Abstract
Single-particle electron microscopy (EM) has been shown to be very powerful for studying structures and associated conformational changes of macromolecular complexes. In the context of analyzing conformational changes of complexes, distinct EM density maps obtained by image analysis and three-dimensional (3D) reconstruction are usually analyzed in 3D for interpretation of structural differences. However, graphic visualization of these differences based on a quantitative analysis of elastic transformations (deformations) among density maps has not been done yet due to a lack of appropriate methods. Here, we present an approach that allows such visualization. This approach is based on statistical analysis of distances among elastically aligned pairs of EM maps (one map is deformed to fit the other map), and results in visualizing EM maps as points in a lower-dimensional distance space. The distances among points in the new space can be analyzed in terms of clusters or trajectories of points related to potential conformational changes. The results of the method are shown with synthetic and experimental EM maps at different resolutions.
Introduction
Single-particle analysis (SPA) of two-dimensional (2D) transmission electron microscopy (EM) images of isolated biological macromolecular complexes is routinely used to compute three-dimensional (3D) density maps of a wide range of complexes (e.g., proteins, ribosomes, or viruses) (1). In this way, EM information, integrated with a large range of other types of data (e.g., from x-ray crystallography, NMR, modeling, etc.), often provides very valuable information on how these macromolecular complexes perform their functions in the cell.
EM density maps are ideally computed from images of complexes that have identical conformation and different, uniformly distributed, random orientations. However, quite often, complexes present some degree of flexibility. Methodological extensions of SPA have thus been proposed to analyze flexible complexes (2, 3, 4, 5, 6, 7, 8, 9, 10, 11). A classical approach to analyzing macromolecular flexibility is to classify a set of particle images into distinct classes composed of particles with similar conformations, and then to reconstruct an EM density map for each class (2, 3, 4, 5, 6, 7). To explain differences between the obtained EM density maps in terms of conformational flexibility, the density maps are analyzed independently as well as with respect to each other (12, 13, 14, 15, 16). Multivariate statistical analysis (MSA), introduced to EM in the 1980s for analyzing mixed populations of images (17, 18) and now an integral part of many image-analysis approaches and software, allows analysis and visualization of mixed populations of 2D or 3D data by analyzing principal axes (eigenvectors) of the total data variance. However, to our knowledge, no method currently allows graph visualization of differences among sets of EM density maps based on conformational modeling by elastic transformations (deformations) among maps and a quantitative analysis of these elastic transformations. To fill this gap, we here propose such a method.
The classical class-based approaches rest on the assumption that flexibility is discrete, which is not true for a large range of biological systems characterized by continuous flexibility. When flexibility is a continuous process, these class-based approaches may lead to a resolution loss in the density maps coming from the classes, because each density map may represent the average conformation of several slightly different conformations that were assigned to the same class. Some recent approaches (8, 9, 10) explicitly consider continuous flexibility and perform a multidimensional analysis of particular conformational variables (specific to each approach), which brings all images from the given set of single-particle images into a common quantitative reference frame. In this common frame, images are shown as points, and distances among points are related to conformational differences among the corresponding complexes. Such approaches make it possible to analyze possible trajectories of conformational changes by exploring the regions of the common frame that are most densely populated. The common frame onto which EM data are mapped will be referred to as the “distance space” or “map of structures.”
Experimental raw individual 2D images of complexes with continuous flexibility can be seen as spread over some axes described by linear combinations of normal modes, as shown in (8). When density maps are reconstructed from such images sorted according to conformational similarity into a few discrete groups along an axis, each of these maps represents the average conformation of slightly different conformations from the same group (8). These density maps can be thus seen as discrete samples of a continuous trajectory. However, the question is whether this trajectory could be extrapolated from its discrete, unordered samples (density maps) obtained by classical class-based approaches. The approach described here was developed to help with a rough “extrapolation” of the original continuous trajectory from such discrete, unordered samples. This approach is based on continuous normal-mode analysis (NMA) of a relatively small set of 3D density maps (usually, 3–10), and it could be used as the first step toward a fine extrapolation of the trajectory by methods such as hybrid EM normal-mode analysis (HEMNMA), which is based on a “continuous” analysis of a large set of 2D images using normal modes of a reference density map (8).
In the proposed approach, each experimental density map is “modeled” by elastic deformation of other density maps from the given set of initially rigid-body aligned maps, to compare the maps in terms of their structural and conformational differences. Such modeling is used to evaluate how much one map (target) can be explained by the other map (reference), which is done by computing cross-correlation (normalized between 0 and 1) between the target map and its model obtained by deforming the reference map until it best fits the target map (through flexible fitting using normal modes). The unexplained part (what is left after fitting) is termed “dissimilarity,” which is computed by subtracting the obtained cross-correlation from 1. The dissimilarity measure characterizes the difference (distance) between maps that cannot be explained by flexible fitting. The smaller the dissimilarity, the better is the characterization of the deformation between the two maps in terms of conformational motions described by normal modes. Indeed, normal modes have been shown to be very successful in predicting large-scale low-frequency conformational motions of complexes that were also observed experimentally (19, 20, 21, 22, 23). The reference map deformed by flexible fitting is locally rigid-body aligned with the target map before computing the dissimilarity to correct for the potential errors in the initial rigid-body alignment that is done without taking into account the deformation. The combined flexible fitting and local rigid-body alignment will be referred to as elastic alignment.
The obtained dissimilarities among the elastically aligned EM density maps are then used to construct a matrix of distances among the density maps. The distance matrix is analyzed using a statistical multivariate analysis method that projects the distances among the density maps onto a lower-dimensional space in which each EM density map is shown as a point. The dimension of the new space is usually 1, 2, or 3, which allows its full visualization. The process of projecting the distance matrix onto a lower-dimensional space is similar to that used in MSA (17, 18), but the meaning of the distance matrix here is different from that in MSA. More precisely, the distance matrix in the proposed approach describes pairwise dissimilarities not for pairs of original EM maps, but for pairs of elastically aligned EM maps (basically, after one map was deformed to fit the other).
In the obtained common quantitative reference frame (map of structures), potential clusters or trajectories of points can be analyzed by the user in terms of conformational changes. The proposed approach will be referred to as “StructMap,” which stands for structure mapping. The results of StructMap are shown with one set of synthetic EM data and three sets of experimental EM data. The synthetic EM density maps were generated using the EM density map of the closed conformation of the rabbit skeletal muscle type 1 ryanodine receptor (RyR1) complex from Samso et al. (24). The experimental EM density maps comprise the eukaryotic primosome DNA polymerase Pol α-B subunit complex (Pol α-B) from Klinge et al. (14), the Escherichia coli 70S ribosome complex from Fischer et al. (15), and the human 80S ribosomal complex from polysomes from Behrmann et al. (25).
Materials and Methods
In this section, we describe the methodology that is being proposed as well as the synthesis of the test data set that is used in Experiment 1, described in Results.
StructMap method
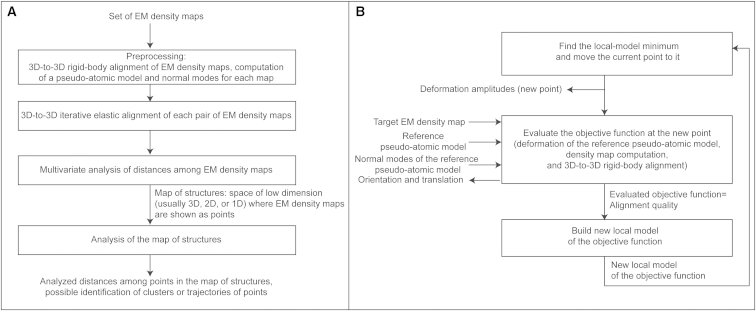
StructMap is comprised of four steps (Fig. 1 A): 1) preprocessing, which consists of 3D-to-3D rigid-body alignment of given EM maps, as well as the computation of pseudoatomic models and the corresponding normal modes of the maps; 2) iterative elastic 3D-to-3D alignment of each pair of EM maps from a given set of EM maps; 3) multivariate analysis of distances among the elastically aligned EM maps; and 4) analysis of the resulting low-dimensional space of distances among EM maps. We here describe each of these steps.
Figure 1.
Flowchart of the proposed StructMap method (A) and the iterative elastic 3D-to-3D alignment step between any two EM density maps (B). The measure of dissimilarity between two finally aligned EM density maps is taken as the distance between the two maps. The distances among all pairs of EM maps are used to construct a distance matrix that is then analyzed with a multivariate analysis method, so as to project all EM maps onto a common distance space (map of structures), in which each EM map is represented as a point. Points may then be analyzed in terms of their positions and mutual distances to potentially identify clusters or trajectories of points.
