Abstract
Diaminopimelate decarboxylase (DAPDC) catalyzes the final step in the diaminopimelate biosynthesis pathway of bacteria. The product of the reaction is the essential amino acid l-lysine, which is an important precursor for the synthesis of the peptidoglycan cell wall, housekeeping proteins, and virulence factors of bacteria. Accordingly, the enzyme is a promising antibacterial target. Previous structural studies demonstrate that DAPDC exists as monomers, dimers, and tetramers in the crystal state. However, the active oligomeric form has not yet been determined. We show using analytical ultracentrifugation, small angle x-ray scattering, and enzyme kinetic analyses in solution that the active form of DAPDC from Bacillus anthracis, Escherichia coli, Mycobacterium tuberculosis, and Vibrio cholerae is a dimer. The importance of dimerization was probed further by generating dimerization interface mutants (N381A and R385A) of V. cholerae DAPDC. Our studies indicate that N381A and R385A are significantly attenuated in catalytic activity, thus confirming that dimerization of DAPDC is essential for function. These findings provide scope for the development of new antibacterial agents that prevent DAPDC dimerization.
Keywords: analytical ultracentrifugation, dimerization, enzyme, enzyme kinetics, small-angle X-ray scattering (SAXS), diaminopimelate decarboxylase, lysine biosynthesis
Introduction
Diaminopimelate decarboxylase (DAPDC)3 (E.C. 4.1.1.20) is a member of the pyridoxal 5′-phosphate (PLP)-dependent decarboxylases (1). It catalyzes the irreversible and stereospecific decarboxylation of meso-diaminopimelate (meso-DAP) in the final step of the diaminopimelate (DAP) biosynthesis pathway of bacteria and plants (2, 3). The product of the reaction, l-lysine, is an important building block for the biosynthesis of the peptidoglycan cell wall, housekeeping proteins, and virulence factors of bacteria. Consequently, DAPDC represents a promising target for the development of novel antibacterial agents (4).
A DAPDC reaction mechanism has been previously proposed (5, 6). The reaction mechanism is thought to be initiated by the formation of a Schiff base between PLP and a conserved active site lysine from the (Y/F)AXKA motif (7). The substrate, meso-DAP, then binds and forms a subsequent Schiff base with PLP. This is suggested to be coordinated by the highly conserved CE(S/T)XD motif provided by an adjacent subunit (7). Decarboxylation of meso-DAP is then mediated by the sulfhydryl nucleophile provided by the conserved cysteine of the CE(S/T)XD motif thereby leaving the substrate cradled between two monomers (8). This proposed mechanism suggests DAPDC functions as a dimer.
To support this assertion, previous structural studies of DAPDC from Methanocaldococcus jannaschii (PDB codes 1TWI and 1TUF) (9), Aquifex aeolicus (PDB code 2P3E), and Brucella melitensis (PDB code 3VAB) show that the enzyme crystallizes as a homodimer, with each monomer comprised of two domains (9). Domain I forms an α/β barrel, whereas domain II is comprised of a mixed β-sheet flanked by α-helices (9). The overall fold is similar to the functionally related enzyme ornithine decarboxylase, which also uses PLP as a cofactor in a decarboxylation of a secondary amine (10). Although the crystal structures of DAPDC from all species are shown to adopt the same tertiary fold, significant differences are observed at the quaternary level in the crystal state. For example, the asymmetric unit of DAPDC from Escherichia coli (PDB codes 1KNW and 1KO0) (Fig. 1A) (11), Thermotoga maritima (PDB code 2YXX), and Helicobacter pylori (PDB code 2QGH) (8) is a monomer; whereas DAPDC from Vibrio cholerae (PDB code 3N2B) adopts a tetramer (Fig. 1B). Interestingly, DAPDC from Mycobacterium tuberculosis has been crystallized as both a dimer (PDB codes 1HKW and 1HKV) (7) and a tetramer (PDB code 2O0T) (Fig. 1, C and D) (12). However, the tetrameric form observed in the asymmetric unit of DAPDC from M. tuberculosis (Fig. 1D) differs from that observed for V. cholerae (Fig. 1B).
FIGURE 1.

Quaternary structure of DAPDC enzymes in the crystal form. The asymmetric units of A, Ec-DAPDC (PDB code 1KO0); B, Vc-DAPDC (PDB code 3N2B); C, Mt-DAPDC dimer (PDB code 1HKV); and D, Mt-DAPDC tetramer (PDB code 2O0T) are displayed. The in solution assemblies calculated by PISA are shown for E, Ec-DAPDC (PDB code 1KO0), F, Mt-DAPDC (PDB code 1HKV and 2O0T); and G, Vc-DAPDC (PDB code 3N2B).
Adding to this confusion are the results of earlier studies aimed at characterizing the quaternary structure of DAPDC in solution. Analytical ultracentrifugation analysis of E. coli DAPDC suggested that the enzyme exists as a 5.4 S tetramer with a molecular mass of 200 kDa (13). Although small-angle x-ray scattering (SAXS) of M. tuberculosis DAPDC at 9 and 18 mg ml−1 found it to be mostly dimeric, with the scattering data best represented by a mixture of 93% dimer and 7% tetramer (12).
Accordingly, we set out in this study to comprehensively characterize the quaternary structure of DAPDC in aqueous solution from different bacterial species. Given that the aforementioned kinetic mechanism proposes that active site residues are contributed by adjacent subunits, we hypothesized that DAPDC functions as a dimer in solution.
Experimental Procedures
Protein Expression and Purification of DAPDC Enzymes
DAPDC from Bacillus anthracis (Ba-DAPDC), E. coli (Ec-DAPDC), and M. tuberculosis (Mt-DAPDC) were expressed and purified as described in Peverelli and Perugini (14). Expression and purification of V. cholerae DAPDC (Vc-DAPDC) was achieved as follows. The nucleotide sequence of the V. cholerae DAPDC gene (lysA) was first identified from the Kyoto Encyclopedia of Genes and Genomes database as entry VC0125. The amino acid sequence was subsequently altered to match the amino acid sequence from PDB code 3N2B. Next, the codon-optimized nucleotide sequence was commercially synthesized by GeneArt (Life Technologies) and subcloned into the expression vector pET11a (Novagen). The vector was then transformed into E. coli BL21(DE3) cells and expression and purification of the recombinant enzyme was performed as previously described (14, 15).
Generation of Vc-DAPDC N381A and R385A Mutants
The QuikChange II XL site-directed mutagenesis kit (Stratagene) was employed to introduce the R385A point mutation into the V. cholerae lysA gene ligated into pET11a using the primer set of 5′-CAATTACAATACCCGTCCGGCTGTTGCCGAAGTTATGGTT-3′ and 5′-AACCATAACTTCGGCAACAGCCGGACGGGTATTGTAATTG-3′ (mismatched nucleotides are underlined). Mutagenesis was performed according to the manufacturer's instructions with successful mutations confirmed by dideoxynucleotide sequencing. By contrast, the N381A mutant was commercially synthesized by Bioneer and subcloned into the expression vector pET11a (Novagen). The recombinant N381A and R385A mutants were expressed and purified using the same protocol adopted for the wild-type enzyme.
Mass Spectrometry
To determine the molar mass of purified recombinant DAPDC enzymes, electrospray ionization time-of-flight mass spectrometry was performed as previously described by Hor et al. (16). Recombinant proteins were also subjected to trypsin digestion and the resulting tryptic fragments were sequenced by tandem mass spectrometry as reported previously (17).
