Abstract
Formyl peptide receptor 3 (Fpr3, also known as Fpr-rs1) is a G protein-coupled receptor expressed in subsets of sensory neurons of the mouse vomeronasal organ, an olfactory substructure essential for social recognition. Fpr3 has been implicated in the sensing of infection-associated olfactory cues, but its expression pattern and function are incompletely understood. To facilitate visualization of Fpr3-expressing cells, we generated and validated two new anti-Fpr3 antibodies enabling us to analyze acute Fpr3 protein expression. Fpr3 is not only expressed in murine vomeronasal sensory neurons but also in bone marrow cells, the primary source for immune cell renewal, and in mature neutrophils. Consistent with the notion that Fpr3 functions as a pathogen sensor, Fpr3 expression in the immune system is up-regulated after stimulation with a bacterial endotoxin (lipopolysaccharide). These results strongly support a dual role for Fpr3 in both vomeronasal sensory neurons and immune cells. We also identify a large panel of mouse strains with severely altered expression and function of Fpr3, thus establishing the existence of natural Fpr3 knock-out strains. We attribute distinct Fpr3 expression in these strains to the presence or absence of a 12-nucleotide in-frame deletion (Fpr3Δ424–435). In vitro calcium imaging and immunofluorescence analyses demonstrate that the lack of four amino acids leads to an unstable, truncated, and non-functional receptor protein. The genome of at least 19 strains encodes a non-functional Fpr3 variant, whereas at least 13 other strains express an intact receptor. These results provide a foundation for understanding the in vivo function of Fpr3.
Keywords: cellular immune response, chemotaxis, gene expression, genetic polymorphism, immunodeficiency, neuroimmunology, pattern recognition receptor (PRR), G protein-coupled receptor, olfaction, olfactory receptor (OR)
Introduction
Social animals benefit from sharing information that promotes fitness and survival (1–3). In rodents, there is extensive evidence that the detection of chemosensory cues emitted by other individuals is a fundamental feature of social recognition and a prerequisite for a wide range of social behaviors (4–6). For example, rodents use such chemical signals to distinguish between parasitized individuals, recognize infected conspecifics, and avoid and display aversive responses to infected individuals (7–9). Defining the cellular and molecular mechanisms and neural circuits underlying the detection of infection- and illness-associated states is not only crucial for understanding the sensing of health status in animals but also for advancing our knowledge of disease-related odors and diagnostic olfactory biomarkers in humans (10, 11).
The vomeronasal organ (VNO)4 is an olfactory substructure of the mammalian nose that is essential for social recognition and chemical communication (12). The VNO houses vomeronasal sensory neurons (VSNs), specialized nerve cells that detect a wide range of volatile and non-volatile molecular cues, including pheromones and predator odors (13–15). Among these cues are major histocompatibility complex class I binding peptides (antigens) and other small urinary peptides (16–18) thus providing initial support for an evolutionary link between recognition mechanisms in immune cells and subsets of VSNs (14, 19). More recently, clear evidence has emerged that the VNO plays an important role in mediating sick conspecific avoidance and thus seems to function as a sensor of infection (20), but the underlying cellular and molecular basis for this function remains unclear. VSNs are characterized by the expression of large families of G protein-coupled receptors, vomeronasal type 1 receptors (Vmn1r) expressed in the apical vomeronasal layer and vomeronasal type 2 receptor (Vmn2r) that function together with non-classical class I major histocompatibility complex receptors of the H2-Mv subtype in the basal layer (21, 22). An important development has been the identification of a third family of G protein-coupled receptors expressed in subsets of VSNs as follows: the formyl peptide receptors (Fprs) (23, 24). Fprs are primarily known from the innate immune system where they are involved in chemoattraction of phagocytic immune cells to sites of infection mediating host defense against invading microorganisms (25–28). Fprs detect a wide range of inflammation markers as well as bacterial and mitochondrial peptides (25–28), and consistent with the results from immune cells, some of these ligands have also been shown to activate specific VSNs (23, 29). Hence, members of the Fpr family are currently prime candidates for mediating VNO pathogen sensing, although it cannot be ruled out that some Vmn1r or Vmn2r receptors or even receptors expressed in other olfactory subsystems such as the main olfactory epithelium (30) could also contribute to this task.
The mouse genome encodes seven full-length Fpr genes (31). Two of these, Fpr1 and Fpr2, are expressed in leukocytes, and like their human orthologues FPR1 and FPR2, they function in innate immune defense (27, 28). The remaining five Fprs are expressed in non-overlapping subsets of VSNs as follows: Fpr-rs3, Fpr-rs4, Fpr-rs6, and Fpr-rs7 all coexpress with the G protein α-subunit Gαi2, whereas Fpr-rs1 (recently renamed to Fpr3, see Mouse Genome Informatics Database gene ID 1194495) coexpresses with Gαo (23, 24). Here, we focus on Fpr3 whose function has been quite unclear. We previously identified a set of formylated bacterial signal peptides as natural agonists for this receptor (32), and we demonstrated that human FPR3 responds to a similar ligand spectrum (32). These in vitro studies, together with the close sequence homology of both receptors (32, 33), indicate that human and mouse Fpr3 could share orthologous roles in pathogen detection. However, the known expression patterns of both receptors seem to contrast with this idea. Human FPR3 is found in immune cells (34), but no evidence for its expression in sensory neurons has been reported, possibly due to the fact that a functional VNO is missing in humans (4). Conversely, careful quantitative PCR and in situ hybridization studies clearly showed that mouse Fpr3 mRNA is present in VSNs, but both studies found no evidence for an expression outside the olfactory system (23, 24). So far, only a single report for the presence of low amounts of Fpr3 mRNA in Northern blots from murine leukocytes exists (31), but several other studies could not detect Fpr3 from blood samples (23), bone marrow (35), dendritic cells (36), or neutrophils (37), despite performing highly sensitive reverse transcriptase-polymerase chain reaction (RT-PCR) experiments. However, it is conceivable that Fpr3 protein is present in immune cells and that the failure to detect its expression can be attributed to low mRNA levels.
To address these questions, we generated two specific Fpr3 antibodies that enabled the direct detection of Fpr3 protein. We report the expression of Fpr3 not only in murine VSNs but also in bone marrow cells, the primary source for immune cell renewal, and in mature neutrophils. Importantly, we find that Fpr3 expression in the immune system can be up-regulated by stimulation with a bacterial endotoxin (lipopolysaccharide, LPS) that mimics bacterial infection. These findings strongly support a dual role for Fpr3 in both VSNs and immune cells. We also identify and characterize a natural Fpr3 gene variant that leads to a non-functional receptor. This variant is expressed in a wide range of mouse strains, thus identifying the existence of natural Fpr3 knock-out strains.
Experimental Procedures
Peptide-spot Assay for Antibody Characterization
Peptides (15 amino acid residues with an overlap of 10 residues) covering the whole length of Fpr3 were synthesized on acid-hardened cellulose membranes derivatized with a polyethylene glycol spacer. Membranes were equilibrated in 150 mm NaCl, 50 mm Tris/HCl (pH 7.5) for 30 min at room temperature. Each antibody was solved at 4 μg/ml in phosphate-buffered saline (PBS) with 5% milk powder, then added to the membrane, and incubated overnight at 4 °C. After washing with PBS, the membrane was incubated with the corresponding peroxidase-coupled secondary antibody overnight at 4 °C. Thereafter, the membrane was washed twice with PBS for 10 min, incubated with enhanced chemiluminescence solution, and analyzed using a Fusion SL (Peqlab) luminescence imaging system.
Antibody Generation
The polyclonal rabbit antibody Fpr3-ECL1 was generated by an epitope that was determined by epitope mapping of a commercially available antibody (sc-18195; M-20; Santa Cruz Biotechnology, Inc.) that weakly detected overexpressed Fpr3 in human embryonic kidney cells containing the simian vacuolating virus 40 T-antigen (HEK293T) at a concentration of 2 μg/ml. The mapping revealed two epitopes, AMKEKWPFGWFLCKL and MQFSGSYKIIGRLVN. Each peptide was synthesized and used to immunize a rabbit. Immunocytochemical tests on HEK293T cells expressing Fpr3 revealed that serum from the AMKEKWPFGWFLCKL-injected rabbit showed strong Fpr3 immunoreactivity, whereas serum from the MQFSGSYKIIGRLVN-injected rabbit did not. After 12 weeks, the animals were sacrificed; whole blood was collected, and Fpr3-ELC1 was purified by affinity chromatography with the sulfo-linked AMKEKWPFGWFLCKL peptide. Fpr3-ELC1 was adjusted to a stock concentration of 2 mg/ml.
The monoclonal mouse antibody Fpr3-ECL2 was developed in cooperation with the Abmart Co. Epitope scoring of the Fpr3 sequence was used to determine four peptide sequences (WGNSVEERLNTA, LSEDSGHISDTR, HSLSSRLQRALS, and ITTKIHKKAFV) spanning intracellular, extracellular, and C-terminal epitopes on Fpr3. For immunogen production, these epitopes were overexpressed in Escherichia coli and purified by nickel-affinity chromatography. Each immunogen was then injected into three female 8–12-week-old BALB/C mice. For hybridoma cell generation spleen cells from the best responding mice were fused to SP2/0 myeloma cells. Single parent cell colonies were obtained and tested by ELISA for their ability to bind the antigen. The nine most productive and stable clones were injected into the peritoneal cavity of mice. After 10–14 days ascites fluid was obtained and tested by immunocytochemistry experiments with Fpr3-expressing HEK293T cells. Antibodies produced by a hybridoma cell line against WGNSVEERLNTA were the most sensitive ones. They were used for all Fpr3-ECL2 immunochemistry experiments.
Cloning of Murine Fpr Genes
Fpr1, Fpr2, Fpr3 (previously known as Fpr-rs1), Fpr-rs3, Fpr-rs4, Fpr-rs6, and Fpr-rs7 were amplified from C57BL/6NJ mice by PCR and subcloned into pcDNA3.1(+) (Thermo Scientific) as described previously (33). Fpr3Δ424–435 is identical to Fpr3 except for a 12-nucleotide in-frame deletion occurring from base pair 424 to 435. Fpr3Δ424–435 was amplified and subcloned into pcDNA3.1(+)from genomic DNA of 129X1/Sv mice using the Phusion HF Master Mix (Thermo Scientific) according to the manufacturer's protocol with the primer and reaction conditions as described (33). The Fpr3Δ424–435 fusion construct was obtained by in-frame subcloning of an Fpr3 PCR product from genomic DNA of 129X1/Sv into a modified pcDNA5/FRT/TO vector containing an N-terminal rhodopsin epitope (Rho tag) and a C-terminal herpes simplex virus epitope (HSV tag). Complete coding sequences of all receptor genes were validated by sequencing.
