Abstract
Export of the Escherichia coli serotype O9a O-antigenic polysaccharides (O-PS) involves an ATP-binding cassette (ABC) transporter. The process requires a non-reducing terminal residue, which is recognized by a carbohydrate-binding module (CBM) appended to the C terminus of the nucleotide-binding domain of the transporter. Here, we investigate the process in Klebsiella pneumoniae serotype O12 (and Raoultella terrigena ATCC 33257). The O12 polysaccharide is terminated at the non-reducing end by a β-linked 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) residue. The O12 ABC transporter also binds its cognate O-PS via a CBM, and export is dependent on the presence of the terminal β-Kdo residue. The overall structural architecture of the O12 CBM resembles the O9a prototype, but they share only weak sequence similarity, and the putative binding pocket for the O12 glycan is different. Removal of the CBM abrogated O-PS transport, but export was restored when the CBM was expressed in trans with the mutant CBM-deficient ABC transporter. These results demonstrate that the CBM-mediated substrate-recognition mechanism is evolutionarily conserved and can operate with glycans of widely differing structures.
Keywords: ABC transporter, bacteria, cell surface, glycobiology, glycoconjugate, glycosylation, Klebsiella pneumonia, lipopolysaccharide (LPS), membrane transport, polysaccharide
Introduction
Cell-surface carbohydrates are essential for the viability and pathogenicity of bacteria. Biosynthesis of these molecules is initiated in the cytoplasm and so requires an export system to deliver the completed glycan (or assembly intermediates) to the external face of the cytoplasmic membrane. In the periplasm, further structural modifications can be accomplished prior to the translocation of the finished product to its final site on the cell surface. ATP-binding cassette (ABC)3 transporters provide one strategy used in the export of bacterial glycans, with examples acting on substrates, including capsular polysaccharide, teichoic acids, N- and O-linked oligosaccharides, and O-antigenic polysaccharide (O-PS) components of lipopolysaccharides (LPS) (1). ABC transporters are widespread in nature, being found in all organisms where they carry out a variety of import and export functions. They are built around a conserved core architecture, with a dimeric transmembrane domain (TMD) that forms the translocation channel, and a pair of nucleotide binding domains (NBDs), which hydrolyze ATP to drive the transport cycle (2, 3). However, the details of ABC transporter architecture vary between systems; the two halves of the TMD and NBD domains can either be identical or encoded by different genes, and the various domains can be present as separate polypeptides or fused. In many bacterial exporters, the NBD and TMD domains are fused to form half-transporters. A typical O-PS ABC transporter is composed of two identical TMDs (Wzm proteins) and two identical NBDs (Wzt proteins). They export undecaprenyl diphosphate (Und-PP)-linked completed O-PS glycans across the cytoplasmic membrane. In the periplasm, the OPS is attached to lipid A-core, which is synthesized separately, and the completed LPS molecule is translocated to the cell surface by the Lpt complex (4–7).
Two ABC transporter-dependent O-PS synthesis strategies have been identified. In the case of the Klebsiella pneumoniae O2a (polygalactose O-PS), cytosolic glycan synthesis and export are obligatorily coupled. Competition between the assembly machinery and ABC transporter dictate glycan chain length (8). In an alternative assembly strategy, described in detail in Escherichia coli O9a, the Und-PP-linked polymannose O-PS is subject to a non-reducing terminal modification that terminates chain extension. The terminator dictates glycan chain length and serves as an export signal recognized by the ABC transporter. In this mechanism, synthesis and export are not obligatorily coupled (8). The terminator residue is a phosphomethyl group added by the WbdD protein, which possesses methyltransferase and kinase domains (9–11). WbdD recruits the O-PS polymerase, WbdA, to active biosynthesis complexes in the membrane (12). The termination reaction occurs when the chain reaches a certain length and is affected by the stoichiometry of the WbdD-WbdA complex (13). In addition, WbdD possesses an extended coiled-coil region separating its membrane-anchoring amphipathic helix from the catalytic domains; this serves as the molecular ruler to fine-tune glycan chain length (11, 14). The terminated O-PS is recognized by a carbohydrate binding module (CBM) appended to the C terminus of the NBD of the cognate ABC transporter (15, 16). As their name implies, CBMs are non-catalytic polysaccharide/oligosaccharide-recognizing subdomains of many glycosidic enzymes and lectins. Currently, there are 71 defined CBM families, based on primary sequence similarity, classified in the carbohydrate-active enzyme (CAZy) database. The main purpose of CBMs is to potentiate long term saccharide-protein interactions. For example, CBMs increase the relative concentrations of a glycosidic hydrolase on its polysaccharide substrate (17), in turn increasing the efficiency of degradation (18). In addition, the CBMs of some glycosyltransferases assist in substrate recruitment (19). Type A CBMs bind crystalline glycan surfaces (e.g. cellulose), whereas type B CBMs recognize internal components of a glycan chain, and type C CBMs bind terminal units (20, 21). The structure of Wzt-C from E. coli O9a reveals a type C CBM with an immunoglobulin-like fold (16). However, it is not classified on the CAZy server because Wzt has no modifying activity directed against a carbohydrate substrate.
The Wzt CBM prevents non-terminated O-PS from being transported. This creates a quality control mechanism that ensures the exporter only delivers mature O-PS of appropriate chain length; chain length is a factor in the ability of O-PS to confer resistance to complement-mediated serum killing (22). Additionally, the CBM may ensure the presence of a non-reducing terminal acidic group, although the advantages of this are unknown. Because of the interactions they mediate with the saccharide terminus, these ABC transporters are specific for their substrates, unlike the K. pneumoniae O2a that can export structurally diverse O-PS (including O9a) (8). The mechanistic steps of Und-PP-glycan export have been established for the CBM-lacking ABC half-transporter (PglK) from the N-linked protein glycosylation pathway from Campylobacter (23). However, it is unclear whether the fine details of this model also apply to the heterotetrameric Und-PP-O-PS transporters.
Sequence analyses identify C-terminal domains, potentially corresponding to CBMs, in ABC transporters other than the closely related family of polymannose O-PSs represented by E. coli O9a (1). However, there is currently no evidence for a conserved mechanism. To address this, we investigated export of an O-PS structure composed of a disaccharide repeat unit [→4)-α-l-Rha-(1→3)-d-GlcNAc-(1→] produced by Raoultella terrigena ATCC 33257 and K. pneumoniae O12 (Fig. 1) (24, 25). These closely related species possess the same O-PS biosynthesis gene locus (25), presumably reflecting horizontal gene transfer. Hereafter, the genes and proteins are identified by the K. pneumoniae serotype (O12). The O12 O-PS is terminated with a β-linked 3-deoxy-d-manno-oct-2-ulosonic acid (β-Kdo) residue (24). Here, we test the hypothesis that the C-terminal domain of WztO12 specifically recognizes the β-Kdo residue at the non-reducing terminus of its cognate O-PS to regulate export.
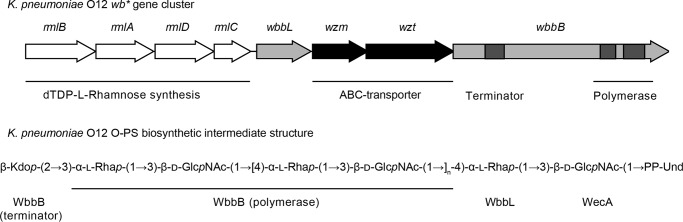
FIGURE 1.
Organization of the O-antigen biosynthesis gene cluster (wb*) from K. pneumoniae O12 and R. terrigena ATCC 33257 and the corresponding O-antigenic polysaccharide biosynthesis product. WecA is encoded at a separate site within the chromosome. The rmlBADC genes encode enzymes for the production of dTDP-l-Rha precursor and wzm-wzt encode the ABC transporter TMD and NBDs, respectively. The system requires two proteins with glycosyltransferase activities. WbbL is a monofunctional rhamnosyltransferase, whereas WbbB contains three glycosyltransferase domains; two are required for O12 polymerization and one for addition of the terminal β-Kdo residue. The component sugars are 2-acetylamino-2-deoxyglucose (N-acetylglucosamine; GlcNAc), 6-deoxymannose (rhamnose; Rha), and 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo).
Experimental Procedures
Bacterial Strains and Growth Conditions
Bacterial cultures were grown with aeration at 37 °C in Luria Broth Base (LB) (Invitrogen), supplemented with appropriate antibiotics (100 μg ml−1 ampicillin; 34 μg ml−1 chloramphenicol; 50 μg/ml kanamycin). R. terrigena ATCC 33257 and E. coli transformed with plasmid pKM114 (25), containing the O-PS-biosynthesis gene locus from R. terrigena ATCC 33257, were gifts from U. Mamat (Research Centre Borstel, Leibnitz Centre for Medicine and Biosciences, Borstel, Germany). E. coli CWG1217 [F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1nupG[pPA261]Δwzx-wbbK KanR] (26) was used to express protein for purifications. CWG1219 (CWG1217 ΔgtrA) (26) was used for in vivo experiments. For protein purification for crystallography, plasmids were used to transform either E. coli BL21 (DE3) [F− ompT hsdSB (rB−mB−) gal dcm] (Invitrogen) or the methionine auxotroph B834 (DE3) [F− ompT hsdSB (rB−mB−) gal dcm met] (Novagen).
DNA Methods
Custom oligonucleotide primers were obtained from Sigma. PCR amplification was performed using KOD DNA polymerase (EMD Millipore). The PureLink PCR purification kit (Invitrogen) was used to clean PCR products. Restriction digestions and ligation reactions were performed according to the manufacturer's instructions. The PureLink Quick Plasmid Miniprep kit (Invitrogen) was use to isolate plasmid DNA from overnight cultures. DNA sequencing was performed in the Genomics Facility of the Advanced Analysis Center at the University of Guelph.
Plasmid Constructs
The plasmids used in this study are summarized in Table 1, and the primers used are described in Table 2.
TABLE 1.
