Abstract
The phylum Ciliophora plays important roles in a wide range of biological studies. However, the evolutionary relationships of many groups remain unclear due to a lack of sufficient molecular data. In this study, molecular dataset was expanded with representatives from 55 orders and all major lineages. The main findings are: (1) 14 classes were recovered including one new class, Protocruziea n. cl.; (2) in addition to the two main branches, Postciliodesmatophora and Intramacronucleata, a third branch, the Mesodiniea, is identified as being basal to the other two subphyla; (3) the newly defined order Discocephalida is revealed to be a sister clade to the euplotids, strongly suggesting the separation of discocephalids from the hypotrichs; (4) the separation of mobilids from the peritrichs is not supported; (5) Loxocephalida is basal to the main scuticociliate assemblage, whereas the thigmotrichs are placed within the order Pleuronematida; (6) the monophyly of classes Phyllopharyngea, Karyorelictea, Armophorea, Prostomatea, Plagiopylea, Colpodea and Heterotrichea are confirmed; (7) ambiguous genera Askenasia, CyclotrichiumParaspathidium and Plagiocampa show close affiliation to the well known plagiopyleans; (8) validity of the subclass Rhynchostomatia is supported, and (9) the systematic positions of Halteriida and Linconophoria remain unresolved and are thus regarded as incertae sedis within Spirotrichea.
The ciliated protists are a large and diverse group of microbial eukaryotes that are of central importance in the functioning of microbial food webs by mediating the transfer of organic matter and energy between different trophic levels1,2. Due to their short lifespan and unique biological characters (e.g., nuclear dimorphism, chromosomal fragmentation), they have attracted the attention of researchers in a wide range of disciplines including cytology, evolutionary biology and genetics3. Despite this attention, however, no broadly accepted hypothesis exists for the phylogenetic relationships within the Ciliophora. For example, two widely used classification systems, i.e. those of Corliss4 based mainly on morphological characters, and Lynn5 inferred mainly from ultrastructural characters and small subunit ribosomal DNA (SSU rDNA) sequences, are broadly consistent at class-level but differ widely at order- or family- levels. In recent years, investigations based on gene trees have been increasingly used to reconstruct ciliate phylogeny and have helped to resolve a number of phylogenetic problems6,7,8,9,10,11. Unfortunately, most of these investigations are based on sequence data from a single gene, namely SSU rDNA, although a few multi-gene phylogenetic studies have been performed on far more limited numbers of species6,9,10,11. To date, no molecular phylogenetic analysis has focused on a full-scale classification of the Ciliophora at the level of order/family.
Comprehensive taxon sampling coupled with gene-rich analyses are critical for resolving accurate phylogenies12. However, sampling and identifying targeted ciliate species/groups is very difficult and time-consuming due to their microscopic size. According to Lynn5, there are about 300 families and 57 orders of ciliates. In the present study, phylogenetic analyses were carried out based on sequence data from up to four gene markers in a total of 152 species representing 110 families and 55 orders. The main aim of this investigation was to resolve phylogenetic relationships among the principal ciliate groups including all available orders (excluding Cariacotrichida for which the SSU rDNA sequence is short and in vivo characters have yet to be reported).
Results
Concatenated, SSU rDNA, and LSU rDNA trees are topologically similar, and most of their nodes are well supported (Figs 1 and 2). By contrast, ITS1-5.8S-ITS2 rDNA and alpha-tubulin genes produce poorly supported topologies in both deep and crown nodes (Additional file 2: Figs S1–S6), so we do not describe these two trees in detail.
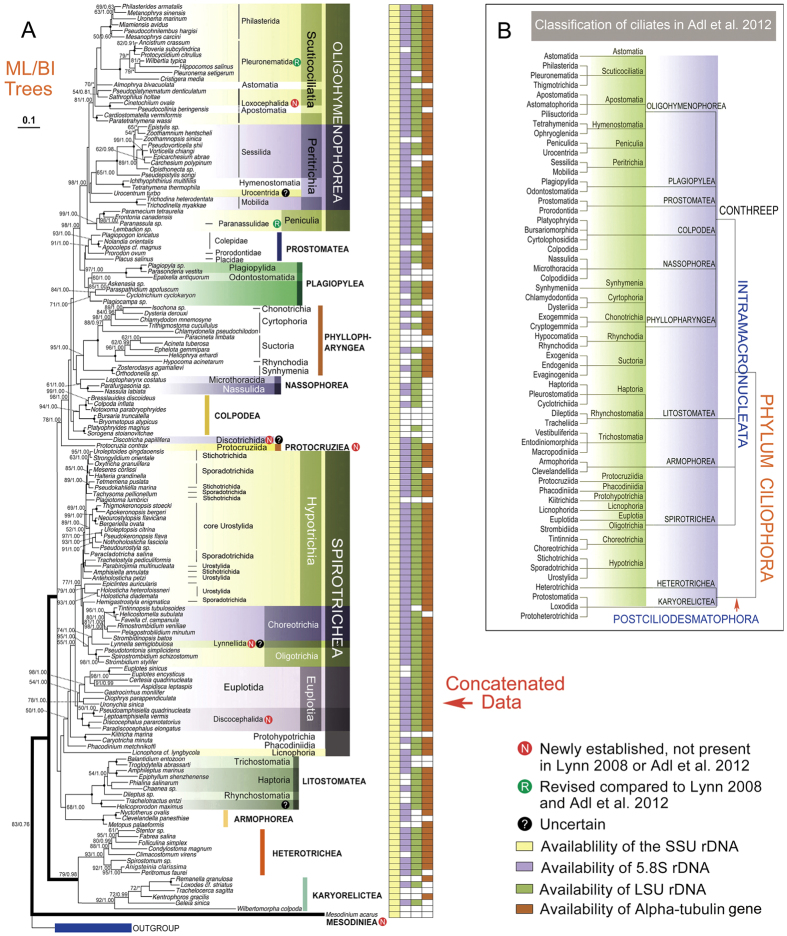
Figure 1.
Phylogenetic tree (A) and classification (B) of the phylum Ciliophora. (A) Maximum likelihood (ML) tree reconstructed using 152 ciliates and concatenated genes (the SSU rDNA sequence is available for all the taxa whereas the 5.8S rDNA, LSU rDNA and alpha-tubulin gene sequences are available for only a subset of these taxa, Additional file 1: Table S2). Numbers at nodes represent the bootstrap values of maximum likelihood (ML) out of 1000 replicates and the posterior probability of Bayesian analysis (BI). Only bootstraps above 50% are shown. Fully supported (100%/1.00) branches are marked with solid circles. Asterisk (*) indicates disagreement between ML and BI analyses. The three main branches of ciliates are in bold. The scale bar corresponds to 10 substitutions per 100 nucleotide positions. (B) Classification scheme of phylum Ciliophora according to Lynn5 and Adl et al.44.
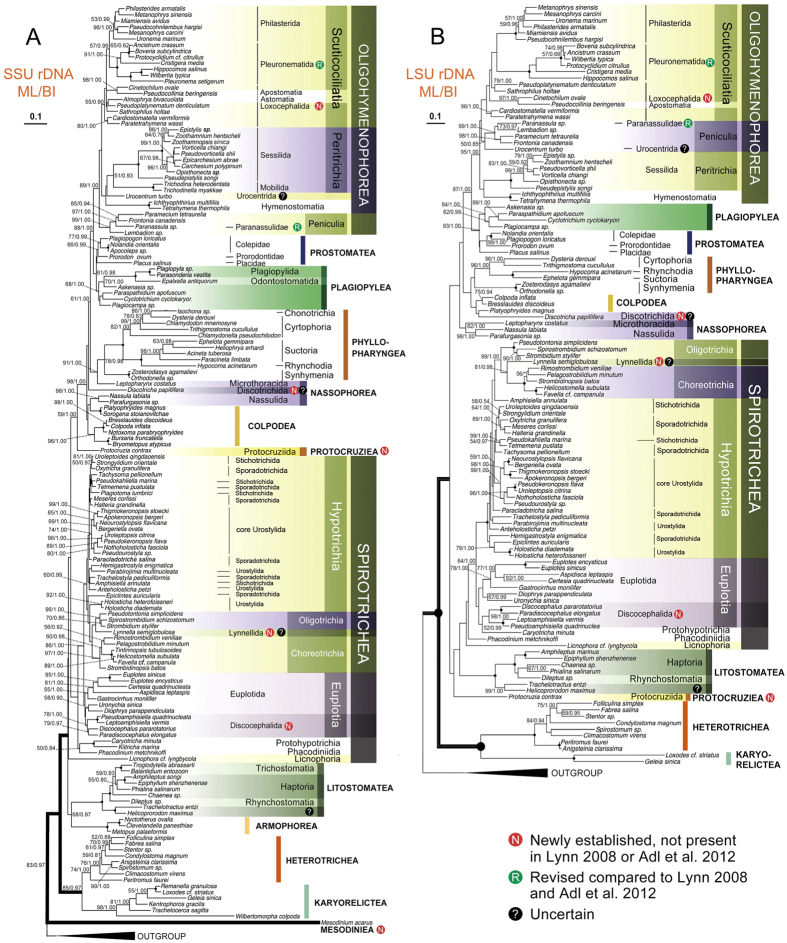
Figure 2.
Maximum likelihood (ML) trees of the phylum Ciliophora based on the SSU rDNA ((A), 152 taxa) and LSU rDNA ((B), 118 taxa). Numbers at nodes represent the bootstrap values of maximum likelihood (ML) out of 1000 replicates and the posterior probability of Bayesian analysis (BI). Only bootstraps above 50% are shown. Fully supported (100%/1.00) branches are marked with solid circles. Asterisk (*) indicates disagreement between ML and BI analyses. The three main branches of ciliates are in bold. The scale bar corresponds to 10 substitutions per 100 nucleotide positions.
Concatenated Tree
Both ML and BI analyses show a similar topology in the concatenated tree, that is, with three main groups are recognizable (Fig. 1A): (1) Mesodiniea, represented by the type genus Mesodinium; (2) subphylum Postciliodesmatophora, which comprises Karyorelictea and Heterotrichea, and; (3) subphylum Intramacronucleata, which comprises all other classes. Within the Postciliodesmatophora, the classes Heterotrichea and Karyorelictea is each recovered as a monophyletic assemblage. Within the Intramacronucleata, two major superclades are recovered, one comprising the classes Colpodea + Oligohymenophorea + Nassophorea + Plagiopylea + Prostomatea + Phyllopharyngea (CONthreeP), and the other comprising the classes: Litostomatea + Armophorea + Spirotrichea (SAL) (Fig. 1A).
