Abstract
The nucleosome core particle (NCP) is the basic building block of chromatin. Nucleosome-nucleosome interactions are instrumental in chromatin compaction, and understanding NCP self-assembly is important for understanding chromatin structure and dynamics. Recombinant NCPs aggregated by multivalent cations form various ordered phases that can be studied by x-ray diffraction (small-angle x-ray scattering). In this work, the effects on the supramolecular structure of aggregated NCPs due to lysine histone H4 tail acetylations, histone H2A mutations (neutralizing the acidic patch of the histone octamer), and the removal of histone tails were investigated. The formation of ordered mainly hexagonal columnar NCP phases is in agreement with earlier studies; however, the highly homogeneous recombinant NCP systems used in this work display a more compact packing. The long-range order of the NCP columnar phase was found to be abolished or reduced by acetylation of the H4 tails, acidic patch neutralization, and removal of the H3 and H2B tails. Loss of nucleosome stacking upon removal of the H3 tails in combination with other tails was observed. In the absence of the H2A tails, the formation of an unknown highly ordered phase was observed.
Introduction
In most species of eukaryotes, genomic DNA is packed by nuclear proteins, forming chromatin. Chromatin consists of regular linear arrays of DNA-histone complexes, the nucleosomes. A central part of the nucleosome is the nucleosome core particle (NCP), which is formed by the wrapping of 145–147 bp DNA as a 1.70–1.75-turn superhelix around an octamer of four core histone proteins, two H2A/H2B dimers, and an (H3/H4)2 tetramer (1, 2). The NCPs are connected by linker DNA of variable length (10–80 bp), forming a beads-on-a-string chromatin fiber that is further folded into various dynamic secondary and tertiary structures that are still not fully understood. In higher organisms, an additional protein, the linker histone H1, is also regularly present and contributes to chromatin folding, but cells are remarkably tolerant to H1 depletion (3) and it has been shown that fibers with short nucleosome repeat lengths can fold to their natural degree of compaction even in the absence of the H1 histone (4).
NCPs show aggregation behavior similar to that of chromatin fibers, and this phase separation is sensitive to the ionic conditions, with the presence of multivalent cations with charge ≥ +2 provoking self-association and aggregation (5, 6, 7, 8, 9, 10, 11, 12, 13, 14). This sensitivity is due to the polyelectrolyte nature of the NCP, which is a polycation-polyanion complex between the histone octamer (HO) and the DNA (15). Nucleosomal DNA carries a large negative charge (−290e to −294e), which is only partially neutralized by the histones (the total net charge of the HO is +148e). The charge of the eight core histones is unevenly distributed between the octamer globular domain (+60e) and the eight flexible, unstructured N-termini (the histone tails) (1, 16), each of which carries a charge of +9e to +14e due to a varying number of positively charged lysine and arginine residues. Consequently, the NCP, which has a significant net negative charge (∼−148e), can be visualized as a central particle (−236e) to which the positive flexible tails are attached (15, 17). The tails can interact with the DNA of their own NCP, with the linker DNA, and with the other NCPs and proteins present in chromatin. They can also perform important roles in chromatin folding (4, 18) and are essential for primary nucleosome-nucleosome interactions (19). Folding and aggregation of chromatin are facilitated and regulated by tail bridging between adjacent NCPs (8, 11, 20).
Amino acids in the histone tails are subject to numerous posttranslational modifications, with charge quenching acetylations of the ε-amino groups of the lysines being the most frequent and dynamic. Acetylated chromatin is more easily digested by nucleases (21, 22) and shows higher solubility in monovalent salt (NaCl and KCl) and in the presence of millimolar concentrations of Mg2+ (22, 23, 24). The histone tails also play important roles in transcription, replication, repair, and recombination, both as direct modulators of chromatin folding and as a recognition platform for regulatory proteins (25, 26). These functions of the tails are regulated through histone modifications, and the presence of tail acetylations typically correlates with transcriptionally active chromatin (27, 28, 29, 30).
The cation-induced aggregation of NCPs is similar to the behavior exhibited by other polyelectrolyte systems such as DNA (15, 31); however, DNA condensation normally can only be induced by cations with a charge of +3 or higher (32), whereas NCP and chromatin systems can be condensed by divalent cations such as Mg2+ and Ca2+ (15). NCP-NCP attraction is mediated by the histone tails and is observed in solutions of monovalent salt (8, 11) as well as in the presence of Mg2+, whereas under similar conditions, DNA, as well as NCPs and nucleosome arrays lacking the tails, do not show aggregation (4, 11) (and references cited in (4)). The close NCP-NCP contact between the flat surfaces of the HO core on both sides of the cylindrical wedge-shaped NCP, called nucleosome stacking, is a common structural feature of the condensed state of chromatin. This stacking has been observed in NCP crystals (1, 2, 33, 34, 35), in NCP liquid crystalline phases (9, 10, 13, 36, 37), in the crystal of the tetranucleosome (38), in folded nucleosome arrays (39, 40, 41), and in cryo-electron microscopy (cryo-EM) images of frozen isolated native chromatin (42, 43).
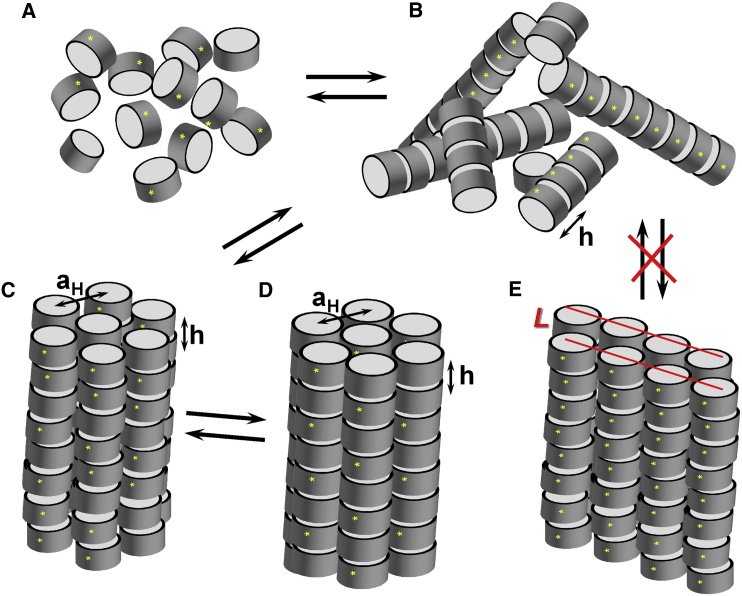
Various aggregates of NCPs phase-separated in the presence of multivalent cations (or a high concentration of monovalent salt and osmotic pressure) have been systematically characterized by Bertin et al. (13) and Livolant et al. (44) (and references cited therein). These phases include isotropic NCPs similar to the solution-state, NCP columnar phase with no long-range order between columns, as well as the lamello-columnar and highly ordered columnar hexagonal phases (44) (schematically shown in Fig. 1). The phenomenon of supramolecular ordering in NCP aggregates is determined by a complex combination of variables in the mixture and by intrinsic NCP properties. The former include NCP concentration, buffer conditions, cation type, concentration, and osmotic pressure. It was previously shown that the cation-induced formation of condensed NCP phases follows a polyelectrolyte behavior similar to DNA condensation (5). Aggregation of NCPs is dependent on the charge and nature of the cations present in solution (5, 6, 7, 13), and the histone tails mediate NCP-NCP contacts in a salt-dependent manner (7, 8, 11, 12, 14). However, despite all of these observations, knowledge about the molecular structure and thermodynamics of the NCP-NCP contacts, and the precise roles of the tails and tail modifications in the modulation of NCP-NCP interactions and the formation of ordered phases is still limited. Even less is known about the contribution of intrinsic NCP features, such as the cylindrical wedge shape, inhomogeneous surface charge distribution, dyadic symmetry, and chirality (45, 46, 47). Most of the earlier studies used either cell-extracted NCPs or NCPs reconstituted using HOs from chicken blood. These NCPs have intrinsic features with a certain degree of heterogeneity in DNA length, and unknown degrees and positions of histone posttranslational modifications. One can vary the NCP tail and surface charge distribution in a controlled way using recombinant NCPs to address several aspects of nucleosome interactions, in particular modification effects and the roles of histone tails. The synchrotron x-ray diffraction method (using small-angle x-ray scattering (SAXS)) is well suited to address the influence of NCP charge distribution through the detection and characterization of order in the NCP phases (9, 10, 44, 45, 46, 47, 48). The effects of dyadic symmetry and NCP chirality on the order in NCP phases, however, can only be addressed by cryo-EM and optical microscopy methods.