Plasmid summary
| Plasmid name | Description | Ref. |
|---|---|---|
| pKM114 | A pMBL19 derivative containing the 10.3-kb R. terrigena wb* gene cluster | 25 |
| pWQ284 | Plasmid vector with l-arabinose-inducible promoter; Cmr | 48 |
| pWQ811 | pMBL19 derivative containing tetracycline-inducible promoter; Apr | 26 |
| pWQ552 | Protein expression vector under control of Ptet; Apr | 49 |
| pBAD18-Kn | Plasmid vector with l-arabinose-inducible promoter; Knr | 34 |
| pWQ674 | pWQ284 derivative containing wztO12; Cmr | This study |
| pWQ114 | pWQ284 derivative containing FLAG-wztO12; Cmr | 26 |
| pWQ840 | pWQ284 derivative containing FLAG-wztO12 (1–265); Cmr | This study |
| pWQ841 | pWQ284 derivative containing wzm-wztO2a; Cmr | This study |
| pWQ842 | pWQ284 derivative containing wzm-wztO12; Cmr | This study |
| pWQ675 | pWQ284 derivative containing wztO12 with an internal NdeI site; Cmr | This study |
| pWQ689 | pWQ284 derivative containing wztO12-His10; Cmr | This study |
| pWQ843 | pWQ284 derivative containing wztO12-C-His10; Cmr | This study |
| pWQ844 | pWQ284 derivative containing His6-TEV-wzt-CO12; Cmr | This study |
| pWQ845 | pWQ552 derivative containing wbbB (401–1103)-rbs-wbbL; Apr | This study |
| pWQ703 | pWQ552 derivative containing wbbL-rbs-wbbB; Apr | This study |
| pWQ847 | pBAD18-Kan derivative containing wztO12-C; Knr | This study |
| pWQ677 | pWQ811 derivative containing wbbL-wzm-rbs-wbbB; Apr | 26 |
| pWQ672 | pWQ811 derivative containing wbbL-wzm-wzt-wbbB; Apr | This study |
| pWQ848 | pWQ284 derivative containing wztO12 F298A; Cmr | This study |
| pWQ849 | pWQ284 derivative containing wztO12 G348Q; Cmr | This study |
| pWQ850 | pWQ284 derivative containing wztO12 Q355A; Cmr | This study |
| pWQ851 | pWQ284 derivative containing wztO12 D358A; Cmr | This study |
| pWQ852 | pWQ284 derivative containing wztO12 G361Q; Cmr | This study |
| pWQ853 | pWQ284 derivative containing wztO12 R407A; Cmr | This study |
| pWQ854 | pWQ284 derivative containing wztO12 Q412A; Cmr | This study |
| pWQ855 | pWQ284 derivative containing wztO12 R414A; Cmr | This study |
| pWQ856 | pWQ284 derivative containing FLAG-wztO12 G348Q; Cmr | This study |
| pWQ857 | pWQ284 derivative containing FLAG-wztO12 E183Q; Cmr | This study |
| pWQ866 | pWQ284 derivative containing FLAG-wztO12 E183Q G348Q; Cmr | This study |
| pWQ858 | pWQ284 derivative containing wztO12-C-His10 F298A; Cmr | This study |
| pWQ859 | pWQ284 derivative containing wztO12-C-His10 G348Q; Cmr | This study |
| pWQ860 | pWQ284 derivative containing wztO12-C-His10 Q355A; Cmr | This study |
| pWQ861 | pWQ284 derivative containing wztO12-C-His10 D358A; Cmr | This study |
| pWQ862 | pWQ284 derivative containing wztO12-C-His10 G361Q; Cmr | This study |
| pWQ863 | pWQ284 derivative containing wztO12-C-His10 R407A; Cmr | This study |
| pWQ864 | pWQ284 derivative containing wztO12-C-His10 Q412A; Cmr | This study |
| pWQ865 | pWQ284 derivative containing wztO12-C-His10 R414A; Cmr | This study |
TABLE 2.
Primers used in the generation of plasmids
Capital letters are complementary to the template DNA. Restriction endonuclease recognition sequences are underlined. His and FLAG-tag sequences are italicized. Boldface sequences are rbs sites. Fwd is forward, and Rev is reverse.
| Primer name | Sequence | Construction of plasmid |
|---|---|---|
| 674 Fwd | 5′-aaaaaagaattccatATGTCCTCTAATGAAATTGCTATCC-3′ | pWQ674 |
| pWQ848 | ||
| pWQ851 | ||
| 674 Rev | 5′-aaaaaaaaaagcttggtaccTTACTCTCCATTCGAAATAATTTTC-3′ | pWQ674 |
| pWQ848 | ||
| pWQ851 | ||
| 689 Fwd | caactaggaggtacccatatgGCGGACCAGGGTTCAGCTG | pWQ689 |
| 689 Rev | aaaaaaaaaagcttggtaccttagtgatggtgatggtgatggtgatggtgatgtgACTCTCCATTCGAAATAATTTTC | pWQ689 |
| 672 Fwd | 5′-aaaaaaggtaccaggaggtgaattctATGACTTATGAAGCAATGAAGCC-3′ | pWQ672 |
| 672 Rev | 5′-aaaaaatctagaCTTCCGGCCTTTTGCTATG-3′ | pWQ672 |
| 841 Fwd | 5′-gactactagtATGAGTATAAAGATGAAGTACAATTTAGG-3′ | pWQ841 |
| 841 Rev | 5′-gatcaagcttTTAAGCCATTGCTTGCTTGTAC-3′ | pWQ841 |
| 842 Fwd | 5′-gactactagtATGAAAAACCCGCATCAAAAACTTTC-3′ | pWQ842 |
| 842 Rev | 5′-gatcaagcttTTACTCTCCATTCGAAATAATTTTCAAC-3′ | pWQ842 |
| 675-1 Fwd | 5′-aaaaaagaattccatATGTCCTCTAATGAAATTGCTATCC-3′ | pWQ675 |
| 675-1 Rev | 5′-CGCcatatgtggtacctcctaGTTTGCGGCTGGCTTCTCG-3′ | pWQ675 |
| 675-2 Fwd | 5′-CAACtaggaggtaccacatatgGCGGACCAGGGTTCAGCTG-3 | pWQ675 |
| 675-2 Rev | 5′-aaaaaaaaaagcttggtaccTTACTCTCCATTCGAAATAATTTTC-3′ | pWQ675 |
| 843 Fwd | 5′-aaaaaagaattccatATGTCCTCTAATGAAATTGCTATCC-3′ | pWQ843 |
| 843 Rev | 5′-gaaaattatttcgaatggagagtcacatcaccatcaccatcaccatcaccatcacTAAGGTACCAAGCtttttttttt-3′ | pWQ843 |
| 844 Fwd | 5′-ggaggaattcaccatgcatcatcatcatcatcatGAGAACCTATATTTTCAAGGCGCGGACCAGGGTTCAGCTGAAGAAGTTAGCG-3′ | pWQ844 |
| 844 Rev | 5′-gatcaagcttTTATGACTCTCCATTCGAAATAATTTTCAACGG-3′ | pWQ844 |
| 845-1 Fwd | 5′-tctatcagtgatagaggATGCTGGCTGTATTTTTACC-3′ | pWQ845 |
| 845-1 Rev | 5′-CTTCATTGCTTCATAAGTCATTATTTACCTCCTTTATGATCCTAGCGGTTGCGCTTAAACTC-3′ | pWQ845 |
| 845-2 Fwd | 5′-gagtttaagcgcaaccgctaggatcATAAAGGAGGTAAATA ATGACTTATGAAGCAATGAAG-3′ | pWQ845 |
| 845-2 Rev | 5′-ccgggtaccatggtgCAACCTAACTTCCGGGGAATTTC-3′ | pWQ845 |
| 703 Fwd | 5′-aaaaaagctagcaggaggttggtaccATGCTGGCTGTATTTTTACCTCCC-3′ | pWQ703 |
| 703 Rev | 5′-aaaaaaaagcttccatggaCTAGCGGTTGCGCTTAAACTCC-3′ | pWQ703 |
| 840 Fwd | 5′-gatcgaattcATGgattacaaggatgacgacgataagTCCTCTAATGAAATTGC- TATCCAGGTCAC-3′ | pWQ840 |
| 840 Rev | 5′-gatcaagcttTTAGCGGTTTGCGGCTG-3′ | pWQ840 |
| 847 Fwd | 5′-gatcggtaccaggaggatctcatATGGCGGACCAGGGTTC-3′ | pWQ847 |
| 847 Rev | 5′-gatctctagaTTACTCTCCATTCGAAATAATTTTCAAC-3′ | pWQ847 |
| 856 Fwd | 5′-gatcgaattcATGgattacaaggatgacgacgataagTCCTCTAATGAAATTGC- TATCCAGGTCAC-3′ | pWQ856 |
| 856 Rev | 5′-gatcaagcttTTACTCTCCATTCGAAATAATTTTCAACGGAG-3′ | pWQ856 |
| 858 Fwd | 5′-gatcgaattcATGTCCTCTAATGAAATTGC-3′ | pWQ858–865 |
| 858 Rev | 5′-gaaaattatttcgaatggagagtcacatcaccatcaccatcaccatcaccatcacTAAGGTACCAAGCtttttttttt-3′ | pWQ858–865 |
| wztF298A Fwd | 5′-CTGGTGAGAAGCGCggCGGTAGCGCGCGTG-3′pWQ | pWQ848 |
| wztF298A Rev | 5′-CACGCGCGCTACCGccGCGCTTCTCACCAG-3′ | pWQ848 |
| wztG348Q Fwd | 5′-CCTGCGCTTTCGACATGGAGATctgTAACGCGATATCTTTAATTTCTTC-3′ | pWQ849 |
| wztG348Q Rev | 5′-GAAGAAATTAAAGATATCGCGTTAcagATCTCCATGTCGAAAGCGCAGG-3′ | pWQ849 |
| wztQ355A Fwd | 5′-TCCATGTCGAAAGCGgcGGGGGGCGATATCTG-3′ | pWQ850 |
| wztQ355A Rev | 5′-CAGATATCGCCCCCCgcCGCTTTCGACATGGA-3′ | pWQ850 |
| wztD358A Fwd | 5′-TCTCCCCAGATAgCGCCCCCCTGCG-3′ | pWQ851 |
| wztD358A Rev | 5′-CGCAGGGGGGCGcTATCTGGGGAGA-3′ | pWQ851 |
| wztG361Q Fwd | 5′-TCCGATATTACTGTCtgGCCAGATATCGCCCCCC-3′ | pWQ852 |
| wztG361Q Rev | 5′-GGGGGGCGATATCTGGCcaGACAGTAATATCGGA-3′ | pWQ852 |
| wztR407A Fwd | 5′-GATCGAGTTCTTCCgcATCGCCGTTGCCAACCT-3′ | pWQ853 |
| wztR407A Rev | 5′-AGGTTGGCAACGGCGATgcGGAAGAACTCGATC-3′ | pWQ853 |
| wztQ412A Fwd | 5′-CGGACGGCGCgcATCGAGTTCTTCCCGATCG-3′ | pWQ854 |
| wztQ412A Rev | 5′-CGATCGGGAAGAACTCGATgcGCGCCGTCCG-3′ | pWQ854 |
| wztR414A Fwd | 5′-TTTAACTTTCATCATCGGAgcGCGCTGATCGAGTTCTTCC-3′ | pWQ855 |
| wztR414A Rev | 5′-GGAAGAACTCGATCAGCGCgcTCCGATGATGAAAGTTAAA-3′ | pWQ855 |
| wztE183Q Fwd | 5′-GATCTTAATTGTCGACcAGGCGTTGGCGGTTG-3′ | pWQ857 |
| wztE183Q Rev | 5′-CAACCGCCAACGCCTgGTCGACAATTAAGATC-3′ | pWQ857 |
The wztO12 gene was PCR-amplified from pKM114 (25). The primer sequences incorporated EcoRI and HindIII restriction sites for cloning. The PCR product was digested with these enzymes and ligated into pWQ284 (16) to generate pWQ674.