Step 1. Preprocessing: rigid-body alignment of EM maps and computing pseudoatomic models and corresponding normal modes of EM maps
Before starting the elastic alignment of EM maps in step 2, these maps must be rigid-body aligned as well as possible. More precisely, given two EM density maps, the elastic alignment (step 2) is done by flexible and rigid-body alignments of one density map, referred to as the reference density map, until it matches the other density map, referred to as the target density map. However, the rigid-body alignment involved in that step is only local, not global (the initial orientation and position of the deformed reference map with respect to the target map are refined). In this way, the initial rigid-body alignment is corrected at each iteration of the elastic alignment, taking into account the deformation estimated at that iteration.
Rigid-body alignment of EM maps is a part of many common data processing workflows. Before using the approach proposed here, the EM maps will thus, most likely, be already globally rigid-body aligned in some way, which was also the case with the experimental EM maps used in this article. Thus, in the preprocessing step of StructMap, in all experiments in this article, the given maps were only locally rigid-body aligned, meaning that they were aligned around the orientations and positions in which the maps were available before our work. This rigid-body local alignment was performed using the xmipp_volume_align program in Xmipp 3.1. This method was used to refine the alignment of EM maps around the orientation and position in which the maps were available before this work, as aligned by the authors of the maps. However, note that the xmipp_volume_align method can optionally be used for a global rigid-body alignment of EM maps, meaning over all rotations and translations, independent of the currently available ones.
In the elastic alignment procedure (step 2), the flexible alignment is based on deforming the reference density map. The density map deformation is realized by displacing a set of 3D Gaussian functions along vectors that are linear combinations of normal modes of the given density map, where the 3D Gaussian functions are used to model the map densities using the method proposed in (27). More precisely, the reference density map is converted into a collection of 3D Gaussian functions of different amplitudes and positions over the map so that the sum of these Gaussian functions over a voxel approximates the reference-map density at that voxel, with a given mean approximation error over all voxels (27). The 3D Gaussian functions are referred to as “pseudoatoms,” though their positions do not have to coincide with true atomic positions. The density map representation by pseudoatoms (PDB-format structure) will be referred to here as the “pseudoatomic model” to distinguish it from a true atomic-resolution structure.
Normal modes of the given EM map are often computed using the elastic network model of the potential energy function of the complex around a minimum energy conformation (8, 19, 20, 21, 22, 27, 28), as was the case here. In the elastic network approach, the network is composed of nodes that are 3D point particles connected by springs, where the springs represent harmonic restraints on displacements from the equilibrium conformation (28). Here, the elastic network nodes are 3D Gaussian functions (pseudoatoms). The elastic network approach based on nodes determined by 3D Gaussian functions was shown to result in computation of normal modes that approximate atomic normal modes with high accuracy (8, 27). Additionally, as displacing pseudoatoms (nodes) along normal modes results in deformation of the pseudoatomic model, such Gaussian-based nodes allow easy computation of deformed density maps (by summing 3D Gaussian functions) and a comparable resolution of these maps to the resolution of the original, nondeformed density map. The resolution of the pseudoatomic models (reference and deformed) and the resolution of the deformed density maps can be controlled by controlling the error of pseudoatomic approximation of the given density map (27). Such deformed density maps are compared with the target density maps in the elastic alignment procedure (step 2) to estimate the deformation.
The user-friendly HEMNMA graphical interface (29) was used here for computing both pseudoatomic models and normal modes. More precisely, we used a previously developed software (developed by Tama et al. (22)) for converting EM maps into pseudoatoms (27) and for NMA, which were also used in our previous work based on normal modes of pseudoatomic models (8, 27) before being integrated in HEMNMA (29). More details on the use of HEMNMA for these two tasks are given below (see Use of HEMNMA for Computing Pseudoatomic Models and Normal Modes).
Step 2. Iterative elastic 3D-to-3D alignment of EM density maps
The proposed method involves elastic 3D-to-3D alignment in the continuous NMA framework, which was realized by extending and modifying the elastic 3D-to-2D alignment method of (8), available via HEMNMA interface (29). The elastic alignment here means a combined flexible and rigid-body local alignment of one map with the other. The flexible alignment of two maps is performed by deforming one map using normal modes until it fits the other map, and the deformation is realized by displacing pseudoatoms with a linear combination of normal modes, as in (22, 27). Pseudoatoms (3D Gaussian functions modeling the original, nondeformed EM map) are displaced, using normal modes, with respect to their location in the original map, meaning that the flexible alignment is local around the conformation given by the original map. However, recall that normal modes describe low-frequency large-scale conformational changes, which means a relatively large range of global deformation amplitudes (several nanometers). The rigid-body alignment of two maps is performed by rotating and translating one map until it fits the other. The elastic alignment procedure assumes that maps were initially rigid-body aligned (before computing pseudoatoms and normal modes in the preprocessing step), so that it only performs local rigid-body alignment (refinement of current orientations and translations). The transformation of the map by elastic alignment can thus be mainly described as map deformation, because the rigid-body-alignment part of transformation will be small for initially rigid-body-aligned maps.
The iterative elastic alignment method consists of refining the amplitude of displacement in each normal mode (elastic parameters) as well as the orientation and position (rigid-body parameters) of the reference density map until it matches the target density maps. This is done by minimizing dissimilarity between the two maps, and this dissimilarity (the objective function to be minimized) is here defined as S = 1 − CC, where CC is the cross-correlation between the elastically aligned reference and target density maps. The optimization of the objective function is done with Powell’s UOBYQA method, which uses a quadratic-approximation local model of the objective function subject to a trust region (30). The UOBYQA method takes into account the curvature of the objective function by constructing interpolant quadratic models. A typical iteration of the algorithm generates a new vector of variables (new point) by minimizing the quadratic model. The objective function is then evaluated at the new point and one of the interpolation points is replaced by this new point (30).
The trust-region radius is related to the length of steps used in the objective-function optimization (30). Its meaning here is the norm of the vector of normal-mode displacement-amplitude differences between points at which the objective function is evaluated, where points are vectors of the displacement (deformation) amplitudes and the vector dimension is the dimension of the search space given by the number of normal modes. The trust region is a ball around the best point found currently, with the radius given by the trust-region radius. Like the deformation amplitudes, the trust-region radius has no physical units, as the NMA software used here computes the coordinates of normal-mode vectors in angstroms, whereas it displaces pseudoatoms using normal-mode displacement amplitudes without units. Trust-region-based approaches are known to be more robust to noise thanks to the regularization effect of minimizing objective-function models (usually quadratic) around current iterates over regions of controlled size. The trust-region radius is adapted iteratively to optimize the trust-region size. Actually, the UOBYQA method uses two radii; one of these (ρ) is not allowed to increase, because this would necessitate expensive decrease later, whereas the other (Δ) satisfies Δ ≥ ρ and allows the length of steps to exceed ρ, which improves the efficiency of the algorithm (30). The method requires setting the initial vector of displacement amplitudes (initial point), the maximum number of iterations, the initial and final values of the trust-region radius ρ, and sets the initial value of Δ to be equal to the initial value of ρ.
The developed 3D-to-3D elastic alignment software uses an implementation of the UOBYQA method from CONDOR libraries (31). The 3D-to-3D elastic alignment method is available in Xmipp (32, 33, 34) (version 3.1) as a program under the name xmipp_nma_alignment_vol. This program aligns a reference pseudoatomic model (obtained from a reference EM density map) with a set of target EM density maps, and requires that all density maps have the same size (in voxels) and the same voxel size (in angstroms). For each target EM density map, the reference pseudoatomic model is deformed and its orientation and position refined so that the corresponding density map (after the displaced pseudoatoms are converted into the density-map format) best matches the target EM density map.
In the proposed approach, the initial displacement amplitudes are equal to zero, meaning that the nondeformed reference pseudoatomic model is used to initialize the first iteration. In each UOBYQA iteration (Fig. 1 B), the pseudoatomic model is displaced with the new guess of the normal-mode displacement amplitudes (new point) obtained by minimizing the quadratic model, and the modified pseudoatomic coordinates are converted into a density map. Then, this density map is locally rigid-body aligned with the target density map. The rigid-body alignment is done with a fast rotational matching method that maximizes the cross-correlation between two density maps (26), and the objective function S is evaluated after this alignment. The local model of the objective function is then reconstructed around the new point, and a new iteration is started by finding the minimum of the current local model of the objective function and moving the current vector of deformation amplitudes (current point) to this minimum. The iterations are repeated until the final value of the trust-region radius ρ or the maximum number of iterations is reached. All experiments in this article were performed using the initial and final values of the trust-region radius ρ and the maximum number of iterations of 250, 50, and 10,000, respectively. Within a similar objective-function optimization procedure (in HEMNMA), these values have been shown to produce good results with several complexes of different amplitudes of conformational changes (70S ribosome, Pol α-B complex, and tomato bushy stunt virus) (8). They are set as default values in xmipp_nma_alignment_vol. The use of trust-region radii improves the optimization convergence by improving robustness to noise, but UOBYQA does not guarantee reaching the global optimum. Though reaching the global optimum cannot be guaranteed, the method provides rather good solutions, as shown in this article.