Cell Culture and Transient Transfections
HEK293T PEAKrapid cells (ATCC) were cultivated in Dulbecco's modified Eagle's medium (DMEM, Sigma) supplemented with 10% (v/v) heat-inactivated fetal calf serum (FCS, Sigma), 100 units/ml penicillin (Sigma), 0.1 mg/ml streptomycin (Sigma), 2 mm l-glutamine (Sigma) until 80–90% confluency. For transfection, cells were seeded at 20–30% confluence on poly-d-lysine-coated (10 μg/ml in PBS) black 96-well μCLEAR-Plates (Greiner Bio-One). Cells were transfected 24 h later using jetPEI® (Polyplus-transfection SA) according to the manufacturer's protocol for 96-well transfections. For immunocytochemical analyses, 0.25 μg of plasmid DNA encoding a receptor were transfected. For calcium imaging experiments, 0.125 μg of DNA plasmid encoding a receptor were cotransfected with equal amounts of a plasmid encoding the G protein α-subunit Gα16.
Mouse Strains
For strain names, we used the nomenclature employed by the Mouse Genome Informatics database. C57Bl/6NCrl (MGI:2683688), FVB/NCrl (MGI:2165215), and BALB/cJ (MGI:2159737) mice were acquired from Charles River; 129X1/Sv (MGI:2164536) mice were purchased from The Jackson Laboratory, and NZB/Ola (MGI:2160555) mice were provided by Reinhart Kluge (Max Rubner Laboratorium, German Institute of Human Nutrition, Potsdam Rehbrücke, Germany). Ear stamps of Mus musculus musculus (Kazakhstan, Czech Republic), Mus musculus domesticus (Germany, France),Mus musculus castaneus (Thailand), and Mus musculus spretus (Spain) were kindly provided by Diethard Tautz (Max-Planck-Institut für Evolutionsbiologie, Plön, Germany).
Blood Cells
Blood samples were obtained from 8- to 12-week-old mice that were euthanized by CO2. Blood (10 μl) was deposited on a microscope slide (Superfrost Plus; Menzel-Gläser) and smeared immediately.
Bone Marrow Cells
Isolation was performed as described (38). 8–12- Week-old mice were euthanized with CO2 and decapitated. Femoral bones were isolated and stored in ice-cold calcium/magnesium-free Hanks' balanced salt solution buffer (Gibco) containing 10 mm HEPES for 10 min. The epiphysis was removed from both ends, and 1–2 ml of Hanks' balanced salt solution with HEPES was forced through the bone shaft with a syringe (20-gauge needle) to flush out the bone marrow. The resulting solution was counted with a MOXI Z cell counter (Orflo) using Type S Moxi Z Cassettes (Orflo) and centrifuged at 300 × g (brake on lowest level) for 8 min at 4 °C. Supernatant was discarded, and the cell pellet was resuspended in RPMI 1640 medium (Gibco) containing 100 units/ml penicillin, 0.1 mg/ml streptomycin, 5 mm l-glutamine, and 10% (v/v) FCS at a concentration of 4 × 106 cells/ml. Cells were seeded on Petri dishes (∼1 × 107 cells/dish) and incubated for 2 h at 37 °C and 5% CO2. Stimulation of bone marrow cells was achieved by adding 150 μg/ml lipopolysaccharide of Salmonella enteriditis (Sigma) into the culture medium of seeded cells. Stimulated and unstimulated samples were incubated for additional 8 h at 37 °C and 5% CO2 before their use in experiments.
Dissociation of Vomeronasal Tissue
VNO epithelium of 8–12-week-old mice was detached from the cartilage and minced in PBS at 4 °C (29, 39). The pooled tissue from three to five mice was incubated for 20 min at 37 °C in 1 ml of PBS supplemented with papain (0.22 units/ml; Worthington), 1.1 mm EDTA (Thermo Scientific), and 5.5 mm l-cysteine hydrochloride (Sigma). Subsequently, cells were kept on ice for 5 min in 1 ml of DNase buffer (600 μl of PBS with 400 μl of 5× Colorless GoTaq® reaction buffer (Promega) and 50 units of DNase I (Thermo Scientific)). Thereafter, the reaction was stopped by adding 10 ml of DMEM (Invitrogen) supplemented with 10% (v/v) FCS and centrifuged for 5 min at 1000 × g at 4 °C. After removal of the supernatant, cells were resuspended in 200 μl of DMEM supplemented with 10% (v/v) FCS and gently extruded by pipetting. The supernatant containing the dissociated cells was seeded on coverslips coated with concanavalin-A (0.5 mg/ml, overnight at 4 °C; Sigma) and incubated for 1 h at 37 °C and 5% CO2.
Immunocytochemistry
Cells were fixed for 4 min in 4% (m/v) methanol-free paraformaldehyde (Polyscience), rinsed in PBS, and treated with a blocking solution (PBS supplemented with 5% (v/v) FCS) and 0.25% (v/v) Triton X-100 for 30 min at room temperature. Thereafter, cells were incubated overnight at 4 °C with the primary antibody in blocking solution. After rinsing with PBS, staining was obtained by sample incubation for 60 min at room temperature with fluorescence-conjugated secondary antibodies in blocking solution containing Hoechst 33342 (1 μg/ml; Hoechst) to counterstain the cell nuclei. The same immunocytochemistry protocol was used for HEK293T cells, dissociated VNO cells, and leukocytes from blood and bone marrow.
Antibodies
The commercial Fpr3 antibodies M-20 (sc-18195; Santa Cruz Biotechnology, Inc., 0.2 mg/ml) and N-20 (orb100776; Biorbyt, 0.5 mg/ml) showed insufficient detection of Fpr3 in receptor-transfected HEK cells. We therefore generated two new antibodies. Both antibodies had a stock concentration of 2 mg/ml. All secondary antibodies had a stock concentration of 2 mg/ml. Working concentrations are given in parentheses. For Fpr3 detection, an affinity-purified polyclonal rabbit anti-Fpr3 IgG (Fpr3-ECL1; 2 μg/ml) and a monoclonal mouse anti-Fpr3 IgG (Fpr3-ECL2; 0.2 μg/ml) were used. Fpr3-ECL1 was stained with a polyclonal goat anti-rabbit Alexa Fluor 488 IgG (2 μg/ml; A-11034; Invitrogen). In HEK293T cells, Fpr3-ECL2 was stained with a polyclonal goat anti-mouse Alexa Fluor 546 IgG (0.5 μg/ml; A-11003; Invitrogen). For VNO and blood samples, we used either a polyclonal goat anti-mouse Alexa Fluor 488 IgG (2 μg/ml; A-11029; Molecular Probes) or a polyclonal donkey anti-mouse Alexa 647 IgG (2 μg/ml; A-31571; Invitrogen). OMP detection was obtained using a combination of a polyclonal goat anti-OMP IgG (2 μg/ml; 544–10001; Wako Chemicals USA, Inc.) and a polyclonal donkey anti-goat Alexa Fluor 555 IgG (2 μg/ml; A-21432; Invitrogen). For Gαo a polyclonal rabbit anti-Gαo IgG (0.2 μg/ml; sc-387; Santa Cruz Biotechnology, Inc.), for PDE4A a polyclonal rabbit anti-PDE4A IgG (0.5 μg/ml; PD4-112AP; FabGennix), and for V2R2 (vomeronasal type 2 receptor 2) a polyclonal rabbit anti-V2R2 IgG (1:10,000; provided by R. Tirindelli, University of Parma, Parma, Italy) were applied as primary antibodies. Staining for all three markers was done with a polyclonal donkey anti-rabbit Alexa Fluor 555 IgG (2 μg/ml; A-31572; Invitrogen). CD45R (cluster of differentiation molecule 45R) was detected by a polyclonal FITC rat anti-mouse CD45R/B220 IgG (0.25 μg/ml; clone RA3-6B; 2553087; BD Pharmingen) and stained with a polyclonal donkey anti-rat Alexa Fluor 488 IgG (2 μg/ml; A-21208; Invitrogen). For Ly6G (lymphocyte antigen 6G) a monoclonal rat anti-mouse Ly6G IgG (0.25 μg/ml; clone 1A8; 127601; BioLegend) was used as primary antibody. For colocalization experiments with Fpr3-ECL1, staining was obtained by a polyclonal donkey anti-rat CF633 IgG (2 μg/ml; 20137; Biotium), and for colocalization with Fpr3-ECL2, a polyclonal donkey anti-rat Alexa Fluor 488 IgG (2 μg/ml; A-21208; Invitrogen) was applied for staining. The Rho and HSV tags were detected with a monoclonal anti-rhodopsin mouse IgG of anti-4D2 (1:500; R. Molday, Centre for Macular Research, University of British Columbia, Canada) and a monoclonal mouse anti-HSV IgG (0.1 μg/ml; 69171; Novagen), respectively. Both antibodies were stained with a polyclonal goat anti-mouse Alexa Fluor 546 IgG (0.25 μg/ml; A-11003; Invitrogen).
Antibody Specificity
Several independent lines of evidence indicate high specificity of the newly developed Fpr3-ECL1 and Fpr3-ECL2 antibodies. First, epitope mapping of both antibodies shows that the linear motif of Fpr3-ECL1 is only present in 17 mouse proteins, that the Fpr3-ECL2 motif is present in 119 mouse proteins (supplemental Table S1, A and B), and that both antibodies can only recognize their motif in a specific conformation (Fig. 1, A and B). Second, both antibodies do not recognize any other mouse Fpr (Fig. 2, A and B). Third, we performed blocking peptide controls that are well suited to monitor unspecific binding such as binding via the Fc-region (Fc-region is the fragment crystallizable domain of immunoglobulins) of the antibodies. In these reactions, ECL1 or ECL2 antibodies were pre-incubated for 1 h at room temperature with 10 μg/ml of the peptide that was used for antibody generation. Thus, the specific binding sites of the antibodies are blocked. Subsequently, all steps of the normal immunostaining protocol were performed. Control reactions showed no staining beyond background level (Figs. 3B and 4C). Fourth, both antibodies were found to label the same cells, although they were independently generated to detect different domains of Fpr3 (Fig. 4D). Fifth, the molecular characteristics of VSNs identified by our Fpr3 antibodies are fully consistent with those VSNs previously reported to express Fpr3 mRNA. Sixth, the staining of both Fpr3 antibodies is diminished in mouse lines that express a non-functional Fpr3 variant (Figs. 5, A and B and 6).
