shanghai SLAC Experimental Animal Co., Shanghai, China). At the same time, a construct of RNF6 small hairpin RNA targeting 5′-TCAGTGAATTTCAATGGTA-3′ was made in an shRNA expression vector (GV248, GeneChem Biotechnology Inc.). When the tumors were palpable, 10 μg of shRNF6 or scramble shRNA (shNC) in 100 μl of in vivo-jetPEI® Delivery Reagent (nitrogen/phosphorus = 6) (Polyplus-transfection Inc., New York, NY) was injected into tumors every week and continued for 3 weeks according to the manufacturer's instructions. Tumor sizes and mice body weights were monitored every 3 days. This xenograft study was approved by the Review Board of Animal Care and Use of Soochow University.
Statistical Analysis
Student's t test was used for comparisons of two groups in the studies. All statistical tests were two-sided, and a p value < 0.05 was considered statistically significant.
Results
RNF6 Is Highly Expressed in Blood Cancer Cells but Not in the Normal Counterparts
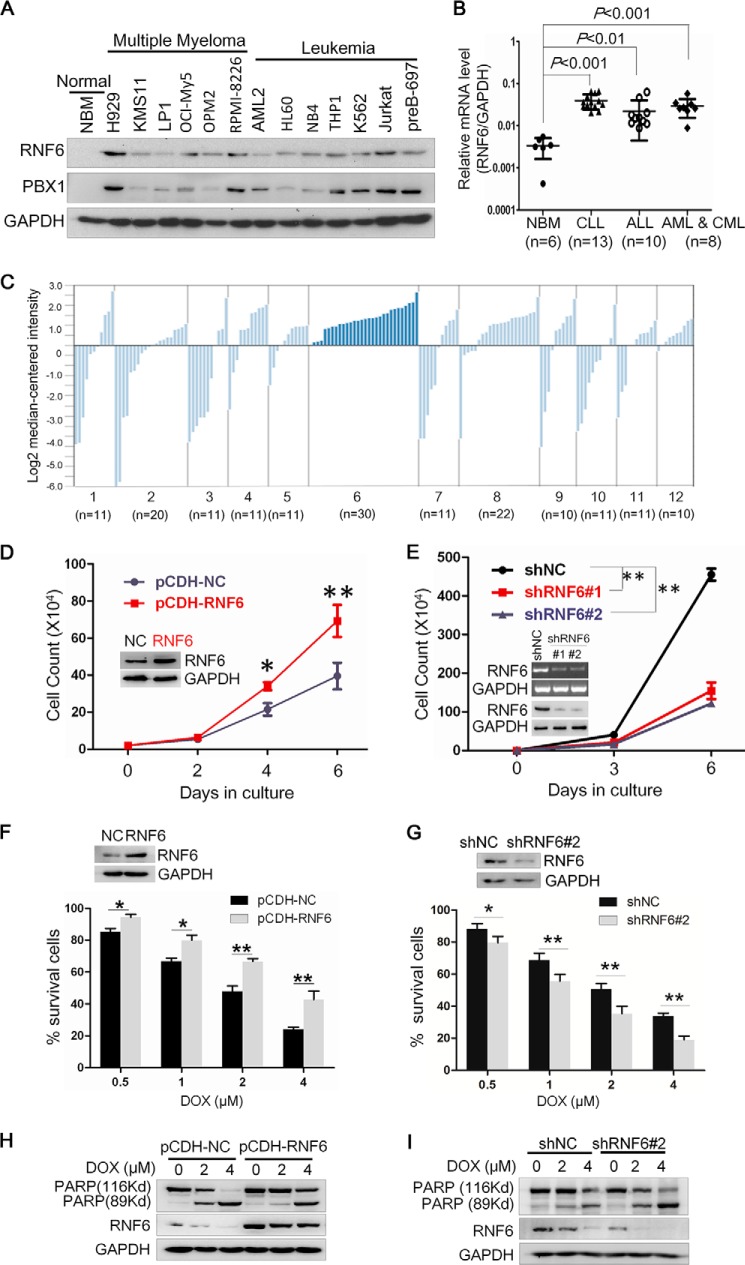
Previous studies show that RNF6 is overexpressed in prostate cancer tissues and cell lines but that it is not expressed or only slightly expressed in normal prostate tissues (3). To find out the expression profile of RNF6 in blood cancers, an immunoblotting assay was performed to measure RNF6 in a panel of leukemia and multiple myeloma cell lines and normal bone marrow cells. As shown in Fig. 1A, RNF6 was not detected in normal blood cells but expressed at a high level in most of the leukemia cell lines. To confirm this finding, primary bone marrow species from 31 leukemia patients (containing chronic and acute lymphoid leukemia and chronic and acute myelogenous leukemia) and 5 healthy donors were analyzed with qRT-PCR. As shown in Fig. 1B, the mRNA level of RNF6 was very low in normal bone marrow cells, but it was markedly elevated in the bone marrow samples from all leukemia patients, which was consistent with the result from leukemia cell lines (Fig. 1A). To further verify this finding, the Oncomine mRNA database was further analyzed. Oncomine Research Edition houses 715 datasets from 86,733 various cancer cell and tissue samples. By searching this public database, we found RNF6 in 12 types of cancers. As shown in Fig. 1C, the mRNA level of RNF6 varied in different types of cancers, but it was universally up-regulated in leukemia, lymphoma, and prostate cancer, which was consistent with our finding (Fig. 1, A and B) and the previous study on prostate cancers (3).
FIGURE 1.