The main difference between the objective-function optimizations in the elastic alignment procedure in HEMNMA and that in StructMap is the different nature of the target data (2D images in HEMNMA versus 3D density maps in StructMap). As the reference data are in both cases 3D density maps, HEMNMA is based on elastic 2D-to-3D data alignment, whereas StructMap is based on elastic 3D-to-3D data alignment. This means that the elastic alignment procedure in HEMNMA requires computation of 2D projections of a 3D density map (to perform its matching with target images), which is not the case for StructMap. Note here that this comparison is made with regard only to technical details of the two elastic alignment procedures, and not to the results that the entire StructMap and HEMNMA methodologies can produce. Actually, StructMap is not meant to replace HEMNMA, because the two methodologies are complementary and can be combined, as shown in the Results (Experiment 2) and treated further in the Discussion.
Step 3: Multivariate distance analysis
The optimized value of the dissimilarity measure, S, obtained for each aligned pair of EM density maps is used to construct an N × N symmetrical distance matrix, D, where N is the number of given EM maps. As the alignment of the ith EM map, Vi, with respect to the jth EM map, Vj, is done through elastic geometric transformations of Vi until it matches Vj, this alignment will result in dissimilarity Sij that will generally be different from Sji, representing the dissimilarity of Vj aligned with respect to Vi. Thus, we set the ijth and jith elements of the distance matrix, D (Dij and Dji, respectively) to be the average between Sij and Sji (i.e., Dij = Dji = (Sij +Sji)/2). Note that the differences between Sij and Sji are usually small (a strong asymmetry of the distance matrix would indicate that the underlying matching of one volume to the other is ill defined, which was not observed here). We set Dii = 0, since there is no distance from an EM density map to itself.
To perform multivariate analysis of the distance matrix, we use a nonmetric multidimensional scaling method (35), which is available in MATLAB (MATLAB and Statistics Toolbox Release 2011b, The MathWorks, Natick, MA) under the name mdscale. The method returns N points in p dimensions, where the number of dimensions, p (p ≤ N), is an input parameter (in normal practice, p = 1, p = 2, or, at most, p = 3), such that the Euclidean distances between the obtained points approximate a monotonic transformation of distances in the distance matrix. The mdscale method optimizes the so-called Kruskal’s normalized stress (35) function that measures the degree of correspondence between the input distance matrix and the output points. The points are plotted in a p-dimensional distance space (map of structures). In this article, results of the mdscale method are deterministic, as the method was used with no randomness option.
Step 4: Analysis of the map of structures
The pattern that the points make in the new, p-dimensional space, together with their relative distances, can be used to identify clusters of points that correspond to similar EM maps. In some cases, the disposition of points in the p-dimensional space can also be used to explore potential sequences of conformational changes that we will then refer to as “trajectories of points.”
Data generation for the experiment with synthetic EM maps
For one of the experiments performed using the proposed method, we discretized two distinct synthetic trajectories of continuous conformational changes of the same complex. The two synthesized trajectories are completely fictional and may not exist or coexist in the experimental case. They were synthesized by displacing the EM map of the closed conformation of RyR1 (from Samso et al. (24)) using two different normal modes from the same set of normal modes. The two normal modes used for the displacement were selected to produce opening-closing movements of RyR1. The displacement in one of the two modes (mode 8) produces symmetrical conformational changes that correspond to those usually reported in the literature. By symmetrical changes, we mean that the object remains symmetrical after the displacement using normal modes. The displacement in the other mode (mode 9), in turn, produces asymmetrical conformational changes. By asymmetrical changes, we mean that the object becomes asymmetrical after the displacement using normal modes. To the best of our knowledge, those asymmetrical changes have not been previously reported. The synthesis of this test data set is fully described in this section. The results of the experiment with this data set are given in the Results section (Experiment 1).
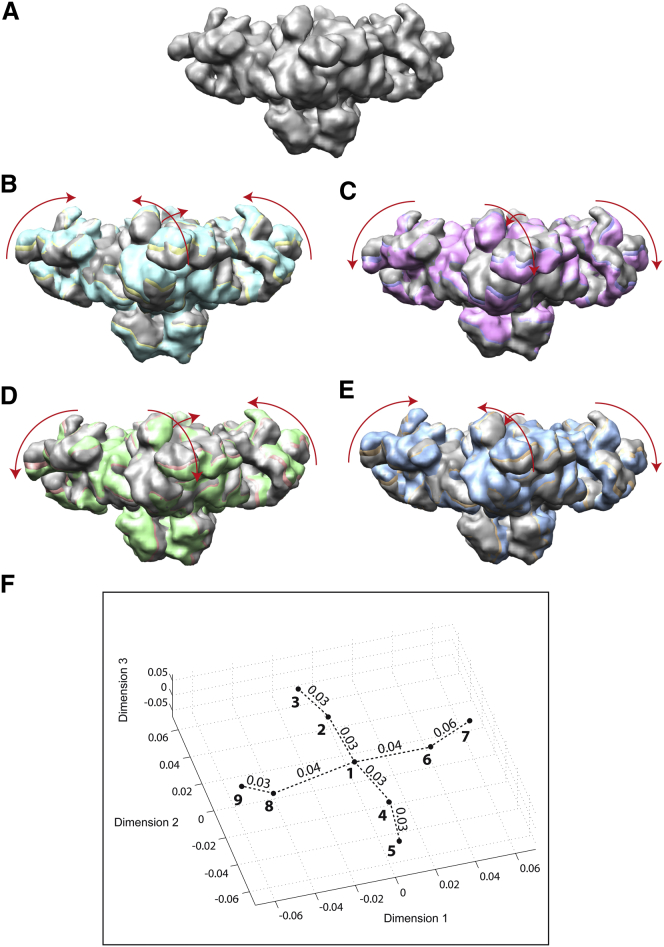
We used the EM density map of the closed conformation of RyR1 deposited at the Electron Microscopy Data Bank (EMDB) with code EMD-1606 (24) (resolution, 10.2 Å; size, 180 × 180 × 180 voxels; voxel size, 2.8 Å × 2.8 Å × 2.8 Å). The pseudoatomic model of the EM density map and its normal modes 8 and 9 were used to compute 8 additional pseudoatomic models. The density maps obtained from the nine pseudoatomic models were projected to obtain 500 randomly oriented projections for each density map. The projections had a uniform angular distribution, a size of 128 × 128 pixels, and a pixel size of 3.94 Å × 3.94 Å. Note here that the projection resolution is lower than the resolution that could be obtained by projecting the obtained density maps onto the image planes whose size is given by the original density map size (i.e., image size of 180 × 180 pixels and pixel size of 2.8 Å × 2.8 Å), which was chosen to simulate a real experiment as, in practice, the object’s 2D projection images are collected at a limited resolution due to image formation and detection limitations. Each projection was modified by adding simulated experimental noise and contrast transfer function (CTF). The CTF and noise were simulated for a 200 kV microscope with a spherical aberration of 2 mm, defocus of 1 μm and signal/noise ratio of 0.2, using the method of Velazquez-Muriel et al. (36). Finally, each of the nine sets of simulated images was used to compute the corresponding 3D reconstruction (Fig. 2, A–E). Note here that resolution of the reconstructed density maps is low (∼20 Å), as a relatively small number (500) of noisy and CTF-affected images was used for the reconstruction.
Figure 2.
Synthetic RyR1 data experiment using synthetic density maps representing symmetrical (B and C) and asymmetrical (D and E) structural changes of the complex (A). (A) Synthetic density map 1. (B) Map 1 (gray) overlapped with maps 2 (yellow) and 3 (cyan). (C) Map 1 (gray) overlapped with maps 4 (violet) and 5 (magenta). (D) Map 1 (gray) overlapped with maps 6 (pink) and 7 (green). (E) Map 1 (gray) overlapped with maps 8 (brown) and 9 (blue). (F) Projection of density maps onto a 3D distance space. In (A)–(E), all density maps are viewed from the same direction. In each image, arrows show the main directions of deformation of map 1 (gray) when fitting the other two maps (overlapped), and the arrow scale shows the largest distance among these three maps after the deformation according to Table S1. In (F), the density maps are marked with their indexes and circles, and the length of each dotted line segment is the distance between two conformations that is shown above the segment in arbitrary units. See also Fig. 3. To see this figure in color, go online.
The reconstructed density map referred to as 1 corresponds to the nondisplaced initial pseudoatomic model (i.e., zero displacement amplitudes in all modes) (Fig. 2 A). Four of the other reconstructed density maps, referred to as 2, 3, 4, and 5 (Fig. 2, B and C), correspond to the displacement of the initial pseudoatomic model in mode 8 using displacement amplitudes of 300, 500, −300, and −500 (no units), respectively, and zero displacement amplitudes in other modes. Four additional reconstructed density maps, referred to as 6, 7, 8, and 9 (Fig. 2, D and E), correspond to the displacement of the initial pseudoatomic model in mode 9 using displacement amplitudes of 300, 500, −300, and −500 (no units), respectively, and zero displacement amplitudes in other modes.
