Abstract
After reanalyzing simulations of NuG2—a designed mutant of protein G—generated by Lindorff-Larsen et al. with time structure-based independent components analysis and Markov state models as well as performing 1.5 ms of additional sampling on Folding@home, we found an intermediate with a register-shift in one of the β-sheets that was visited along a minor folding pathway. The minor folding pathway was initiated by the register-shifted sheet, which is composed of solely nonnative contacts, suggesting that for some peptides, nonnative contacts can lead to productive folding events. To confirm this experimentally, we suggest a mutational strategy for stabilizing the register shift, as well as an infrared experiment that could observe the nonnative folding nucleus.
Main Text
There are many important questions surrounding the physics of protein folding that remain unanswered (1). Why do proteins fold quickly? What is the role of nonnative interactions in the folding process? Are there multiple pathways to the folded state? Molecular dynamics (MD) simulation has proven to be a powerful tool that can provide an atomic level answer to these and other biophysical questions (2, 3, 4) as well as a way for interpreting and predicting new experiments (5). Nonetheless, making sense of large and high-dimensional MD datasets can be difficult, but the analysis can be made simpler by employing Markov state models (MSMs). An MSM consists of a set of states (groups of peptide conformations) and the probabilities of interconversion between those states (6, 7, 8, 9). There are two main steps in the MSM construction process. The first is clustering the data into some (preferably small) set of states and the second is estimating the probabilities of transitioning between states. Recent work has highlighted the importance of the state decomposition: the general features of the model depend significantly on the choice of the state space and basic structural metrics, such as root mean-square deviation (RMSD) in atom positions or dihedral angles may not be the best choice for clustering protein conformations (10). In addition, recent improvements have illustrated that distance metrics designed to ignore fast degrees of freedom can produce a superior state decomposition and provide better estimates of kinetic and thermodynamic observables (11, 12). Alternative approaches, which use an energy-minimization during the clustering step, can also be useful for avoiding pathologies of strictly structural metrics (13).
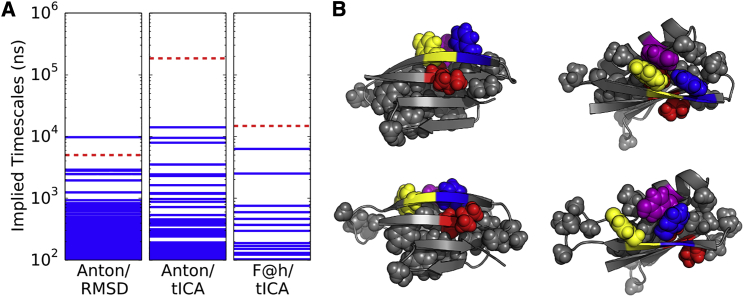
In this Letter, we discuss the time structure-based independent components analysis (tICA) method applied to simulations of the peptide NuG2. This protein is a mutant of protein G, which was computationally designed to fold faster via mutations that stabilized a β-sheet between strands 1 and 2 (14). Previously, Beauchamp et al. (15) built an MSM on the dataset generated by Lindorff-Larsen et al. (16) utilizing the RMSD of atom positions as the distance metric during the clustering step. We reanalyzed the same dataset, but used tICA to build the new MSM (see MSM Construction in the Supporting Material). When we compared the RMSD-based model to one built using the tICA metric, we found that there was a new slow timescale (∼180 μs) in the tICA model that was absent in the RMSD-based model (Fig. 1). This slow process corresponded to a near-native state, which had a two-residue register shift in the sheet formed between strands 1 and 2. In fact, this process was also observed in the RMSD-based model (see Fig. S1 in the Supporting Material), but the corresponding timescale was two orders-of-magnitude faster in the original model (Fig. 1). The timescale’s sensitivity to the choice of state decomposition illustrated that this eigenprocess was not adequately sampled in the original simulation. In fact, this register-shifted state was only visited at the very end of a single trajectory. To improve our estimate of the register-shifted state’s kinetic and thermodynamic properties, we used Folding@home to generate 1.5 ms of aggregate sampling (17) (see Folding@home Sampling in the Supporting Material). The folding timescale of the new MSM was slightly faster (∼6 μs) than previous models, likely due to using one order-of-magnitude fewer states. However, recent results have illustrated that models built with greater than a few hundred states may be overfit to the observed data (18), and we are therefore more confident in this model than the previous ones.
