Abstract
Peptidylglycine α-amidating monooxygenase (PAM) (EC 1.14.17.3) catalyzes peptide amidation, a crucial post-translational modification, through the sequential actions of its monooxygenase (peptidylglycine α-hydroxylating monooxygenase) and lyase (peptidyl-α-hydroxyglycine α-amidating lyase (PAL)) domains. Alternative splicing generates two different regions that connect the protease-resistant catalytic domains. Inclusion of exon 16 introduces a pair of Lys residues, providing a site for controlled endoproteolytic cleavage of PAM and the separation of soluble peptidylglycine α-hydroxylating monooxygenase from membrane-associated PAL. Exon 16 also includes two O-glycosylation sites. PAM-1 lacking both glycosylation sites (PAM-1/OSX; where OSX is O-glycan-depleted mutant of PAM-1) was stably expressed in AtT-20 corticotrope tumor cells. In PAM-1/OSX, a cleavage site for furin-like convertases was exposed, generating a shorter form of membrane-associated PAL. The endocytic trafficking of PAM-1/OSX differed dramatically from that of PAM-1. A soluble fragment of the cytosolic domain of PAM-1 was produced in the endocytic pathway and entered the nucleus; very little soluble fragment of the cytosolic domain was produced from PAM-1/OSX. Internalized PAM-1/OSX was rapidly degraded; unlike PAM-1, very little internalized PAM-1/OSX was detected in multivesicular bodies. Blue native PAGE analysis identified high molecular weight complexes containing PAM-1; the ability of PAM-1/OSX to form similar complexes was markedly diminished. By promoting the formation of high molecular weight complexes, O-glycans may facilitate the recycling of PAM-1 through the endocytic compartment.
Keywords: electron microscopy (EM), furin, lysosome, neuropeptide, protein secretion, AtT-20 cell, multivesicular body
Introduction
Many of the enzymes involved in prohormone post-translational processing are subjected to both glycosylation and endoproteolytic cleavage as they travel through the secretory pathway. Peptidylglycine α-amidating monooxygenase (PAM3; EC 1.14.17.3), which catalyzes a late step in the generation of bioactive peptides (1), is a good example of this. Tissue-specific and developmentally regulated alternative splicing (1) generates integral membrane isoforms that differ only by the presence of a longer non-catalytic region between its two catalytic domains, peptidylglycine α-hydroxylating monooxygenase (PHM) and peptidyl-α-hydroxyglycine α-amidating lyase (PAL) (2, 3). This additional region, which is encoded by a single exon (exon 16), includes a prohormone convertase cleavage site (Lys436–Lys437) and multiple O-glycosylation sites (4–6).
Differences observed between the behavior of PAM-1, which includes exon 16 and is both O-glycosylated and cleaved, and PAM-2, which does not include exon 16 and is neither O-glycosylated nor extensively cleaved, encouraged us to explore the contribution of O-sugars to PAM-1 cleavage, trafficking, and transmembrane signaling. The single N-glycosylation site in PAM is glycosylated, but its elimination had no effect on PAM localization or function (7). Although PHM and PAL act sequentially, there is no evidence that their presence in the same protein facilitates the conversion of peptidylglycine substrates into amidated products (1).
Prohormone convertase-mediated cleavages regulate many fundamental biological pathways, and O-glycans in or immediately adjacent to cleavage sites affect the ability of these subtilisin-like endoproteases to recognize and cleave their substrates (8). Recent studies identified site-specific GalNAc-type O-glycosylation as an important regulator of prohormone convertase cleavage. When Tango1, a transport and Golgi organization protein required for protein secretion, cannot be O-glycosylated, it is subjected to furin-mediated proteolysis, and apical secretion and secretory vesicle formation are disrupted (9). The cleavage and inactivation of fibroblast growth factor 23 can be blocked by O-glycosylation of a Thr located in the protease recognition motif; phosphorylation of a nearby Ser prevents O-glycosylation at this site, allowing growth factor degradation (10). Cleavage of human MUC1 by immunoproteasomes is largely blocked when O-GalNAc is added near the preferred cleavage sites (11).
Glycosylation is one of the most abundant protein modifications (12, 13). Although N-glycosylation occurs at predictable sites (-Asn-Xaa-Ser/Thr-, where Xaa is not Pro) and must be initiated in the endoplasmic reticulum, O-glycosylation sites cannot be predicted based on primary sequence (14, 15). Based on earlier studies using [3H]galactose, [35SO42−Met], and [35S]Met, sulfated galactose-containing O-linked sugars are found on at least two Ser/Thr residues in the exon 16 region of PAM-1 (3, 15). A prohormone convertase 1-mediated cleavage at the Lys-Lys site in exon 16 separates soluble PHM from membrane PAL, and at least one O-glycosylation site is found in each product.
Mucin-type O-glycosylation is initiated by a family of 20 UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases (GalNAc-transferases) (EC. 2.4.1.41). Members of this highly conserved family catalyze the addition of monosaccharides to the side chains of Ser and Thr residues in target proteins (16). These O-linked oligosaccharides are generally short, often containing only one to four sugar residues; the O-linked oligosaccharides identified in PAM-1 share these properties (3). O-Glycosylation governs many aspects of cellular homeostasis, including vascular integrity (17), notch signaling (12), and intestinal homeostasis (18). Many congenital diseases have been attributed to deficits in O-glycosylation (19, 20), including autoimmune disorders (21), heterotaxy (22), hyperphosphatemia (23, 24), nephropathy, and breast adenocarcinoma (25).
Materials and Methods
Sample Generation and Identification of O-Glycosylation Sites in PAM
A stably transfected AtT-20 cell line secreting rat PAM-4 (rPAM-4) (1) was grown in T-75 flasks. Spent medium was collected daily and stored frozen after addition of protease inhibitors (30 μg/ml phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin, 50 μg/ml lima bean trypsin inhibitor, 2 μg/ml pepstatin, and 16 μg/ml benzamidine). Secreted proteins were concentrated by precipitation with 60% (NH4)2SO4. Pellets were resuspended and equilibrated with 20 mm NaTES, pH 7.0. After SDS-PAGE (Bio-Rad Criterion TGX gels, 4–15% gradient) and visualization with SilverSNAP (Pierce), rPAM-4 was excised and processed for mass spectroscopy as described (26). Trypsin digestion was carried out overnight at 37 °C; for LC MS/MS, peptides were separated using an RP C18 nanoACQUITY UPLC column with a Symmetry C18 trap column (27). Mass spectral data were acquired in the LTQ Orbitrap XL as described. Peaks targeted for MS/MS fragmentation by collision-induced dissociation were first isolated with a 2-Da window followed by normalized collision energy of 35%. Resulting LC MS/MS data were analyzed and searched utilizingMASCOT distiller and search engine (version 2.3). Manual analyses and verification of peptide masses were further carried out for additional molecular masses corresponding to the carbohydrate attachment.
Generation and Expression of Mutant PAM-1 and PAM-4
Mass spectroscopy identified three potential O-glycosylation sites in PAM-4; site-directed mutagenesis using the Stratagene QuikChange protocol (La Jolla, CA) was used to construct expression vectors encoding mutant PAM-4 proteins lacking one site: T417A, S442A, and S471G/T472A. The DNA sequence of each vector was verified. AtT-20 and pEAK Rapid cells (Edge Biosystems, Gaithersburg, MD) were fed with serum-free medium 2 h before transfection and were transiently transfected using Lipofectamine 2000 (Invitrogen) in Opti-MEM (Life Technologies, Inc.). Spent medium was collected after 24–48 h, and cells were harvested into 20 mm NaTES, 10 mm mannitol, 1% Triton X-100, pH 7.4 (TMT), for Western blot analysis.
Based on our analysis of the PAM-4 mutants, a vector encoding PAM-1 that lacked both O-glycosylation sites in exon 16 (pCI-Neo-KrPAM-1/OSX; where OSX is O-glycan-depleted mutant of PAM-1) was created by introducing the T417A and the S471G/T472A mutations into PAM-1 using the Stratagene QuikChange protocol; the DNA sequence of the vector was verified. Stable AtT-20 cell lines expressing PAM-1/OSX, were established using drug selection (0.5 mg/ml G-418) and subcloned by limiting dilution. Drug-resistant lines were selected based on their ability to secrete active PHM and were again subcloned by limiting dilution. Two stable clonal cell lines were chosen for further analysis; results for both lines were similar.
Analysis of Cells
Adenoviral Infection
To infect AtT-20 cells stably expressing PAM-1/OSX with α1-PDX-encoding adenovirus (28), cells were seeded into 12-well plates at 1 × 105/well. The α1-PDX adenovirus was diluted in PBS to achieve a multiplicity of infection of 1 in a final volume of 500 μl/well. The diluted adenovirus solution was added directly to cells in growth medium, mixed by swirling, and incubated for 30 min to permit infection. After this incubation period, 0.5 ml of high glucose DMEM containing 2% fetal bovine serum was added to each well, and the cells were incubated for 24 h at 37 °C in a 5% CO2 incubator. The medium was then supplemented with an additional 1 ml of DMEM containing 2% fetal bovine serum, and the cells were incubated for another 24 h. Forty eight hours after infection, cells were scraped into TMT supplemented with protease inhibitors; clarified lysates were subjected to Western blot analysis.
Secretion
To assess basal secretion, AtT-20 cells in 6-well plates were incubated in complete serum-free medium (CSFM) with 0.05 mg/ml bovine serum albumin (3 ml/well) for 24 h; spent medium was centrifuged to remove non-adherent cells. To measure secretagogue stimulation, cells were first rinsed with pre-warmed CSFM and then incubated in 2 ml of CSFM for two sequential 1-h basal secretion periods. Cells were then incubated in CSFM containing no secretagogue (basal) or 2 mm BaCl2 for 1 h; media were collected, and cells were extracted into TMT. PAM activity was quantified using 125I-labeled acetyl-Tyr-Val-Gly; optimal amounts of copper and ascorbate were supplied for the assay (29).
Immunofluorescence
Cells grown on glass chamber slides were fixed using 4% paraformaldehyde and processed as described; primary antibody was applied for 2 h (30). Rabbit polyclonal antiserum to rat TGN38(155–249) (JH1479 (5)) was diluted 1:1000, and spent media from a mouse hybridoma secreting monoclonal antibody to recombinant PAM C-terminal domain (6E6 (5)) was diluted 1:50. The PAM antibody was visualized using Cy3-conjugated donkey anti-mouse IgG (715-166-151; Jackson ImmunoResearch), and TGN38 antibody was visualized using fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (711-096-152; Jackson ImmunoResearch). Slides were cover-slipped using ProLong Gold anti-fade mounting agent (Invitrogen).
Antibody internalization studies were carried out as described (31) using a PAM ectodomain antibody (JH629 or JH471, 1:1000) (3) and Alexa Fluor 488-conjugated wheat germ agglutinin (WGA) (1 μg/ml Molecular Probes) for 5 or 10 min. After a 5–35-min chase, cells were fixed with 4% paraformaldehyde, permeabilized using 0.125% Triton X-100, and incubated with monoclonal antibody to syntaxin 6 (1:100; BD Transduction Laboratories) for 1 h at room temperature. Secondary antibodies were Alexa Fluor 488- or 555-conjugated F(ab′)2 goat anti-rabbit or anti-mouse IgG (A11070, A21430, A10684, and A21425; Molecular Probes) (31). Cells were viewed and photographed with a Leica TCS SP8 MP CARS microscope with a glycerol immersion ×63 objective (NA 1.3).