To generate plasmid pWQ841, K. pneumoniae O2a wzm-wzt was PCR-amplified from pWQ391 (8). The primers incorporated SpeI and HindIII sites for cloning into pWQ284. Plasmid pWQ842 was produced by amplifying R. terrigena wzm-wzt from pKM114 and cloned in pWQ284.
pWQ675 contains wztO12 with an internal NdeI site located immediately upstream of the codon encoding amino acid 267 to facilitate removal of bases encoding the N-terminal domain of Wzt (amino acids 1–266) from the construct for later experiments. Fragments of wztO12 were PCR-amplified from pKM114, and the primers incorporated EcoRI, HindIII, and NdeI sites. The fragments were joined using overlap PCR, cleaved with EcoRI and HindIII, and ligated into pWQ284. To express WztO12-His10, the gene was amplified from pWQ675 with primers containing a sequence encoding a C-terminal His10 tag. The PCR fragment was digested with KpnI and HindIII and ligated into pWQ284 to generate pWQ689. To produce the C-terminal CBM (WztO12-C-His10), plasmid pWQ675 was digested with NdeI and religated to remove the 798 bp from wztO12 and generating pWQ843.
For expression of the WztO12 CBM for crystallization, a DNA fragment encoding residues 267–442 from R. terrigena (gi AY376146.1) was amplified by PCR from pKM114 and ligated into pWQ284 to generate pWQ844. The cloning strategy added 14 residues (MHHHHHHENLYFQG) at the N terminus of the protein, as well as a C-terminal serine derived from the vector.
Plasmid pWQ840 contains a gene encoding the first 265 residues of WztO12. Primers used to amplify the fragment from pKM114 introduced an N-terminal FLAG tag (DYKDDDDK) and flanking restriction sites to facilitate cloning into pWQ284.
Plasmid pWQ845 contains a DNA fragment encoding residues 401–1103 of WbbB (WbbB(401–1103)) followed by an engineered ribosome-binding site (rbs) upstream of the wbbL open reading frame. The construct was made using vector pWQ811 (linearized with EcoRI), and the two PCR fragments were ligated together using the Gibson assembly kit (New England Biolabs). The wbbB fragment was PCR-amplified using primers that incorporated a 5′ region overlapping the vector (pWQ811, linearized with EcoRI) and a 3′ region encoding an rbs, as well as sequence overlapping gene wbbL. The wbbL gene was amplified with primers incorporating a 5′ region overlapping wbbB and the engineered rbs and a 3′ region overlapping the vector.
Plasmid pWQ703 contains wbbL and wbbB. The primers were used to amplify wbbL from pKM114 restriction enzyme sites for cloning. The sequence encoding wbbB was amplified from pKM114 with primers introducing a preceding rbs and flanking restriction sites. The PCR products were digested and ligated into pWQ811 in successive steps.
For genetic complementation studies, plasmid pWQ847 was constructed. This plasmid contains a PCR fragment amplified from pKM114 encoding an rbs followed by sequence encoding Wzt-C (residues 267–442). Introduced XbaI and KpnI restriction sites facilitated cloning into pBAD18-kan.
Most WztO12 mutants were generated by site-directed mutagenesis using pWQ674 as a template. The protocol from the QuikChange mutagenesis kit (Agilent) was followed, but the cycling conditions were altered to adhere to the directions provided for the KOD polymerase (Table 1). Mutants F298A and D358A were generated by overlap PCR of fragments containing wzt bases 1–907 and bases 880–1329 introducing the F298A mutation and fragments 1–1083 and 1061–1329 introducing the D358A mutation. These were digested with EcoRI and HindIII and ligated into pWQ284. WztO12 mutations were verified by DNA sequencing.
WztO12-C mutants for in vitro binding analysis were generated by PCR amplification using the corresponding Wzt-mutant plasmid as a template with primers (858 forward and 858 reverse), which incorporated a C-terminal His10 tag and EcoRI and HindIII restriction sites to facilitate cloning into pWQ284. Plasmid pWQ672 was generated by PCR amplification of wbbL-wzm-wzt-wbbB from pKM114 with primers 672 forward and 672 reverse, digestion with KpnI and XbaI, and ligation into pWQ811. Plasmid pWQ856 encoding FLAG-WztO12 G348Q was produced by PCR amplification of WztO12 G348Q from pWQ849 with primers that introduced an N-terminal FLAG tag along with EcoRI and HindIII sites. The insert was digested and ligated into pWQ284.
Protein Expression and Purification
For apoprotein expression, E. coli BL21(pWQ844) cells were grown in 1 liter of LB cultures (with 34 μg/ml chloramphenicol) at 37 °C until an A600 nm 0.6 was reached. Recombinant protein expression was induced by adding 0.1% l-(+)-arabinose and continuing incubation for 2 h at 37 °C. For selenomethionyl protein expression, B834 pWQ844 cells were grown in 5-ml LB cultures (with 34 μg/ml chloramphenicol) at 37 °C overnight. Cells from the overnight culture were harvested by centrifugation and resuspended in selenomethionine-supplemented minimal media (SSMM) (0.25 mm l-selenomethionine, 0.4% (w/v) glucose, 1 mm magnesium sulfate, 0.3 mm calcium chloride, 4 μm biotin, 3.8 μm thiamine, 56 mm sodium phosphate dibasic, 29 mm potassium phosphate monobasic, 8.6 mm sodium chloride, 9.3 mm ammonium chloride, 0.17 mm EDTA, 65 μm iron(II) chloride, 6.2 μm zinc chloride, 0.76 μm copper(II) chloride dehydrate, 0.42 μm cobalt(II) chloride hexahydrate, 1.6 μm boric acid, 80 nm manganese (II) chloride tetrahydrate). The culture was incubated for 1 h at 37 °C and then used to inoculate 90 ml of SSMM. This culture was grown until an A600 nm ∼0.8 was achieved and then used to inoculate 900 ml of SSMM, which was again grown to A600 nm ∼0.8. The culture was then transferred to 16 °C, and recombinant protein expression was induced using 0.1% l-(+)-arabinose for 20 h. For both native and selenomethionine preparations, cells were harvested by centrifugation at 5,000 × g for 15 min at 4 °C, resuspended in buffer A (50 mm BisTris, pH 7.0, and 150 mm NaCl), and frozen at −20 °C. Cells were lysed using an EmulsiFlex-C3 (Avestin) at 15,000–17,000 p.s.i. The cell lysate was cleared using successive centrifugation steps at 5,000 × g for 15 min and 74,000 × g for 1 h. Cell- and membrane-free supernatant was passed through a 2-ml nickel-nitrilotriacetic acid-agarose IMAC gravity column (Bio-Rad). The matrix was washed in successive steps with 10 ml of buffer A with 50 mm imidazole, and then 100 mm imidazole was added before the protein was eluted with 20 ml of buffer A supplemented with 500 mm imidazole. Eluted protein was dialyzed using a 3500-Da molecular mass cutoff Slidealyzer Dialyzer Cassette (ThermoScientific) against storage buffer (150 mm Tris-HCl, pH 7.4, containing 150 mm NaCl) to remove the imidazole and then concentrated using a VivaSpin500 3-kDa molecular mass cutoff column (General Electric Healthcare). Protein folding of mutants was confirmed with differential scanning fluorimetry using the Protein Thermal Shift dye kit (ThermoFisher Scientific), according to the manufacturer's instructions. A StepOnePlus Real Time PCR system (ThermoFisher Scientific) was used in this protocol (27).
Structure Determination
Form 1 crystals were grown in a sitting drop configuration by mixing 5 mg/ml protein (selenomethionyl or native) at a ratio of 1:1 with 0.1 m Na-HEPES, pH 7.5, containing 0.8 m sodium phosphate and 0.8 m potassium phosphate. Crystals formed small plates (50 mm) after 5 days at room temperature. Form 2 crystals grew in a sitting drop configuration by mixing 23 mg/ml protein at a ratio of 1:1 with 0.1 m Tris-Cl, pH 8.5, containing 0.3 m sodium acetate and 20% w/v PEG 2000. Crystals formed large (600 mm) prisms. Crystals were cryoprotected with paratone-N prior to freezing with liquid nitrogen. Form 1 crystals were of the monoclinic space group C2 and diffracted to 1.85 Å with selenomethionyl protein and to 1.7 Å with native protein. Form 2 crystals were of the orthorhombic space group P212121 and diffracted to 2.2 Å. The structure of WztO12-C was initially determined using single anomalous scattering from the form 1 selenomethionyl crystals. Anomalous substructure searching with Phenix autosol found all nine selenium atoms. Despite relatively weak phases (overall figure of merit 0.239), the presence of 3-fold non-crystallographic symmetry allowed autotracing to correctly trace over half of the structure. Manual rebuilding in Coot (28) and refinement in Phenix (29) were used to complete the structure. The higher resolution native monoclinic dataset was refined in the same way. The orthorhombic structure was then determined using molecular replacement in Phenix. Data collection and refinement statistics are shown in Table 3. Structural figures were prepared in PyMOL.
TABLE 3.
Data collection, model refinement and final structure statistics
| Crystallographic data collection statistics |
|||
|---|---|---|---|
| Selmet (peak) | Monoclinic | Orthorhombic | |
| Protein Data Bank | 5HNO | 5HNP | |
| Space group | C2 | C2 | P212121 |
| Cell dimensions | |||
| a | 141.65 | 141.75 | 47.78 |
| b | 53.60 | 53.55 | 104.07 |
| c | 87.71 | 87.47 | 106.33 |
| β | 125.38 | 125.77 | 90.0 |
| Wavelength (Å) | 0.978635 | 1.03322 | 1.03321 |
| Resolution range (Å) | 50–1.85 | 40–1.7 | 45–2.2 |
| Redundancy | 2.9 | 3.7 | 7.6 |
| Completeness (last shell)a | 0.993 (0.998) | 0.995 (0.994) | 0.998 (1.0) |
| 〈I/σ(I)〉 (last shell)a | 10.5 (1.4) | 19.8 (1.87) | 20.1 (2.35) |
| Rsym (last shell)a | 0.073 (0.953) | 0.035 (0.88) | 0.054 (0.95) |
| CC½ highest shell | 67 | 80 | 80 |
| X-ray structure refinement statistics | |||
| Rcryst | 0.1831 | 0.1928 | |
| Rfreeb | 0.2155 | 0.2189 | |
| Asymmetric unit contents | |||
| Protein chains | 3 | 2 | |
| Water molecules | 276 | 201 | |
| Other molecules | 2 | 1 Cl− | |
| Average ADPs (Å2) | |||
| Protein | 49.35 | 41.2 | |
| Water | 45.66 | 40.1 | |
| r.m.s.d. bond lengths (Å) | 0.0043 | 0.0037 | |
| r.m.s.d. bond angles (°) | 0.921 | 0.889 | |
| Ramachandran favored (%) | 97.4 | 97.8 | |
| Outliers (%) | 0 | 0 | |
a The last shell includes all reflections between 1.90 and 1.85 Å for the Selmet dataset, between 1.74 and 1.70 Å for the native monoclinic dataset and between 2.26 and 2.20 Å for the native.
b Rfree calculated using 5% of the data, which were chosen randomly.