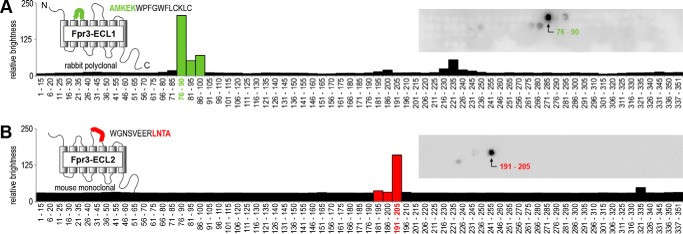
FIGURE 1.
Generation of two novel specific Fpr3 antibodies. Peptide spot array analyses of the polyclonal rabbit antibody Fpr3-ECL1 (A, green) and the monoclonal mouse antibody Fpr3-ECL2 (B, red) are shown. Models indicate the positions and sequences of the immunization peptides used for antibody generation. The amino acid motifs that are recognized by the antibodies are indicated in the respective colors. Both antibodies were tested against peptides comprising the complete Fpr3 sequence. Each spot consists of a 15-amino acid peptide overlapping by 10 residues with its predecessor. Insets show original arrays visualized by enhanced chemiluminescence. The charts show the quantification of staining intensities; bars containing the AMKEK or LNTA motif are highlighted in green or red, respectively. Numbers denote the peptide positions in the receptor protein.
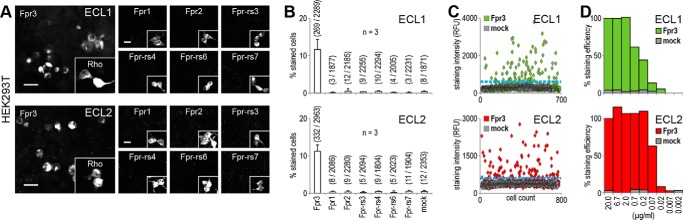
FIGURE 2.
Both antibodies detect Fpr3 and do not recognize other Fpr family members. A, immunostaining of HEK293T cells to test the cross-reactivity of Fpr3-ECL1 (upper panel) and Fpr3-ECL2 (lower panel) for the indicated receptors. Insets show stainings with an anti-rhodopsin-epitope (Rho) antibody as a positive control for expression of the indicated receptors. Scale bars, 20 μm. B, average percentage of stained cells from three independent experiments. Numbers in parentheses denote positive versus total cells. Error bars, S.D. C, representative scatter plots of staining intensities for both antibodies in relative fluorescence units (RFU). Each dot represents an individual cell. Threshold (blue line) was set to maximal intensity level observed for mock-transfected cells. D, antibody dilution experiment examining the sensitivity of both antibodies tested in decreasing concentrations. Signals were normalized to 1:100 dilution. Immunoreactivity was compared with mock-transfected cells. Error bars, S.D.
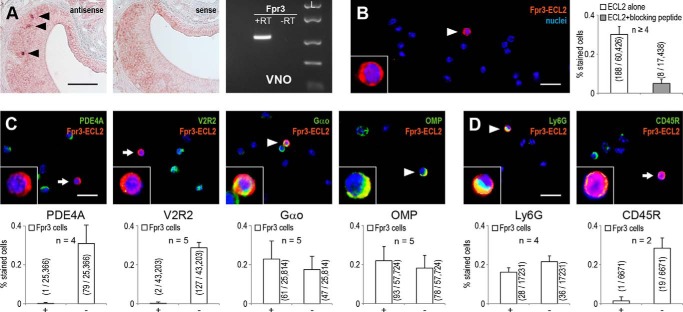
FIGURE 3.
Fpr3 is expressed in vomeronasal sensory neurons and immune cells. A, left, in situ hybridization with Fpr3 sense and antisense probes on coronal slices of the vomeronasal organ. Black triangles mark Fpr3-positive cells. Scale bar, 100 μm. Right, RT-PCR analysis of Fpr3 expression in the vomeronasal organ. A band of the correct size and sequence was observed in VNO cDNA (+RT) but not in the negative control without reverse transcriptase (−RT). Similar results were obtained in five independent experiments. Size marker (L) FastRuler Middle Range DNA Ladder. B, left, representative immunostaining with Fpr3-ECL2 on dissociated vomeronasal cells of C57Bl/6NCrl mice. The white triangle marks a Fpr3-positive cell (red). Right, quantification of Fpr3 immunoreactivity. Average frequency of Fpr3 expression was analyzed in a total of 60,426 cells from six independent experiments. Antibody specificity was demonstrated by blocking the specific binding site through preincubation with 10 μg/ml of the peptide used for antibody generation. C, colocalization of Fpr3 (red) with different cellular markers (green) on dissociated vomeronasal cells from C57Bl/6NCrl mice using phosphodiesterase 4A (PDE4A) as a marker for the apical zone of the VNO, vomeronasal type 2 receptor 2 (V2R2) as a marker for Vmn2r-expressing cells, G protein subunit Gαo as a marker for the basal zone, and OMP as a marker for mature VSNs. D, colocalization of Fpr3 (red) with immune cell markers (green) on dissociated cells. Fpr3 staining colocalized with Ly6G (lymphocyte antigen 6G), a neutrophil marker. No colocalization was seen for CD45R (cluster of differentiation molecule 45R) that is expressed by most immune cells but absent in neutrophils. In all colocalization experiments, the colocalizing cells are marked with a white triangle and non-colocalizing with a white arrow. All cells were counterstained with the nuclear dye Hoechst33342 (blue). Insets show Fpr3-positive cells in a ×3 magnification. Scale bars, 20 μm. The bar chart below each picture denotes the quantification from two to five experiments, analyzing the colocalizations in a total of 7000 to 57,000 cells. Precise numbers are given in the graphs. Fpr3-positive cells that coexpress the marker are labeled by +, and cells not coexpressing the marker are labeled with −. Values in parentheses denote positive versus total cells. Error bars, S.D.
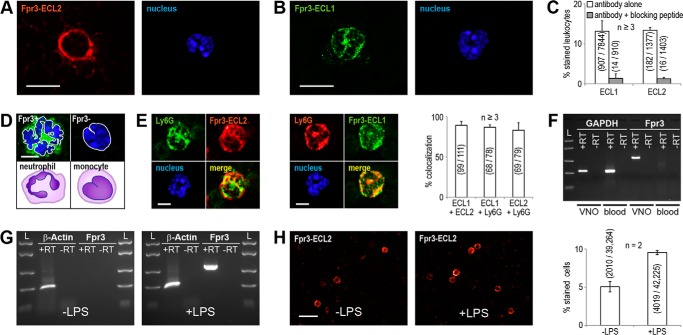
FIGURE 4.
Fpr3 expression in neutrophils is enhanced by LPS stimulation. Representative immunostainings of blood cells from C57Bl/6NCrl mice with the monoclonal mouse antibody Fpr3-ECL2 (A) or the polyclonal rabbit antibody Fpr3-ECL1 (B). For nuclear staining Hoechst33342 was used (right). Scale bars, 10 μm. C, quantification and specificity of Fpr3-ECL2 and Fpr3-ECL1 immunoreactivity in leukocytes. Antibody specificity was demonstrated by blocking the specific binding site through preincubation with 10 μg/ml of the peptide used for antibody generation. D, comparison of nuclear morphologies for Fpr3-positive and -negative leukocytes. Fpr3 was stained with Fpr3-ECL1 (green), and for nuclear staining Hoechst33342 (blue) was used. Fpr3-positive cell (upper left) showing a clearly multiple lobed nucleus, typical for neutrophil granulocytes (lower left). Fpr3-negative cell (upper right) showing a horseshoe-shaped nucleus, typical for monocytes (lower right). Scale bar, 5 μm. E, colocalization between the monoclonal mouse antibody Fpr3-ECL2 (left) and the polyclonal rabbit antibody Fpr3-ECL1 (middle) with the neutrophil marker Ly6G (lymphocyte antigen 6G). Cell nuclei are shown in blue. Scale bar, 5 μm. Right, colocalization between Fpr3 and Ly6G immunoreactivity in leukocytes. Bars show average percentage of colocalizing cells from at least three independent experiments. Numbers in parentheses denote positive versus total cells. Error bars, S.D. F, RT-PCR analysis of Fpr3 expression in mouse blood. Fpr3 expression was only detected in the positive control reaction from vomeronasal cDNA. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh, reverse transcription control) could be amplified from vomeronasal and blood cDNA. Similar results were obtained in three independent experiments. Size marker (L) FastRuler Middle Range DNA Ladder. G, RT-PCR analysis of Fpr3 expression in bone marrow. Fpr3 could not be detected from unstimulated mouse bone marrow cDNA (left). Fpr3 expression was observed upon stimulation with 150 μg/ml lipopolysaccharide (LPS) from S. enteriditis (right). Actb (actin, beta) could be amplified from all cDNAs. Similar results were obtained in two independent experiments. H, representative immunostainings for Fpr3 in unstimulated (left) and LPS-stimulated (middle) bone marrow cells from C57Bl/6NCrl mice and their quantification (right). Scale bar, 20 μm. Bar chart shows mean increase of Fpr3-expressing cells upon LPS stimulation from two independent experiments carried out as triplicate. Numbers in parentheses denote positive versus total cells. Error bars, S.D.
FIGURE 5.
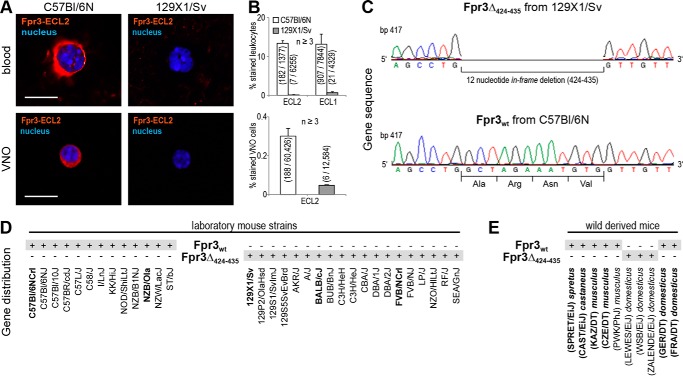
Strain-specific loss of Fpr3 expression in mice. A, representative immunostainings of Fpr3 expression in leukocytes and vomeronasal cells of C57Bl/6NCrl mice (left) versus 129X1/Sv mice (right). Fpr3 is only detectable in cells from C57Bl/6NCrl mice. Scale bars, 10 μm. B, quantifications of Fpr3 expression in leukocytes (upper panel) and vomeronasal cells (lower panel). Bar charts show average percentage of stained cells from at least three independent experiments. Numbers in parentheses denote positive versus total cells. Error bars, S.D. C, genotyping of Fpr3 in 129/Sv and C57Bl/6NCrl mice revealed two receptor variants Fpr3wt and Fpr3Δ424–435. A 12 nucleotide in-frame deletion from base pair 424 to 435 was observed in 129X1/Sv mice resulting in a loss of an alanine, arginine, asparagine, and valine at the end of the second intracellular loop. Identical results were obtained from three individuals of each strain. D, distribution of both Fpr3 gene variants in different laboratory mouse strains. Gray shading denotes the genomically encoded variants. Bold letters annotate in house sequenced strains. All other data were obtained from the Mouse Genomes Project. E, distribution of both Fpr3 gene variants in wild type-derived mouse strains from different subspecies and geographical origins. M. musculus castaneus were from Thailand (CAST/EiJ). M. musculus domesticus were from Germany (GER/DT), France (FRA/DT), the United States of America (LEWES/EiJ, WSB/EiJ), and Switzerland (ZALENDE/EiJ). M. musculus musculus were from Kazakhstan (KAZ/DT) and Czech Republic (CZE/DT, PWK/PhJ), M. musculus spretus were from Spain (SPRET/EiJ).