RNF6 is overexpressed in leukemia cells and promotes leukemia cell proliferation. A, the whole-cell lysates from normal bone marrow (NBM) stem cells and multiple myeloma and leukemia cell lines were extracted, and RNF6 and PBX1 protein levels were measured by immunoblotting analyses. GAPDH was used as a loading control. B, bone marrow species from 31 leukemia patients and 6 healthy donors were applied to extract total RNA, followed by qRT-PCR analysis to measure the mRNA level of RNF6. C, RNF6 expression levels in various types of cancers were retrieved from the Oncomine mRNA database. The numbers 1–12 represent different types of cancers: 1, bladder cancer; 2, brain and CNS cancer; 3, breast cancer; 4, colorectal cancer; 5, kidney cancer; 6, leukemia; 7, lung cancer; 8, lymphoma; 9, melanoma; 10, ovarian cancer; 11, pancreatic cancer; and 12, prostate cancer. n means the dataset number for each type of cancer. D, K562 cells were infected with lentiviral RNF6 or control, followed by trypan blue exclusion assay at days 0, 2, 4, and 6. NC, negative control. E, K562 cells were infected with lentiviral shRNF6 (shRNF6#1 or shRNF6#2) or control shRNA (shNC) followed by trypan blue exclusion assay at days 0, 3, and 6. F and G, K562 cells were infected with lentiviral RNF6 (F) or shRNF6 (G) for 48 h, followed by DOX treatment at the indicated concentrations for 24 h and cell viability assay using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method. H and I, K562 cells infected with lentiviral RNF6 (H) or shRNF6 (I) for 48 h were treated with DOX at the indicated concentrations for 24 h, followed by immunoblotting assay to evaluate PARP and RNF6. GAPDH was used as an internal control. *, p < 0.05, **, p < 0.01, as compared with control. Error bars indicate ± S.E.
RNF6 Promotes Leukemia Cell Proliferation
Because RNF6 was not detectable in healthy blood cells but was highly expressed in leukemia cells, we wondered whether RNF6 was important for leukemia cell proliferation and survival. To this end, the chronic myelogenous leukemia-derived K562 cell line was infected with lentiviral RNF6, followed by cell viability assay. As shown in Fig. 1D, RNF6 promoted K562 cell proliferation and there were more cells in the RNF6-infected group than in the mock one. In contrast, knockdown of RNF6 with shRNA led to decreased cell proliferation (Fig. 1E).
RNF6 Is Decreased by Doxorubicin
To find out whether RNF6 could be affected by any anti-leukemia drugs, K562 cells were infected with lentiviral RNF6 or lentiviral shRNF6 followed by treatment with DOX, a major anti-leukemia drug. These cells were then subjected to cell proliferation and cell death evaluation. As shown in Fig. 1F, ectopic expression of RNF6 significantly attenuated DOX-inhibited cell survival. Consistent with this finding, knockdown of endogenous RNF6 by shRNA sensitized DOX-inhibited cell survival (Fig. 1G). Moreover, RNF6 rescued K562 cells from DOX-induced apoptosis. DOX induced more than 40 and 90% of PARP1 cleavage at 2 and 4 μm, respectively. When RNF6 was ectopically expressed, PARP1 cleavage was markedly abolished. The PARP1 cleavage levels were reduced to 5 and 40% at 2 and 4 μm DOX, respectively (Fig. 1H). In contrast, knockdown of RNF6 enhanced DOX-induced apoptosis in terms of PARP expression (Fig. 1I). These results indicated that RNF6 could be developed as a therapeutic target.
RNF6 Knockdown Delays Chronic Myelogenous Leukemia-derived Tumor Growth in Nude Mice
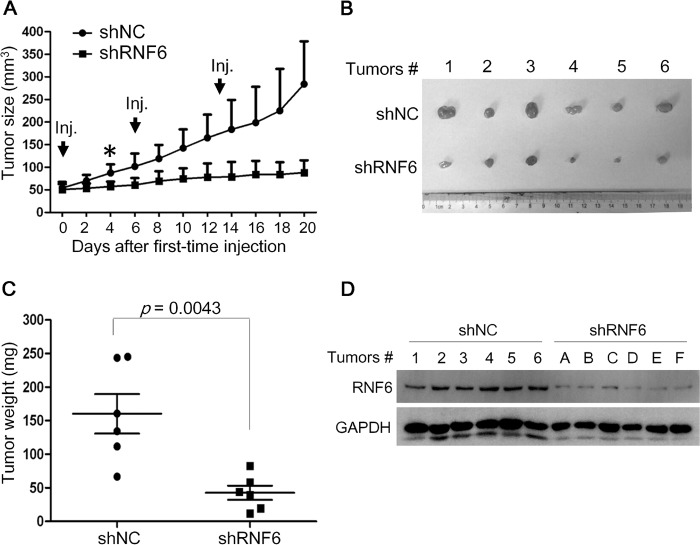
The above findings showed that RNF6 promoted leukemia cell growth and that RNF6 could be down-regulated by an anti-leukemia drug. To find out its effects on leukemia in vivo, K562 cells were inoculated subcutaneously into the right flanks of nude mice to establish a xenograft model. When tumors reached 50–60 mm3, shRNF6 or mock expressing vectors carried by the in vivo-jetPEI® Delivery Reagent were intratumorally injected once a week for 3 continuous weeks. Introduction of shRNF6 suppressed tumor growth in 4 days (Fig. 2A), and at the end of the experiment, tumors injected with shRNF6 were significantly smaller than controls (Fig. 2, B and C). Analyses on the tumor tissues showed that RNF6 was also decreased in the tumors with shRNF6 (Fig. 2D). These results thus further demonstrated that RNF6 was oncogenic in leukemia and promoted leukemia growth.
FIGURE 2.
RNF6 knockdown delays tumor growth in K562-derived xenografts in nude mice. A, K562 cells were subcutaneously injected (Inj.) into the right flanks of each mouse. When tumors were palpable, tumors were injected with 10 μg of shRNF6 or control shRNA (shNC) at days 0, 6, and 13 as indicated by arrows. Tumor sizes were monitored every other day. Error bars indicate ± S.E. B, tumors were excised from nude mice. C, tumor weight was measured at the end of the experiment. D, tumor tissues were applied for total protein preparation and immunoblotting assay against RNF6 and GAPDH. Error bars indicate ± S.E.