Within the class Phyllopharyngea, the subgroup Subkinetalia (subclasses Cyrtophoria, Chonotrichia, Rhynchodia, and Suctoria), and the subclass Synhymenia, are both monophyletic and strongly supported (Fig. 1A).
In the class Nassophorea, the order Microthoracida clusters with the order Nassulida in the concatenated tree (61%ML, 1.00BI) (Fig. 1A). Discotricha is consistently placed as an early branching lineage within the ConThreeP superclade with high support (98% ML, 1.00 BI).
Protocruziidia, represented by the well-known genus Protocruzia and traditionally classified as a member of Spirotrichea5, groups with CONthreeP. With Protocruzia excluded, the class Spirotrichea is monophyletic (Fig. 1A), albeit with low support (<50% ML, <0.5 BI). Licnophorida occupies the basal position within the spirotrichean clade. Each of the other major subgroups, i.e. Euplotia, Protohypotrichia, Phacodiniida, Oligotrichia and Choreotrichia, forms a separate clade within the spirotrichean assemblage. By contrast, the subclass Hypotrichia is non-monophyletic with Epiclintes, Holosticha and Hemigastrostyla grouping with the choreotrichs and oligotrichs. Furthermore, groupings within the three hypotrich orders (Sporadotrichida, Stichotrichida, and Urostylida) are ambiguous. For example, six species of the order Stichotrichida occupy five separated clades; seven species of the order Sporadotrichida occupy six different clades; and although eight out of 13 species of Urostylida form a moderately supported clade (89% ML, 1.00 BI), the other five species occupy three separate clades. Within the Euplotia, the order Euplotida is monophyletic whereas the order Discocephalida clusters with Pseudoamphisiella.
As expected, the subclasses Oligotrichia and Choreotrichia, are both monophyletic with high support (Fig. 1A). The newly proposed order Lynnellida clusters with Choreotrichia, forming a group that is sister to the subclass Oligotrichia. The subclass Choreotrichia is comprised of two groups, the tintinnids and the aloricate choreotrichians.
The class Litostomatea consists of three subclasses, Haptoria, Trichostomatia and Rhynchostomatia. Haptoria is paraphyletic in all the gene trees (Figs 1 and 2), with the Trichostomatia (represented by Balantidium and Troglodytella) nesting within it and Helicoprorodon clustering with Trachelotractus, forming an early-branching lineage of litostomateans.
The Oligohymenophorea forms a maximally supported clade and comprises six subclasses (Fig. 1A). Both the subclasses Apostomatia and Astomatia, each of which is represented by a single sequence, nest within the scuticociliate assemblage rendering the Scuticociliatia polyphyletic. The scuticociliate order Philasterida is monophyletic. By contrast, the traditional scuticociliate order Pleuronematida sensu Lynn 2008, represented by five genera and four families, is not monophyletic with the thigmotrichids consistently nesting within it (100% ML, 1.00 BI). The order Loxocephalida, represented by five genera and two families, is paraphyletic with Pseudoplatynematum and Sathrophilus clustering with the Philasterida/Pleuronematida + Astomatia assemblage, Cardiostomatella and Paratetrahymena forming a separate clade that is basal to rest of the scuticociliates (100% ML, 1.00 BI), and Cinetochilum clustering with the subclass Apostomatia (100% ML, 1.00 BI).
The subclass Peritrichia comprises two orders, Sessilida and Mobilida. Although each order is monophyletic, the Sessilida groups with the subclass Hymenostomatia and the family Urocentridae (subclass Peniculia), rather than with the Mobilida, rendering the Peritrichia non-monophyletic.
The subclass Peniculia is represented by five families and five genera. Four of these genera (Paramecium, Frontonia, Lambadion, and Paranassula) form a well-supported clade that is basal to the rest of the Oligohymenophorea (Fig. 1A). The fifth genus, Urocentrum (family Urocentridae), occupies a position between the hymenostomes and the peritrich order Mobilida.
A close relationship between the classes Prostomatea and Plagiopylea is well supported (97% ML, 1.00 BI). The Prostomatea-Plagiopylea group (hereafter referred to as the PP clade) is sister to the class Oligohymenophorea with high support (98% ML, 1.00 BI), forming a three-class metaclade (Fig. 1A).
The class Prostomatea is represented by one order, namely Prorodontida, which is paraphyletic. The monophyletic family Colepidae (represented by Apocoleps, Nolandia and Plagiopogon) is closely related to Prorodontidae (represented by Prorodon) with variable support (91%ML, 1.00BI). The family Placidae (represented by Placus) is basal to the Colepidae-Prorodontidae. The prostome family Plagiocampidae (represented by Plagiocampa), however, clusters with two genera of uncertain taxonomic affiliation, Cyclotrichium and Paraspathidium (see below).
The class Plagiopylea is formed by the well-known plagiopyleans and four ambiguous genera, Plagiocampa, Paraspathidium, Cyclotrichium and Askenasia. The former three form a moderately supported sister group (71%ML, 1.00BI) to Askenasia and the well-known plagiopyleans (85% ML, 1.00 BI).
Topological Difference between SSU rDNA and Concatenated Trees
Within the class Spirotrichea, the genus Hemigastrostyla is not sister to oligotrichs in the SSU rDNA tree (Fig. 2A), whereas this is the case in the concatenated tree (Fig. 1A), and the euplotid family Uronychiidae clusters with Discocephalida rather than Euplotida (Fig. 2A).
Within the class Oligohymenophorea, Cristigera groups with other cyclidiids and thigmotrichids with moderate to high support in the SSU rDNA tree (91% ML, 1.00 BI), rather than branching before all the other pleuronematids and thigmotrichids. Furthermore, in contrast to the concatenated tree, the monophyly of the subclass Peritrichia is supported in the SSU rDNA tree, with the sessilids and mobilids clustering together.
The subclass Licnophoria, represented by Licnophora, branches relatively early in the SSU rDNA tree and is basal to all groups except the assemblages Litostomatea-Armophorea and the Heterotrichea-Karyorelictea (Fig. 2A) vs. sister to Spirotrichea in the concatenated tree (Fig. 1A).
The two orders, Microthoracida and Nassulida, which are believed to be the members of the class Nassophorea, are placed in separate clades: the former branches in a position that is sister to the class Phyllopharyngea whereas the latter clusters with the colpodean genera Platyophrides and Sorogena (Fig. 2A) vs. form one clade in the concatenated tree (Fig. 1A).
Topological Difference between LSU rDNA and Concatenated Trees
Various taxa are located in different positions in the LSU rDNA tree compared to the concatenated tree (Figs 1A and 2B). These include: Protocruzia, which branches within the SAL clade (vs. with the CONthreeP clade); the genera Amphisiella, Trachelostyla, Parabirojimia and Anteholosticha, all of which are located outside (vs. within) the main hypotrich clade; Caryotricha, which is nested within the Discocephalida (vs. basal to Spirotrichea); the order Lynnellida, which is more closely related to the subclass Oligotrichia than to the subclass Choreotrichia; Wilbertia, which clusters with the thigmotrichids (vs. with the pleuronematid Hippocomos); and Urocentrum, which clusters with the peniculids (vs. with the peritrichs and hymenostomes). Furthermore, in the SSU rDNA tree the basal clade within the class Oligohymenophorea is that comprising Peritrichia and Hymenostomatia whereas in the concatenated tree the Peniculia is basal.
Discussion
Relationships and Arrangement within the Phylum Ciliophora
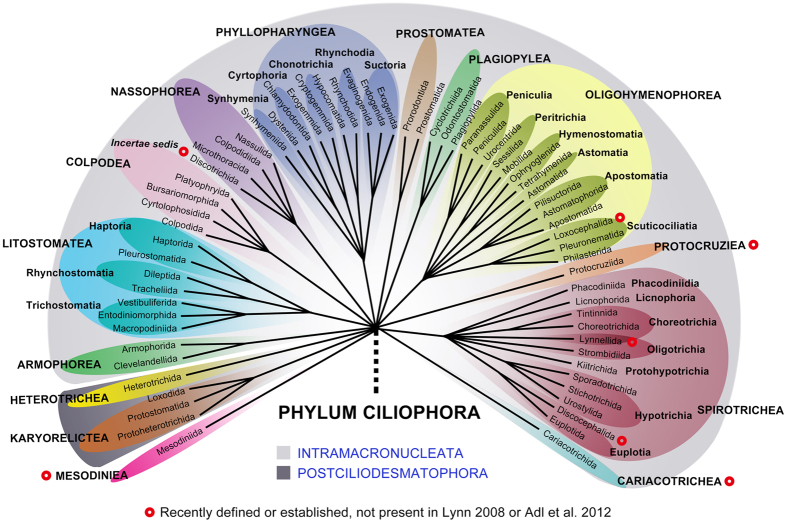
It is widely accepted that the phylum Ciliophora comprises two main groups, the subphyla Intramacronucleata and Postciliodesmatophora (Figs 3 and 4), although relationships among the constituent classes are generally not well resolved due to low support values in gene trees11,13,14,15. In the concatenated and LSU rDNA trees (Figs 1A and 2B), two main groups could be recognized within the subphylum Intramacronucleata: one group has six classes, namely Colpodea, Oligohymenophorea, Nassophorea, Phyllopharyngea, Plagiopylea and Prostomatea (CONthreeP); the other main group comprises three morphologically diverse classes, Spirotrichea, Armophorea and Litostomatea (SAL). It is noteworthy, however, that whereas both Intramacronucleata and Postciliodesmatophora could be defined by morphological differences5, there are no strong morphological synapomorphies for either CONthreeP or SAL. Nevertheless, the clade comprising Armophorea and Litostomatea is supported by some morphological and morphogenetic synapomorphies, which unite them into a new infraphylum, Lamellicorticata16.
Figure 3. Systematic scheme for the phylum Ciliophora suggested by the present and previous works.
The order Discocephalida was established by Wicklow47, and revised to contain two suborders Discocephalina and Pseudoamphisiellina by Miao et al.45. The order Discotrichida was established in Fan et al.103. The order Loxocephalida was originally proposed by Jankowski18 and was confirmed by Li et al.66, Gao et al.60, Zhang et al.62, etc. The order Lynnellida was established by Liu et al.55. The class Cariacotrichea was established by Orsi et al.122. The order Mesodiniida was resurrected and the class Mesodiniea was established by Chen et al.17. The classes Protocruziea and Licnophoriea are defined in the present study.