Figure 1.
(A–E) Schematic presentation of the phases formed by NCPs: (A) isotropic NCP phase, (B) columnar NCP phase, (C) 2D columnar hexagonal phase, and (D) 3D orthorhombic quasi-hexagonal phase. The distance between stacked NCPs is marked as h, the intercolumnar distance is aH, and stars indicate the direction of the dyad axis. (E) A single bilayer of the lamello-columnar NCP phase observed by Mangenot et al. (9, 10) and Livolant et al. (44) in systems where NCP condensation was caused by osmotic stress and addition of monovalent cations. Lines indicate NCP layers in the bilayer. Bilayers in turn formed higher-order structures, with the bilayer-bilayer distance dependent on conditions. To see this figure in color, go online.
Large quantities of homogeneous NCPs with well-defined DNA sequences can be obtained using recombinant histones (49). Such well-defined systems allow one to determine the contributions of individual tails and specific alterations within them to the association behavior of mononucleosomes. As noted above, much of the previously published data on NCP systems was obtained from cell-extracted chromatin or in solutions of NCPs obtained by nuclease treatment of the chromatin (4, 6, 9, 10, 13, 50) (and references cited therein). A growing number of studies are using well-defined chromatin and NCPs prepared by recombinant methods (12, 14, 40, 51, 52, 53, 54, 55, 56, 57, 58). However, a limited amount of data are available for systems with modifications of the individual histones, such as mutants lacking tails, containing only the globular part of the histone molecule (12), or with selected tail lysines replaced by glutamine (K→Q mutation), mimicking acetylation (40, 51, 52, 53, 54, 55, 56, 59). Published results indicate that acetylation or the absence of tails stabilizes NCPs or arrays in the solution state against self-association and promotes the unfolding of compact nucleosome arrays (12, 51, 52, 53, 58, 60). The H4 tail (12, 52, 61, 62) and specifically H4-K16 (51, 55, 58) have been found to be critical for intramolecular folding of individual nucleosome arrays. Of note, one of the H4 K16-R23 regions (of the two H4 tails) is involved in NCP stacking, since it is located between the two NCPs in practically all reported NCP crystal structures, interacting with the acidic patch located on the globular domain of the H2A histone of the neighboring NCP (1, 63).
In this work, we characterized the x-ray diffraction spectra of various precipitated NCP samples and their Bragg peak patterns (using the synchrotron SAXS beamline) to investigate the supramolecular structure in NCP aggregates, complemented by a SAXS dilute-solution investigation for selected samples. We studied the effects caused by H4 tail acetylations or deletions of tails on the supramolecular structure of NCPs aggregated by Mg2+ and cobalt(III)hexammine (CoHex3+) cations. Wild-type (WT) and mutant NCPs were reconstituted from recombinant Xenopus laevis histones with a 145 bp DNA fragment of the 601 nucleosome positioning sequence (64). Specifically, NCPs containing H4 histones with lysines acetylated at the K16 position (H4-K16Ac) and at the K5, K8, K12, and K16 positions (H4-QuadAc) were investigated. To compare the effects of acetylations with K→Q mutations (both quenching the lysine charge), NCPs containing H4 tails mutated at the same K5, K8, K12, and K16 positions (H4-K16Q and H4-QuadQ) were studied. The effect of H2A histone acidic patch neutralization was investigated. Additionally, the effects of deletion of individual tails and combinations thereof were investigated. In a recent work using well-defined recombinant NCPs, we investigated the effects of H4 tail acetylation and deletion of tails on cation-induced NCP aggregation (20), and the work presented here complements that study.
Materials and Methods
A detailed description of the materials and methods applied in this work is given in Supporting Materials and Methods in the Supporting Material. Below, a brief account of the applied techniques is presented.
Preparation of NCPs
Recombinant WT, mutated, and truncated X. laevis core histone proteins H2A, H2B, H3, and H4 were expressed in the Escherichia coli [BL21 (DE3) pLysS] strain and purified by gel-filtration and cation-exchange chromatography. H4 histones containing acetylated lysine, H4-K16Ac, and H4-QuadAc (H4 histone with lysine residues acetylated at positions 5, 8, 12, and 16) were synthesized by natural chemical ligation and purified as described previously (58).
The HOs were refolded by dialysis from a mixture of WT or modified core histones H2A, H2B, H3, and H4, and purified by gel-filtration chromatography. The purity and completeness of the HO refolding was checked by 18% SDS-PAGE.
The 145 bp DNA of the 601 Widom nucleosome positioning sequence (64) was used in this work to reconstitute NCPs. In a few measurements, NCPs reconstituted with the 147 bp palindromic human α-satellite DNA fragment (1, 2) were also used. The 145 bp 601 DNA construct in the pUC19 plasmid was amplified using the E. coli HB101 strain, extracted by the alkaline lysis method, released by EcoRV, and separated by PEG 6000.
The NCPs were reconstituted from DNA and HO by a stepwise dialysis in a high- to low-salt buffer (49, 65, 66). The best yield was obtained for the WT NCPs; reconstitution of the NCPs with mutated and deleted tails was less efficient, but still was notably better than that described by Bertin et al. (12, 14) for tailless NCP constructs. The quality of the reconstitution was assayed by 5% PAGE (see Fig. S3 C), yielding an estimated presence of <5% of free DNA. After NCP precipitation from solution by multivalent cations for the x-ray diffraction measurements, any free DNA will remain in the supernatant and this stage acts as a further purification.
Sample preparation, data collection, and analysis
We investigated the structures of cation-induced NCP aggregates using SAXS, where ordered phases can be identified from the way they diffract x-rays. Table S1 contains a summary of more than 200 analyzed samples. Along with WT-NCPs assembled with HO consisting of recombinant, fully charged histones, we reconstituted four variants of the NCP using various H4 histones with single- or tetra-acetylated or mutated lysine residues in the histone tail: H4-K16Ac, H4-QuadAc, H4-K16Q, and H4-QuadQ. Additionally, one HO was prepared with three charge-neutralizing mutations in the acidic patch of the H2A histone (the mutations were D90S+E91T+E92T, abbreviated as H2A-STT). The other NCP samples contained various combinations of tailless histones. NCP aggregation was induced by addition of divalent Mg2+ or trivalent CoHex3+ cations. Aggregation of a few samples of WT-NCP was also induced by the polyamines spermidine3+ (Spd3+) and spermine4+ (Spm4+). The final concentration of the NCP was 8 mg/mL. For comparison and reference purposes, solution SAXS spectra for WT-NCP solutions in a concentration range of 1.25–17 mg/mL NCP and two concentrations of KCl (10 and 100 mM) were obtained. Furthermore, solution spectra of the H4K16Q, H4-QuadQ, gH2A, and gH2BgH2B samples were collected for two concentrations of KCl.
High-energy synchrotron radiated x-rays were used to compensate for the weak scattering intensity of the biological samples. The samples were prepared as described in Supporting Materials and Methods. Measurements were made at the beamline 23A SWAXS end-station at the National Synchrotron Radiation Research Center (Hsinchu, Taiwan) (67). Some samples were repeatedly measured at intervals of several months with no apparent changes in the spectral characteristics (data not shown).