Circular Dichroism (CD) Spectroscopy
CD spectroscopy was performed using an Aviv Biomedical Model 410-SF CD spectrophotometer as reported previously (16, 18–20). Samples typically consisted of 0.15 mg ml−1 enzyme in a buffer of 20 mm Tris, 150 mm NaCl, 1 mm Tris(2-carboxyethyl)phosphine (TCEP), pH 8.0. Spectra were measured in a 1-mm quartz cuvette between 190 and 250 nm employing a step size of 0.5 nm and 4-s averaging time. Data were analyzed using the CONTINLL algorithm and SP43 database from the CDPro software package (21).
Absorbance Detected Analytical Ultracentrifugation
Sedimentation velocity and sedimentation equilibrium experiments were performed in a Beckman XL-I analytical ultracentrifuge with a four-hole An-60 Ti or eight-hole An-50 Ti rotor at 20 °C as described previously (16, 18–20, 22, 23). The partial specific volume (v̄) of the DAPDC samples together with the density and viscosity of the buffers were computed using the program SEDNTERP (24). For sedimentation velocity experiments, double sector quartz cells were loaded with 400 μl of reference (20 mm Tris, 150 mm NaCl, 1 mm TCEP, pH 8.0) and 380 μl of sample (3 μm), and data were collected at 40,000 rpm from 5.8 to 7.3 cm using a step size of 0.003 cm without averaging. Sedimentation velocity data were analyzed using the continuous size-distribution model (25, 26) employing the program SEDFIT. For sedimentation equilibrium experiments of R385A and N381A, double sector quartz cells were loaded with 140 μl of reference and 100 μl of sample (0.4–1.0 μm) with the sample sector also containing 20 μl of heavy (FC43) oil. Experiments were performed at 9,600 rpm and 14,300 rpm. When sedimentation equilibrium was attained (∼24 h at each speed), data were collected from 6.7 to 7.3 cm using a 0.001-cm step size with 10 averages. The resulting dual speed sedimentation equilibrium data at multiple protein concentrations were fitted globally to various self-association models, including a monomer-dimer model (16, 23), using SEDPHAT software (27).
Fluorophore Labeling of Vc-DAPDC
Prior to labeling, 1.0 ml of Vc-DAPDC (7 mg ml−1) was dialyzed overnight at 4 °C against 5 liters of 100 mm sodium bicarbonate, pH 8.3, 1 mm TCEP and containing 0.1 mm PLP to protect the active site lysine (Lys72) from modification. 160 nmol of Alexa Fluor 488 5-tetrafluorophenyl ester (dissolved in 60 μl of dimethyl sulfoxide) was then added to the dialyzed Vc-DAPDC sample and the mixture was incubated at room temperature for 80 min with continuous mixing on a rocking platform. Unincorporated fluorophore was removed by Superose 12 size exclusion liquid chromatography on a 1.0 cm × 30-cm column previously equilibrated with 20 mm Tris, 150 mm NaCl, 1 mm TCEP, 0.1 mm PLP, pH 8.0. The optimized labeling procedure yielded a ratio of 1.2:1 fluorophore to Vc-DAPDC monomer (determined using manufacturer's instructions).
Fluorescence Detected Analytical Ultracentrifugation (AU-FDS)
Sedimentation velocity and sedimentation equilibrium experiments were performed in a Beckman XL-A analytical ultracentrifuge equipped with a fluorescence detection system (Aviv Biomedical) using Alexa Fluor 488-labeled Vc-DAPDC at nanomolar concentrations. For sedimentation velocity experiments, double sector cells containing sapphire windows were loaded with 400 μl of sample. Data were collected at 40,000 rpm from 5.8 to 7.3 cm at multiple photomultiplier voltages to yield ∼3,000 fluorescence counts for the highest concentration in each cell. The resulting fluorescence versus radial data were analyzed using a continuous size distribution model (25, 26) employing SEDFIT software. For sedimentation equilibrium experiments, double sector sapphire cells were loaded with 120 μl of sample and 40 μl of heavy oil. Samples were centrifuged at 9,000 and 14,000 rpm until sedimentation equilibrium was attained (∼24 h for each speed). At equilibrium, data were collected from 6.7 to 7.3 cm at multiple photomultiplier voltages to optimize the signal-to-noise ratio and initial fluorescence counts to ∼1,000. Resulting data were corrected for photomultiplier voltage differences and normalized to pseudo-absorbance values. The pseudo-absorbance profiles were then fitted to various self-association models, including a monomer-dimer model (16, 23), using SEDPHAT software (27).
Small-angle X-ray Scattering
SAXS experiments were performed on the SAXS/WAXS beamline at the Australian Synchrotron (Melbourne, Australia). Buffer and enzyme samples at multiple concentrations were loaded into 96-well plates and incubated on a temperature-controlled sample platform at 20 °C for the duration of the experiment. Scattering data of the x-ray beam were collected using a Pilatus-1 M pixel-array detector (Dectris, Switzerland). The sample-to-detector distance was 1.6 m, covering a range of momentum transfer 0.01 Å−1 < q < 0.6 Å−1 (q = 4πsinθ/λ, where 2θ is the scattering angle and λ = 1.03 Å is the x-ray wavelength). Data sets of ∼20 frames covering a 2-s exposure time were collected from a sample exposed to an x-ray beam through a 1.5-mm quartz capillary. Frames were converged and buffer scattering subtracted using ScatterBrain software (Australian Synchrotron). Guinier plots and P(r) distributions were produced using PRIMUS (28–31), whereas scattering fits to crystal structures were performed using CRYSOL (32). Data generated for Ec-DAPDC and Vc-DAPDC were fitted against their respective crystal structures (PDB codes 1KNW and 3N2B), whereas the data for Mt-DAPDC were fitted against its dimeric crystal structure (PDB code 1HKV) or the tetramer generated from the T. maritima DAPDC (PDB code 2YXX). Data collected on Ba-DAPDC were fitted against PDB code 1HKV, as M. tuberculosis DAPDC is the closest homolog available in the PDB. Ab initio models of each DAPDC enzyme were generated using DAMMIN software (available from ATSAS online) (33).
Enzyme Kinetics Assay
DAPDC enzyme kinetic assays were performed as described in Peverelli and Perugini (14). Briefly, pre-initiation mixtures comprised of 200 mm Tris buffer, pH 8.0, 0.25–40 mm DAP, 2.5 μm saccharopine dehydrogenase, 0.16 mm NADH, 1 mm TCEP, 25 mm α-ketoglutarate, and 0.1 mm PLP were incubated for 10 min at 37 °C, DAPDC (final concentration of 5–500 nm) was then added to initiate the reaction, which was monitored by measuring the decrease in absorbance at 340 nm as a function of time using a temperature-controlled Varian Cary 4000 UV-visible spectrophotometer at 37 °C. Assays were performed in triplicate and analyzed using a Michaelis-Menten kinetic model to determine the Km for meso-DAP and kcat as reported previously (14).