Electron Microscopy
For PAM antibody/colloidal gold labeling, cells were incubated with antibody JH629 (diluted 250-fold in DMEM/HEPES/BSA) for 30 min at 4 °C. Cells were then rinsed at 4 °C with DMEM/HEPES/BSA and incubated for 30 min at 4 °C with protein A/15-nm colloidal gold (diluted 50-fold in DMEM/HEPES; University of Utrecht, Utrecht, Netherlands) (31). After rinsing in DMEM/HEPES at 4 °C, cells were incubated (chased) in culture medium at 37 °C for the times indicated. Cells were then fixed with 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer, pH 7.2, post-fixed with 1% osmium tetroxide and 1.5% potassium ferrocyanide, dehydrated, and embedded in an Epon resin (32). Ultrathin sections were post-stained with uranyl acetate and lead citrate and viewed and photographed at ×8000 with a Jeol JEM-1400 electron microscope equipped with a Gatan Orius SC 1000B bottom-mounted CCD camera. For quantification of gold particles, specimens were systematically scanned until 20 labeled cells had been imaged for each cell line in each experiment. Gold particle data were subjected to the Friedman Repeated Measures ANOVA on Ranks; data for each organelle were compared using the Wilcoxon Signed Rank Test in SigmaPlot.
For simultaneous detection of PAM antibody and WGA uptake, the 30-min 4 °C incubation with protein A/15-nm gold included peroxidase-conjugated WGA (30 μg/ml; Sigma). The cells were then rinsed in DMEM/HEPES, chased in culture medium for 5 min, fixed with 1.5% glutaraldehyde in 0.1 m phosphate buffer, and incubated with 0.25 mg/ml diaminobenzidine and 0.6 mg/ml hydrogen peroxide for 15 min on ice. The cells were then rinsed, post-fixed, dehydrated, and embedded as described above. To improve visualization of the diaminobenzidine precipitate, ultrathin sections were post-stained only with uranyl acetate.
Protease Inhibitor Treatment
PAM-1 and PAM-1/OSX AtT-20 cells were fed with CSFM containing 0.05 mg/ml BSA. Cells were incubated with MG132 (2 μm; added from a 10 mg/ml stock in DMSO; Sigma) for 4.5 h. Cells were then extracted into ice-cold TMT containing protease inhibitors (33); lysates were allowed to tumble for 30 min at 4 °C and then clarified. Cells were also incubated with NH4Cl (20 mm; added from a 160 mm isotonic stock in water), concanamycin A (1 nm, added from a 10 μm stock in DMSO; Sigma), lactacystin (10 μm, added from a 10 mm stock in water; Sigma), or leupeptin(20 μm, added from a 2 mm stock in water; Sigma). After these treatments, cells were extracted into SDS-lysis buffer containing the protease inhibitor mixture described above.
Surface Biotinylation
Cells were incubated for 30 min in CSFM/air and then rinsed with HSG (15 mm HEPES, 120 mm NaCl, 2 mm CaCl2, 4 mm KCl, 25 mm glucose, pH 7.5). For assessing steady state plasma membrane localization, surface biotinylation was carried out on ice, and all solutions used were pre-chilled. For assessment of endocytic trafficking, surface biotinylation was carried out for 10 min at 37 °C. Sulfo-NHS-LC-biotin (1.25 mm in HSG; Pierce) was applied on ice or at 37 °C. The reaction was quenched by incubation in CSFM/air, 2 mg/ml BSA for 5 min, followed by rinsing in CSFM/air. Cells were either extracted immediately or chased for up to 4 h in CSFM/air, 1 mg/ml BSA. Following extraction into TMT supplemented with protease inhibitors and centrifugation at 14,000 × g for 20 min, clarified lysates and media were incubated with Neutravidin beads (25 μl of slurry; Pierce) for 1 h at 4 °C. Beads were rinsed twice with TMT containing protease inhibitors and once with buffer lacking detergent before eluting the bound proteins into Laemmli sample buffer by heating for 5 min at 95 °C. Input samples and eluates were analyzed on 4–15% gradient gels (Bio-Rad). Band intensities were quantified using GeneTools software and exposures in the linear range. For assessing the contribution of lysosomal proteases to the degradation of internalized PAM, the chase medium contained leupeptin alone (20 μm) or in combination with MG132 (2 μm).
Biochemical Analyses
Trypsin Sensitivity Assay
AtT-20 cells were extracted into TM buffer (20 mm NaTES, 10 mm mannitol, pH 7.4) containing protease inhibitor mixture. Membranes were prepared by differential centrifugation (33). Pellets were suspended in 20 mm Tris-HCl, pH 8.0, 0.2 m NaCl, 0.02% Nonidet P-40. Samples (1 μg of protein) were digested at 37 °C for 5 min with 0.01–0.2 μg of l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (Sigma) added from a 5 mg/ml stock (in 1 mm HCl, 1 mm CaCl2); digestion was terminated by adding 0.5 μl of phenylmethylsulfonyl fluoride (30 mg/ml) (34). Samples were assayed for PHM and PAL activity (29), fractionated by SDS-PAGE, and analyzed by Western blotting with region-specific PAM antibodies.
Western Blotting
Samples were subjected to SDS-PAGE and Western blot analysis as described (30). Antigen-antibody complexes were detected using horseradish peroxidase-conjugated secondary antibody and Super Signal West Pico chemiluminescent substrate (Pierce). Affinity-purified rabbit polyclonal antibodies (1:1000 dilution for each) used to visualize PAM included the following: PHM antibody JH1761 (raised to rPAM-1(37–382)) (5); exon 16 antibody JH629 (raised to rPAM-1(394–498)) (3); PAL antibody JH471 (raised to rPAM-1(463–864)) (5); and CD antibody CT267 (raised to rPAM-1(965–976)) (33).
Biosynthetic Labeling
To assess the effects of O-glycosylation on stability, AtT-20 cells expressing PAM-1 or PAM-1/OSX were incubated in medium containing [35S]Met/[35S]Cys (NEG772; PerkinElmer Life Sciences) for 30 min and harvested immediately or chased for 1, 2, or 4 h. Immunoprecipitation and analysis were carried out as described using affinity-purified antibody to PHM (30).
Blue Native-PAGE (BN-PAGE) Analysis
Cells grown to confluence in 15-cm dishes were rinsed once with CSFM and harvested into fresh CSFM. Cells were pelleted and suspended in 10 volumes of ice-cold 25 mm imidazole, 250 mm sucrose, pH 7.4 supplemented with protease inhibitors. Cells were homogenized using a motor-driven Potter-Elvehjem homogenizer with a Teflon pestle at 4 °C, and the cell debris was pelleted (1000 × g for 5 min). Supernatants were centrifuged for 15 min at 100,000 × g, and pellets were solubilized in native-PAGE sample buffer (50 mm BisTris, 50 mm NaCl, 10% glycerol, Ponceau; Life Technologies, Inc.) containing 0.5% n-dodecyl-β-d-maltoside and protease inhibitors for 30–45 min at 4 °C. Insoluble material was removed by centrifugation.
Supernatants (10 μg of protein) were subjected to one-dimensional BN-PAGE after addition of 0.05% Coomassie G-250. Samples were separated on linear 3–12% (w/v) BisTris gels (Life Technologies, Inc.) in 15 mm BisTris, 50 mm Tricine, and 0.05% Coomassie G-250 running buffer with 15 mm BisTris, pH 7.0, as the anode buffer. Native Mark unstained protein standards (Life Technologies, Inc.) were used to calibrate the first dimension. For Western blot analysis, complexes were transferred onto PVDF membranes using NuPAGE transfer buffer (Life Technologies, Inc.) with 10% methanol and a constant current of 200 mA for 2.5 h.
For two-dimensional BN-PAGE, single lanes were excised (∼5 × 1 cm) and reduced by incubation with gentle shaking in 5 ml of 1× NuPAGE sample buffer (10% glycerol and 2.5% lithium dodecyl sulfate) with 50 mm dithiothreitol for 15–30 min at room temperature. Gel strips were then incubated in NuPAGE sample buffer with 20% ethanol and 5 mm dithiothreitol for 15 min at room temperature. Single lanes were placed on top of 4–20% (w/v) Tris-glycine Zoom SDS-polyacrylamide gels and electrophoresed in parallel with molecular mass markers (Precision Plus Dual Color Protein Standards, Bio-Rad). Proteins were then transferred onto PVDF membranes and subjected to Western blot analysis.
Results
Identification of O-Glycosylation Sites in Exon 16
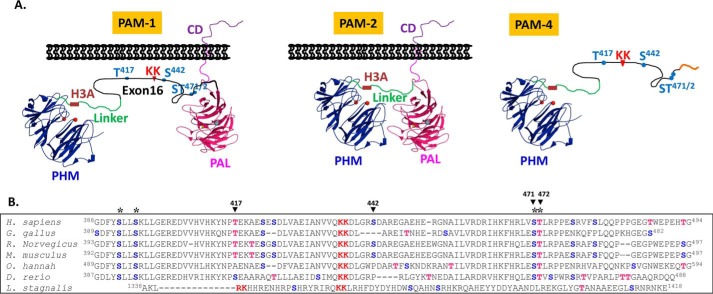
To determine whether the O-glycosylation of PAM was of functional significance, we first identified the sites at which this modification occurred. Tissue-specific and developmentally regulated alternative splicing generates two major forms of vertebrate PAM (Fig. 1A). In the shorter isoform (PAM-2), PHM and PAL are separated by a 36-amino acid linker that includes a pH-sensitive His cluster (H3A) essential for efficient endocytic trafficking (30). In the longer isoform (PAM-1), a 105-amino acid intrinsically unstructured region (encoded by exon 16) is inserted between PHM and PAL (Fig. 1A); the pair of basic amino acids in exon 16 can be cleaved by prohormone convertase 1, separating soluble PHM from membrane PAL (35). We previously reported the presence of at least two O-linked sugars in this region (3). Based on the assumption that functionally important Ser/Thr residues would be conserved, a ClustalW2 alignment of this region of several vertebrate PAM proteins was examined (Fig. 1B); four Ser/Thr residues were conserved from human to zebrafish.
FIGURE 1.
Vertebrate PAM exon 16 has well conserved potential O-glycosylation sites. A, two major PAM isoforms in rodents and human are shown, rodent and human PAM-1 include exon 16 (105 and 107 amino acids, respectively; black line). The protease-resistant PHMcc (blue) is followed by a protease-sensitive 36-amino acid linker (green) that includes a pH-sensitive His cluster (H3A). In PAM-4, a minor splice variant, transcripts terminate in the intron following exon 16 (1). The PALcc (red) is shown, along with the cleavage site in exon 16 (KK) and the Ser/Thr sites explored by mutagenesis. B, ClustalW alignment of the alternatively spliced exon from several vertebrate PAM proteins; the conserved PC cleavage site (KK, in red) and conserved potential O-glycosylation sites (*) are shown: Ser (blue); Thr (red). Accession numbers for the sequences shown: Homo sapiens, NP_000910.2; Gallus gallus, XP_424857.3; Rattus norvegicus, NP_037132.2; Mus musculus, NP_038654.2; Ophiophagus hannah, ETE72045.1; Danio rerio, XP_699436.4; Lymnaea stagnalis, AAD42258.1. Although L. stagnalis PAM included an alternatively spliced exon with a prohormone convertase cleavage site, potential O-glycosylation sites were not conserved.