PBX1 Is an Essential Modulator of RNF6 Transcription
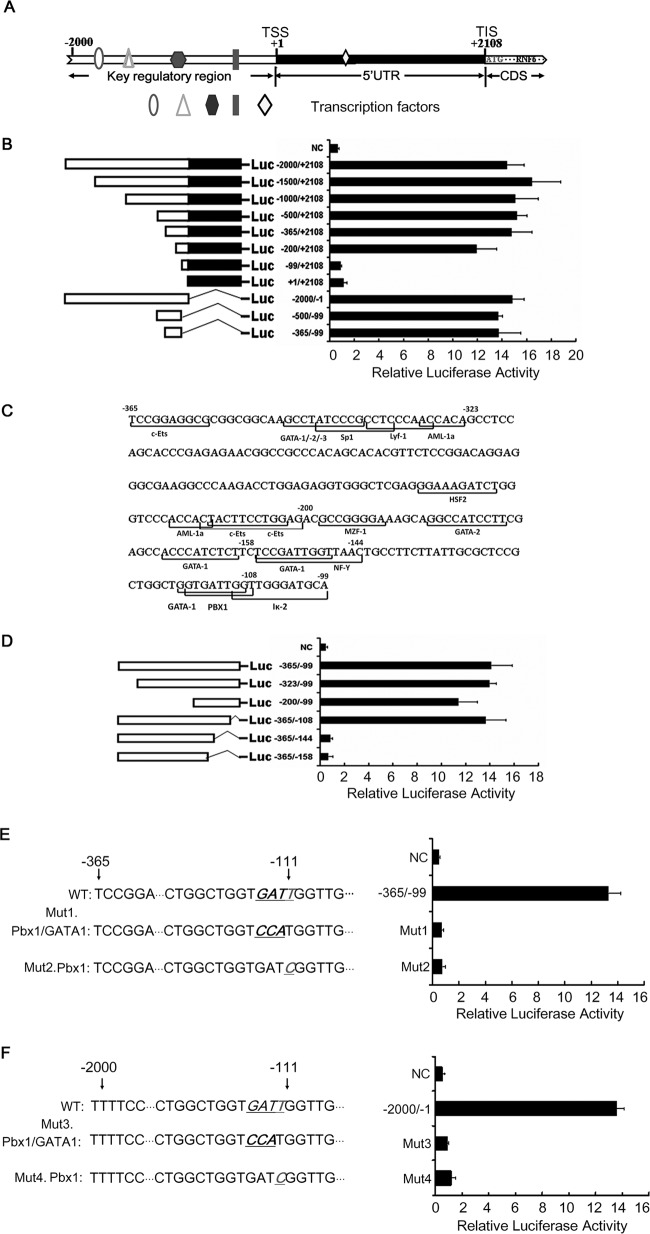
The above studies showed that RNF6 was overexpressed in various blood cancer cells and contributed to leukemia cell growth. To find out the key modulator for RNF6 expression, we analyzed the activity of various RNF6 regulatory sequences using luciferase as a reporter. The schematic structure of the RNF6 regulatory region is shown in Fig. 3A, and a series of truncated sequences was constructed as shown in Fig. 3B. Based on the activity of the truncated sequences, the fragment of −365 to +2108 retained a high level of promoter activity, but the luciferase activity was completely lost when it was driven by the fragment of −99 to 2108 (Fig. 3B), which suggested that the −365 to −99 region was essential for RNF6 transcription. Next we constructed a reporter system driven by the fragment of the RNF6 regulatory fragment from −365 to −99; the result showed that the transcriptional activity was restored (Fig. 3B, bottom). Therefore, these results suggested that the key regulatory sequence recognized by a transcription factor was probably present in the region of −365 to −99.
FIGURE 3.
The PBX1 binding site is essential for RNF6 transcription. A, the schematic diagram of RNF6 gene regulatory sequence was predicted by the UCSC Genome Browser website. TSS: transcription start site; TIS: translation initial site. B, the left side shows diagrammatic representations of the luciferase reporter gene constructs, and the right side shows the luciferase activity mediated by those constructs after transient transfection into HEK293 cells. C, the predicted binding sites of transcription factors in the core regulatory region of RNF6 were obtained by the TFSearch software. D, the constructs containing the RNF6 core regulatory region were transfected into HEK293 cells. A Dual-Luciferase reporter assay system was used to analyze the transcription activity of different RNF6 regulatory constructs. E and F, PBX1 and GATA1 transcription binding sites were mutated in the −365 to −99 fragment (E) or in the −2000 to −1 fragment (F) of RNF6 regulatory region. Mut1 and Mut3 were acquired by mutating the GAT to CCA. Mut2 and Mut4 were obtained by mutating T to C. The luciferase activities of these mutants were measured. Error bars indicate ± S.E.
To understand the specific modulator of RNF6 expression, we next predicted the binding sites of transcription factors using the software TFSearch and found that there were several recognition elements of transcription factors including c-ETS, SP1, AML-1, GATA, and PBX1 (Fig. 3C); therefore, the regulatory region of −365 to −99 was further truncated down as shown in Fig. 3D. The luciferase assay indicated that the region of −144 to −108 was essential for the RNF6 transcription because deletion of the −144 to −108 region led to loss of the promoter activity (Fig. 3D).
The TFSearch software predicted that the region of −144 to −108 harbors two transcription factor binding sites: 5′-GGTGATTGG-3′ for GATA1 and 5′-GTGATTGGT-3′ for PBX1. To differentiate the crucial effects of GATA1 and PBX1 on RNF6 promoter activity, we next mutated GATA1 and PBX1 binding elements, respectively. When both the GATA1 and the PBX1 binding sites were mutated in the −365 to −104 region, the regulatory sequence lost its transcriptional activity (Fig. 3E). A similar result was seen in the construct with mutated PBX1 and intact GATA1 binding sites (Fig. 3E). These results indicated that the PBX1 binding site was critical. To confirm this hypothesis, we mutated the core nucleotides (GATT) of both the GATA1 and the PBX1 binding sites to CCAT in the region of −2000 to −1 in the RNF6 regulatory region; the luciferase assay indicated that this region lost its activity (Fig. 3F). When the PBX1 binding site was mutated, whereas the one for GATA1 was intact in the same region of −2000 to −1, the fragment failed to modulate luciferase expression (Fig. 3F). Therefore, these results demonstrated that the PBX1 binding site was essential for RNF6 transcription. The RNF6 gene was probably a direct target of PBX1.