Results
PISA Analyses of DAPDC Crystal Structures
To support our hypothesis that DAPDC functions as a dimer, we first performed computational analyses of DAPDC crystal structures from E. coli (PDB code 1KNW), M. tuberculosis (PDB code 1HKV), and V. cholerae (PDB code 3N2B). As reported earlier, these orthologs crystallize as monomers, dimers/tetramers, and tetramers, respectively. The in silico analyses were performed using the Protein Interfaces, Surfaces and Assemblies (PISA) web-based server (34), which calculates the nature of interactions stabilizing protein-protein interfaces in crystal structures. The PISA algorithm can therefore aid in discriminating between interfaces resulting from crystal artifacts versus those that are structurally relevant (34, 35). The PISA results conducted on 1KNW, 1HKW, and 3N2B suggest the enzymes exist as dimers (Fig. 1, E–G). We were therefore interested in confirming these predictions by studying the quaternary structure of the enzymes in aqueous solution.
Recombinant DAPDC Enzymes Are Folded and Enzymatically Active
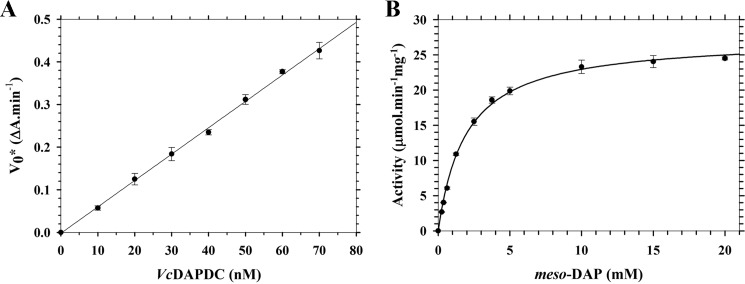
Before assessing and comparing the quaternary structure of Ec-DAPDC, Mt-DAPDC, and Vc-DAPDC, we first needed to express and purify milligram quantities of the recombinant enzymes. To expand the diversity of orthologs to also include a DAPDC from a Gram-positive bacterium, we also expressed and purified Ba-DAPDC. All four enzymes were produced in milligram quantities (1.69–132 mg) with >95% purity as assessed by SDS-PAGE. Mass spectrometric analyses showed that the recombinant products were comprised of the correct primary structure. CD spectroscopy studies were also performed to confirm that Ba-DAPDC, Ec-DAPDC, Mt-DAPDC, and Vc-DAPDC adopt α/β structures in solution (Table 1), consistent with x-ray crystal structure analyses (7–9, 11, 12). The enzymatic activity of the folded recombinant enzymes were next determined and compared using a recently optimized coupled assay employing saccharopine dehydrogenase (14, 36). The enzyme kinetic data for Vc-DAPDC is shown in Fig. 2. The resultant data fitted well to the Michaelis-Menten equation (R2 = 0.99) thereby imparting a Kmmeso-DAP of 1.9 mm, a Vmax of 27 μmol min−1 mg−1, and a catalytic rate constant (kcat) of 22 s−1 (Table 2). This is similar to the kinetic parameters determined for Ba-DAPDC, Ec-DAPDC, and Mt-DAPDC (Table 2). We were next interested in determining the quaternary structure of the active recombinant enzymes in solution.
TABLE 1.
Secondary structure of recombinant and mutant DAPDC enzymes
CD data were analyzed using CDPro (21) with the CONTINLL algorithm and the SP43 reference database.
| DAPDC | α-Helix | β-Sheet | Turn | Random | R.m.s. deviationa |
|---|---|---|---|---|---|
| % | |||||
| B. anthracis | 26 | 21 | 21 | 32 | 0.09 |
| E. coli | 25 | 24 | 22 | 29 | 0.05 |
| M. tuberculosis | 24 | 25 | 21 | 30 | 0.10 |
| V. cholerae | 27 | 24 | 21 | 28 | 0.08 |
| N381A | 26 | 24 | 20 | 30 | 0.04 |
| R385A | 30 | 20 | 21 | 29 | 0.07 |
a R.m.s. deviation, root mean square deviation.
FIGURE 2.
Enzyme kinetic analysis of Vc-DAPDC. The catalytic activity of Vc-DAPDC was investigated using the DAPDC-SDH coupled assay (14, 36). Solid circles represent the mean of triplicate measurements and error bars represent the standard deviation. A, a rate versus enzyme concentration profile. The solid line represents the fit to a simple linear regression model. B, activity of Vc-DAPDC plotted as a function of substrate (meso-DAP) concentration. The solid line represents the data fitted to the Michaelis-Menten equation.
TABLE 2.
Catalytic parameters of recombinant DAPDC enzymes
| DAPDC | Kmmeso-DAP | kcat | Vmax | Specific activity |
|---|---|---|---|---|
| mm | s−1 | units mg−1 | ||
| B. anthracis | 0.68 | 58 | 72 | 69 |
| E. coli | 0.97 | 55 | 70 | 66 |
| M. tuberculosis | 1.6 | 28 | 34 | 32 |
| V. cholerae | 1.9 | 22 | 27 | 24 |
| V. cholerae (FDS) | NDa | ND | ND | 22 |
| N381A | ND | ND | ND | 3.2 |
| R385A | ND | ND | ND | 1.9 |
a ND, not determined.
DAPDC Exists as a Dimer in Solution
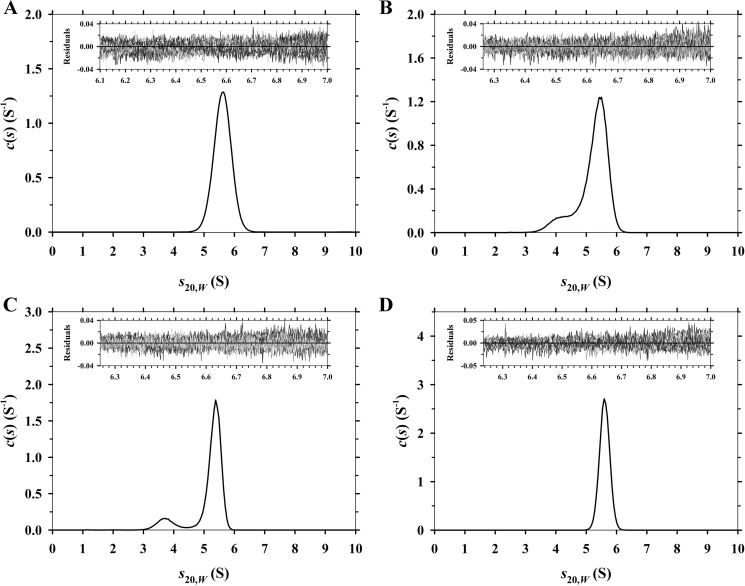
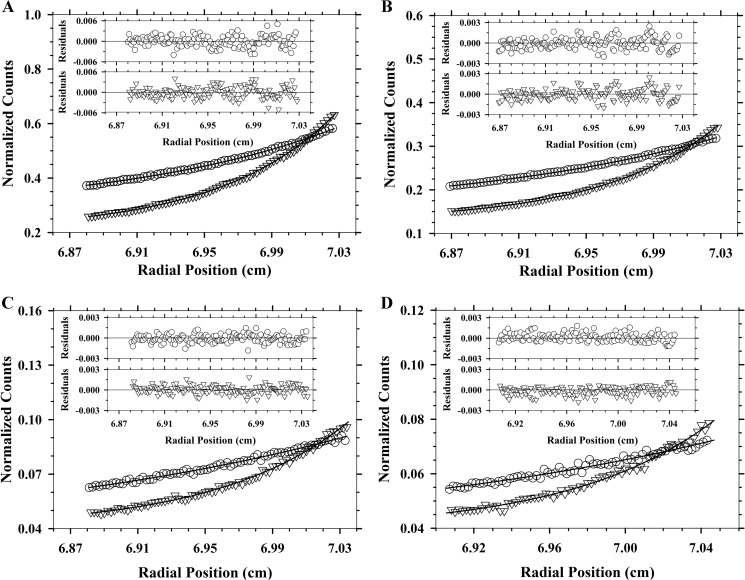
To characterize and compare the quaternary structure of Ba-DAPDC, Ec-DAPDC, Mt-DAPDC, and Vc-DAPDC, absorbance-detected sedimentation velocity experiments were initially conducted in the analytical ultracentrifuge at a protein concentration of 150 μg ml−1 (3 μm). The raw absorbance versus radial position data were fitted to a continuous size-distribution model yielding the c(s) distributions depicted in Fig. 3. The results show that Ba-DAPDC and Vc-DAPDC exist as a monodisperse 5.6 S species (Fig. 3, A and D; Table 3). The equivalent c(M) distribution results in a single species with a modal molecular mass of 83 kDa for Ba-DAPDC and 92 kDa for Vc-DAPDC (Table 3). This suggests that the 5.6 S species is a dimer. Similarly, Ec-DAPDC and Mt-DAPDC exist primarily as a ∼5.6 S species, but the c(s) distributions also show a small proportion of a ∼4 S species (Fig. 3, B and C; Table 3). The c(M) distribution results for Ec-DAPDC and Mt-DAPDC indicate that the ∼4 S species has a molecular mass of ∼50 kDa (Table 3), suggesting that this species is a monomer. Nevertheless, the results of the sedimentation velocity experiments demonstrate that DAPDC from all four bacterial species primarily exist as dimers in aqueous solution. We therefore set out to validate these results using small-angle x-ray scattering.