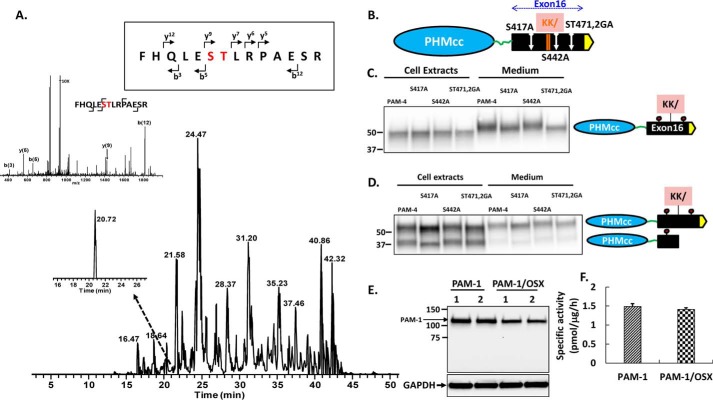
To facilitate site identification, we used AtT-20 cells stably expressing rat PAM-4, a natural isoform that includes PHM but terminates shortly after the end of exon 16 (Fig. 1A) (1). Secreted 52-kDa PAM-4 isolated from spent medium was subjected to in-gel trypsin digestion. A search for peaks that could correspond to O-glycosylated peptides identified one with an additional mass of 406 Da, which corresponds to two GalNAc molecules (GalNAc = 203.1 Da). MS/MS spectra confirmed the presence of GalNAc residues on Ser471 and/or Thr472, because the y9 and b12 peptide fragments both showed the additional mass (Fig. 2A). The identification of tryptic peptides containing unmodified Ser/Thr residues left only two Ser/Thr residues as candidate sites, Thr417 and Ser442. To determine whether they were O-glycosylated, we mutated Thr417 to Ala (T417A) and Ser442 to Ala (S442A); to confirm our identification of Ser471–Thr472 as a site of O-glycosylation, both residues were mutated (S471G/T472A) (Fig. 2B).
FIGURE 2.
Identification of O-glycosylation sites in exon 16. A, AtT-20 PAM-4 (52 kDa) was subjected to in-gel trypsin digestion. The MS/MS fragment spectrum, along with fragment assignments for O-glycosylated rPAM(466–479), are shown; brackets indicate the b and y ion assignments. Both the y(9) and b(12) fragment ions with the GalNAc modification were detected. B, diagram showing the location of each site-directed mutation relative to the cleavage site in exon 16. PAM-4 and mutant PAM-4 were expressed transiently in pEAK and AtT-20 cells. Using an antibody to the exon 16 region, the apparent molecular weights of the PAM-4 proteins in the media and in cell lysates were compared. For pEAK cells (C), a decrease in the mass of PAM-4/S417A and PAM-4/S471G/T472A versus PAM-4 was apparent in the medium. For AtT-20 cells (D), a decrease in the mass of PAM-4/S417A and S471G/T472A (ST471,2GA) was apparent in intact PAM-4 in both cells and media. The decreased mass of PAM-4/S417A was also apparent in the cleaved protein, which no longer contains S471G/T472A. E, PAM-1 and PAM-1/OSX were transiently expressed in duplicate wells of pEAK cells; lysates (5 μg of protein) were fractionated by SDS-PAGE and visualized using antibody to PHM. F, specific activity of PHM in lysates prepared from pEAK cells transiently expressing PAM-1 or PAM-1/OSX (n = 3).
Vectors encoding PAM-4 and the three mutants were transiently expressed in pEAK cells and in AtT-20 cells. In pEAK cells, which do not store secreted proteins, a mobility shift was only apparent in the medium: PAM-4/T417A and PAM-4/S471G/T472A were slightly smaller than PAM-4 (Fig. 2C). In AtT-20 cells, PAM-4 is subjected to endoproteolytic cleavage; although the 52-kDa precursor is not stored in secretory granules, the smaller product is stored (5). In AtT-20 cell medium, as in pEAK cell medium, PAM-4/T417A and PAM-4/S471G/T472A were slightly smaller than PAM-4 (Fig. 2D); a similar shift in size was observed when the cell extracts were examined. The PAM-4 cleavage product includes T417A, but not S471G/T472A (Fig. 2B); consistent with this, the PAM-4/T417A cleavage product was smaller than any of the other cleavage products (Fig. 2D, lane 2). Taken together, our data indicated that O-glycosylation occurred at Thr417 and Ser471Thr472, but not at Ser442, in both pEAK and AtT-20 cells.
Both mutations (T417A and S471G/T472A) were introduced into PAM-1, creating PAM-1/OSX, which differed from native PAM-1 only by its inability to undergo O-glycosylation at these two sites in exon 16. Transient expression of PAM-1/OSX in pEAK cells demonstrated that the protein was stable and as active as PAM-1 (Fig. 2, E and F). Because PAM-1 lacking these O-glycosylation sites appeared to fold successfully, AtT-20 lines stably expressing PAM-1/OSX were generated; two independent lines were selected so that we could determine whether the O-glycosylation of PAM-1 played a role in its cleavage or trafficking.
PAM-1/OSX Is More Susceptible to Endoproteolytic Cleavages
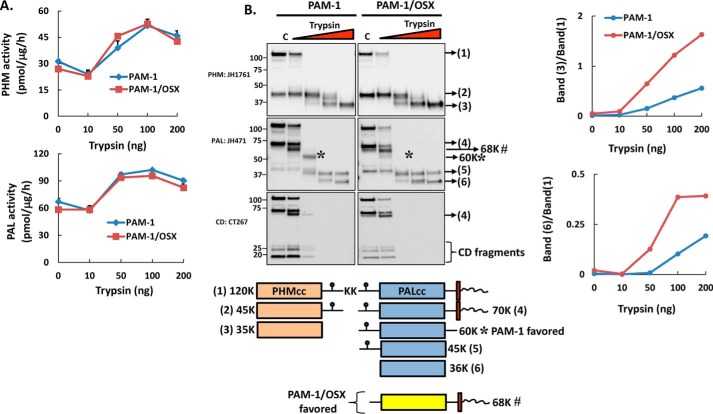
We next asked whether the absence of O-glycans made PAM-1/OSX more sensitive to proteolytic digestion (Fig. 3). Trypsin was used previously to define the catalytic cores of PHM (PHMcc) and PAL (PALcc) and to reveal the inhibitory effect of its cytosolic domain on enzyme activity (34). Membranes from AtT-20 cells expressing PAM-1 or PAM-1/OSX were digested with increasing amounts of trypsin. As seen previously for PAM-3, an increase in PHM activity was observed after digestion of PAM-1 with trypsin (34); digestion of PAM-1/OSX produced an increase of the same magnitude (Fig. 3A). PAL activity was similarly increased by trypsin digestion of PAM-1 and PAM-1/OSX.
FIGURE 3.
PAM-1/OSX is more sensitive to trypsin digestion than PAM-1. Membranes from PAM-1 and PAM-1/OSX AtT-20 cells were digested with increasing amounts of trypsin for 5 min at 37 °C. A, samples were assayed for PHM and PAL activity. B, after fractionation by SDS-PAGE, antibodies specific for PHM, PAL, and the cytosolic domain (CD) were used to identify major tryptic fragments. Representative blots are shown; the proteins that correspond to each numbered band are illustrated. 60K*, unique proteolytic product seen in PAM-1 cells; 68K#, cleavage product seen more in PAM-1/OSX cells. Graphs show the appearance of PHMcc and PALcc (band#3 and band#6, respectively) relative to PAM-1 in the control as a function of trypsin added; the experiment was repeated twice, with similar results.
The protein products generated by trypsin were separated by SDS-PAGE; antisera to PHM, PAL, and the cytosolic domain were used to compare the cleavage products (Fig. 3B). As expected, the cytosolic domains of both proteins were protease-sensitive, and the major intermediates observed (45-kDa PHM and 45-kDa PAL) were consistent with cleavage at the Lys–Lys437 site in exon 16 and the Lys–Lys822 site at the C terminus of PALcc. For the most part, bands of similar mass were detected in digests of PAM-1 and PAM-1/OSX. The 68-kDa product recognized by the PAL and CD antibodies was more prevalent in PAM-1/OSX digests than in PAM-1 digests (68K#, Fig. 3B). Although prominent in PAM-1 digests, 60-kDa PAL (60K*) was not a major PAM-1/OSX product (Fig. 3B); this product was not recognized by the CD antibody. Consistent with the hypothesis that PAM-1/OSX would be more susceptible to cleavage by trypsin, conversion of PAM-1/OSX into PHMcc (band#3) and PALcc (band#6) proceeded more rapidly than observed with PAM-1 (Fig. 3B, graph).
O-Glycosylation of PAM-1 Limits Its Endoproteolytic Cleavage in AtT-20 Cells
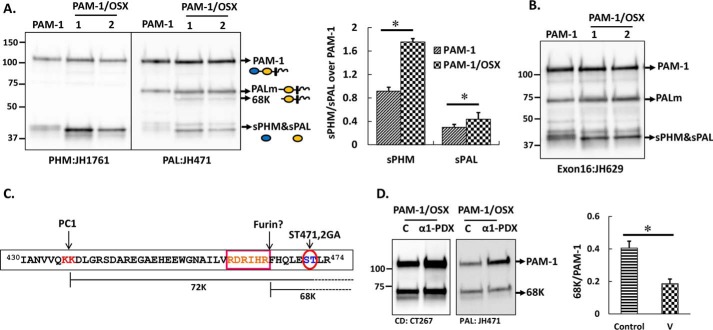
We next asked whether the proteolytic processing of PAM-1 and PAM-1/OSX in AtT-20 cells differed. Although the same major products, 45-kDa sPHM and 70-kDa PALm (where PALm is the cleavage product of PAM-1 lacking sPHM), were detected, the steady state ratio of cleaved products to intact PAM was higher in PAM-1/OSX cells (Fig. 4A, graph). In addition, a new 68-kDa PALm fragment was detected in PAM-1/OSX lysates (Fig. 4A). Because this product was recognized by an antibody specific for the C terminus of PAM-1, it must lack more of exon 16 than the 70-kDa PALm product found in PAM-1 cells. Consistent with this, an antibody raised to exon 16 (JH629) failed to recognize the 68-kDa fragment (Fig. 4B). These observations suggested that the absence of O-linked glycans exposed an additional proteolytic cleavage site in PAM-1/OSX.
FIGURE 4.