Purification of LPS
LPS with non-terminated O-PS was generated in E. coli CWG1219 cells co-expressing pWQ841(wzm-wztO2a) and pWQ845 (wbbB(401–1103)-rbs-wbbL) with 100 ng/ml anhydrotetracycline inducer for pWQ845. Expression of the contents of pWQ841 was reliant on the leaky pBAD promoter. LPS was isolated from R. terrigena ATCC 33257 and E. coli using the hot phenol method of extraction (30). Five g of cell pellet was resuspended in 50 ml of 50 mm Tris-Cl, pH 7.5, containing 5 mm EDTA, and the cells were disrupted by sonication for 3 min in 15-s pulses. Hen egg white lysozyme (Sigma) was added to a final concentration of 2 mg/ml, and the solution was stirred for 16 h at 4 °C. The solution was diluted to a final volume of 100 ml by adding the same buffer, and MgCl2 was added to a final concentration of 10 mm. DNA and RNA removal was performed by adding 125 units of Benzonase® endonuclease (Novagen) and incubating at room temperature for 30 min on a rotary shaker. The solution was warmed to 70 °C in a water bath. An equal volume of 90% phenol (prewarmed to 70 °C) was added to the suspension. The mixture was stirred by hand for 20 min until a single phase was evident. The mixture was then cooled on ice to <15 °C, and the resulting phases were separated by centrifugation at 10,000 × g for 15 min. The aqueous phase was collected and dialyzed against water until no phenol odor was detectable. The retentate was lyophilized under vacuum (Labonco) and dissolved in 25 ml of 20 mm sodium acetate, pH 7.0. LPS was collected as a pellet after ultracentrifugation at 105,000 × g for 16 h at 4 °C, resuspended in MilliQ water, and lyophilized.
In Vitro LPS Binding Assays
Binding assays were performed in 1-ml reactions comprising buffer B (25 mm BisTris, pH 7.0, containing 250 mm NaCl), 200 μg of native LPS, or 8.9 mg of LPS with non-terminated O-PS and 200 μg of WztO12-C-His10. Reaction mixtures were incubated on a rotary shaker for 30 min at room temperature. Each reaction mixture was added to 50 μl of PureProteome nickel magnetic beads (Millipore) equilibrated with buffer B and incubated for 30 min at room temperature on a rotary shaker. The beads were collected with a magnet and washed three times with 500 ml of buffer B. Protein was eluted stepwise from the beads using three washes with 100 μl of buffer B containing 500 mm imidazole. The samples were then examined for protein and LPS contents by PAGE.
Protein Detection
Protein samples were boiled in SDS-PAGE loading buffer for 10 min and separated on a 12% acrylamide resolving gel by SDS-PAGE using Tris-glycine buffer (31). Purified protein was detected by SimplyBlue SafeStain (Life Technologies, Inc.). Western blots were performed with polyclonal rabbit antibodies directed against the C-terminal domain of WztO12 on nitrocellulose membranes (Protran, PerkinElmer Life Sciences). Alkaline phosphatase-conjugated goat anti-rabbit secondary antibodies (Cedar Lane) were used, and the immunoblot was developed with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (Roche Applied Science). Mouse anti-FLAG antibodies were obtained from Sigma and were detected with alkaline phosphatase-conjugated goat anti-mouse secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) and developed as described previously. To generate antibodies specific for WztO12-C, serum was collected from rabbits immunized with purified His-tagged WztO12-C, and antibodies were purified by affinity chromatography using WztO12-C protein conjugated to CNBr-activated Sepharose. Antibodies specific for WztO12-C were eluted with 200 mm glycine, pH 2.8.
LPS Detection
Whole-cell lysates were prepared by solubilizing equivalent amounts of cells (determined by A600 nm) in SDS-PAGE loading buffer, heating to 100 °C for 10 min, and treating with proteinase K (32). The resulting lysates were separated using SDS-PAGE in Tris-glycine buffer on a 12% acrylamide resolving gel (31). LPS was visualized with silver staining (33). Immunoblot detection of O-antigens was performed by transferring lysates separated by SDS-PAGE to nitrocellulose membranes and probing with rabbit antiserum specific for the R. terrigena ATCC 33257 O-antigen structure (26). Alkaline phosphatase-conjugated goat anti-rabbit secondary antibodies (Cedar Lane) were used, and the immunoblot was developed with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (Roche Applied Science).
Results
Terminating β-Kdo Residue Is Required for O-PS Export
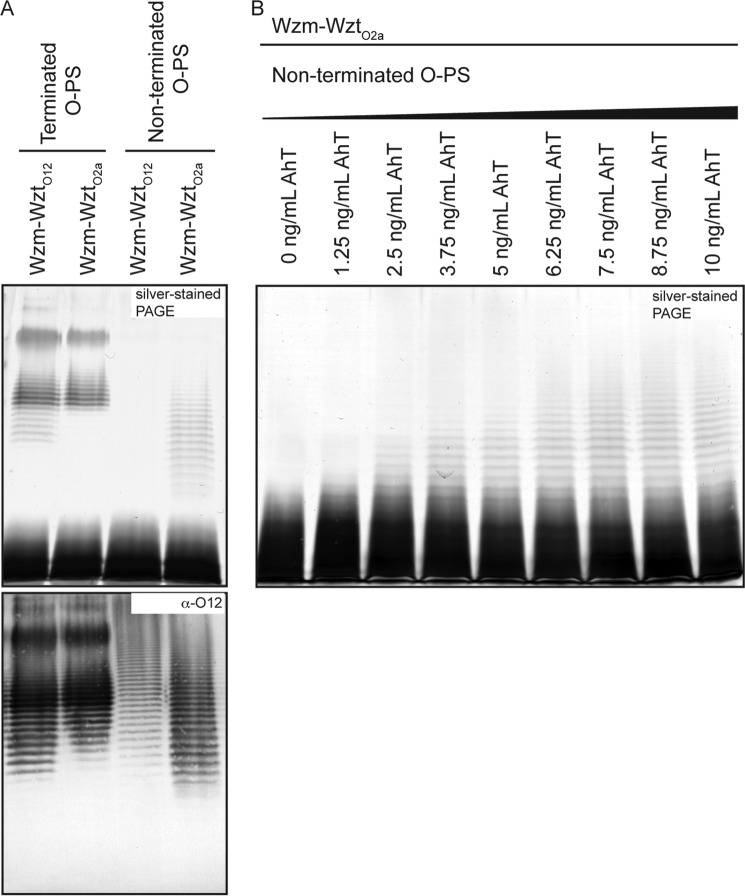
Biosynthesis of the O12 OPS requires glycosyltransferase (GT) activity provided by the WbbL and WbbB proteins (26). WbbB contains three predicted GT catalytic domains. Expression of WbbL and WbbB in E. coli CWG1219 (pWQ703) generates O12 glycan that can be exported by the native ABC transporter (Wzm-WztO12) and assembled into LPS (Fig. 2A). The promiscuous ABC transporter (8) from K. pneumoniae serotype O2a (Wzm-WztO2a), also transports the O12 glycan with a very similar size profile; a slight reduction in the amount of shorter polymers was evident (Fig. 2A). An N-terminal domain (residues 1–401) of WbbB encodes a β-Kdo transferase domain responsible for chain termination.4 Expression of WbbB(401–1103) with WbbL in E. coli CWG1219 (pWQ845) resulted in synthesis of O-PS with wild-type repeat-unit structure, as determined by reactivity with antibodies directed against K. pneumoniae O12 antigen, but the native chain length regulation is lost as expected (Fig. 2A). The Western immunoblot detects both LPS and unexported Und-PP-linked glycan, whereas the silver-stained gel only reports O-PS that is exported and incorporated into LPS (26). The uncapped O12 glycan was still exported by the O2a ABC transporter, as evidenced in the silver-stained SDS-PAGE, but it was no longer a substrate for the native O12 ABC transporter. Titration of the expression of the GTs (WbbL and WbbB(401–1103)) by increasing the amount of anhydrotetracycline inducer resulted in enhanced synthesis and higher average chain lengths in the non-terminated LPS (Fig. 2B). This is consistent with the operation of the native O2a system where the stoichiometry of the export:biosynthesis components controls chain length (8).
FIGURE 2.
β-Kdo reducing terminal residue is required for recognition of O-PS by the ABC transporter in K. pneumoniae O12. The figure shows representative SDS-PAGE of whole-cell lysates stained with silver (A, upper panel), which highlights all LPS molecules (but not unexported O-PS), as well as the corresponding immunoblot with antibodies specific for the O12 O-PS, which identifies intracellular undecaprenyl diphosphate-linked glycan, as well as O-PS exported and ligated to lipid A-core (A, lower panel). Properly terminated O-PS is synthesized in E. coli CWG1219 transformed with pWQ703 encoding WbbL and WbbB. This material is exported to the surface by either the native O12 ABC transporter (pWQ842) or the transporter from K. pneumoniae O2a (pWQ841). The absence of chain termination in CWG1219 (pWQ845) encoding WbbB(401–1103) and WbbL generates glycan that is a substrate for the O2a transporter but is no longer recognized by the O12 transporter. B shows representative SDS-PAGE of whole-cell lysates stained with silver, demonstrating the change in non-terminated O-antigen chain length due to differential expression of the biosynthesis enzymes under conditions of constant transporter production. The expression of WbbL and the C-terminal domains of WbbB (pWQ845) was elevated using increasing amounts of anhydrotetracycline (AhT), whereas the levels of O2a transporter remain constant (pWQ841).
C-terminal Domain of WztO12 Is Required for Export
The NBD protein (WztO12) has a size consistent with the presence of a functional C-terminal CBM (1). WztO12 includes 440 residues compared with the WztO2a protein at 246 residues. The additional C-terminal sequence shares weak homology (21% identity, E = 3e−8) with the WztO9a CBM. To investigate this further, WztO12 was truncated within a region predicted to be weakly ordered by JPred4 (35). The resulting N-terminal domain (residues 1–265) is referred to as Wzt-N. No export occurred in E. coli CWG1219 transformants expressing WbbL and WbbB, Wzt-N, and the corresponding TMD protein (Wzm) (Fig. 3A), despite the presence of abundant Und-PP-linked O-PS detected in immunoblots with O12-specific antiserum (Fig. 3B). The absence of export was not due to the absence of protein expression because Wzt-N-FLAG was detected (Fig. 3C). Introduction of WztO12-C in trans restored transport of O-PS, with longer O-PS structures favored.