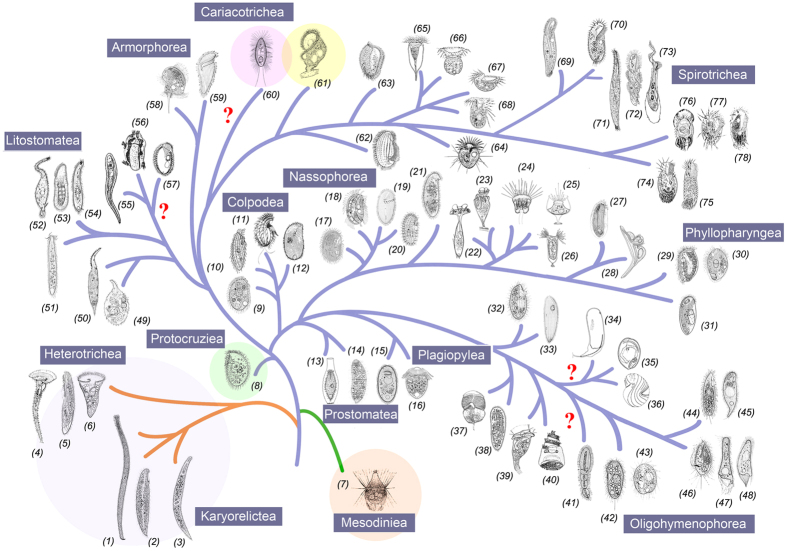
Figure 4. Hypothetical evolution of ciliated protozoa based on both morphological and molecular data to show the relationship and the positions of the taxa at order level.
(1–8), (14–17), (21), (24–26), (29–32), (36), (38), (39), (42–54), (58), (61), and (63–78) are from the present authors. (9) is from Bardele et al.123. (10–13), (19), (22–23), (27–28), (33–35), (37), (40–41), (55–56), (59), and (62) are from Corliss4. (18) is from Foissner et al.124, (20) is from Foissner et al.125. (57) is from Dehority126. (60) is from Orsi et al.122.
The subphylum Postciliodesmatophora comprises two classes, the Heterotrichea and Karyorelictea, both of which were recovered as monophyletic groups in our analyses. This is consistent both with previous phylogenetic analyses15,17 and with morphological data5.
The systematics of the family Mesodiniidae Jankowski in Small & Lynn, 1985, represented in the present study by the genus Mesodinium, has long been problematic. Traditionally it has been assigned to one of two orders within the subclass Haptoria, i.e. Haptorida4,5,18,19,20 or Cyclotrichida5. Puytorac19 established the order Mesodiniida for the family Mesodiniidae, however this taxon has not been widely accepted. Nevertheless, the systematic placement of the mesodiniids is ambiguous given that they invariably branch very deep in a range of gene trees and, based on their unique morphological features, they are usually completely isolated from other groups17,21,22. It has recently been suggested that the order Mesodiniida should be resurrected and that the new class Mesodiniea, be established for this taxon17. In the present study, Mesodiniea is consistently basal to all other ciliate lineages thus supporting its validity as a separate class and suggesting that it may even be separated at subphylum level (Figs 1A and 2A). However, the class Mesodiniea was not monophyletic in phylogenomic analyses based on 127 genes9. More data are therefore needed in order to validate this class.
Relationships within Heterotrichs and Traditionally Related Groups
Ciliates with a non-specialized somatic ciliature and an adoral zone of membranelles have long been classified as heterotrichs18,20. In recent decades, some “traditional” heterotrichs such as armorphoreans, Licnophora, Phacodinium, Protocruzia, and Plagiotoma have been excluded based on ultrastructural and/or molecular data23,24,25. As a result of these findings, Lynn5 assigned the “true” heterotrichs to the class Heterotrichea, a decision that is firmly supported by the current analyses.
According to Lynn5, Phacodiniidia Small and Lynn, 1985 and Protocruziidia Puytorac et al.25 are subclasses of the class Spirotrichea. Each contains only a single genus. Hitherto, the systematic positions of these two subclasses have never been satisfactorily resolved. Both are usually placed within the class Spirotrichea despite the fact that neither has a replication band during macronuclear DNA replication, an important apomorphy for the Spirotrichea5,20,26,27. In the case of Protocruziidia, other evidence casting doubt on its affiliation to the Spirotrichea includes its infraciliature (non-specialized somatic ciliature, non-differentiation of the ventral-dorsal sides, etc)27 and its process of ontogenesis which is a mixture of parakinetal and scuticokinetal modes28. Although some early molecular studies suggested that Protocruzia could be related to Spirotrichea24,29,30, this finding has been repeatedly rejected by recent studies which invariably conclude that it is not a member of the class Spirotrichea11,31. Li et al.31 suggested that Protocruzia represents a separate class, although they failed to define this taxon. Thus, the new class Protocruziea n. cl. is formally established here, based on the subclass Protocruziidia which contains a single order, Protocruziida Jankowski in Small & Lynn, 1985, a single family, Protocruziidae Jankowski, 1980, and a single genus, Protocruzia de Faria, da Cunha and Pinto, 1922, with the characters diagnosed for its subclass and the order5. The current analyses reveal that Protocruziea n. cl. belongs to neither of the main sub-groups of Intramacronucleata (CONthreeP or SAL) but instead occupies a position between the two as sister to the CONthreeP/Discotrichida assemblage (Figs 1A and 2A).
Protocruziea de Puytorac et al. 1987 n. cl
Diagnosis
Body small-sized, bilaterally flattened; somatic ciliature typically with dikinetids on both left and right sides; extrusomes present; adoral zone with several membranelles on left of dominant oral region; paroral membrane composed of dikinetids; stomatogenesis in mixokinetal mode; nuclear apparatus as a cluster of similar-sized nuclei with paradiploid macronuclei surrounding one or more micronuclei; free-swimming in marine and brackish water habitats; one order.
Type order
Protocruziida Jankowski, 1980 The genus Phacodinium is another highly questionable taxon regarding its phylogenetic position having long been regarded as a heterotrich, then as having affiliations to the hypotrichs and finally as being an intermediate form between the heterotrichs and hypotrichs32. In Lynn & Small’s system33, it was treated as a sedis mutabilis and assigned to the order Phacodiniida. The first molecular investigation based on SSU rDNA sequence data concluded that Phacodiniida is basal within Spirotrichea and suggested the establishment of a new subclass Phacodiniidia24. The present study supports this finding with Phacodiniidia occupying a basal position within the Spirotrichea.
Licnophora, for which the subclass Licnophoria Corliss, 1957 was established, is characterized by its highly specialized ciliature, unique morphology (e.g. presence of the adhesive disc with concentric kinetal rings) and the unusual pattern in ontogenesis with the cell undergoing a highly modified form of homothetogenic fission, i.e., a type of parallel division producing two daughter cells that develop alongside each other, similar to that in peritrichs4,28. Lynn5 assigned Licnophoria to the class Spirotrichea, although this placement has been repeatedly contradicted by molecular studies which indicate that it does not consistently cluster with spirotricheans15,17,34. Regarding its unique morphology that is unlike that of any spirotrichean, Licnophoria may represent a separate class that is sister to Spirotrichea. However, as molecular data are available for only a few species and complete information on morphogenesis remain unclear, we treat Licnophoria as incertae sedis within the SAL group.
Phylogeny of Hypotrichia sensu str
The subclass Hypotrichia s.l. (formerly Stichotrichia) is one of the most morphologically diverse group of ciliates and has been subject to numerous phylogenetic analyses6,35,36,37,38. This has resulted in the development of at least ten systematic systems4,5,19,20,39. In the Lynn’s system5, three orders of hypotrichs were recognized, namely Stichotrichida, Urostylida and Sporadotrichida, based mainly on the ventral ciliary pattern and partly on morphogenetic features. Until now, however, classification within this subclass remained unclear6,35,36,37,38.
In common with previous investigations, the multi-gene analyses reported here failed to separate the Stichotrichida and Sporadotrichida, the clustering patterns of these two orders being ambiguous due to low support values in all trees. This indicates that the pattern of the ventral ciliature, i.e. in specific, localized frontal and ventral groups in Sporadotrichida vs. in one or more linear longitudinal files in Stichotrichida, which is used as a main apomorphy at order level5, might be a result of convergent evolution. As in previous phylogenetic analyses6,35,36, the order Urostylida is non-monophyletic, consisting of monophyletic “core urostylids” and others (Figs 1 and 2). It is also noteworthy that classifications of hypotrichous orders based mainly on morphological characters are distinct from each other and none is completely consistent with trees based on molecular data4,5,39,40. Thus, it seems likely that some morphological characters regarded as apomorphies at order level might be plesiomorphies.
Systematic Position of the Subclass Protohypotrichia
The subclass Protohypotrichia, which contains a single order Kiitrichida, was established in 2009 based on both ontogenetic and molecular information41. Historically, the members of this group were believed to be a primordial assemblage within the Hypotrichia or ancestral forms of euplotids4,19,20,33. This hypothesis has received increasing support, both by the addition of more gene sequence data34,41 and by the recognition of various unique morphological and morphogenetic characters42,43. In the present study, the addition of newly sequenced LSU rDNA, 5.8S rDNA, and alpha-tubulin genes invariably resulted in the protohypotricheans clustering in a well-supported clade that is basal to the hypotrichs and euplotids (Figs 1A and 2). Thus all available evidence, molecular, morphological and ontogenetic, indicates that the Protohypotrichia should be recognized as a distinct group at subclass rank that is ancestral both to the hypotrichs and to the euplotids.