PBX1 Binds to the RNF6 Core Regulatory Region and Promotes RNF6 Expression
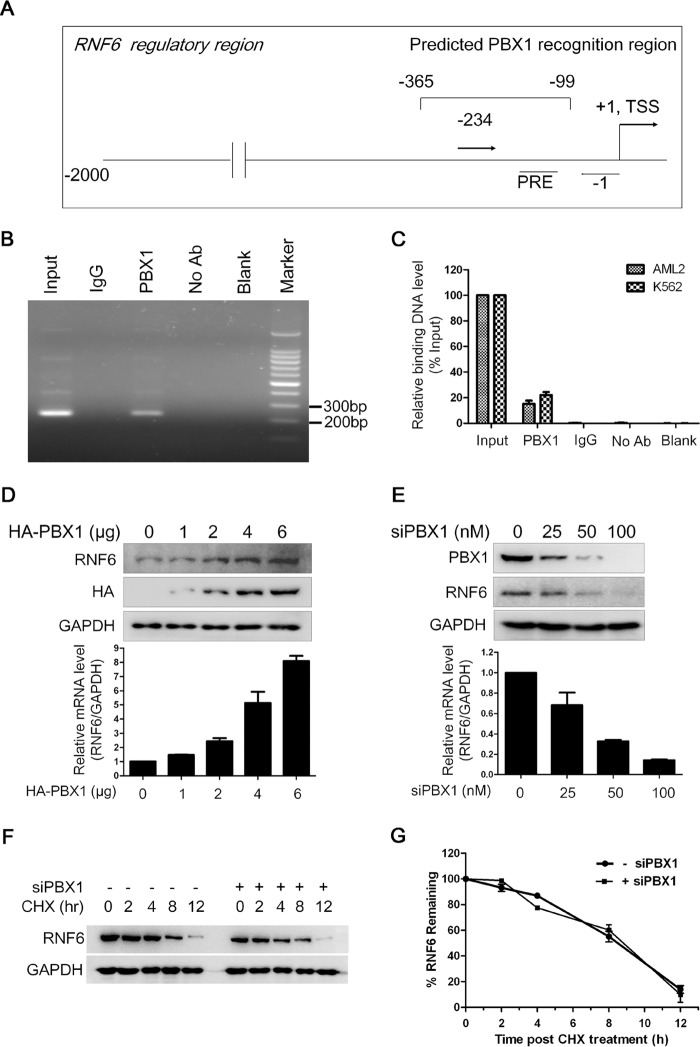
To further verify our hypothesis, we wondered whether PBX1 could recognize and bind to the RNF6 promoter in vivo. To this end, we performed a ChIP assay in HEK293 cells that express both RNF6 and PBX1 (data not shown). The PBX1 specific antibody was incubated with sonicated genomic DNA and precipitated by protein-A beads. Primers designed for the region of −234 and −1 (Fig. 4A) were used for the semiquantitative RT-PCR. As shown in Fig. 4B, the RT-PCR result showed that the fragment of −234 to −1 was precipitated with an anti-PBX1 antibody, suggesting that PBX1 recognized and bound to the regulatory region containing the PBX1 recognition element. To confirm this finding in leukemia cells, we analyzed the binding of PBX1 to RNF6 core regulatory region in two leukemia cell lines, OCI-AML2 and K562. As shown in Fig. 4C, qRT-PCR results showed that PBX1 bound to RNF6 core regulatory sequence. All these results thus suggested that PBX1 regulated RNF6 transcription.
FIGURE 4.
PBX1 binds to the specific RNF6 regulatory sequence and modulates RNF6 expression. A, the schematic illustration of primer design. +1 represents the transcription start site (TSS). B, ChIP assay was applied to determine the binding between PBX1 protein and PBX1 recognition region in the RNF6 regulatory sequence in HEK293 cells. The RNF6 regulatory sequence in the ChIP precipitates with PBX1 antibody was analyzed by RT-PCR. No Ab, no antibody. C, the binding between PBX1 protein and the RNF6 regulatory sequence in leukemia cell lines OCI-AML2 and K562 were analyzed by qRT-PCR after ChIP. D and E, K562 cells were transfected with PBX1 plasmids (D) or PBX1 siRNA (E) for 72 h, followed by whole cell lysates and total RNA preparation. The protein and mRNA levels of RNF6 were measured by immunoblotting (IB) and qRT-PCR, respectively. F, K562 cells were transfected with 25 nm siPBX1 for 60 h, followed by cycloheximide (CHX) chase assay to evaluate RNF6 stability. G, the statistical analysis for RNF6 in F. Error bars indicate ± S.E.
To further analyze the modulation of PBX1 on RNF6 expression, we first evaluated the expression profiles of both PBX1 and RNF6 in leukemia cells. Immunoblotting assay showed that both PBX1 and RNF6 were seen in most of the blood cancer cell lines but not in normal cells. The expression pattern of PBX1 was very similar to RNF6 (Fig. 1A). To find out whether PBX1 promoted RNF6 expression, the expression level of RNF6 was evaluated in the presence or absence of PBX1. As shown in Fig. 4D, enforced expression of PBX1 up-regulated RNF6 in K562 cells at both mRNA and protein levels. Consistent with this finding, when PBX1 was knocked down by siPBX1, RNF6 was decreased at both mRNA and protein levels (Fig. 4E). However, PBX1 knockdown had no effects on the RNF6 stability (Fig. 4, F and G). Therefore, these results collectively suggested that PBX1 bound to the RNF6 promoter and modulated its expression.
