Abstract
Acquisition and distribution of metal ions support a number of biological processes. Here we show that respiratory growth of and iron acquisition by the yeast Saccharomyces cerevisiae relies on potassium (K+) compartmentalization to the trans-Golgi network via Kha1p, a K+/H+ exchanger. K+ in the trans-Golgi network facilitates binding of copper to the Fet3p multi-copper ferroxidase. The effect of K+ is not dependent on stable binding with Fet3p or alteration of the characteristics of the secretory pathway. The data suggest that K+ acts as a chemical factor in Fet3p maturation, a role similar to that of cations in folding of nucleic acids. Up-regulation of KHA1 gene in response to iron limitation via iron-specific transcription factors indicates that K+ compartmentalization is linked to cellular iron homeostasis. Our study reveals a novel functional role of K+ in the binding of copper to apoFet3p and identifies a K+/H+ exchanger at the secretory pathway as a new molecular factor associated with iron uptake in yeast.
Keywords: copper, iron, iron response element (IRE), potassium transport, protein secretion, ferroxidase
Introduction
Diverse biological pathways, such as energy generation, transcription regulation, and neurotransmission, are dependent on ionic concentration gradients across the membranes and on metal-containing proteins. Organisms have evolved delicate mechanisms for uptake, distribution, utilization, and detoxification of such ions (1–6). Defect in homeostasis of these species is implicated in numerous disorders, such as growth retardation, neurodegeneration, and cancer (7–12).
Channels, transporters, and ion pumps at the plasma membrane and in subcellular organelles determine ionic cellular distribution, which is critical for maintenance of membrane potential, pH control, osmolality, transport of nutrients, and protein activity (1, 13–16). For example, cations, such as K+, Na+, and Mg2+, interact with negatively charged phosphate in DNA and RNA and thus affect folding, dynamics, and protein-nucleic acid interactions (17–19). This illustrates an example of how ions can affect macromolecular structure and function independent of site-specific binding. Comparable effects of ions on proteins have not been reported, however.
It is estimated that one-third of the proteome contains at least one metal ion or metal-containing prosthetic group as the catalytic and/or structural cofactor (2). For instance, oxidative phosphorylation and many other mitochondrial functions rely on metalloproteins containing iron, heme, Fe-S cluster, copper, manganese, and/or zinc. Iron acquisition, synthesis of heme and Fe-S clusters, and incorporation of these cofactors into the subunits of the respiratory chain complexes is vital for normal growth and development (8, 20–22). Insufficient iron acquisition is a widespread problem in virtually all organisms (8–9). Ferroxidases in fungi, algae, and humans are multi-copper-containing enzymes that cooperate with cell surface iron transporters to translocate iron across the plasma membrane (23, 24). This indicates that copper is required for respiration via maintaining iron homeostasis as well as functioning as a cofactor of the complex IV. Despite significant research progress in identification of molecular factors involved in iron and copper homeostasis, there are gaps in our understanding of the pathways and mechanisms.
To gain greater insight into copper and iron homeostasis in support of mitochondrial function, we selected and characterized a yeast strain displaying respiration deficiency that can be rescued by surplus copper or iron. Thorough characterization of the metal metabolism in this strain indicates that potassium (K+) compartmentalization to the trans-Golgi network (TGN)2 via a K+/H+ exchanger promotes binding of copper to apoFet3p ferroxidase. This occurs without stable binding of K+ to Fet3p or alteration of normal characteristics of the TGN. KHA1 transcription is up-regulated under iron deficiency, indicating active control by iron status of K+ compartmentalization. These results suggest a novel function for K+ in copper enzyme activation and identify Kha1p, a K+ transporter in the TGN, as a new molecular factor involved in copper and iron homeostasis.
Experimental Procedures
Yeast Strains, Culture Conditions, and Growth Assays
BY4741 wild-type (WT) control Saccharomyces cerevisiae strain and isogenic strains possessing indicated gene deletion (25) were obtained from Open Biosystems. Cells were cultured at 30 °C in the synthetic complete (SC) media (2% (w/v) dextrose, 0.2% (w/v) amino acid mixture, 0.67% (w/v) yeast nitrogen base) lacking uracil for plasmid selection (SC-ura), YPD media (1% (w/v) yeast extract, 2% (w/v) Bacto-peptone, 2% (w/v) dextrose)), and non-fermentable YPEG media (1% (w/v) yeast extract, 2% (w/v) Bacto-peptone, 2% (w/v) ethanol, 3% (w/v) glycerol) as indicated at each experiment. Solid media contains 1.5% (w/v) agar.
Selection of Yeast Mutants Displaying Copper and Iron-rescued Respiratory Deficiency
A collection of BY4741 strains possessing individual gene deletion by homologous recombination of the KanMX4 cassette (25) (Open Biosystems) was replica plated on non-fermentable YPEG media supplemented with the copper chelator bathocuproine disulfonate (BCS, 10 μm). This allowed us to identify strains displaying complete or subtle defect in respiratory growth in conjunction with copper metabolism. Copper- and iron-dependent respiration deficiency was determined by culturing cells on plates with additional supplementation of CuSO4 (10 μm final concentration) or FeSO4 (20 μm final concentration). Fitness of the strains under a fermentable growth condition was determined using the media containing glucose. The deleted gene of each strain was identified by PCR amplification of the region containing a gene-specific barcode (25) followed by sequencing of the PCR products.
Plasmids
KHA1 coding sequence obtained by PCR was inserted into the HindIII and XhoI sites in the p416-TEF vector (26) for TEF2 gene promoter-mediated constitutive expression in yeast. For construction of a C-terminal fusion of an epitope or fluorescent protein, a NotI restriction enzyme site was generated in the PCR primer before the stop codon. A DNA fragment encoding triple hemagglutinin epitope (HA), enhanced yellow fluorescent protein (YFP), or red fluorescent protein (RFP) was inserted into the NotI site. The same approach was employed to express YFP-fused Fet3p. For C-terminal c-myc epitope tagging of Ccc2p (Ccc2-myc), a reverse PCR primer included the c-myc sequence before the stop codon. FET3 fused with the sequence containing c-myc epitopes (Fet3p-myc) was integrated into its genomic locus by homologous recombination (27), which allowed expression of c-myc tagged FET3 by its own promoter. Functional integrity of these proteins fused with an epitope or fluorescent protein was assessed by functional complementation assays using yeast strains possessing knock-out of corresponding gene.
For β-galactosidase reporter assays, PCR-amplified KHA1 promoter (700 bp) was inserted into the EcoRI and PstI sites of the pCM64-lacZ vector (28). pCM64-FET3-lacZ and pCM64-CTR1-lacZ reporter plasmids (29, 30) determined FET3 and CTR1 gene expression, respectively. A reporter plasmid containing unfolded protein response elements (UPRE) was described previously (31). p316GALlcc1 plasmid contains a gene encoding laccase of Pycnoporus coccineus (32).
Fet3p and Glutathione S-Transferase (GST) Purification
Methods for purification of Fet3p lacking its C-terminal transmembrane domain was prepared as described (33) as was apoFet3p (34). GST was expressed in BY4741 yeast strain using p415-GPD vector (26) and purified using glutathione (GSH)-agarose (Thermo Scientific).
Oxidase Activity Assays of Fet3p
Fet3p oxidase activities were measured by in-gel and spectrophotometric assays using p-phenylenediamine dihydrochloride as a substrate (35, 36). Cells expressing c-myc-tagged Fet3p under the control of its own promoter were cultured to mid-log phase in YPD or SC media with and without supplementation of iron chelator bathophenanthroline disulfonate (BPS) (Sigma) (80 μm) and copper chelator BCS (Sigma) (50 μm). BPS enhances expression of Fet3p to easily detectable levels. BCS-induced copper limitation allowed us assess of the roles of cellular factors in assembling copper into apoFet3p. Cells were washed twice with K+-free buffer and broken by vortexing (8 × 1 min) with glass beads in Tris-HCl buffer (50 mm, pH 7.4) containing protease inhibitor mixture (Roche Applied Science), phenylmethanesulfonyl fluoride (PMSF, 1 mm) (Sigma), and BCS (50 μm). BCS was added to prevent copper loading to apoFet3p during cell lysis. After removing unbroken cells and glass beads by centrifugation at 300 × g for 3 min, membrane fractions were obtained by centrifugation (21,000 × g, 15 min). The samples were resuspended in the same buffer containing Triton X-100 (1%, v/v) on ice for 30 min with vortexing every 5 min and then cleared by centrifugation (21,000 × g, 20 min). Protein concentration was measured by the bicinchoninic acid (BCA) assay method using a kit (Pierce). Samples obtained from a FET3 gene knock-out yeast strain and copper-deficient cells producing apoFet3p were used as negative controls.