Proteolytic processing of PAM-1 and PAM-1/OSX in AtT-20 cells differs. An AtT-20 line expressing PAM-1 and two lines expressing PAM-1/OSX were compared. A, equal amounts of protein (5 μg) fractionated by SDS-PAGE were visualized using affinity-purified antibodies against PHM or PAL. The major products of PAM-1 cleavage are indicated by arrows. A 68-kDa band was detected with the PAL antibody only in PAM-1/OSX cells. Bar graph showing steady state ratio of sPHM or soluble PAL over PAM-1 in both cell lines was obtained by quantifying blots from five independent experiments (*, p < 0.05). B, when the same samples were probed with an exon 16-specific antibody, the 68-kDa fragment was not recognized. C, schematic identifies a potential furin cleavage site, which could generate 68-kDa PAL. D, PAM-1/OSX cells were infected with adenovirus expressing α1-PDX 48 h before harvest. Cell lysates from control (C) and virus-infected (α1-PDX) cells were prepared in 1% Triton X-100 and fractionated by SDS-PAGE. Western blotting using a CD antibody (left) or a PAL antibody (right) showed a decrease in the amount of 68-kDa PAL. Production of 68-kDa PAL in triplicate samples of control and virus-infected cells from two independent experiments using both antibodies was quantified (*, p < 0.05). v, virus.
Furin, a secretory pathway subtilisin-like convertase, is expressed in AtT-20 cells (37). A potential furin cleavage site (38) precedes the S471G/T472A O-glycan site in exon 16 (Fig. 4C). To test the hypothesis that the absence of an O-glycan at this site in PAM-1/OSX made this additional cleavage site accessible to a furin-like endoprotease, leading to the production of both 68- and 70-kDa PALm, we used α1-PDX Portland, a bioengineered serpin that serves as a highly selective furin inhibitor (28). Cells stably expressing PAM-1/OSX were infected with adenovirus-expressing α1-PDX. Cell lysates prepared 48 h later were analyzed for the presence of 68-kDa PALm; its levels were reduced in cells expressing α1-PDX (Fig. 4D, graph). This allowed us to conclude that O-glycosylation of exon 16 limited its endoproteolytic cleavage by masking a site that could otherwise be cleaved by furin.
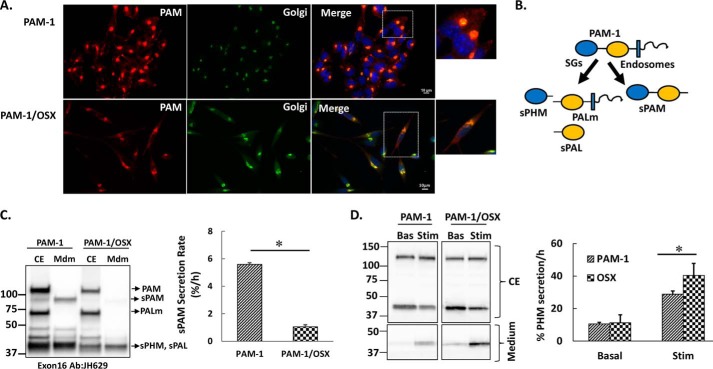
Secretion of PAM-1 Products Is Altered in PAM-1/OSX Cells
AtT-20 cells expressing PAM-1 or PAM-1/OSX were fixed and stained simultaneously for PAM and a Golgi marker. In both cell lines, PAM staining was largely localized to the perinuclear region in a complex reticular pattern that overlapped extensively with TGN38 staining (Fig. 5A). The two PAM-1/OSX cell lines examined both adopted a more polarized morphology than the PAM-1 cell line.
FIGURE 5.
Secretion of PAM-1 products is altered in PAM-1/OSX cells. A, steady state localization of PAM was assessed in AtT-20 lines expressing PAM-1 or PAM-1/OSX; PAM was visualized using an antibody to its cytosolic domain (6E6; Cy3 anti-mouse), and the Golgi complex was visualized using an antibody to TGN38 (FITC anti-rabbit). Scale bar, 10 μm. B, schematic illustrating soluble products of secretory granule (SG) and endosomal processing of PAM-1. C, PAM-1 and PAM-1/OSX cells were incubated for 1 h in basal medium; cell extracts (CE) and media (Mdm) were subjected to Western blot analysis using an antibody to exon 16 (JH629). The basal secretion rate of sPAM, which is produced only in the endocytic pathway, was calculated from these Western blots (% = 100 × sPAMmdm/PAM-1cell) and was diminished in PAM-1/OSX cells (*, p < 0.05, n = 6). D, duplicate wells of AtT-20 cells expressing PAM-1 or PAM-1/OSX were either incubated in basal medium (Bas) or medium containing 2 mm BaCl2 (Stim) for 30 min; cell extracts (CE, upper) and spent medium (lower) were subjected to Western blot analysis using an antibody specific to PHM (JH1761). Secretion rates (% cell content/h) were calculated by taking the ratio of sPHM in the medium to sPHM in the cell extract, although its basal secretion did not differ, the stimulated secretion of sPHM was increased in PAM-1/OSX cells (*, p < 0.05, n = 3).
To compare the secretory pathways in PAM-1 and PAM-1/OSX cells, basally secreted PAM products and cell extracts were compared. Under basal conditions, PAM-1 AtT-20 cells secreted sPAM, a product of the endocytic pathway (30), along with sPHM; strikingly, very little sPAM was found in the basal medium of PAM-1/OSX AtT-20 cells (Fig. 5, B and C). sPHM and soluble PAL, products of the regulated secretory pathway, are secreted basally, but the rate at which they are secreted increases in response to secretagogues like BaCl2 (Fig. 5D). Basal secretion of sPHM was indistinguishable in PAM-1 and PAM-1/OSX cells. However, the response of PAM-1/OSX cells to BaCl2 was more robust than the response in PAM-1 cells (30). Western blots from three independent experiments were quantified, showing significantly greater release of sPHM in response to BaCl2 stimulation of PAM-1/OSX cells. Taken together, our data indicated that the secretion of products of the endocytic pathway differed dramatically in PAM-1 and PAM-1/OSX cells.
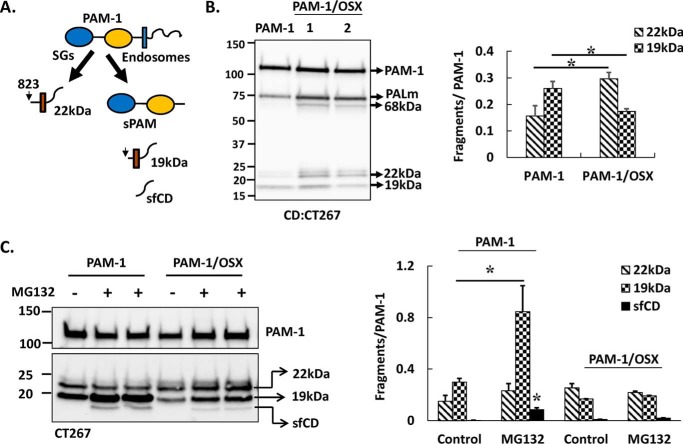
Endocytic Pathway Processing of PAM-1/OSX Is Altered
The cleavages that allow secretion of soluble fragments of PAM generate integral membrane fragments that remain in the cell (Fig. 6A) Although secretory granule cleavages between PAL and the transmembrane domain produce a 22-kDa TMD/CD fragment, production of sPAM yields a 19-kDa TMD/CD fragment (Fig. 6A) (33). Both TMD/CD products can serve as substrates for regulated intramembrane proteolysis by γ-secretase, releasing sfCD, a rapidly degraded cytosolic protein that accumulates in the nucleus in a phosphorylation-dependent manner (36). The PAM-1 products produced in the endocytic pathway are best distinguished using a CD antibody (Fig. 6B). As observed using a PAL antibody, PAM-1/OSX was more extensively processed to PALm than PAM-1. Expressed as a ratio to intact PAM-1, levels of 22-kDa TMD/CD rose in lysates of PAM-1/OSX cells, whereas levels of 19-kDa TMD/CD fell. This observation was also consistent with diminished endocytic processing of PAM-1/OSX.
FIGURE 6.
Endocytic processing of PAM-1/OSX is altered. A, schematic shows membrane-associated C-terminal fragments of secretory granule (SG) and endosomal processing of PAM-1 in AtT-20 cells. B, samples from Fig. 4A were analyzed using an affinity-purified antibody specific for the C terminus of PAM (CT267). Signals for 19- and 22-kDa intermediates in the two cell types from two independent experiments were quantified and expressed as a percentage of intact 120-kDa PAM-1 (*, p < 0.05, n = 6). C, PAM-1 and PAM-1/OSX cells (duplicate wells) were incubated in basal medium without (−) or with (+) MG-132 for 4.5 h. Cell lysates were analyzed using the PAM-CD antibody. Signals for sfCD, 19- and 22-kDa intermediates in MG-132-treated versus control wells from two independent experiments were quantified and expressed as a percentage of intact 120-kDa PAM-1 (* for the effect of MG-132, p < 0.05, n = 6).
To further explore the endocytic processing of PAM-1/OSX, cells were treated with a peptide aldehyde, MG-132, a protease inhibitor (33). SDS lysates prepared from control and MG-132-treated AtT-20 cells expressing PAM-1 or PAM-1/OSX were examined using CD antibody. As expected, MG-132 treatment of PAM-1 cells increased the amount of 19-kDa TMD/CD and sfCD (Fig. 6C). In contrast, MG-132 failed to produce an increase in levels of 19-kDa TMD/CD and sfCD in PAM-1/OSX cells. The decreased levels of 19-kDa TMD/CD, a substrate for γ-secretase, in PAM-1/OSX cells would be expected to contribute to decreased production of sfCD and support the conclusion that O-glycans play an essential role in the endocytic trafficking of PAM-1.
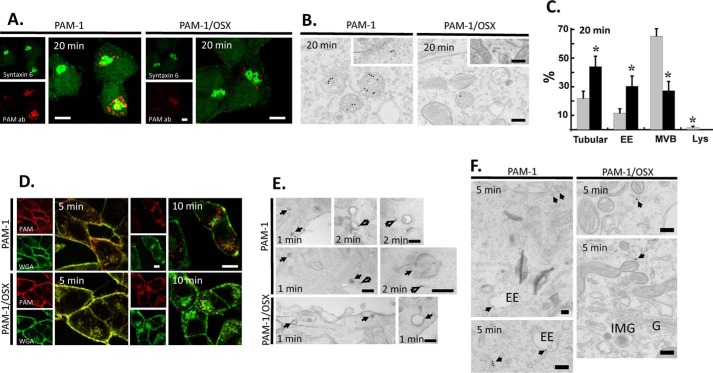
Internalized PAM-1/OSX Is in Tubular Endosomes
Because our biochemical studies indicated that PAM-1/OSX mostly failed to reach the endocytic compartments generating sPAM and 19-kDa TMD/CD, we next sought to establish the step at which the endocytic trafficking of PAM-1 and PAM-1/OSX deviated. Previous surface biotinylation and antibody uptake experiments have provided a detailed description of the pathway traveled by PAM-1 internalized from the plasma membrane; after traversing multiple endosomal compartments and returning to the TGN, it can again be incorporated into nascent secretory granules (31). As expected, after 20 min of uptake, PAM antibody internalized by PAM-1 AtT-20 cells was concentrated in the TGN area, partially overlapping the distribution of syntaxin 6, a TGN marker; internalized antibody was also in round structures collected around the TGN (Fig. 7A, left). When PAM antibody was internalized by PAM-1/OSX AtT-20 cells, the fluorescently labeled endocytic structures were smaller, much less prevalent, and not as effectively aggregated in the perinuclear area, showing less colocalization with syntaxin 6 (Fig. 7A, right). Similar results were obtained using antibodies for exon 16 (JH629) or PAL (JH471).