Systematics of the Subclass Euplotia and Related Groups
Based on previous studies and the results of the present work, we accept the system proposed by Adl et al.44 that the assemblage comprising the euplotids and discocephalids represents a distinct taxon at subclass level, namely Euplotia. Previously these two subgroups were treated as two orders (Euplotida and Discocephalida) in different subclasses33, or as suborders within the order Euplotida5. In the present study, the monophyly of Euplotida is recovered in trees inferred both from LSU rDNA gene sequence data alone (Fig. 2B) and from concatenated data of four genes (Fig. 1A), although the support values are not high. In the SSU rDNA tree the discocephalids nest within the Euplotida assemblage. The discocephalines are found only in marine biotopes and are characterized by their cephalized body shape. The group most closely related to Discocephalina is generally thought to be Pseudoamphisiellina, this conclusion being based both on morphological characters (e.g., cephalized body shape, highly developed fiber system connecting the cirri, generally two clearly separated ventral rows, and highly developed transverse cirri) and ontogenetic characters (e.g., the unique formation of the ventral rows during morphogenesis)45,46,47,48. This finding is supported by the present study, the sister relationship between these two groups being consistently recovered in all trees (Figs 1 and 2). We also propose to resurrect the order Discocephalida, originally established by Wicklow47, to contain two subgroups, namely Discocephalina and Pseudoamphisiellina45. Discocephalids have been assigned to a range of different groups4,19,25,33,47. Previous studies based both on ontogenetic46,47 and molecular data45,49 indicate that the discocephalids are clearly separated from the hypotrichs s. str., and probably represent an independent lineage at order level that is intermediate between the euplotids and other groups within the Spirotrichea. This hypothesis is firmly supported by the present study.
New Understanding of the Oligotrichs s.l
Traditionally, the loricate (tintinnids) and non-loricate (oligotrichids) oligotrichs were considered to be sister groups50. Only in last three decades have they been assigned to separate subclasses, the Choreotrichia and Oligotrichia, respectively. Choreotrichians are characterized by having a closed AZM and the group includes both tintinnids and some non-loricate forms, e.g. the strobilidids and related taxa. By contrast, oligotrichians have an open AZM5,33,44,51,52. The separation of these two subclasses is supported by the present study.
The family Lynnellidae was established by Liu et al.53 for the genus Lynnella, which shares some morphological features with both oligotrichians and choreotrichians. In a previous study based on analyses of all available data, i.e. morphological and molecular characters, Li et al.54 concluded that members of the Lynnellidae are intermediate forms between the Choreotrichia and Oligotrichia. Recently, Liu et al.55 established the order, Lynnellida for the Lynnellidae. The present study broadly supports these findings with Lynellida either sister to the oligotrichians (Fig. 2A) or sister to choreotrichians (Figs 1A and 2B).
The order Halteriida, represented by the genus Halteria, is a unique group that has long puzzled taxonomists4,5,19,33,44. Based on their morphology and pattern of morphogenesis, halteriids share similarities with the oligotrichs sensu lato but differ from the hypotrichs56,57. However, the SSU rDNA gene sequence data suggest that Halteriida might be a member of the hypotrichs/stichotrichs, possibly belonging to the oxytrichids, a highly specialized group of hypotrichs with very conservative modes of stomatogenesis24,58. Given this conflicting evidence, we believe that the Halteriida should be regarded as incertae sedis within Spirotrichea.
Phylogeny of Scuticociliates and Closely Related Taxa
Due to their small size and similar morphologies and ciliary patterns, scuticociliates are one of the most ambiguous groups of ciliates4,5,59,60,61,62. According to Lynn5, the subclass Scuticociliatia contains three orders: Philasterida, Pleuronematida, and Thigmotrichida. Although Philasterida is a well-outlined lineage, analyses of gene sequences data have challenged the monophyly of both Pleuronematida and Thigmotrichida63,64. Indeed as more data have accumulated, the thigmotrichids are often nested within the Pleuronematida, close to the cyclidiids60,65. This is broadly consistent with Puytorac19 who regarded the thigmotrichids as a suborder within the order Pleuronematida. A fourth order of scuticociliates, Loxocephalida, was originally proposed by Jankowski18 to contain certain Cinetochilum-like taxa which were previously assigned in the order Philasterida. Loxocephalida has been repeatedly recovered as a basal group to the core scuticociliates66. Present analyses indicate that Loxocephalida is a polyphyletic assemblage that is most closely related to Astomatia and Apostomatia, and clearly support its separation from Philasterida (Figs 1 and 2). Nevertheless, further studies are needed in order to clarify the systematics of the loxocephalids.
The Astomatia and Apostomatia are two specialized subclasses within the class Oligohymenophorea5,67,68. Astomes are endosymbionts typically found in the digestive tract of annelids, especially oligochaetes, and entirely lack an oral apparatus19. By contrast, apostomes are usually found as epibionts of marine and brackish water crustaceans, and have highly modified oral structures and polymorphic life cycles69. Analyses based on molecular data consistently reveal both groups to be closely related to the scuticociliates60,62. Present analyses show that either astomes or apostomes are more closely related to the loxocephalids than to the core scuticociliates, which is consistent with previous studies60,62,70. A reasonable hypothesis could be that they may be derived from loxocephalid-like ancestral lineages, their highly specialized morphologies being a result of adaptation to their symbiotic life styles60,62.
Phylogeny of Peniculia and Hymenostomatia
The peniculians are characterized by their three oral polykinetids aligned longitudinally in the oral cavit 5,19,33,71,72,73. The present phylogenetic analyses are consistent with previous studies in recovering the peniculians as a group that occupies a basal position within the olighymenophorean assemblage and is most closely related to the hymenostomatians and peritrichs74,75.
The hymenostomatians are characterized by having a well-defined buccal cavity with a paroral membrane, which may be unciliated and reduced, and typically three oral polykinetids5. In keeping with previous arrangements19,33, Lynn5 divided the subclass Hymenostomatia into two orders, Tetrahymenida and Ophryoglenida, represented by the well-known model organisms Tetrahymena and Ichthyophthirius, respectively. The findings of the present study are consistent with the above assignments and with previous studies76, supporting the monophyly of Hymenostomatia and its two orders.
Urocentrids have traditionally been regarded as a family within the subclass Peniculia4, although Puytorac et al.25 elevated them to the rank of order, which was accepted in later classification schemes5,19. However, both morphological (e.g. distinctive girdle of somatic cilia, somatic kinetids only as monokinetids with broad, tangential transverse ribbon; somatic extrusomes as mucocysts, etc.) and molecular evidence reveal that the urocentrids are divergent from all other typical peniculines and the position of the urocentrid assemblage is unstable in gene trees4,74,77. Based on the present findings and morphological data, classifying the urocentrids as an order is acceptable, however the phylogenetic position of the order Urocentrida is uncertain.
The Monophyly of the Traditional Peritrichs
Historically, peritrich ciliates were considered to be a well-defined group comprising two orders: Sessilida and Mobilida4,33. Recent molecular analyses, however, have challenged this arrangement suggesting that the molecular and morphological information are not always congruent78,79,80,81,82,83,84,85. Based on SSU rDNA sequence data, the monophyly of the Peritrichia s. l. was considered sufficiently doubtful for the recognition of the mobilids as a separate subclass from an entirely sessilid subclass Peritrichia s. str., a decision subsequently supported by alpha-tubulin gene trees86. It has recently been suggested that support for and against monophyly of the peritrichs s.l. depends on methods of alignment, and methods of masking ambiguously aligned nucleotide positions87. In the present study, with the inclusion of additional sequence data, the concatenated tree and single-gene trees result in conflicting findings concerning the relationship between the sessilids and mobilids. As shown in Figs 1 and 2, species of the two lineages were recovered as a monophyletic group in the SSU rDNA tree whereas they are separated in the concatenated tree, albeit with very low support values. However, sessilids and mobilids show a close relationship based on the morphological and the morphogenetic evidence which exclude them from all other non-peritrich groups5,28,86,88,89,90,91. Therefore, we do not believe there is sufficient evidence for the non-monophyly of the peritrichs s. l. or for the separation of the sessilids and mobilids at subclass level. Consequently we support the continued recognition of the subclass Peritrichia sensu Lynn 2008 and its two orders, Sessilida and Mobilida.
Phylogenetic Relationship within Litostomatea
The class Litostomatea has been traditionally rather poorly defined as having an apically positioned cytostome, uniform somatic ciliation and a non-distinct oral apparatus4,5,33,92. Lynn5 recognized two subclasses, Haptoria and Trichostomatia. A third subclass, Rhynchostomatia, was recently established by Vd’ačný et al.93. Recent molecular phylogenetic analyses, however, do not provide unambiguous support for the monophyly of Haptoria, with several of its members grouping with Trichostomatia, and the haptorid genera Helicoprorodon and/or Trachelotractus occupying a basal position within the class Litostomatea94. Our analyses with additional sequences also failed to recover the subclass Haptoria as a monophyletic group. In addition, for the first time, we reveal the close phylogenetic relationship between Helicoprorodon and Trachelotractus, and confirm their basal position within the Litostomatea. Our findings support a previous suggestion based on morphological features that the helicoprorodonids represent an independent group, possibly at the rank of order within the subclass Haptoria or even as a subclass within the class Litostomatea95,96,97. Helicoprorodon and Trachelotractus differ from other haptorids in having a peribuccal ridge with extrusomes, and specialized ciliary rows curving around the pharyngeal opening98.
Further Insights into Phyllopharyngea and Nassophorea
Members of classes Phyllopharyngean and Nassophorean have a basket-like, ventrally opening oral apparatus or cyrtos99 as result of which it has long been assumed that these two groups are closely related5,33. The class Nassophorea sensu Lynn, 2008 comprises three orders, Synhymeniida, Nassulida and Microthoracida5. However, SSU rDNA-based phylogenies have shown that the order Synhymeniida clusters strongly with the class Phyllopharyngea rather than with the other two orders of Nassophorea. Based on these findings, Gong et al.100 revised the higher classification of these groups, regarding the synhymeniids as a subclass of the class Phyllopharyngea. In the present study, trees based on multi-gene data (Fig. 1A) also recover the synhymeniids as a distinct lineage within the Phyllopharyngea.
The group-name “Subkinetalia” was coined for phyllopharyngean super-clade comprising the subclasses Cyrtophoria, Chonotrichia, Rhynchodia and Suctoria, the synapomorphy of which is the possession of subkinetal microtubules100. The multi-gene based phylogeny in the present study is consistent with this finding100,101,102 and reveals a highly supported monophyletic Phyllopharyngea comprising two subgroups, the Subkinetalia (cyrtophorians, rhynchodians, chonotrichians, suctorians) and Synhymenia. Since the Subkinetalia represents a taxon between the ranks of subclass and class we believe it is more biological meaningful to refer to this as a superclade sharing same synapomorphic character.
Discotrichids are traditionally classified as a family within the nassophorean order Microthoracida5. However, Fan et al.103 established the order Discotrichida since discotrichids do not group with other microthoracids and are even distinct from all other nassophorean lineages. The present study also recovers the discotrichids (represented by Discotricha) as a distinct lineage that occupies a basal position within CONthreeP. However, evolutionary relationships between the discotrichids and other nassophoreans (microthoracids and nassulids) remain uncertain, probably due to undersampling within these groups.