pH Measurement of Subcellular Compartment
pH luorin, a pH-sensitive fluorescent protein, was used to measure the pH of the lumen of the TGN and cytosol (37, 38). Yeast strains were transformed with p416Met25, p416Met25-pHluorin, and p416Met25-pH-Gef1E230A plasmids (38), expressing empty vector, a cytosolic pH-sensitive fluorescent protein, and a pH-sensitive fluorescent protein fused with non-functional Gef1p to target it to the lumen of the TGN, respectively. Validation of pH sensitivity, subcellular localization, and detail protocols for pH measurement using these proteins were published previously (37, 38). Cells at mid-log phase were collected by centrifugation (1000 × g, 5 min) and resuspended in PBS (50 mm, pH 7.0). Fluorescent signals of the cells in response to two excitations (407 nm, 488 nm) were measured using a fluorescence-activated cell sorter (FACS) (Beckman). After background correction using empty vector transformed cells, the ratio of fluorescent emissions at the indicated excitations were determined. The pH was calculated using a calibration curve of each pH-sensitive protein using the methods described previously (37, 38).
In Vitro Copper Binding to ApoFet3p
Membrane protein extracts containing apoFet3p were obtained using a CCC2 deleted strain; CCC2 encodes a copper transporter that delivers copper to the secretory pathway. CuSO4 preincubated with 1 mm ascorbate was added to the samples (30 μg of yeast membrane protein in Tris-HCl buffer (50 mm, pH 7.4)) for 30 min on ice with gentle agitation every 5 min. Fet3p oxidase activities were measured as described above. To determine the effects of K+ and/or Na+ on copper binding to apoFet3p, samples were resuspended in the buffer supplemented with K2SO4 or Na2SO4. pH effects on copper metallation of apoFet3p were measured using MES buffer (50 mm, pH 5.5 and 6.0) and phosphate buffer (50 mm, pH 6.0, 6.5, 7.0, and 7.5).
Western Blotting
After washing cells twice with Tris-HCl buffer (50 mm Tris, pH 7.4) by centrifugation, total and membrane protein extracts were prepared by breaking cells with glass beads in the same buffer containing protease inhibitor mixture (Complete Mini, Roche Applied Science), PMSF (1 mm), and Triton X-100 (1%, v/v). Protein concentrations were measured using a BCA kit (Pierce). Cell lysates were denatured in a SDS sample buffer containing dithiothreitol (DTT) (100 mm) at 60 °C for 15 min, separated by SDS-PAGE, and then transferred to a nitrocellulose membrane. HA, c-myc, FLAG, YFP, or RFP-fused proteins were detected by chemiluminescence using corresponding antibodies (Rockland, 600-401-384, 400-406, 381, 200-301-383, 600-401-215, 200-301-379, respectively) and horseradish peroxidase-conjugated anti-rabbit IgG (Santa Cruz, J2915) or anti-mouse IgG (Santa Cruz, H0415) antibodies. Phosphoglycerate kinase 1 (Pgk1p) was probed using anti-Pgk1p antibodies (Molecular Probes, 459250) to determine equal loading.
Subcellular Fractionation
Cell lysates were loaded onto a linear sucrose gradient prepared by subsequently layering 10, 20, 35, or 60% (v/v) sucrose to form a 9-l step gradient (39). Samples were subjected to centrifugation using a Beckman SW41 Ti Rotor (200,000 × g, 14 h). Fractions (500 μl each) were collected from the top of the gradient.
Limited Trypsin Proteolysis
Purified holo-Fet3p, apoFet3p, and GST in the Tris-HCl buffer (50 mm, pH 7.4) were co-incubated with or without CuSO4 (10 μm, preincubated with 1 mm ascorbate) and 50 mm K2SO4 on ice for 30 min. Proteolysis by trypsin (1–5 μg/ml, Sigma) was conducted at 37 °C for 30 min. The addition of soybean trypsin inhibitor (Fluka BioChemika, 0.2 μg/ml) for 15 min on ice stopped the reaction. Proteolysis patterns were visualized by SDS-PAGE followed by silver staining.
Prediction of K+ Binding Sites in Fet3p
Putative K+ ion binding sites were identified in the crystal structure of Fet3p (PDB code 1ZPU) using the ion-binding site prediction program Vale (40). Default parameters for K+, derived from small molecule crystal structures, were used for the ion radius (1.33 Å), a grid spacing of 0.1 Å and selecting sites with either 4 or 5 ligating atoms (the number typically observed in structures with K+ binding sites). The output from Vale is a list of putative binding sites in Protein Data Bank format; this list was superimposed onto the input structure allowing for visualization of the predicted binding sites. Structural models of K+ binding to Fet3p were generated using UCSF Chimera software (41).
Site-directed Mutagenesis
The FET3 coding sequence lacking the transmembrane domain (amino acids 1–555) was amplified by PCR using genomic DNA of BY4741 strain and cloned into the p416-GPD vector (26) for TDH3 gene promoter-mediated constitutive expression of Fet3p(1–555) in a fet3Δ strain. One FLAG epitope sequence was inserted before the stop codon. The residues Glu-487, Asn-49, Asp-501, or Ser-161/Glu-162 was substituted to alanine by the primer overlap extension method (39). Total protein extracts were prepared by breaking cells with glass beads in the Tris-HCl buffer (50 mm, pH 7.4) containing protease inhibitor mixture (Roche Applied Science), and phenylmethanesulfonyl fluoride (PMSF, 1 mm) (Sigma). Secreted Fet3(1–555) species were collected by concentrating the culture media using spin columns (Amicon-Ultra-15) followed by washing the columns using Tris-HCl buffer (50 mm, pH 7.4).
Measurement of Metal Levels
The levels of major physiological metals were measured by inductively coupled plasma mass spectrometry (Agilent Model 7500cs, Santa Clara, CA) (39). Data were normalized to cell number or protein concentration.
Measurement of 86Rb+
86Rb+ (PerkinElmer Life Sciences) uptake into subcellular compartments was measured using the method described previously (42). 86Rb+ uptake was normalized to cell number.
Determination of the Effects of K+ on Cu+ Solubilization
The BCS-Cu+ complex exhibits an absorbance at 490 nm (43). To determine if K+ can affect the binding of Cu+ to BCS, K2SO4 (0–200 mm) was added in Tris-HCl buffer (50 mm, pH 7.4) along with CuSO4 (25 μm) and dihydroascorbic acid (0.1–1 mm). The reaction was initiated by adding BCS (0–200 mm). Samples were incubated at room temperature for 10 min and the appearance of the Cu+-BCS complex was measured at 490 nm.
Quantitative PCR
WT and fet3Δ cells were co-cultured with BPS (80 μm) in the SC media overnight. Cells were then re-inoculated (A600 0.1) into the fresh media containing iron chelator BPS (80 μm) for two consecutive 12-h cultures. This iron limitation induced known iron deficiency-responsive genes, such as FET3. Total RNA was subjected to cDNA synthesis followed by quantitative PCR using primer sets that are specific for KHA1 and PGK1.
β-Galactosidase Assays
β-Galactosidase activities were measured as described previously (44). Enzyme activities were converted to Miller units [(A420)/(reaction time, min) (assay volume, ml) (A600 of cells in the culture)].
Laccase Assays
A yeast S. cerevisiae plasmid containing a laccase gene of P. coccineus (32) was transformed into wild type, kha1Δ, and vma1Δ strains. Laccase activities were measured as described previously (32).
Superoxide Dismutase (SOD) Assay
Activities of yeast Sod1p were determined by an in-gel assay measuring nitrite formation from hydroxylamine in the presence of a superoxide anion generation system (45).
Gel Filtration Chromatography of Fet3p
Purified apoFet3p (500 μg) in the Tris-HCl buffer (50 mm, pH 7.4) was mixed with K2SO4 (100 mm final concentration) and ascorbate (1 mm final concentration) and incubated on ice for 30 min, which is the same condition for in vitro copper reconstitution assays and limited trypsin proteolysis of Fet3p. The samples were loaded onto a Sephadex G-100 column (1.5 × 30 cm) and then eluted (0.5 ml/min) with Tris-HCl buffer. Fractions (1 ml each) were subjected to a protein assay and inductively coupled plasma mass spectrometry for detecting Fet3p and K+, respectively.