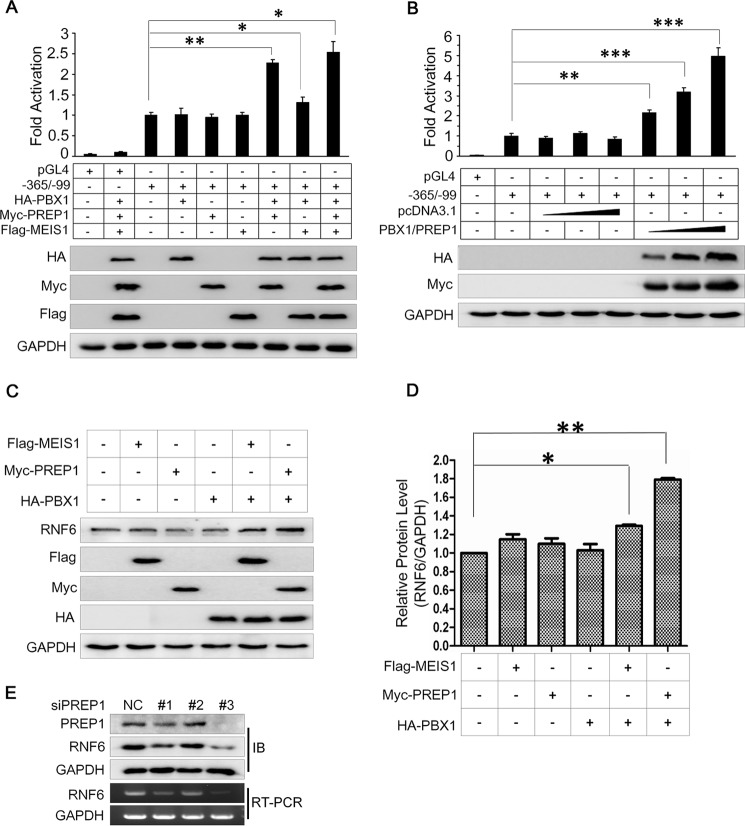
PBX1 Partners with PREP1 but Not MEIS1 to Modulate RNF6 Expression
To perform its regulatory activity as a transcription factor, PBX1 usually forms a heterodimer with the three-amino acid loop extension (TALE) family of transcription factors PREP1 or myeloid ecotropic viral integration site-1 (MEIS1) (8). To find out which TALE member is important for PBX1 in regulating RNF6 expression, we analyzed the RNF6 promoter (−365 to −99)-driven luciferase activity modulated by PBX1 in the presence of PREP1 or MEIS1. As shown in Fig. 5A, PBX1 alone did not significantly increase the luciferase activity; however, co-transfection with PBX1 and PREP1 but not MEIS1 markedly increased RNF6 promoter activity (Fig. 5A). Co-transfection of PBX1/PREP1 induced RNF6 promoter activity in a concentration-dependent manner (Fig. 5B), suggesting that PREP1 was important for PBX1 transcriptional activity in regulating RNF6 expression. To confirm this hypothesis, PBX1 was co-transfected with PREP1 or MEIS1 into HeLa cells that express endogenous RNF6 (data not shown). Immunoblotting analysis revealed that PREP1 but not MEIS1 markedly up-regulated RNF6 expression (Fig. 5, C and D). To find out whether PREP1 down-regulated RNF6 expression, K562 cells were transfected with siPREP1. The result showed that siPREP1 decreased the RNF6 transcription and protein expression (Fig. 5E), suggesting that PREP1 is also important for RNF6 transcription. These results thus solidified the finding that PBX1 modulated RNF6 expression by heterodimerizing with PREP1.
FIGURE 5.
PBX1 requires PREP1 to regulate RNF6 transcription. A, PBX1, PREP1, MEIS1, and the core regulatory sequence of RNF6 were co-transfected into HEK293 cells. The RNF6 transcription level was analyzed by a Dual-Luciferase reporter assay system. The same lysates were applied to measure the RNF6 protein level using immunoblotting. B, PBX1 and PREP1 plasmids were co-transfected into HEK293 cells with increasing concentrations followed by measurement of RNF6 transcription in a Dual-Luciferase reporter assay system. C, PBX1, PREP1, and MEIS1 plasmids were co-transfected in HeLa cells, and cell lysates were then prepared for immunoblotting against specific proteins as indicated. D, statistical analysis of C. E, K562 cells were transfected with PREP1 siRNAs for 72 h, and then whole cell lysates and total RNA were extracted for immunoblotting and RT-PCR as indicated. *, p < 0.05; **, p < 0.01; ***, p < 0.001, as compared with control. Error bars indicate ± S.E.