Fig. 2, B and C, shows that the sequences of discrete states 3-2-1-4-5 and 5-4-1-2-3 describe symmetrical opening and closing of the complex, respectively, whereas Fig. 2, D and E, shows that sequence 7-6-1-8-9 (or 9-8-1-6-7) describes asymmetrical opening or closing of the complex (one side of the complex opens while the other side closes).
Use of HEMNMA for computing pseudoatomic models and normal modes
As mentioned earlier in this section, the pseudoatomic models and their normal modes were computed using the HEMNMA graphical interface (29). In this subsection, we give more details on the HEMNMA parameter settings used for this purpose.
The HEMNMA graphical interface (29) allows use of a 3D binary mask and adjustment of the desired EM-map approximation error and the standard deviation of Gaussian functions (for computing pseudoatomic models), an interaction cutoff distance between pseudoatoms (for computing normal modes), and adjustment of the collectivity threshold and the number of lowest-frequency normal modes that will be analyzed in terms of their collectivity (for selecting the modes that will be used in the further analysis that is here the elastic alignment of EM density maps). The 3D binary mask can be used to assure that some parts of the EM density map are not represented by Gaussian functions (e.g., a noisy background of the complex). Here, the 3D binary mask was obtained by a combination of EM-map thresholding and several morphological operations. The desired error of the EM-map representation by Gaussian functions determines the end of the iterative process by which Gaussian functions are added to the pseudoatomic representation, and here, it was set to a value of 5%, which usually results in good representations (29). The Gaussian-function standard deviation was adjusted to a value between 1 and 2 voxels, as typically done to better optimize the EM-map approximation error (29). The elastic network model for the computation of normal modes has the interaction cut-off distance parameter that determines the distance between the pseudoatoms above which they do not interact, and its value is usually adjusted between 10 Å and 30 Å, according to the size of the complex (in voxels) (8, 22). Here, the interaction cutoff distance parameter was set to 10 Å for Pol α-B, 25 Å for 70S and 80S, and 30 Å for RyR1. It has been shown that highly collective low-frequency modes are relevant to functional conformational changes (19, 20) and that such changes can usually be described by a few modes among the first (lowest-frequency) 20–50 pseudoatomic normal modes (8, 22). The collectivity degree (37) measures the collectivity of motions in a normal mode by counting the number of pseudoatoms affected by the mode. The collectivity degree approaches 1 for maximally collective motions and 0 for localized motions (37). Here, the collectivity degree was computed and analyzed for the first 20 normal modes in the case of lower-resolution density maps (synthetic maps of RyR1 and experimental maps of Pol α-B), whereas the first 30 normal modes were analyzed in terms of collectivity in the case of higher-resolution density maps (experimental maps of 70S and 80S). The modes collectivity threshold determines a subset of the total set of normal modes that will be used for further analysis. The collectivity threshold of 0.15 was used here, which means that only modes with a collectivity degree >0.15 were used for the elastic alignment of EM density maps. The collectivity threshold of 0.15 has been shown to successfully reject poorly collective modes that are very likely unrelated to functional conformational changes (8, 29). The six lowest-frequency modes were not used either, as they are related to rigid-body movements (8, 22, 29).
Results
In this section, we show the performance of the StructMap method with synthetic and experimental EM density maps. The most important information gained by using StructMap is the graphical and numerical distance-space results i.e., the overall graphical view of differences (distances) among a set of elastically aligned density maps (represented by points in the distance space), as well as all pairwise distances among these maps. In the distance space, some pairs of points are connected by lines to facilitate discussion of distances among points and a potential disposition (configuration) of points in clusters or trajectories in the distance space. In the experiments with experimental EM maps (Experiments 2–4), lines are drawn to connect points representing subsequent states according to the previously published work in which these maps were obtained, which was done to visualize the patterns that these previously published results (sequences) make in the distance spaces obtained in this work. In the experiment with synthetic EM maps (Experiment 1), lines are drawn to connect points following their ground-truth order (i.e., the synthesized sequence of states 5-4-1-2-3 and 9-8-1-6-7), which was done to visualize the pattern that this ground-truth order makes in the obtained distance space. The summary of all pairwise point distances obtained by analyzing the sets with more than three EM maps is given in Tables S1–S3 in the Supporting Material.
Figures in this section also show rigid-body-aligned density maps (from the preprocessing step) that are complemented with arrows indicating the movements that mainly contribute to the elastic transformation (deformation) between maps, and the arrow scale shows the distance between maps in the distance space. This is additional information gained by using StructMap.
Experiment 1: Analysis of synthetic EM density maps
Here, we present the results of StructMap using the synthetic data set described in Materials and Methods. The goal of the experiment was to analyze the pattern created by the synthetic EM density maps in the resulting map of structures, to see how the synthesized conformational trajectories look in this map (e.g., as two distinct trajectories or not). Recall that the synthesized conformational trajectories are completely imaginary, which means that the results presented here should not be interpreted in terms of experimental RyR1conformational changes. A pseudoatomic model and its 20 normal modes were computed for each given synthetic density map. Then, each density map (via its pseudoatomic model) was elastically aligned with all other density maps and the obtained 9 × 9 distance matrix was projected onto a space of three, two, and one dimensions (Figs. 2 F and 3, A and B, respectively).
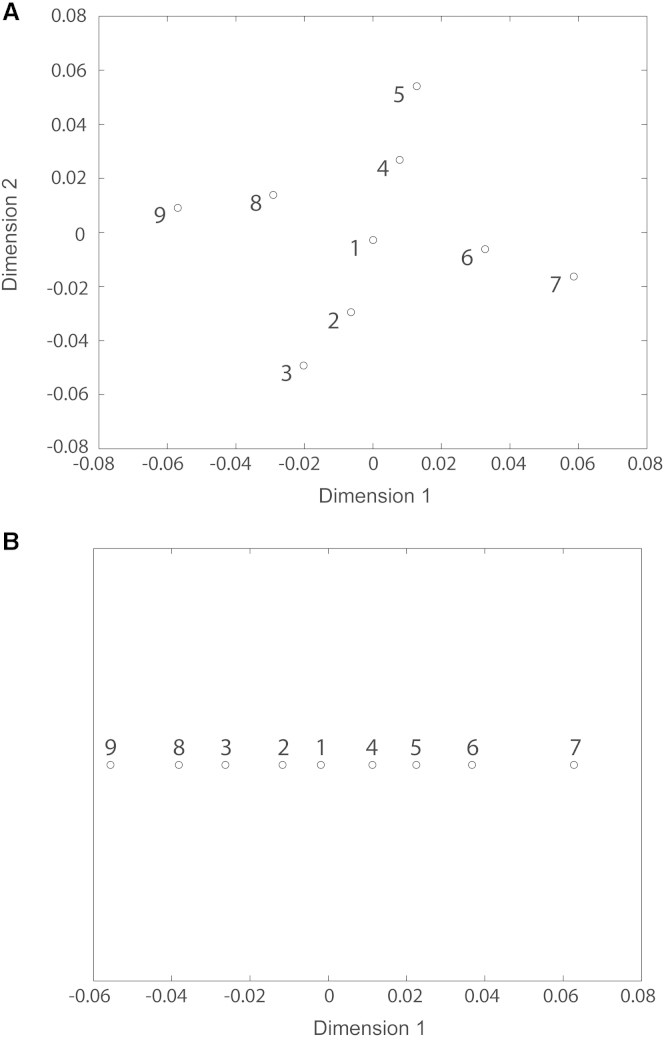
Figure 3.
Projection of synthetic RyR1 density maps onto a distance space of a lower dimension than 3. (A) The 2D distance space. (B) The 1D distance space. The density maps are marked with their indexes and circles. See also Fig. 2.
In the 3D distance space, one can note a two-trajectory configuration of points 1–9 that correspond to synthetic density maps 1–9, respectively (Fig. 2 F). This two-trajectory configuration is visually emphasized by connecting points according to their ground-truth order (i.e., 5-4-1-2-3 and 9-8-1-6-7). The different distances among neighboring points on the two trajectories mean that the same deformation amplitude has a different impact on the two types of deformation, from the point of view of the mapped dissimilarity measure. More precisely, the larger distances on trajectory 9-8-1-6-7 compared to trajectory 5-4-1-2-3 suggest that the same deformation amplitude has a larger impact on the asymmetrical deformation than on the symmetrical deformation. All pairwise point distances obtained in this experiment are summarized in Table S1.
As 2D and 1D maps of structures could also be useful in practice, especially when analyzing a small number of EM maps, we projected the same set of synthetic density maps onto these two spaces as well. The results of projecting density maps onto a 2D distance space are consistent with those obtained by projecting the same density maps onto a 3D distance space (Fig. 3 A). Regarding the projection of the same density maps onto a 1D distance space, there is a nonsurprising loss of information about the two-trajectory configuration (Fig. 3 B); however, the 1D distance mapping still allows sorting out of similar and dissimilar density maps. For instance, Fig. 3 B clearly shows that density maps 1, 2, and 4 are the most similar, whereas maps 7 and 9 are the most dissimilar. Thus, 1D distance mapping may still be useful when analyzing a small number of EM density maps (e.g., three to four density maps), as in the case shown in Experiment 2.
