Abstract
ARID1A is a tumor suppressor gene that belongs to the switch/sucrose non-fermentable chromatin remodeling gene family. It is mutated in many types of human cancer with the highest frequency in endometrium-related ovarian and uterine neoplasms including ovarian clear cell, ovarian endometrioid, and uterine endometrioid carcinomas. We have previously reported that mutations in the promoter of human telomerase reverse transcriptase (TERT) rarely co-occur with the loss of ARID1A protein expression, suggesting a potential role of ARID1A in telomere biology. In this study, we demonstrate that ARID1A negatively regulates TERT transcriptional regulation and activity via binding to the regulatory element of TERT and promotes a repressive histone mode. Induction of ARID1A expression was associated with increased occupancy of SIN3A and H3K9me3, known transcription repressor and histone repressor marks, respectively. Thus, loss of ARID1A protein expression caused by inactivating mutations reactivates TERT transcriptional activity and confers a survival advantage of tumor cells by maintaining their telomeres.
Keywords: chromatin remodeling, ovarian cancer, telomerase reverse transcriptase (TERT), telomere, tumor suppressor gene, ARID1A
Introduction
ARID1A3 encodes BAF250 (also known as p270), which interacts with the ATPase subunit, BRG1 or BRM, and a set of core subunits (BAF47, BAF155, and BAF170) to form the switch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex. By utilizing the energy from ATP hydrolysis, these complexes rearrange the distribution of nucleosomes, thereby modifying chromatin configuration and DNA accessibility to cellular machineries involved in transcription, DNA replication, DNA methylation, and DNA repair (1–3).
The discovery of molecular genetic aberrations in chromatin remodelers converged genetic and epigenetic dysregulation, which contributes to the development and progression of numerous diseases, including cancer. Somatic ARID1A mutations were first discovered in ovarian clear cell carcinoma (4, 5) and subsequently reported in endometrium-derived carcinomas and several other cancer types (6, 7). Mutations of ARID1A occur throughout the entire coding sequence, mostly frameshift and nonsense mutations, thereby resulting in a loss of ARID1A protein expression. Hence, ARID1A is implicated as a tumor suppressor, and indeed, functional studies have demonstrated that ARID1A loss can promote tumorigenesis by affecting proliferation, differentiation, and apoptosis (8–10). At the molecular level, our previous study demonstrated that the ARID1A complex interacted and collaborated with p53 to regulate transcription of several effectors including CDKN1A encoding p21 (8). In genetically engineered mice, whereas Arid1a deletion by itself is insufficient to transform cells, co-deletion of Arid1a with either Pten or Pik3ca is required to drive the formation of ovarian endometrioid and clear cell-like carcinomas, respectively (11, 12).
To further elucidate molecular mechanisms of ARID1A in preventing tumorigenesis, we sought to identify additional molecular genetic alterations that tend to be absent in ARID1A mutated tumors. The nature of “mutual exclusiveness” between two molecular genetic events typically implicates their involvement in similar or overlapping biological functions. Therefore, alteration in one of the molecular genetic events will be sufficient to achieve pathway activation. Somatic mutations of KRAS and BRAF represent a quintessential example of mutual exclusiveness because KRAS mutations rarely co-occur with BRAF mutations in many types of human cancers (13–15). We previously reported that known activating mutations in the promoter of human TERT rarely co-occurred with loss of ARID1A protein expression, which is a surrogate marker for its inactivating mutation (16). Somatic activating mutations at the TERT promoter have recently been reported in melanoma (17, 18) and other malignancies including ovarian clear cell carcinoma (19–22). The majority of reported mutations are located at two hot spots with each mutation creating a new 11-bp motif essential for the recruitment of the multimeric GABP transcription factor, known to enhance transcriptional activity of the TERT promoter in vitro and increase TERT mRNA expression in cancer cells including those derived from ovarian clear cell carcinoma (16–18, 22–24). Considering the above findings and that several tumor suppressor genes have been reported to act as TERT transcriptional repressors (25), it raises the possibility that similar to TERT promoter (activating) mutation, loss of ARID1A expression may lead to up-regulation of TERT expression in certain types of tumor. In this study, we employed both reverse (silencing and gene knockout of ARID1A) and forward (induction of ARID1A) approaches to determine whether ARID1A directly regulates TERT transcription and telomerase activity.
Experimental Procedures
Cell Culture
The cell lines used in this study included human embryonic kidney epithelial cells (HEK293FT), human osteosarcoma cells (U2OS), human endometrial epithelial cells (hEM3), and human ovarian clear cell carcinoma cell lines (OVISE and OV207). HEK293FT cells were cultured in DMEM (Life Technologies) supplemented with 10% FBS (Atlanta Biologicals, Norcross, GA) and 1% penicillin/streptomycin (Life Technologies). In this study, we generated the human endometrial epithelial cell line, hEM3, which was established from a primary culture of normal endometrial epithelial cells. hEM3 cells expressed epithelial cell markers including cytokeratin-8, Ep-CAM, and E-cadherin, as well as estrogen receptor. More than 99% of these cells were positive for Ep-CAM using flow cytometry. This cell line harbored wild-type ARID1A and expressed BAF250. Because endometrium-related neoplasms are derived from endometrial epithelial cells, hEM3 offers an appropriate cell model to study these carcinomas. hEM3 cells were cultured in RPMI 1640 medium (Life Technologies) supplemented with 15% FBS, 1% nonessential amino acids (Life Technologies), and 1% penicillin/streptomycin. All other cell lines were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin. Mouse primary fibroblast cells were established from the ear of Arid1aflox/flox adult mice using an established protocol (11, 26) and were cultured in tissue culture dishes coated with 1% gelatin (Sigma-Aldrich) using DMEM (Life Technologies) supplemented with 15% FBS, 1% nonessential amino acids (Life Technologies), and 1% penicillin/streptomycin. OVISE and OV207 ARID1A-inducible cells were established previously (8). Ectopic expression of ARID1A in U2OS and hEM3 ARID1A KO CRISPR/Cas9 cells was achieved by transfection of pcDNA6-V5/HisB plasmid containing wild-type full-length ARID1A (27).
Gene Silencing by RNA Interference
ARID1A expression was suppressed using the Stealth RNAiTM siRNAs HSS112108, HSS112109, and HSS112110 (Life Technologies). HSS112109 was used in all subsequent studies because of its ability to produce the highest inhibition of ARID1A expression in hEM3 and HEK293FT cells. TERT expression was suppressed using Silencer® Select siRNA s372 (Life Technologies). Stealth RNAiTM siRNA negative control medium GC (Life Technologies) and Silencer® Select Negative Control No. 1 siRNA (Life Technologies) were used as negative controls for ARID1A and TERT siRNAs, respectively. siRNAs were transfected by Lipofectamine® RNAiMAX (Life Technologies), and cells were harvested 72 h after transfection and subjected to subsequent experiments.