FIGURE 3.
C-terminal domain of O12 Wzt is essential for export activity. A and B show representative SDS-PAGE and Western immunoblots (as indicated) of proteinase K-treated whole-cell lysates showing LPS and O12 glycan, respectively. C and D show representative immunoblots with primary antibodies directed against N-terminal FLAG tag on Wzt and the native C-terminal domain of Wzt, respectively. N-terminally FLAG-tagged Wzt (pWQ114) or Wzt-N (Wzt(1–265); pWQ840) together with the TMD (Wzm) and the O12 biosynthesis GTs (pWQ677) are shown. Induction of gene expression was accomplished using 0.1% arabinose (pWQ114) and 2.5 ng/ml anhydrotetracycline (pWQ840), and cell cultures were grown to an A600 nm of ∼0.8. In the 3rd lane, Wzt-C was introduced in trans with pWQ847, and the 1st two lanes contain empty pBAD18-Kan vector controls. The calculated sizes of the proteins are as follows: FLAG-WztO12 49.2 kDa; FLAG-WztO12-N 29.1 kDa; and WztO12-C 19.2 kDa. Positions of protein bands are denoted with an asterisk.
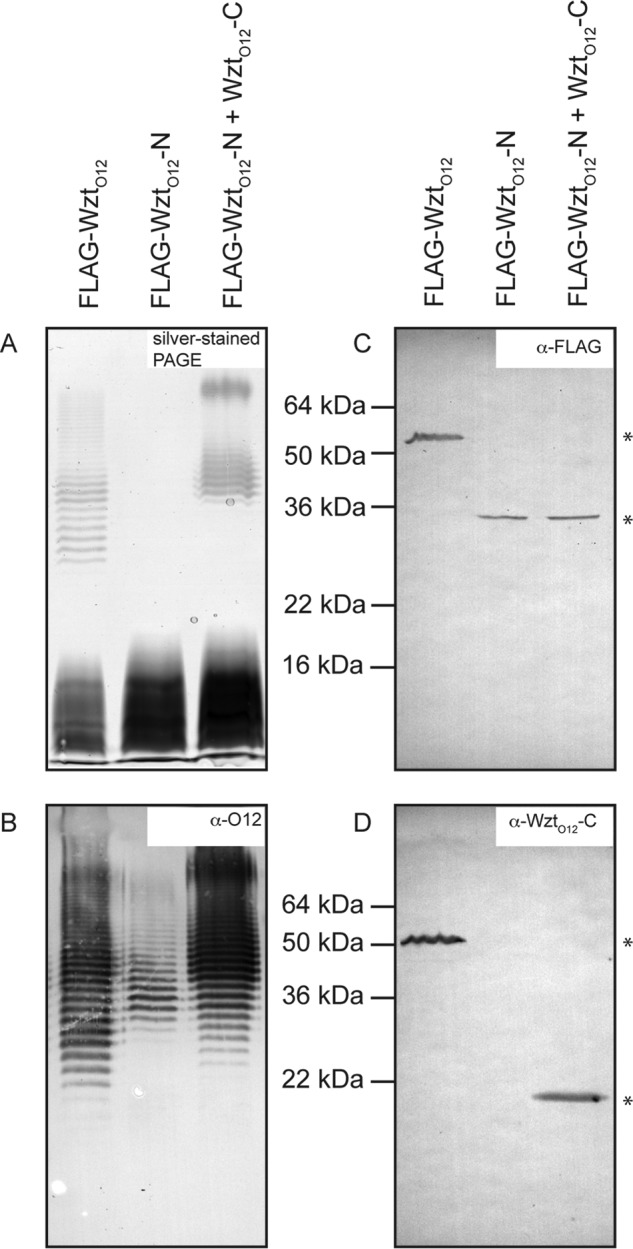
WztO12-C Binds Specifically Its Cognate β-Kdo-terminated LPS in Vitro
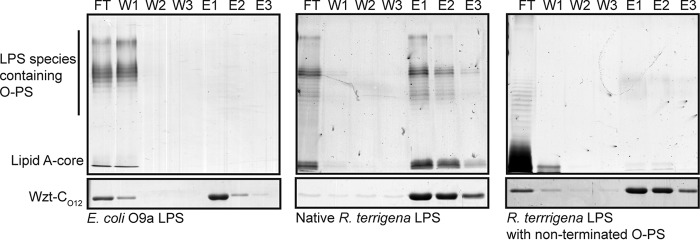
Purified E. coli O9a LPS or R. terrigena LPS was incubated with WztO12-C-His10. Protein-LPS complexes were bound to magnetic nickel-nitrilotriacetic acid beads. Protein was eluted with imidazole, and the fractions were analyzed by SDS-PAGE to detect LPS and protein (Fig. 4). Elution fractions from reactions containing WztO12-C-His10 and (negative control) E. coli O9a LPS contained only protein; LPS was confined to the flow-through fraction. Conversely, when WztO12-C-His10 was incubated with its cognate LPS, LPS and protein species co-eluted.
FIGURE 4.
C-terminal domain of O12 Wzt binds specifically to its non-terminated LPS. Pulldown experiments were performed using purified Wzt-CO12-His10 and three different LPS species obtained from E. coli O9a: R. terrigena ATCC 33257, a recombinant E. coli TOP10 derivative expressing the wzm-wzt genes from K. pneumoniae O2a, and the glycosyltransferases wbbL and wbbB(401–1103), encoding the O-PS polymerization domains from R. terrigena ATCC 33257 to produce non-terminated LPS. Reaction mixtures containing WztO12-C-His10 and the identified LPS were mixed with magnetic nickel beads. The supernatant (flow-through; FT) was collected. The beads were washed with buffer three times (W1–W3) and protein was eluted with buffer containing 500 mm imidazole (E1–E3). Protein samples were detected by SDS-PAGE (lower) and LPS was detected by SDS-PAGE and silver staining after proteinase-K treatment (upper). Representative PAGE are displayed.
LPS with O-PS lacking the terminal β-Kdo residue was purified from E. coli CWG1219 cells co-expressing WbbB(401–1103) and WbbL and the promiscuous Wzm-Wzt transporter from K. pneumoniae O2a. The elution profile of reactions containing WztO12-C-His10 and this LPS showed no binding. The LPS was confined to the flow-through, whereas protein was eluted in the elution steps (Fig. 4).
Structure of WztO12 C-terminal Domain
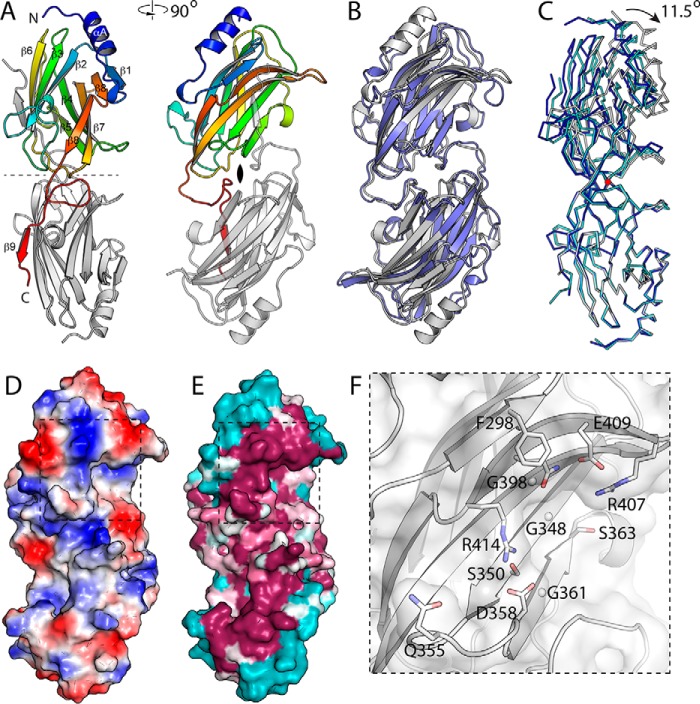
We determined the structure of the WztO12 C-terminal domain (WztO12-C) by single anomalous diffraction phasing of a selenomethionine derivative. This crystal form proved monoclinic, with three molecules in the asymmetric unit. A high resolution native structure was also determined for this crystal form. This structure was then used to determine the structure of the native protein using a dataset from an orthorhombic crystal with two molecules per asymmetric unit. Despite being from substantially smaller crystals (less than 0.1% of the volume of the native crystals), the monoclinic crystal form diffracted considerably more strongly, and our analysis will focus primarily on this structure. The three protomers in the monoclinic structure are arranged as a dimer and a half-dimer that is completed by a second copy of the same molecule related by a molecular 2-fold axis (Fig. 5, A and B). WztO12-C adopts an immunoglobulin fold and is organized as two antiparallel β-sheets, one five-stranded (order 5, 4, 7, 8, and 1) and the other four-stranded (order 2, 3, 6, and 9′, with 9′ from the other half of the dimer). A single three-turn α-helix is found at the N terminus. This domain forms a dimer, with extensive interactions afforded by the C-terminal β-strand, which is domain swapped. The first ordered residue is 274, indicating that residues immediately N-terminal to this may form a linker.
FIGURE 5.
Structure of WztO12-C. A, structure of the WztO12-C dimer, with two orthogonal views. In the left panel, the 2-fold symmetry axis (dashed line) is in plane of the page. In the right panel, the view is down the 2-fold axis. Secondary structure elements as marked. The structure is organized as a pair of back-to-back β-sheets. The second sheet is completed by the last β-strand that is provided by its dimeric partner. B, superposition of Wzt-CO12 (white) on WztO9a-C (blue). The two structures are overall very similar, with an r.m.s.d. of 2.2 Å. The major difference is in the N terminus, especially αA, which is longer and better defined in Wzt-CO12 and also displaced. C, domain flexibility in Wzt-CO12. The three independent dimers in the two structures were superposed using the lower protomer. The apo-dimer (white) is at one extreme of the range of motion, and the C-C′ dimer of the Selmet crystal is at the other end (blue); the A-B dimer of the Selmet crystal is intermediate in conformation (cyan) and on the trajectory linking the two extremes. The red circle marks the location of the rotation axis, which is orthogonal to the page and at right angles to the 2-fold symmetry axis of the dimers. Note that the orientation of this panel is as in A. D, surface of WztO12-C colored by electrostatic potential. Blue represents positive potential; red indicates negative. The dashed line shows the area of the inset in F. E, surface of WztO12-C colored by sequence conservation. Sequences with E values for the C-domain of greater than e-24 were used to construct the alignment, which was then mapped onto the structure using Consurf. Magenta represents conserved residues on the surface, cyan indicates highly variable. Note the ridge of conserved residues that is well positioned to interact with the NBD domain. F, key residues in the potential binding pocket, with side chains shown as sticks (glycine Cα as spheres). These residues line an extended pocket on the surface of WztO12-C and likely represent the terminal residue binding site.