Statistical Analysis
Data are presented as the means ± S.D., and statistical comparisons of control and experimental groups were performed using Student's t test. p < 0.05 was considered to be significant.
Results
Kha1p Is a New Molecular Factor Involved in Respiratory Growth and Iron and Copper Homeostasis in Yeast
A collection of S. cerevisiae strains carrying individual gene knockouts (25) was screened by monitoring a growth defect on non-fermentable carbon media (YPEG) supplemented with the copper chelator, BCS (10 μm). In addition to identifying known genes involved in copper and iron homeostasis, this approach also identified the KHA1 gene, which encodes a predicted monovalent cation-H+ exchanger (Fig. 1A). Although KHA1 does not have any previously identified role in respiration or the metabolism of copper or iron, either copper or iron supplementation in the growth media rescued respiration deficiency of kha1Δ cells (Fig. 1A). The similar degree of growth between WT control and kha1Δ cells on media containing glucose indicated a specific role for KHA1 in respiratory growth (Fig. 1A). KHA1 deletion leads to reduction in steady state cellular levels of iron without a significant change of copper, zinc, or magnesium (Fig. 1B). Expression of FET3 was higher in kha1Δ cells relative to WT cells (Fig. 1C), reflecting the cellular iron deficiency followed by up-regulation of the iron uptake system. The similar copper levels (Fig. 1B) coupled to no significant change in expression of CTR1 (Fig. 1C), which encodes the major high affinity copper importer (5), suggested that copper uptake into kha1Δ cells was at wild type levels. Nevertheless, overexpression of KHA1 partially rescued the growth defect of a strain lacking CTR1 but not one lacking the FTR1 iron importer (Fig. 1D). These results collectively suggest that Kha1p is involved in acquisition of iron and the utilization of copper in support of respiratory growth.
FIGURE 1.
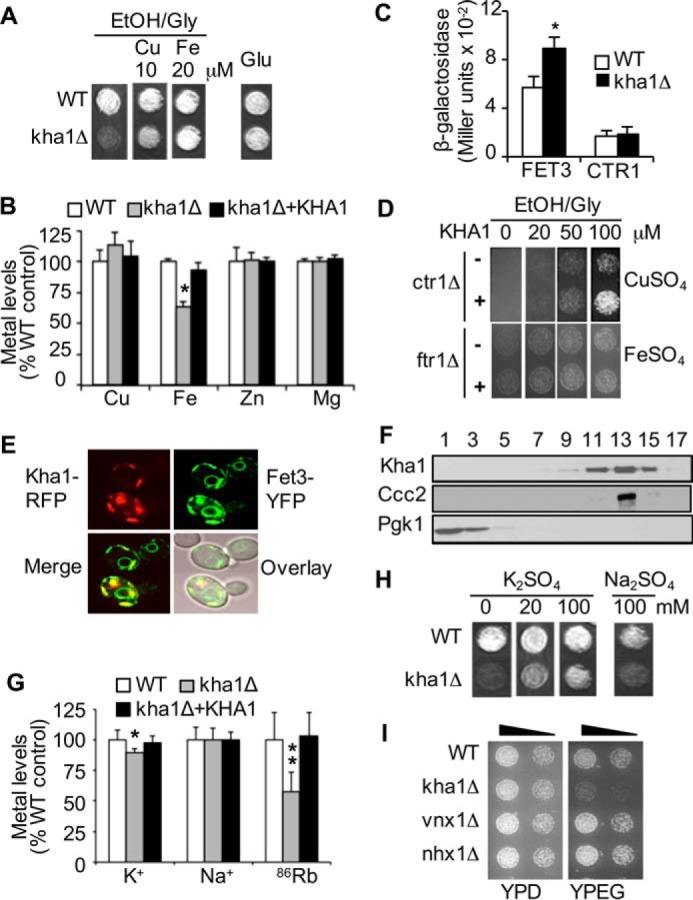
Kha1p is a new molecular factor involved in respiratory growth and homeostasis of copper, iron, and potassium. A, growth of WT control and kha1Δ strains on non-fermentable YPEG media (EtOH/Gly) containing ethanol and glycerol as carbon sources and fermentable media (Glu) containing glucose with and without CuSO4 or FeSO4 supplementation at the indicated concentration. B, cellular metal levels were measured, normalized to cell number, and presented as % of those in WT cells. C, WT and kha1Δ cells expressing iron or copper-responsive reporter (FET3-lacZ and CTR1-lacZ, respectively) were subjected to β-galactosidase assays. D, growth of ctr1Δ and ftr1Δ cells with and without KHA1 overexpression was monitored on EtOH/Gly media supplemented with CuSO4 or FeSO4. E, RFP-fused Kha1p (Kha1-RFP) and YFP-fused Fet3p (Fet3-YFP) were expressed in ftr1Δ cells to visualize Kha1p and Fet3p. F, subcellular fractionation of cells expressing Kha1-RFP and c-myc epitope tagged Ccc2p. Samples were subjected to Western blotting. G, cellular K+ and Na+ were measured using inductively coupled plasma mass spectrometry. Cells were permeabilized with saponin and incubated with 86Rb for 10 min. Endomembrane compartmentalized 86Rb was measured using a γ-counter. Metal levels were normalized to cell number. H, cell growth was monitored on the YPEG media supplemented with K2SO4 or Na2SO4. I, growth of yeast strains on YPEG and fermentable YPD media. All growth assays were conducted at least twice with two different clones. Average ± S.D. (n = 6) is presented. One (*) and two (**) asterisks indicate p < 0.05 and p < 0.01, respectively.
K+ Accumulation in the Secretory Pathway via Kha1p
KHA1 encodes a predicted monovalent cation (K+ and/or Na+) and H+ exchanger (46). Although this family of proteins has been identified from many organisms ranging from bacteria to humans (13, 15–16, 46), the physiological role of Kha1p has not been defined. Previous studies suggested that Kha1p might serve as a K+ efflux transporter (47), which localized to the Golgi (48, 49) or mitochondria (50). Cells expressing Kha1p fused with RFP (Kha1p-RFP) display fluorescence associated with vesicle-like compartments (Fig. 1E). We examined whether Kha1p co-localized with Fet3p and Ccc2p, two known molecular factors involved in copper metabolism. The majority of Fet3p fused with YFP (Fet3p-YFP) is detected at the cell surface in WT cells; however, Fet3p accumulates in the secretory pathway in cells lacking Ftr1p due the lack of Fet3p-Ftr1p complex formation (51). A significant overlap in subcellular distribution between Fet3p-YFP and Kha1p-RFP was observed in ftr1Δ cells (Fig. 1E). Localization of Kha1p-RFP in the secretory pathway was further confirmed by subcellular fractionation followed by Western blotting of Kha1p and Ccc2p fused with RFP and c-myc epitope, respectively (Fig. 1F). Kha1-RFP, Fet3-YFP, and Ccc2p-Myc are fully functional as determined by complementation of phenotypes of cells carrying deletions of the corresponding genes (data not shown).
KHA1 deletion decreased cellular K+ levels; in contrast, Na+ levels were not affected (Fig. 1G). Uptake into subcellular membrane-bound compartments of 86Rb+ as a tracer for K+ (42) was significantly reduced in kha1Δ cells relative to control WT cells (Fig. 1G). In addition, K+ but not Na+ rescued the respiratory deficiency of kha1Δ cells (Fig. 1H). These results collectively suggest that Kha1p plays a role in K+ accumulation in the secretory pathway. S. cerevisiae expresses other intracellular monovalent cation transporters, including Vnx1p and Nhx1p in the vacuole and late endosome, respectively (16); however, deletion of VNX1 and NHX1 did not impair respiratory growth (Fig. 1I).
pH of the Secretory Pathway and Fet3p Secretion Are Not Affected by KHA1 Gene Deletion
Given that Kha1p is a predicted K+-H+ exchanger, Kha1p deficiency might lead to changes in the physicochemical characteristics of the secretory pathway (e.g. pH, vesicle trafficking, and fusion). This could affect copper metabolism, Fet3p maturation, and/or cell surface expression of the Fet3p-Ftr1p iron uptake complex. To assess these possibilities, we examined the pH of the secretory pathway and cytosol using a pH-sensitive green fluorescent protein (GFP) as reporter (37) (Fig. 2A). In-frame fusion of this GFP to a non-functional Gef1p chloride channel delivered this reporter to the lumen of the TGN (38). Calibration curves for determining pH (Fig. 2B) were obtained as previously described (37, 38). The pH of the cytosol and TGN of the cells were found to be 7.00 ± 0.82 and 6.22 ± 0.67, respectively (Fig. 2C), both within the normal range (52). No pH difference between WT and kha1Δ cells was observed, suggesting that Kha1p is not a major factor in maintenance of the pH in the secretory compartments. In addition, there was no significant change in subcellular distribution of Fet3p in kha1Δ cells (Fig. 2D), suggesting that secretion of Fet3p was not altered. This data were supported also by subcellular fractionation of Fet3p (data not shown).