FIGURE 7.
Endocytic trafficking of PAM-1/OSX is altered. The endocytic trafficking of PAM-1 and PAM-1/OSX was monitored by incubating live cells with ectodomain antibody to PAM. A, cells incubated with PAM antibody for 5 min were chased for 15 min, fixed, and permeabilized; endogenous syntaxin 6 and internalized ectodomain antibody were visualized. Scale bars, 10 μm. B, cells incubated with PAM antibody-gold complexes at 4 °C were visualized after a 20-min chase at 37 °C. Representative images are shown as follows: three labeled multivesicular bodies are seen in the PAM-1 cell and one in the PAM-1/OSX cell; insets show labeled tubular structures. Scale bars, 200 nm. C, graph shows the percentage of total gold particles in tubular structures, early endosomes (EE), multivesicular bodies (MVB), and lysosomes (Lys) in PAM-1 (gray bars) and PAM-1/OSX (black bars) cells after the 20-min chase (mean ± S.E.; *, p < 0.001). D, PAM-1 and PAM-1/OSX cells exposed to PAM antibody and to fluorescently tagged WGA for 5 min were rinsed and chased for 5 or 10 min before fixation; internalized antibody was visualized after permeabilization. Scale bars, 10 μm. E and F, PAM-1 and PAM-1/OSX cells kept on ice were incubated with PAM antibody-gold complexes and WGA-HRP; cells were fixed after a chase incubation at 37 °C for 1, 2, or 5 min. E, after a 1- or 2-min chase, co-localized PAM antibody-gold complexes and peroxidase product are indicated by arrows; open arrows mark PAM-1 separated from WGA and in tubules. Scale bars, 200 nm. F, after the 5-min chase, antibody/gold particles were found in tubular structures and early endosomes (EE) in PAM-1 cells; in PAM-1/OSX cells, antibody/gold particles were confined to tubular structures localized peripherally or near the Golgi complex (G, Golgi stack; IMG, immature secretory granule). Scale bars, 200 nm.
We turned to electron microscopy and antibody-gold complexes to better define the endocytic structures containing antibody internalized by PAM-1 or PAM-1/OSX (Fig. 7B). As expected, antibody-gold complexes internalized by PAM-1 during a 20-min incubation at 37 °C were largely localized to multivesicular bodies; in contrast, the far scarcer antibody-gold complexes internalized by PAM-1/OSX were most commonly found in tubular structures. Although the multivesicular bodies in PAM-1/OSX cells contained only one or two antibody-gold complexes, multiple complexes were often seen in PAM-1 cell multivesicular bodies. Quantification of the percentage of gold particles associated with different endosomal compartments revealed clear differences (Fig. 7C), although 75% of the gold particles in PAM-1 cells were in multivesicular bodies, this number dropped to 27% in PAM-1/OSX cells. Almost half (44%) of the antibody-gold complexes in PAM-1/OSX cells were in tubular structures; this number dropped to 21% in PAM-1 cells. The lack of PAM-1/OSX in late endosomal structures was consistent with its diminished endocytic processing.
We next examined earlier stages of endocytic trafficking using WGA, a lectin, and a general marker for endocytic trafficking (Fig. 7D). As expected (31), antibody internalized by PAM-1 cells quickly separated from internalized WGA. In contrast, antibody internalized by PAM-1/OSX cells colocalized with WGA after a 5- or 10-min chase (Fig. 7D).
Because differences in the endocytic trafficking of PAM-1 and PAM-1/OSX were apparent within 5 min of uptake, we further examined the early stages of endocytosis using a mixture of PAM antibody-gold complexes and WGA linked to HRP. For a short period of time (1–2 min), an endosomal compartment that contained both gold particles and peroxidase product was seen in PAM-1 and PAM-1/OSX cells. Electron microscopy revealed an early sorting process that separated antibody internalized by PAM-1 into tubules lacking WGA-HRP. In contrast, antibody internalized by PAM-1/OSX and WGA-HRP remained largely co-localized (Fig. 7E). After the 5-min chase, antibody internalized by PAM accumulated in early endosomes and tubular structures, although antibody internalized by PAM-1/OSX was confined to tubular structures, which could be located peripherally or near the Golgi complex (Fig. 7F). To directly follow the fate of internalized PAM-1/OSX, we turned to surface biotinylation.
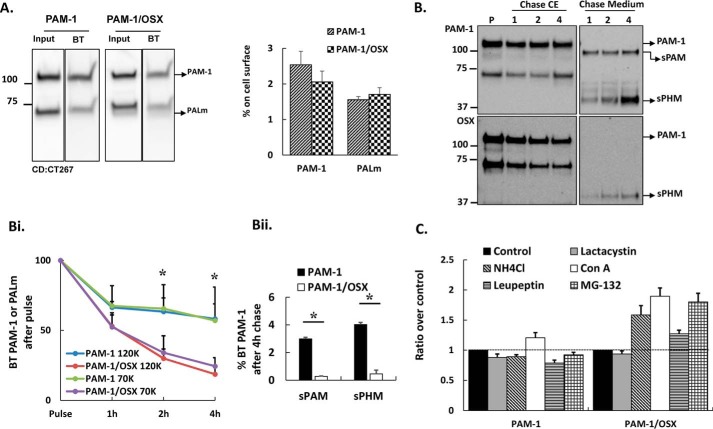
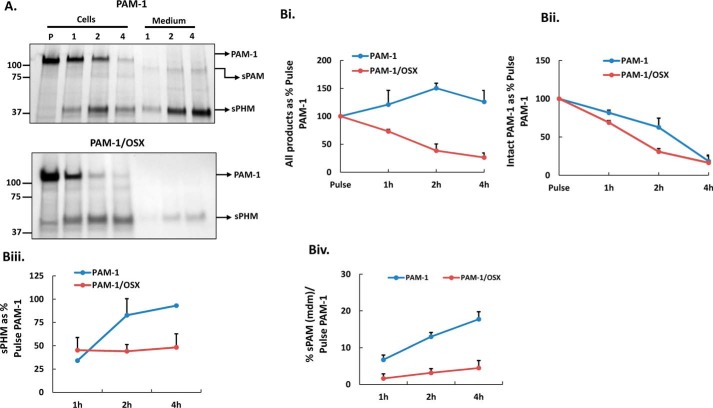
Internalized PAM-1/OSX Is Degraded, Not Recycled
The greatly diminished antibody internalization observed in PAM-1/OSX cells could reflect a decrease in the amount of PAM-1/OSX on the cell surface or its rapid degradation after internalization. After reaching the plasma membrane, PAM-1 is internalized; whether internalized PAM-1 is returned to the secretory pathway or degraded is governed by signals in its cytosolic domain (37). Surface biotinylation at 4 °C revealed a similar percentage of the total PAM-1 and PALm on the plasma membrane in PAM-1 and PAM-1/OSX cells (Fig. 8A).
FIGURE 8.
Internalized PAM-1/OSX is degraded more rapidly than internalized PAM-1. A, biotinylated proteins were isolated from PAM-1 and PAM-1/OSX AtT-20 cells exposed to cell-impermeant activated biotin for 10 min at 4 °C. Input and biotinylated proteins (BT) fractionated by SDS-PAGE were visualized using a C-terminal antibody. Input lanes contained 5% as much sample as biotinylated lanes. Data for PAM-1 and PALm from two separate experiments were quantified. B, cells exposed to cell impermeant-activated biotin for 10 min at 37 °C were analyzed immediately (pulse, P) or chased for 1, 2, or 4 h. Biotinylated proteins isolated from cell extracts (CE) and chase media were fractionated and probed for PAM using an exon 16 antibody. Arrows indicate products produced from biotinylated PAM-1. B, panel i, line graph shows biotinylated PAM-1 and PALm recovered at each chase point plotted as % biotinylated PAM-1 or PALm after the pulse (*, p < 0.05; PAM-1 and PALm, n = 3). B, panel ii, biotinylated sPAM and sPHM recovered from the media after the 4-h chase are plotted as % biotinylated PAM-1 after the pulse. Values are averages of two experiments with duplicate samples. C, PAM-1 or PAM-1/OSX AtT-20 cells were treated with vehicle (Control) or other pharmacological agents to inactivate proteases; PAM-1 in cell lysates was visualized using the PHM antibody. Non-saturated images were quantified; each bar represents the mean from two experiments performed in triplicate ± S.D. Con A, concanamycin A.
Surface biotinylation for 10 min at 37 °C followed by incubation for 0, 1, 2, or 4 h at 37 °C was used to assess recycling versus degradation of internalized PAM (Fig. 8B). As observed previously, the amount of biotinylated PAM-1 and PALm declined substantially during a 1-h chase; more than half of the biotinylated PAM-1 and PALm remained intact after a 4-h chase. In contrast, biotinylated PAM-1/OSX and PALm/OSX disappeared more rapidly, with only 18–25% remaining intact after a 4-h chase (Fig. 8B, panel i). Recycled PAM-1 can be converted into sPAM in the endocytic pathway and into sPHM after it is returned to the secretory pathway. Medium samples from PAM-1/OSX cells contained very little biotinylated sPAM, although sPHM was detectable, the percentage of the biotinylated PAM-1/OSX converted into sPHM was much lower than the percentage of biotinylated PAM-1 converted into sPHM (Fig. 8B, panel ii). Our surface biotinylation studies indicated that PAM-1/OSX was rapidly degraded after internalization.
When treating PAM-1/OSX AtT-20 cells with MG-132, we noted that the levels of intact PAM-1 rose (Fig. 6C); the same treatment of PAM-1 AtT-20 cells did not produce an increase in PAM-1 levels. In addition to acting as a potent proteasomal inhibitor, MG-132 inhibits cathepsins A and B, major lysosomal proteases (38–41). To distinguish between the role of lysosomes and the proteasome in the degradation of internalized PAM-1/OSX, AtT-20 cells were treated with lactacystin, a specific proteasomal inhibitor (41), for 5 h. Lactacystin treatment was without effect on levels of PAM-1 or PAM-1/OSX (Fig. 8C), leading us to focus our efforts on lysosomal degradation.
Lysosomal proteases were inhibited by raising lysosomal pH using 20 mm NH4Cl, an alkalinizing agent, or concanamycin, a V-ATPase inhibitor (Fig. 8C) (42). Both treatments resulted in the accumulation of PAM-1/OSX but had little or no effect on levels of PAM-1. In addition, the effect of leupeptin, a designated lysosomal protease inhibitor (43), was assessed; an increase in intact PAM protein was again observed only in PAM-1/OSX cells. Data for MG-132 are shown for comparison; like alkalinization, MG132 had a greater effect on PAM-1/OSX levels than leupeptin. At steady state, our data indicated that O-glycosylation of PAM-1 protected it from degradation (Fig. 8C).