FIGURE 3.
Sedimentation velocity analysis of DAPDC enzymes. Continuous sedimentation coefficient (c(s)) distributions plotted as a function of standardized sedimentation coefficient (s20,w) in units of Svedberg (S). A, Ba-DAPDC; B, Ec-DAPDC; C, Mt-DAPDC; and D, Vc-DAPDC. Residuals for each experiment are displayed as insets.
TABLE 3.
Summary of hydrodynamic properties of DAPDC enzymes
| DAPDC | Oligomer | Theoretical molecular massa | s20,w | Experimental molecular massb | f/f0c | KD2→1d |
|---|---|---|---|---|---|---|
| kDa | S | kDa | nm | |||
| B. anthracis | Dimer | 98 | 5.6 | 83 | 1.3 | NDe |
| E. coli | Monomer | 47 | 4.2 | 52 | 1.1 | ND |
| Dimer | 94 | 5.5 | 81 | 1.4 | ||
| M. tuberculosis | Monomer | 50 | 3.7 | 50 | 1.3 | ND |
| Dimer | 100 | 5.4 | 86 | 1.4 | ||
| V. cholerae | Dimer | 98 | 5.6 | 92 | 1.3 | ND |
| V. cholerae labeled (Abs) | Dimer | 100 | 5.8 | 98 | 1.3 | ND |
| V. cholerae | Monomer | 50 | 3.6 | 41 | 1.3 | 0.06 |
| labeled (FDS) | Dimer | 100 | 5.6 | 91 | 1.3 | |
| N381A | Monomer | 49 | 3.6 | 46 | 1.2 | 17.3 |
| Dimer | 98 | 5.1 | 76 | 1.4 | ||
| R385A | Monomer | 49 | 3.7 | 51 | 1.2 | 410 |
| dimer | 98 | 5.2 | 77 | 1.4 |
a Molecular masses derived from amino acid sequence.
b Molecular masses obtained from the ordinate maximum of peaks in the c(M) distribution.
c Frictional ratio derived using the v̄ bar method (24).
d Determined from global nonlinear best fit to a monomer-dimer equilibrium model using sedimentation equilibrium data at multiple concentrations and rotor speeds.
e ND, not determined.
SAXS Confirms DAPDC Is Dimeric
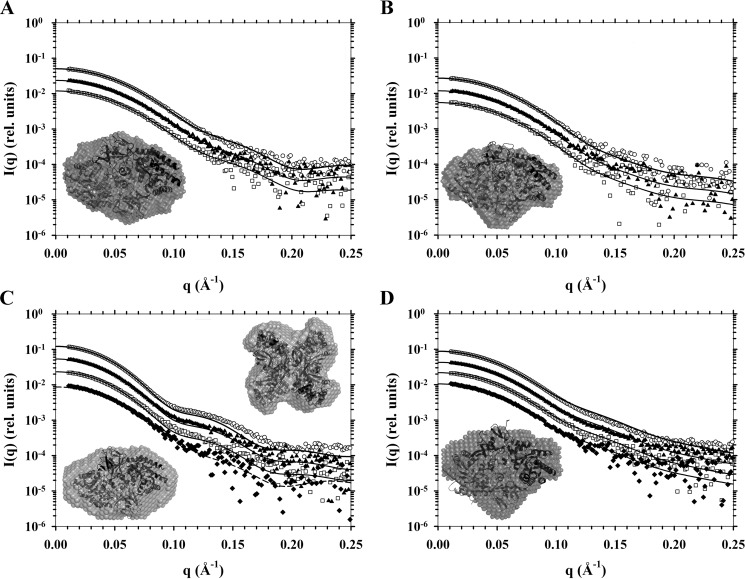
To support the initial analytical ultracentrifugation results, SAXS experiments were performed using a synchrotron x-ray source at DAPDC concentrations of 3–30 μm. The resulting scattering data were subjected to Guinier analysis (28, 29) to determine the radius of gyration (Rg) (Table 4). The maximum dimension of the scattering particle (Dmax) for each DAPDC enzyme (Table 4) were determined from the pairwise distance-distribution function (P(r)) calculated using the indirect Fourier transform method (30). Scattering data were also evaluated using CRYSOL (32) incorporating appropriate PDB entries of DAPDC enzymes (Fig. 4; Table 4). These analyses showed that the experimental scattering profiles for Ba-DAPDC, Ec-DAPDC, and Vc-DAPDC best fit to the theoretical scattering profile of a dimer (Fig. 4, A, B, and D). However, the scattering data for Mt-DAPDC fit best to a dimer at 3 μm, but to a tetramer at 30 μm (Fig. 4C), which is consistent with previous studies (12). Furthermore, ab initio bead models were created using DAMMIN (33) to establish shape envelopes from the scattering data of each DAPDC sample. Crystal structures were overlaid with the bead models as shown in Fig. 4 (insets). Consistent with the CRYSOL results, the ab initio models exhibit shape envelopes that correspond to dimers for Ba-DAPDC, Ec-DAPDC, and Vc-DAPDC (Fig. 4, A, B, and D). Whereas, the bead models for Mt-DAPDC generated from the lowest (3 μm) and highest (30 μm) concentration datasets displayed a dimeric and tetrameric shape envelope, respectively (Fig. 4C). The bead models provide further evidence that the structurally relevant quaternary structure of DAPDC is a dimer. We were thus interested in further probing the relationship between the dimeric structure and enzyme function. To do this, we decided to create mutant forms of Vc-DAPDC with attenuated dimerization propensity and compare the catalytic activity of the mutants to the wild-type dimer. Nevertheless, it was important to first determine the dimerization affinity of wild-type Vc-DAPDC to enable comparison with dimer attenuated mutant forms.