Generation of ARID1A Knocked Out hEM3 Cells by CRISPR/Cas9
The pSpCas9n(BB)-2A-Puro (PX462) CRISPR/Cas9 vector used in this study was obtained from the Addgene (plasmid no. 48141). Cloning was performed as previously described (28) using a pair of CRISPR single-guide RNA (sgRNA) specifically targeting exon 15 of ARID1A, namely top strand-nicking sgRNA (AACGGCGGGATGGGTGACCC) and bottom strand-nicking sgRNA (TACAGTCGTGCTGCCGGCCC). hEM3 cells were transiently transfected using Lipofectamine® 3000 (Life Technologies), and positive cells were selected in the presence of 1 μg/ml puromycin. Two weeks after transfection, single clones were isolated. Sequencing revealed deletion of 16 bp (chr1: 26,773,393–26,773,408) and combination of deletion 16 of bp with a 2-bp insertion (chr1: 26,773,390–26,773,407) on each allele, respectively. The efficiency of ARID1A knock-out was verified by immunoblotting.
RNA Extraction, Reverse Transcriptase, and Quantitative RT-PCR
Total RNA from cultured cells was isolated using the RNeasy Plus mini kit (Qiagen). In addition, a total of 30 anonymous formalin-fixed paraffin-embedded clear cell carcinoma tissues were obtained from the archival files in the Department of Pathology, the Johns Hopkins Hospital under appropriate approval of institutional review boards. Hematoxylin and eosin stained sections were reviewed by a pathologist (I.-M. S.) to confirm the diagnosis before experiments. Total RNA from formalin-fixed paraffin-embedded dissected tumor tissues was then extracted using the RNeasy formalin-fixed paraffin-embedded kit (Qiagen). First strand cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR was performed using OneTaq® Hot Start DNA polymerase (New England Biolabs, Ipswich, MA) and SYBR Green I (Life Technologies). The primers used for Quantitative RT-PCR are listed in Table 1.
TABLE 1.
Primer pairs used in this study
| Application | Primers Name | Forward | Reverse |
|---|---|---|---|
| qRT-PCR | ARID1A | CAGTACCTGCCTCGCACATA | GCCAGGAGACCAGACTTGAG |
| TERT | TGGGCACGTCCGCAAG | GAGCTCTGCTCGATGACGAC | |
| β-actin | GTTGTCGACGACGAGCG | GCACAGAGCCTCGCCTT | |
| Telomerase activity | TS (F)/ACX (R) | AATCCGTCGAGCAGAGTT | GCGCGGCTTACCCTTACCCTTACCCTAACC |
| Telomere length | Tel1b (F)/Tel2b (R) | CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT | GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT |
| 36B4u (F)/36B4d (R) | CAGCAAGTGGGAAGGTGTAATCC | CCCATTCTATCATCAACGGGTACAA | |
| ChIP qPCR | CDKN1A (p21) | CTGTGGCTCTGATTGGCTTT | CCCTTCCTCACCTGAAAACA |
| CHGA | AGAGAGAAGCCTCACTCAGACAG | CACCCCGTGCTATTTTTCCTA | |
| TERT TSS | AGCCCCTCCCCTTCCTTTCC | AGCGCACGGCTCGGCAGC | |
| TERT-500 | CTCCGTCCTCCCCTTCAC | GAGGCCCTGGGAACAGGT | |
| TERT-700 | CCGGTGGGTGATTAACAGAT | ACTTGGGCTCCTTGACACAG | |
| TERT-1200 | AATTTCCTCCGGCAGTTTCT | GGGATGCTGTAGCTGAGGTC | |
| TERT-1700 | CCCACTCAAGTGTTGTGGTG | CCCCAGTATTATGGGTGCAG |
Immunoblot Analysis
The cells were homogenized in protein lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, 0.1% SDS) supplemented with Halt protease inhibitor mixture (Thermo Scientific, Waltham, MA). Protein were separated using 4–15% Mini-PROTEAN® TGXTM Precast protein gels (Bio-Rad), and immunoblot analyses were performed using standard procedures. The blots were developed using the ClarityTM Western ECL blotting substrate (Bio-Rad). The following antibodies were used for immunoblot analysis: anti-ARID1A (1:1000, D2A8U; Cell Signaling, Danvers, MA), TERT (1:2000, ab183105; Abcam, Cambridge, MA), and GAPDH (1:5000, G9545; Sigma-Aldrich).
SYBR Green Real Time Telomeric Repeat Amplification Protocol (SYBR-TRAP) Telomerase Activity Assay
Samples for telomerase activity assays were extracted by suspending cell pellets in CHAPS lysis buffer (1,000 cells/μl), incubated for 30 min on ice, and centrifuged at 12,000 × g at 4 °C for 20 min. Protein concentration of extracts was determined with the DC protein assay (Bio-Rad). The SYBR Green RQ-TRAP assay was conducted using Platinum Taq DNA polymerase (Life Technologies) and SYBR Green I with 375 ng of protein, 1 μm of telomerase primer TS, and 0.5 μm of anchored return primer ACX. Primer sequences are listed in Table 1. Reaction mixes were incubated for 30 min at 30 °C and amplified in 35 cycles of two-step PCR with 30 s at 95 °C and 90 s at 60 °C. Samples after heat inactivation at 85 °C for 10 min serve as negative controls.
Telomere Length Analysis by qPCR
Quantitative telomere length assay was performed using a modified protocol (29, 30). Briefly, genomic DNA was extracted from cells using the DNeasy blood and tissue kit (Qiagen). Triplicate PCRs using 5 ng of each DNA, 1 μm telomere primer Tel, and 1 μm single gene copy primer 36B4 were carried out in a 12.5-μl reaction volume using the Maxima SYBR Green qPCR Master Mixes (Thermo Scientific). Thermocycling conditions were 3 min of initial denaturation step at 95 °C, followed by 35 cycles of denaturation for 1 min at 95 °C, annealing for 10 s at 57 °C, and extension for 45 s at 70 °C. Relative T/S ratios reflecting relative length differences in telomeric DNA (29, 30) were determined by generating standard curves for telomere lengths and the single gene copy from a reference DNA sample. Primer sequences used in this experiment are listed in Table 1.
Telomere Length Analysis by Fluorescence in Situ Hybridization
The method for telomere-specific fluorescence in situ hybridization was previously described (31). Briefly, cells on chamber slides were hybridized with a Cy3-labeled peptide nucleic acid probe complementary to the mammalian telomere repeat sequence (N terminus to C terminus, CCCTAACCCTAACCCTAA), and nuclei were counterstained using DAPI. A fluorescein isothiocyanate-labeled peptide nucleic acid probe specific for human centromeric DNA repeats (ATTCGTTGGAAACGGGA; CENP-B binding sequence) was also included as a positive control. The slides were then imaged with a Nikon 50i epifluorescence microscope equipped with X-Cite series 120 illuminator (EXFO Photonics Solutions Inc., Ontario, Canada), and gray scale images were captured using the Photometrics CoolsnapEZ digital camera. The images were analyzed using Telometer, a custom software plugin created for the open source image analysis program ImageJ.