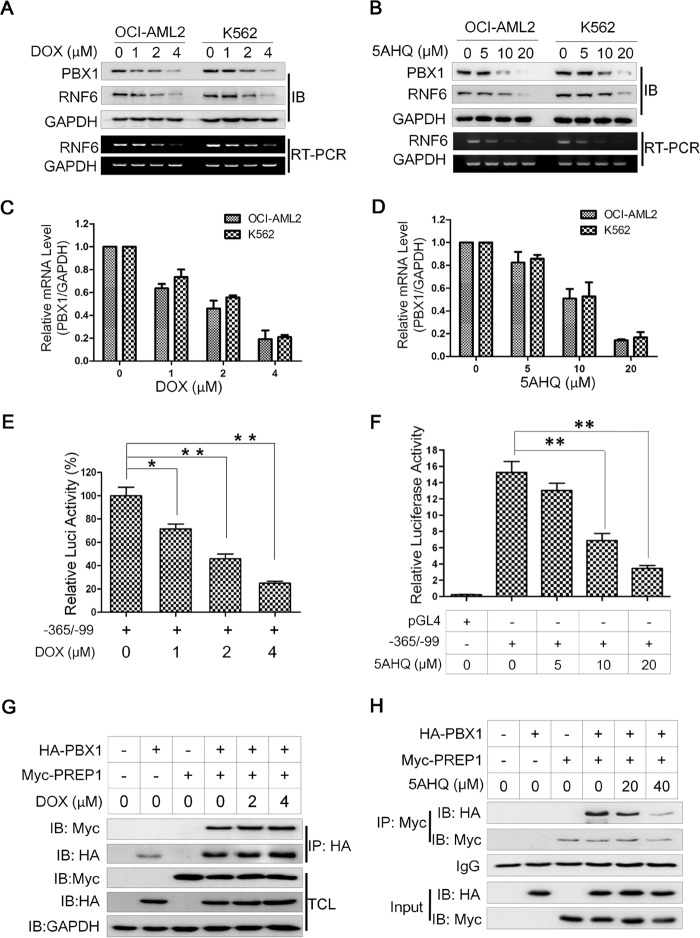
Anti-leukemia Agents Down-regulate RNF6 by Decreasing PBX1 Expression
PBX1 has long been considered as an important transcription factor in lymphoid leukemia (9). The above studies suggested that RNF6 was critical for leukemia cell growth, as well as for PBX1-directed RNF6 transcription; therefore, we wondered whether any drugs could target this PBX1/RNF6 axis and induce leukemia cell death. To this end, doxorubicin, a major anti-leukemia drug, and 5AHQ, a novel potential leukemia treatment (10), were chosen for the study. Two leukemia cell lines, OCI-AML2 and K562, were treated with DOX or 5AHQ for 24 h followed by immunoblotting analyses against RNF6 and PBX1. Our results showed that both DOX and 5AHQ down-regulated the expression of PBX1 and RNF6 in two leukemia cell lines in a concentration-dependent manner at both RNA and protein levels (Fig. 6, A–D). These two agents also suppressed the luciferase activity under direction of the RNF6 core regulatory region (−365/−99) (Fig. 6, E and F), suggesting that PBX1/PREP1 transcriptional activity could be modulated by anti-leukemia agents. Because the PBX1/PREP1 dimer was critical for RNF6 transcription (Fig. 5), we wondered whether these agents could display any effects on PBX1/PREP1 association. To this end, PBX1 and PREP1 plasmids were co-transfected into HEK293 cells, followed by DOX or 5AHQ treatment. These cells were then subjected to co-immunoprecipitation and subsequent immunoblotting analysis. As shown in Fig. 6G, PBX1 was identified at a similar level in the PREP1 co-immunoprecipitates in the concentrations of DOX to down-regulate PBX1 and RNF6, which suggested that DOX could not disrupt the association of PBX1/PREP1. However, 5AHQ might act in a different manner. As shown in Fig. 6H, 5AHQ decreased the PBX1 level in the immunoprecipitates of PREP1, especially at higher concentrations. Therefore, these findings suggested that these anti-leukemia agents might suppress RNF6 transcription mainly by suppressing PBX1 expression. However, 5AHQ probably also disrupted the association of the PBX1/PREP1 complex.
FIGURE 6.
Anti-leukemia agents down-regulate RNF6 expression in association with PBX1 inhibition. A and B, leukemia cells (K562 and OCI-AML2) were treated with increasing concentrations of DOX (A) or 5AHQ (B) for 24 h, followed by measurement PBX1 and RNF6 expression. IB, immunoblotting. E and F, the RNF6 core regulatory region (−365/−99) was transfected into HEK293 cells for 24 h, followed by DOX (C) or 5AHQ (D) treatment with increasing concentrations for 12 h. The transcriptional activity of this regulatory sequence was analyzed by a Dual-Luciferase reporter assay system. Luci, luciferase. G and H, HA-PBX1 and Myc-PREP1 plasmids were co-transfected into HEK293 cells for 24 h, followed by DOX (G) or 5AHQ (H) treatment for 12 h. Whole cell lysates were prepared for co-immunoprecipitation (IP) and immunoblotting against specific antibodies as indicated. *, p < 0.05, **, p < 0.01, as compared with control. Error bars indicate ± S.E.
Discussion
The present study shows that RNF6 is selectively expressed in leukemia cells but not in normal blood cells and that RNF6 probably acts as an oncogene in promoting leukemia cell growth in both in vitro and in vivo models. This finding is different from a previous proposal that RNF6 is believed to be a potential tumor suppressor because multiple mutations in the RNF6 gene were identified in human esophageal squamous cell carcinoma (1). However, our finding suggests that RNF6 is oncogenic in leukemia, which is consistent with a study that shows RNF6 is an oncogene in prostate cancer because RNF6 is associated with prostate cancer progression and tumor growth, whereas silence of RNF6 abolishes prostate cancer cell growth and delays tumor growth (3). Together with these previous studies, we believe that, at least, RNF6 is probably a double-faceted gene dependent on specific cell types and tissue contexts. The general expression profile of RNF6 in different types of cancers also demonstrates this because RNF6 is up-regulated in some cancers, and it is down-regulated in others.
Our present study demonstrates that RNF6 is highly expressed in leukemia cell lines and primary patients' tissues modulated by PBX1, an important transcription factor extensively studied in pre-B-cell leukemia, although many transcription factors have been reported in leukemia cells and are responsible for leukemogenesis (11), such as CCAAT/enhancer-binding protein α (C/EBPA) (12), SP1 (12), NF-κB (13), STAT3 (14), PU.1 (15), HOX9 (16), PBX1 (9), and many others (11). PBX1 is frequently reported as a fusion protein in association with a chromosomal translocation t(1;19) involving itself and TCF3/E2A (17). For example, the E2A-PBX1 fusion leads to multiple second genomic aberrations involved in the tumor suppressor PAX5 and a key cell signaling pathway of JAK2/STAT3 in acute lymphoblastic leukemia (17). The present study demonstrates that RNF6 is a direct downstream gene because there is a specific PBX1 recognition site in the RNF6 regulatory region. By binding to the recognition element, PBX1 triggers the transcription of RNF6. When the PBX1 recognition element in the RNF6 regulatory region is deleted, the promoter activity will be completely lost. In addition, PBX1 induced endogenous RNF6 expression, and when PBX1 was knocked down, RNF6 expression was also decreased.