FIGURE 2.
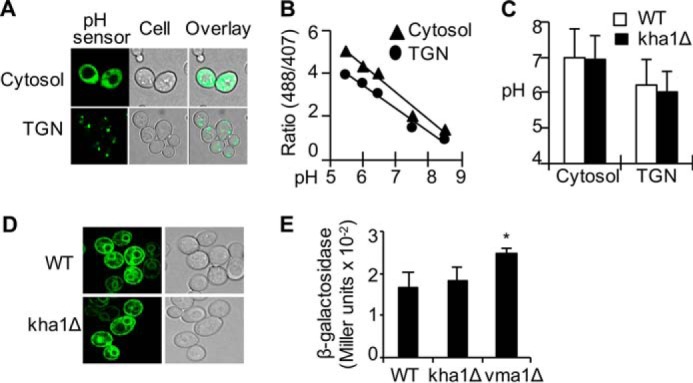
No significant change in the functions of the secretory pathway in Kha1p-dificient cells. A–C, measurement of pH of the Golgi vesicles and cytosol. Cells were transformed with a plasmid expressing a pH-sensitive fluorescent protein at the cytosol or at the lumen of the TGN. A, visualization of subcellular distribution of the pH sensors by fluorescent microscopy. B, a calibration curve reflecting the ratio of emission intensity at the indicated pH. Data represent the average of cells in A600 = 1. C, pH at the cytosolic or lumen of the TGN was determined by using the calibration curve. D, no difference in subcellular distribution of Fet3p fused with YFP (Fet3-YFP) in WT control and kha1Δ cells. E, KHA1 gene knock-out did not lead to unfolded protein response. WT, kha1Δ, and vma1Δ cells were transformed with a reporter plasmid of unfolded protein response. Cells cultured in SC media were subjected to β-galactosidase assays. Data represent the average ± S.D. (n = 4). The asterisk (*) indicates p < 0.05 compared with WT control cells.
We next examined whether in kha1Δ cells the secretory pathway exhibited stress characteristics. Protein folding or maturation defects lead to protein retention and a subsequent unfolded protein response (UPR) (53). Fet3p in ftr1Δ cells is a target of this quality control system (51). Deletion of subunit A of vacuolar H+-ATPase (vma1Δ) for instance induces UPR (54), which was confirmed by UPR reporter assays (Fig. 2E). The kha1Δ cells did not manifest UPR, however (Fig. 2E). These results indicate that kha1Δ cells do not display any defect in the basic function and physiochemical characteristics of the secretory pathway.
Kha1p Is Required for Expression of Active Fet3p
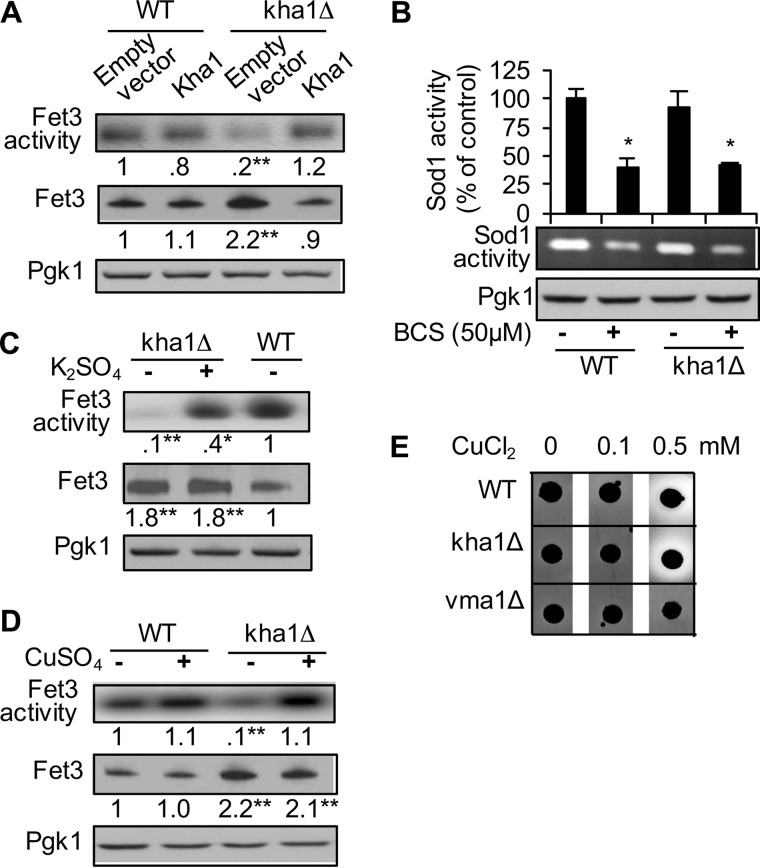
In line with iron deficiency (Fig. 1B) and the up-regulation of FET3 expression (Fig. 1C), Fet3p levels in kha1Δ cells were >2-fold higher than that in WT cells (Fig. 3A, middle panel, third lane). However, Fet3p oxidase activities were ∼20% that of the activity expressed by WT (Fig. 3A, upper panel, third lane). This indicated that most of the Fet3p in kha1Δ cells is nonfunctional. However, kha1Δ cells had normal activities of the cytosolic, copper-requiring enzyme, Sod1p (Fig. 3B). Surplus K+ (100 mm K2SO4) or copper (25 μm CuCl2) in the growth media resulted in dramatic recovery of Fet3p activity and returned expression levels to normal (Fig. 3, C, second lane, and D, fourth lane), suggesting the hypothesis that K+ deficiency in kha1Δ cells leads to inefficient copper incorporation into apoFet3p.
FIGURE 3.
Reduced Fet3p activity in kha1Δ cells and its recovery by surplus copper or K+ in the growth media. A, C, and D, chromosomal FET3 in WT control and kha1Δ cells was fused with c-myc epitope. Whole cell extracts were subjected to Fet3p oxidase assays and Western blotting using anti-myc antibodies. The numbers indicate relative oxidase activities (n = 4). The experiments were conducted without and with supplementation of K2SO4 (100 mm) (C) or CuSO4 (25 μm) (D) in the media for 30 min before collecting cells. B, KHA1 knock-out does not affect Sod1p activities. WT and kha1Δ strains were cultured at mid-log phase in YPD media with and without supplementation of copper chelator, BCS. Total cell lysates were subjected to in-gel Sod1p activity assays. E, enzymatic activities of a laccase of P. coccineus in the yeast S. cerevisiae. Representative figures of at least four colonies are presented. The white hollow around the growing cells reflects laccase activities. The data represent the average ± S.D. (n = 4). One (*) and two (**) asterisks indicate p < 0.05 and p < 0.01, respectively, compared with WT cells expressing empty vector or without K+ and copper co-culture.
Although >10 proteins in mammals acquire copper in the secretory pathway, Fet3p is the only known secreted cuproprotein in the S. cerevisiae. We thought to determine the specificity of K+ by expressing secretory cuproproteins of humans in kha1Δ yeast. However, neither active human superoxide dismutase 3 (hSOD3) nor ceruloplasmin was expressed in WT cells (data not shown). On the other hand, a gene encoding P. coccineus laccase was expressed in WT and kha1Δ cells. Copper supplementation to the growth media was necessary for visualization of laccase activities as has been reported (32). The color change around the cells growing on substrate-containing solid media reflected this activity (Fig. 3E). The similar size of the activity zone associated with WT cells in comparison to kha1Δ ones indicated that KHA1 gene deletion did not affect laccase maturation. As expected a strain lacking VMA1, which encodes a subunit of the V-type ATPase that plays a critical role secretory pathway pH regulation, manifested a defect in laccase maturation (Fig. 3E). These results suggest that Kha1p and K+ play specific roles in the maturation of Fet3p.