Experiment 2: EM density maps of Pol α-B
In this experiment, we used three EM density maps of Pol α-B from Klinge et al. (14). They correspond to different states of bending of the flexible linker between two lobes of the complex. The EM density maps have a size of 64 × 64 × 64 voxels, with voxel size 3.8 Å × 3.8 Å × 3.8 Å, and a resolution between 23 Å and 25 Å. They are referenced by indexes 1, 2, and 3, as obtained from the authors (14). A pseudoatomic model and its 20 normal modes were first computed for each EM density map. Then, each density map (via its pseudoatomic model) was elastically aligned with all other density maps, and the obtained 3 × 3 distance matrix was projected onto a 1D distance space (Fig. 4 A).
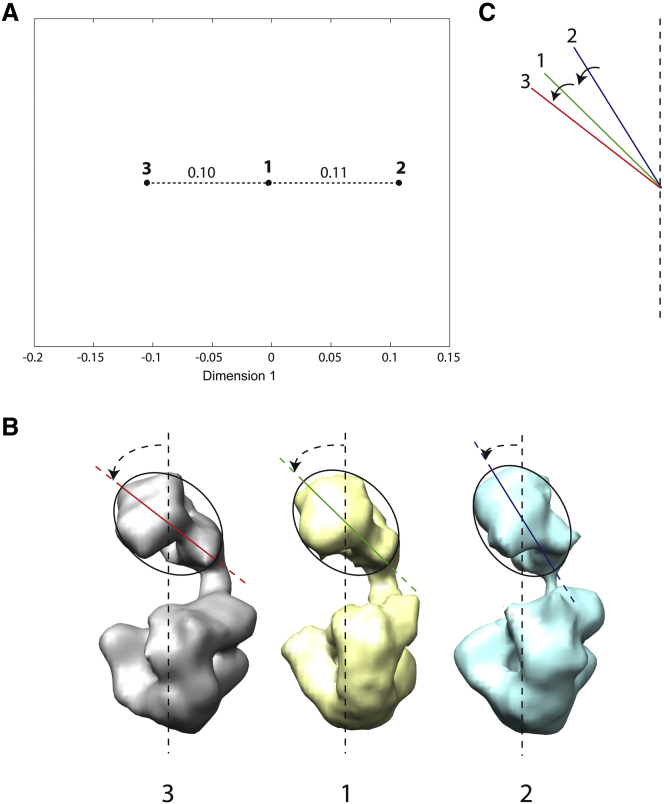
Figure 4.
Projection of three EM density maps of DNA polymerase Pol α-B complex onto a 1D distance space. (A) The 1D distance space (EM maps are marked with circles and the length of each dotted line segment is the distance between two EM maps that is shown above the segment in arbitrary units). (B) Rigid-body-aligned EM density maps (3-1-2) ordered according to their disposition in the distance space shown in (A), with dashed arrows showing different degrees of bending of the complex, i.e., rotation of the upper lobe toward the lower lobe (the elastic transformation among these maps is mainly described by the bending movement). (C) Upper-lobe axis of overlapped maps 1–3, shown with the same color as the color of the upper-lobe axis in (B) (red, map 3; green, map 1; blue, map 2), with solid arrows showing the deformation direction (the rotation of the upper lobe of map 2 toward map 1 and the rotation of the upper lobe of map 1 toward map 3). In (C), the arrows do not show the amount of deformation, but they show (by the arrow scale) the distance between maps in the map pairs (1,2) and (1,3) after the deformation according to (A). Note that the vertical lines shown in (B) and (C) are not the same but they are parallel to each other. To see this figure in color, go online.
The 1D distance mapping results (Fig. 4 A) show that EM density map 1 is almost equally distant from the other two EM density maps (the distance of map 1 to the other two maps is ∼0.1). Thus, this sequence could be interpreted as a movement around conformation 1, in the order 3-1-2 or 2-1-3 (Fig. 4 B). EM maps in the order 3-1-2 correspond to the unbending of the complex from conformation 3 to conformation 2 (Fig. 4, B and C). These results are coherent with previously published results (Fig. 6 of Klinge et al. (14)) but, contrary to the previous study, they are here based on a quantitative distance analysis.
These results could be used in combination with HEMNMA to explore conformational changes of Pol α-B at a finer level of detail, by a continuous analysis of 2D images using normal modes of a reference density map, as was done in (8). In this context, the results of StructMap could be used to select the reference density map for analyzing images by HEMNMA. Actually, in (8), 12,000 Pol α-B single-particle images (used for computing maps 1–3) were analyzed using HEMNMA, and map 3 was used as the reference map. The reference map was chosen to be map 3 because this map was obtained from the most populated class of images (maps 3, 1, and 2 come from classes with 42%, 30%, and 28% of the total number of images, respectively (14)). Although map 3 was reconstructed from the largest number of images, or exactly because of that, it is tempting to think that some heterogeneity could have been incorporated in that map. The fact that it is one of the two end points in the distance map (Fig. 4 A) could support this hypothesis, meaning that some other conformations could exist even farther to the left of map 3 in the distance map, but they could not be identified (they were somehow hidden) because images were sorted into only three classes. Thus, the results of StructMap (distance map, Fig. 4 A) show that it would be interesting to use map 3 for the image analysis with HEMNMA to identify conformations that could not be identified by splitting images into only three classes (e.g., those to the left of map 3 in the distance map), which was actually done by Jin et al. (8). The second end point in the distance map, map 2, would be less interesting to use for the continuous analysis of images (as the reference map for HEMNMA), because it lacks some mass at the level of the linker between the lobes, which was explained by a strong heterogeneity of conformations in the corresponding class of images (14).
Experiment 3: EM density maps of E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex
The EM density maps of different pre- and posttranslocational states of E. coli 70S complex published by Fischer et al. (15) were downloaded from the EMDB database. Among them, we have analyzed those that had the same size (128 × 128 × 128 voxels) and voxel size (2.8 Å × 2.8 Å × 2.8 Å) and contained the entire 70S complex. The analyzed data set contained seven EM density maps of resolution between 12 Å and 20 Å (15). These EM maps correspond to pretranslocational states pre2 to pre5 (EMD-1717 to EMD-1720, respectively) and posttranslocational states post1 to post3 (EMD-1721 to EMD-1723, respectively). A pseudoatomic model and its 30 normal modes were computed for each EM density map, each density map (via its pseudoatomic model) was elastically aligned with all other density maps, and the resulting 7 × 7 distance matrix was projected onto a 3D distance space (Fig. 5 A).
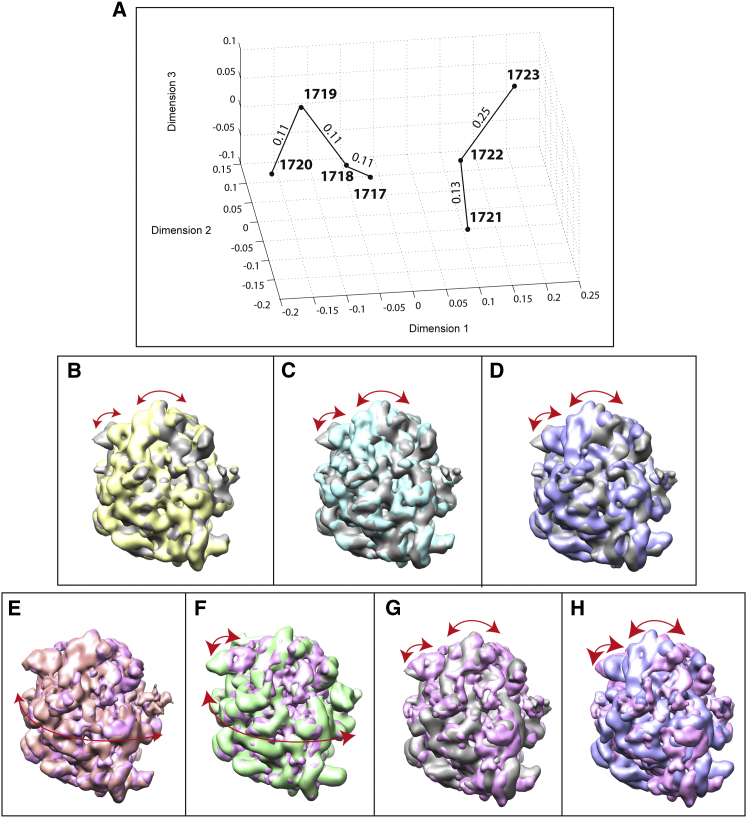
Figure 5.