Figure 1.
(A) A slow eigenprocess (red, dashed line) was found in the both the tICA and RMSD models built with the Anton dataset, but with two very different timescales indicating a large uncertainty in the model. After producing a new dataset on Folding@home, we found that the timescale was best estimated to be ∼15 μs. (B) The slow timescale corresponded to a two-residue register shift in strand two. The native state is shown in the top two images, and the register shift in the bottom two. There is a corresponding two-residue shift in the hydrophobic core, with Tyr16 (yellow) contacting Tyr33 (purple) in the native state, while in the register-shifted state, Phe14 (blue) stacks with Tyr33 (purple). To see this figure in color, go online.
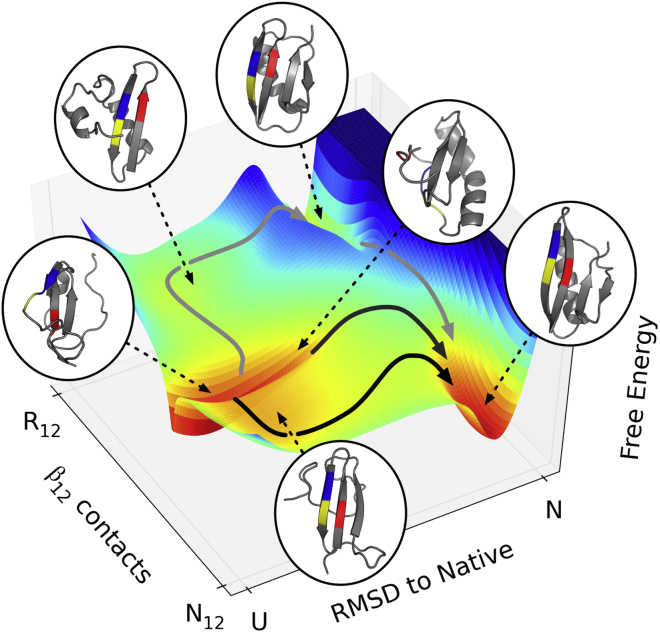
Using transition path theory (19), we found that most of the reactive flux went through a pathway that first forms the sheet between strands 1 and 2, and then forms the remainder of the native secondary structure (Fig. 2). However, ∼1% of the flux flowed through the register-shifted state along a pathway that again first forms the sheet between strands 1 and 2, but with a two-residue register shift. After the formation of this sheet, the remainder of the secondary structure forms as it is in the native state and then in a final step, strand 2 shifts to the native register. Because the register shift contacts are nonnative, this pathway is nucleated by a state that is made up of entirely nonnative secondary structure.
Figure 2.
Folding occurred along three main pathways. The largest flux pathway (∼55%) proceeded through the native sheet between strands 1 and 2 (N12) and then to the folded state. The second largest pathway (∼10%) proceeded by first forming the sheet between strands 3 and 4, and then folding to the native state. Additionally, a low-flux (∼1%) pathway proceeded through the register-shifted state (R12). To see this figure in color, go online.
The register-shifted state is fairly stable, having an equilibrium free energy (ΔG) of ∼2 kcal/mol above the native state. Interestingly, the two-residue shift in secondary structure is also accompanied by a two-residue shift in the hydrophobic contacts in the core (Fig. 1). A similar shift in the hydrophobic contacts was observed for the register-shifted states found in NTL91–39 (11), suggesting a larger trend that register-shifted states can be stabilized by favorable hydrophobic packing. In fact, these results suggest that register shifts are more probable when the shift in register does not significantly disrupt the hydrophobic core.