Searching with DALI (36) shows that WztO12-C is structurally most similar to WztO9a-C (2r5o), with an r.m.s.d. of 2.2 Å and a Z-score of 17.5. Sequence identity between these domains is 21%. The structures show a common overall fold, differing mainly in that WztO12-C has an extended N-terminal α-helix; the corresponding residues are substantially displaced in WztO9a. WztO12-C also shows weak similarity to a variety of other immunoglobulin fold proteins, with the more similar examples including Rho-GDP dissociation inhibitor 1 (2jhy, 2.3 Å r.ms.d., Z-score 10.1) and β-mannosidase (2vqu; r.m.s.d. 5.7 Å, Z-score 9.4).
Structural Flexibility of WztO12-C
The independent observation of multiple copies of a given structure in one or more crystal forms can be revealing, as crystal packing interactions contribute a small amount of interaction energy that can weakly stabilize one conformer from the envelope of conformations sampled by the protein in solution. Comparison of the structures of the five independently determined protomers between the two crystal forms shows that the structures superpose well, with r.m.s.d. values of 0.25 Å or less for each pairwise comparison. More flexible regions include the β7-β8 loop, which is poorly ordered in all protomers, and the β2-β3 loop, which adopts two distinct conformations. Comparison of the three independent dimers in the two crystal forms (A-B and C-C′ in the monoclinic structure, and A-B in the orthorhombic structure) (Fig. 5C) reveals flexibility in the dimer interface, with protomers flexing along a hinge that is orthogonal to the 2-fold symmetry axis. Residues in β9 as well as the preceding loop (423 to end) are more closely associated with the second protomer and move with it. The A-B dimer from the orthorhombic and monoclinic structures represent the two extremes of this motion, with an 11.5° rotation between them; the C-C′ dimer from the monoclinic structure occupies an intermediate state, although closer to the other monoclinic dimer. Of note, the presence of this third conformation between the two extremes suggests that there is a preferred rotation axis between the protomers, rather than the region simply being generally flexible. The significance of this flexibility is not clear, but it could possibly play a role in the functional cycle of this protein. For example, oligosaccharide binding may induce inter-subunit hinge bending in WztO12-C, which in turn triggers export by NBD-WztO12. It is also worth noting that the observed flexibility probably represents three random sampling points of the trajectory in the absence of ligand. As such, it may not represent the full range of motion available over the course of the protein's functional cycle.
Candidate Binding Pocket
WztO12-C has a candidate substrate-binding pocket located on the exposed surface of the five-stranded β-sheet. This region is the most conserved surface on the structure, and this is also the location of the pocket identified in WztO9a. This sheet curves to create a distinctly concave surface, with the candidate carbohydrate-binding site located in its approximate center. Here, a deep pocket is created by the presence of three glycine residues, Gly-348, Gly-361, and Gly-398, which combined with small adjacent residues Ala-346 and Ser-363 create an extensive pocket with the backbone of the β-sheet as its floor. Although all three glycine residues are conserved in WztO9a, the rest of the flanking residues are very different, consistent with the different terminal residue specificities. Arg-407, Asp-409, Gln-412, and Arg-414 on β8 form the back of the pocket, providing two Arg residues that may be important for forming favorable electrostatic interactions with the carboxylate group of Kdo. Ser-350 and Asp-358 are conserved residues that line the pocket, contributed by adjacent strands. All of these residues are highly conserved, although Arg-107 and Arg-414 are relatively mobile in the absence of ligand (with high atomic displacement parameters and large differences in conformation between protomers). This pocket is larger than required to accommodate Kdo, with space for the next two sugar residues available in the direction of Ile-365. The equivalent pocket in WztO9a-C was confirmed to be important for O-antigen binding and export (16). Unfortunately, we were unable to obtain substrate complexes by soaking with millimolar concentrations of either Kdo or a trisaccharide product of WbbB.
Mutation of the Putative Binding Pocket Alters LPS Chain Length
Candidate amino acids from the proposed binding pocket were selected for site-directed mutagenesis. Mutant versions of WztO12 were expressed alongside WbbL-Wzm-WbbB. Western blots with α-WztO12-C primary antibodies demonstrate similar levels of wild-type and mutant WztO12 proteins (Fig. 6C). The LPS profile produced with expression of the F298A, Q355A, and D358A proteins were very similar to the wild type. WztO12G348Q and G361Q completely eliminated transport under experimental conditions (Fig. 6A), even though O-PS was present (Fig. 6B). Three mutations, R407A, Q412A, and R414A, altered the chain length distribution of LPS species, with a loss of species with shorter chain lengths. Interestingly, the chain length distribution seen in Western immunoblots of cells harboring each of these two mutants resembled more closely that seen with the export-null mutants.
FIGURE 6.
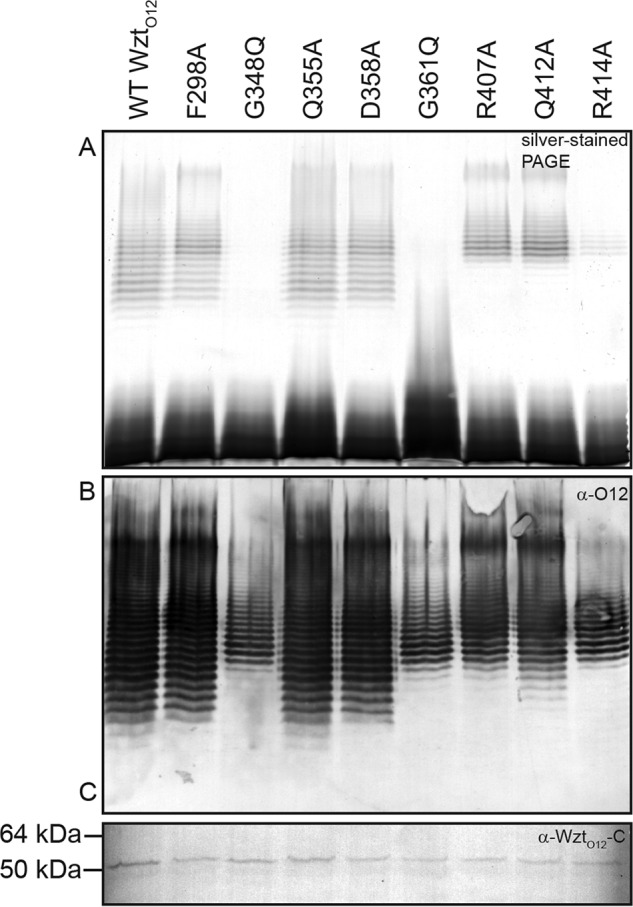
Mutagenesis of key residues in the Wzt-C putative binding pocket alter O-antigen export activity. Size distribution of LPS species is altered by the mutation of candidate O-PS-binding site residues in R. terrigena ATCC 33257 Wzt-C. Samples were proteinase K-treated whole-cell lysates that were subject to SDS-PAGE and silver staining. A and B are representative silver-stained SDS-PAGE and an α-O12 immunoblot of proteinase K-treated samples, respectively. C is an α-Wzt-CO12 Western blot that demonstrates expression of Wzt mutants.
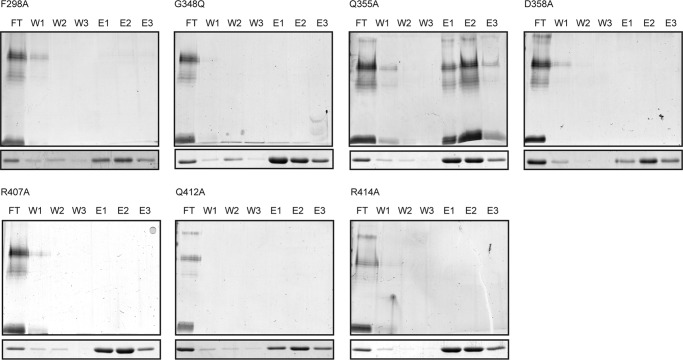
Mutation of the Putative Binding Pocket Impairs LPS Binding in Vitro
The point mutations were introduced into the WztO12-C-His10 construct to facilitate in vitro binding studies, as described above. Under the experimental conditions used here, only the Q355A mutant retained binding activity and co-eluted with LPS (Fig. 7). In all other cases, the LPS was detected only in the flow-through. These results implicate the pocket on the five β-strand face of the protein in binding the O-PS chain. As well, the results suggest that polar and electrostatic interactions from Arg-407 and Arg-414 as well as Asp-358 with the terminal Kdo sugar are important for binding. Gln-412 possibly contributes to binding through polar interactions with its amide group. The sole aromatic residue in the pocket, Phe-298, may contribute to ring-stacking.
FIGURE 7.
Mutagenesis of key residues in the Wzt-C putative binding pocket reduces LPS affinity. Wzt-CO12-binding site mutants have reduced O-PS affinity compared with wild type. Pulldown experiments were performed using purified WztO12-C-His10 mutants and LPS obtained from R. terrigena ATCC 33257. WztO12-C-His10 was incubated with LPS and magnetic nickel beads. Beads were pulled down with a magnet, and the supernatant (flow-through; FT) was collected. The beads were washed with buffer three times (W1–W3), and protein was eluted with buffer containing 500 mm imidazole (E1–E3). Protein samples were detected by SimplyBlue staining SDS-12% polyacrylamide gels (lower). LPS samples were proteinase K-treated fractions, and LPS was detected by SDS-PAGE and silver staining (upper).