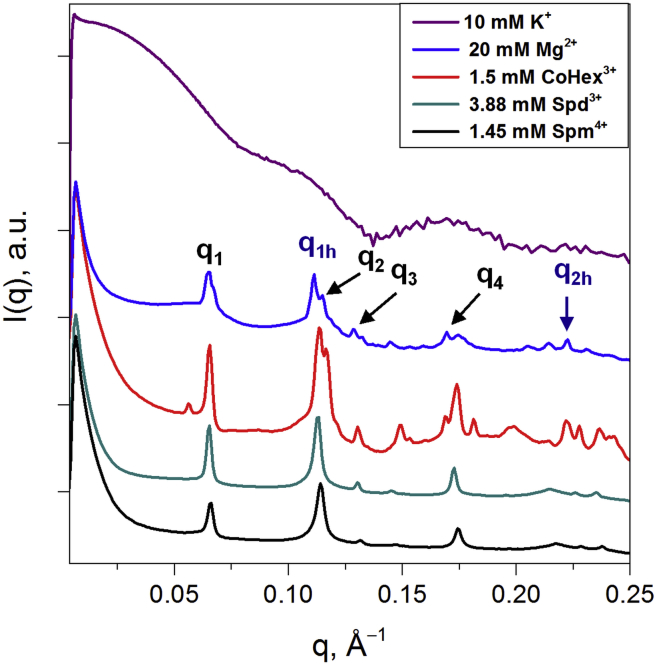
The phases of the NCP aggregates were identified from the periodicity of the Bragg peaks as a function of the scattering vector q. NCP stacking and hexagonally ordered columns generate sets of peaks with a periodicity of 1:2:3:4 and 1:√3:√4:√7:√9, respectively (9, 44, 68). The NCP stacking distance h and the intercolumnar distance aH in columnar hexagonal phases (Fig. 1, C and D) were calculated from the position of the first peaks, q1h and q1, using the equations h = , and aH = . Fig. 2 displays an example of an x-ray diffraction spectrum with annotated peaks.
Figure 2.
SAXS spectra of the solution and most-ordered phases of WT-NCP with cations (the nature and concentration of the ions are indicated in the graph). The solution SAXS spectrum of WT-NCP at 1.25 mg/mL in 10 mM KCl (top spectrum) largely overlaps with the profile calculated from the atomic coordinates of the NCP crystal structure 1KX5 (2) (see Fig. S2). In the presence of multivalent cations (second to last spectra from the top), NCPs precipitated from solution form ordered columnar hexagonal structures. On the spectrum in the presence of 20 mM Mg2+, the peak characteristics of the 3D orthorhombic order (q1, q2, q3, and q4) due to NCP-NCP stacking within the columns (q1h and q2h) are indicated. To see this figure in color, go online.
Results and Discussion
We used SAXS to study the effects of lysine acetylations in the H4 histone tails, neutralization of the acidic patch on the H2A globular domain, and removal of histone tails in the recombinant NCP aggregated by Mg2+ and CoHex3+ cations (Table S1). In addition, H4-K16Q, H2A-STT, gH2A, and gH2AgH2B samples in solution at two concentrations of monovalent KCl were investigated.
Solution SAXS spectra
The diffraction spectrum intensity I(q) originates from x-rays scattered by the atoms of individual NCPs (by the form factor, P(q)) and by the structures formed by interacting NCPs (by the structure factor, S(q)), hence I(q) ∼ P(q) × S(q)). Fig. 2 shows spectra of the WT-NCP recorded in the presence of five cations, namely, K+, Mg2+, CoHex3+, and the polyamines Spd3+ and Spm4+. It is well established that at low concentrations of monovalent cations (K+ or Na+), the interaction between nucleosomes is weak and repulsive (7, 48). Therefore, the SAXS spectrum obtained from the solution of homogeneous and isotropic NCPs is primarily defined by the form factor P(q) of the NCPs (top magenta curve in Fig. 2). For the WT-NCP, we recorded SAXS spectra of NCP solutions with 10 and 100 mM KCl in a range of NCP concentrations from 1.25 to 17 mg/mL (see Fig. S1). Our results agree with the solution SAXS data reported by others (7, 14, 48). At low NCP concentrations (CNCP = 1.25 and 2.5 mg/mL) in 10 mM KCl, the experimental SAXS profiles show an almost perfect overlap with the simulated spectrum calculated from the atomic structure of the NCP obtained from molecular-dynamics simulations (69), which compared with the NCP structure in the crystal (2) exhibits collapse of the histone tails on the globular part of the NCP. According to physicochemical principles (70), oligocationic tails should be bound to polyanionic DNA under low-salt conditions, and this most realistic NCP structure shows the best agreement with the measured SAXS spectrum. NCP structures with the tails extended outside of the globular part or with the tails removed showed worse agreement with experimental data. We used two programs to simulate SAXS spectra from atomic coordinates, CRYSOL (71) and FoXS (72) (Fig. S2), which produced very similar results.
We used the experimental form factor of the WT-NCP obtained at CNCP = 1.25 mg/mL in 10 mM KCl to estimate the structure factors of the WT-NCP in solution. The scattering profiles together with calculated structure factors (Fig. S1, C and D) allow a better assessment of the NCP ordering when broad maxima in scattering vector ranges of 0.09–0.10 and 0.16–0.17 Å−1 of the P(q) profile contribute significantly and overlap with peaks from weak NCP-NCP interactions. Samples of WT-NCPs show little interaction in both 10 and 100 mM KCl. At 10 mM KCl, an increase in the NCP concentration results in a progressive drop of the intensity of the S(q) profile at low q-values (Fig. S1 C), indicating repulsive NCP-NCP interactions. In contrast, at 100 mM KCl, weak maxima observed at 0.012–0.016 Å−1 (Fig. S1 D) are likely to indicate the weak attractive NCP-NCP interactions as reported by others for NCP solutions with increased concentrations of NaCl and NCP (7, 11, 48).
The scattering from solutions of selected mutated and tailless NCPs was recorded to assess the form factor that could indicate the change in shape of the particles caused by modifications. Selected SAXS profiles recorded for the WT-NCP, H4-K16Q, and H2A-STT NCPs are compared in Fig. S3 A. Fig. S3 B shows spectra in solutions of the WT-NCP, gH2A, and gH2AgH2B NCPs. Figs. S4 and S5 present all solution SAXS spectra collected for the selected mutated and tailless NCPs. H4-K16Q, and H2A-STT samples produced scattering spectra with features almost identical to the WT-NCPs (Fig. S3 A). The results in Fig. S3 A demonstrate (together with the corresponding gels in Fig. S3 C and other assays, data not shown) that modifications/mutations have only minor effect on the particle shape and size and that our systems are homogeneous, which is known from previous studies with these constructs. The tailless particles on the other hand, produced featureless spectra, distinct from the WT-NCP (Fig. S3 B). These spectra are in agreement with the spectra reported by Livolant and colleagues (14). The absence of the minima as the most pronounced effect of tail deletion was suggested to originate from a change in DNA wrapping around the octamer core and may also indicate that DNA wrapping is more dynamic and flexible.
Cation-induced condensed phases of WT-NCP
The formation of ordered phases by WT-NCP (i.e., for histones without any modifications or tail deletions) in a range of concentrations of Mg2+, CoHex3+, Spd3+, and Spm4+ was investigated (Figs. 2, 3, S6, and S7; Tables S1 and S2). Upon addition of multivalent cations, the highly homogeneous recombinant NCPs instantaneously aggregate, and precipitate upon gentle centrifugation (see Supporting Materials and Methods). In a certain cation concentration range, highly ordered phases were identified from SAXS spectra (Fig. 2). The spectral features are reproducible without significant changes for periods from immediate measurements within days of preparation up to several months (data not shown).