K+ Promotes Binding of Copper to ApoFet3p
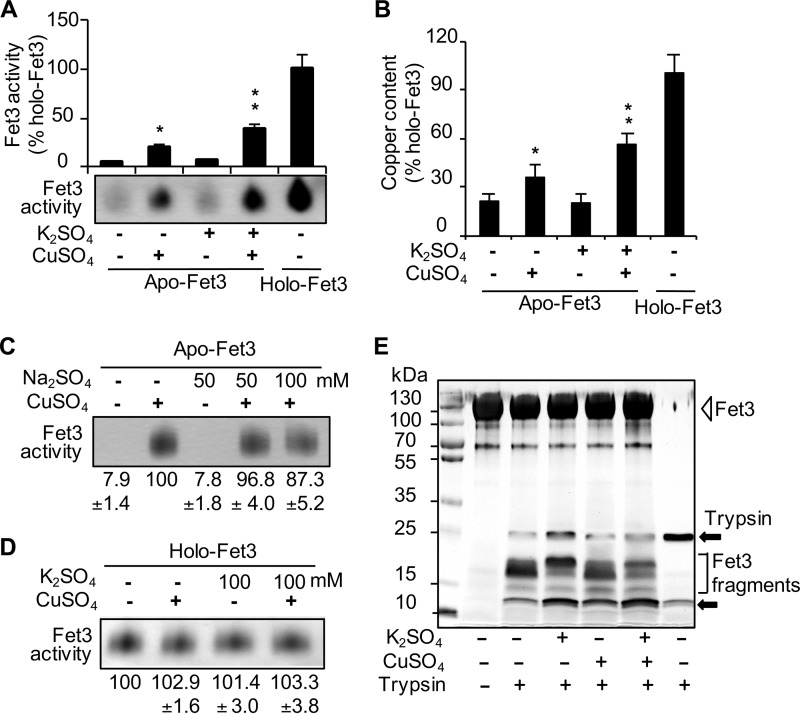
We hypothesized that K+ transported into the lumen of the secretory pathway may facilitate insertion of copper ions into apoFet3p. The catalytic domain of Fet3p was expressed in S. cerevisiae and purified (33). ApoFet3p was obtained by extracting copper ions from the purified Fet3p (34) to the level below one copper per Fet3p. Given that Fet3p contains four copper atoms (24), the apoFet3p samples displayed negligible oxidase activity (Fig. 4A, first lane). Co-incubation of purified apoFet3 with CuSO4 (5 μm, reduced to Cu+ by ascorbate) followed by oxidase assays showed that K+ is not essential for copper insertion into Fet3p under this experimental condition (Fig. 4A, second lane). K+ concentration in the cytosol is ∼150 mm. Given the fact that Kha1p serves to support a K+ compartmentalization, we surmise that the K+ concentration in the TGN should be similar or higher than it is in the cytoplasm. Although K+ up to 200 mm did not affect holo-Fet3p activity (Fig. 4D), K+ significantly enhanced the copper-dependent apoFet3p activation (Fig. 4A, fourth lane). Measurement of copper in Fet3p displayed an anticipated correlation between copper incorporation and oxidase activities of Fet3p (Fig. 4B). In contrast to the effect of K+, Na+ did not stimulate the copper-dependent Fet3p activation (Fig. 4C). The K+ effect was independent of pH (data not shown).
FIGURE 4.
K+ promotes binding of copper cofactors to apoFet3p. A, purified apo- and holo-Fet3p (5 μg) was incubated with or without K2SO4 (100 mm) and CuSO4 (5 μm). Fet3p oxidase activities were measured using an in-gel assay method. B, purified apo- and holo-Fet3p (120 μg) was processed as described in A, and loosely bound or unbound copper was removed by dialysis. Total copper content of Fet3p was measured. Single (*) and double (**) asterisks indicate p < 0.05 and p < 0.01, respectively, relative to the control condition (no CuSO4 and K2SO4). C, purified apoFet3p (5 μg) and CuSO4 (5 μm) were incubated with Na2SO4 at the indicated concentrations. Fet3p oxidase activities were measured. D, purified holo-Fet3p (5 μg) was incubated with and without K2SO4 at the indicated concentrations and CuSO4 (5 μm). Fet3p oxidase activities were measured. E, apoFet3p co-incubated with K2SO4 (100 mm) and CuSO4 (5 μm) was subjected to limited trypsin proteolysis. Samples were separated by SDS-PAGE and visualized by silver staining. Data represent the average ± S.D. and/or representative figure of at least three independent experiments.
We thought that Fet3p might require K+ for its activity similar to other K+-containing enzymes (14). However, the crystal structure of Fet3p gives no evidence of stably bound K+ (55). Also, no significant level of K+ could be detected in purified holo-Fet3p by inductively coupled plasma mass spectrometry (data not shown). Similarly, gel filtration of mixtures of Fet3p and K+ as in Fig. 4A could not detect K+ in the fractions containing Fet3p (data not shown). These results collectively indicate that K+ does not stably bind to Fet3p nor serve as a cofactor for Fet3p activity.
We hypothesized that K+ might affect the conformation of apoFet3p in a manner that promoted copper binding. To address this, purified apoFet3p was subjected to partial trypsin proteolysis after incubating with and without K+ and/or copper. SDS-PAGE of the samples followed by silver staining showed different Fet3p fragmentation patterns when the protein was co-incubated with K+ (Fig. 4E, third lane). However, copper without K+ did not appear to have a significant effect (Fig. 4E, fourth lane). The fragmentation patterns in the presence of both K+ and copper (Fig. 4E, fifth lane) likely reflect maturation of a portion of apoFet3 to the holo-form as shown by activity assay (Fig. 4A). The same experiments conducted for GST (data not shown) indicated that the K+ effects observed in Fet3p are specific, and K+ does not change trypsin activities. Therefore, although K+ is neither stably associated with Fet3p nor required for holo-Fet3p activities, K+ appears to promote incorporation of copper into apoFet3p by functioning as a structural modifier through a transient and/or weak interaction(s).
Site-directed Substitution of Specific Residues in Fet3p Abrogates the K+ Effect
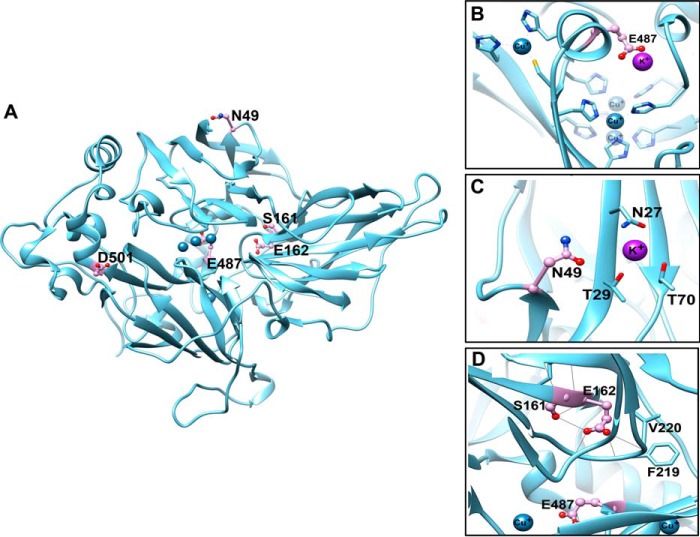
Although our data did not support stable binding of K+ to Fet3p, we predicted potential K+ sites in Fet3p (40) and modeled K+ binding based on the Fet3p structure (PDB code 1ZPU) (41) (Fig. 5).
FIGURE 5.
Prediction of K+ binding sites in Fet3p. Fet3p (PDB code 1ZPU) residues predicted to be involved in K+ binding are shown in ball and stick representation. Carbon atoms are colored in pink, nitrogen atoms are in dark blue, and oxygen atoms are in red. Bound copper atoms are represented as light blue spheres. The figures were generated using UCSF Chimera software. A, four predicted K+ sites, containing Glu-487, Asn-49, Asp-501, and Ser-161/Glu-662, are indicated in the catalytic domain of Fet3p. The K+ coordination sites containing Glu-487 (B), Asn-49 (C), or Ser-161/Glu-662 (D) are enlarged. The solid gray lines (D) indicate hydrogen bond networks.