TABLE 4.
Summary of SAXS results of recombinant DAPDC enzymes
| DAPDC | Concentration | Data Rg | Dmax | χ vs PDBa | Fit Rg | PDB Dmax |
|---|---|---|---|---|---|---|
| mg ml−1 | Å | Å | ||||
| B. anthracis | 0.84 | 29.7 ± 0.1b | 100 | 0.715 | 29.5 | 102 |
| 0.42 | 30.0 ± 0.2 | 100 | 0.599 | 29.5 | ||
| 0.21 | 29.9 ± 0.3 | 103 | 0.553 | 29.5 | ||
| E. coli | 0.50 | 29.5 ± 0.2 | 103 | 0.677 | 28.9 | 96.8 |
| 0.25 | 29.6 ± 0.2 | 104 | 0.615 | 28.8 | ||
| 0.13 | 29.6 ± 0.8 | 104 | 0.549 | 28.6 | ||
| M. tuberculosis | 1.4 | 36.5 ± 0.1 | 117 | 2.14 | 35.8 | 111 |
| 0.70 | 35.8 ± 0.2 | 120 | 0.848 | 35.0 | ||
| 0.35 | 35.1 ± 0.3 | 115 | 0.878 | 34.7 | ||
| 0.17 | 34.9 ± 0.6 | 117 | 0.855 | 34.7 | ||
| V. cholerae | 1.5 | 30.5 ± 0.1 | 107 | 1.70 | 30.6 | 98.5 |
| 0.74 | 30.5 ± 0.1 | 107 | 0.947 | 30.5 | ||
| 0.37 | 30.4 ± 0.3 | 104 | 0.667 | 30.6 | ||
| 0.19 | 30.9 ± 1.0 | 113 | 0.601 | 30.6 | ||
FIGURE 4.
SAXS analyses of DAPDC enzymes. Scattering intensity in relative units is plotted as a function of momentum transfer in Å−1. Buffer corrected scattering data were fitted to theoretical scattering profiles (solid or dashed lines) generated from atomic coordinates from PDB files. Data from highest to lowest concentrations are, respectively, represented by open circles (○), solid triangles (▴), open squares (□), and if so required closed diamonds (♦). Included as insets are DAMMIN ab initio bead models generated from scattering data with overlaid cartoon crystals structures. A, Ba-DAPDC scattering data from 0.84, 0.42, and 0.21 mg ml−1 samples and a DAMMIN bead model fitted to the Mt-DAPDC dimer (PDB code 1HKV). B, Ec-DAPDC scattering data from 0.50, 0.25, and 0.13 mg ml−1 samples and a DAMMIN bead model fitted to the dimer assembly calculated by PISA of the E. coli DAPDC crystal structure (PDB code 1KO0). C, Mt-DAPDC scattering data from 1.40, 0.70, 0.35, and 0.17 mg ml−1 samples and DAMMIN bead models fitted to either its dimer (PDB code 1HKV) or the PISA tetramer assembly generated from the T. maritima DAPDC crystal structure (PDB code 2YXX). Solid lines are fits to the 2YXX tetramer and the dashed line is the fit to the 1HKV dimer. D, Vc-DAPDC scattering data from 1.48, 0.74, 0.37, and 0.19 mg ml−1 samples and a DAMMIN bead model fitted to the dimer of PDB code 3N2B (i.e. chains A and B).
AU-FDS Studies of Vc-DAPDC
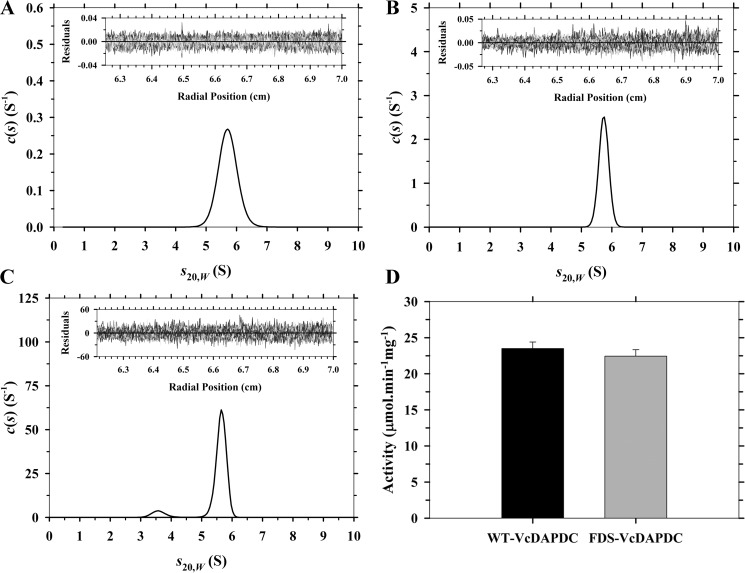
Although the initial sedimentation velocity studies outlined earlier indicate that Vc-DAPDC exists as a stable dimer at a concentration of 3 μm (Fig. 3D); we set out to examine the concentration dependence of Vc-DAPDC dimerization by diluting the enzyme concentration by 5-fold to 30 μg ml−1 (0.6 μm) (Fig. 5A). However, no significant dissociation of the dimer was observed. This indicated that the dimerization dissociation constant was considerably tighter than 0.6 μm. Accordingly, a more sensitive method for detection was required and thus Vc-DAPDC was labeled with Alexa Fluor 488 to facilitate sedimentation studies via AU-FDS at submicromolar concentrations. Initially, absorbance-detected sedimentation velocity studies were performed on the fluorescently labeled enzyme to ensure that labeling did not perturb dimerization. At 150 μg ml−1, the labeled enzyme sedimented as a single species with an s20,w value of 5.8 S (Fig. 5B) and a molecular mass of 98 kDa, consistent with the unlabeled enzyme (Fig. 3D; Table 3). Sedimentation velocity experiments were thus performed in AU-FDS using 5 nm fluorescently labeled enzyme. The resulting continuous size distribution shows evidence of dissociation of the 5.6 S dimer to a 3.6 S monomer (Fig. 5C; Table 3). Importantly, we also determined that the specific activity of the Alexa Fluor 488-labeled enzyme was 22.4 units mg−1, which is similar to the unlabeled enzyme (Fig. 5D; Table 2). This demonstrates that labeling did not significantly alter the catalytic function of Vc-DAPDC.
FIGURE 5.
Comparison of fluorescently labeled and unlabeled Vc-DAPDC. A–C, continuous sedimentation coefficient (c(s)) distributions plotted as a function of standardized sedimentation coefficients (s20,w) with residuals of fits as insets. A, Vc-DAPDC at 30 μg ml−1 (∼600 nm); B, fluorescently labeled Vc-DAPDC at 150 μg ml−1 (∼3,010 nm); and C, fluorescently labeled Vc-DAPDC at 0.25 μg ml−1 (5 nm). AU-FDS technique was used for C. D, comparison of wild-type Vc-DAPDC (WT-VcDAPDC) and fluorescently tagged Vc-DAPDC (FDS-VcDAPDC) specific activity at a fixed concentration of 100 nm. Error bars represent the standard deviation of triplicate measurements.