To quantify differences in the turnover of newly synthesized PAM-1 and PAM-1/OSX, pulse-chase labeling experiments were performed. PAM-1 and PAM-1/OSX AtT-20 cells were incubated in medium containing [35S]Met/[35S]Cys for 30 min and extracted or further incubated in chase medium for 1, 2, or 4 h; PHM was recovered from cell extracts and media by immunoprecipitation (Fig. 9A). After traversing the Golgi complex, newly synthesized PAM-1 can travel directly to the cell surface (constitutive pathway) or enter the regulated secretory pathway, where it is subject to AP-1-mediated return to the endocytic pathway or to cleavage by secretory granule proteases (44). Exocytosis places any granule resident PAM-1 and PALm on the cell surface, after which it can undergo endocytosis.
FIGURE 9.
Biosynthetic labeling demonstrates extensive degradation of PAM-1/OSX. A, AtT-20 cells expressing PAM-1 or PAM-1/OSX were incubated with [35S]Met/[35S]Cys for 30 min and either harvested immediately (pulse, P) or chased for 1, 2, or 4 h; aliquots representing 1/3rd of each cell extract or medium sample were immunoprecipitated with a PHM antibody. Immunoprecipitates were fractionated by SDS-PAGE and visualized by fluorography; major PAM products are indicated by arrows on the right. Data for PAM-1, sPAM, and sPHM in cell extracts and media were quantified using films exposed for different times; the amount of PAM-1 or PAM-1/OSX present after the pulse was taken as 100%. B, panel i, total recovery of intact PAM-1 (B, panel ii) and sPHM (B, panel iii) was calculated by taking 120-kDa PAM present after the pulse as 100%. B, panel iv, secretion of newly synthesized sPAM was plotted as a percent of pulse PAM-1 at each chase point in both cell types. Graphs were constructed from mean values of two independent experiments.
Analysis of cell extracts revealed a rapid decline in levels of newly synthesized PAM-1 and PAM-1/OSX (Fig. 9A). Although newly synthesized sPHM accumulated in the cells and then in the media of PAM-1 cells, production of newly synthesized sPHM from PAM-1/OSX ceased after 2 h. We first quantified the total recovery of labeled PAM proteins during the chase (Fig. 9B, panel i). As expected, almost all of the newly synthesized PAM-1 could be accounted for in the cells or in the medium. In contrast, only 25% of the newly synthesized PAM-1/OSX was recovered after the 4-h chase.
An examination of the amount of intact PAM-1 remaining at each chase time revealed a more significant loss of PAM-1/OSX after the 2-h chase than after the 1-h chase (Fig. 9B, panel ii), as expected if its increased degradation occurred in the endocytic pathway. Further support for this conclusion comes from the fact that production of sPHM from PAM-1 increased over time, while production of sPHM from PAM-1/OSX ceased after 1 h (Fig. 9B, panel iii). Taken together, our analyses demonstrated that loss of its O-glycans subjected PAM-1/OSX to increased degradation following endocytosis, eliminating its re-entry into the regulated secretory pathway.
Bifunctional sPAM, which is produced in the endocytic compartment (Fig. 6A), was readily detected in the medium of PAM-1 cells after the 2-h chase (Fig. 9A); in contrast, very little sPAM was produced by PAM-1/OSX cells. After the 4-h chase, 16–18% of the newly synthesized intact PAM-1 was recovered from spent medium as sPAM, whereas less than 5% of the newly synthesized PAM-1/OSX was recovered as sPAM (Fig. 9B, panel iv).
PAM-1/OSX Shows Diminished Ability to Form Complexes
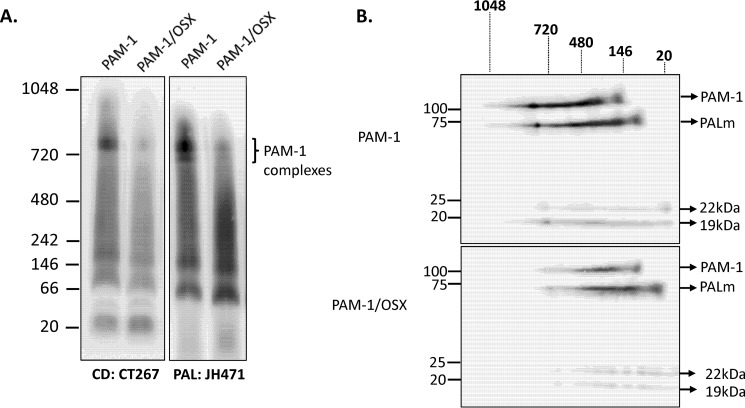
The alterations observed in the trafficking and processing of PAM-1/OSX closely resembled the alterations observed in PAM-1/H3A, which lacks a pH-sensitive His cluster in the linker regions separating PHMcc from exon 16 (30). Because the altered behavior of PAM-1/H3A suggested a role for pH-dependent interactions of PAM-1 with other proteins, we turned to blue native-PAGE to compare the ability of PAM-1 and PAM-1/OSX to participate in multiprotein complexes. Membranes prepared from AtT-20 cells expressing PAM-1 or PAM-1/OSX were solubilized in 0.5% n-dodecyl β-d-maltopyranoside, a mild non-ionic detergent. More than 90% of the membrane-associated PAM was soluble in 0.5% n-dodecyl β-d-maltopyranoside.
After electrophoretic separation at pH 7.5, native complexes were electroblotted onto a PVDF membrane and visualized using a CD antibody. Complexes ranging in mass from 780 to 66 kDa were consistently observed in PAM-1 AtT-20 samples (Fig. 10A). In some samples, three distinct complexes at 790 ± 25, 770 ± 0, and 720 ± 10 kDa could be resolved. These high molecular weight complexes were either absent or much less prevalent in samples prepared from PAM-1/OSX cells. Our data suggest that O-glycosylation of the exon 16 region of PAM-1 played an essential role in the ability of PAM-1 to interact with other proteins.
FIGURE 10.
Blue native-PAGE analysis reveals diminished ability of PAM-1/OSX to form complexes in AtT-20 cells. A, solubilized membrane proteins prepared from PAM-1 and PAM-1/OSX AtT-20 cells were separated by blue native-PAGE. Molecular weight markers are indicated to the left. After transfer to a PVDF membrane under native conditions, duplicate blots were probed with antibodies to PAL or PAM-CD. Multiple high molecular weight complexes containing PAM were apparent. B, single gel lanes from PAM-1 or PAM-1/OSX AtT-20 cells were excised, reduced, and denatured and then fractionated by SDS-PAGE. After transfer to a PVDF membrane, PAM proteins were visualized using a CD antibody; PAM products are indicated on the right and native molecular weight markers are shown above the blots. This experiment was repeated three times with separate lysates, with similar results.
To explore this hypothesis, gel lanes containing samples from PAM-1 and PAM-1/OSX AtT-20 cells fractionated in one dimension were reduced and denatured before separation in the second dimension by SDS-PAGE. After transferring the denatured proteins to a PVDF membrane, PAM proteins were visualized using a CD antibody (Fig. 10B). As expected, Western blot signals for PAM-1/OSX failed to align with those for PAM-1, confirming the diminished ability of PAM-1/OSX to participate in the higher order complexes formed with PAM-1 (Fig. 10B). After two-dimensional BN-PAGE/SDS-PAGE, proteins belonging to the same complex aligned vertically in the second dimension; the three largest PAM complexes differed in their content of PAM-1, PALm, and TMD/CD.
Discussion
Two Sites in PAM Exon 16 Are Subjected to Mucin-type O-Glycosylation
Our earlier studies suggested that the sulfated O-glycans detected in PAM-1 attached to Ser/Thr residues at two sites in exon 16 could be labeled with [3H]galactose and that at least one was also sialylated (3). The smaller tryptic glycopeptide was mapped to a site preceding the endoproteolytic cleavage site in exon 16, and the larger more heavily glycosylated tryptic glycopeptide was mapped to a site beyond this cleavage site. Mass spectroscopic analysis of the tryptic peptides of PAM-4 allowed us to identify Ser471Thr472 as the attachment site(s) beyond the cleavage site. Because fragmentation did not occur between Ser471 and Thr472, both residues were modified to preclude O-glycosylation at either position. Two Ser/Thr-containing exon 16 tryptic peptides were not identified in our mass spectroscopic analysis of rat PAM-4, identifying them as potential O-glycosylation sites; the single Ser/Thr in each peptide was mutated to Ala. The decreased mass of PAM-4 bearing a T417A mutation identified this as a second O-glycosylation site in exon 16; mutation of Ser442 was without effect on the mass of PAM-4. Mutations at both sites (Thr417 and Ser471Thr472) were combined to create PAM-1/OSX, an O-glycan depleted version of PAM-1, which retained full catalytic activity and was largely localized to post-endoplasmic reticulum compartments when stably expressed in AtT-20 cells (Fig. 5A).
The synthesis of O-linked oligosaccharides commences in the Golgi apparatus; in many proteins, their presence influences protein localization, stability, activity, and interactions with other biomolecules (45). The transfer of GalNAc from UDP-N-acetylgalactosamine onto Ser and Thr residues in target proteins is catalyzed by a large family of polypeptide GalNAc transferases (8). Transcriptomic analysis demonstrated significant levels of expression of seven GalNAc transferases in AtT-20 cells (1, 2, 9–12, 18) (data not shown). Different expression patterns and different substrate specificities mean that mutations in a single GalNAc transferase can affect leukocyte rolling (GALNT1), left-right development (GALNT11), or the development of colon cancer (GALNT12). Sulfated O-glycans in pituitary glycoprotein hormones such as thyrotropin and luteinizing hormone influence multiple functions, including subunit combination, intracellular proteolysis, and subunit aggregation (46–49).
Glycosyltransferase specificity relies on higher order protein structure rather than on the primary sequence (50), meaning that a consensus sequence motif for this type of modification has not been identified. These enzymes prefer long and disordered regions, and Pro residues are commonly found near O-glycosylation sites (at −1 and/or +3) (51). Consistent with these observations, exon 16 is an intrinsically disordered region, Thr417 is preceded by Pro416, and Pro475 is separated from Ser471Thr472 by two amino acids.
In both the cytosolic and luminal compartments, Ser and Thr are subject to both glycosylation and phosphorylation. In the cytosol, both modifications are reversible, and dynamic O-linked monosaccharide addition (GlcNAc-β-Ser/Thr) and phosphorylation are key regulators of protein-protein interactions and protein function (52). The addition of mucin-type O-glycans is not reversible, and our knowledge of interactions between secretory pathway protein O-GalNAc addition and phosphorylation is still quite limited.