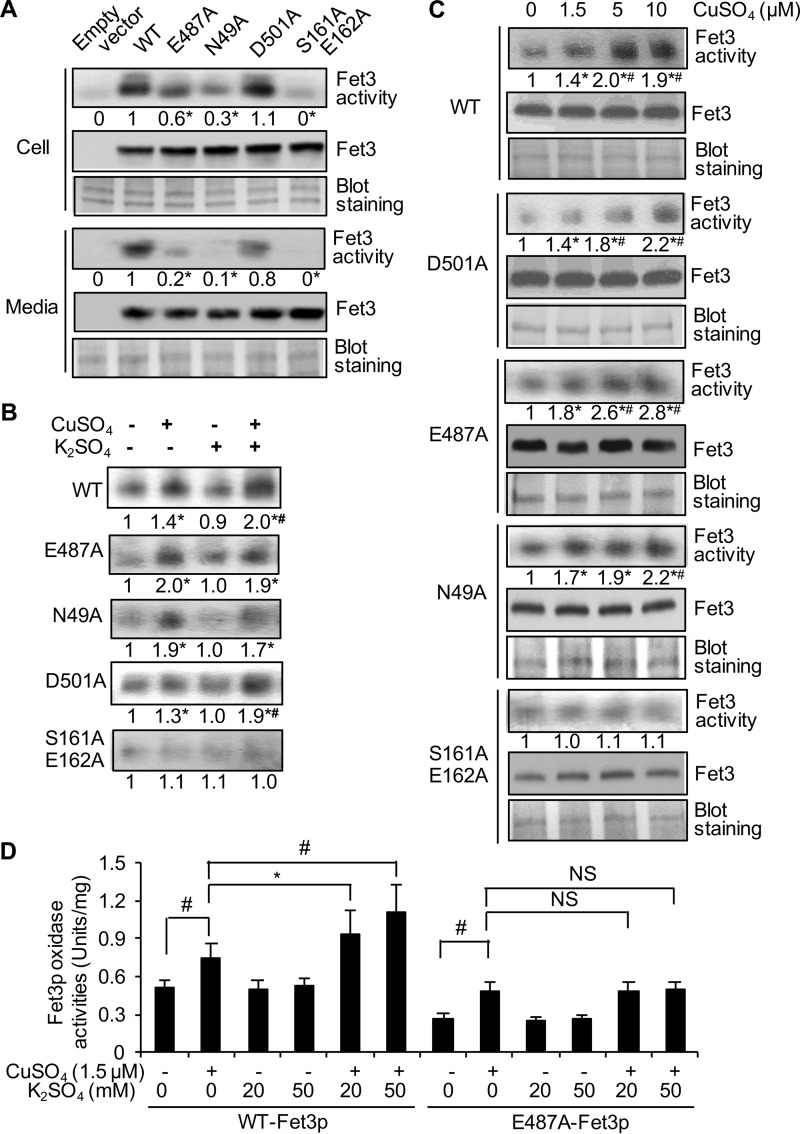
Site-directed substitution of residues Glu-487, Asn-49, Asp-501, and Ser-161/Glu-162 of each potential K+ site revealed that substitution of Glu-487 or Asn-49 with alanine (Ala) abolished the K+ effect on Fet3p maturation (Fig. 6). Expression levels of Fet3p possessing the mutation(s) were comparable with those of wild type control; however, all except Fet3p(D501A) showed significantly lower oxidase activities (Fig. 6A). In vitro maturation assays using secreted Fet3p showed further activation of wild-type Fet3p in a copper- and K+-dependent manner, indicating that a significant portion of Fet3p is in the apo-form (Fig. 6B, top panel). Copper activated Fet3p(E487A) and Fet3p(N49A) (Fig. 6, B, second and third panels, C, and D); however, K+ did not promote copper-induced activation of these Fet3p mutants (Fig. 6, B, second and third panels, and D). This suggests that Glu-487 and Asn-49 are important residues for K+-dependent Fet3p maturation. Glu-487 is the outer sphere to the trinuclear copper cluster (Fig. 5, A and B) and donates a proton during O—O bond cleavage in the enzyme's reduction of dioxygen to water (24). Asn-49 is at the surface (Fig. 5A). Fet3p(D501A) showed a similar pattern to that of WT (Fig. 6, A–C). Fet3p(S161A/E162A) displayed near non-detectable activity under all experimental conditions (Fig. 6, A–C), indicating inactivation by the mutations. These two residues are involved in an extensive hydrogen bond network (Fig. 5D). The substitutions may disrupt the local structure. Collectively, these results suggest that specific residues of Fet3p mediate the K+ effects on facilitating copper binding to Fet3p.
FIGURE 6.
Specific residues in Fet3p are responsible for K+ effects on biogenesis of functional Fet3p. The catalytic domain of Fet3p tagged with a flag epitope was constitutively expressed in fet3Δ yeast cells. Glu-487, Asn-49, Asp-501, or Ser-161/Glu-162 were substituted to alanine (Ala). A, total cell lysates (Cell) and secreted Fet3p (Media) were subjected to in-gel Fet3p oxidase assays and Western blotting using anti-FLAG antibodies. Blots were stained to determine equal loading. Signal intensities reflecting Fet3p oxidase activities were normalized to Fet3p expression levels and then presented as relative levels of WT control. B, equal amounts of culture media containing secreted Fet3p were incubated with CuSO4 (1.5 μm) and/or K2SO4 (50 mm) to induce Fet3p maturation in vitro. C, equal amounts of collected culture media were incubated with CuSO4 to induce holo-Fet3p formation in vitro. Fet3p oxidase activities were normalized to Fet3p protein levels and then presented as relative activities of those without copper addition. The numbers underneath the figures (average of four experiments) indicate the relative levels of Fet3p oxidase activities (A–C). The asterisks (*) indicate p < 0.05 relative to the result of WT (A) or control (no copper and/or K+) (B and C). Pound symbol (#) indicate p < 0.05 relative to samples co-incubated with 1.5 μm CuSO4 (B) or samples containing copper without K+ (C) (second lane of each panel). D, oxidase activities of the catalytic domain of WT control and E487A Fet3p in cell lysates were measured using a spectrophotometry method. Background activities for p-phenylenediamine oxidation in cells expressing an empty vector were subtracted. The average ± S.D. (n = 4) is presented. Asterisks (*), pounds symbols (#), and NS indicate p < 0.05, p < 0.01, and no significant difference, respectively.
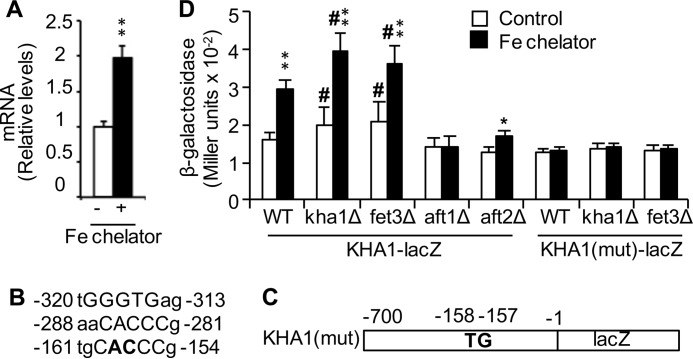
Iron Deficiency Up-regulates KHA1 Expression
Given the role of Kha1p in maturation of Fet3p, iron deficiency might induce KHA1 expression to promote Fet3p maturation followed by iron uptake. Aft1p and Aft2p are major transcription factors involved in expression control of genes involved in iron acquisition, including FET3 (29, 56). Indeed, KHA1 mRNA levels are higher in cells co-cultured with an iron chelator (Fig. 7A). Its promoter region (within −700 bp) contains three predicted Aft1/2p response elements (Fig. 7B) similar to those found in the promoters of other iron-regulated genes (29). Construction of β-galactosidase reporters with and without site-directed mutation of AC(−158 and −157) to TG (Fig. 7C) followed by reporter assays showed up-regulation of KHA1 in iron deficiency induced by growth in the presence of an iron chelator or deletion of either KHA1 or FET3 (Fig. 7D). This iron-dependent induction of KHA1 was dependent on Aft1p and the Aft1/2p response elements (Fig. 7D). KHA1 gene induction is severely compromised (∼15% of WT cells) in the absence of Aft2p (Fig. 7D). These results are consistent with the conclusion that KHA1 is a target of Aft1/2p and support the hypothesis that Kha1p-mediated K+ compartmentalization is a component of iron homeostasis in yeast.
FIGURE 7.
Iron deficiency up-regulated KHA1 expression. A, WT control cells cultured with (+) and without (−) iron (Fe) chelator BPS in the SC media were subjected to quantitation of KHA1 mRNA. B, three predicted Aft1p and Aft2p response elements at the promoter (within −700 bp). GGGTG core sequences are indicated along with one 5′ and two 3′ nucleotides. C, schematic presentation of lacZ reporter of KHA1 with site-directed mutations of AC (−158 and −157) to TG. D, The indicated strains expressing KHA1-lacZ reporter were cultured with or without BPS iron chelator and subjected to β-galactosidase assays. The average ± S.D. (n = 4) is presented. Single (*) and double (**) asterisks indicate p < 0.05 and p < 0.01, respectively, relative to the results of the same strain cultured in control media (no iron chelator). The pound symbol (#) indicates p < 0.05 relative to WT strain cultured in the same media.