Luciferase Reporter Assay
Cells were plated onto 6-well plates 24 h before transfection. The human TERT promoter luciferase reporter gene constructs (German Cancer Research Center, Dr. Rajiv Kumar), covering 2209 bp of the TERT promoter, followed by 219 bp of exon 1 and 60 bp of intron 1 region (17), were transfected using Lipofectamine® 3000 reagent according to the manufacturer's protocol. pRL-Renilla reporter plasmid (Promega, Fitchburg, WI) was co-transfected as an internal control. After another 24-h incubation with DMSO (control) and doxycycline (to induce ARID1A expression), cells were lysed and subjected to a luciferase activity assay using the Dual-Glo luciferase assay system (Promega). The reporter assay was performed according to Promega's instructions. Each transfectant was assayed in triplicate. The activity of firefly luciferase was normalized to Renilla luciferase to obtain final reporter activity.
Chromatin Immunoprecipitation Assay
The ChIP assay was performed as previously described (8) with the following modification. The cells were grown in 15-cm dishes, and ARID1A expression was induced by incubating the OVISE cells with 1 μg/ml doxycycline for 48 h, with an equal amount of DMSO used as a control on a separate dish. Approximately 1.2 × 107 cells were treated with 5 mm dimethyl-3–3′-dithiobispropionimidate for 30 min at 4 °C. The cells were washed cells twice with cold Dulbecco's PBS (Life Technologies) and incubated with quenching buffer (100 mm Tris-HCl, 150 mm NaCl) for 5 min. The cells were cross-linked with 1% formaldehyde for 10 min, quenched with 125 mm glycine for 5 min, and washed in PBS. Nuclear content was then extracted by using cytoplasmic lysis buffer (5 mm PIPES, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40), followed by nuclear lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, 0.5% Nonidet P-40). The chromatins were sheared by three cycles of 30 s on/30 s off sonication for 8 min with the Bioruptor (Diagenode, Denville, NJ) to generate DNA fragments ranging from 300 to 600 bp. The sonicated lysates were diluted with ChIP dilution buffer (1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, 167 mm NaCl) and immunoprecipitated with rotation overnight at 4 °C with 2 μg of antibodies. The antibody-chromatin complex was then precipitated for 6 h by protein G DYNAL magnetic beads (Life Technologies). Antibody-protein complexes bound to beads were washed once with high salt buffer (0.5 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.0), once with low salt buffer (0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.0), and twice with Tris-EDTA buffer, pH 8.0. After reverse cross-linking with 1% SDS and 0.1 m NaHCO3 at 65 °C overnight, DNA fragments were purified using a QIAquick PCR purification kit (Qiagen). Binding to promoter regions was analyzed by quantitative real time PCR, and data were normalized to qPCR signals obtained from an input sample. Primer sets for the CDKN1A, CHGA, and TERT are described in Table 1.
Statistical Considerations
Two-tailed unpaired t test was performed in all studies to assess differences between the control and ARID1A-modified cells, unless otherwise stated.
Results
ARID1A Negatively Regulates TERT Transcription
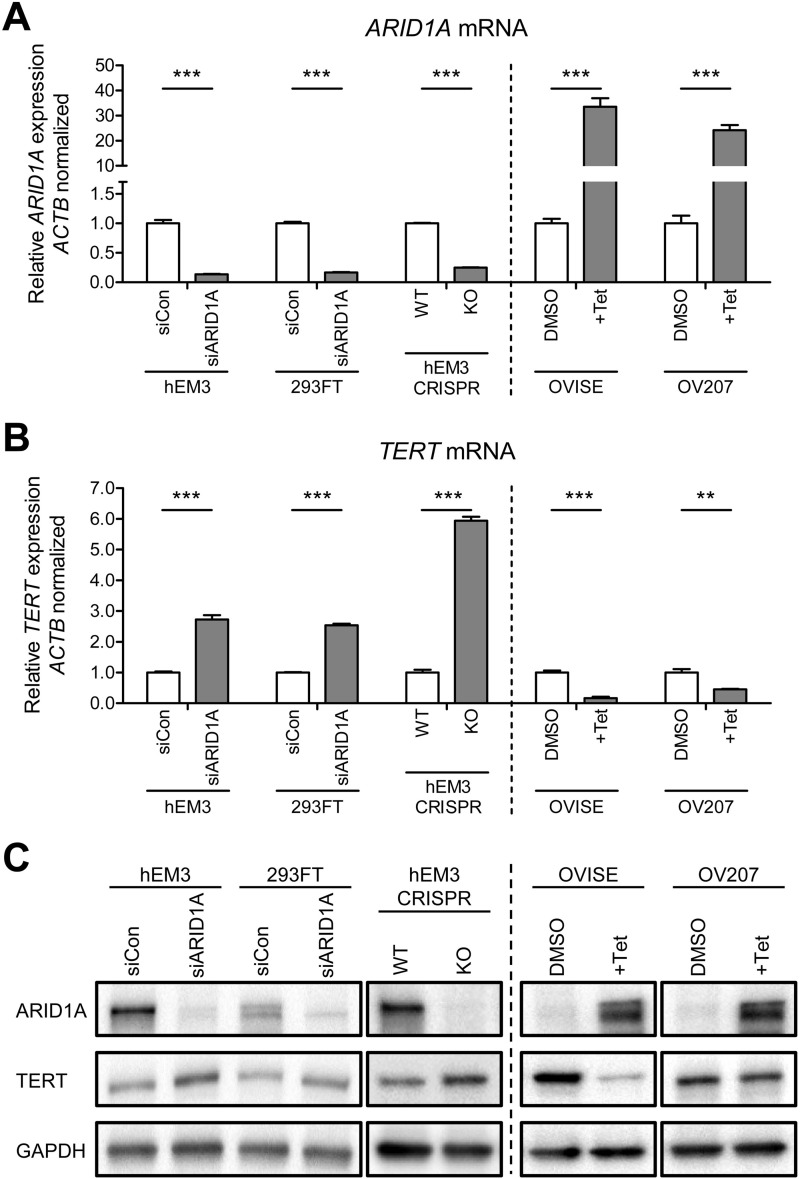
To determine whether loss or down-regulation of ARID1A expression as occurs in many endometrium-derived carcinomas is sufficient to activate TERT transcription, we first silenced ARID1A expression using siRNA in nontransformed HEK293FT and hEM3 human endometrial epithelial cells, both containing wild-type ARID1A. Although HEK293FT is a well established human embryonic kidney epithelial cell line, hEM3 cell line was derived from normal premenopausal endometrium where the endometrium-related gynecologic carcinomas arise. Quantitative real time PCR showed the robustness of ARID1A knockdown (83–87%) by ARID1A-targeting siRNAs as compared with scrambled control siRNA in both cell lines (p < 0.001; Fig. 1A). Reduction in ARID1A expression was associated with a significant increase in TERT mRNA by 2.73- (p < 0.001) and 2.53-fold (p < 0.001), respectively (Fig. 1B). To determine whether these changes at the mRNA levels were reflected at the protein levels, we performed immunoblot and observed fold changes that were similar to those detected at the mRNA level (Fig. 1C). The specificity of TERT antibody for immunoblotting was confirmed by reduced TERT protein expression in TERT siRNA knockdown 293FT cells (supplemental Fig. S1). Thus, a decrease in ARID1A protein expression was associated with an increase of TERT protein expression.