PBX1 is a member of the TALE family in which it usually acts as a heterodimer with other transcription factors MEIS1 or PREP1 dependent on its function (18). Generally speaking, PBX1 acts as a tumor suppressor in association with PREP1, but it promotes tumor progression when heterodimerizing with MEIS1 (8). For example, PBX1 collaborates with MEIS1 in the induction of acute myelogenous leukemia cells by accelerating leukemic transformation (19). However, in the present study, we found that PBX1 prefers to partner with PREP1 to modulate RNF6 transcription because the RNF6 promoter activity is markedly increased only when PBX1 and PREP1 are both present. Therefore, this study also suggests that PREP1 probably also acts as an oncoprotein. This is consistent with recent studies in which PREP1 enhances the transcription of murine leukemia virus (Moloney MLV) (20) and its overexpression induces epithelia-mesenchymal transition and metastasis in non-small-cell lung cancer by regulating the TGF-β-SMAD3 pathway (21). Therefore, it is reasonable that PBX1 promotes leukemia cell proliferation in association with PREP1.
RNF6 has been found to be responsible for prostate cancer progression, and it appears to be a promising target for prostate cancer treatment, but direct evidence is lacking (3). In the present study, we further found that the PBX1/RNF6 axis could be developed as a therapeutic target for leukemia. This conclusion is based on several factors: 1) both RNF6 and PBX1 are overexpressed in leukemia cells; 2) enforced expression of RNF6 promotes leukemia cell proliferation; 3) knockdown of RNF6 inhibits leukemia cell proliferation and delays leukemia-derived tumor growth in vivo; and 4) RNF6 can be decreased by the anti-leukemia agents DOX and 5AHQ, whereas enforced RNF6 expression can rescue leukemia cells from apoptosis induced by DOX. Based on the present study, there are at least two strategies to develop PBX1/RNF6-targeted drugs. One is to down-regulate PBX1 and or PREP1 expression, and the other one is to disrupt the PBX1/PREP1 association. As demonstrated in Figs. 5 and 6, both knockdown of PBX1/PREP1 and dissociation of the PBX1/PREP1 result in decreased expression of RNF6, which further leads to leukemia cell apoptosis. For example, 5AHQ decreases PBX1 and RNF6 at lower concentrations, but it can also interfere with the formation of the PBX1 and PREP1 heterodimer at higher concentrations. Because 5AHQ has been established as a proteasomal inhibitor (10), the question arises: what is the mechanism through which 5AHQ decreases PBX1 expression? Current studies have demonstrated that proteasomal inhibitors can modulate gene transcription in addition to modulating protein stability. Proteasomal inhibitors such as bortezomib and clioquinol have been demonstrated to regulate gene transcription by inhibiting histone deacetylases, thus regulating gene expression in an epigenetic manner (22, 23).
However, several questions about RNF6 in leukemia are still open. The most important one is: how does RNF6 promote leukemia cell proliferation? How does RNF6 promote leukemia cell line-derived tumor growth in vivo? Previous studies show that RNF6 is associated with progression of prostate cancer by modulating androgen receptor activity as a ubiquitin ligase (3). However, this mechanism seems unreasonable for leukemia. To understand RNF6 in leukemia, a specific protein substrate of RNF6 should be identified from leukemia cells. Another study proposed that RNF6 is probably a transcription regulatory protein because it can bind to the Inha promoter, thus up-regulating Inha expression and being involved in germinal differentiation (24). Whether RNF6 also acts as a transcription factor in leukemia deserves further studies.
In summary, our present study focused on the regulatory mechanism of RNF6 expression in leukemia and demonstrated that RNF6 is critical for leukemia cell proliferation as a direct target gene of PBX1. This study forms a rationale for PBX1/RNF6-based drug development toward leukemia therapy.
Author Contributions
X. M. designed the study; X. X., K. H., Y. Z., X. L., B. C., and Z. Z. conducted experiments; X. T., D. W., Y. Z., and X. M. analyzed data; X. M. wrote the manuscript.
This work was partly supported by National Natural Science Foundation of China Grants 81320108023 (to X. M.), 81272632 (to X. M.), 81270645 (to X. T.), by Natural Science Foundation of Jiangsu Province Grant BE2014630 (to X. M.), by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), by Jiangsu Key Laboratory for Translational Research and Therapeutics of Neuro-Psycho-Diseases Grant BK2013003 (to X. M.), and by Suzhou City Science and Technology Programs Grant SYS201457 (to X. T.). The authors declare that they have no conflicts of interest with the contents of this article.
- AR
- androgen receptor
- 5AHQ
- 5-amino-8-hydroxyquinoline
- DOX
- doxorubicin
- PRE
- PBX1 recognition element
- PARP
- poly(ADP-ribose) polymerase
- TALE
- three-amino acid loop extension
- qRT-PCR
- quantitative real-time PCR.