Discussion
Identification of a K+/H+ exchanger as a molecular factor involved in respiratory growth of yeast and characterization of its function and regulation provides new insights into the mechanisms underlying iron acquisition. K+-promoted activation of the Fet3p ferroxidase by copper illustrates a previously unrecognized role of K+ and its organellar transporter in the maturation of this enzyme, which is essential to high affinity iron uptake in yeast.
Alkali cations and metal ions affect protein function and stability through site-specific binding (14, 34). However, our elemental analysis of purified holo-Fet3p did not detect stably bound K+, which is consistent with a previous report on crystal structure of Fet3p (55). K+ did not affect the oxidase activities of holo-Fet3p either. Nevertheless, apoFet3p manifests conformational differences in the presence of K+ as determined by limited trypsin proteolysis, and specific residues in Fet3p mediate the effect of K+ on copper activation of apoFet3p. In this regard, the roles of cellular cations in the folding and dynamics of nucleic acids is likely relevant (17–19). Because DNA and RNA are negatively charged polyelectrolytes, cations function as counter ions to reduce the electrostatic repulsion that might interfere with their folding. The possibility of the similar effects of ions on proteins has not yet been ascertained. Fet3p possesses Glu and Asp residues 8.0% and 5.5%, respectively; thus, Fet3p is not particularly abundant in negatively charged residues. However, the copper sites in Fet3p are found among clusters with negative charged residues, including Asp-94, Glu-185, Asp-409, Asp-458, and Glu-487, along with copper binding His residues (24). K+ may preset the copper sites for copper binding via electrostatic interactions. Determination of K+-induced conformational dynamics of Fet3p would shed new light on K+-protein interactions and the roles for the identified residues in maturation and activities of Fet3p.
Our study reveals a novel role of subcellular K+ compartmentalization in Fet3p maturation; this mechanism adds to the previously reported correlation between the proton (H+) gradient in the secretory pathway and iron uptake in yeast. Thus, the V-type ATPase and 2Cl−/1H+ antiporters (Gef1p in yeast, and CLC4 in mammals) are required for pH control of the secretory pathway and iron uptake as well (38, 57–60). The acidic pH and H+ gradient of the secretory compartment(s) is critical for supporting the functions of this organelle (54, 61). The positive electrical potential across the membrane, which is generated by H+ accumulation due to V-type ATPase function, could be neutralized by Cl− transported by a 2Cl−/1H+ exchanger. The significance of organellar acidification and HCl formation has been linked to diverse cellular processes, including iron homeostasis, vesicular trafficking, protein degradation, and bone resorption (61–63). This H+ gradient is required for Kha1p-mediated K+ transport into the lumen of the TGN via the K+/H+ exchange mechanism. Therefore, perturbation of iron metabolism in cells lacking V-type ATPase or Gef1p chloride channel could be attributed, at least partially, to the failure of K+ transport into the TGN.
In addition to Kha1p, yeast expresses two other organellar transporters of monovalent cations (K+ and/or Na+). Nhx1p plays a role in the regulation of cellular pH, vesicle trafficking, and vacuole fusion (46). Vnx1p also is involved in the regulation of cellular pH (16). Despite the similar modes of action and functions of these transporters, our results indicate that Kha1p does not affect secretory pathway pH, nor the pH of the cytosol. However, Kha1p function relies on a proton gradient across the secretory compartment membrane. The specific role for Kha1p-mediated compartmentalization of K+ in iron homeostasis is further supported by the regulation of KHA1 expression via iron-specific transcription regulators. Thus, Kha1p could be a unique member of this protein family that might be conserved in other organisms. Identification and characterization of the counterpart(s) of Kha1p in humans, animals, and plants is ongoing in the hope of gaining a better understanding of iron, copper, and K+ homeostasis and their functional interaction(s) in higher eukaryotes.
Our data provide no evidence in support of the hypothesis that K+ promotes the binding of copper to other copper-containing enzymes. It is worth emphasizing that iron deficiency induces KHA1 expression via iron-responsive transcription regulators. The system for K+ compartmentalization to the secretory pathway, therefore, could be evolved specifically for efficient acquisition of iron. No significant growth defect of kha1Δ cells under conditions of surplus media iron also supports this argument. Nevertheless, further experiments could reveal possible K+ effects on the maturation of proteins in other organisms.
Author Contributions
X. W. and J. L. designed the experiments. X. W., H. K., J. S., J. J. B., D. W. G., W. H. J., and D. J. K. conducted the experiments. X. W., P. J. H., E. D. C., D. J. K., and J. L. interpreted the data. X. W., D. J. K., and J. L. wrote the paper.
Acknowledgments
Plasmids containing pH-sensitive GFP, FET3 and CTR1-lacZ reporters, and UPR reporter, an expression construct of a laccase gene of P. coccineus, were kindly provided by Drs. Blanche Schwappach, Dennis Winge, Peter Walter, and Hisashi Hoshida, respectively. We thank Danielle Shea for FACS analysis. We also thank Nathan Smith and Eric Shuman for technical assistance and Matt O'Dell and Sunho Lee for editing this manuscript.
This work was supported by National Institutes of Health Grants DK79209 (to J. L.), HL49413, HL73813, and HL112303 (to E. D. C.), and P30GM103335 (to the Nebraska Redox Biology Center). This work was also supported by the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (A-600115007202; to X. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- TGN
- trans-Golgi network
- SC
- synthetic complete
- BCS
- bathocuproine disulfonate
- BPS
- bathophenanthroline disulfonate
- RFP
- red fluorescent protein
- UPR
- unfolded protein response.