FIGURE 1.
Regulation of TERT mRNA expression levels by ARID1A. A and B, quantitative real time PCR demonstrates (A) silencing of ARID1A and (B) up-regulation of TERT mRNA expression after knockdown by siARID1A in hEM3 endometrial epithelial cells and HEK293FT embryonic kidney cells. CRISPR-Cas9 genome editing of ARID1A increases TERT mRNA expressions expression in hEM3 ARID1A KO cells. Reintroduction of ARID1A in ARID1A-null ovarian clear cells (OVISE and OV207) leads to (A) up-regulation of ARID1A and (B) down-regulation of TERT mRNA expression. β-actin (ACTB) was used as an internal loading control. C, immunoblot analysis demonstrating changes in TERT expression following modification of ARID1A expression using siRNA, CRISPR/Cas9, and Tet-on inducible systems. GAPDH was used as a protein loading control. The data are expressed as means ± S.E.; three independent experiments were performed. **, p < 0.01; ***, p < 0.001.
We next employed the CRISPR/Cas9 genome editing to generate cells lacking endogenous ARID1A (KO) in hEM3 cells (Fig. 1, A and C). The level of ARID1A protein was clearly ablated (Fig. 1C), whereas the levels of TERT mRNA and protein were increased substantially in hEM3 ARID1A KO cells as compared with ARID1A-expressing parental hEM3 cells (5.94-fold, p < 0.001; Fig. 1, B and C). We then used a forward approach by overexpressing ARID1A in OVISE and OV207 ovarian clear cell carcinoma lines that harbor deleterious ARID1A mutations and, as a result, do not express ARID1A. Using the Tet-On inducible system (8), we observed a significant 55–83% reduction of TERT mRNA (Fig. 1B), and protein expression after ARID1A expression was restored by exposure of cells to tetracycline (p < 0.001; Fig. 1, A and C). These findings suggested a negative role of ARID1A in regulating TERT transcriptional activation.
ARID1A Inhibits Enzymatic Activity of Telomerase and Shortens Telomeres
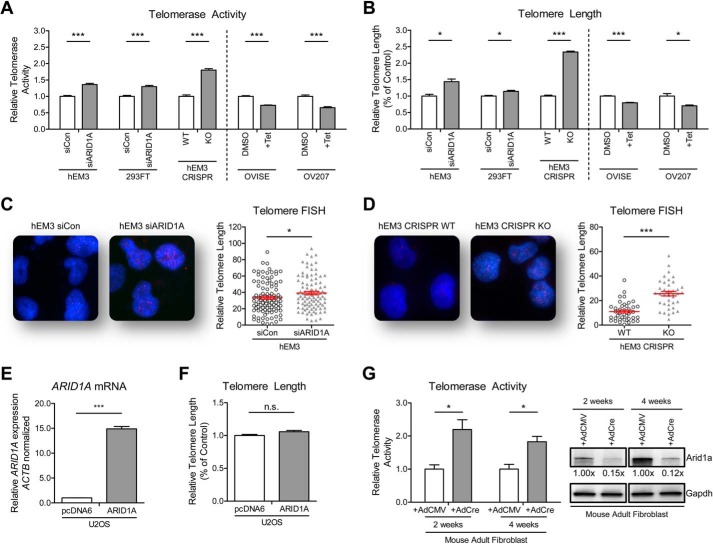
Because TERT expression generally correlates with enzyme activity of telomerase (32), we employed a real time quantitative method based on telomeric repeat amplification protocol (SYBR-TRAP) (33) and demonstrated that ARID1A silencing increased telomerase activity by1.30–1.80-fold (p < 0.001; Fig. 2A). Next, we applied qPCR to semiquantify the telomere length in the cells population. We found significantly longer telomeres (2.34-fold increase, p < 0.001; Fig. 2B) in hEM3 cells after ARID1A knock-out using CRISPR/Cas9 than in ARID1A wild-type cells, with a lesser extent in ARID1A siRNA knockdown models of hEM3 and HEK293FT cells. Telomere length was also analyzed by fluorescence in situ hybridization on hEM3 cells in which ARID1A has been silenced using siRNA for an extended period of 36 days, and hEM3 cells with CRISPR/Cas9-mediated ARID1A knock-out. In both models, depletion of ARID1A significantly increased the telomere-specific fluorescence in situ hybridization signals (p < 0.05; Fig. 2, C and D). On the contrary, overexpression of ARID1A in OVISE and OV207 ovarian clear cell carcinoma cell lines significantly decreased telomerase activities by 27–34% (p < 0.001; Fig. 2A) and significantly shortened the telomeres by 20–30% as compared with control cells without ARID1A overexpression (p < 0.05; Fig. 2B).
FIGURE 2.
ARID1A regulates telomerase activity and telomere length. A and B, silencing of ARID1A resulted in an increase of telomerase activity (A) and average telomere length (B) as determined by the SYBR-TRAP assay and qPCR, respectively. Expression of ARID1A in ARID1A-null ovarian clear cells (OVISE and OV207) leads to suppression of (A) telomerase activity and (B) telomere length. C, representative images and quantitation of telomere-fluorescence in situ hybridization (FISH) on hEM3 cells that has been treated continuously for 36 days with control and ARID1A siRNA revealed shorter telomeres in ARID1A knockdown than their control cells (n = 100). D, ARID1A knock-out using CRISPR/Cas9 resulted in significant telomere shortening in the hEM3 cells (n = 40). E and F, ectopic expression of ARID1A in U2OS telomerase-negative cells (E) resulted in no significant alteration in telomere length (F). G, increase in telomerase activities were observed in mouse primary fibroblast cells following induction of Arid1a knockdown by exposure to adeno-Cre virus. Gapdh was used as a protein loading control. The data were obtained from triplicate experiments and are shown as means ± S.E. *, p < 0.05; ***, p < 0.001.
To further support that the observed telomere phenotypes were specifically due to ARID1A, we ectopically re-expressed ARID1A in hEM3 ARID1A KO cells. We confirmed that transient expression of ARID1A resulted in a decrease of TERT mRNA expression, a reduction of telomerase activity, and telomere shortening (supplemental Fig. S2). To elucidate the role of telomerase in ARID1A-regulated telomere length, we transiently overexpressed ARID1A in the U2OS telomerase-negative human cell line. Overexpression of ARID1A in U2OS cells did not affect telomere length (Fig. 2, E and F). This is in contrast to the telomere shortening observed in ARID1A-inducible telomerase-positive OVISE and OV207 cells (Fig. 2B), suggesting that ARID1A negatively regulates telomeres through a telomerase-dependent mechanism.
In addition to the cell line study, we also examined the effect of ARID1A loss in primary cells. Considering that it is unfeasible to perform ARID1A genetic deletion in human primary cells, we applied an alternative approach by using primary fibroblast cultures derived from an inducible Arid1a knock-out mouse model (11). Induction of Arid1a deletion using adeno-Cre virus resulted in a 1.83–2.20-fold increase of telomerase activities in these mouse primary fibroblasts (Fig. 2G). Together, the above results suggest that ARID1A suppresses TERT expression and activity, thus compromising telomerase-dependent telomere length maintenance, leading to decreased average telomere length.