To quantify the dimerization affinity of Alexa Fluor 488-labeled Vc-DAPDC, sedimentation equilibrium experiments were conducted at two rotor speeds (9,000 and 14,000 rpm) and four enzyme concentrations (2, 4, 20, and 40 nm). The resulting data were globally fitted to various self-association models, including monomer-dimer, monomer-trimer, monomer-tetramer, and dimer-tetramer. The global nonlinear best fit resulted to a monomer-dimer model, which yielded a dimer-monomer dissociation constant (KD2→1) of 0.061 nm (Fig. 6; Table 3). This is very tight, therefore supporting the hypothesis that dimerization is critical for DAPDC function. We next set out to design dimerization attenuation mutants to compare with the wild-type Vc-DAPDC dimer.
FIGURE 6.
AU-FDS sedimentation equilibrium analysis of Vc-DAPDC. Once sedimentation of samples were at equilibrium at 9,000 (○) and 14,000 (▿) rpm, fluorescence emission readings of Alexa Fluor 488 from 519 nm were collected, corrected for photomultiplier voltage differences, and finally normalized to pseudo-absorbance values (y axes) for use in the program SEDPHAT. Overlaid to the experimental data are the global nonlinear regression best fit to a monomer-dimer self-association model (global reduced χ2 = 0.039). Residuals are displayed above each plot for 9,000 (○) and 14,000 (▿) rpm data. The KD2→1 of 0.061 nm was yielded from experimental Vc-DAPDC concentrations of A, 40; B, 20; C, 4; and D, 2 nm.
Dimer Interface Mutants Are Attenuated in Dimerization Propensity and Catalytic Activity
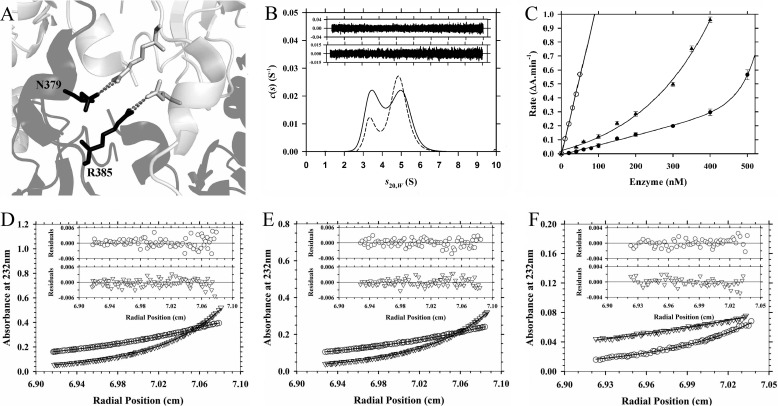
There are several residues that facilitate subunit interactions at the dimerization interface of Vc-DAPDC. This includes Arg385, which forms a hydrogen bond with Asn379 on the adjacent subunit (Fig. 7A). We therefore decided to mutate Arg385 to an Ala in an attempt to create a dimerization attenuated mutant. Successful implementation of the point mutation R385A was confirmed by dideoxynucleotide sequencing and mass spectrometry. The structure of R385A was analyzed by CD spectroscopy, which demonstrates that the point mutation did not perturb the secondary structure of the enzyme relative to wild-type Vc-DAPDC (Table 1).
FIGURE 7.
Characterization of Vc-DAPDC dimer interface point mutants. A, adjacent schematic chains of Vc-DAPDC are shown in black and light gray. In stick representation are the residues Arg385 (targeted for the R385A mutation) and Asp379, with gray dashed lines representing hydrogen bond formation across the dimer interface. B, continuous sedimentation coefficient (c(s)) distribution plotted as a function of standardized sedimentation coefficient (s20,w) for R385A (solid line) and N381A (dashed line) at an initial concentration of 400 nm. The residuals for the fits are displayed as an inset for N381A (top) and R385A (bottom). C, a rate versus enzyme concentration profile was generated for wild-type Vc-DAPDC (○), N381A (▴), and R385A (●). Vc-DAPDC data fits to a simple linear regression model. N381A and R385A data fit to a nonlinear regression model encompassing exponential growth. Error bars represent the standard deviation of triplicate measurements. D–F, absorbance at 232 nm from sedimentation equilibrium experiments were collected at 9,600 (○) and 14,300 (▿) rpm for R385A. The global nonlinear regression best fit (solid lines) to the monomer-dimer self-association model (global reduced χ2 = 0.079) are shown. Residuals are displayed above each plot for 9,600 (○) and 14,300 (▿) rpm data. The KD2→1 of 0.41 μm was yielded from experimental R385A concentrations of D, 1,000; E, 600; and F, 400 nm.
Next, the quaternary structure of R385A was investigated using analytical ultracentrifugation. Sedimentation velocity experiments were performed over a concentration range of 0.4–20 μm. At 0.4 μm, the continuous size distribution analysis showed that the enzyme exists in a monomer-dimer equilibrium with similar proportions of each species (Fig. 7B; Table 3). This suggests the dimerization dissociation constant of R385A is ∼400 nm. To better quantify the dimerization affinity of R385A, sedimentation equilibrium experiments were performed at two rotor speeds (9,600 and 14,300 rpm) and multiple protein concentrations (400, 600, and 1,000 nm). The resulting data were fitted to a monomer-dimer self-association model, which yielded a KD2→1 of 0.41 μm consistent with the sedimentation velocity results (Fig. 7, D–F; Table 3). This indicates that dimerization of the R385A mutant is ∼6,660-fold weaker than wild-type Vc-DAPDC.
The catalytic activity of R385A was next examined and compared with the wild-type enzyme. At a fixed concentration of 100 nm, the specific activity of R385A is 1.9 units mg−1, which is ∼8% of the wild-type enzyme. These results demonstrate that the wild-type Vc-DAPDC dimer is considerably more active than the dimer attenuated R385A mutant. Furthermore, the point mutant does not display a linear relationship between rate and enzyme concentrations (Fig. 7C). A distinct upward curvature is observed between 5 and 500 nm, which is indicative of a self-associating enzyme where the lower order species is significantly less active than the higher order oligomer (16, 20, 23). By contrast, the wild-type enzyme displays a linear rate versus concentration relationship at concentrations (5–50 nm) well above the KD2→1 (Fig. 7C).
To further validate that loss of catalytic function correlates with attenuation in dimerization, a second dimer interface mutant (N381A) was cloned, expressed, purified, and characterized in solution (Tables 1–3; Fig. 7, B and C). Asn381 is positioned at the dimer interface ∼10 Å from Arg385, and forms an inter-chain hydrogen bond with Glu86. Although less pronounced than the R385A mutant, the results show that N381A is also attenuated in dimerization propensity (Fig. 7B; Table 3) and catalytic activity (Fig. 7C). Indeed, comparison of the structure-activity properties of wild-type Vc-DAPDC, N381A, and R385A shows that specific activity (Table 2) correlates with dimerization affinity (i.e. decreasing KD2→1) (Table 3).
Discussion
Given that the catalytic mechanism proposed for DAPDC assumes the enzyme exists as a dimer (7–9, 12), it is interesting to note that previous studies have not yet established whether the enzyme functions as a dimer in solution. Although the crystal structure of DAPDC from some species suggests the enzyme exists as a dimer, monomeric and tetrameric asymmetric units are also observed in the crystal state for several DAPDC structures. Nevertheless, we hypothesized that DAPDC functions as a dimer and therefore non-dimeric forms observed in the crystalline state are artifacts of crystallization or simply represent the asymmetric unit. Accordingly, the overall aim of this study was to characterize the quaternary structure of DAPDC from different bacterial species in aqueous solution.