FIGURE 6.
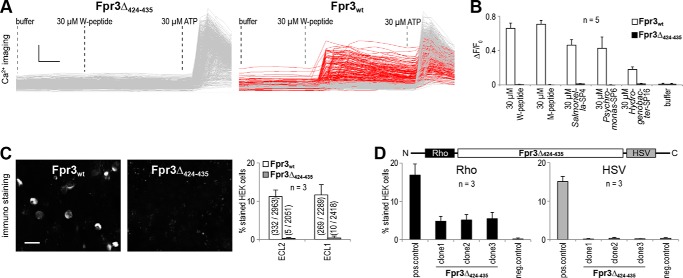
Variant-specific loss of Fpr3 function. A, single-cell calcium responses obtained in HEK293T cells transfected with Fpr3Δ424–435 or Fpr3wt and stimulated with W-peptide. Each trace represents an individual cell. Left, Fpr3Δ424–435-transfected cells. None of 686 cells responded to 30 μm W-peptide. Right, Fpr3wt-transfected cells. 177 of 475 cells (red) responded to 30 μm W-peptide. Buffer was used to exclude mechanical activation; 30 μm ATP that activates endogenous receptors was used as a positive control for cell viability. Scale bars, vertical 0.5 340 nm/380 nm and horizontal 10 s. B, mean calcium responses to various agonists of HEK293T cells transfected with Fpr3wt or Fpr3Δ424–435. Buffer was used as a negative control. Bars denote average percentage of responding cells from five independent experiments, measured in duplicates. Error bars, S.D. C, immunostainings of HEK293T cells expressing Fpr3wt (left) or Fpr3Δ424–435 (middle) and their quantifications (right). Only Fpr3wt was detectable. Bar chart shows average percentage of stained cells from three independent experiments. Numbers in parentheses denote positive versus total cells. Error bars, S.D. Scale bars, 20 μm. D, immunostainings of HEK293T cells transfected with an independent plasmid copy of Rho-Fpr3Δ424–435-HSV designed to produce a fusion protein of Fpr3Δ424–435 containing an N-terminal rhodopsin-epitope (Rho) and a C-terminal herpes simplex virus-epitope (HSV). Left, quantification of stainings with an anti-Rho antibody. Right, quantification of stainings with an anti-HSV antibody. Bars show average percentage of stained cells from three independent experiments. Two plasmids were transfected as controls as follows: Rho-Fpr3wt, to produce a fusion protein of Fpr3wt with an N-terminal Rho-epitope; and hT2R16-HSV, to produce a fusion protein of hT2R16 with a C-terminal HSV epitope. Error bars, S.D.
In Situ Hybridization
Fpr3 sense and antisense probes (nucleotide 672–1056) from National Center for Biotechnology Information database, accession number NM_008042.2 plus 153 bp of the 3′-untranslated region were synthesized from a full-length Fpr3 DNA template using a digoxigenin-labeling procedure during in vitro transcription. Tissue was obtained from anesthetized mice that were perfused with ice-cold 4% (m/v) paraformaldehyde (pH 7.4). The VNO was dissected and incubated overnight in 4% (m/v) paraformaldehyde at 4 °C. Subsequently, it was incubated in increasing sucrose gradients (m/v) of 10% (2 h), 20% (2 h), and 30% (overnight) at 4 °C. Next, the VNO was embedded in Tissue-Tek O.C.T. (Sakura Finetek), frozen in 2-methylbutane that was cooled by dry ice, and stored at −80 °C. For in situ hybridization, 12-μm thick slices were cut and gathered on microscope slides (Superfrost Plus, Menzel-Gläser). In situ hybridization was performed according to a published protocol (40), except that the treatments with H2O2 and proteinase K and the drying of the slices in increasing ethanol concentrations were omitted. For detection of the digoxigenin-labeled probes, we used an anti-digoxigenin antibody (Roche Applied Science, 0.15 units/ml). For detection of hybridized RNA, slices were incubated with detection buffer (175 μg/ml 5-bromo-4-chloro-3-indolyl phosphate, 300 μg/ml nitro blue tetrazolium, 1 mm levamisole in alkaline phosphatase buffer) at room temperature. Staining reaction was stopped at 4 °C, and slices were mounted with fluorescence mounting medium (DAKO).
Image Acquisition and Data Analysis
All representative images from blood and vomeronasal cells were taken with an Olympus BX61 fluorescence microscope with an X-Cite® series 120PC (EXFO) light source. Representative pictures of HEK293T cells and montage pictures for quantifications were taken with the BD Pathway 855 bioimaging system (BD Biosciences). Quantifications were evaluated with the BD-image Explorer software (BD Biosciences).
RT-PCR Analysis
RNA from VNO and bone marrow of mice was obtained with the innuPREP RNA mini kit (Analytik Jena AG) according to the manufacturer's protocol. Mouse whole blood RNA was purchased from Zyagen. Quality was assessed by gel electrophoresis and photometric measurements. cDNA was synthesized from 0.5 μg of total RNA using Smart cDNA Synthesis protocol (Clontech) and Superscript II Reverse Transcriptase (Invitrogen). PCR was performed with Phusion High Fidelity DNA polymerase (New England Biolabs Inc.) according to the manufacturer's protocol. Conditions for all primers were 98 °C for 15 s followed by 35 cycles (denaturation, 98 °C for 10 s; annealing, 64 °C for 10 s; elongation, 72 °C for 40 s) and 72 °C for 20 s. Primer sequences for Fpr3 and Fpr3Δ424–435 were GATCAGATGTGGTGATCTATGATTCTAC and AGGAAGTGAAGCCAAATTGGT. GAPDH was amplified with ACCACAGTCCATGCCATCAC and TCCCACCACCCTGTTGCTGTA. For Actb (actin, beta), we used CTGGAACGGTGAAGGTGACA and AAGGGACTTCCTGTAACAATGCA.
Calcium Imaging
Cell population responses of transfected HEK293T cells (∼50,000 cells per well) were recorded using a fluorometric imaging plate reader system (Molecular Devices) as described previously (32, 33, 41). Briefly, 48 h after transfection, cells were incubated with 2 μm Fluo-4 AM (Molecular Probes) and 50 μm probenicid (Sigma) for 2 h at room temperature in a C1 bath solution (130 mm NaCl, 10 mm HEPES, 5 mm KCl, 2 mm CaCl2, 5 mm glucose (pH 7.4)). Before each experiment cells were rinsed three times with C1. Response amplitudes (ΔF/F0) were calculated by dividing the maximal change in fluorescence after ligand application by baseline fluorescence. All experiments were performed in duplicate wells using at least three independent transfections. For automated single cell calcium imaging, we used the BD Pathway 855 bioimaging system (BD Biosciences). Cells were loaded with Fura-2/AM (Molecular Probes) using trypan red plus (1/20; AAT Bioquest). They were cotransfected with a receptor and Gα16. Calcium-dependent fluorescence signals were recorded at 0.5 Hz. 30 μm ATP served as a positive control to monitor cell viability. Cells that responded to C1 buffer or did not respond to ATP were excluded from the analysis. Images were taken with the Bioimaging system and quantified using Attovision software (BD Biosciences).
Ligands
Purity of all ligands was >95%. W-peptide was obtained from Innovagen. D- and L-M-peptide were purchased from GenScript. Salmonella-SP4 and Psychromonas-SP6 were acquired from VCPBIO. Hydrogenobacter-SP16 was purchased from United Biosystems. All ligands were dissolved in the assay buffer solution C1.
Results
Generation and Validation of Two Novel Fpr3 Antibodies
To gain new insight into the function of Fpr3, we first used immunofluorescence analysis. In initial experiments, we tested two commercially available antibodies for their capability to detect murine Fpr3 in transfected HEK cells but found them to produce insufficient results. We therefore generated two novel antibodies binding to non-overlapping sites on Fpr3 and carefully characterized their epitopes (Fig. 1) and cross-reactivity to other Fprs (Fig. 2). We focused on the detection of the extracellular loops of Fpr3 because these possess the highest sequence divergence among different members of the Fpr family. By peptide immunization of rabbits with AMKEKWPFGWFLCKL, we obtained the polyclonal antibody ECL1 that is directed against the first extracellular loop of Fpr3. Furthermore, we used a mouse hybridoma cell line to produce the monoclonal antibody ECL2 that is directed against the WGNSVEERLNTA sequence present in the second extracellular loop of Fpr3.
Antibodies sometimes recognize more than one domain within a protein. Multiple recognition sites and unfavorable epitopes can increase the chance for unspecific binding or cross-reactivity. We used peptide spot array analysis to precisely determine the recognition sites of both antibodies (Fig. 1). We generated a set of 69 peptides that covered all 351 amino acid residues of Fpr3 (supplemental Table S1A) and spotted them on a membrane. Each peptide had a length of 15 amino acids and overlapped by 10 residues with the preceding peptide. Antibodies were tested for their reactivity at 4 μg/ml dilution on the synthesized peptides. Both antibodies showed strong immunoreactivity to specific sequences that were used for their generation (Fig. 1, A and B; indicated by the green and red bars, respectively). There was very little cross-reactivity to a few peptides from other domains of the receptor (Fig. 1, A and B). Secondary antibody controls showed no interaction with any domain. Comparison between signal intensities and peptide sequences revealed the key binding residues of ECL1 and ECL2 that are contained in AMKEK and LNTA, respectively (Fig. 1, A and B). Interestingly, both motifs are also contained in two neighboring peptides to which the antibodies showed no strong immunoreactivity. This suggests that both antibodies recognize only a specific conformation of their respective peptide epitopes. Database analysis of ∼20,000 mouse genes (42) revealed the presence of the AMKEK and LNTA motifs in 0.08 and 0.6% of the mouse genes, respectively (supplemental Table S1, B and C). Thus, both motifs are reasonably specific for Fpr3. Moreover, they are not found in most murine Fprs except for Fpr2, which has a sequence identity of 88.5% to Fpr3.