ARID1A Regulates TERT Promoter Activity via Binding to the TERT Regulatory Region
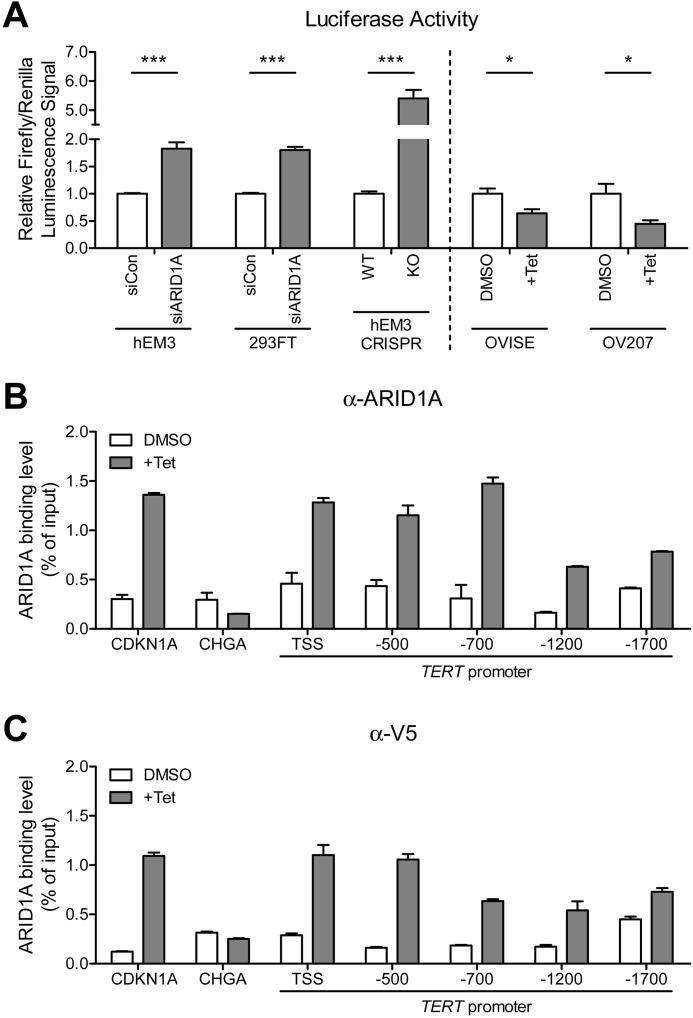
The SWI/SNF chromatin remodeling complex has been shown to alter the compactness of chromatin structure, and therefore, ARID1A may negatively or positively affect the accessibility of transcription complexes such as RNA polymerase II (Pol II) and transcription repressors or activators in gene promoters or enhancers (34–36). To investigate whether ARID1A directly regulates TERT promoter, we performed luciferase activity assay by transfecting cells with TERT promoter luciferase reporter plasmids, followed by modification of ARID1A expression via ectopic overexpression and/or siRNA-mediated silencing. Our data demonstrated that ARID1A knockdown and knock-out significantly increased TERT promoter activity by 1.80–5.40-fold in both hEM3 and HEK293FT cells (p < 0.001; Fig. 3A), whereas ectopic expression of ARID1A in ARID1A-null OVISE and OV207 clear cell carcinoma cell lines significantly suppressed its promoter activity (p < 0.05; Fig. 3A). These results support that ARID1A negatively regulates TERT promoter activity.
FIGURE 3.
ARID1A affects TERT promoter activity by direct binding to the TERT promoter region. A, the human TERT promoter luciferase reporter plasmid was co-transfected with pRL-Renilla construct into nontransformed human epithelial cells (hEM3 and 293FT) with ARID1A knockdown, and ARID1A mutated ovarian clear cells (OVISE and OV207) with ARID1A induction. B and C, ChIP-qPCR was performed using anti-ARID1A antibody (HPA005456, Sigma-Aldrich) (B) and anti-V5 antibody (ab15828, Abcam) (C) in both ARID1A-induced (+ Tet) and noninduced (DMSO) cells. CDKN1A (p21) and CHGA promoters were included as a positive and a negative control, respectively. The data were obtained from triplicate experiments and are expressed as means ± S.E. *, p < 0.05; ***, p < 0.001.
Next, we determined whether ARID1A bound to TERT promoter region using ChIP-qPCR in OVISE ARID1A-V5-inducible cells. Using an anti-ARID1A antibody for pulldown, we observed that ARID1A protein was enriched in the TERT promoter from the transcription starting site (TSS) to a −1700-bp region upstream of the TSS in the ARID1A-induced but not in the noninduced cells (Fig. 3B). ARID1A enrichment at the TERT promoter was confirmed by another antibody against the V5 epitope tag (Fig. 3C). ChIP-qPCR was also performed at the regulatory locus of CHGA gene that was not expressed in human epithelial cells, serving as a negative control (Fig. 3, B and C). Indeed, there was no appreciable enrichment of ARID1A in this locus, further confirming the specificity of the applied ChIP methodology.
ARID1A Associates with SIN3A Co-repressor and Repressive Histone Modifications at the TERT Promoter Site
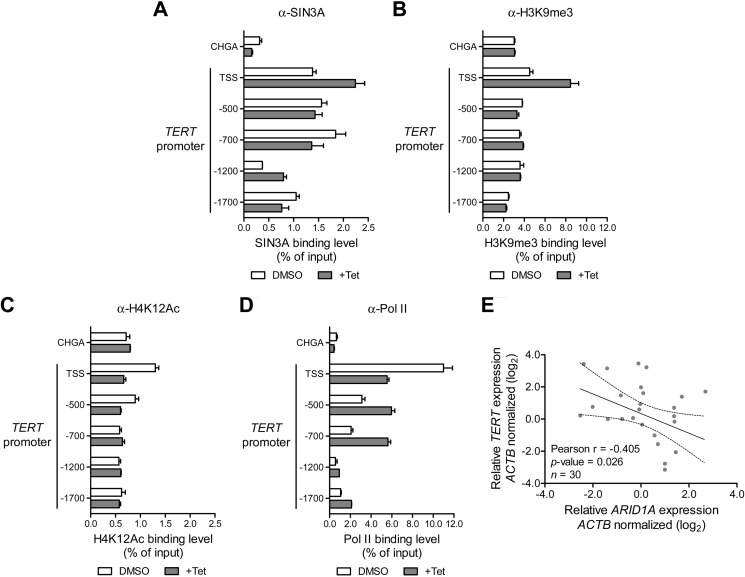
SIN3A is a critical component of multifunctional repressor protein complexes that regulate gene transcription (37). ARID1A has been reported to interact with SIN3A to suppress transcription of genes involved in cell cycle regulation and tumor progression such as c-Myc, cdc2, cyclin E, cyclin A, and IL-6 (12, 35, 38). Here, our ChIP analysis demonstrated an increase of promoter occupancy by SIN3A in ARID1A-induced cells as compared with noninduced cells (Fig. 4A), indicating the presence of ARID1A-SIN3A co-repressor complex at the TSS and the −1200-bp region of the TERT promoter. Because SIN3A-containing complex modifies histones, we also examined alterations of histones acetylation and methylation by ARID1A. Promoter occupancy experiments were performed using two repressor marks, H3K9me3 and H3K27me3, as well as two activation marks, H3K4me3 and H4K12Ac. Increased H3K9me3 was observed at the TSS of TERT promoter of ARID1A-induced cells, whereas the H4K12Ac activation mark was decreased (Fig. 4, B and C). Of note, no significant difference was observed for the levels of H3K4me3 and H3K27me3 at the TSS of TERT (data not shown). These data suggest that ARID1A induces a complex mechanism of histone modifications at the TSS of TERT promoter. The observed methylation and deacetylation of histones are consistent with the presence of the SIN3A complex in the same region.