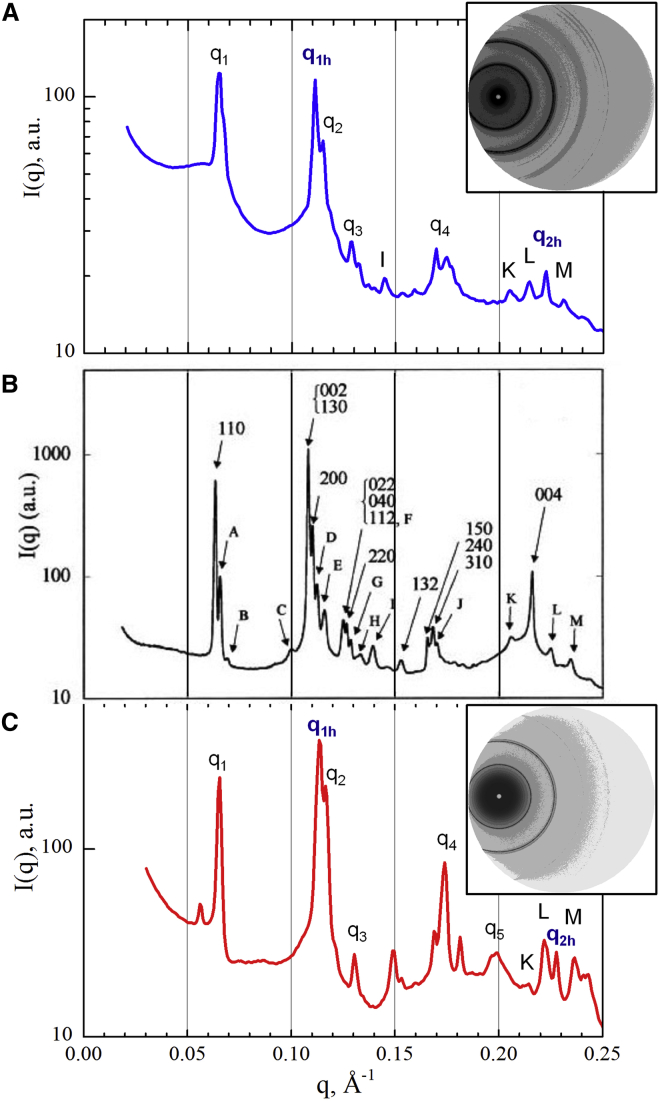
Figure 3.
(A and C) Spectra of highly ordered phases formed by WT-NCP in the presence of 20 mM Mg2+ (A) and 1.5 mM CoHex3+ (C). The insets show corresponding SAXS diffraction patterns. (B) The spectrum for NCPs obtained from nuclease-digested calf thymus chromatin in 160 mM NaCl under an osmotic pressure of 4.7 × 105 Pa after 4 months of equilibration and after partial orientation in a 10 T magnetic field for 15 days (9) is shown for comparison. In (B), peaks are marked according to the periodicity of the orthorhombic lattice (a, b, c) with dimensions (aH, √3 × aH, ∼h). Peaks indicated by letters from A to M are suggested to correspond to a superstructure along the c-axis of a periodicity close to 6c: A (1, 1, 1/3), B (1, 1, 1/2), C (0, 2, 1 + 1/2), D (1, 3, 1/2), E (0, 0, 2 + 1/6), F (0, 0, 2 + 1/3), G (0, 4, 1/2) and (2, 2, 1/2), H (0, 0, 2 + 1/2), I (2, 1, 1 + 1/2), J (3, 1, 1/2), K (3, 3, 1 + 1/2), L (0, 0, 4 + 1/6), M (0, 0, 4 + 1/3). Panel (B) is reprinted with permission from the Biophysical Society. To see this figure in color, go online.
Columnar hexagonal phases are formed in solutions of WT-NCP in the presence of millimolar concentrations of Mg2+, CoHex3+, Spd3+ and Spm4+ as indicated by a clear appearance of the characteristic hexagonal order peaks, q1, q2, q3, and q4 at ∼0.065, 0.113, 0.13, and 0.172 Å−1, with the expected ratio of 1:√3:√4:√7 (Fig. 2). The set of peaks with a 1:2 ratio at ∼0.11 and 0.22 Å−1, assigned respectively as q1h and q2h indicate columnar stacking of the NCPs. The most ordered WT-NCP aggregates were observed at 20 mM Mg2+, 1.5 mM CoHex3+, 3.88 mM Spd3+, and 1.45 mM Spm4+ (Fig. 2; Table S2). The presence of only q1,2,3 and q1h,2h peaks, illustrated by spectra for WT-NCP with Spd3+ and Spm4+ indicates a two-dimensional (2D) columnar hexagonal phase. The presence of peaks in addition to the columnar hexagonal phase, illustrated in spectra from the WT-NCP with Mg2+ and CoHex3+, indicates the formation of a three-dimensional (3D) orthorhombic quasi-hexagonal phase. The additional peaks indicate correlation between columnar and hexagonal orders like in a crystal. This interpretation was developed by Mangenot et al. (9) and Livolant et al. (44), who used the intercolumnar distance aH and NCP stacking distance h to characterize the hexagonal phase, and a, b, c indices for the orthorhombic quasi-hexagonal phase. The orthorhombic quasi-hexagonal phase has a slightly distorted hexagonal order, with peak positions deviating by ∼2% from the hexagonal periodicity. In this work, although we distinguish between phases with 2D columnar hexagonal and 3D orthorhombic quasi-hexagonal order, we use the 2D notation for both.
The NCP stacking distances h = 55–57 Å and the intercolumnar distances aH = 110–112 Å calculated from the SAXS spectra of the WT-NCP are inversely related to the cation charge, being the shortest for the Spm4+ and largest for the Mg2+ ions, indicating the relative compactness of the NCP packing (Table S2). Previous computational studies have also shown more condensed internucleosome packing induced by CoHex3+ compared with Mg2+ (20, 57, 58). The characteristic distances observed for recombinant WT-NCPs that were precipitated by multivalent cations were generally comparable or less than the values reported for native cell-extracted NCPs condensed by osmotic shock in the presence of NaCl (h = 57.7–60 Å and aH = 110–116 Å) (44).
The 3D orthorhombic quasi-hexagonal symmetry in NCP aggregates is not unexpected given the orthorhombic packing obtained for single crystals of NCPs (1, 2). In Fig. 3, we compare our spectra of ordered WT-NCP phases condensed by Mg2+ and CoHex3+ with a spectrum previously obtained for highly ordered NCPs prepared from nuclease-digested calf thymus chromatin, aggregated by osmotic pressure at a high monovalent salt concentration, oriented in a magnetic field for 15 days and measured after 4 months of equilibration (9, 44). The comparison allows us to index some of the correlation peaks in our x-ray diffraction spectra. However, our assignment is incomplete and should be considered tentative, and furthermore, our samples were not oriented and thus were more disordered. It is challenging to assign all diffraction peaks from the averaged one-dimensional x-ray diffraction spectrum from a polycrystalline sample. 2D spectra with the phase information from single crystals are needed to unequivocally assign all peaks and determine the structure. However, it is clear that the recombinant WT-NCP samples aggregated by Mg2+ or CoHex3+ are indicative of a degree of ordering similar to that previously obtained under conditions that favor high order (9, 44). This shows that the formation of highly ordered columnar hexagonal phases is a natural characteristic of mononucleosomes that likely corresponds to the equilibrium phase under physiological conditions.
The polyelectrolyte behavior of NCP interactions in a wide range of Mg2+ concentrations is noteworthy. A gradual loss of hexagonal order is observed at cation concentrations significantly lower or higher than 20 mM (Fig. S6 A). In the range of 8–14 and 24–50 mM Mg2+, an NCP columnar phase with no long-range order is observed. The broad peaks arise from the average intercolumnar distance (q1 ∼0.065 Å−1), the average NCP stacking distance within columns, and the columnar form factor from the local arrangement (q2 and q1h ∼0.11 Å−1 and q3 ∼0.17 Å−1) (44). At 3 and 80 mM Mg2+, an isotropic NCP phase is observed, and the spectra are similar to the form factor obtained from a solution of WT-NCP. At Mg2+ concentrations lower than 3 mM or higher than 80 mM, the NCPs remain soluble. In contrast, a stable columnar hexagonal phase is observed in a broad range of CoHex3+ concentrations (1.5–30 mM), with an isotropic NCP phase being observed at 1.2 mM CoHex3+ (see Fig. S6).
We also obtained x-ray diffraction spectra for a few samples of NCPs reconstituted with a different DNA template, namely, the 147 bp palindromic human α-satellite DNA fragment. The spectra are shown in Fig. S8 and the pattern shows a 2D hexagonal columnar phase, displaying sharp peaks, in addition to some weak additional peaks indicating distortion from the 2D hexagonal arrangement. Consequently, the columnar hexagonal phase was obtained for mononucleosomes extracted from cells that likely contain an unknown amount of posttranslational modifications in the histone tails (Livolant et al. (44)).