The genus Paranassula in the nassophorean order Nassulida possesses distinct morphological characters that separates it from other nassulids, e.g. two polykinetids that are restricted to a shallow oral cavity and the presence of a paroral kinety92. Furthermore, phylogenetic analyses based on SSU rDNA and LSU rDNA sequence data suggest that Paranassula is related to Peniculia (class Oligohymeophorea)102. Our analyses with newly added gene sequence data support the most recent assignment for this taxon, i.e. Paranassula should be assigned to subclass Peniculia, class Oligohymenophorea. However, in contrast to the conclusion of Zhang et al.102 which suggested a resurrection of order Paranassulida Deroux in de Puytorac et al. (1993), we propose that Paranassula should be assigned to the order Peniculida, as a member of the family Paranassulidae Fauré-Fremiet, 1962. The phylogenetic positions of other genera in the family Paranassulidae, e.g. Enneameron, Gullmarella, remain uncertain due to undersampling.
Systematically Ambiguous Taxa Find Their Close Relatives in the Class Plagiopylea
The systematic positions of the litostomatean genera Askenasia, Cyclotrichium, Paraspathidium and the prostome genus Plagiocampa have long been ambiguous. The former three have been variously assigned to the Cyclotrichida and Haptorida, or even as incertae sedis in the phylum Ciliophora5,93,95. Jankowski104 established the family Cyclotrichiidae and the order Cyclotrichida for Cyclotrichium which he assigned to the class Litostomatea, but failed to define these taxa. In the absence of any molecular data, Lynn5,33 regarded Cyclotrichiidae as a junior synonym of Didiniidae within the order Haptorida while retaining the order Cyclotrichida to include Mesodiniidae, e.g. Askenasia, Mesodinium and Myrionecta. Recent studies based on rDNA and the alpha-tubulin protein gene sequence data suggested that the two cyclotrichid genera Askenasia, Cyclotrichium, and the prostome genus Paraspathidium, should be removed from the class Litostomatea but failed to give their exact positions within the classes Plagiopylea and Prostomatea, respectively94,102. Lynn5 assigned the prostome genus Plagiocampa to the order Prorodontida. However, recent phylogenetic analyses recognized a distinct subclade comprising Plagiocampidae and two closely related genera Urotricha and Cryptocaryon7,102, which have a closer relationship to Cyclotrichium and Paraspathidium than to the prostomes102.
In the present study, based on multi-gene analyses, the concatenated gene trees robustly show that these four genera with uncertain taxonomic affiliations, i.e., Askenasia, Cyclotrichium, Paraspathidium and Plagiocampa, are most closely related to the plagiopyleans. Because of the highly specialized infraciliature and anaerobic life style of the plagiopyleans, their morphological synapomorphies are not well understood5. It is possible, for example, that a combination of the following features could define this clade (class Plagiopylea + Askenasia + Paraspathidium-Cyclotrichium + various prostomatean morphospecies represented by Plagiocampa): somatic monokinetids and an oralized somatic ciliature around a dominant cytostome consisting of densely ciliated dikinetids105,106,107. In addition, a brosse structure composed of dikinetids on the border of the oral slit has been commonly observed in Paraspathidium, Urotricha and Plagiocampa106,107. Based on the information above, we tentatively place Askenasia Paraspathidium, Cyclotrichium and various prostomatean morphospecies represented by Plagiocampa in the class Plagiopylea, pending further evidence including increased taxon sampling, ultrastructural studies and phylogenomic analyses.
A Highly Supported Metaclade of Oligohymenophorea, Prostomatea and Plagiopylea within the CONthreeP
Several studies based on SSU rDNA sequence data have revealed a close evolutionary relationship among the classes Oligohymenophorea, Prostomatea and Plagiopylea5,7,108. Following the inclusion of sequences of additional genes and increased taxon sampling, the metaclade containing Oligohymenophorea, Prostomatea and Plagiopylea was recovered with high support. A close relationship between prostomes and oligohymenophoreans has been suggested based on similarities of their patterns of morphogenesis revealed by electron microscopy102,109,110,111. However, the phenotypic features that unite the riboclass Plagiopylea with Oligohymenophorea and Prostomatea remain unknown.
Material and Methods
DNA Extraction, Gene Sequencing, Dataset Assembly and Alignments
Gene sequence data were obtained for a total of 104 species representing almost all the main ciliate lineages. Genomic DNA extraction, PCR amplifications and sequencing were performed as described in previous studies59,112 for the following genes: completed sequence (~1800 bp) of the SSU rDNA; a partial sequence (~500 bp) of the ITS1-5.8S-ITS2; a partial sequence (~1800 bp) of the LSU rDNA; and, a partial sequence (~1000 bp) of the alpha-tubulin gene.
In total, 232 sequences were submitted to the GenBank database (Additional file 1: Table S1). In order to maximize the taxonomic diversity of ciliates included in our analyses, newly characterized sequences were combined with relevant sequences obtained from GenBank (Additional file 1: Table S2). Six datasets were evaluated: (1) concatenation of the aligned SSU rDNA, 5.8S DNA, LSU rDNA and alpha-tubulin amino acid sequences from datasets 2–5; (2) SSU rDNA sequences including all 152 group representatives; (3) 5.8S rDNA sequences of 113 taxa; (4) LSU rDNA sequences of 118 taxa; (5) alpha-tubulin amino acid sequences of 116 taxa; (6) alpha-tubulin nucleotide sequences, including the first two codon positions, of 116 taxa. Orthologs of alpha-tubulin for concatenations were selected according to Gao and Katz15.
Sequences were aligned using the GUIDANCE algorithm with default parameters in GUIDANCE web server113. Regions that could not be unambiguously aligned were excluded from the phylogenetic analyses. Because the ITS regions are too divergent to be aligned, only the 5.8S rDNA of the ITS1-5.8S-ITS2 region was used. The lengths of the final alignments of datasets (1)-(6) were 3794, 1661, 164, 1612, 357, 714 positions, respectively.
Phylogenetic Analyses
Three apicomplexans and three dinoflagellates were used as outgroups (Additional file 1: Table S2)44. Maximum likelihood (ML) analyses were carried out using RAxML-HPC2 v7.6.6114 on CIPRES Science Gateway115. The DNA partition was analyzed with GTR + gamma. ProtTest 2.4116 selected the MtArt + I + G + F amino acid replacement matrix as the best-fitting model for alpha-tubulin amino acid sequences. The alpha-tubulin amino acid partition was run under the MtArt + gamma model as this was the best-fitting model available in RAxML. Support for the best-scoring ML tree came from 1000 bootstrap replicates. Bayesian inference (BI) analysis was performed with MrBayes 3.2.2117 on CIPRES Science Gateway using the GTR + I + G model for the DNA partition as selected by MrModeltest v.2.2118 and using mixed model for the alpha-tubulin amino acid partition. Markov chain Monte Carlo (MCMC) simulations were run with two sets of four chains for 4,000,000 generations with a sample frequency of 100 generations. The first 10,000 trees were discarded as burn-in. All remaining trees were used to calculate posterior probabilities using a majority rule consensus. Systematic schemes are mainly based on Lynn5 and Adl et al.44, except for some revisions made in the present study.
The approximately unbiased (AU) test119 was used to test the monophyly of the focal group against competing phylogenetic hypotheses (Table 1). Constrained ML trees enforcing the monophyly of the respective focal groups were generated based on SSU rDNA data. For all constraints, internal relationships within the constrained groups and among the remaining taxa were unspecified. The site-wise likelihoods for the resulting constrained topologies and the non-constrained ML topology were calculated using PAUP120 and were then analyzed in CONSEL121 with standard parameters to obtain p-values.
Table 1. Approximately Unbiased test (AU) results based on SSU rDNA data.
| Topology constraints | AU test |
|---|---|
| Climacostomidae | 0.011 |
| Urostylida + Pseudoamphisiellidae | 0.007 |
| Oligotrichia + Lynnella | 0.682 |
| Choreotrichia + Lynnella | 0.397 |
| Scuticociliatia | 0.065 |
| Philasterida + loxocephalids | 0.028 |
| Loxocephalida | 0.021 |
| Pleuronematida | 7.00E-06 |
| Peniculia | 0.218 |
| Peritrichia | 0.657 |
| Colpodea | 0.365 |
| Nassophorea | 0.069 |
| Cyclotrichida | 0.088 |
| Plagiopylea | 0.695 |
| Prostomatea | 0.253 |
Notes: The topology constraints column refers to proposed taxonomic groups that were tested for monophyly through the approximately unbiased test (AU). Rejected monophyly (p < 0.05) is highlighted in gray.
Additional Information
How to cite this article: Gao, F. et al. The All-Data-Based Evolutionary Hypothesis of Ciliated Protists with a Revised Classification of the Phylum Ciliophora (Eukaryota, Alveolata). Sci. Rep. 6, 24874; doi: 10.1038/srep24874 (2016).
Supplementary Material
Acknowledgments
This work is supported by the Natural Science Foundation of China (grant numbers 31430077 to W. Song, and 31471973 to Z. Yi), the Applied Basic Research Plan of Qingdao (15-12-1-1-jch). International Exchanges Scheme supported by the Royal Society/Natural Science Foundation of China (31411130122), and the BBSRC China Partnering Award Scheme, provided support for training in data analyses. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Many thanks are due to Dr. Yan Zhao, Dr. Jiamei Li, Ms. Lijuan Huang, Pu Wang, and Mr. Weibo Zheng, graduate students at OUC or SCNU, for their help with gene sequencing.
Footnotes
Author Contributions W.S., F.G. and Z.Y. designed the study. F.G. and Z.Y. undertook experiments and performed all analysis; F.G., A.W., Q.Z., J.G., M.M., P.S., D.X., J.H., Z.Y. and W.S. drafted and wrote the manuscript. All authors contributed to and approved the final manuscript.