FIGURE 4.
ARID1A-SIN3A co-repressor complex is localized at the TSS region of TERT promoter. A–D, ARID1A expression was induced by incubating the OVISE ARID1A-inducible cells with 1 μg/ml doxycycline for 48 h (+Tet), and DMSO was used as a control on a separate group. ChIP assays were performed to examine the occupancy of SIN3A (NB600-1263, Novus Biologicals) (A), histone 3 lysine 9 trimethylation (H3K9me3, ab8898, Abcam) (B), histone 4 lysine 12 acetylation (H4K12Ac, 07-595, Millipore) (C), and RNA Pol II (ab5095, Abcam) (D). Immunoprecipitated DNA samples were quantified by qPCR, and their enrichment were expressed as percentage of the input. E, Pearson's correlation analysis demonstrated a significant negative relationship between ARID1A and TERT mRNA expression in ovarian clear cell carcinoma tissues.
Furthermore, we examined the effect of ARID1A induction on TERT promoter occupancy by Pol II, which is critical for transcription initiation. We showed that induced expression of ARID1A significantly inhibited the recruitment of Pol II to the TSS of the TERT promoter (Fig. 4D). This suggests that the ARID1A-containing SWI/SNF chromatin remodeling complex in collaboration with SIN3A can modify histone methylation and acetylation to compromise Pol II occupancy, resulting in a reduction of TERT transcription activity. Thus, loss of such interaction caused by ARID1A inactivating mutation releases its negative regulation on transcription and enhances TERT expression.
Inverse Correlation between ARID1A and TERT mRNA in Ovarian Clear Cell Carcinomas
The above results support the view that chromatin remodeling regulated by ARID1A is one of the mechanisms controlling TERT expression activity. To further investigate the contribution of ARID1A genetic alterations to the TERT expression at the tissue levels, we assessed the mRNA expression of ARID1A and TERT in 30 ovarian clear cell carcinoma tissues. Pearson's correlation analysis demonstrated a significant inverse relationship between ARID1A and TERT transcript levels (r = −0.405, p = 0.026; Fig. 4E).
Discussion
Telomeres are DNA-protein complexes located at the chromosomal termini and protect the free ends from degradation and subsequent end to end fusion. Following each cellular division, telomeres shorten because of the end replication problem, ultimately leading to telomere dysfunction and induction of replicative senescence (39). To ensure unlimited proliferative potential, cancer cells evolve to develop mechanisms to maintain telomere length. This study demonstrates for the first time a critical function of a SWI/SNF chromatin remodeler protein, ARID1A, in negatively regulating telomerase activity by direct promoter occupancy and transcriptional repression of TERT. These results are of substantial significance from the perspective of cancer biology because avoiding telomere crisis is essential for mammalian cells to achieve replicative immortality, a critical step in tumorigenesis (39, 40). Hence, loss of ARID1A caused by either inactivating mutations or epigenetic silencing may offer a new mechanism to avoid cellular senescence triggered by progressive telomere shortening. This finding, together with previous reports showing other chromatin remodeling complex proteins, including ATRX and mINO80, are required for efficient telomere replication and maintenance of genome stability (41, 42), points to the important role of chromatin remodeling in telomere biology.
Telomeres are maintained in a dynamic manner through two major mechanisms including the telomerase/TERT pathway (43, 44) and the telomerase-independent recombination-based ALT pathway (39, 45). We did not detect marked telomere length heterogeneity, a hallmark of the ALT pathway, in all of the cell lines studied. Moreover, a previous report on the prevalence of ALT in human cancers demonstrated that the ALT pathway was rarely used by ovarian neoplasms including clear cell and endometrioid carcinomas (46). Because ARID1A expression did not have an effect on telomere length in telomerase negative U2OS cells (Fig. 2F), it is highly likely that ARID1A-mediated telomere dynamics can be attributed to TERT expression and activity rather than modulating the ALT pathway, highlighting the significance of telomerase regulation by ARID1A in ovarian neoplasms.
It has been established that SIN3A is an accessory factor of the transcription repressor complex that regulates the transcriptional activity of TERT (38). Our current study demonstrated that re-expression of ARID1A promoted recruitment of the SIN3A co-repressor to the TERT regulatory element. Given the known association of SIN3A and HDAC activity (38), recruitment of SIN3A to the TERT promoter region may facilitate conversion of histones into a repressive mode (heterochromatin) through deacetylation of histone H4K12Ac and methylation of histone H3K9me3. qPCR analysis from these ChIP experiments indicated that Pol II had a strong preference for occupying the TERT promoter in the ARID1A noninduced cells. Indeed, the removal of Pol II from DNA frequently correlates with a closed chromatin state, which prevents target gene transcription. Therefore, loss of ARID1A, which frequently occurs in endometrium-related and other carcinomas, may induce an open euchromatin configuration, allowing the transcription of TERT to start. In support of the above observations, a recent study demonstrated that mutation in TERT promoter can epigenetically switch chromatin into an active mode (24). Furthermore, Wu et al. (47) demonstrated the ability of BRG1, the catalytic core subunit of the SWI/SNF chromatin remodeling complex, to promote histone deacetylase at the TERT TSS and thus suppresses its transcription. ARID1A is known as one of the core components that facilitates the targeting and binding of the complexes with AT-rich DNA region within chromatin (48). This suggests that ARID1A plays an important role in repressing TERT, because the BRG1-containing SWI/SNF complex may lose the ability to bind chromatin DNA in the absence of ARID1A. Thus, it is likely that the nucleosome remodeling activity regulated by the SWI/SNF ATP-dependent complex provides accessibility of SIN3A-containing complexes to the TERT promoter region, leading to deacetylation and subsequent repression of target chromatin (49, 50). Future studies are warranted to investigate the relative importance of ARID1A and/or BRG1 in modulating chromatin structure to regulate TERT and whether inhibition of telomerase can be exploited as a therapeutic potential in patients with ovarian clear cell and endometrioid carcinomas.