By comparing the hydrophobic contacts in the register-shifted and native folds, Tyr16 and Phe14 appear to be in competition for forming a contact with Tyr33 (Fig. 1). Therefore, by mutating one of these residues we believe it is possible to stabilize (or destabilize) the register-shifted state relative to the native fold. For instance, the Y16T mutation (which reverses one of the mutations made by Nauli et al. (14)) would remove the Tyr16-Tyr33 contact formed in the native fold and possibly force the protein to adopt a register-shifted conformation with Phe14 contacting Tyr33. The stacking of two benzenes has been estimated to be between 1 and 2 kcal/mol (20), and so, this mutation could shift the population significantly. However, we note that other mutations, which disrupt the contacts with Tyr33 or the hydrophobic packing, could also be useful. Because the register-shifted conformation has a significantly different backbone hydrogen-bonding network, infrared (IR) spectroscopy with carefully placed 13C=18O probes provides a powerful tool to confirm our observations. The stability of the register-shifted state puts observing this conformation at the cusp of what is possible with a conventional IR experiment. Therefore, we suggest leveraging the between-strand coupling of two heavy carbonyl labels to observe the state. Briefly, when two labeled carbonyls are on adjacent strands and within one or two residues, an anomalously large IR absorption occurs (21). We suggest labeling Leu5 and Thr12, which will be adjacent in the register-shifted conformation but separated by two residues in the native fold (see Fig. S6).
Our results provide compelling evidence for the necessity of kinetically informed distance metrics, and shed light on the limits of simple structural metrics such as RMSD. In addition, our new dataset indicates that the folding of NuG2 can proceed through a minor pathway that is nucleated entirely by nonnative secondary structure. Many have assumed that nonnative contacts can only give rise to so-called glassy free-energy landscapes that will slow the folding process (22). However, the mutations made by Nauli et al. (14) that introduced these stable nonnative contacts actually increased the rate of folding by several orders of magnitude. These observations are consistent with previous work from Clementi and Plotkin (23), who found that favorable nonnative interactions can speed up the folding reaction in simplified models, but because we studied a real protein, we can go further to suggest specific experiments that can verify (or refute) our conclusions. In fact, register shifts have been observed in many other MD studies (10, 24), and these nonnative folding nuclei may be quite prevalent. So long as the nonnative structure can correct itself without completely unfolding, then nonnative contacts may lead to productive folding events.
Author Contributions
C.R.S designed and performed the research, analyzed the data, and wrote the article; D.S. ran simulations and wrote the article; and V.S.P. designed the research and wrote the article.
Acknowledgments
We thank Robert T. McGibbon, Thomas J. Lane, and Kyle Beauchamp for discussions regarding MSMs as well as Caitlin Davis, Yu-Shan Lin, and Stephen Fried for useful discussions regarding IR spectroscopy.
The V.S.P. group gratefully acknowledges support from grants No. NIH-R01-GM062868 and No. NSF-MCB-0954714 and the SIMBIOS NIH Center for Biomedical Computation through the National Institutes of Health Roadmap for Medical Research Grant (No. U54-GM07297).
Editor: Nathan Baker.
Footnotes
Supporting Materials and Methods, Supporting Results, and eight figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30107-2.
Supporting Citations
References (25, 26, 27, 28, 29) appear in the Supporting Material.