Supramolecular structures for NCPs with modified and tailless histones
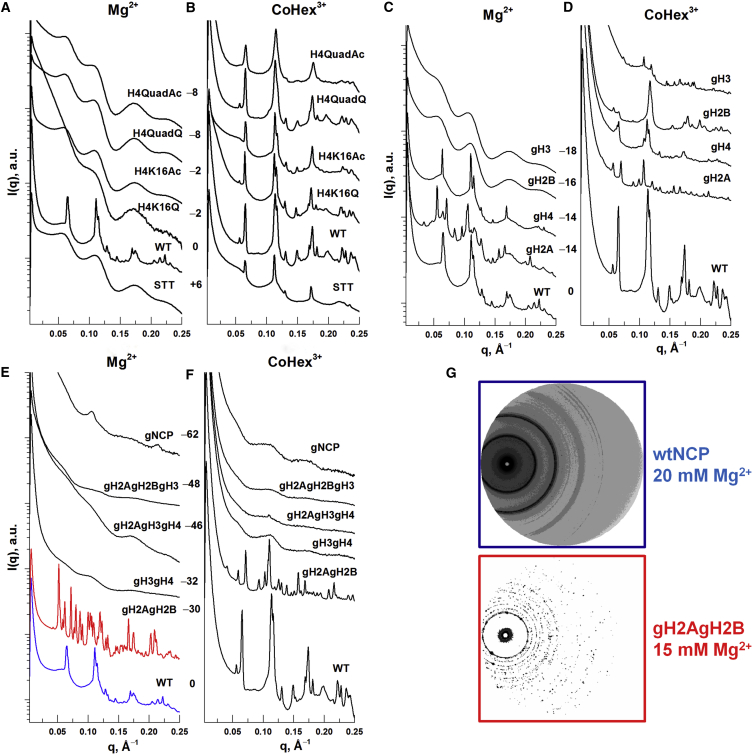
Spectra representing the most ordered phases for NCPs with various histone modifications obtained in presence of Mg2+ or CoHex3+ are summarized in Fig. 4. The concentration ranges of Mg2+ and CoHex3+ investigated for each variant of NCP and the type of the most ordered phase are schematically presented in Fig. 5 (see also Table S2 and Figs. S9–S22). In Fig. 4 the spectra are divided into three groups: 1) NCPs with mutated and acetylated histones (H2A-STT, H4-K16Q, H4-K16Ac, H4-QuadQ, and H4-QuadAc; Fig. 4, A and B); 2) NCPs with one of the four histone tails truncated (gH2A, gH4, gH2B, and gH3; Fig. 4, C and D), and 3) NCPs lacking two or more histone tails (gH2AgH2B, gH3gH4, gH2AgH3gH4, gH2AgH2BgH3, and gNCP; Fig. 4, E and F). Within each graph the spectra are aligned in the order that reflects the change of the charge in the HO relative to the charge of the WT-NCP (numbers are shown between the Mg2+ and CoHex3+ panels). All modifications, excluding H2A-STT, increase the net negative charge of NCPs. Changes in the NCP charge and charge distribution alone may alter the NCP structure and affect NCP-NCP interactions, which in turn may influence the degree of order within the NCP aggregates in a cation-dependent manner. In addition, acetylation or tail deletion may affect the NCP structure and NCP-NCP interactions in a cation-independent manner, combining electrostatic and steric contributions.
Figure 4.
(A–F) SAXS spectra for NCPs with various histone modifications and with added Mg2+ (A, C, and E) or CoHex3+ (B, D, and F). (A and B) NCPs with octamers containing mutated or H4-acetylated histones. (C and D) NCPs with HOs lacking tails in one of the four histones (gH2A, gH2B, gH3, and gH4). (E and F) NCPs with octamers lacking tails in two or more of the four histones. For each NCP modification, the concentrations of Mg2+ and CoHex3+ were varied and the SAXS spectrum corresponding to the most-organized NCP phase is shown. All spectra are given in Figs. S9–S22. In each panel, the WT-NCP SAXS profile is given for comparison; the change of the net NCP charge relative to the WT-NCP is indicated next to the sample identifiers. (G) Two types of NCPs containing tailless histone H2A (gH2A and gH2AgH2B) form highly ordered structures that are different from all other NCP variants. Shown is a comparison of SAXS patterns recorded in the presence of Mg2+ for WT-NCP (top frame) and gH2AgH2B (bottom frame). Corresponding 1D spectra are shown in panel (E). To see this figure in color, go online.
Figure 5.
NCP samples with different variants of core histones studied in a range of concentrations of Mg2+ (left) and CoHex3+ (right) salts. Numbers show the change of the net NCP charge relative to the WT-NCP. Circles show the studied Mg2+ or CoHex3+ concentrations, and the concentration that resulted in the most-ordered NCP phase for a given sample is indicated by a symbol. The corresponding NCP phases are listed below the chart. To see this figure in color, go online.
All of our results are subject to the assumption that we were able to detect the most ordered state in the range of measured concentrations of counterions.
One noticeable feature of the SAXS spectra shown in Fig. 4 is that NCP samples with CoHex3+ generally display a columnar hexagonal order, except for the NCPs lacking two or more histone tails. Samples with the corresponding NCPs aggregated by Mg2+, in contrast, display columnar hexagonal phases, columnar disordered phases, and the isotropic NCP phase.
The H4-K16Q, H4-QuadQ, and H4-K16Ac NCPs show a degree of order and phase behavior rather similar to the WT-NCP in the presence of CoHex3+. The H2A-STT NCP, in contrast, shows a columnar hexagonal order only at 2 mM CoHex3+ and a disordered columnar phase for CoHex3+ concentrations in the 1.2–4 mM CoHex3+ range measured. The limited measurements obtained with H4-QuadAc show a columnar hexagonal phase in the presence of CoHex+3, and based on the similarity in the effects of K16Q and K16Ac modifications, we assume that the H4-QuadAc modification effect is similar to the H4-QuadQ one. The characteristic NCP stacking and intercolumnar distances in the columnar hexagonal phases of NCPs with modified histones are in the range of h = 54.6–55.9 Å and aH = 109.9–111.8 Å. Interestingly, no apparent dependence on the nature (acetylation versus K→Q mutation) and number (single H4-K16 or tetra H4-K5, K8, K12, or K16 change) of lysine modifications was observed.
With the more biologically relevant Mg2+, the columnar hexagonal phase was not achieved for all five mutated NCPs. The absence of hexagonal order appears to be cation dependent, because well-ordered columnar hexagonal phases were observed for the NCP variants with mutated or modified histones in the presence of CoHex+3 (Fig. 4 B). The NCP columnar phase was observed for all but H4-K16Q. The latter showed an isotropic NCP phase. The modifications in the H4 histone tails, as well as the STT mutation in the H2A histone, led to significantly reduced order compared with the WT-NCP in the presence of Mg2+ (Fig. 4 A). The investigated modifications in the presence of Mg2+ seemed to prevent the ordering of NCP columns.
We observed two specific effects of the mutations: 1) the H2A acidic patch neutralization greatly reduced the CoHex3+ range for columnar hexagonal phase formation, and 2) H4-K16Q prevented NCP stacking in the presence of Mg2+. The neutralization of three negative charges on the NCP stacking interface had a destabilizing effect on the columnar hexagonal phase in comparison with neutralization of one or four positive charges on H4 tail. The observed difference may be due to the nature or the location of the neutralized charges and is analogous to a similar disruption of the NCP-NCP contacts in folding of nucleosome arrays reported for the H2A.Bbd variant of the histone H2A with charge-quenching alteration of the acidic patch (73).
The removal of histone tails from one core histone shows that not all tails are equal (Fig. 4, C and D). The absence of H2B and H3 tails led to formation of the columnar NCP phase in the presence of Mg2+ and columnar hexagonal phases in the presence of CoHex3+ in a cation-dependent manner. For both Mg2+ and CoHex3+, removal of the H4 tails did not affect the formation of columnar hexagonal phases, which is similar to the behavior exhibited by the WT-NCP. Surprisingly, in the presence of either of the two cations (Mg2+ and CoHex3+), the NCP with truncation of N-terminal as well as C-terminal tails in the H2A histones produced x-ray diffraction spectra with a large number of peaks that were not previously observed for NCPs here or in the literature. The gH3 samples displayed spectra with peak patterns similar to those observed for gH2A with 2 mM CoHex3+. A possible interpretation of these spectra is that there is a mixture of more than one phase, including one of hexagonal order. The removal of tails has no effect on phase structure in the case of the histone H4, is cation dependent for the histones H3 and H2B, and leads to structural changes for the H2A histone.