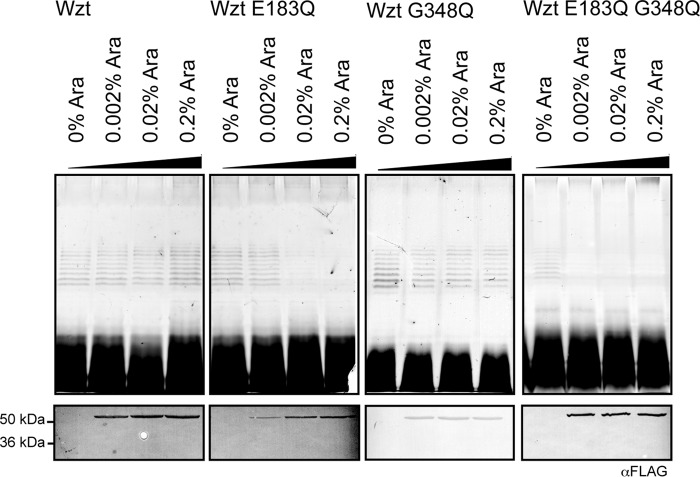
WztO12 Mutants Defective in Glycan Binding Do Not Display a “Dominant-negative” Phenotype
Studies with the maltose ABC importer (MalK) demonstrated that transport was severely affected by an ATPase defect in a single NBD (37). The same is true for the vitamin B12 (BtuD) transporter (38) where a single functional ATPase supported just 5% of wild-type transport levels. We used a dominant-negative approach as a simple preliminary method for investigating the transport response to increasing levels of NBDs with glycan-binding defects (Fig. 8). The WztO12-binding site mutants G348Q change a residue essential for the export of Und-PP-O-PS (Fig. 6) and in vitro LPS binding (Fig. 7). This protein has no detectable folding defects, and the mutation is not in a position that should influence dimerization. FLAG-tagged WztO12 G348Q was overexpressed along with low levels of WbbL, Wzm, WbbB, and wild-type WztO12. Surprisingly, export was not affected, even with high levels of G348Q expression; O-PS-substituted LPS was still observed on silver-stained SDS-polyacrylamide gels. The lack of any detectable dominant-negative effect by the mutant was surprising. One interpretation is that this particular system is refractory to dominant-negative effects for some reason, so we tested this possibility using a catalytically null variant. A glutamic acid residue within the Walker box motif that is conserved across ABC transporters is generally critical for function (39). The corresponding WztO12 mutant (E183Q) was generated. High level expression of the E183Q mutant with wild-type Wzt, WbbL, WbbB, and Wzm results in a transition of LPS to a form devoid of O-PS (as observed by silver-stained SDS-polyacrylamide gels) demonstrating its inability to support transport. A similar dominant-negative phenotype was also observed when a Wzt double mutant containing both the CBM mutation, G348Q, and the Walker box mutation, E183Q, was overexpressed. From the precedent established with the MalK and BtuD systems, these results are consistent with a scenario where, as levels of the mutant protein rise, there is a transition to ABC transporter complexes with heterodimers and homodimers of inactive NBD proteins.
FIGURE 8.
Half-site binding by Wzt-CO12 permits export of O-antigen. Samples were whole-cell lysates. The top panels display representative silver-stained SDS-polyacrylamide gels, and the samples were proteinase K-treated. The lower panels are representative Western immunoblots with primary antibodies directed against the N-terminal FLAG tag. N-terminally FLAG-tagged WztO12 (pWQ114), WztO12 E183Q (pWQ857), WztO12 G348Q (pWQ856), or WztO12 E183QG348Q (pWQ866) were induced with arabinose (0–0.2%) in cells expressing wild-type Wzt, the TMD (Wzm), and O12 biosynthesis GTs (pWQ672). Expression of genes cloned in pWQ674 was performed without inducer (leaky expression) and cells were grown to an A600-nm of ∼0.8.
Discussion
A wide variety of Gram-positive and Gram-negative bacteria are predicted to contain glyco-transporters with an extended C-terminal domain on the NBD (1). Examples include members of the genera Clostridium, Pseudomonas, Vibrio, Yersinia, and Rhizobium. These ABC transporters play essential roles in the production of O-PS and glycoproteins, but the structures and functions of the C-terminal domains are unknown. Prior to this study, the only fully characterized example was from E. coli O9a, where the C-terminal domain is a CBM (15, 16). The R. terrigena/K. pneumoniae O12 Wzt CBM provides a second example and reveals a conserved process for two O-PSs with significantly different carbohydrate structures. The C-terminal extension of WztO12 encodes a lectin-like CBM, which recognizes its cognate O-PS with a conserved binding pocket. Binding is dependent on the presence of a β-Kdo residue at the non-reducing terminus of the O-PS substrate and is a requisite step in the transport mechanism. The general similarity to the E. coli O9a model increases our confidence in the widespread applicability of the export strategy in other systems.
Our analyses of the role of the CBM took advantage of both in vivo activity and in vitro LPS-binding tests. The in vitro system was much more susceptible to mutations in the CBM. Several mutants were still able to export sufficient O-PS for abundant O-PS-substituted LPS molecules, despite showing no binding of LPS in vitro. Possibly this implies that carbohydrate binding has additional determinants in the holo-complex. Aromatic residues played a critical role in O-PS binding in E. coli O9a, potentially by ring stacking. In contrast, electrostatic and polar interactions dominate in O12 CBM recognition. In either case, a pair of glycine residues form the base of the binding pocket. Replacement with bulkier glutamine residues appears to completely abrogate carbohydrate binding and export, presumably by steric exclusion. The activities of several other bacterial CBMs recognizing terminal residues have been characterized, although they are in the minority in comparison with CBMs recognizing features within the glycan chain. The family 66, type C CBM of the Bacillus subtilis β-fructosidase (SacC) contains a β-sandwich fold with the binding pocket on the concave face. It binds strongly to the non-reducing terminal fructose with only moderate hydrogen bonding to the penultimate residue, allowing it to recognize a broader range of substrates with various linkages (40). This increased hydrolytic activity by ∼100-fold. Likewise, the Bacillus halodurans laminarinase and Saccharophagus degradans β-agarase family 6 CBMs bind the non-reducing terminus of their substrates, albeit using loop inter-strand loop regions (41, 42). Conversely, Thermotoga maritima xylanase 10A identifies residues located at the reducing terminus of xylans and cellulose (43).
The longer saccharide chain length phenotype observed with most binding pocket mutants is interesting. Precedent from the E. coli O9a chain length regulation systems (14) suggests that chain length should be dictated primarily by the action of the capping Kdo transferase domain of WbbB, with the Wzt-Wzm export system acting downstream. However, the accumulation of capped saccharides in the cytosol by slowed transport may indirectly affect the relative activities of the chain elongation and termination activities of WbbB, for example by product inhibition of the Kdo transferase domain.
The route of Und-PP-glycan flipping has been described recently in PglK from the N-linked protein glycosylation pathway in Campylobacter jejuni (23). In this case, PglK is a half-transporter, and the substrate is a lipid-linked heptasaccharide, rather than a longer polysaccharide. The conserved pyrophosphate-sugar moiety interacts with the interior of the transmembrane channel to stimulate ATP binding. Conformational changes in the NBDs, induced by nucleotide binding, are translated to opening of the TMDs to the periplasm and translocation of the oligosaccharide through the lumen of the transporter, whereas the lipid component remains in the membrane. ATP hydrolysis and nucleotide release facilitate release of the Und-PP-glycan substrate and resets the transporter to the resting state for a new round of transport. Sequence and organizational differences between Wzm-WztO12 and PglK are large enough to preclude confident modeling of the subtle features important in the PglK mechanism. However, integration of a CBM into this model offers intriguing regulatory possibilities. One possible analog of the O12 system is the NBD of the E. coli maltose importer, MalK, which contains a C-terminal extension involved in trans-inhibition. In particular, the phosphorylated enzyme IIAGlc suppresses transport by binding to the NBD-regulatory domain interfaces, locking the transporter in an ATPase-deficient conformation (44); MalT, in contrast, binds in the absence of intracellular maltose to stimulate maltose intake. By analogy, the binding of the cognate O-PS by WztO12 could induce a conformational change that is a necessary precondition of dimerization, nucleotide binding, or nucleotide hydrolysis by the NBD.
Although the functional transporter possesses two CBMs, we consider it unlikely that both are essential to initiate transport. The model proposed for PglK invokes flipping of a single Und-PP-glycan at a time, implying half-site binding (23); given that the O12 glycan substrate is much larger, half-site binding would seem a reasonable assumption in this system, too. This is consistent with the lack of a dominant-negative phenotype with a Wzt mutant defective in glycan binding. Although it is conceivable that mutant Wzt never reaches levels high enough to prevent wild-type Wzt dimers from forming, this is not consistent with the sensitivity of the system to the overexpression of the E183Q mutant (which was able to integrate and inactivate the complex at expression levels comparable with those evident with G348Q (Fig. 8)).
More interesting is the observation that no dominant-negative effects were observed even at the highest levels of CBM-defective protein expression when expressed in conjunction with the chromosomal Wzt. This suggests that the finite available Wzm somehow avoids being locked into transport-incompetent complexes by excess transport-incompetent mutant homodimer Wzt. One hypothesis explaining this observation is that the abrogation of carbohydrate binding interferes with the assembly of a functional ABC transporter complex. In some transporters, NBDs exist as monomers in the absence of nucleotide, and only dimerize in vitro in the presence of ATP (45–47). In Wzt, the CBM domains likely form a strongly dimerized platform (implied by the extensive interactions between the subunits) that may then control the interaction geometry of the NBD domains in a carbohydrate binding-dependent manner. In this model, binding of carbohydrate to a single CBM site drives NBD dimerization, followed by association with Wzm and ultimately transport. This hypothesis is consistent with the lack of a dominant-negative phenotype demonstrated by highly overexpressing the glycan binding-deficient G348Q mutant in the presence of chromosomal Wzt; without the ability to bind carbohydrate, the G348Q homodimers fail to form a stable complex with Wzm, but the G348Q/WT heterodimers still mediate transport. In contrast, the E183Q G348Q/WT heterodimers can bind carbohydrate and therefore initiate NBD dimerization and Wzm binding; however, this complex is unable to hydrolyze ATP. This leaves these heterodimers trapped in a state incapable of completing transport, leading to the observed dominant-negative phenotype. Confirming this hypothesis will require detailed biochemical analysis that is beyond the scope of this work. Now that the overall conservation of the CBM role is established in two systems with different glycan structures, reaching an understanding of how carbohydrate binding at the CBM can trigger transport becomes an important research priority for this field.
Author Contributions
E. Mann, B. R. C., M. S. K., and C. W. conceived the study. E. Mann and M. S. K. performed the experiments. E. Mann, E. Mallette, M. S. K., and C. W. analyzed and interpreted the data. E. Mann, M. S. K., and C. W. wrote the manuscript and B. R. C. provided input for the final draft.
Acknowledgment
We thank S. Labiuk at Canadian Light Source for collecting the selenomethionine dataset.
This work was supported by Canadian Institutes of Health Research Grant MOP 62877 (to C. W.) and Discovery Grants 327280 (to M. S. K.) and 2015-04622 (to C. W.) from the National Science and Engineering Research Council of Canada. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5HNO and 5HNP) have been deposited in the Protein Data Bank (http://wwpdb.org/).
O. G. Ovchinnikova, E. Mallette, A. Koizumi, T. L. Lowary, M. S. Kimber, and C. Whitfield, submitted for publication.
- ABC
- ATP-binding cassette
- OPS
- O-antigenic polysaccharide
- TMD
- transmembrane domain
- NBD
- nucleotide-binding domain
- Und-P
- undecaprenolphosphate
- GT
- glycosyltransferase
- CBM
- carbohydrate-binding module
- Kdo
- 3-deoxy-d-manno-oct-2-ulosonic acid
- SSMM
- selenomethionine-supplemented minimal medium
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- rbs
- ribosome-binding site
- r.m.s.d.