Consistent with our hypothesis, we show using analytical ultracentrifugation, SAXS, and enzyme kinetics that DAPDC from B. anthracis, E. coli, M. tuberculosis, and V. cholerae are functional dimers in solution. These results offer strong support to the proposed catalytic mechanism implicating an essential role for dimerization for DAPDC function (7–9, 12). Moreover, the critical CE(S/T)XD motif required for catalysis is provided by the adjacent subunit in the dimeric complex. Dimerization has also been shown to be critical for other family members of the PLP-dependent decarboxylases, such as eukaryotic ornithine decarboxylase, where the active site is completed by the contribution of Lys69 and Cys360 from opposing subunits (10, 37). These are equivalent to the DAPDC Lys and Cys residues in the respective (Y/F)AXKA and CE(S/T)XD motifs. Mutation of these residues in mouse ornithine decarboxylase leads to heavily attenuated activity (38, 39). Clearly, these cross-interface interactions are highly important.
Interestingly, these interactions are also observed for several other enzymes, including dihydrodipicolinate synthase, which catalyzes the first committed and rate-limiting step in the DAP biosynthesis pathway (2, 3). For dihydrodipicolinate synthase, dimerization has been shown to be important for completing the active site of each monomer through the interdigitation of a key catalytic triad residue (18, 20, 22, 23, 35, 40). Similarly, recent studies demonstrate that dimerization is required for the catalytic activity of diaminopimelate epimerase, which also functions in the DAP pathway (16). Additionally, dimerization has been proposed to be required for reorientating the active site in the activation of apoptotic initiator caspase 9 (41). Indeed, it is predicted that oligomerization is critical for providing key active site determinants in ∼1 in 6 enzymes (42). The importance of enzyme oligomerization is therefore not always pertinent for evolving allosteric regulation sites, because more and more examples of the role of quaternary structure for the completion of active sites are being discovered. One such example is reported in this study.
Protein-protein interfaces represent highly specific drug targets, as conservation is much higher in active sites relative to interfaces. Given the increase in drug-resistant bacteria associated with the overuse and misuse of broad spectrum antibacterials, exploiting the dimerization interface of DAPDC may provide a means to negate the incidence of broad spectrum drug resistance (2, 3, 43–45).
In conclusion, we show that DAPDC from multiple bacterial species form dimers in solution and that self-association is required for activity. Consequently, preventing dimerization of DAPDC may provide a viable approach for inhibition and subsequent discovery of new antibacterial agents.
Author Contributions
M. A. P. conceived the project; M. G. P. and M. A. P. designed the majority of the experiments and co-wrote the manuscript; M. G. P. and T. P. S. C. performed the experiments and analyzed the data; N. K. co-designed, co-performed, and assisted M. G. P. with the analysis of the SAXS experiments.
Acknowledgments
We thank Dr. Manfred S. Weiss (Helmholtz Centre for Materials and Energy, Berlin) for generously providing us with the plasmid for M. tuberculosis DAPDC and Prof. Paul F. Cook (University of Oklahoma) for providing us with the plasmid for Saccharomyces cerevisiae saccharopine dehydrogenase. Part of this research was undertaken on the SAXS/WAXS beamline at the Australian Synchrotron, Victoria, Australia, and we acknowledge the La Trobe University-Comprehensive Proteomics Platform (LTU-CPP) for access to essential equipment employed in this study.
This work was supported in part by Australian Research Council (ARC) Discovery Project DP150103313. The authors declare that they have no conflicts of interest with the contents of this article.
- DAPDC
- diaminopimelate decarboxylase
- AU-FDS
- fluorescence-detected analytical ultracentrifugation
- DAP
- diaminopimelate
- Dmax
- maximum particle size
- FDS
- fluorescence detection system
- P(r)
- pairwise distance-distribution function
- PISA
- protein interfaces, surfaces and assemblies
- PLP
- pyridoxal 5′-phosphate
- SAXS
- small-angle x-ray scattering
- TCEP
- tris(2-carboxyethyl)phosphine
- PDB
- Protein Data Bank.
References
- 1. Dewey D. L., and Work E. (1952) Diaminopimelic acid and lysine: diaminopimelic acid decarboxylase. Nature 169, 533–534 [DOI] [PubMed] [Google Scholar]
- 2. Dogovski C., Atkinson S. C., Dommaraju S. R., Hor L., Dobson R. C. J., Hutton C. A., Gerrard J. A., and Perugini M. A. (2009) in Biotechnology (Doelle H. W., and Rokem S., eds) pp. 116–136, Eolss Publishers, Oxford, UK [Google Scholar]
- 3. Dogovski C., Atkinson S. C., Dommaraju S. R., Downton M., Hor L., Moore S., Paxman J. J., Peverelli M. G., Qiu T. W., Reumann M., Siddiqui T., Taylor N. L., Wagner J., Wubben J. M., and Perugini M. A. (2012) in Biochemistry (Ekinci D., ed) pp. 225–262, InTech Open Access Publisher, Rijeka, Croatia [Google Scholar]
- 4. Hutton C. A., Southwood T. J., and Turner J. J. (2003) Inhibitors of lysine biosynthesis as antibacterial agents. Mini Rev. Med. Chem. 3, 115–127 [DOI] [PubMed] [Google Scholar]
- 5. Asada Y., Tanizawa K., Sawada S., Suzuki T., Misono H., and Soda K. (1981) Stereochemistry of meso-α,ϵ-diaminopimelate decarboxylase reaction: the first evidence for pyridoxal 5′-phosphate dependent decarboxylation with inversion of configuration. Biochemistry 20, 6881–6886 [DOI] [PubMed] [Google Scholar]
- 6. Kelland J. G., Palcic M. M., Pickard M. A., and Vederas J. C. (1985) Stereochemistry of lysine formation by meso-diaminopimelate decarboxylase from wheat germ: use of 1H-13C NMR shift correlation to detect stereospecific deuterium labeling. Biochemistry 24, 3263–3267 [DOI] [PubMed] [Google Scholar]
- 7. Gokulan K., Rupp B., Pavelka M. S. Jr., Jacobs W. R. Jr., and Sacchettini J. C. (2003) Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis. J. Biol. Chem. 278, 18588–18596 [DOI] [PubMed] [Google Scholar]
- 8. Hu T., Wu D., Chen J., Ding J., Jiang H., and Shen X. (2008) The catalytic intermediate stabilized by a “down” active site loop for diaminopimelate decarboxylase from Helicobacter pylori. J. Biol. Chem. 283, 21284–21293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ray S. S., Bonanno J. B., Rajashankar K. R., Pinho M. G., He G., De Lencastre H., Tomasz A., and Burley S. K. (2002) Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor. Structure 10, 1499–1508 [DOI] [PubMed] [Google Scholar]
- 10. Osterman A., Grishin N. V., Kinch L. N., and Phillips M. A. (1994) Formation of functional cross-species heterodimers of ornithine decarboxylase. Biochemistry 33, 13662–13667 [DOI] [PubMed] [Google Scholar]