NCP systems lacking tails in two (gH3gH4), three (gH2AgH3gH4, gH2AgH2BgH3), or all four histones (gNCP) did not form columnar hexagonal phases with Mg2+ and CoHex3+ (Fig. 4, E and F). The gNCP samples displayed peaks characteristic of NCP stacking at 80 and 100 mM Mg2+, and the isotropic NCP phase was observed in the 12–60 mM Mg2+ range. The NCP samples gH3gH4, gH2AgH2BgH3, and gH2AgH3gH4 resembled gNCP in the 10–30 mM Mg2+ range, forming an isotropic NCP phase. All NCP samples lacking more than one tail mentioned above formed isotropic NCP phases in the range of 1.5–5 mM CoHex3+ (gNCPs were measured in a broader range of 1.5–27 mM CoHex3+).
In the presence of Mg2+, the gH2AgH2B samples display unusual spectra with sharp peaks, indicating long-range order such as that found in a crystal (see the spectrum highlighted by red in Fig. 4 E). These are similar to the spectra of the gH2A sample, and the diffraction pattern reveals a crystal-like organization (Fig. 4 G, bottom spectrum in the red frame compared with the spectrum from the WT-NCP). It seems likely that these spectra are due to the formation of at least two phases, one of which could be the commonly observed columnar hexagonal structure (the expected hexagonal peaks can be tentatively assigned). It appears that the removal of the H2A and H2B tails resulted in the formation of highly ordered phases of yet unknown structure. The spectra of gH2AgH2B at 1.5 or 5 mM CoHex3+ are very similar to the gH2A spectra at 2 mM CoHex3+. The same set of peaks of higher intensity for the former is shifted to a higher q range, indicating tighter packing. Thus, we suggest that the same phase of unknown structure and long-range order is formed by NCPs lacking H2A tails and by NCPs lacking H2A and H2B tails in the presence of Mg2+ and CoHex3+. The formation of the columnar hexagonal phase by gH2AgH2B NCPs was also observed in the presence of CoHex3+. Currently, it is not possible to suggest what sort of structure is formed, and additional experiments are needed to gain insight into this effect.
In general, it was observed that the ordering of the NCP aggregates decreased as the number of tailless histones increased (with the notable exceptions of the gH2A and gH2AgH2B systems). The order decreased in both the NCP stacking and the hexagonal columnar association.
Two broad diffraction peaks with a 1:2 positional ratio observed in the spectra of gNCPs at 80 and 100 mM Mg2+ (Figs. 4 E and S22) indicate a possible NCP stacking of short-range order with a mean distance h = 59.4 Å (Table S2). This distance is significantly larger than that observed for the other NCP variants, suggesting very loosely stacked NCP aggregates. It may be noted that the electrophoretic mobility of the gNCP is slower than that of the WT and the mutated/acetylated NCPs (20), suggesting that the gNCPs may have an extended size, probably due to detachment of the entry/exit parts of the DNA from the histone core. A similar DNA detachment effect may apply to the gH2AgH3gH4 sample, where a weak peak corresponding to h = 57.1 Å was observed at 3 mM CoHex3+ (Fig. 4 F; Table S2). The absence of columnar ordering in these spectra may suggest a smectic NCP organization with layers of NCP that lack proper stacking and are laterally disordered due to the extended DNA.
Studies of NCPs concentrated by variation of the osmotic pressure in the presence of monovalent salt have identified the formation of bilayers of NCPs stacked in columns that are also visible in EM images and show a scattering peak q1L at low q corresponding to a distance of ∼350 Å (9, 10, 44). However, in our study, as well as in other studies of ordered phases in the presence of multivalent cations (13, 74), the bilayer phase was never observed. Such a loose bilayer arrangement was seen in NCP systems with monovalent salt and under osmotic pressure (44) (and references therein). In general, multication-induced condensation of DNA (31, 75) and nucleosome arrays (76) proceeds as a cooperative process, an all-or-nothing transition. It is reasonable to expect that NCP aggregation would follow the same process. Therefore, under the conditions applied here, one would expect the existence of either NCPs in solution or fully condensed phases of NCPs without bilayer intermediates.
Estimation of the size of the columnar hexagonal NCP domains
A lower boundary of the linear domain size s of the ordered aggregates can be estimated from the fitted full width at half-maximum (FWHM) of the first diffraction peak, namely, s = 2π/FWHM. In the columnar hexagonal NCP phase, the length across the columnar hexagonal domain s(aH) and the length of the NCP columns s(h) can be estimated using the FWHM of q1 and q1h peaks, respectively. The domain sizes of columnar hexagonal phases are listed in Table S3. The number of columns and stacked NCPs in the column may be calculated by dividing the domain size s(aH) and s(h) by aH and h, respectively. A WT-NCP columnar hexagonal phase made of 20–30 columns, each consisting of 40–50 NCPs as estimated from the SAXS data, agrees with previous observations made using cryo-EM (44). A reduction in domain size in comparison with the WT-NCP was observed for H4-QuadAc and gH2B with CoHex3+, whereas the opposite effect was observed for H4-K16Q with CoHex3+ and for gH4 with Mg2+. The general observation was a larger number of NCPs stacked in the column compared with the number of arranged columns.
Conclusions
In solutions with low (<50 mM) monovalent salt, interactions between NCPs are repulsive. The addition of millimolar concentrations of Mg2+ or CoHex3+ results in phase separation and the formation of NCP aggregates with different arrangements of particles, ranging from the isotropic NCP phase to NCP columnar and columnar hexagonal phases. The major contribution to free energy that drives phase separation comes from the entropic gain of the release of monovalent counterions (15).
The x-ray diffraction method enabled us to quantitatively assess the short- and long-range order in many NCP aggregates, and to identify and distinguish a number of known phases with NCP stacking and columnar arrangement order. These advantages compensate for the limitations of polycrystalline samples of higher disorder, incomplete sampling that assumes detection of the most ordered phases, and the inability to solve the structure with regard to the order of dyad symmetry and NCP chirality in general, and for some of the tailless NCPs in this work in particular.
Our results are in general agreement with earlier x-ray diffraction studies (9, 10, 44) in which a variety of NCP phases were observed. Compared with previous studies performed on inhomogeneous nucleosomes extracted from native chromatin (9, 10, 44), we observed more compact packing and faster equilibration of the highly homogeneous recombinant NCPs. The WT-NCP stacking distances in the range of h = 55–57 Å and intercolumnar distances of aH = 110–112 Å are shorter and more uniform than those observed for cell-extracted NCPs.
In the 3–80 mM Mg2+ range, the phase behavior of the WT-NCP aggregates is dependent on the cation concentration. With increasing Mg2+ concentration, the order changes from isotropic NCP to columnar disordered and finally columnar hexagonal. A further increase in Mg2+ concentration leads to the same set of phases in reverse order. Our observations suggest a model of columnar hexagonal phase formation in which column formation by NCP stacking is followed by intercolumnar interactions. For CoHex3+, the isotropic NCP to 3D to 2D columnar hexagonal phase transition in the narrow range of 1.2–2.5 mM CoHex3+ was observed. The columnar hexagonal phase persisted in the 2.5–30 mM CoHex3+ range. The long-range order of the NCP columnar phase disappeared or was reduced by acetylation of the H4 tails, acidic patch neutralization.