O-Glycosylation Affects the Stability of PAM
Regulated secretory pathway proteins like PAM, which enter immature secretory granules upon exiting the Golgi complex, become prohormone convertase substrates as luminal pH drops and calcium levels rise (53–55). Prohormone convertase 1-mediated cleavage at the Lys-Lys site in PAM-1 (35) increases the specific activity of PHM and allows the secretion of soluble PHM; PAM-2, which lacks exon 16 and this Lys-Lys site, remains largely intact. Our metabolic labeling study provided no evidence for enhanced cleavage at this Lys-Lys site in PAM-1/OSX. Soluble PHM accumulates at the tips of the cellular processes in PAM-1/OSX cells, which release more sPHM in response to secretagogue stimulation.
Recent studies have demonstrated a role for O-glycans in controlling prohormone convertase processing of FGF 23 (8, 10), Tango-1 (9), pro-brain natriuretic peptide (56), and angiopoietin-like protein-3 (ANGPTL3) (57). Lacking O-glycans in exon 16, a 68-kDa product that contained part of exon 16, PAL, the transmembrane and cytosolic domains of PAM, was generated from PAM-1/OSX. Based on the charge complementarity of its catalytic pocket, furin, a widely expressed prohormone convertase whose crystal structure has been determined (58), shows a preference for RXXR. Among furin substrates, the frequency of Arg at P4 is >90%, but a less favorable amino acid residue at P4 can be compensated for by Arg or Lys at P2 and P6. The ability of α1-antitrypsin Portland to diminish 68-kDa PALm production from PAM-1/OSX is consistent with the conclusion that attachment of an O-glycan to Ser-Thr472 blocks the ability of a furin-like enzyme to cleave after Arg465.
Differences in the protease sensitivity of PAM-1/OSX were also apparent in the in vitro studies conducted with trypsin. Lower amounts of trypsin were required to produce soluble PHM (band#3) and soluble PAL (band#6) from PAM-1/OSX than from PAM-1. In addition, the 60-kDa fragment of PAL generated from PAM-1 was not observed when PAM-1/OSX was exposed to trypsin. This product is thought to represent cleavage at the Lys-Lys site in exon 16 and at a site between the C terminus of PAL and the transmembrane domain. The β-propeller structure of PALcc positions its N and C termini in close proximity (59); as a result, the O-glycans attached to Ser471–Thr472 could interact with the stalk region that separates PALcc from the transmembrane domain, affecting its susceptibility to proteolysis.
O-Glycosylation of PAM-1 Is Essential for Endocytic Trafficking and for Complex Formation
At steady state, only about 2% of the intact PAM-1 in AtT-20 corticotrope tumor cells is on the cell surface (Fig. 8) (5, 37). PAM-1 that reaches the plasma membrane as a result of basal or regulated secretion is rapidly internalized, after which it can be returned to the regulated secretory pathway or subjected to lysosomal degradation. PAM trafficking through multivesicular bodies is controlled by the phosphorylation and dephosphorylation of multiple sites in its intrinsically disordered cytosolic domain and by the interaction of this region with multiple cytosolic proteins, such as Kalirin and Uhmk1 (31, 36). Based on our analysis of the endocytic trafficking of PAM-1/OSX, pH-dependent, O-glycan-mediated luminal interactions play an essential role in the endocytic trafficking of PAM-1.
The alterations observed within the first 2 min of the removal of PAM-1/OSX from the cell surface may account for its altered stability, processing, and secretion (60, 61). Ectodomain antibody internalized by PAM-1 could later be found in early endosomes and multivesicular bodies; the antibody internalized by PAM-1/OSX remained in WGA-positive tubular endosomes. All of the other differences observed can be explained as a consequence of this deficit. First, its impaired ability to exit the tubular endosomes would preclude the production of sPAM/OSX and 19-kDa TMD/CD. Lacking its normal substrate, the γ-secretase-mediated cleavage that generates sfCD could not occur (33); the altered morphology of both PAM-1/OSX AtT-20 lines is consistent with this conclusion. The increased sensitivity of PAM-1 OSX to proteolytic degradation in test tube experiments and in intact cells could make it more susceptible to lysosomal degradation, but the subcellular location at which internalized PAM-1/OSX is degraded remains unclear. Exposure to leupeptin was less effective at stabilizing PAM-1/OSX than elevating luminal pH with concanamycin or NH4Cl. Strikingly, PAM-1 lacking the His cluster that renders it pH-sensitive (PAM-1/H3A) was also much more sensitive to degradation in the endocytic pathway (30).
The aberrant endocytic trafficking of PAM-1/H3A provided the first evidence that the luminal domains of PAM-1 played an essential role in its endocytic trafficking (30). Our current studies demonstrate an essential role for the O-glycans attached to exon 16 in both endocytic trafficking and the formation of high molecular weight complexes. It is tempting to speculate that the ability of PAM-1 to form these complexes is linked to its ability to exit from tubular endosomes and traverse the multivesicular bodies. The mild detergents used in BN-PAGE, which was developed for the analysis of mitochondrial protein complexes, appear well suited to the study of endocytic trafficking (62); at steady state, most of the PAM-1 in cell lysates is located in endocytic compartments (5, 63). Aberrant O-glycosylation is known to alter early steps in the endocytic trafficking of MUC1, a type 1 transmembrane protein with multiple mucin-type O-linked glycans (64), and Notch 1 (65). In addition, lectins are well recognized contributors to the endocytic trafficking so essential to immune cell functioning.
Author Contributions
K. S. V., R. E. M., and B. A. E. designed and performed experiments and wrote the paper; N. B. did the electron microscopy and ultrastructural image analysis; T. K. T. L. performed the mass spectrometric analyses.
Acknowledgments
We thank Darlene D'Amato and Yanping Wang for their many contributions to the experiments and Drs. Iris Lindberg and Gary Thomas for the α1-PDX-encoding adenovirus and advice on its use. We also thank Jean Kanyo for assistance in collecting and interpreting the mass spectroscopy data. We thank the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki, for providing laboratory facilities.
This work was supported by National Institutes of Health Grants DK-032949 and DA-018343, the Janice and Rodney Reynolds Endowment, the Scoville Endowment, the Liv och Hälsa Foundation, Finska Läkaresällskapet, and the Perklén Foundation. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PAM
- peptidylglycine α-amidating monooxygenase
- CSFM
- complete serum-free medium
- PAL
- peptidyl-α-hydroxyglycine α-amidating lyase
- PALcc
- catalytic core of PAL
- PHM
- peptidylglycine α-hydroxylating monooxygenase
- PHMcc
- catalytic core of PHM
- sPAM
- soluble PAM
- sfCD
- soluble fragment of PAM cytosolic domain
- TGN
- trans-Golgi network
- TMD/CD
- trans-membrane domain/cytoplasmic domain
- WGA
- wheat germ agglutinin
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- BN-PAGE
- blue native-PAGE
- r
- rat
- α1-PDX
- α1-antitrypsin Portland.
References
- 1. Eipper B. A., Milgram S. L., Husten E. J., Yun H. Y., and Mains R. E. (1993) Peptidylglycine α-amidating monooxygenase: a multifunctional protein with catalytic, processing, and routing domains. Protein Sci. 2, 489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ouafik L. H., Stoffers D. A., Campbell T. A., Johnson R. C., Bloomquist B. T., Mains R. E., and Eipper B. A. (1992) The multifunctional peptidylglycine α-amidating monooxygenase gene: exon/intron organization of catalytic, processing, and routing domains. Mol. Endocrinol. 6, 1571–1584 [DOI] [PubMed] [Google Scholar]
- 3. Yun H. Y., Milgram S. L., Keutmann H. T., and Eipper B. A. (1995) Phosphorylation of the cytosolic domain of peptidylglycine α-amidating monooxygenase. J. Biol. Chem. 270, 30075–30083 [DOI] [PubMed] [Google Scholar]
- 4. Stoffers D. A., Ouafik L., and Eipper B. A. (1991) Characterization of novel mRNAs encoding enzymes involved in peptide α-amidation. J. Biol. Chem. 266, 1701–1707 [PubMed] [Google Scholar]
- 5. Milgram S. L., Kho S. T., Martin G. V., Mains R. E., and Eipper B. A. (1997) Localization of integral membrane peptidylglycine α-amidating monooxygenase in neuroendocrine cells. J. Cell Sci. 110, 695–706 [DOI] [PubMed] [Google Scholar]
- 6. Ciccotosto G. D., Schiller M. R., Eipper B. A., and Mains R. E. (1999) Induction of integral membrane PAM expression in AtT-20 cells alters the storage and trafficking of POMC and PC1. J. Cell Biol. 144, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolhekar A. S., Bell J., Shiozaki E. N., Jin L., Keutmann H. T., Hand T. A., Mains R. E., and Eipper B. A. (2002) Essential features of the catalytic core of peptidyl-α-hydroxyglycine α-amidating lyase. Biochemistry 41, 12384–12394 [DOI] [PubMed] [Google Scholar]
- 8. Schjoldager K. T., Vester-Christensen M. B., Goth C. K., Petersen T. N., Brunak S., Bennett E. P., Levery S. B., and Clausen H. (2011) A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J. Biol. Chem. 286, 40122–40132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang L., Syed Z. A., van Dijk Härd I., Lim J. M., Wells L., and Ten Hagen K. G. (2014) O-Glycosylation regulates polarized secretion by modulating Tango1 stability. Proc. Natl. Acad. Sci. U.S.A. 111, 7296–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tagliabracci V. S., Engel J. L., Wiley S. E., Xiao J., Gonzalez D. J., Nidumanda Appaiah H., Koller A., Nizet V., White K. E., and Dixon J. E. (2014) Dynamic regulation of FGF23 by Fam20C phosphorylation, GalNAc-T3 glycosylation, and furin proteolysis. Proc. Natl. Acad. Sci. U.S.A. 111, 5520–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ninkovic T., and Hanisch F. G. (2007) O-Glycosylated human MUC1 repeats are processed in vitro by immunoproteasomes. J. Immunol. 179, 2380–2388 [DOI] [PubMed] [Google Scholar]
- 12. Takeuchi H., and Haltiwanger R. S. (2014) Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 453, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lowe J. B., and Marth J. D. (2003) A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 72, 643–691 [DOI] [PubMed] [Google Scholar]
- 14. Gill D. J., Chia J., Senewiratne J., and Bard F. (2010) Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J. Cell Biol. 189, 843–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanisch F. G., Müller S., Hassan H., Clausen H., Zachara N., Gooley A. A., Paulsen H., Alving K., and Peter-Katalinic J. (1999) Dynamic epigenetic regulation of initial O-glycosylation by UDP-N-acetylgalactosamine:peptide N-acetylgalactosaminyltransferases. site-specific glycosylation of MUC1 repeat peptide influences the substrate qualities at adjacent or distant Ser/Thr positions. J. Biol. Chem. 274, 9946–9954 [DOI] [PubMed] [Google Scholar]
- 16. Levery S. B., Steentoft C., Halim A., Narimatsu Y., Clausen H., and Vakhrushev S. Y. (2015) Advances in mass spectrometry driven O-glycoproteomics. Biochim. Biophys. Acta 1850, 33–42 [DOI] [PubMed] [Google Scholar]