References
- Kathol M., Norf H., Arndt H. & Weitere M. Effects of temperature increase on the grazing of planktonic bacteria by biofilm-dwelling consumers. Aquat. Microb. Ecol. 55, 65–79 (2009). [Google Scholar]
- Xu H., Zhang W. & Jiang Y. Do early colonization patterns of periphytic ciliate fauna reveal environmental quality status in coastal waters? Environ. Sci. Pollut. Res. Int. 21, 7097–7112 (2014). [DOI] [PubMed] [Google Scholar]
- Hausmann K. & Bradbury P. Ciliates: cells as organisms. (Gustav Fischer Verlag, 1996). [Google Scholar]
- Corliss J. O. The ciliated protozoa: characterization, classification and guide to the literature. (Pergamon Press, 1979). [Google Scholar]
- Lynn D. H. The ciliated protozoa: characterization, classification and guide to the literature. 3rd edition edn, (Springer Verlag, 2008). [Google Scholar]
- Huang J., Chen Z., Song W. & Berger H. Three-gene based phylogeny of the Urostyloidea (Protista, Ciliophora, Hypotricha), with notes on classification of some core taxa. Mol. Phylogenet. Evol. 70, 337–347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Z., Dunthorn M., Song W. & Stoeck T. Increasing taxon sampling using both unidentified environmental sequences and identified cultures improves phylogenetic inference in the Prorodontida (Ciliophora, Prostomatea). Mol. Phylogenet. Evol. 57, 937–941 (2010). [DOI] [PubMed] [Google Scholar]
- Foissner W., Kusuoka Y. & Shimano S. Morphology and gene sequence of Levicoleps biwae n. gen., n. sp. (Ciliophora, Prostomatida), a proposed endemic from the ancient Lake Biwa, Japan. J. Eukaryot. Microbiol. 55, 185–200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. Phylogenomics of non-model ciliates based on transcriptomic analyses. Protein Cell 6, 373–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J. M. et al. Phylogenomic analyses reveal subclass Scuticociliatia as the sister group of subclass Hymenostomatia within class Oligohymenophorea. Mol. Phylogenet. Evol. 90, 104–111 (2015). [DOI] [PubMed] [Google Scholar]
- Gentekaki E. et al. Large-scale phylogenomic analysis reveals the phylogenetic position of the problematic taxon Protocruzia and unravels the deep phylogenetic affinities of the ciliate lineages. Mol. Phylogenet. Evol. 78, 36–42 (2014). [DOI] [PubMed] [Google Scholar]
- Parfrey L. W. et al. Evaluating support for the current classification of eukaryotic diversity. PloS Genet. 2, e220 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunthorn M., Hall M., Foissner W., Stoeck T. & Katz L. A. Broad taxon sampling of ciliates using mitochondrial small subunit ribosomal DNA. Acta Protozool. 53, 207–213 (2014). [Google Scholar]
- Dunthorn M. et al. Placing environmental next-generation sequencing amplicons from microbial eukaryotes into a phylogenetic context. Mol. Biol. Evol. 31, 993–1009 (2014). [DOI] [PubMed] [Google Scholar]
- Gao F. & Katz L. A. Phylogenomic analyses support the bifurcation of ciliates into two major clades that differ in properties of nuclear division. Mol. Phylogenet. Evol. 70, 240–243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vdacny P., Orsi W. & Foissner W. Molecular and morphological evidence for a sister group relationship of the classes Armophorea and Litostomatea (Ciliophora, Intramacronucleata, Lamellicorticata infraphyl. nov.), with an account on basal litostomateans. Eur. J. Protistol. 46, 298–309 (2010). [DOI] [PubMed] [Google Scholar]
- Chen X., Ma H., Al-Rasheid K. A. S. & Miao M. Molecular data suggests the ciliate Mesodinium (Protista: Ciliophora) might represent an undescribed taxon at class level. Zool. Syst. 40, 31–40 (2015). [Google Scholar]
- Jankowski A. W. Conspectus of a new system of the phylum Ciliophora. Trudy Zool. Inst., Leningr. 94, 103–121 (1980). [Google Scholar]
- Puytorac P. d. In Traité de zoologie, infusoires ciliés. Vol. 2 (ed de Puytorac P. ) 621–679 (Masson, 1994). [Google Scholar]
- Small E. B. & Lynn D. H. In An illustrated guide to the Protozoa (eds Lee J. J., Hutner S. H., & Bovee E. D. ) 393–575 (Soc. Protozool. Special Publ., Allen Press, 1985). [Google Scholar]
- Johnson M. D., Tengs T., Oldach D. W., Delwiche C. F. & Stoecker D. K. Highly divergent SSU rRNA genes found in the marine ciliates Myrionecta rubra and Mesodinium pulex. Protist 155, 347–359 (2004). [DOI] [PubMed] [Google Scholar]
- Strüder-Kypke M. C., Wright A. D., Foissner W., Chatzinotas A. & Lynn D. H. Molecular phylogeny of litostome ciliates (Ciliophora, Litostomatea) with emphasis on free-living haptorian genera. Protist 157, 261–278 (2006). [DOI] [PubMed] [Google Scholar]
- Affa’a F. M., Hickey D. A., Struder-Kypke M. & Lynn D. H. Phylogenetic position of species in the genera Anoplophrya, Plagiotoma, and Nyctotheroides (Phylum Ciliophora), endosymbiotic ciliates of annelids and anurans. J. Eukaryot. Microbiol. 51, 301–306 (2004). [DOI] [PubMed] [Google Scholar]
- Shin M. K. et al. Phylogenetic position of the ciliates Phacodinium (Order Phacodiniida) and Protocruzia (Subclass Protocruziidia) and systematics of the spirotrich ciliates examined by small subunit ribosomal RNA gene sequences. Eur. J. Protistol. 36, 293–302 (2000). [Google Scholar]
- Puytorac P., Grain J. & Mignot J. P. Prècis de protistologie. Boubèe èd. (1987). [Google Scholar]
- Grolière C. A., de Puytorac P. & Grain J. Observations de quelques espèces de cilies endocommensaux d’echinides du Golfe du Mexique et de la Mer des Antilles. Protistologica 16, 233–239 (1980). [Google Scholar]
- Song W. & Wilbert N. Morphological investigation on some free living ciliates (Protozoa, Ciliophora) from China sea with description of a new hypotrichous genus, Hemigastrostyla nov. gen. Arch. Protistenkd 148, 413–444 (1997). [Google Scholar]
- Foissner W. In Ciliates cell as organisms. (eds Hausmann K. & Bradbur P. C. ) (Gustav Fischer, 1996). [Google Scholar]
- Hammerschmidt B. et al. Insights into the evolution of nuclear dualism in the ciliates revealed by phylogenetic analysis of rRNA sequences. J. Eukaryot. Microbiol. 43, 225–230 (1996). [DOI] [PubMed] [Google Scholar]
- Lynn D. H. My journey in ciliate systematics. J. Eukaryot. Microbiol. 43, 253–260 (1996). [Google Scholar]
- Li L. et al. Protocruzia, a highly ambiguous ciliate (Protozoa; Ciliophora): very likely an ancestral form for Heterotrichea, Colpodea or Spirotrichea? With reevaluation of its evolutionary position based on multigene analyses. Sci. China Life Sci. 53, 131–138 (2010). [DOI] [PubMed] [Google Scholar]
- da Silva Neto I. D. Structural and ultrastructural observations of the ciliate Phacodinium metchnicoffi Certes, 1891 (Heterotrichea, Phacodiniida). Eur. J. Protistol. 29, 209–218 (1993). [DOI] [PubMed] [Google Scholar]
- Lynn D. H. & Small E. B. In An illustrated guide to the protozoa (eds Lee J. J., Leedale G. F. & Bradbury P. ) (Allen Press Inc., 2002). [Google Scholar]
- Miao M. et al. Caryotricha minuta (Xu et al., 2008) nov. comb., a unique marine ciliate (Protista, Ciliophora, Spirotrichea), with phylogenetic analysis of the ambiguous genus Caryotricha inferred from the small-subunit rRNA gene sequence. Int. J. Syst. Evol. Microbiol. 59, 430–438 (2009). [DOI] [PubMed] [Google Scholar]
- Schmidt S. L., Bernhard D., Schlegel M. & Foissner W. Phylogeny of the Stichotrichia (Ciliophora; Spirotrichea) reconstructed with nuclear small subunit rRNA gene sequences: discrepancies and accordances with morphological data. J. Eukaryot. Microbiol. 54, 201–209 (2007). [DOI] [PubMed] [Google Scholar]
- Yi Z. & Song W. Evolution of the order Urostylida (Protozoa, Ciliophora): new hypotheses based on multi-gene information and identification of localized incongruence. PloS one 6, e17471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt E. A. et al. Phylogenetic relationships among 28 spirotrichous ciliates documented by rDNA. Mol. Phylogenet. Evol. 29, 258–267 (2003). [DOI] [PubMed] [Google Scholar]
- Foissner W. et al. Reconciling classical and molecular phylogenies in the stichotrichines (Ciliophora, Spirotrichea), including new sequences from some rare species. Eur. J. Protistol. 4, 265–281 (2004). [Google Scholar]
- Berger H. Monograph of the Urostyloidea (Ciliophora, Hypotricha). Monographiae Biol. 85, 1–1304 (2006). [Google Scholar]
- Song W. et al. Progress in Protozoology. (Qingdao Ocean University Press, 1999). [Google Scholar]
- Li L. et al. Does Kiitricha (Protista, Ciliophora, Spirotrichea) belong to Euplotida or represent a primordial spirotrichous taxon? With suggestion to establish a new subclass Protohypotrichia. Int. J. Syst. Evol. Microbiol. 59, 439–446 (2009). [DOI] [PubMed] [Google Scholar]
- Jiang J. et al. Two new marine ciliates, Caryotricha rariseta n. sp. and Discocephalus pararotatorius n. sp. (Ciliophora, Spirotrichea), with phylogenetic analyses inferred from the small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 60, 388–398 (2013). [DOI] [PubMed] [Google Scholar]
- Shao C., Song W., Warren A. & Al-Rasheid K. A. Morphogenesis of Kiitricha marina Nozawa, 1941 (Ciliophora, Spirotrichea), a possible model for the ancestor of hypotrichs s. l. Eur. J. Protistol. 45, 292–304 (2009). [DOI] [PubMed] [Google Scholar]
- Adl S. M. et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M., Shao C., Chen X. & Song W. Evolution of discocephalid ciliates: molecular, morphological and ontogenetic data support a sister group of discocephalids and pseudoamphisiellids (Protozoa, Ciliophora) with establishment of a new suborder Pseudoamphisiellina subord. n. Sci. China Life Sci. 54, 634–641 (2011). [DOI] [PubMed] [Google Scholar]