- root mean square deviation
- Selmet
- selenomethionine.
References
- 1. Cuthbertson L., Kos V., and Whitfield C. (2010) ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 74, 341–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Higgins C. F., and Linton K. J. (2004) The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11 918–926 [DOI] [PubMed] [Google Scholar]
- 3. Beis K. (2015) Structural basis for the mechanism of ABC transporters. Biochem. Soc. Trans. 43, 889–893 [DOI] [PubMed] [Google Scholar]
- 4. Polissi A., and Sperandeo P. (2014) The lipopolysaccharide export pathway in Escherichia coli: structure, organization and regulated assembly of the Lpt machinery. Mar. Drugs. 12, 1023–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simpson B. W., May J. M., Sherman D. J., Kahne D., and Ruiz N. (2015) Lipopolysaccharide transport to the cell surface: biosynthesis and extraction from the inner membrane. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. May J. M., Sherman D. J., Simpson B. W., Ruiz N., and Kahne D. (2015) Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whitfield C., and Trent M. S. (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 [DOI] [PubMed] [Google Scholar]
- 8. Kos V., Cuthbertson L., and Whitfield C. (2009) The Klebsiella pneumoniae O2a antigen defines a second mechanism for O-antigen ATP-binding cassette transporters. J. Biol. Chem. 284, 2947–2956 [DOI] [PubMed] [Google Scholar]
- 9. Clarke B. R., Cuthbertson L., and Whitfield C. (2004) Nonreducing terminal modifications determine the chain length of polymannose O-antigens of Escherichia coli and couple chain termination to polymer export via an ATP-binding cassette transporter. J. Biol. Chem. 279, 35709–35718 [DOI] [PubMed] [Google Scholar]
- 10. Clarke B. R., Richards M. R., Greenfield L. K., Hou D., Lowary T. L., and Whitfield C. (2011) In vitro reconstruction of the chain termination reaction in biosynthesis of the Escherichia coli O9a O-polysaccharide: the chain length regulator, WbdD, catalyzes the addition of methyl phosphate to the non-reducing terminus of the growing glycan. J. Biol. Chem. 286, 41391–41401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hagelueken G., Huang H., Clarke B. R., Lebl T., Whitfield C., and Naismith J. H. (2012) Structure of WbdD: a bifunctional kinase and methyltransferase that regulates the chain length of the O-antigen in Escherichia coli O9a. Mol. Microbiol. 86, 730–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clarke B. R., Greenfield L. K., Bouwman C., and Whitfield C. (2009) Coordination of polymerization, chain termination, and export in assembly of the Escherichia coli lipopolysaccharide O9a antigen in an ATP-binding cassette transporter-dependent pathway. J. Biol. Chem. 284, 30662–30672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. King J. D., Berry S., Clarke B. R., Morris R. J., and Whitfield C. (2014) Lipopolysaccharide O-antigen size distribution is determined by a chain extension complex of variable stoichiometry in Escherichia coli O9a. Proc. Natl. Acad. Sci. U.S.A. 111, 6407–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hagelueken G., Clarke B. R., Huang H., Tuukkanen A., Danciu I., Svergun D. I., Hussain R., Liu H., Whitfield C., and Naismith J. H. (2015) A coiled-coil domain acts as a molecular ruler to regulate O-antigen chain length in lipopolysaccharide. Nat. Struct. Mol. Biol. 22, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cuthbertson L., Powers J., and Whitfield C. (2005) The C-terminal domain of the nucleotide-binding domain protein Wzt determines substrate specificity in the ATP-binding cassette transporter for the lipopolysaccharide O-antigens in Escherichia coli serotypes O8 and O9a. J. Biol. Chem. 280, 30310–30319 [DOI] [PubMed] [Google Scholar]
- 16. Cuthbertson L., Kimber M. S., and Whitfield C. (2007) Substrate binding by a bacterial ABC transporter involved in polysaccharide export. Proc. Natl. Acad. Sci. U.S.A. 104, 19529–19534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reinikainen T., Ruohonen L., Nevanen T., Laaksonen L., Kraulis P., Jones T. A., Knowles J. K., and Teeri T. T. (1992) Investigation of the function of mutated cellulose-binding domains of Trichoderma reesei cellobiohydrolase I. Proteins 14, 475–482 [DOI] [PubMed] [Google Scholar]
- 18. Várnai A., Siika-Aho M., and Viikari L. (2013) Carbohydrate-binding modules (CBMs) revisited: reduced amount of water counterbalances the need for CBMs. Biotechnol. Biofuels. 6, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alderwick L. J., Lloyd G. S., Ghadbane H., May J. W., Bhatt A., Eggeling L., Fütterer K., and Besra G. S. (2011) The C-terminal domain of the arabinosyltransferase Mycobacterium tuberculosis EmbC is a lectin-like carbohydrate binding module. PLoS Pathog. 7, e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilbert H. J., Knox J. P., and Boraston A. B. (2013) Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 23, 669–677 [DOI] [PubMed] [Google Scholar]
- 21. Boraston A. B., Bolam D. N., Gilbert H. J., and Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murray G. L., Attridge S. R., and Morona R. (2006) Altering the length of the lipopolysaccharide O-antigen has an impact on the interaction of Salmonella enterica serovar typhimurium with macrophages and complement. J. Bacteriol. 188, 2735–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perez C., Gerber S., Boilevin J., Bucher M., Darbre T., Aebi M., Reymond J.-L., and Locher K. P. (2015) Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 524, 433–438 [DOI] [PubMed] [Google Scholar]
- 24. Vinogradov E., Frirdich E., MacLean L. L., Perry M. B., Petersen B. O., Duus J. Ø., and Whitfield C. (2002) Structures of lipopolysaccharides from Klebsiella pneumoniae. Elucidation of the structure of the linkage region between core and polysaccharide O chain and identification of the residues at the non-reducing termini of the O chains. J. Biol. Chem. 277, 25070–25081 [DOI] [PubMed] [Google Scholar]
- 25. Mertens K., Müller-Loennies S., Stengel P., Podschun R., Hansen D. S., and Mamat U. (2010) Antiserum against Raoultella terrigena ATCC 33257 identifies a large number of Raoultella and Klebsiella clinical isolates as serotype O12. Innate Immun. 16, 366–380 [DOI] [PubMed] [Google Scholar]
- 26. Mann E., Ovchinnikova O. G., King J. D., and Whitfield C. (2015) Bacteriophage-mediated glucosylation can modify lipopolysaccharide O-antigens synthesized by an ATP-binding cassette (ABC) transporter-dependent assembly mechanism. J. Biol. Chem. 290, 25561–25570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Phillips K., and de la Peña A. H. (2011) The combined use of the Thermofluor assay and ThermoQ analytical software for the determination of protein stability and buffer optimization as an aid in protein crystallization. Curr. Protoc. Mol. Biol. Chapter 10, Unit 10.28 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Adams P. D., Grosse-Kunstleve R. W., Hung L.-W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 30. Westphal O. (1965) Bacterial lipopolysaccharide-extraction with phenol water and further application of procedure. Methods Carbohydr. Chem. 1, 83–91 [Google Scholar]
- 31. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 32. Hitchcock P. J., and Brown T. M. (1983) Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsai C. M., and Frasch C. E. (1982) A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119, 115–119 [DOI] [PubMed] [Google Scholar]
- 34. Guzman L. M., Belin D., Carson M. J., and Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose P(BAD) promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drozdetskiy A., Cole C., Procter J., and Barton G. J. (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davidson A. L., and Sharma S. (1997) Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J. Bacteriol. 179, 5458–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tal N., Ovcharenko E., and Lewinson O. (2013) A single intact ATPase site of the ABC transporter BtuCD drives 5% transport activity yet supports full in vivo vitamin B12 utilization. Proc. Natl. Acad. Sci. U.S.A. 110, 5434–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Orelle C., Dalmas O., Gros P., Di Pietro A., and Jault J.-M. (2003) The conserved glutamate residue adjacent to the Walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J. Biol. Chem. 278, 47002–47008 [DOI] [PubMed] [Google Scholar]
- 40. Cuskin F., Flint J. E., Gloster T. M., Morland C., Baslé A., Henrissat B., Coutinho P. M., Strazzulli A., Solovyova A. S., Davies G. J., and Gilbert H. J. (2012) How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 20889–20894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Bueren A. L., Morland C., Gilbert H. J., and Boraston A. B. (2005) Family 6 carbohydrate binding modules recognize the non-reducing end of β-1,3-linked glucans by presenting a unique ligand binding surface. J. Biol. Chem. 280, 530–537 [DOI] [PubMed] [Google Scholar]
- 42. Henshaw J., Horne-Bitschy A., van Bueren A. L., Money V. A., Bolam D. N., Czjzek M., Ekborg N. A., Weiner R. M., Hutcheson S. W., Davies G. J., Boraston A. B., and Gilbert H. J. (2006) Family 6 carbohydrate binding modules in β-agarases display exquisite selectivity for the non-reducing termini of agarose chains. J. Biol. Chem. 281, 17099–17107 [DOI] [PubMed] [Google Scholar]
- 43. Boraston A. B., Creagh A. L., Alam M. M., Kormos J. M., Tomme P., Haynes C. A., Warren R. A., and Kilburn D. G. (2001) Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry 40, 6240–6247 [DOI] [PubMed] [Google Scholar]
- 44. Boos W., and Shuman H. (1998) Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62, 204–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., and Hunt J. F. (2002) ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Z., Xiang Q., Zhu X., Dong H., He C., Wang H., Zhang Y., Wang W., and Dong C. (2014) Structural and functional studies of conserved nucleotide-binding protein LptB in lipopolysaccharide transport. Biochem. Biophys. Res. Commun. 452, 443–449 [DOI] [PubMed] [Google Scholar]
- 47. Nsahlai C. J., and Silver R. P. (2003) Purification and characterization of KpsT, the ATP-binding component of the ABC-capsule exporter of Escherichia coli K1. FEMS Microbiol. Lett. 224, 113–118 [DOI] [PubMed] [Google Scholar]
- 48. Greenfield L. K., Richards M. R., Vinogradov E., Wakarchuk W. W., Lowary T. L., and Whitfield C. (2012) Domain organization of the polymerizing mannosyltransferases involved in synthesis of the Escherichia coli O8 and O9a lipopolysaccharide O-antigens. J. Biol. Chem. 287, 38135–38149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Willis L. M., Stupak J., Richards M. R., Lowary T. L., Li J., and Whitfield C. (2013) Conserved glycolipid termini in capsular polysaccharides synthesized by ATP-binding cassette transporter-dependent pathways in Gram-negative pathogens. Proc. Natl. Acad. Sci. U.S.A. 110, 7868–7873 [DOI] [PMC free article] [PubMed] [Google Scholar]