References
- 1. Macdonald D. H., Lahiri D., Sampath A., Chase A., Sohal J., and Cross N. C. (1999) Cloning and characterization of RNF6, a novel RING finger gene mapping to 13q12. Genomics 58, 94–97 [DOI] [PubMed] [Google Scholar]
- 2. Lo H. S., Hu N., Gere S., Lu N., Su H., Goldstein A. M., Taylor P. R., and Lee M. P. (2002) Identification of somatic mutations of the RNF6 gene in human esophageal squamous cell carcinoma. Cancer Res. 62, 4191–4193 [PubMed] [Google Scholar]
- 3. Xu K., Shimelis H., Linn D. E., Jiang R., Yang X., Sun F., Guo Z., Chen H., Li W., Chen H., Kong X., Melamed J., Fang S., Xiao Z., Veenstra T. D., and Qiu Y. (2009) Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell 15, 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qin X., Chen S., Qiu Z., Zhang Y., and Qiu F. (2012) Proteomic analysis of ubiquitination-associated proteins in a cisplatin-resistant human lung adenocarcinoma cell line. Int. J. Mol. Med. 29, 791–800 [DOI] [PubMed] [Google Scholar]
- 5. Mao X., Hou T., Cao B., Wang W., Li Z., Chen S., Fei M., Hurren R., Gronda M., Wu D., Trudel S., and Schimmer A. D. (2011) The tricyclic antidepressant amitriptyline inhibits D-cyclin transactivation and induces myeloma cell apoptosis by inhibiting histone deacetylases: in vitro and in silico evidence. Mol. Pharmacol. 79, 672–680 [DOI] [PubMed] [Google Scholar]
- 6. Shi M., Zhou X., Zhang Z., Wang M., Chen G., Han K., Cao B., Liu Z., and Mao X. (2014) A novel PI3K inhibitor displays potent preclinical activity against an androgen-independent and PTEN-deficient prostate cancer model established from the cell line PC3. Toxicol. Lett. 228, 133–139 [DOI] [PubMed] [Google Scholar]
- 7. Mao X., Cao B., Wood T. E., Hurren R., Tong J., Wang X., Wang W., Li J., Jin Y., Sun W., Spagnuolo P. A., MacLean N., Moran M. F., Datti A., Wrana J., et al. (2011) A small-molecule inhibitor of D-cyclin transactivation displays preclinical efficacy in myeloma and leukemia via phosphoinositide 3-kinase pathway. Blood 117, 1986–1997 [DOI] [PubMed] [Google Scholar]
- 8. Dardaei L., Longobardi E., and Blasi F. (2014) Prep1 and Meis1 competition for Pbx1 binding regulates protein stability and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 111, E896–E905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aspland S. E., Bendall H. H., and Murre C. (2001) The role of E2A-PBX1 in leukemogenesis. Oncogene 20, 5708–5717 [DOI] [PubMed] [Google Scholar]
- 10. Li X., Wood T. E., Sprangers R., Jansen G., Franke N. E., Mao X., Wang X., Zhang Y., Verbrugge S. E., Adomat H., Li Z. H., Trudel S., Chen C., Religa T. L., Jamal N., et al. (2010) Effect of noncompetitive proteasome inhibition on bortezomib resistance. J. Natl. Cancer Inst. 102, 1069–1082 [DOI] [PubMed] [Google Scholar]
- 11. Somasundaram R., Prasad M. A., Ungerbäck J., and Sigvardsson M. (2015) Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood 126, 144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mao X., Liang S. B., Hurren R., Gronda M., Chow S., Xu G. W., Wang X., Beheshti Zavareh R., Jamal N., Messner H., Hedley D. W., Datti A., Wrana J. L., Zhu Y., Shi C. X., et al. (2008) Cyproheptadine displays preclinical activity in myeloma and leukemia. Blood 112, 760–769 [DOI] [PubMed] [Google Scholar]
- 13. Chen G., Han K., Xu X., Du X., Zhang Z., Tang J., Shi M., Wang M., Li J., Cao B., and Mao X. (2014) An anti-leishmanial thiadiazine agent induces multiple myeloma cell apoptosis by suppressing the nuclear factor κB signalling pathway. Br. J Cancer 110, 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bruserud Ø., Nepstad I., Hauge M., Hatfield K. J., and Reikvam H. (2015) STAT3 as a possible therapeutic target in human malignancies: lessons from acute myeloid leukemia. Expert Rev. Hematol. 8, 29–41 [DOI] [PubMed] [Google Scholar]
- 15. Imperato M. R., Cauchy P., Obier N., and Bonifer C. (2015) The RUNX1-PU.1 axis in the control of hematopoiesis. Int. J. Hematol. 101, 319–329 [DOI] [PubMed] [Google Scholar]
- 16. Collins C. T., and Hess J. L. (2016) Role of HOXA9 in leukemia: dysregulation, cofactors and essential targets. Oncogene 35, 1090–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duque-Afonso J., Feng J., Scherer F., Lin C. H., Wong S. H., Wang Z., Iwasaki M., and Cleary M. L. (2015) Comparative genomics reveals multistep pathogenesis of E2A-PBX1 acute lymphoblastic leukemia. J. Clin. Invest. 125, 3667–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calvo K. R., Knoepfler P., McGrath S., and Kamps M. P. (1999) An inhibitory switch derepressed by Pbx, Hox, and Meis/Prep1 partners regulates DNA-binding by Pbx1 and E2a-Pbx1 and is dispensable for myeloid immortalization by E2a-Pbx1. Oncogene 18, 8033–8043 [DOI] [PubMed] [Google Scholar]
- 19. Thorsteinsdottir U., Kroon E., Jerome L., Blasi F., and Sauvageau G. (2001) Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell. Biol. 21, 224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chao S. H., Walker J. R., Chanda S. K., Gray N. S., and Caldwell J. S. (2003) Identification of homeodomain proteins, PBX1 and PREP1, involved in the transcription of murine leukemia virus. Mol. Cell. Biol. 23, 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Risolino M., Mandia N., Iavarone F., Dardaei L., Longobardi E., Fernandez S., Talotta F., Bianchi F., Pisati F., Spaggiari L., Harter P. N., Mittelbronn M., Schulte D., Incoronato M., Di Fiore P. P., et al. (2014) Transcription factor PREP1 induces EMT and metastasis by controlling the TGF-β-SMAD3 pathway in non-small cell lung adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 111, E3775–E3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cao B., Li J., Zhu J., Shen M., Han K., Zhang Z., Yu Y., Wang Y., Wu D., Chen S., Sun A., Tang X., Zhao Y., Qiao C., Hou T., and Mao X. (2013) The antiparasitic clioquinol induces apoptosis in leukemia and myeloma cells by inhibiting histone deacetylase activity. J. Biol. Chem. 288, 34181–34189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kikuchi J., Wada T., Shimizu R., Izumi T., Akutsu M., Mitsunaga K., Noborio-Hatano K., Nobuyoshi M., Ozawa K., Kano Y., and Furukawa Y. (2010) Histone deacetylases are critical targets of bortezomib-induced cytotoxicity in multiple myeloma. Blood 116, 406–417 [DOI] [PubMed] [Google Scholar]
- 24. Lopez P., Vidal F., Martin L., Lopez-Fernandez L. A., Rual J. F., Rosen B. S., Cuzin F., and Rassoulzadegan M. (2002) Gene control in germinal differentiation: RNF6, a transcription regulatory protein in the mouse Sertoli cell. Mol. Cell. Biol. 22, 3488–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]