Although the observations discussed above favor our view regarding the role of ARID1A in suppression of tumor development through multiple mechanisms including decreasing TERT transcription and compromising telomere maintenance in cancer cells, other alternative explanations and limitations of this study should also be highlighted. It has been recently reported that activation of TERT expression confers a survival advantage of neoplastic cells by both telomere-dependent and -independent mechanisms. In the latter process, TERT is required to stabilize the binding of MYC to its target gene promoters, thus affecting MYC-dependent transcriptional programs and oncogenic properties (51). It has also been recently reported that elevated TERT mRNA levels correlates with reduced disease-specific survival in urothelial carcinoma patients (22). In support of our in vitro study, we found clinical relevance in patient samples by which a significant inverse correlation was observed between ARID1A and TERT mRNA expression in ovarian clear cell carcinoma tissues. These data were obtained from a relatively small cohort of specimens and warrant validation in a larger multisite cohort. Moreover, it would be interesting to compare telomere length in ovarian cancer patient samples with and without ARID1A loss. Loss of ARID1A has been suggested as an early molecular event that contributes to tumor development. As an example, the absence of ARID1A immunoreactivity is observed not only in ovarian clear cell and endometrioid carcinomas but also in their adjacent endometriotic cyst epithelium that is contiguous to the carcinoma (52). Because endometriotic cyst epithelium is the cell of origin in many ovarian clear cell and endometrioid carcinomas, future studies should address whether the cyst epithelium with ARID1A loss has higher levels of TERT expression and longer telomere than those cysts with ARID1A retention.
In summary, the results from this study demonstrate that ARID1A suppresses TERT transcription by modulating the chromatin structure at the TERT regulatory/promoter region. Loss of ARID1A expression caused by inactivating mutations that are common in endometrium-associated neoplasms reactivates TERT transcriptional activity and confers survival advantage of cancer cells by maintaining their telomeres and preventing them from progressively and critically shortening to a state that is incompatible with tumor cell survival (46). Thus, this study establishes a new caretaker role for ARID1A, whereby it contributes to tumor suppression by modulating TERT expression and telomere maintenance.
Author Contributions
Y. S. R. designed and conducted a majority of the experiments outlined in the paper, analyzed the results, and wrote the paper. J.-G. J. generated the human endometrial epithelial cell line and performed part of the chromatin immunoprecipitation experiments. R.-C. W. provided critical feedback and assisted with qPCR telomere length experiments. Y. K. performed formalin-fixed paraffin-embedded tissues experiments. C. M. H. and A. K. M. provided critical feedback on experimental design and conducted experiments on telomere fluorescence in situ hybridization. T.-L. W. and I.-M. S. conceived the idea, designed experiments, and wrote the paper with Y. S. R.
Supplementary Material
Acknowledgments
We acknowledge and thank Dr. Rajiv Kumar from German Cancer Research Center (Heidelberg, Germany) for providing us with the human TERT promoter luciferase reporter gene constructs.
This work was supported by NCI, National Institutes of Health grants CA165807 and CA129080 (to I. M. S.), CA148826 (to T. L. W.), CA187512 (to T. L. W.); DoD grants W81XWH-11–2-0230 (to I. M. S. and T. L. W.) and W81XWH-14–1-0221 (to T. L. W.); Katie Oppo Research Fund (to I. M. S.); The Ephraim and Wilma Shaw Roseman Foundation (to I. M. S.); HERA Women's Cancer Foundation OSB Grant (to Y. S. R.); and Research Fellowship from the Uehara Memorial Foundation (to Y. K.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1 and S2.
- ARID1A
- AT-rich interactive domain 1A
- TERT
- telomerase reverse transcriptase
- SWI/SNF
- switch/sucrose non-fermentable
- ALT
- alternative lengthening of telomeres
- Pol II
- RNA polymerase II
- qPCR
- quantitative PCR
- TSS
- transcription starting site.
References
- 1. Watanabe R., Ui A., Kanno S., Ogiwara H., Nagase T., Kohno T., and Yasui A. (2014) SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 74, 2465–2475 [DOI] [PubMed] [Google Scholar]
- 2. Clapier C. R., and Cairns B. R. (2009) The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 [DOI] [PubMed] [Google Scholar]
- 3. Shen J., Peng Y., Wei L., Zhang W., Yang L., Lan L., Kapoor P., Ju Z., Mo Q., Shih I. M., Uray I. P., Wu X., Brown P. H., Shen X., Mills G. B., and Peng G. (2015) ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 5, 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones S., Wang T. L., Shih I. M., Mao T. L., Nakayama K., Roden R., Glas R., Slamon D., Diaz L. A. Jr., Vogelstein B., Kinzler K. W., Velculescu V. E., and Papadopoulos N. (2010) Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiegand K. C., Shah S. P., Al-Agha O. M., Zhao Y., Tse K., Zeng T., Senz J., McConechy M. K., Anglesio M. S., Kalloger S. E., Yang W., Heravi-Moussavi A., Giuliany R., Chow C., Fee J., et al. (2010) ARID1A mutations in endometriosis-associated ovarian carcinomas. New Engl. J. Med. 363, 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilson B. G., and Roberts C. W. (2011) SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 [DOI] [PubMed] [Google Scholar]
- 7. Wu R. C., Wang T. L., and Shih I. M. (2014) The emerging roles of ARID1A in tumor suppression. Cancer Biol. Ther. 15, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guan B., Wang T. L., and Shih I. M. (2011) ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 71, 6718–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mamo A., Cavallone L., Tuzmen S., Chabot C., Ferrario C., Hassan S., Edgren H., Kallioniemi O., Aleynikova O., Przybytkowski E., Malcolm K., Mousses S., Tonin P. N., and Basik M. (2012) An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene 31, 2090–2100 [DOI] [PubMed] [Google Scholar]
- 10. Wu J. N., and Roberts C. W. (2013) ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 3, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guan B., Rahmanto Y. S., Wu R. C., Wang Y., Wang Z., Wang T. L., and Shih Ie-M. (2014) Roles of deletion of Arid1a, a tumor suppressor, in mouse ovarian tumorigenesis. J. Natl. Cancer Inst. 106, dju146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chandler R. L., Damrauer J. S., Raab J. R., Schisler J. C., Wilkerson M. D., Didion J. P., Starmer J., Serber D., Yee D., Xiong J., Darr D. B., Pardo-Manuel de Villena F., Kim W. Y., and Magnuson T. (2015) Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat. Commun. 6, 6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan T. L., Zhao W., Leung S. Y., and Yuen S. T. (2003) BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 63, 4878–4881 [PubMed] [Google Scholar]
- 14. Singer G., Oldt R. 3rd, Cohen Y., Wang B. G., Sidransky D., Kurman R. J., and Shih I. M. (2003) Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J. Natl. Cancer Inst. 95, 484–486 [DOI] [PubMed] [Google Scholar]