Single tail deletion effects result in diversity of the spectra. The H4 tail deletion results in spectra similar to those of WT-NCP for both Mg2+ and CoHex3+, with the formation of columnar hexagonal phases. Deletion of gH2A tails in the presence of Mg2+ or CoHex3+ produces spectra with multiple peaks, indicating highly ordered structures with peak positions that are not in agreement with the previously reported columnar hexagonal phase (9). Deletion of H2B and H3 tails results in the NCP columnar phase in the presence of Mg2+. In the presence of CoHex3+, multiple peaks at high q (>0.15 Å−1) indicate ordering in the gH2B spectrum, but the low intensity of q1 with the high-intensity q2 peak with a shoulder and the absence of regular NCP stacking peak make phase identification difficult. The gH3 spectrum in the presence of CoHex3+ is similar to that of gH2A. The electrostatic contribution of the tail deletion effect may be due to the fact that NCP columns lacking H2B and H3 tails are more electronegative than columns lacking H2A and H4 tails that form columnar hexagonal phases. Recent coarse-grained simulations from our laboratory, using an advanced coarse-grained NCP model that reproduces the detailed shape of the NCP core and geometry and the flexibility of the tails, confirmed that in phases of aggregated NCPs, the H2A and H4 tails stabilize the NCP-NCP stacking contact, whereas the H3 and H2B tails are responsible for screening and bridging interactions between the lateral surfaces of the contacting NCPs (17). The gH2A and gH3 samples are special cases because an unknown structure is observed. The tail deletion also changes the shape of the NCPs with a different and probably more dynamic DNA wrapping around the HO, besides changing the charge and charge distribution, thereby allowing the formation of new contacts. One particular effect of the tail removal is the unpeeling of the DNA ends, which makes the NCP resemble the chromatosome, a complex of NCP with the linker histone H1 (77).
In the case of four samples with more than one tail removed (namely, gH3gH4, gH2AgH3gH4, gH2AgH2BgH3, and gNCP), the aggregates produced featureless spectra, indicating the formation of an isotropic NCP phase with no long-range order in the presence of Mg2+ or CoHex3+. We conclude that the removal of the H3 tail in combination with any other tail is sufficient to prevent the formation of NCP columns, possibly by inhibiting NCP stacking. The H3 tail is the largest of the four tails, and its essential role in long-range NCP order is not surprising.
Our observation that the effects of the absence of H2A and H2B tails resulted in novel (to our knowledge), highly ordered structures in the NCP aggregates is intriguing. These effects may be biologically relevant, because H2A/H2B dimers are more dynamic than (H3/H4)2 tetramers, and the H2A histone family has the largest number of variants. The highly ordered phases observed in this work deserve to be characterized using high-resolution methods such as cryo-EM and crystallography. The role of individual tails may be addressed by NMR. In addition, studies of aggregates of NCP reconstituted with (H3/H4)2 tetramers only or with H2A histone variants may contribute to our understanding of the role of histone tails.
Finally, it must be strongly emphasized that the reconstitution of tailless constructs results in NCPs with different particle shapes, which likely is caused by a different, more dynamic DNA wrapping around the HO with unwrapping at the DNA ends, resulting in a more dynamic, heterogeneous system. For this reason, the effects observed for the tailless systems could be due both to the change in NCP particle structure and to the absence of the tails, which is expected to result in decreased inter-NCP interactions and diminished order. Therefore, the above conclusions and discussions regarding tailless systems should be considered tentative, and further characterization of these systems is warranted.
Author Contributions
N.K. and L.N. designed research. N.V.B., Y.L., C.-J.S., and N.K. performed research. A.A., C.-F.L., and R.Y. contributed analytical tools. N.V.B., Y.L., and N.K. analyzed data. N.V.B., N.K., and L.N. wrote the manuscript.
Acknowledgments
We thank Dr. Curt Davey and Prof. Timothy Richmond for gifts of the plasmids encoding DNA and core histones, and Prof. Martin Zacharias for sharing coordinates of the NCPs obtained in molecular dynamics simulations.
This work was supported by the Singapore Agency for Science Technology and Research (A∗STAR) through the Biomedical Research Council (grant 10/1/22/19/666) and by the Singapore Ministry of Education Academic Research Fund (AcRF) through a Tier 3 grant (MOE2012-T3-1-001).
Editor: Lois Pollack.
Footnotes
Nikolay V. Berezhnoy and Ying Liu contributed equally to this work.
Supporting Materials and Methods, Supporting Results, three tables, and 22 figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30067-4.
Supporting Material
References
- 1.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Davey C.A., Sargent D.F., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 3.Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 4.Hansen J.C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 5.Raspaud E., Chaperon I., Livolant F. Spermine-induced aggregation of DNA, nucleosome, and chromatin. Biophys. J. 1999;77:1547–1555. doi: 10.1016/S0006-3495(99)77002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Frutos M., Raspaud E., Livolant F. Aggregation of nucleosomes by divalent cations. Biophys. J. 2001;81:1127–1132. doi: 10.1016/S0006-3495(01)75769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangenot S., Leforestier A., Livolant F. Salt-induced conformation and interaction changes of nucleosome core particles. Biophys. J. 2002;82:345–356. doi: 10.1016/S0006-3495(02)75399-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangenot S., Raspaud E., Livolant F. Interactions between isolated nucleosome core particles. A tail bridging effect? Eur. Phys. J. E. 2002;7:221–231. [Google Scholar]
- 9.Mangenot S., Leforestier A., Livolant F. X-ray diffraction characterization of the dense phases formed by nucleosome core particles. Biophys. J. 2003;84:2570–2584. doi: 10.1016/S0006-3495(03)75062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangenot S., Leforestier A., Livolant F. Phase diagram of nucleosome core particles. J. Mol. Biol. 2003;333:907–916. doi: 10.1016/j.jmb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Bertin A., Leforestier A., Livolant F. Role of histone tails in the conformation and interactions of nucleosome core particles. Biochemistry. 2004;43:4773–4780. doi: 10.1021/bi036210g. [DOI] [PubMed] [Google Scholar]
- 12.Bertin A., Renouard M., Durand D. H3 and H4 histone tails play a central role in the interactions of recombinant NCPs. Biophys. J. 2007;92:2633–2645. doi: 10.1529/biophysj.106.093815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertin A., Mangenot S., Livolant F. Structure and phase diagram of nucleosome core particles aggregated by multivalent cations. Biophys. J. 2007;93:3652–3663. doi: 10.1529/biophysj.107.108365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertin A., Durand D., Mangenot S. H2A and H2B tails are essential to properly reconstitute nucleosome core particles. Eur. Biophys. J. 2007;36:1083–1094. doi: 10.1007/s00249-007-0212-9. [DOI] [PubMed] [Google Scholar]
- 15.Korolev N., Allahverdi A., Nordenskiöld L. The polyelectrolyte properties of chromatin. Soft Matter. 2012;8:9322–9333. [Google Scholar]
- 16.Luger K., Richmond T.J. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 17.Fan Y., Korolev N., Nordenskiöld L. An advanced coarse-grained nucleosome core particle model for computer simulations of nucleosome-nucleosome interactions under varying ionic conditions. PLoS One. 2013;8:e54228. doi: 10.1371/journal.pone.0054228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodcock C.L., Dimitrov S. Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev. 2001;11:130–135. doi: 10.1016/s0959-437x(00)00169-6. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe A.P., Hayes J.J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Lu C., Nordenskiöld L. Influence of histone tails and H4 tail acetylations on nucleosome-nucleosome interactions. J. Mol. Biol. 2011;414:749–764. doi: 10.1016/j.jmb.2011.10.031. [DOI] [PubMed] [Google Scholar]
- 21.Lewis P.N., Guillemette J.G., Chan S. Histone accessibility determined by lysine-specific acetylation in chicken erythrocyte nuclei. Eur. J. Biochem. 1988;172:135–145. doi: 10.1111/j.1432-1033.1988.tb13865.x. [DOI] [PubMed] [Google Scholar]
- 22.Perry M., Chalkley R. The effect of histone hyperacetylation on the nuclease sensitivity and the solubility of chromatin. J. Biol. Chem. 1981;256:3313–3318. [PubMed] [Google Scholar]
- 23.Perry M., Chalkley R. Histone acetylation increases the solubility of chromatin and occurs sequentially over most of the chromatin. A novel model for the biological role of histone acetylation. J. Biol. Chem. 1982;257:7336–7347. [PubMed] [Google Scholar]