- Shao C. et al. Systematic position of Discocephalus-like ciliates (Ciliophora: Spirotrichea) inferred from SSU rDNA and ontogenetic information. Int. J. Syst. Evol. Microbiol. 58, 2962–2972 (2008). [DOI] [PubMed] [Google Scholar]
- Wicklow B. J. The Discocephalina (n. subord.): ultrastructure, morphogenesis and evolutionary implications of a group of endemic marine interstital hypotrichs (Ciliophora, Protozoa). Protistologica 18, 299–330 (1982). [Google Scholar]
- Shao C. et al. Morphogenesis of the marine ciliate, Pseudoamphisiella alveolata (Kahl, 1932) Song & Warren, 2000 (Ciliophora, Stichotrichia, Urostylida) during binary fission. J. Eukaryot. Microbiol. 53, 388–396 (2006). [DOI] [PubMed] [Google Scholar]
- Yi Z. et al. A molecular phylogenetic investigation of Pseudoamphisiella and Parabirojimia (Protozoa, Ciliophora, Spirotrichea), two genera with ambiguous systematic positions. Eur. J. Protistol. 44, 45–53 (2008). [DOI] [PubMed] [Google Scholar]
- Agatha S. Global diversity of aloricate Oligotrichea (Protista, Ciliophora, Spirotricha) in marine and brackish sea water. PloS one 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Gong J., Lynn D., Lin X. F. & Song W. B. An updated phylogeny of oligotrich and choreotrich ciliates (Protozoa, Ciliophora, Spirotrichea) with representative taxa collected from Chinese coastal waters. Syst. Biodivers. 7, 235–242 (2009). [Google Scholar]
- Snoeyenbos-West O. L., Salcedo T., McManus G. B. & Katz L. A. Insights into the diversity of choreotrich and oligotrich ciliates (Class: Spirotrichea) based on genealogical analyses of multiple loci. Int. J. Syst. Evol. Microbiol. 52, 1901–1913 (2002). [DOI] [PubMed] [Google Scholar]
- Liu W., Yi Z., Lin X. & Al-Rasheid K. A. Morphologic and molecular data suggest that Lynnella semiglobulosa n. g., n. sp. represents a new family within the subclass Choreotrichia (Ciliophora, Spirotrichea). J. Eukaryot. Microbiol. 58, 43–49 (2011). [DOI] [PubMed] [Google Scholar]
- Li J., Liu W., Gao S., Warren A. & Song W. Multigene-based analyses of the phylogenetic evolution of oligotrich ciliates, with consideration of the internal transcribed spacer 2 secondary structure of three systematically ambiguous genera. Eukaryot. Cell 12, 430–437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. W. et al. Two new genera of planktonic ciliates and insights into the evolution of the family Strombidiidae (Protista, Ciliophora, Oligotrichia). PloS one 10, e0131726 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agatha S. A cladistic approach for the classification of oligotrichid ciliates (Ciliophora: Spirotricha). Acta Protozool. 43, 201–217 (2004). [PMC free article] [PubMed] [Google Scholar]
- Agatha S. & Foissner W. Conjugation in the spirotrich ciliate Halteria grandinella (Muller, 1773) Dujardin, 1841 (Protozoa, Ciliophora) and its phylogenetic implications. Eur. J. Protistol. 45, 51–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D. C. & Prescott D. M. Phylogenetic relationships among hypotrichous ciliates determined with the macronuclear gene encoding the large, catalytic subunit of DNA polymerase alpha. J. Mol. Evol. 45, 301–310 (1997). [DOI] [PubMed] [Google Scholar]
- Gao F., Katz L. A. & Song W. Insights into the phylogenetic and taxonomy of philasterid ciliates (Protozoa, Ciliophora, Scuticociliatia) based on analyses of multiple molecular markers. Mol. Phylogenet. Evol. 64, 308–317 (2012). [DOI] [PubMed] [Google Scholar]
- Gao F., Katz L. A. & Song W. Multigene-based analyses on evolutionary phylogeny of two controversial ciliate orders: Pleuronematida and Loxocephalida (Protista, Ciliophora, Oligohymenophorea). Mol. Phylogenet. Evol. 68, 55–63 (2013). [DOI] [PubMed] [Google Scholar]
- Lynn D. H. & Strüder-Kypke M. Scuticociliate endosymbionts of echinoids (phylum Echinodermata): phylogenetic relationships among species in the genera Entodiscus, Plagiopyliella, Thyrophylax, and Entorhipidium (phylum Ciliophora). J. Parasitol. 91, 1190–1199 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al. Molecular evolution of Cinetochilum and Sathrophilus (Protozoa, Ciliophora, Oligohymenophorea), two genera of ciliates with morphological affinities to scuticociliates. Zool. Scr. 40, 317–325 (2011). [Google Scholar]
- Miao M., Wang Y., Li L., Al-Rasheid K. A. S. & Song W. Molecular phylogeny of the scuticociliate Philaster (Protozoa, Ciliophora) based on SSU rRNA gene sequences information, with description of a new species P. apodigitiformis sp. n. Syst. Biodivers. 7, 381–388 (2009). [Google Scholar]
- Miao M., Wang Y., Song W., Clamp J. C. & Al-Rasheid K. A. S. Description of Eurystomatella sinica n. gen., n. sp., with establishment of a new family Eurystomatellidae n. fam. (Protista, Ciliophora, Scuticociliatia) and analyses of its phylogeny inferred from sequences of the small-subunit rRNA gene. Int. J. Syst. Evol. Microbiol. 60, 460–468 (2010). [DOI] [PubMed] [Google Scholar]
- Gao F., Gao S., Wang P., Katz L. A. & Song W. Phylogenetic analyses of cyclidiids (Protista, Ciliophora, Scuticociliatia) based on multiple genes suggest their close relationship with thigmotrichids. Mol. Phylogenet. Evol. 75, 219–226 (2014). [DOI] [PubMed] [Google Scholar]
- Li L. et al. Phylogenetic position of the marine ciliate, Cardiostomatella vermiforme (Kahl, 1928) Corliss, 1960 inferred from the complete SSrRNA gene sequence, with establishment of a new order Loxocephalida n. ord. (Ciliophora, Oligohymenophorea). Eur. J. Protistol. 42, 107–114 (2006). [DOI] [PubMed] [Google Scholar]
- Fokam Z., Ngassam P., Strüder-Kypke M. & Lynn D. Genetic diversity and phylogenetic position of the subclass Astomatia (Ciliophora) based on a sampling of six genera from West African oligochaetes (Glossoscolecidae, Megascolecidae), including description of the new genus Paraclausilocola n. gen. Eur. J. Protistol. 47, 161–171 (2011). [DOI] [PubMed] [Google Scholar]
- Grain J. & Puytorac P. d. Particularités ultrastructurales des cinétosomes et de leurs annexes fibrillaires chez certains ciliés astomes Hoplitophryidae. J. Microsc. 19, 231–246 (1974). [Google Scholar]
- Bradbury P. C., Song W. & Zhang L. Stomatogenesis during the formation of the tomite of Hyalophysa chattoni (Hymenostomatida: Ciliophora). Eur. J. Protistol. 33, 409–419 (1997). [Google Scholar]
- Zhang Q., Fan X., Clamp J. C., Al-Rasheid K. A. S. & Song W. Description of Paratetrahymena parawassi n. sp. using morphological and molecular evidence and a phylogenetic analysis of Paratetrahymena and other taxonomically ambiguous genera in the order Loxocephalida (Ciliophora, Oligohymenophorea). J. Eukaryot. Microbiol. 57, 483–493 (2010). [DOI] [PubMed] [Google Scholar]
- Fan X., Chen X., Song W., Al-Rasheid K. A. S. & Warren A. Two new marine Frontonia species, F. mengi spec. nov. and F. magna spec. nov. (Protozoa; Ciliophora), with notes on their phylogeny based on SSU rRNA gene sequence data. Int. J. Syst. Evol. Microbiol. 61, 1476–1486 (2011). [DOI] [PubMed] [Google Scholar]
- Fan X. et al. Morphology of three new marine Frontonia species (Ciliophora; Peniculida) with note on the phylogeny of this genus. Eur. J. Protistol. 49, 312–323 (2013). [DOI] [PubMed] [Google Scholar]
- Pan X. et al. Studies on three diverse Frontonia species (Ciliophora, Peniculida), with brief notes on 14 marine or brackish congeners. Acta Protozool. 52, 35–49 (2013). [Google Scholar]
- Strüder-Kypke M. C., Wright A. D., Fokin S. I. & Lynn D. H. Phylogenetic relationships of the Subclass Peniculia (Oligohymenophorea, Ciliophora) inferred from small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 47, 419–429 (2000). [DOI] [PubMed] [Google Scholar]
- Yi Z. et al. Phylogeny of six oligohymenophoreans (Protozoa, Ciliophora) inferred from small subunit rRNA gene sequences. Zool. Scr. 38, 323–331 (2009). [Google Scholar]
- Bourland W. A. & Struder-Kypke M. C. Agolohymena aspidocauda nov. gen., nov. spec., a histophagous freshwater tetrahymenid ciliate in the family Deltopylidae (Ciliophora, Hymenostomatia), from Idaho (northwest USA): morphology, ontogenesis and molecular phylogeny. Eur. J. Protistol. 46, 221–242 (2010). [DOI] [PubMed] [Google Scholar]
- Strüder-Kypke M. C. & Lynn D. H. Comparative analysis of the mitochondrial cytochrome c oxidase subunit I (COI) gene in ciliates (Alveolata, Ciliophora) and evaluation of its suitability as a biodiversity marker. Syst. Biodivers. 8, 131–148 (2010). [Google Scholar]
- Miao W., Yu Y. & Shen Y. Phylogenetic relationships of the subclass Peritrichia (Oligohymenophorea, Ciliophora) with emphasis on the genus Epistylis, inferred from small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 48, 583–587 (2001). [DOI] [PubMed] [Google Scholar]
- Miao W., Yu Y., Shen Y. & Zhang X. Intraspecific phylogeography of Carchesium polypinum (Peritrichia, Ciliophora) from China, inferred from 18S-ITS1-5.8S ribosomal DNA. Sci. China 47, 11–17 (2004). [DOI] [PubMed] [Google Scholar]
- Sun P., Clamp J., Xu D., Kusuoka Y. & Miao W. Vorticella Linnaeus, 1767 (Ciliophora, Oligohymenophora, Peritrichia) is a grade not a clade: redefinition of Vorticella and the families Vorticellidae and Astylozoidae using molecular characters derived from the gene coding for small subunit ribosomal RNA. Protist 163, 129–142 (2012). [DOI] [PubMed] [Google Scholar]