To test whether these antibodies cross-react with Fpr2 or any other murine Fpr, we next used the antibodies on HEK cells that were transiently transfected with either Fpr3 or another member of the murine Fpr family (Fig. 2, A–C). Both antibodies recognized Fpr3-expressing cells in a nearly identical manner. ECL1 stained 11.6 ± 2.7% and ECL2 stained 11.2 ± 1.7% of the Fpr3-transfected cells. All Fprs are well expressed in HEK293T cells (33), and we did not observe any specific reactivity to Fpr2 or any other human or murine Fpr. Finally, we determined the sensitivity of both antibodies by testing them in serial dilutions on Fpr3-expressing cells (Fig. 2D). The polyclonal ECL1 showed no loss in reactivity up to a 1:1000 dilution; the monoclonal ECL2 was even more sensitive and showed no activity loss until 1:10,000. These results demonstrate high specificity of the newly developed Fpr3 antibodies.
Fpr3 Protein Is Expressed in VSNs and in Immune Cells
Reports on Fpr3 expression patterns in immune cells are inconsistent (31, 35–37), but two independent studies (23, 24) raised clear evidence for Fpr3 expression in a small subpopulation (<1%) of VSNs by quantitative PCR and in situ hybridization. However, direct detection of the Fpr3 protein is still missing. Because the expression pattern of a given gene can vary in different mouse strains, we first used in situ hybridization and RT-PCR to confirm the expression of Fpr3 mRNA in a small subset of VSNs from C57Bl/6NCrl mice (Fig. 3A). Next, we examined the expression of Fpr3 protein in individual, dissociated VNO cells. For these experiments, we used the mouse monoclonal ECL2 antibody because of its superior sensitivity over the polyclonal ECL1 antibody. Quantitative analysis of 60,426 cells from seven independent experiments revealed Fpr3 labeling in 188 cells, which corresponds to an average expression rate of 0.3%. Specificity of the staining was established through the blocking peptide control (Fig. 3B). Fpr3 mRNA has been found in sensory neurons of the basal VNO layer that also express the G protein α-subunit Gαo (24) but are negative for type 2 vomeronasal receptors (Vmn2r) and markers of apical VSNs such as PDE4A (24). The basic expression pattern for Fpr3 protein in our experiments was consistent with these results as there was virtually no colocalization between Fpr3 and PDE4A or V2R2, a marker for Vmn2r-positive VSNs (43) (Fig. 3C).
With respect to Gαo expression, we only observed colabeling of Fpr3 with Gαo in 61/108 cells (56%) (Fig. 3C). When we tested an antibody against OMP, which is present in all mature VSNs (44), only 54% of the Fpr3-positive cells also expressed this protein (Fig. 3C). Thus, nearly half of the cells positive for Fpr3 in this preparation do not express specific markers of mature VSNs, indicating that they could include non-olfactory cell types. As our cell preparation was likely to contain trace contaminations from white blood cells, we hypothesized that the Fpr3 antibody detected some leukocytes. To test this, we examined two immune cell markers for their coexpression with Fpr3 (Fig. 3D). We used the lineage-specific R-isoform of the cluster of differentiation molecule (CD45R), a plasma membrane phosphatase, which is expressed in most leukocytes but absent in neutrophils, and the lymphocyte antigen 6G (Ly6G), a marker for neutrophils (45). We did not observe CD45R expression in Fpr3-positive cells, but we detected coexpression of Ly6G and Fpr3 in 28 of 64 (44%) analyzed cells. Hence, these results provide evidence for the expression of Fpr3 protein in neurons of the VNO as well as in specific subsets of immune cells.
Fpr3 Expression in Neutrophils Is Enhanced by LPS Stimulation
To characterize Fpr3-positive immune cells in greater detail, we performed combined immunocytochemical and RT-PCR analyses in mouse blood cells (Fig. 4). We first used the new antibodies to directly test leukocytes from murine blood smears for Fpr3 expression. We observed a staining in 182/1377 leukocytes with ECL2 (Fig. 4A) and in 907/7844 leukocytes with ECL1 (Fig. 4B). The mean percentages of leukocyte stainings for ECL2 and ECL1 were 13.4 ± 0.6 and 13.2 ± 2.6%, respectively (Fig. 4C). Furthermore, the signals of both antibodies colocalized to 89.0 ± 4.8% (Fig. 4E). Both stainings could be abolished by blocking of the antibody-binding sites with the peptides that were used for antibody generation (Fig. 4C). The number of Fpr3-expressing leukocytes corresponded well to the typical range of neutrophils in mouse blood that is usually between 9 to 18% (46). Moreover, Fpr3-positive leukocytes showed a multi-lobed nucleus, typical for mature neutrophils (Fig. 4D). Colocalization experiments between Fpr3 and the neutrophil marker Ly6G revealed colabeling of 86.7 ± 3.1% and 83.1 ± 9.3% for ECL1 and ECL2, respectively (Fig. 4E). These results unambiguously demonstrate the presence of Fpr3 protein in mouse neutrophils.
Next, we performed RT-PCR to investigate Fpr3 mRNA expression in blood cells. Surprisingly, we could not detect Fpr3 in blood RNA, although we readily obtained PCR products in control reactions such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a control for cDNA quality or amplification of Fpr3 from vomeronasal cDNA as a control for correct reaction conditions (Fig. 4F). The RNA amount in mature neutrophils is relatively low (47) and in blood, only mature neutrophils circulate whereas their maturation and proliferation takes place in bone marrow. Hence, it is possible that the mRNA levels for Fpr3 are much higher in maturing neutrophils. Moreover, it is well known that mRNA levels of specific genes in leukocytes rise upon stimulation with bacterial stimuli (48–50). To further increase RNA levels, we stimulated bone marrow cells with lipopolysaccharide (LPS) from S. enteriditis prior to mRNA isolation and examined both preparations for Fpr3 expression. We did not observe Fpr3 in unstimulated bone marrow cells but readily detected its expression in all LPS-stimulated samples (Fig. 4G). Thus, Fpr3 mRNA levels in unstimulated cells are relatively low and LPS stimulation induces an increase in Fpr3 expression, which would explain why most previous RT-PCR studies failed to detect it. The rise in Fpr3 RNA should correspond with an increased level of the Fpr3 protein. Indeed, we found that the number of Fpr3-positive cells almost doubled, from 5.1 ± 0.7 to 9.5 ± 0.3%, after stimulation with LPS (Fig. 4H). Thus, expression of Fpr3 protein in mouse neutrophils can be induced by a powerful bacterial virulence factor with proinflammatory properties.
Fpr3 Protein Expression Occurs in a Strain-specific Manner
Our experiments described thus far were performed in C57Bl/6NCrl mice. Two previous studies (31, 51) reported divergent sequences for Fpr3 between BALB/c and 129/S6 mice, with a main difference of four amino acids missing in the fourth transmembrane region. We hypothesized that similar variations could also occur in other mouse strains, possibly leading to strain-specific variations in the Fpr3 expression pattern.
To test this, we performed a systematic expression and genotype analysis of Fpr3 in five mouse strains and combined these results with sequence data from 32 laboratory and nine wild type-derived strains (Fig. 5 and Table 1). First, we analyzed 129X1/Sv mice. Remarkably, this strain showed no Fpr3 expression in leukocytes and VNO cells, a result that was highly reproducible in multiple animals (n = 9) using both Fpr3 antibodies (Fig. 5, A and B). We next performed additional immunostainings in leukocytes from C57Bl/6NCrl, 129X1/Sv, BALB/cJ, FVB/N, and NZB/Ola mice. We observed Fpr3 protein expression in NZB/Ola and C57Bl/6NCrl mice. By contrast, BALB/cJ, FVB/N, and 129X1/Sv mice showed no Fpr3 expression despite the fact that we could produce Fpr3 mRNA from VNO cDNA in all negatively tested strains. Possibly, variations in the Fpr3 gene alter the Fpr3 protein structure and thus prevent its detection by the antibodies. We therefore amplified and sequenced Fpr3 from genomic DNA of C57Bl/6NCrl and 129X1/Sv mice and compared the results. This analysis indeed revealed two distinct Fpr3 variants (Fig. 5C) as follows: the Fpr3 sequence from C57Bl/6NCrl mice perfectly matched the annotated NCBI reference sequence NM_008042.2, whereas the Fpr3 sequence from 129X1/Sv mice showed a 12-nucleotide in-frame deletion. This deletion includes the nucleotides 424–435 of the coding region and results in the loss of alanine 142, arginine 143, asparagine 144, and valine 145 but leaves the open reading frame intact. We named the version carrying the 12-nucleotide deletion Fpr3Δ424–435 and the full-length version Fpr3wt. Next, we examined Fpr3 in BALB/cJ, FVB/N, and NZB/Ola and found a clear correlation between specific gene variants and the presence or absence of Fpr3 antibody staining. Fpr3Δ424–435 is present in the genome of 129X1/Sv, BALB/cJ, and FVB/N mice that have no detectable Fpr3 protein, whereas C57Bl/6NCrl and NZB/Ola, in which we could readily detect Fpr3 protein, carry the Fpr3wt gene. To obtain a more comprehensive view of the distribution of both Fpr3 variants in different inbred strains, we analyzed the Mouse Genomes Project database and found Fpr3wt present in C57, C58, I, KK NOD, NZB, NZW, and ST mice, whereas 129S, 129P, AKR, A, BALB, BUB, C3H, CBA, DBA, FVB, LP, NZO, RF, and SEA all carry the Fpr3Δ424–435 variant (Fig. 5D).
TABLE 1.