Projection of seven EM density maps of 70S ribosome (EMD-1717 to EMD-1723) onto a 3D distance space. (A) The 3D distance space. (B–H) Overlap of rigid-body-aligned EM density maps, with the same view for all overlapped maps for 1718 vs. 1717 (gray) (B), 1719 vs. 1717 (gray) (C), 1720 vs. 1717 (gray) (D), 1722 vs. 1721 (magenta) (E), 1723 vs. 1721 (magenta) (F), 1717 vs. 1721 (magenta) (G), and 1720 vs. 1721 (magenta) (H). In (A), the EM maps are marked with circles and the corresponding EMDB entry codes. Straight lines are used to connect subsequent states according to the proposal by Fischer et al (15), and the length of each line segment is the distance between two maps that is shown above the segment in arbitrary units. In (B)–(H), arrows show the movements that mainly contribute to the elastic transformation (deformation) between two overlapped maps, and the arrow scale shows the distance between these maps in the distance space. Bidirectional arrows were used to recall that StructMap computes the elastic transformation between two maps in both directions. Note that the arrow scale shows the dissimilarity between maps after the deformation, but the amount of deformation (movement) is not represented by arrows. However, note that the type of movement is represented by arrows (i.e., rotation of 30S with respect to 50S and L1 stalk motion in (B)–(D) and (G) and (H), rolling/unrolling of the complex in (E), and rolling/unrolling of the complex and L1 stalk motion in (F)). To see this figure in color, go online.
The 3D map of structures and pairwise map distances (Fig. 5 A and Table S2) show that most distances among maps 1717–1720 (four distances of 0.11, one distance of 0.2, and one distance of 0.21 (arbitrary units)) are smaller than the distances from maps 1717–1720 to maps 1721–1723 (three distances of 0.2 and nine distances ≥0.25 (arbitrary units)). This means that maps 1717–1720 correspond to similar conformational states and that these states are generally less different with respect to each other than with respect to states 1721–1723, which is also visible when the maps are overlapped. For instance, the orientation of the 30S subunit is generally more similar among maps 1717–1720 (Fig. 5, B–D) than is the 30S orientation between these maps and maps 1721–1723 (Fig. 5, G and H). This is consistent with the findings of Fischer et al. (15), indicating that maps 1717–1720 correspond to similar pretranslocational states, whereas maps 1721–1723 correspond to similar posttranslocational states. Our results complement the results of Fischer et al. (15) by providing a quantitative analysis of the conformational differences. For instance, the results (Fig. 5 A and Table S2) additionally show that maps 1721 and 1722 are more similar (distance of 0.13) than maps 1722 and 1723 (distance of 0.25) or maps 1721 and 1723 (distance of 0.25), which is also visible in Fig. 5, E and F.
Experiment 4: EM density maps of native human 80S ribosomal complex from polysomes
In this experiment, we used 11 EM density maps of native human 80S ribosomal complexes from polysomes, deposited in the EMDB by Behrmann et al. (25). They correspond to states along the elongation cycle of 80S (25). In the clockwise direction along the elongation cycle shown in Fig. 1 of Behrmann et al. (25), the EM density maps correspond to the classical iPRE (EMD-2907), classical-1 PRE (EMD-2909), PRE∗ (EMD-2906), rotated-1 PRE (EMD-2904), rotated-2 PRE (EMD-2905), POST-i3 (EMD-2903), POST-i2 (EMD-2902), POST (EMD-2875), prerecycling (EMD-2910), postdecoding/posthydrolysis (EMD-2908), and postdecoding/postdissociation (EMD-2911) states. Note that the density maps of translocation and decoding-sampling/-recognition complexes were not used in our experiment, as these complexes were not observed experimentally in the original work (25). The density map EMD-2875 has a size of 400 × 400 × 400 voxels, with voxel size 0.945 Å × 0.945 Å × 0.945 Å and a resolution of 3.5 Å according to the FSC0.143 resolution criterion (25). All other EM density maps have a size of 200 × 200 × 200 voxels, a voxel size of 1.89 Å × 1.89 Å × 1.89 Å and resolution between 5 Å and 10 Å according to the FSC0.5 resolution criterion (25). We thus resized map EMD-2875 so that it has the same number of voxels and the same voxel size as the other EM density maps, which is required by StructMap (see Materials and Methods). A pseudoatomic model and its 30 normal modes were computed for each density map, each density map (via its pseudoatomic model) was elastically aligned with all other density maps, and the obtained 11 × 11 distance matrix was projected onto a 3D distance space (Fig. 6 A).
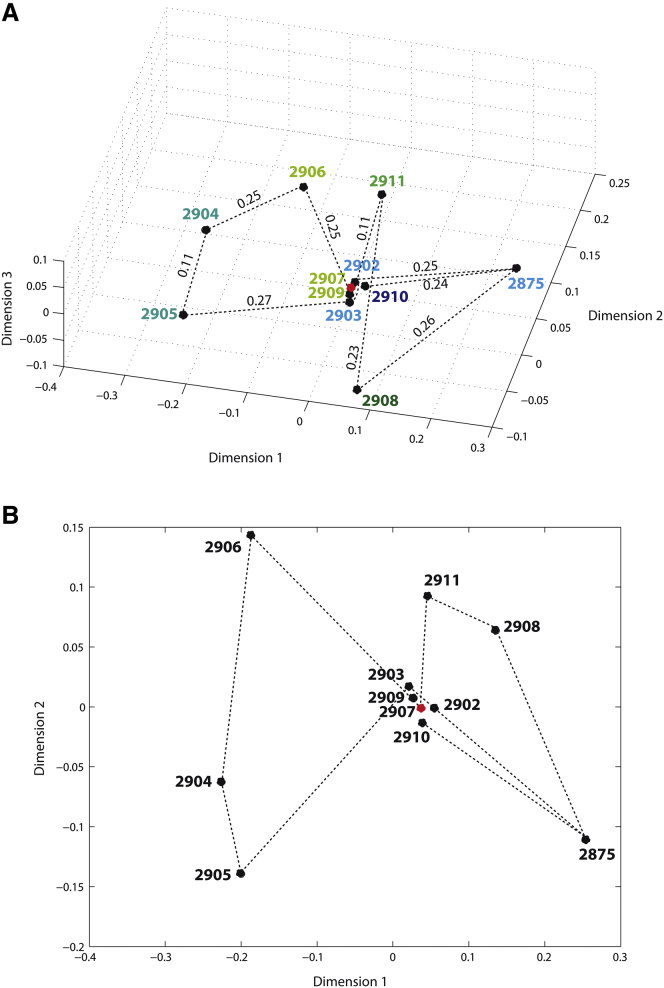
Figure 6.
Projection of 11 EM density maps of 80S ribosome (EMD-2875 and from EMD-2902 to EMD-2911) onto the 3D distance space (A) and the 2D distance space (B). The density maps are marked with circles and the corresponding EMDB entry codes. The length of each dotted line segment is the distance between two density maps that is, in (A), shown above the segment in arbitrary units. The dotted lines are used to connect subsequent states according to the proposal by Behrmann et al. (25). In (A), different colors are used for map numbers to represent complexes with different compositions, according to Behrmann et al. (25). The same colors are attributed to the maps with the same or similar composition, which resulted in the use of six different colors for the following types of composition: 1) 2904 and 2905 (two tRNAs but slightly different, P/E, A/P in 2905 and P/E, A/A in 2904); 2) 2906–2909 (three tRNAs: E/E, P/P, and A/A); 3) 2902, 2903, and 2875 (two tRNAs: E/E, P/P); 4) 2911 (three tRNAs: E/E, P/P, and A/T); 5) 2908 (eEF1A and three tRNAs: E/E, P/P, and A/T); and 6) 2910 (eRF1, ABCE1, and two tRNAs: E/E,P/P). The classical iPRE state (EMD-2907) is marked with a red point in (A) and (B). See also Fig. 7. To see this figure in color, go online.
The 3D map of structures shows that conformations 2902, 2903, 2907, 2909, and 2910 are similar to each other and different from other conformations (Fig. 6 A). Also, in a very simplified form, it seems that the other conformations are projected so that they belong to two different groups that are similarly distant from the central group (2902, 2903, 2907, 2909, and 2910). One group comprises conformations 2904–2906, whereas the other group is made of conformations 2875, 2908, and 2911 (Fig. 6 A). The overlapped density maps (Fig. 7 and Figs. S1 and S2) show that the three different coarse groups can be explained by the existence of at least two different types of changes in conformation and composition of the complex. One of these is a rotation of the small subunit (40S) with respect to the large subunit (60S), which could explain the groups 2904–2906 (40S is rotated in these maps, with different amounts of rotation, as shown in Fig. 7, L–O, and Fig. S1). The other is rolling of the complex, which could explain the central group of maps. Indeed, different degrees of rolling of the complex can be noticed in maps 2902, 2903, 2907, 2909, and 2910 (Fig. 7, A–H, and Fig. S1). The remaining group is comprised of maps of complexes with slightly different degrees of unrolling and different compositions (2875, 2908, and 2911) (Fig. 7, I and J, and Fig. S1). More precisely, in map 2875, the A and F sites are empty; in map 2908, they are occupied by A/T tRNA (A site) and eEF1A (F site); finally, in map 2911, the F site is empty and the A site is occupied by A/T tRNA (Fig. 7, I–K). Note here that we used a mask that suits the shape of map 2875 to compute pseudoatomic models, implying that computations were done using density maps with partly removed additional mass from the A and F sites (Fig. S2, A–E). The contribution of masking in this case thus was not only in removing the background noise, but also in focusing the analysis onto the conformational changes (by minimizing compositional differences while preserving the associated conformational differences).