- Utz L. R. & Eizirik E. Molecular phylogenetics of subclass Peritrichia (Ciliophora: Oligohymenophorea) based on expanded analyses of 18S rRNA sequences. J. Eukaryot. Microbiol. 54, 303–305 (2007). [DOI] [PubMed] [Google Scholar]
- Williams D. & Clamp J. C. A molecular phylogenetic investigation of Opisthonecta and related genera (Ciliophora, Peritrichia, Sessilida). J. Eukaryot. Microbiol. 54, 317–323 (2007). [DOI] [PubMed] [Google Scholar]
- Sun P., Clamp J. C. & Xu D. P. Analysis of the secondary structure of ITS transcripts in peritrich ciliates (Ciliophora, Oligohymenophorea): Implications for structural evolution and phylogenetic reconstruction. Mol. Phylogenet. Evol. 56, 242–251 (2010). [DOI] [PubMed] [Google Scholar]
- Sun P., Clamp J. C., Xu D. P., Kusuoka Y. & Hori M. Molecular phylogeny of the family Vorticellidae (Ciliophora, Peritrichia) using combined datasets with a special emphasis on the three morphologically similar genera Carchesium, Epicarchesium and Apocarchesium. Int. J. Syst. Evol. Microbiol. 61, 1001–1010 (2011). [DOI] [PubMed] [Google Scholar]
- Sun P. et al. An ITS-based phylogenetic framework for the genus Vorticella: finding the molecular and morphological gaps in a taxonomically difficult group. Proc. R. Soc. B 280(1771), 20131177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y. et al. Alpha-tubulin and small subunit rRNA phylogenies of peritrichs are congruent and do not support the clustering of mobilids and sessilids (Ciliophora, Oligohymenophorea). J. Eukaryot. Microbiol. 57, 265–272 (2010). [DOI] [PubMed] [Google Scholar]
- Zhan Z., Xu K. & Dunthorn M. Evaluating molecular support for and against the monophyly of the Peritrichia and phylogenetic relationships within the Mobilida (Ciliophora, Oligohymenophorea). Zool. Scr. 42, 213–226 (2013). [Google Scholar]
- Gong Y., Yu Y., Villalobo E., Zhu F. & Miao W. Reevaluation of the phylogenetic relationship between mobilid and sessilid peritrichs (Ciliophora, Oligohymenophorea) based on small subunit rRNA genes sequences. J. Eukaryot. Microbiol. 53, 397–403 (2006). [DOI] [PubMed] [Google Scholar]
- Zhan Z., Xu K., Warren A. & Gong Y. Reconsideration of phylogenetic relationships of the subclass Peritrichia (Ciliophora, Oligohymenophorea) based on small subunit ribosomal RNA gene sequences, with the establishment of a new subclass Mobilia Kahl, 1933. J. Eukaryot. Microbiol. 56, 552–558 (2009). [DOI] [PubMed] [Google Scholar]
- Wang H., Clamp J. C., Shi X., Utz L. R. & Liu G. Evolution of variations in the common pattern of stomatogenesis in peritrich ciliates: evidence from a comparative study including a new description of stomatogenesis in Pseudepistylis songi Peng et al., 2007. J. Eukaryot. Microbiol. 59, 300–324 (2012). [DOI] [PubMed] [Google Scholar]
- Xu K. & Song W. In Pathogenic Protozoa in Mariculture (eds Song W. et al. ) 291–314 (Science Press, 2003). [Google Scholar]
- Carey P. G. Marine interstitial ciliates. An illustrated key. (Chapman and Hall, 1992). [Google Scholar]
- Vd’ačný P., Bourland W. A., Orsi W., Epstein S. S. & Foissner W. Phylogeny and classification of the Litostomatea (Protista, Ciliophora), with emphasis on free-living taxa and the 18S rRNA gene. Mol. Phylogenet. Evol. 59, 510–522 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang Q., Simpson A. & Song W. Insights into the phylogeny of systematically controversial haptorian ciliates (Ciliophora, Litostomatea) based on multigene analyses. Proc. R. Soc. B 279, 2625–2635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner W. & Foissner I. The fine structure of Fuscheria terricola Berger et al., 1983 and a proposed new classification of the subclass Haptoria Corliss, 1974 (Ciliophora, Litostomatea). Arch. Protistenkd. 135, 213–235 (1988). [Google Scholar]
- Grain J. In Infusoires Ciliés - Systematique. Traité de Zoologie. II Fasc. 2. (ed de Puytorac P. ) 267–310 (Masson, 1994). [Google Scholar]
- Lipscomb D. L. & Riordan G. P. The ultrastructure of Chaenea teres and an analysis of the phylogeny of the haptorid ciliates. J. Protozool. 37, 287–300 (1990). [Google Scholar]
- Foissner W. Updating the Trachelocercids (Ciliophora, Karyorelictea). IV. Transfer of Trachelocerca entzi Kahl, 1927 to the Gymnostomatea as a new genus, Trachelotractus gen. n. (Helicoprorodontidae). Acta Protozool. 36, 63–74 (1997). [Google Scholar]
- Kahl A. Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria). 2. Holotricha. Tierwelt Dtl. 21, 181–398 (1931). [Google Scholar]
- Gong J. et al. Small subunit rRNA phylogenies show that the class Nassophorea is not monophyletic (Phylum Ciliophora). J. Eukaryot. Microbiol. 56, 339–347 (2009). [DOI] [PubMed] [Google Scholar]
- Kivimaki K. L., Bowditch B. M., Riordan G. P. & Lipscomb D. L. Phylogeny and systematic position of Zosterodasys (Ciliophora, Synhymeniida): a combined analysis of ciliate relationships using morphological and molecular data. J. Eukaryot. Microbiol. 56, 323–338 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al. Further insights into the phylogeny of two ciliate classes Nassophorea and Prostomatea (Protista, Ciliophora). Mol. Phylogenet. Evol. 70, 162–170 (2014). [DOI] [PubMed] [Google Scholar]
- Fan X. et al. Phylogeny of the poorly known ciliates, microthoracida, a systematically confused taxon (ciliophora), with morphological reports of three species. J. Eukaryot. Microbiol. 61, 227–237 (2014). [DOI] [PubMed] [Google Scholar]
- Jankowski A. W. In Protista: Handbook on Zoology, Part 2. (eds Krylow M. V. & Frolov A. O. ) 415–976 (St. Petersburg, 2007). [Google Scholar]
- Diggles B. K. Some information on the morphology of Cryptocaryon irritans from south-east Queensland, Australia. Eur. J. Protistol. 33, 200–210 (1997). [Google Scholar]
- Foissner W. Infraciliature and systematic position of the marine interstitial ciliates (Protozoa, Ciliophora) Lopezoterenia torpens (Kahl, 1931) nov. gen., nov. comb., Discotricha papillifera Tuffrau, 1954, and Paraspathidium fuscum (Kahl, 1928) Fjeld, 1955. Rev. Soc. Mex. Hist Nat. 47, 41–63 (1997). [Google Scholar]
- Foissner W. A compilation of soil and moss ciliates (Protozoa, Ciliophora) from Germany, with new records and descriptions of new and insufficiently known species. Eur. J. Protistol. 36, 253–283 (2000). [Google Scholar]
- Lipscomb D. L. & Riordan G. The ultrastructure of Placus striatus and a revision of the family Placidae(Ciliophora). J. Eukaryot. Microbiol. 59, 407–422 (2012). [DOI] [PubMed] [Google Scholar]
- Fleury A., Delgado P., Iftode F. & Adoutte A. A molecular phylogeny of ciliates: What does it tell us about the evolutionary of the cytoskeleton and of developmental strategies? Dev. Genet. 13, 247–254 (1992). [Google Scholar]
- Baroin-Tourancheau A., Delgado P., Perasso R. & Adoutte A. A broad molecular phylogeny of ciliates: identification of major evolutionary trends and radiations within the phylum. Proc. Natl. Acad. Sci. USA 89, 9764–9768 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardele C. F. Ultrastructure, morphogenetics, and a discussion of the phylogeny of the prostome ciliate Balanion planctonicum. Eur. J. Protistol. 35, 17–33 (1999). [Google Scholar]
- Yi Z., Katz L. A. & Song W. Assessing whether alpha-tubulin sequences are suitable for phylogenetic reconstruction of Ciliophora with insights into its evolution in euplotids. PloS one 7, e40635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O. et al. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38, W23–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P. & Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771 (2008). [DOI] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W. & Schwartz T. In Proceedings of the Gateway Computing Environments Workshop (GCE) 1–8 (New Orleans, LA, 2010). [Google Scholar]
- Abascal F., Zardoya R. & Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Nylander J. A. (2004). MrModeltest v2. Uppsala University.
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002). [DOI] [PubMed] [Google Scholar]
- Swofford D. L. (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. URL http://www.paup.csit.fsu.edu/.
- Shimodaira H. & Hasegawa M. Consel: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247 (2001). [DOI] [PubMed] [Google Scholar]
- Orsi W. et al. Class Cariacotrichea, a novel ciliate taxon from the anoxic Cariaco Basin, Venezuela. Int J Syst Evol Microbiol 62, 1425–1433 (2012). [DOI] [PubMed] [Google Scholar]
- Bardele C. F., Foissner W. & Blanton R. L. Morphology, morphogenesis and systematic position of the sorocarp forming ciliate Sorogena stoianovitchae Bradbury-and-Olive, 1980. J. Protozool. 38, 7–17 (1991). [Google Scholar]
- Foissner W., Skogstad A. & Putt J. R. Morphology and infraciliature of Trochiliopsis australis n. sp., Pelagohalteria viridis (Fromentel, 1876) n. g., n. comb., and Strobilidium lacustris n. sp. (Protozoa, Ciliophora). J. Protozool. 35, 489–497 (1988). [Google Scholar]
- Foissner W., Agatha S. & Berger H. Soil ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with emphasis on two contrasting environments, the Etosha region and the Namib desert. Part I: Text and line drawings. Part II: Photographs. Denisia 5, 494 (2002). [Google Scholar]
- Dehority B. A. A new family of entodiniomorph protozoa from the marsupial forestomach, with descriptions of a new genus and five new species. J. Eukaryot. Microbiol. 43, 285–295 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.