Supporting Material
References
- 1.Dill K.A., MacCallum J.L. The protein-folding problem, 50 years on. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 2.Lane T.J., Shukla D., Pande V.S. To milliseconds and beyond: challenges in the simulation of protein folding. Curr. Opin. Struct. Biol. 2013;23:58–65. doi: 10.1016/j.sbi.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buch I., Giorgino T., De Fabritiis G. Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2011;108:10184–10189. doi: 10.1073/pnas.1103547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelley N.W., Vishal V., Pande V.S. Simulating oligomerization at experimental concentrations and long timescales: a Markov state model approach. J. Chem. Phys. 2008;129:214707. doi: 10.1063/1.3010881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voelz V.A., Jäger M., Pande V.S. Slow unfolded-state structuring in Acyl-CoA binding protein folding revealed by simulation and experiment. J. Am. Chem. Soc. 2012;134:12565–12577. doi: 10.1021/ja302528z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senne M., Trendelkamp-Schroer B., Noé F. EMMA: a software package for Markov model building and analysis. J. Chem. Theory Comput. 2012;8:2223–2238. doi: 10.1021/ct300274u. [DOI] [PubMed] [Google Scholar]
- 7.Prinz J.-H., Wu H., Noé F. Markov models of molecular kinetics: generation and validation. J. Chem. Phys. 2011;134:174105. doi: 10.1063/1.3565032. [DOI] [PubMed] [Google Scholar]
- 8.Schütte C., Noé F., Vanden-Eijnden E. Markov state models based on milestoning. J. Chem. Phys. 2011;134:204105. doi: 10.1063/1.3590108. [DOI] [PubMed] [Google Scholar]
- 9.Bowman G.R., Huang X., Pande V.S. Using generalized ensemble simulations and Markov state models to identify conformational states. Methods. 2009;49:197–201. doi: 10.1016/j.ymeth.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kellogg E.H., Lange O.F., Baker D. Evaluation and optimization of discrete state models of protein folding. J. Phys. Chem. B. 2012;116:11405–11413. doi: 10.1021/jp3044303. [DOI] [PubMed] [Google Scholar]
- 11.Schwantes C.R., Pande V.S. Improvements in Markov state model construction reveal many non-native interactions in the folding of NTL9. J. Chem. Theory Comput. 2013;9:2000–2009. doi: 10.1021/ct300878a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez-Hernández G., Paul F., Noé F. Identification of slow molecular order parameters for Markov model construction. J. Chem. Phys. 2013;139:015102. doi: 10.1063/1.4811489. [DOI] [PubMed] [Google Scholar]
- 13.Wales D.J. Energy landscapes: some new horizons. Curr. Opin. Struct. Biol. 2010;20:3–10. doi: 10.1016/j.sbi.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Nauli S., Kuhlman B., Baker D. Computer-based redesign of a protein folding pathway. Nat. Struct. Biol. 2001;8:602–605. doi: 10.1038/89638. [DOI] [PubMed] [Google Scholar]
- 15.Beauchamp K.A., McGibbon R., Pande V.S. Simple few-state models reveal hidden complexity in protein folding. Proc. Natl. Acad. Sci. USA. 2012;109:17807–17813. doi: 10.1073/pnas.1201810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindorff-Larsen K., Piana S., Shaw D.E. How fast-folding proteins fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 17.Shirts M., Pande V.S. COMPUTING: screen savers of the world unite! Science. 2000;290:1903–1904. doi: 10.1126/science.290.5498.1903. [DOI] [PubMed] [Google Scholar]
- 18.McGibbon R.T., Schwantes C.R., Pande V.S. Statistical model selection for Markov models of biomolecular dynamics. J. Phys. Chem. B. 2014;118:6475–6481. doi: 10.1021/jp411822r. [DOI] [PubMed] [Google Scholar]
- 19.Noé F., Schütte C., Weikl T.R. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc. Natl. Acad. Sci. USA. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgensen W.L., Severance D.L. Aromatic-aromatic interactions: free energy profiles for the benzene dimer in water, chloroform, and liquid benzene. J. Am. Chem. Soc. 1990;112:4768–4774. [Google Scholar]
- 21.Huang R., Wu L., Keiderling T.A. Cross-strand coupling and site-specific unfolding thermodynamics of a trpzip β-hairpin peptide using 13C isotopic labeling and IR spectroscopy. J. Phys. Chem. B. 2009;113:5661–5674. doi: 10.1021/jp9014299. [DOI] [PubMed] [Google Scholar]
- 22.Onuchic J.N., Wolynes P.G. Theory of protein folding. Curr. Opin. Struct. Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Clementi C., Plotkin S.S. The effects of nonnative interactions on protein folding rates: theory and simulation. Protein Sci. 2004;13:1750–1766. doi: 10.1110/ps.03580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baiz C.R., Lin Y.-S., Tokmakoff A. A molecular interpretation of 2D IR protein folding experiments with Markov state models. Biophys. J. 2014;106:1359–1370. doi: 10.1016/j.bpj.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 26.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 27.Piana S., Lindorff-Larsen K., Shaw D.E. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011;100:L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 29.Darden T., York D., Pedersen L. Particle mesh Ewald: an N log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.