- 17. Herzog B. H., Fu J., and Xia L. (2014) Mucin-type O-glycosylation is critical for vascular integrity. Glycobiology 24, 1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bergstrom K. S., and Xia L. (2013) Mucin-type O-glycans and their roles in intestinal homeostasis. Glycobiology 23, 1026–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thiel C., and Körner C. (2013) Therapies and therapeutic approaches in congenital disorders of glycosylation. Glycoconj. J. 30, 77–84 [DOI] [PubMed] [Google Scholar]
- 20. Hennet T. (2012) Diseases of glycosylation beyond classical congenital disorders of glycosylation. Biochim. Biophys. Acta 1820, 1306–1317 [DOI] [PubMed] [Google Scholar]
- 21. Ju T., and Cummings R. D. (2005) Protein glycosylation: chaperone mutation in Tn syndrome. Nature 437, 1252. [DOI] [PubMed] [Google Scholar]
- 22. Tran D. T., and Ten Hagen K. G. (2013) Mucin-type O-glycosylation during development. J. Biol. Chem. 288, 6921–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., and Sprecher E. (2004) Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 36, 579–581 [DOI] [PubMed] [Google Scholar]
- 24. Kato K., Jeanneau C., Tarp M. A., Benet-Pagès A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., and Clausen H. (2006) Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J. Biol. Chem. 281, 18370–18377 [DOI] [PubMed] [Google Scholar]
- 25. Stuchlová H. M., Raška M., Clausen H., and Novak J. (2013) Aberrant O-glycosylation and anti-glycan antibodies in an autoimmune disease IgA nephropathy and breast adenocarcinoma. Cell. Mol. Life Sci. 70, 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma X. M., Miller M. B., Vishwanatha K. S., Gross M. J., Wang Y., Abbott T., Lam T. T., Mains R. E., and Eipper B. A. (2014) Nonenzymatic domains of Kalirin7 contribute to spine morphogenesis through interactions with phosphoinositides and Abl. Mol. Biol. Cell 25, 1458–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krishnan N., Lam T. T., Fritz A., Rempinski D., O'Loughlin K., Minderman H., Berezney R., Marzluff W. F., and Thapar R. (2012) The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol. Cell. Biol. 32, 4306–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jean F., Stella K., Thomas L., Liu G., Xiang Y., Reason A. J., and Thomas G. (1998) α1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. Proc. Natl. Acad. Sci. U.S.A. 95, 7293–7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De M., Ciccotosto G. D., Mains R. E., and Eipper B. A. (2007) Trafficking of a secretory granule membrane protein is sensitive to copper. J. Biol. Chem. 282, 23362–23371 [DOI] [PubMed] [Google Scholar]
- 30. Vishwanatha K., Bäck N., Mains R. E., and Eipper B. A. (2014) A histidine-rich linker region in peptidylglycine α-amidating monooxygenase has the properties of a pH sensor. J. Biol. Chem. 289, 12404–12420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bäck N., Rajagopal C., Mains R. E., and Eipper B. A. (2010) Secretory granule membrane protein recycles through multivesicular bodies. Traffic 11, 972–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bäck N., Litonius E., Mains R. E., and Eipper B. A. (2004) Fluoride causes reversible dispersal of Golgi cisternae and matrix in neuroendocrine cells. Eur. J. Cell Biol. 83, 389–402 [DOI] [PubMed] [Google Scholar]
- 33. Rajagopal C., Stone K. L., Mains R. E., and Eipper B. A. (2010) Secretion stimulates intramembrane proteolysis of a secretory granule membrane enzyme. J. Biol. Chem. 285, 34632–34642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Husten E. J., Tausk F. A., Keutmann H. T., and Eipper B. A. (1993) Use of endoproteases to identify catalytic domains, linker regions, and functional interactions in soluble peptidylglycine α-amidating monooxygenase. J. Biol. Chem. 268, 9709–9717 [PubMed] [Google Scholar]
- 35. Fricker L. D. (2010) Analysis of mouse brain peptides using mass spectrometry-based peptidomics: implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol. Biosyst. 6, 1355–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Francone V. P., Ifrim M. F., Rajagopal C., Leddy C. J., Wang Y., Carson J. H., Mains R. E., and Eipper B. A. (2010) Signaling from the secretory granule to the nucleus: Uhmk1 and PAM. Mol. Endocrinol. 24, 1543–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steveson T. C., Keutmann H. T., Mains R. E., and Eipper B. A. (1999) Phosphorylation of cytosolic domain Ser(937) affects both biosynthetic and endocytic trafficking of peptidylglycine α-amidating monooxygenase. J. Biol. Chem. 274, 21128–21138 [DOI] [PubMed] [Google Scholar]
- 38. Ito H., Watanabe M., Kim Y. T., and Takahashi K. (2009) Inhibition of rat liver cathepsins B and L by the peptide aldehyde benzyloxycarbonyl-leucyl-leucyl-leucinal and its analogues. J. Enzyme Inhib. Med. Chem. 24, 279–286 [DOI] [PubMed] [Google Scholar]
- 39. Longva K. E., Blystad F. D., Stang E., Larsen A. M., Johannessen L. E., and Madshus I. H. (2002) Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shirley R. B., Kaddour-Djebbar I., Patel D. M., Lakshmikanthan V., Lewis R. W., and Kumar M. V. (2005) Combination of proteasomal inhibitors lactacystin and MG132 induced synergistic apoptosis in prostate cancer cells. Neoplasia 7, 1104–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kadlcíková J., Holecek M., Safránek R., Tilser I., and Kessler B. M. (2004) Effects of proteasome inhibitors MG132, ZL3VS and AdaAhx3L3VS on protein metabolism in septic rats. Int. J. Exp. Pathol. 85, 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sobota J. A., Bäck N., Eipper B. A., and Mains R. E. (2009) Inhibitors of the V0 subunit of the vacuolar H+-ATPase prevent segregation of lysosomal- and secretory-pathway proteins. J. Cell Sci. 122, 3542–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qin H., Shao Q., Igdoura S. A., Alaoui-Jamali M. A., and Laird D. W. (2003) Lysosomal and proteasomal degradation play distinct roles in the life cycle of Cx43 in gap junctional intercellular communication-deficient and -competent breast tumor cells. J. Biol. Chem. 278, 30005–30014 [DOI] [PubMed] [Google Scholar]
- 44. Bonnemaison M. L., Bäck N., Duffy M. E., Ralle M., Mains R. E., and Eipper B. A. (2015) Adaptor protein-1 complex affects the endocytic trafficking and function of peptidylglycine α-amidating monooxygenase, a luminal cuproenzyme. J. Biol. Chem. 290, 21264–21279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Janetzko J., and Walker S. (2014) The making of a sweet modification: structure and function of O-GlcNAc transferase. J. Biol. Chem. 289, 34424–34432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Willey K. P. (1999) An elusive role for glycosylation in the structure and function of reproductive hormones. Hum. Reprod. Update 5, 330–355 [DOI] [PubMed] [Google Scholar]
- 47. Taylor T., and Weintraub B. D. (1985) Thyrotropin (TSH)-releasing hormone regulation of TSH subunit biosynthesis and glycosylation in normal and hypothyroid rat pituitaries. Endocrinology 116, 1968–1976 [DOI] [PubMed] [Google Scholar]
- 48. Manzella S. M., Dharmesh S. M., Beranek M. C., Swanson P., and Baenziger J. U. (1995) Evolutionary conservation of the sulfated oligosaccharides on vertebrate glycoprotein hormones that control circulatory half-life. J. Biol. Chem. 270, 21665–21671 [DOI] [PubMed] [Google Scholar]
- 49. Manzella S. M., Hooper L. V., and Baenziger J. U. (1996) Oligosaccharides containing β 1,4-linked N-acetylgalactosamine, a paradigm for protein-specific glycosylation. J. Biol. Chem. 271, 12117–12120 [DOI] [PubMed] [Google Scholar]
- 50. Hansen J. E., Lund O., Engelbrecht J., Bohr H., Nielsen J. O., and Hansen J. E. (1995) Prediction of O-glycosylation of mammalian proteins: specificity patterns of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase. Biochem. J. 308, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thanka Christlet T. H., and Veluraja K. (2001) Database analysis of O-glycosylation sites in proteins. Biophys. J. 80, 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kamemura K., and Hart G. W. (2003) Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: a new paradigm for metabolic control of signal transduction and transcription. Prog. Nucleic Acids Res. Mol. Biol. 73, 107–136 [DOI] [PubMed] [Google Scholar]
- 53. Dillon S. L., Williamson D. M., Elferich J., Radler D., Joshi R., Thomas G., and Shinde U. (2012) Propeptides are sufficient to regulate organelle-specific pH-dependent activation of furin and proprotein convertase 1/3. J. Mol. Biol. 423, 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou A., and Mains R. E. (1994) Endoproteolytic processing of proopiomelanocortin and prohormone convertases 1 and 2 in neuroendocrine cells overexpressing prohormone convertases 1 or 2. J. Biol. Chem. 269, 17440–17447 [PubMed] [Google Scholar]
- 55. Schmidt W. K., and Moore H. P. (1995) Ionic milieu controls the compartment-specific activation of pro-opiomelanocortin processing in AtT-20 cells. Mol. Biol. Cell 6, 1271–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Semenov A. G., Postnikov A. B., Tamm N. N., Seferian K. R., Karpova N. S., Bloshchitsyna M. N., Koshkina E. V., Krasnoselsky M. I., Serebryanaya D. V., and Katrukha A. G. (2009) Processing of pro-brain natriuretic peptide is suppressed by O-glycosylation in the region close to the cleavage site. Clin. Chem. 55, 489–498 [DOI] [PubMed] [Google Scholar]
- 57. Schjoldager K. T., Vester-Christensen M. B., Bennett E. P., Levery S. B., Schwientek T., Yin W., Blixt O., and Clausen H. (2010) O-Glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J. Biol. Chem. 285, 36293–36303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Henrich S., Cameron A., Bourenkov G. P., Kiefersauer R., Huber R., Lindberg I., Bode W., and Than M. E. (2003) The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat. Struct. Biol. 10, 520–526 [DOI] [PubMed] [Google Scholar]
- 59. Chufán E. E., De M., Eipper B. A., Mains R. E., and Amzel L. M. (2009) Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme. Structure 17, 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lakadamyali M., Rust M. J., and Zhuang X. (2006) Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell 124, 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jean-Alphonse F., Bowersox S., Chen S., Beard G., Puthenveedu M. A., and Hanyaloglu A. C. (2014) Spatially restricted G protein-coupled receptor activity via divergent endocytic compartments. J. Biol. Chem. 289, 3960–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schägger H., Cramer W. A., and von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 63. Oyarce A. M., and Eipper B. A. (1995) Identification of subcellular compartments containing peptidylglycine α-amidating monooxygenase in rat anterior pituitary. J. Cell Sci. 108, 287–297 [DOI] [PubMed] [Google Scholar]
- 64. Altschuler Y., Kinlough C. L., Poland P. A., Bruns J. B., Apodaca G., Weisz O. A., and Hughey R. P. (2000) Clathrin-mediated endocytosis of MUC1 is modulated by its glycosylation state. Mol. Biol. Cell 11, 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stanley P. (2007) Regulation of Notch signaling by glycosylation. Curr. Opin. Struct. Biol. 17, 530–535 [DOI] [PMC free article] [PubMed] [Google Scholar]