Laboratory and wild-derived strains exhibiting two distinct Fpr3 variants and their geographic origin
| Strains | Fpr3 Variant | Origin | Inbred Strain Category |
|---|---|---|---|
| Laboratory mouse strains | |||
| C57Bl/6NCrl | Fpr3wt | United Kingdom | Castle's mice |
| C57Bl/6NJ | Fpr3wt | United Kingdom | Castle's mice |
| C57Bl/10J | Fpr3wt | United Kingdom | Castle's mice |
| C57BR/cdJ | Fpr3wt | United Kingdom | Castle's mice |
| C57L/J | Fpr3wt | United Kingdom | Castle's mice |
| C58/J | Fpr3wt | United Kingdom | Castle's mice |
| I/LnJ | Fpr3wt | United Kingdom | Castle's mice |
| NZB/B1NJ | Fpr3wt | United Kingdom | Castle's mice |
| NZB/Ola | Fpr3wt | United Kingdom | Castle's mice |
| NZW/LacJ | Fpr3wt | United Kingdom | Castle's mice |
| KK/HiJa | Fpr3wt | Japan | Colonies from China and Japan |
| NOD/ShiLtJ | Fpr3wt | Switzerland | Swiss mice |
| ST/bJ | Fpr3wt | Unknown | Other inbred strains |
| 129X1/Sv | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| 129P2/OlaHsd | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| 129S1/SvImJ | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| 129S5SvEvBrd | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| AKR/J | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| A/J | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| BALB/cJ | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| C3H/HeH | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| C3H/HeJ | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| CBA/J | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| DBA/1J | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| DBA/2J | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| LP/J | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| NZO/HILtJ | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| RF/J | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| SEA/GnJ | Fpr3Δ424–435 | United Kingdom | Castle's mice |
| FVB/NCrl | Fpr3Δ424–435 | Switzerland | Swiss mice |
| FVB/NJ | Fpr3Δ424–435 | Switzerland | Swiss mice |
| BUB/BnJ | Fpr3Δ424–435 | Unknown | Other inbred strains |
| Wild-derived mouse strains | |||
| SPRET/EiJ | Fpr3wt | Spain | Strains derived from wild mice |
| CZE/DT | Fpr3wt | Czech Republic | Strains derived from wild mice |
| PWK/PhJ | Fpr3wt | Czech Republic | Strains derived from wild mice |
| GER/DT | Fpr3wt | Germany | Strains derived from wild mice |
| FRA/DT | Fpr3wt | France | Strains derived from wild mice |
| CAST/EiJ | Fpr3wt | Thailand | Strains derived from wild mice |
| KAZ/DT | Fpr3wt | Kazakhstan | Strains derived from wild mice |
| ZALENDE/EiJ | Fpr3D424–435 | Switzerland | Strains derived from wild mice |
| LEWES/EiJ | Fpr3D424–435 | United States of America | Strains derived from wild mice |
| WSB/EiJ | Fpr3D424–435 | United States of America | Strains derived from wild mice |
a Inbred strain categories were adopted from Ref. 55.
Because of this high frequency of the variants in laboratory mice, we asked about their frequency in wild type-derived animals. Most laboratory mouse strains are cross-breedings from three Mus musculus subspecies: M. musculus castaneus, M. musculus domesticus, and M. musculus musculus. To elucidate which of these ancestors carry the Fpr3Δ424–435 variant, we investigated the Fpr3 sequence of wild type-derived Mus musculus strains (Fig. 5E). Our panel of six genomic DNAs covered samples of M. musculus castaneus, M. musculus domesticus, M. musculus musculus, and an additional M. musculus spretus sample. The four samples were collected from different locations on the following three continents: North America, Europe, and Asia. The analysis of these DNA samples by direct sequencing of PCR products showed that they all carried Fpr3wt. Additional data mining using Mouse Genomes Project data indicated that three M. musculus domesticus lines carry the Fpr3Δ424–435 variant. Thus, Fpr3Δ424–435 was most likely introduced into inbred mouse lines through breeding with M. musculus domesticus mice that accidentally carried Fpr3Δ424–435.
Fpr3Δ424-435 Loss of Function Because of Diminished Expression
As described above, the deletion in Fpr3Δ424–435 leads to a loss of only four amino acids at the C-terminal end of the second intracellular loop but leaves the open reading frame intact. The presence of Fpr3 mRNA in the absence of a detectable protein strongly argues for structural alterations in Fpr3Δ424–435. We thus hypothesized that these alterations may affect receptor function. To test this, we used an established in vitro calcium imaging assay (33). We first performed calcium imaging in HEK cells transiently transfected with either Fpr3Δ424–435 or Fpr3wt and recorded calcium responses to 30 μm W-peptide, a potent Fpr3 agonist (32, 33). 177/475 cells (37%) transfected with Fpr3wt were robustly activated by the W-peptide, but none of the analyzed 686 cells transfected with Fpr3Δ424–435 responded to this stimulus (Fig. 6A). This dramatic result was unexpected as the deletion only affected four intracellular amino acids leaving the open reading frame intact. A loss of function in this assay was also observed with other Fpr3 agonists s follows: three bacterial signal peptide fragments that we recently identified as Fpr3 activators (32) and the synthetic M-peptide that exhibits high sequence divergence to the W-peptide. Transfected cells were neither activated by M-peptide nor by any of the three sequence divergent bacterial signal peptides (Fig. 6B). Thus, we conclude that Fpr3Δ424–435 is non-functional.
The absence of specific Fpr3 staining in VNO cells and leukocytes from mice carrying the Fpr3Δ424–435 gene could be caused by a diminished expression or lacking expression of the Fpr3 protein. We analyzed the degree of protein expression for both Fpr3 variants by immunostaining with ECL1 and ECL2 in HEK cells. Although Fpr3wt expression could be readily detected with both antibodies in these cells, no staining was visible for Fpr3Δ424–435 (Fig. 6C). This finding, obtained with an overexpression system, closely resembled the results from VSNs and leukocytes. The lack of staining by two independent antibodies that recognize different parts of the receptor strongly suggests a degradation of the Fpr3Δ424–435 variant in HEK cells. However, other consequences of the deletion, e.g. those causing defects in the promoter region or other parts of the plasmid essential for expression of Fpr3Δ424–435, could equally produce such results. To distinguish between these possibilities, we subcloned Fpr3 into a novel vector to create a fusion protein of Fpr3Δ424–435 exhibiting specific N- and C-terminal epitopes. This strategy permitted independent detection of the receptor protein using two different antibodies specific for these epitopes. We attached an N-terminal rhodopsin epitope (Rho tag) and a C-terminal herpes simplex virus epitope (HSV tag) to Fpr3Δ424–435 and named the fusion protein Rho-Fpr3Δ424–435-HSV. To exclude any possible corruption in the vector, we produced, sequenced, and tested three independent copies. In all immunoreactions for the N-terminal Rho tag, all three Rho-Fpr3Δ424–435-HSV plasmids showed a 3-fold diminished expression and a complete loss of signal for the C-terminal HSV tag stainings (Fig. 6D). Together these experiments strongly argue for a structural alteration in Fpr3Δ424–435 that results in the production of an unstable protein, which is C-terminally truncated and subsequently degraded in HEK cells. This mechanism is likely to be responsible for the degradation of Fpr3Δ424–435 in mouse VSNs and leukocytes.
Discussion
This work provides new insight into the function of the formyl peptide receptor Fpr3 in the mouse vomeronasal and immune systems and thus indicates a dual role for this receptor in chemosensory neurons of the olfactory system and in specific immune cells. A key result of this investigation is the identification of a large panel of distinct mouse strains with severely altered expression and function of Fpr3, thus establishing the existence of natural knock-out strains for this receptor. Using two novel Fpr3-specific antibodies, we can clearly attribute distinct Fpr3 expression patterns to the presence or absence of a 12-nucleotide in-frame deletion in the receptor gene. Calcium imaging and immunofluorescence analyses of both gene variants in an in vitro expression system demonstrate the alteration of the receptor structure in one of the Fpr3 variants. The lack of four amino acids leads to an unstable, truncated, and non-functional receptor protein. We find that the genome of at least 19 mouse strains encodes a non-functional Fpr3 variant, whereas at least 13 other strains express an intact receptor. A second key finding is our demonstration that the Fpr3 protein is indeed expressed in mature mouse neutrophils isolated from blood and bone marrow and that exposure to LPS, a powerful bacterial endotoxin, causes pronounced up-regulation of Fpr3 expression in bone marrow cells. These results further substantiate the view that Fpr3 functions as a pathogen sensor.
Fpr3 Expression Is Not Restricted to the Olfactory System
Our newly generated antibodies detected acute Fpr3 protein expression in two different cell types, sensory neurons of the VNO and innate immune cells. The distribution of Fpr3 mRNA in the VNO has been examined by in situ hybridization (23, 24). These reports found Fpr3 in a small subpopulation of VSNs with very specific molecular characteristics, including the expression of G protein α-subunit Gαo, a marker for VSNs of the basal expression zone. Furthermore, tests using a pan V2ra in situ probe that detects 17 of the 122 type 2 vomeronasal receptors indicated a lack of Vmn2r expression in Fpr3-positive VSNs (24). Other formyl peptide receptors expressed in the VNO-Fpr-rs3, Fpr-rs4, Fpr-rs6, and Fpr-rs7 all coexpress with the G protein α-subunit Gαi2 (23, 24). The molecular characteristics of VSNs identified by our Fpr3 antibodies are fully consistent with these studies. The majority of Fpr3-positive cells colocalized with Gαo and OMP, but we found no significant coexpression of Fpr3 with Vmn2rs or PDE4A, which labels Gαi2-positive VSNs (16).
Surprisingly, ∼40% of the Fpr3-positive cells did not colocalize with any VSN-specific marker, thus suggesting the existence of a second cell type in our preparation. Subsequent tests with immune cell markers identified these cells as neutrophils. Fpr3 expression in leukocytes has been discussed somewhat controversially in the past. Although one report detected low amounts of Fpr3 RNA in murine leukocytes (31), other studies failed to confirm this result despite the use of sensitive techniques such as RT-PCR (23, 36, 37). We thus carefully examined blood leukocytes for Fpr3 expression. Our detection of Fpr3 protein in neutrophils by two selective antibodies, the colocalization with the neutrophil marker Ly6G, a clear verification of Fpr3 expression in RT-PCR experiments, and an inducible increase of Fpr3 protein production after LPS exposure all provided clear evidence for the presence of Fpr3 in specific immune cells. These observations thus argue for an orthologous function of Fpr3 in human and mouse immune cells, a result that should help to explain the obvious functional similarities in human and mouse Fpr3 ligand detection (27, 32, 33).
Possible Role for Fpr3 in Pathogen Detection
The primary role of neutrophils is the elimination of microbial infections. Neutrophils are usually the first leukocyte type recruited to an inflammatory site (52), and they respond to a number of bacterial chemoattractants, including classical Fpr activators such as formylated peptides (28, 53). The fact that Fpr3 expression is not limited to the olfactory system but can also be observed in the immune system suggests the detection of similar or even identical chemosensory cues by both systems. Thus, Fpr3 could be used for bacterial pathogen recognition in both systems.