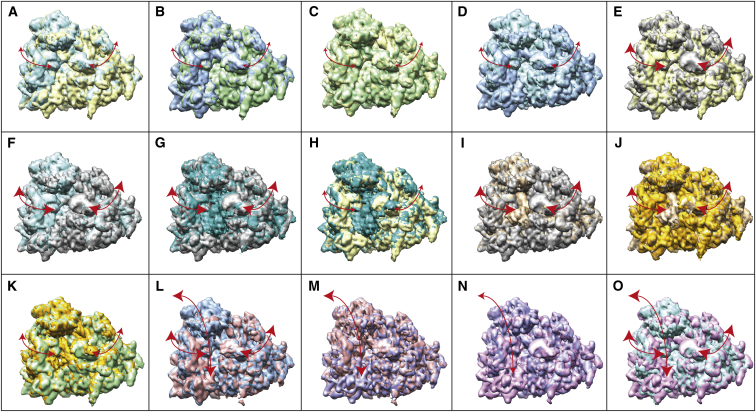
Figure 7.
Overlap of rigid-body-aligned EM density maps of 80S ribosome (EMD-2875 and from EMD-2902 to EMD-2911), with the same view for all overlapped density maps. (A) 2902 (yellow) vs. 2903. (B) 2907 (green) vs. 2909. (C) 2902 (yellow) vs. 2907. (D) 2903 (cyan) vs. 2909. (E) 2875 (gray) vs. 2902. (F) 2875 (gray) vs. 2903. (G) 2875 (gray) vs. 2910. (H) 2902 (yellow) vs. 2910. (I) 2875 (gray) vs. 2908. (J) 2908 (brown) vs. 2911. (K) 2911 (orange) vs. 2907. (L) 2906 (pink) vs. 2909. (M) 2906 (pink) vs. 2904. (N) 2904 (blue) vs. 2905. (O) 2905 (magenta) vs. 2903. In each panel, arrows show the movements that mainly contribute to the elastic transformation (deformation) between two overlapped maps, and the arrow scale shows the distance between these maps in the distance space. Bidirectional arrows were used to recall that StructMap computes the elastic transformation between two maps in both directions. Note that the arrow scale shows the dissimilarity between maps after the deformation, but the amount of deformation (movement) is not represented by arrows. However, note that the type of movement is represented by arrows (i.e., rolling/unrolling of the complex in (A)–(K), rotation of 40S with respect to 60S in (M) and (N), and combination of rolling/unrolling and 40S rotation in (L) and (O). See also Fig. 6 and Figs. S1 and S2. To see this figure in color, go online.
Although a combined conformational and compositional heterogeneity is a very difficult case, the proposed method showed very interesting results, such as the observed grouping of density maps into three coarse but interpretable groups in the same space of distances, as well as a closed trajectory formed by connecting points in the distance space according to the original proposal (Fig. 6). Interestingly enough, the trajectory form could be described as 8-like, particularly in the 2D distance space (Fig. 6 B).
Discussion
In this article, we presented StructMap, which is, to our knowledge, the first methodology that allows visualization of conformational differences among sets of EM density maps in a common and quantitative space and involves elastic alignment of these EM maps for conformational modeling. The elastic alignment of two EM maps is done by flexible deformation, using normal modes, of one map until it fits the other map. The elastic alignment allows the building of a matrix of distances among density maps, which is then analyzed to represent all density maps in the common distance space that is also referred to as the map of structures.
StructMap does not impose any requirements regarding the mass of the complex or the size of its dynamic part. The only requirement is that all EM maps have the same size (number of voxels) and the same voxel size. Also, the work of StructMap will be facilitated if the complex is masked in the density maps to suppress the background noise. Masking of density maps is a part of common data-processing workflows, and we expect that it will be done before starting StructMap. Otherwise, masking can be performed with the HEMNMA graphical interface. Masking can also be used to minimize the impact of compositional differences of complexes (e.g., due to ligand binding) on the analysis of conformational differences among these complexes. The method works with EM maps that are usually available at different resolutions due to the use of different numbers of particles for their reconstruction and other experimental imaging-related issues. In this article, we showed that StructMap yields reliable results (in agreement with previously published results) for EM maps of different resolution, more precisely, different resolution ranges (5–10 Å, 12–20 Å, and 23–25 Å) of EM maps reconstructed from negative stain images (Pol α-B) or cryo images (70S and 80S). A higher-resolution homogeneity of a set of EM maps is expected to yield a better resolution of conformational differences for the average level of detail in that set of maps. Also, a higher resolution of all EM maps in the set is expected to yield a more precise analysis of smaller conformational changes.
Beyond discrete states
Results obtained with synthetic and experimental data show the great potential of the proposed method. Results obtained with experimental data of Pol α-B, 70S, and 80S complexes are fully consistent with previously published results. Moreover, the results presented here complement the previous results with a quantitative analysis of given density maps, which produces an original graphical representation of dissimilarities among the density maps.
The experiment with EM density maps of human 80S ribosomal complexes from polysomes shows a particularly challenging case of heterogeneity, which clearly demonstrates the usefulness of the proposed method. Indeed, the EM density maps used in that experiment represent 11 states of the ribosome during the elongation cycle, and the EM maps differ among each other in both conformation and composition of the complex. With this data set, we observed a grouping of EM density maps in three coarse but interpretable groups, and we obtained a closed trajectory by connecting the EM maps in the obtained distance space according to the sequence proposed by the authors of the maps (25).
Possible trajectories of points in the map of structures
In some cases, trajectories of points can be inferred by connecting points in the map of structures, which suggests the possibility of exploring potential sequences of conformational changes. The points can be connected in many different ways, and here, we would like to discuss possible ad hoc alternatives, or “rules,” to analyze the cloud of points, especially in the absence of a priori information about conformational changes of the studied complex. One such rule could be to identify the shortest trajectory between the two most extreme or distant points, taking into account the assumption that the shortest trajectory corresponds to the lowest “energy” or “effort” required to go from one extreme state to the other (e.g., states 3 and 5 on the 3-2-1-4-5 trajectory in Experiment 1 are the two most distant states and they correspond to maximally closed and maximally open synthetic symmetric conformations of the complex). Another possible rule is to identify the trajectory of points corresponding to those EM density maps that result from the largest numbers of single-particle images, taking into account the assumption that a larger number of images used to reconstruct an EM map means a higher probability that the complex adopts this state. One could also think of combining such rules or even analyzing longer trajectories, taking into account possible random thermal motions of the complex in solution (9).
Network and graph analysis of conformational transitions also has been proposed in the context of analyzing protein conformational ensembles obtained by sampling the conformational space during molecular dynamics simulation of an atomic-resolution (x-ray or NMR) structure, the sampled conformations and transitions being considered as nodes and links of the network, respectively (38, 39, 40). The analysis of potential sequences of conformational changes is particularly difficult in the case of experimental EM maps and can additionally be hindered by a combined conformational and compositional heterogeneity of complexes, as observed in the experiment with the 80S ribosomal complex. Thus, although ad hoc rules could help to start the analysis, additional information about the complex should be employed to ascertain the structural processes that actually take place. That is why we did not use such rules in this article. We instead decided to complement the original work by analyzing similarities and dissimilarities among density maps based on their distances in the obtained distance space, as well as by showing the pattern obtained after connecting the density maps in this distance space according to the sequence proposed in the original publication.
Combination with continuous analysis of images
StructMap could be combined with techniques for continuous analysis of images, such as HEMNMA (8, 29), to explore continuous conformational changes more extensively. In this context, EM maps obtained by discretizing flexibility analysis would be analyzed by StructMap to better understand differences among maps and to select a few of them to use as reference conformations for the continuous image analysis. HEMNMA, which provides an overall view of the conformational distribution based on a 3D-to-2D elastic alignment of images with a given reference EM density map, would be used to perform a fine analysis of the dynamics around the reference conformations identified with the help of StructMap. This approach would be less computationally expensive than performing a fine analysis around every map from the given set of EM density maps. As StructMap performs an automatic analysis and projection of EM density maps onto a common distance space, it would be especially useful if the given set of density maps is large (e.g., containing more than five maps). Though the number of density maps obtained in the same experiment is typically <10, some studies result in much larger numbers of EM density maps (e.g., 50 maps were obtained in (15)). In the same context, StructMap could be combined with other approaches for exhaustive analysis of continuous conformational changes. For instance, StructMap could be used to select reference density maps to rigid-body align images before their analysis by the method proposed by Dashti et al. (9).