- 15. Ardighieri L., Zeppernick F., Hannibal C. G., Vang R., Cope L., Junge J., Kjaer S. K., Kurman R. J., and Shih I. M. (2014) Mutational analysis of BRAF and KRAS in ovarian serous borderline (atypical proliferative) tumours and associated peritoneal implants. J. Pathol. 232, 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu R. C., Ayhan A., Maeda D., Kim K. R., Clarke B. A., Shaw P., Chui M. H., Rosen B., Shih I. M., and Wang T. L. (2014) Frequent somatic mutations of the telomerase reverse transcriptase promoter in ovarian clear cell carcinoma but not in other major types of gynaecological malignancy. J. Pathol. 232, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Horn S., Figl A., Rachakonda P. S., Fischer C., Sucker A., Gast A., Kadel S., Moll I., Nagore E., Hemminki K., Schadendorf D., and Kumar R. (2013) TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 [DOI] [PubMed] [Google Scholar]
- 18. Huang F. W., Hodis E., Xu M. J., Kryukov G. V., Chin L., and Garraway L. A. (2013) Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Killela P. J., Reitman Z. J., Jiao Y., Bettegowda C., Agrawal N., Diaz L. A. Jr., Friedman A. H., Friedman H., Gallia G. L., Giovanella B. C., Grollman A. P., He T. C., He Y., Hruban R. H., Jallo G. I., et al. (2013) TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. U.S.A. 110, 6021–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nault J. C., Mallet M., Pilati C., Calderaro J., Bioulac-Sage P., Laurent C., Laurent A., Cherqui D., Balabaud C., and Zucman Rossi J. (2013) High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 4, 2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vinagre J., Almeida A., Pópulo H., Batista R., Lyra J., Pinto V., Coelho R., Celestino R., Prazeres H., Lima L., Melo M., da Rocha A. G., Preto A., Castro P., Castro L., et al. (2013) Frequency of TERT promoter mutations in human cancers. Nat. Commun. 4, 2185. [DOI] [PubMed] [Google Scholar]
- 22. Borah S., Xi L., Zaug A. J., Powell N. M., Dancik G. M., Cohen S. B., Costello J. C., Theodorescu D., and Cech T. R. (2015) Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 347, 1006–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bell R. J., Rube H. T., Kreig A., Mancini A., Fouse S. D., Nagarajan R. P., Choi S., Hong C., He D., Pekmezci M., Wiencke J. K., Wrensch M. R., Chang S. M., Walsh K. M., Myong S., et al. (2015) Cancer: the transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348, 1036–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stern J. L., Theodorescu D., Vogelstein B., Papadopoulos N., and Cech T. R. (2015) Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 29, 2219–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin S. Y., and Elledge S. J. (2003) Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113, 881–889 [DOI] [PubMed] [Google Scholar]
- 26. Moore C. B., and Allen I. C. (2013) Primary ear fibroblast derivation from mice. Methods Mol. Biol. 1031, 65–70 [DOI] [PubMed] [Google Scholar]
- 27. Guan B., Gao M., Wu C. H., Wang T. L., and Shih I. M. (2012) Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia 14, 986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cawthon R. M. (2002) Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gil M. E., and Coetzer T. L. (2004) Real-time quantitative PCR of telomere length. Mol. Biotechnol. 27, 169–172 [DOI] [PubMed] [Google Scholar]
- 31. Heaphy C. M., de Wilde R. F., Jiao Y., Klein A. P., Edil B. H., Shi C., Bettegowda C., Rodriguez F. J., Eberhart C. G., Hebbar S., Offerhaus G. J., McLendon R., Rasheed B. A., He Y., Yan H., et al. (2011) Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakayama J., Tahara H., Tahara E., Saito M., Ito K., Nakamura H., Nakanishi T., Tahara E., Ide T., and Ishikawa F. (1998) Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat. Genet. 18, 65–68 [DOI] [PubMed] [Google Scholar]
- 33. Wege H., Chui M. S., Le H. T., Tran J. M., and Zern M. A. (2003) SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res. 31, E3–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Euskirchen G., Auerbach R. K., and Snyder M. (2012) SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J. Biol. Chem. 287, 30897–30905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nagl N. G. Jr., Wang X., Patsialou A., Van Scoy M., and Moran E. (2007) Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 26, 752–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wahlström T., Belikov S., and Arsenian Henriksson M. (2013) Chromatin dynamics at the hTERT promoter during transcriptional activation and repression by c-Myc and Mnt in Xenopus leavis oocytes. Exp. Cell Res. 319, 3160–3169 [DOI] [PubMed] [Google Scholar]
- 37. Silverstein R. A., and Ekwall K. (2005) Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47, 1–17 [DOI] [PubMed] [Google Scholar]
- 38. Xu M., Luo W., Elzi D. J., Grandori C., and Galloway D. A. (2008) NFX1 interacts with mSin3A/histone deacetylase to repress hTERT transcription in keratinocytes. Mol. Cell Biol. 28, 4819–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen X., Kamranvar S. A., and Masucci M. G. (2014) Tumor viruses and replicative immortality: avoiding the telomere hurdle. Semin. Cancer Biol. 26, 43–51 [DOI] [PubMed] [Google Scholar]
- 40. Günes C., and Rudolph K. L. (2013) The role of telomeres in stem cells and cancer. Cell 152, 390–393 [DOI] [PubMed] [Google Scholar]
- 41. Clynes D., Jelinska C., Xella B., Ayyub H., Scott C., Mitson M., Taylor S., Higgs D. R., and Gibbons R. J. (2015) Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 6, 7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Min J. N., Tian Y., Xiao Y., Wu L., Li L., and Chang S. (2013) The mINO80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell Res. 23, 1396–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Artandi S. E., and DePinho R. A. (2010) Telomeres and telomerase in cancer. Carcinogenesis 31, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kyo S., and Inoue M. (2002) Complex regulatory mechanisms of telomerase activity in normal and cancer cells: how can we apply them for cancer therapy? Oncogene 21, 688–697 [DOI] [PubMed] [Google Scholar]
- 45. Bryan T. M., Englezou A., Gupta J., Bacchetti S., and Reddel R. R. (1995) Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14, 4240–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heaphy C. M., Subhawong A. P., Hong S. M., Goggins M. G., Montgomery E. A., Gabrielson E., Netto G. J., Epstein J. I., Lotan T. L., Westra W. H., Shih I. M., Iacobuzio-Donahue C. A., Maitra A., Li Q. K., Eberhart C. G., et al. (2011) Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 179, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu S., Ge Y., Huang L., Liu H., Xue Y., and Zhao Y. (2014) BRG1, the ATPase subunit of SWI/SNF chromatin remodeling complex, interacts with HDAC2 to modulate telomerase expression in human cancer cells. Cell Cycle 13, 2869–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kortschak R. D., Tucker P. W., and Saint R. (2000) ARID proteins come in from the desert. Trends Biochem. Sci. 25, 294–299 [DOI] [PubMed] [Google Scholar]
- 49. Nair S. S., and Kumar R. (2012) Chromatin remodeling in cancer: a gateway to regulate gene transcription. Mol. Oncol. 6, 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pal S., Yun R., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., and Sif S. (2003) mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell Biol. 23, 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koh C. M., Khattar E., Leow S. C., Liu C. Y., Muller J., Ang W. X., Li Y., Franzoso G., Li S., Guccione E., and Tergaonkar V. (2015) Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Invest. 125, 2109–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ayhan A., Mao T. L., Seckin T., Wu C. H., Guan B., Ogawa H., Futagami M., Mizukami H., Yokoyama Y., Kurman R. J., and Shih I. M. (2012) Loss of ARID1A expression is an early molecular event in tumor progression from ovarian endometriotic cyst to clear cell and endometrioid carcinoma. Int. J. Gynecol. Cancer 22, 1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.