References
- 1. Cyert M. S., and Philpott C. C. (2013) Regulation of cation balance in Saccharomyces cerevisiae. Genetics 193, 677–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waldron K. J., Rutherford J. C., Ford D., and Robinson N. J. (2009) Metalloproteins and metal sensing. Nature 460, 823–830 [DOI] [PubMed] [Google Scholar]
- 3. Robinson N. J., and Winge D. R. (2010) Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Culotta V. C., Yang M., and O'Halloran T. V. (2006) Activation of superoxide dismutases: putting the metal to the pedal. Biochim. Biophys. Acta 1763, 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim H., Wu X., and Lee J. (2013) SLC31 (CTR) family of copper transporters in health and disease. Mol. Aspects Med. 34, 561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Youn J. H., and McDonough A. A. (2009) Recent advances in understanding integrative control of potassium homeostasis. Annu. Rev. Physiol. 71, 381–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Madsen E., and Gitlin J. D. (2007) Copper and iron disorders of the brain. Annu. Rev. Neurosci. 30, 317–337 [DOI] [PubMed] [Google Scholar]
- 8. Xu W., Barrientos T., and Andrews N. C. (2013) Iron and copper in mitochondrial diseases. Cell Metab. 17, 319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Theil E. C. (2011) Iron homeostasis and nutritional iron deficiency. J. Nutr. 141, 724S–728S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gupta A., and Lutsenko S. (2009) Human copper transporters: mechanism, role in human diseases and therapeutic potential. Future Med. Chem. 1, 1125–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barnham K. J., and Bush A. I. (2008) Metals in Alzheimer's and Parkinson's diseases. Curr. Opin. Chem. Biol. 12, 222–228 [DOI] [PubMed] [Google Scholar]
- 12. Lin S. H., and Halperin M. L. (2007) Hypokalemia: a practical approach to diagnosis and its genetic basis. Curr. Med. Chem. 14, 1551–1565 [DOI] [PubMed] [Google Scholar]
- 13. Brett C. L., Donowitz M., and Rao R. (2005) Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288, C223–C239 [DOI] [PubMed] [Google Scholar]
- 14. Page M. J., and Di Cera E. (2006) Role of Na+ and K+ in enzyme function. Physiol. Rev. 86, 1049–1092 [DOI] [PubMed] [Google Scholar]
- 15. Orlowski J., and Grinstein S. (2007) Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr. Opin. Cell Biol. 19, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ariño J., Ramos J., and Sychrová H. (2010) Alkali metal cation transport and homeostasis in yeasts. Microbiol. Mol. Biol. Rev. 74, 95–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lipfert J., Doniach S., Das R., and Herschlag D. (2014) Understanding nucleic acid-ion interactions. Annu. Rev. Biochem. 83, 813–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wong G. C., and Pollack L. (2010) Electrostatics of strongly charged biological polymers: ion-mediated interactions and self-organization in nucleic acids and proteins. Annu. Rev. Phys. Chem. 61, 171–189 [DOI] [PubMed] [Google Scholar]
- 19. Chen S. J. (2008) RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu. Rev. Biophys. 37, 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lill R., and Mühlenhoff U. (2008) Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 21. Kim H. J., Khalimonchuk O., Smith P. M., and Winge D. R. (2012) Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta 1823, 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maio N., and Rouault T. A. (2015) Iron-sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery. Biochim. Biophys. Acta 1853, 1493–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collins J. F., Prohaska J. R., and Knutson M. D. (2010) Metabolic crossroads of iron and copper. Nutr. Rev. 68, 133–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kosman D. J. (2010) Multicopper oxidases: a workshop on copper coordination chemistry, electron transfer, and metallophysiology. J. Biol. Inorg. Chem. 15, 15–28 [DOI] [PubMed] [Google Scholar]
- 25. Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., and Dow S. W. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- 26. Mumberg D., Müller R., and Funk M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 27. Longtine M. S., McKenzie A. 3rd., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 28. Guarente L. (1983) Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101, 181–191 [DOI] [PubMed] [Google Scholar]
- 29. Rutherford J. C., Jaron S., and Winge D. R. (2003) Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278, 27636–27643 [DOI] [PubMed] [Google Scholar]
- 30. Jensen L. T., Posewitz M. C., Srinivasan C., and Winge D. R. (1998) Mapping of the DNA binding domain of the copper-responsive transcription factor Mac1 from Saccharomyces cerevisiae. J. Biol. Chem. 273, 23805–23811 [DOI] [PubMed] [Google Scholar]
- 31. Cox J. S., and Walter P. (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404 [DOI] [PubMed] [Google Scholar]
- 32. Hoshida H., Fujita T., Murata K., Kubo K., and Akada R. (2005) Copper-dependent production of a Pycnoporus coccineus extracellular laccase in Aspergillus oryzae and Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 69, 1090–1097 [DOI] [PubMed] [Google Scholar]
- 33. Hassett R. F., Yuan D. S., and Kosman D. J. (1998) Spectral and kinetic properties of the Fet3 protein from Saccharomyces cerevisiae, a multinuclear copper ferroxidase enzyme. J. Biol. Chem. 273, 23274–23282 [DOI] [PubMed] [Google Scholar]
- 34. Sedlák E., Ziegler L., Kosman D. J., and Wittung-Stafshede P. (2008) In vitro unfolding of yeast multicopper oxidase Fet3p variants reveals unique role of each metal site. Proc. Natl. Acad. Sci. U.S.A. 105, 19258–19263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Askwith C. C., and Kaplan J. (1998) Site-directed mutagenesis of the yeast multicopper oxidase Fet3p. J. Biol. Chem. 273, 22415–22419 [DOI] [PubMed] [Google Scholar]
- 36. de Silva D., Davis-Kaplan S., Fergestad J., and Kaplan J. (1997) Purification and characterization of Fet3 protein, a yeast homologue of ceruloplasmin. J. Biol. Chem. 272, 14208–14213 [DOI] [PubMed] [Google Scholar]
- 37. Miesenböck G., De Angelis D. A., and Rothman J. E. (1998) Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195 [DOI] [PubMed] [Google Scholar]
- 38. Braun N. A., Morgan B., Dick T. P., and Schwappach B. (2010) The yeast CLC protein counteracts vesicular acidification during iron starvation. J. Cell Sci. 123, 2342–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu X., Sinani D., Kim H., and Lee J. (2009) Copper transport activity of yeast Ctr1 is down-regulated via its C terminus in response to excess copper. J. Biol. Chem. 284, 4112–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nayal M., and Di Cera E. (1994) Predicting Ca2+-binding sites in proteins. Proc. Natl. Acad. Sci. U.S.A. 91, 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 42. Numata M., and Orlowski J. (2001) Molecular cloning and characterization of a novel (Na+,K+)/H+ exchanger localized to the trans-Golgi network. J. Biol. Chem. 276, 17387–17394 [DOI] [PubMed] [Google Scholar]
- 43. Lynch S. M., and Frei B. (1995) Reduction of copper, but not iron, by human low density lipoprotein (LDL). Implications for metal ion-dependent oxidative modification of LDL. J. Biol. Chem. 270, 5158–5163 [DOI] [PubMed] [Google Scholar]
- 44. Rose M. D., Winston F., and Hieter P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, pp. 155–159, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 45. Flohé L., and Otting F. (1984) Superoxide dismutase assays. Methods Enzymol. 105, 93–104 [DOI] [PubMed] [Google Scholar]
- 46. Resch C. T., Winogrodzki J. L., Häse C. C., and Dibrov P. (2011) Insights into the biochemistry of the ubiquitous NhaP family of cation/H+ antiporters. Biochem. Cell Biol. 89, 130–137 [DOI] [PubMed] [Google Scholar]
- 47. Ramírez J., Ramírez O., Saldaña C., Coria R., and Peña A. (1998) A Saccharomyces cerevisiae mutant lacking a K+/H+ exchanger. J. Bacteriol. 180, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Flis K., Hinzpeter A., Edelman A., and Kurlandzka A. (2005) The functioning of mammalian ClC-2 chloride channel in Saccharomyces cerevisiae cells requires an increased level of Kha1p. Biochem. J. 390, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maresova L., and Sychrova H. (2005) Physiological characterization of Saccharomyces cerevisiae kha1 deletion mutants. Mol. Microbiol. 55, 588–600 [DOI] [PubMed] [Google Scholar]
- 50. Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H. E., Schönfisch B., Perschil I., Chacinska A., Guiard B., Rehling P., Pfanner N., and Meisinger C. (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U.S.A. 100, 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sato M., Sato K., and Nakano A. (2004) Endoplasmic reticulum quality control of unassembled iron transporter depends on Rer1p-mediated retrieval from the golgi. Mol. Biol. Cell 15, 1417–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Casey J. R., Grinstein S., and Orlowski J. (2010) Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11, 50–61 [DOI] [PubMed] [Google Scholar]
- 53. Schröder M., and Kaufman R. J. (2005) The mammalian unfolded protein response. Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 54. Kane P. M. (2006) The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70, 177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Taylor A. B., Stoj C. S., Ziegler L., Kosman D. J., and Hart P. J. (2005) The copper-iron connection in biology: structure of the metallo-oxidase Fet3p. Proc. Natl. Acad. Sci. U.S.A. 102, 15459–15464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamaguchi-Iwai Y., Stearman R., Dancis A., and Klausner R. D. (1996) Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15, 3377–3384 [PMC free article] [PubMed] [Google Scholar]
- 57. Davis-Kaplan S. R., Ward D. M., Shiflett S. L., and Kaplan J. (2004) Genome-wide analysis of iron-dependent growth reveals a novel yeast gene required for vacuolar acidification. J. Biol. Chem. 279, 4322–4329 [DOI] [PubMed] [Google Scholar]
- 58. Davis-Kaplan S. R., Askwith C. C., Bengtzen A. C., Radisky D., and Kaplan J. (1998) Chloride is an allosteric effector of copper assembly for the yeast multicopper oxidase Fet3p: an unexpected role for intracellular chloride channels. Proc. Natl. Acad. Sci. U.S.A. 95, 13641–13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gaxiola R. A., Yuan D. S., Klausner R. D., and Fink G. R. (1998) The yeast CLC chloride channel functions in cation homeostasis. Proc. Natl. Acad. Sci. U.S.A. 95, 4046–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang T., and Weinman S. A. (2004) Involvement of chloride channels in hepatic copper metabolism: ClC-4 promotes copper incorporation into ceruloplasmin. Gastroenterology 126, 1157–1166 [DOI] [PubMed] [Google Scholar]
- 61. Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 62. Diab H. I., and Kane P. M. (2013) Loss of vacuolar H+-ATPase (V-ATPase) activity in yeast generates an iron deprivation signal that is moderated by induction of the peroxiredoxin TSA2. J. Biol. Chem. 288, 11366–11377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Supanchart C., and Kornak U. (2008) Ion channels and transporters in osteoclasts. Arch. Biochem. Biophys. 473, 161–165 [DOI] [PubMed] [Google Scholar]