- 11. Momany C., Levdikov V., Blagova L., and Crews K. (2002) Crystallization of diaminopimelate decarboxylase from Escherichia coli, a stereospecific d-amino-acid decarboxylase. Acta Crystallogr. D Biol. Crystallogr. 58, 549–552 [DOI] [PubMed] [Google Scholar]
- 12. Weyand S., Kefala G., Svergun D. I., and Weiss M. S. (2009) The three-dimensional structure of diaminopimelate decarboxylase from Mycobacterium tuberculosis reveals a tetrameric enzyme organization. J. Struct. Funct. Genomics 10, 209–217 [DOI] [PubMed] [Google Scholar]
- 13. White P. J., and Kelly B. (1965) Purification and properties of diaminopimelate decarboxylase from Escherichia coli. Biochem. J. 96, 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peverelli M. G., and Perugini M. A. (2015) An optimized coupled assay for quantifying diaminopimelate decarboxylase activity. Biochimie 115, 78–85 [DOI] [PubMed] [Google Scholar]
- 15. Hor L., Peverelli M. G., Perugini M. A., and Hutton C. A. (2013) A new robust kinetic assay for DAP epimerase activity. Biochimie 95, 1949–1953 [DOI] [PubMed] [Google Scholar]
- 16. Hor L., Dobson R. C., Downton M. T., Wagner J., Hutton C. A., and Perugini M. A. (2013) Dimerization of bacterial DAP epimerase is essential for catalysis. J. Biol. Chem. 288, 9238–9248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brand I. L., Civciristov S., Taylor N. L., Talbo G. H., Pantaki-Eimany D., Levina V., Clem R. J., Perugini M. A., Kvansakul M., and Hawkins C. J. (2012) Caspase inhibitors of the P35 family are more active when purified from yeast than bacteria. PLoS ONE 7, e39248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Atkinson S. C., Dogovski C., Downton M. T., Pearce F. G., Reboul C. F., Buckle A. M., Gerrard J. A., Dobson R. C., Wagner J., and Perugini M. A. (2012) Crystal, solution and in silico structural studies of dihydrodipicolinate synthase from the common grapevine. PLoS ONE 7, e38318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Atkinson S. C., Dogovski C., Downton M. T., Czabotar P. E., Dobson R. C., Gerrard J. A., Wagner J., and Perugini M. A. (2013) Structural, kinetic and computational investigation of Vitis vinifera DHDPS reveals new insight into the mechanism of lysine-mediated allosteric inhibition. Plant Mol. Biol. 81, 431–446 [DOI] [PubMed] [Google Scholar]
- 20. Voss J. E., Scally S. W., Taylor N. L., Atkinson S. C., Griffin M. D., Hutton C. A., Parker M. W., Alderton M. R., Gerrard J. A., Dobson R. C., Dogovski C., and Perugini M. A. (2010) Substrate-mediated stabilization of a tetrameric drug target reveals achilles heel in anthrax. J. Biol. Chem. 285, 5188–5195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sreerama N., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 22. Atkinson S. C., Hor L., Dogovski C., Dobson R. C., and Perugini M. A. (2014) Identification of the bona fide DHDPS from a common plant pathogen. Proteins 82, 1869–1883 [DOI] [PubMed] [Google Scholar]
- 23. Burgess B. R., Dobson R. C., Bailey M. F., Atkinson S. C., Griffin M. D., Jameson G. B., Parker M. W., Gerrard J. A., and Perugini M. A. (2008) Structure and evolution of a novel dimeric enzyme from a clinically-important bacterial pathogen. J. Biol. Chem. 283, 27598–27603 [DOI] [PubMed] [Google Scholar]
- 24. Laue T. M., Shah B. D., Ridgeway T. M., and Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science, pp. 90–125, The Royal Society of Chemistry, Cambridge, United Kingdom [Google Scholar]
- 25. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., and Schubert D. (2002) Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vistica J., Dam J., Balbo A., Yikilmaz E., Mariuzza R. A., Rouault P. A., and Schuck P. (2004) Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 326, 234–256 [DOI] [PubMed] [Google Scholar]
- 28. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 29. Petoukhov M. V., Konarev P. V., Kikhney A. G., and Svergun D. I. (2007) ATSAS 2.1: towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 40, s223–s228 [Google Scholar]
- 30. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 31. Mertens H. D. T., and Svergun D. I. (2010) Structural characterization of proteins and complexes using small-angle x-ray solution scattering. J. Struct. Biol. 172, 128–141 [DOI] [PubMed] [Google Scholar]
- 32. Svergun D. I., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 33. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 35. Dogovski C., Gorman M. A., Ketaren N. E., Praszkier J., Zammit L. M., Mertens H. D., Bryant G., Yang J., Griffin M. D., Pearce F. G., Gerrard J. A., Jameson G. B., Parker M. W., Robins-Browne R. M., and Perugini M. A. (2013) From knock-out phenotype to three-dimensional structure of a promising antibiotic target from Streptococcus pneumoniae. PLoS ONE 8, e83419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laber B., and Amrhein N. (1989) A spectrophotometric assay for meso-diaminopimelate decarboxylase and l-α-amino-ϵ-caprolactam hydrolase. Anal. Biochem. 181, 297–301 [DOI] [PubMed] [Google Scholar]
- 37. Myers D. P., Jackson L. K., Ipe V. G., Murphy G. E., and Phillips M. A. (2001) Long-range interactions in the dimer interface of ornithine decarboxylase are important for enzyme function. Biochemistry 40, 13230–13236 [DOI] [PubMed] [Google Scholar]
- 38. Coleman C. S., Stanley B. A., and Pegg A. E. (1993) Effect of mutations at active site residues on the activity of ornithine decarboxylase and its inhibition by active site-directed irreversible inhibitors. J. Biol. Chem. 268, 24572–24579 [PubMed] [Google Scholar]
- 39. Tsirka S., and Coffino P. (1992) Dominant negative mutants of ornithine decarboxylase. J. Biol. Chem. 267, 23057–23062 [PubMed] [Google Scholar]
- 40. Soares da Costa T. P., Christensen J. B., Desbois S., Gordon S. E., Gupta R., Hogan C. J., Nelson T. G., Downton M. T., Gardhi C. K., Abbott B. M., Wagner J., Panjikar S., Perugini M. A. (2015) Quaternary structure analyses of an essential oligomeric enzyme. Methods Enzymol. 562, 205–223 [DOI] [PubMed] [Google Scholar]
- 41. Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., and Salvesen G. S. (2001) Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. U.S.A. 98, 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ali M. H., and Imperiali B. (2005) Protein oligomerization: how and why. Bioorg. Med. Chem. 13, 5013–5020 [DOI] [PubMed] [Google Scholar]
- 43. Gerrard J. A., Hutton C. A., and Perugini M. A. (2007) Inhibiting protein-protein interactions as an emerging paradigm for drug discovery. Mini Rev. Med. Chem. 7, 151–157 [DOI] [PubMed] [Google Scholar]
- 44. Wells J. A., and McClendon C. L. (2007) Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature 450, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 45. Merabet N., Dumond J., Collinet B., Van Baelinghem L., Boggetto N., Ongeri S., Ressad F., Reboud-Ravaux M., and Sicsic S. (2004) New constrained “molecular tongs” designed to dissociate HIV-1 protease dimer. J. Med. Chem. 47, 6392–6400 [DOI] [PubMed] [Google Scholar]