The work presented in this article opens doors for further development of such combined discrete-continuous approaches. To allow easy and broad use of StructMap, this methodology is currently implemented in Scipion (http://scipion.cnb.csic.es).
Author Contributions
C.O.S.S. and S.J. designed the method and the experiments. C.O.S.S. and M.K. implemented the method. C.O.S.S., S.J., and A.L.A.-C. performed the experiments. C.O.S.S., S.J., A.L.A.-C., and J.M.C. discussed the results. S.J. wrote the manuscript. J.M.C., C.O.S.S., A.L.A.-C., and M.K. reviewed the manuscript.
Acknowledgments
We thank GENCI-CINES/IDRIS (France) for HPC resources (x2013072174, x2014072174, and x2015072174); L. Pellegrini, S. Klinge (Cambridge University, Cambridge, United Kingdom), and O. Llorca (Centro de Investigaciones Biológicas-CSIC, Spain) for generously providing the Pol α-B EM structures; and Dr Isabelle Callebaut (IMPMC, Paris, France) for stimulating discussions.
This work was partially funded by the Centre National de la Recherche Scientifique (CNRS, France) and the Consejo Superior de Investigaciones Cientificas (CSIC, Spain) (Projet International de Coopération Scientifique, PICS 2011); the French National Research Agency (ANR-11-BSV8-010-04); the European Social Fund and the Ministerio de Educación y Ciencia (“Ramón y Cajal” fellowship to C.O.S.S.); the Spanish Ministry of Economy and Competitiveness (AIC-A-2011-0638 and BIO2013-44647-R); and the Comunidad de Madrid (CAM S2010/BMD-2305).
Editor: Andreas Engel.
Footnotes
Two figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30070-4.
Supporting Material
References
- 1.Frank J. Academic Press; San Diego: 1996. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. [Google Scholar]
- 2.Penczek P.A., Frank J., Spahn C.M. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J. Struct. Biol. 2006;154:184–194. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 3.Scheres S.H., Gao H., Carazo J.M. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat. Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 4.Fu J., Gao H., Frank J. Unsupervised classification of single particles by cluster tracking in multi-dimensional space. J. Struct. Biol. 2007;157:226–239. doi: 10.1016/j.jsb.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Elad N., Clare D.K., Orlova E.V. Detection and separation of heterogeneity in molecular complexes by statistical analysis of their two-dimensional projections. J. Struct. Biol. 2008;162:108–120. doi: 10.1016/j.jsb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z., Fang J., Penczek P.A. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyumkis D., Brilot A.F., Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Q., Sorzano C.O., Jonić S. Iterative elastic 3D-to-2D alignment method using normal modes for studying structural dynamics of large macromolecular complexes. Structure. 2014;22:496–506. doi: 10.1016/j.str.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Dashti A., Schwander P., Ourmazd A. Trajectories of the ribosome as a Brownian nanomachine. Proc. Natl. Acad. Sci. USA. 2014;111:17492–17497. doi: 10.1073/pnas.1419276111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katsevich E., Katsevich A., Singer A. Covariance matrix estimation for the cryo-EM heterogeneity problem. SIAM J. Imaging Sci. 2015;8:126–185. doi: 10.1137/130935434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tagare H.D., Kucukelbir A., Rao M. Directly reconstructing principal components of heterogeneous particles from cryo-EM images. J. Struct. Biol. 2015;191:245–262. doi: 10.1016/j.jsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grob P., Cruse M.J., Nogales E. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure. 2006;14:511–520. doi: 10.1016/j.str.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 13.Simonetti A., Marzi S., Klaholz B.P. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 14.Klinge S., Núñez-Ramírez R., Pellegrini L. 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. EMBO J. 2009;28:1978–1987. doi: 10.1038/emboj.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer N., Konevega A.L., Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 16.Lyumkis D., Doamekpor S.K., Joazeiro C.A. Single-particle EM reveals extensive conformational variability of the Ltn1 E3 ligase. Proc. Natl. Acad. Sci. USA. 2013;110:1702–1707. doi: 10.1073/pnas.1210041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Heel M., Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- 18.van Heel M., Frank J. Classification of particles in noisy electron micrographs using correspondence analysis. In: Gelsema I.L.S., Kanal L., editors. Pattern Recognition in Practice. North-Holland; Amsterdam: 1980. pp. 235–243. [Google Scholar]
- 19.Suhre K., Navaza J., Sanejouand Y.H. NORMA: a tool for flexible fitting of high-resolution protein structures into low-resolution electron-microscopy-derived density maps. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1098–1100. doi: 10.1107/S090744490602244X. [DOI] [PubMed] [Google Scholar]
- 20.Tama F., Miyashita O., Brooks C.L., 3rd Flexible multi-scale fitting of atomic structures into low-resolution electron density maps with elastic network normal mode analysis. J. Mol. Biol. 2004;337:985–999. doi: 10.1016/j.jmb.2004.01.048. [DOI] [PubMed] [Google Scholar]
- 21.Chacón P., Tama F., Wriggers W. Mega-Dalton biomolecular motion captured from electron microscopy reconstructions. J. Mol. Biol. 2003;326:485–492. doi: 10.1016/s0022-2836(02)01426-2. [DOI] [PubMed] [Google Scholar]
- 22.Tama F., Wriggers W., Brooks C.L., 3rd Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J. Mol. Biol. 2002;321:297–305. doi: 10.1016/s0022-2836(02)00627-7. [DOI] [PubMed] [Google Scholar]
- 23.Ming D., Kong Y., Ma J. How to describe protein motion without amino acid sequence and atomic coordinates. Proc. Natl. Acad. Sci. USA. 2002;99:8620–8625. doi: 10.1073/pnas.082148899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samsó M., Feng W., Allen P.D. Coordinated movement of cytoplasmic and transmembrane domains of RyR1 upon gating. PLoS Biol. 2009;7:e85. doi: 10.1371/journal.pbio.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behrmann E., Loerke J., Spahn C.M. Structural snapshots of actively translating human ribosomes. Cell. 2015;161:845–857. doi: 10.1016/j.cell.2015.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Pfeffer S., Förster F. Fast and accurate reference-free alignment of subtomograms. J. Struct. Biol. 2013;182:235–245. doi: 10.1016/j.jsb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Nogales-Cadenas R., Jonic S., Sorzano C.O. 3DEM Loupe: analysis of macromolecular dynamics using structures from electron microscopy. Nucleic Acids Res. 2013;41:W363–W367. doi: 10.1093/nar/gkt385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tirion M.M. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 29.Sorzano C.O., de la Rosa-Trevín J.M., Jonić S. Hybrid Electron Microscopy Normal Mode Analysis graphical interface and protocol. J. Struct. Biol. 2014;188:134–141. doi: 10.1016/j.jsb.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Powell M.J.D. UOBYQA: unconstrained optimization by quadratic approximation. Math. Program. 2002;92:555–582. [Google Scholar]
- 31.Berghen F.V., Bersini H. CONDOR, a new parallel, constrained extension of Powell’s UOBYQA algorithm: experimental results and comparison with the DFO algorithm. J. Comput. Appl. Math. 2005;181:157–175. [Google Scholar]
- 32.Sorzano C.O., Marabini R., Pascual-Montano A. XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Scheres S.H., Núñez-Ramírez R., Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Rosa-Trevín J.M., Otón J., Sorzano C.O. Xmipp 3.0: an improved software suite for image processing in electron microscopy. J. Struct. Biol. 2013;184:321–328. doi: 10.1016/j.jsb.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Cox T.F., Cox M.A.A. Chapman & Hall/CRC; Boca Raton, FL: 2001. Multidimensional Scaling. [Google Scholar]
- 36.Velázquez-Muriel J.A., Sorzano C.O., Carazo J.M. A method for estimating the CTF in electron microscopy based on ARMA models and parameter adjustment. Ultramicroscopy. 2003;96:17–35. doi: 10.1016/S0304-3991(02)00377-7. [DOI] [PubMed] [Google Scholar]
- 37.Bruschweiler R. Collective protein dynamics and nuclear spin relaxation. J. Chem. Phys. 1995;102:3396–3403. [Google Scholar]
- 38.Ahlstrom L.S., Baker J.L., Miyashita O. Network visualization of conformational sampling during molecular dynamics simulation. J. Mol. Graph. Model. 2013;46:140–149. doi: 10.1016/j.jmgm.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morcos F., Chatterjee S., Izaguirre J.A. Modeling conformational ensembles of slow functional motions in Pin1-WW. PLOS Comput. Biol. 2010;6:e1001015. doi: 10.1371/journal.pcbi.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caflisch A. Network and graph analyses of folding free energy surfaces. Curr. Opin. Struct. Biol. 2006;16:71–78. doi: 10.1016/j.sbi.2006.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.