Several independent lines of evidence support a role of Fpr3 as a bacterial sensor. First, this receptor has a close structural relationship to Fpr2, which can detect bacterial components such as phenol-soluble modulin peptide toxins and formylated peptides (26, 54). Second, peptide motifs that activate Fpr3 are found far more frequently in bacteria than in other organisms (33). Third, Fpr1 and Fpr2 seem to have evolved as pathogen recognition receptors that recognize structurally conserved export motifs of bacterial signal peptides (32). Fpr3 fits into this mechanistic concept because it is also a bacterial signal peptide receptor, although with a more specific detection range (32). Fourth, our observation that stimulation with LPS, a prototypical bacterial stimulus, evokes a pronounced up-regulation of Fpr3 strongly argues for a role of Fpr3 as a pathogen sensor. LPS is a principal component in the outer membrane of Gram-negative bacteria. Immune cells possess high sensitivity to LPS with the activation of various intracellular cascades that induce the expression of defense systems. Interestingly, LPS-induced increased expression has also been observed for Fpr1 and Fpr2 in microglial cells (48, 49) and for Fpr1 in neutrophils (50). We now show that Fpr3 belongs to the group of genes that are up-regulated after LPS exposure in immune cells and thus likely contributes to immune defense against bacteria.
Strain-specific Loss of Fpr3 Function
Our analyses employing a variety of mouse strains revealed pronounced and unexpected differences in the Fpr3 expression pattern. We did not detect Fpr3 protein in VSNs and neutrophils of a number of mouse strains, despite clear evidence for the presence of vomeronasal Fpr3 mRNA in these strains. Subsequent genotyping of both Fpr3-positive and -negative strains revealed a clear correlation between antibody staining patterns and the presence or absence of a 12-nucleotide in-frame deletion in Fpr3 that leaves the open reading frame intact. This deletion results in a loss of four amino acids in the second intracellular loop. The relatively small change in receptor sequence at an intracellular site could argue for modest effects on the receptor function. However, calcium imaging experiments and subsequent analysis of receptor expression clearly demonstrated a loss of function for this receptor, caused by a lack of expression. Fpr3Δ424–435 was not detectable using ECL1, ECL2, or an HSV tag antibody in both HEK and native cells. This result suggests that the 12-nucleotide in-frame deletion in Fpr3Δ424–435 causes no or extremely diminished protein expression. Another conceivable explanation would be an altered protein structure that masks the antibody-binding site. However, experiments with the Rho-Fpr3Δ424–435-HSV construct revealed a clear but diminished staining for the Rho tag antibody. Together with a lack of staining using the HSV tag antibody, our results indicate a truncated expression of Fpr3Δ424–435. We did not determine the extent to which the receptor was truncated, but the lack of cellular staining with ECL1, which recognizes an epitope localized in front of the in-frame deletion, argues for a severe misfolding of the resulting protein. The strongly reduced staining using the Rho antibody would argue for inefficient receptor synthesis, transport, and/or degradation via the proteasome.
Distribution and Origin of Fpr3Δ424–435
Our database analyses of the frequency of both receptor variants in laboratory mouse strains revealed an astonishing number of mouse lines carrying the non-functional Fpr3Δ424–435 variant. Thus far, we counted 19 different strains with this Fpr3 defect, but the actual number is likely to be much higher because the genome of many strains is not yet sequenced. Fpr3Δ424–435 is widely distributed among different mouse lines. An analysis of mouse strain genealogies (55) revealed the presence of Fpr3Δ424–435 in numerous Castle's mouse founder lines but also in other unrelated strains that originated in Asia or North America (Table 1). We did not identify a consistent heredity transmission pattern that would explain the distribution of Fpr3Δ424–435, but the distribution of Fpr3Δ424–435 among diverse strains argues for an independent introduction into different founder lines.
What is the origin of this Fpr3 variation? A frequent occurrence of Fpr3Δ424–435 in many wild type mice could explain the high frequency of Fpr3Δ424–435 in laboratory mice. The genomes of most laboratory mouse strains include a mixed genetic background derived from M. musculus castaneus, M. musculus domesticus, and M. musculus musculus (56). Our analysis of corresponding wild type-derived mouse strains showed that nearly all of them carry the intact Fpr3wt gene suggesting Fpr3wt to be the naturally occurring Fpr3 variant in wild mice. However, we observed Fpr3Δ424–435 in three substrains of M. musculus domesticus, whereas two other substrains of M. musculus domesticus carried the Fpr3wt variant. Thus M. musculus domesticus is the most likely source for the Fpr3Δ424–435 variant in laboratory strains. The distribution of Fpr3Δ424–435 in wild type M. musculus domesticus is highly unusual because we did not observe a clear geographic origin. One strain from Switzerland and two others from North America carried Fpr3Δ424–435, whereas two other lines from Germany and France carried Fpr3wt. Moreover, our database analyses revealed that the wild type-derived strain representing M. musculus molossinus, which is believed to have solely originated from M. musculus castaneus and M. musculus musculus in Japan (56), also expresses Fpr3Δ424–435 despite a geographic barrier that should prevent a cross-breeding with M. musculus domesticus mice from North America or Europe. This argues for an independent emergence of Fpr3Δ424–435 in several founder mice. An alternative explanation for the high frequency of Fpr3Δ424–435 among laboratory animals could be the loss of Fpr3 function leading to a positive selection of Fpr3Δ424–435 during the breeding process by a phenotype that is favorable under laboratory conditions. It remains to be seen which of these concepts proves to be true. In any case, the existence of a multitude of natural Fpr3 receptor knock-out lines provides a unique opportunity to determine the precise function of Fpr3 in the olfactory and immune system in the context of various genetic backgrounds.
Author Contributions
H. S. performed and analyzed the majority of experiments. B. B. and F. Z. conceived and coordinated the study. H. S., M. J., A. P.-G., T. L.-Z., F. Z., and B. B. designed the research; H. S., M. J., and A. P.-G. performed the research; H. S. and B. B. analyzed the data and contributed to the preparation of the figures; H. S., F. Z., and B. B. wrote the paper. All authors edited the manuscript.
Supplementary Material
Acknowledgments
We thank S. Plant for expert technical and G. Mörschbächer for editorial assistance; M. Pyrski and B. Stein for technical advice; R. Tirindelli and R. Molday for gifts of specific antibodies; F. Kirchhoff, D. Bruns, and R. Kluge for providing mice; and D. Tautz for providing ear stamps from several mouse strains.
This work was supported by Deutsche Forschungsgemeinschaft Grant INST 256/271-1 FUGG (to F. Z. and B. B.), Sonderforschungsbereich 894 Projects A17 (to F. Z.) and P3 (to M. J.), Schwerpunktprogramm 1392 (to F. Z. and T. L.-Z.), the Volkswagen Foundation (to T. L.-Z.), and by the University of Saarland through HOMFORexcellence Grant 2011 (to B. B.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- VNO
- vomeronasal organ
- VSN
- vomeronasal sensory neuron
- ECL
- extracellular loop
- Fpr
- formyl peptide receptor
- HSV
- herpes simplex virus-epitope; major histocompatibility complex
- PDE4A
- phosphodiesterase 4A
- OMP
- olfactory marker protein
- Rho
- rhodopsin epitope
- m/v
- mass/volume.
References
- 1. Arakawa H., Cruz S., and Deak T. (2012) Attractiveness of illness-associated odorant cues in female rats is modulated by ovarian hormones, but not associated with pro-inflammatory cytokine levels. Brain Behav. Immun. 26, 40–49 [DOI] [PubMed] [Google Scholar]
- 2. Hart B. L. (1990) Behavioral adaptations to pathogens and parasites: five strategies. Neurosci. Biobehav. Rev. 14, 273–294 [DOI] [PubMed] [Google Scholar]
- 3. Altizer S., Nunn C. L., Thrall P. H., Gittleman J. L., Antonovics J., Cunningham A. A., Dobson A. P., Ezenwa V., Jones K. E., Pedersen A. B., Poss M., and Pulliam J. R. (2003) Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu. Rev. Ecol. Evol. Syst. 34, 517–547 [Google Scholar]
- 4. Brennan P. A., and Zufall F. (2006) Pheromonal communication in vertebrates. Nature 444, 308–315 [DOI] [PubMed] [Google Scholar]
- 5. Stowers L., and Logan D. W. (2010) Olfactory mechanisms of stereotyped behavior: on the scent of specialized circuits. Curr. Opin. Neurobiol. 20, 274–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Q., and Liberles S. D. (2015) Aversion and attraction through olfaction. Curr. Biol. 25, R120–R129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kavaliers M., Choleris E., and Pfaff D. W. (2005) Recognition and avoidance of the odors of parasitized conspecifics and predators: differential genomic correlates. Neurosci. Biobehav. Rev. 29, 1347–1359 [DOI] [PubMed] [Google Scholar]
- 8. Kavaliers M., Choleris E., and Pfaff D. W. (2005) Genes, odours and the recognition of parasitized individuals by rodents. Trends Parasitol. 21, 423–429 [DOI] [PubMed] [Google Scholar]
- 9. Arakawa H., Cruz S., and Deak T. (2011) From models to mechanisms: odorant communication as a key determinant of social behavior in rodents during illness-associated states. Neurosci. Biobehav. Rev. 35, 1916–1928 [DOI] [PubMed] [Google Scholar]
- 10. Shirasu M., and Touhara K. (2011) The scent of disease: volatile organic compounds of the human body related to disease and disorder. J. Biochem. 150, 257–266 [DOI] [PubMed] [Google Scholar]
- 11. Olsson M. J., Lundström J. N., Kimball B. A., Gordon A. R., Karshikoff B., Hosseini N., Sorjonen K., Olgart Höglund C., Solares C., Soop A., Axelsson J., and Lekander M. (2014) The scent of disease: human body odor contains an early chemosensory cue of sickness. Psychol. Sci. 25, 817–823 [DOI] [PubMed] [Google Scholar]
- 12. Halpern M., and Martínez-Marcos A. (2003) Structure and function of the vomeronasal system: an update. Prog. Neurobiol. 70, 245–318 [DOI] [PubMed] [Google Scholar]