- 24.Tse C., Sera T., Hansen J.C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horn P.J., Peterson C.L. Molecular biology. Chromatin higher order folding—wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 26.Wolffe A.P. Academic Press; San Diego, CA: 1998. Chromatin: Structure and Function. [Google Scholar]
- 27.Turner B.M. Histone acetylation and control of gene expression. J. Cell Sci. 1991;99:13–20. doi: 10.1242/jcs.99.1.13. [DOI] [PubMed] [Google Scholar]
- 28.Calestagne-Morelli A., Ausió J. Long-range histone acetylation: biological significance, structural implications, and mechanisms. Biochem. Cell Biol. 2006;84:518–527. doi: 10.1139/o06-067. [DOI] [PubMed] [Google Scholar]
- 29.Kurdistani S.K., Tavazoie S., Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Szerlong H.J., Prenni J.E., Hansen J.C. Activator-dependent p300 acetylation of chromatin in vitro: enhancement of transcription by disruption of repressive nucleosome-nucleosome interactions. J. Biol. Chem. 2010;285:31954–31964. doi: 10.1074/jbc.M110.148718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korolev N., Berezhnoy N.V., Nordenskiöld L. A universal description for the experimental behavior of salt-(in)dependent oligocation-induced DNA condensation. Nucleic Acids Res. 2012;40:2808–2821. doi: 10.1093/nar/gks214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloomfield V.A. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Finch J.T., Lutter L.C., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- 34.Finch J.T., Brown R.S., Klug A. X-ray diffraction study of a new crystal form of the nucleosome core showing higher resolution. J. Mol. Biol. 1981;145:757–769. doi: 10.1016/0022-2836(81)90313-2. [DOI] [PubMed] [Google Scholar]
- 35.Vasudevan D., Chua E.Y., Davey C.A. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 36.Leforestier A., Dubochet J., Livolant F. Bilayers of nucleosome core particles. Biophys. J. 2001;81:2414–2421. doi: 10.1016/S0006-3495(01)75888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leforestier A., Livolant F. Liquid crystalline ordering of nucleosome core particles under macromolecular crowding conditions: evidence for a discotic columnar hexagonal phase. Biophys. J. 1997;73:1771–1776. doi: 10.1016/S0006-3495(97)78207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schalch T., Duda S., Richmond T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 39.Dorigo B., Schalch T., Richmond T.J. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 40.Robinson P.J.J., Fairall L., Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl. Acad. Sci. USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song F., Chen P., Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 42.Scheffer M.P., Eltsov M., Frangakis A.S. Nucleosomes stacked with aligned dyad axes are found in native compact chromatin in vitro. J. Struct. Biol. 2012;178:207–214. doi: 10.1016/j.jsb.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 43.Castro-Hartmann P., Milla M., Daban J.-R. Irregular orientation of nucleosomes in the well-defined chromatin plates of metaphase chromosomes. Biochemistry. 2010;49:4043–4050. doi: 10.1021/bi100125f. [DOI] [PubMed] [Google Scholar]
- 44.Livolant F., Mangenot S., Durand D. Are liquid crystalline properties of nucleosomes involved in chromosome structure and dynamics? Philos. Trans. A Math. Phys. Eng. Sci. 2007;364:2615–2633. doi: 10.1098/rsta.2006.1843. [DOI] [PubMed] [Google Scholar]
- 45.Garcés R., Podgornik R., Lorman V. Antipolar and anticlinic mesophase order in chromatin induced by nucleosome polarity and chirality correlations. Phys. Rev. Lett. 2015;114:238102. doi: 10.1103/PhysRevLett.114.238102. [DOI] [PubMed] [Google Scholar]
- 46.Manna F.A., Lorman V.L., Žekš B. Polarity and chirality in NCP mesophases and chromatin fibers. Mol. Cryst. Liq. Cryst. 2007;478:83–97. [Google Scholar]
- 47.Lorman V., Podgornik R., Žekš B. Correlated and decorrelated positional and orientational order in the nucleosomal core particle mesophases. Europhys. Lett. 2005;69:1017–1023. [Google Scholar]
- 48.Howell S.C., Andresen K., Qiu X. Elucidating internucleosome interactions and the roles of histone tails. Biophys. J. 2013;105:194–199. doi: 10.1016/j.bpj.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luger K., Rechsteiner T.J., Richmond T.J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 50.Wang X., He C., Ausio J. Effects of histone acetylation on the solubility and folding of the chromatin fiber. J. Biol. Chem. 2001;276:12764–12768. doi: 10.1074/jbc.M100501200. [DOI] [PubMed] [Google Scholar]
- 51.Shogren-Knaak M., Ishii H., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 52.Kan P.-Y., Lu X., Hayes J.J. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol. Cell. Biol. 2007;27:2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Hayes J.J. Site-specific binding affinities within the H2B tail domain indicate specific effects of lysine acetylation. J. Biol. Chem. 2007;282:32867–32876. doi: 10.1074/jbc.M706035200. [DOI] [PubMed] [Google Scholar]
- 54.Routh A., Sandin S., Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson P.J.J., An W., Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J. Mol. Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X., Hayes J.J. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol. Cell. Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korolev N., Allahverdi A., Nordenskiöld L. Electrostatic origin of salt-induced nucleosome array compaction. Biophys. J. 2010;99:1896–1905. doi: 10.1016/j.bpj.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allahverdi A., Yang R., Nordenskiöld L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou B.R., Feng H., Bai Y. Histone H4 K16Q mutation, an acetylation mimic, causes structural disorder of its N-terminal basic patch in the nucleosome. J. Mol. Biol. 2012;421:30–37. doi: 10.1016/j.jmb.2012.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pollard K.J., Samuels M.L., Peterson C.L. Functional interaction between GCN5 and polyamines: a new role for core histone acetylation. EMBO J. 1999;18:5622–5633. doi: 10.1093/emboj/18.20.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dorigo B., Schalch T., Richmond T.J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 62.McBryant S.J., Klonoski J., Hansen J.C. Determinants of histone H4 N-terminal domain function during nucleosomal array oligomerization: roles of amino acid sequence, domain length, and charge density. J. Biol. Chem. 2009;284:16716–16722. doi: 10.1074/jbc.M109.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalashnikova A.A., Porter-Goff M.E., Hansen J.C. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lowary P.T., Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 65.Luger K., Rechsteiner T.J., Richmond T.J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 66.Dyer P.N., Edayathumangalam R.S., Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 67.Jeng U.-S., Su C.H., Liang K.S. A small/wide-angle X-ray scattering instrument for structural characterization of air–liquid interfaces, thin films and bulk specimens. J. Appl. Cryst. 2010;43:110–121. [Google Scholar]
- 68.Safinya C.R. Structures of lipid-DNA complexes: supramolecular assembly and gene delivery. Curr. Opin. Struct. Biol. 2001;11:440–448. doi: 10.1016/s0959-440x(00)00230-x. [DOI] [PubMed] [Google Scholar]
- 69.Roccatano D., Barthel A., Zacharias M. Structural flexibility of the nucleosome core particle at atomic resolution studied by molecular dynamics simulation. Biopolymers. 2007;85:407–421. doi: 10.1002/bip.20690. [DOI] [PubMed] [Google Scholar]
- 70.Korolev N., Vorontsova O.V., Nordenskiöld L. Physicochemical analysis of electrostatic foundation for DNA-protein interactions in chromatin transformations. Prog. Biophys. Mol. Biol. 2007;95:23–49. doi: 10.1016/j.pbiomolbio.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 71.Svergun D.I., Barberato C., Koch M.H.J. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 72.Schneidman-Duhovny D., Hammel M., Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou J., Fan J.Y., Tremethick D.J. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 74.Leforestier A., Fudaley S., Livolant F. Spermidine-induced aggregation of nucleosome core particles: evidence for multiple liquid crystalline phases. J. Mol. Biol. 1999;290:481–494. doi: 10.1006/jmbi.1999.2895. [DOI] [PubMed] [Google Scholar]
- 75.Matulis D., Rouzina I., Bloomfield V.A. Thermodynamics of DNA binding and condensation: isothermal titration calorimetry and electrostatic mechanism. J. Mol. Biol. 2000;296:1053–1063. doi: 10.1006/jmbi.1999.3470. [DOI] [PubMed] [Google Scholar]
- 76.Korolev N., Zhao Y., Nordenskiöld L. The effect of salt on oligocation-induced chromatin condensation. Biochem. Biophys. Res. Commun. 2012;418:205–210. doi: 10.1016/j.bbrc.2011.12.112. [DOI] [PubMed] [Google Scholar]
- 77.Zhou B.R., Jiang J., Bai Y. Structural mechanisms of nucleosome recognition by linker histones. Mol. Cell. 2015;59:628–638. doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.