- 13. Tirindelli R., Dibattista M., Pifferi S., and Menini A. (2009) From pheromones to behavior. Physiol. Rev. 89, 921–956 [DOI] [PubMed] [Google Scholar]
- 14. Chamero P., Leinders-Zufall T., and Zufall F. (2012) From genes to social communication: molecular sensing by the vomeronasal organ. Trends Neurosci. 35, 597–606 [DOI] [PubMed] [Google Scholar]
- 15. Liberles S. D. (2014) Mammalian pheromones. Annu. Rev. Physiol. 76, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leinders-Zufall T., Brennan P., Widmayer P., S P. C., Maul-Pavicic A., Jäger M., Li X. H., Breer H., Zufall F., and Boehm T. (2004) MHC class I peptides as chemosensory signals in the vomeronasal organ. Science 306, 1033–1037 [DOI] [PubMed] [Google Scholar]
- 17. Leinders-Zufall T., Ishii T., Mombaerts P., Zufall F., and Boehm T. (2009) Structural requirements for the activation of vomeronasal sensory neurons by MHC peptides. Nat. Neurosci. 12, 1551–1558 [DOI] [PubMed] [Google Scholar]
- 18. Sturm T., Leinders-Zufall T., Maček B., Walzer M., Jung S., Pömmerl B., Stevanović S., Zufall F., Overath P., and Rammensee H. G. (2013) Mouse urinary peptides provide a molecular basis for genotype discrimination by nasal sensory neurons. Nat. Commun. 4, 1616. [DOI] [PubMed] [Google Scholar]
- 19. Boehm T., and Zufall F. (2006) MHC peptides and the sensory evaluation of genotype. Trends Neurosci. 29, 100–107 [DOI] [PubMed] [Google Scholar]
- 20. Boillat M., Challet L., Rossier D., Kan C., Carleton A., and Rodriguez I. (2015) The vomeronasal system mediates sick conspecific avoidance. Curr. Biol. 25, 251–255 [DOI] [PubMed] [Google Scholar]
- 21. Mombaerts P. (2004) Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 5, 263–278 [DOI] [PubMed] [Google Scholar]
- 22. Leinders-Zufall T., Ishii T., Chamero P., Hendrix P., Oboti L., Schmid A., Kircher S., Pyrski M., Akiyoshi S., Khan M., Vaes E., Zufall F., and Mombaerts P. (2014) A family of nonclassical class I MHC genes contributes to ultrasensitive chemodetection by mouse vomeronasal sensory neurons. J. Neurosci. 34, 5121–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rivière S., Challet L., Fluegge D., Spehr M., and Rodriguez I. (2009) Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature 459, 574–577 [DOI] [PubMed] [Google Scholar]
- 24. Liberles S. D., Horowitz L. F., Kuang D., Contos J. J., Wilson K. L., Siltberg-Liberles J., Liberles D. A., and Buck L. B. (2009) Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ. Proc. Natl. Acad. Sci. U.S.A. 106, 9842–9847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fu H., Karlsson J., Bylund J., Movitz C., Karlsson A., and Dahlgren C. (2006) Ligand recognition and activation of formyl peptide receptors in neutrophils. J. Leukocyte Biol. 79, 247–256 [DOI] [PubMed] [Google Scholar]
- 26. Bloes D. A., Kretschmer D., and Peschel A. (2015) Enemy attraction: bacterial agonists for leukocyte chemotaxis receptors. Nat. Rev. Microbiol. 13, 95–104 [DOI] [PubMed] [Google Scholar]
- 27. Migeotte I., Communi D., and Parmentier M. (2006) Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 17, 501–519 [DOI] [PubMed] [Google Scholar]
- 28. Ye R. D., Boulay F., Wang J. M., Dahlgren C., Gerard C., Parmentier M., Serhan C. N., and Murphy P. M. (2009) International union of basic and clinical pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol. Rev. 61, 119–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chamero P., Katsoulidou V., Hendrix P., Bufe B., Roberts R., Matsunami H., Abramowitz J., Birnbaumer L., Zufall F., and Leinders-Zufall T. (2011) G protein Gαo is essential for vomeronasal function and aggressive behavior in mice. Proc. Natl. Acad. Sci. U.S.A. 108, 12898–12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Munger S. D., Leinders-Zufall T., and Zufall F. (2009) Subsystem organization of the mammalian sense of smell. Annu. Rev. Physiol. 71, 115–140 [DOI] [PubMed] [Google Scholar]
- 31. Gao J. L., Chen H., Filie J. D., Kozak C. A., and Murphy P. M. (1998) Differential expansion of the N-formylpeptide receptor gene cluster in human and mouse. Genomics 51, 270–276 [DOI] [PubMed] [Google Scholar]
- 32. Bufe B., Schumann T., Kappl R., Bogeski I., Kummerow C., Podgórska M., Smola S., Hoth M., and Zufall F. (2015) Recognition of bacterial signal peptides by mammalian formyl peptide receptors: a new mechanism for sensing pathogens. J. Biol. Chem. 290, 7369–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bufe B., Schumann T., and Zufall F. (2012) Formyl peptide receptors from immune and vomeronasal system exhibit distinct agonist properties. J. Biol. Chem. 287, 33644–33655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Devosse T., Guillabert A., D'Haene N., Berton A., De Nadai P., Noel S., Brait M., Franssen J. D., Sozzani S., Salmon I., and Parmentier M. (2009) Formyl peptide receptor-like 2 is expressed and functional in plasmacytoid dendritic cells, tissue-specific macrophage subpopulations, and eosinophils. J. Immunol. 182, 4974–4984 [DOI] [PubMed] [Google Scholar]
- 35. Chiu I. M., Heesters B. A., Ghasemlou N., Von Hehn C. A., Zhao F., Tran J., Wainger B., Strominger A., Muralidharan S., Horswill A. R., Bubeck Wardenburg J., Hwang S. W., Carroll M. C., and Woolf C. J. (2013) Bacteria activate sensory neurons that modulate pain and inflammation. Nature 501, 52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee H. Y., Kang H. K., Jo E. J., Kim J. I., Lee Y. N., Lee S. H., Park Y. M., Ryu S. H., Kwak J. Y., and Bae Y. S. (2004) Trp-Lys-Tyr-Met-Val-Met stimulates phagocytosis via phospholipase D-dependent signaling in mouse dendritic cells. Exp. Mol. Med. 36, 135–144 [DOI] [PubMed] [Google Scholar]
- 37. Southgate E. L., He R. L., Gao J. L., Murphy P. M., Nanamori M., and Ye R. D. (2008) Identification of formyl peptides from Listeria monocytogenes and Staphylococcus aureus as potent chemoattractants for mouse neutrophils. J. Immunol. 181, 1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boxio R., Bossenmeyer-Pourié C., Steinckwich N., Dournon C., and Nüsse O. (2004) Mouse bone marrow contains large numbers of functionally competent neutrophils. J. Leukoc. Biol. 75, 604–611 [DOI] [PubMed] [Google Scholar]
- 39. Pérez-Gómez A., Bleymehl K., Stein B., Pyrski M., Birnbaumer L., Munger S. D., Leinders-Zufall T., Zufall F., and Chamero P. (2015) Innate predator odor aversion driven by parallel olfactory subsystems that converge in the ventromedial hypothalamus. Curr. Biol. 25, 1340–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ishii T., Omura M., and Mombaerts P. (2004) Protocols for two- and three-color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J. Neurocytol. 33, 657–669 [DOI] [PubMed] [Google Scholar]
- 41. Bufe B., Hofmann T., Krautwurst D., Raguse J. D., and Meyerhof W. (2002) The human TAS2R16 receptor mediates bitter taste in response to β-glucopyranosides. Nat. Genet. 32, 397–401 [DOI] [PubMed] [Google Scholar]
- 42. Church D. M., Goodstadt L., Hillier L. W., Zody M. C., Goldstein S., She X., Bult C. J., Agarwala R., Cherry J. L., DiCuccio M., Hlavina W., Kapustin Y., Meric P., Maglott D., Birtle Z., et al. (2009) Lineage-specific biology revealed by a finished genome assembly of the mouse. PLos Biol. 7, e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martini S., Silvotti L., Shirazi A., Ryba N. J., and Tirindelli R. (2001) Co-expression of putative pheromone receptors in the sensory neurons of the vomeronasal organ. J. Neurosci. 21, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Margolis F. L. (1982) Olfactory marker protein (OMP). Scand. J. Immunol. Suppl. 9, 181–199 [DOI] [PubMed] [Google Scholar]
- 45. Lai L., Alaverdi N., Maltais L., and Morse H. C. 3rd. (1998) Mouse cell surface antigens: nomenclature and immunophenotyping. J. Immunol. 160, 3861–3868 [PubMed] [Google Scholar]
- 46. Gowen J. W., and Calhoun M. L. (1943) On the physical basis for genetic resistance to mouse typhoid, Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 29, 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cassatella M. A. (1999) Neutrophil-derived proteins: selling cytokines by the pound. Adv. Immunol. 73, 369–509 [DOI] [PubMed] [Google Scholar]
- 48. Iribarren P., Cui Y. H., Le Y., Ying G., Zhang X., Gong W., and Wang J. M. (2003) IL-4 down-regulates lipopolysaccharide-induced formyl peptide receptor 2 in murine microglial cells by inhibiting the activation of mitogen-activated protein kinases. J. Immunol. 171, 5482–5488 [DOI] [PubMed] [Google Scholar]
- 49. Cui Y. H., Le Y., Gong W., Proost P., Van Damme J., Murphy W. J., and Wang J. M. (2002) Bacterial lipopolysaccharide selectively up-regulates the function of the chemotactic peptide receptor formyl peptide receptor 2 in murine microglial cells. J. Immunol. 168, 434–442 [DOI] [PubMed] [Google Scholar]
- 50. Mandal P., Novotny M., and Hamilton T. A. (2005) Lipopolysaccharide induces formyl peptide receptor 1 gene expression in macrophages and neutrophils via transcriptional and posttranscriptional mechanisms. J. Immunol. 175, 6085–6091 [DOI] [PubMed] [Google Scholar]
- 51. Wang Z. G., and Ye R. D. (2002) Characterization of two new members of the formyl peptide receptor gene family from 129S6 mice. Gene 299, 57–63 [DOI] [PubMed] [Google Scholar]
- 52. Kolaczkowska E., and Kubes P. (2013) Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 [DOI] [PubMed] [Google Scholar]
- 53. Mantovani A., Cassatella M. A., Costantini C., and Jaillon S. (2011) Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 11, 519–531 [DOI] [PubMed] [Google Scholar]
- 54. Wang R., Braughton K. R., Kretschmer D., Bach T. H., Queck S. Y., Li M., Kennedy A. D., Dorward D. W., Klebanoff S. J., Peschel A., DeLeo F. R., and Otto M. (2007) Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514 [DOI] [PubMed] [Google Scholar]
- 55. Beck J. A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J. T., Festing M. F., and Fisher E. M. (2000) Genealogies of mouse inbred strains. Nat. Genet. 24, 23–25 [DOI] [PubMed] [Google Scholar]
- 56. Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., Welsh C. E., Bonhomme F., Yu A. H., Nachman M. W., Pialek J., Tucker P., Boursot P., McMillan L., Churchill G. A., and de Villena F. P. (2011) Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43, 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.